Abstract
The biosynthesis of wax esters in bacteria is accomplished by a unique pathway that combines a fatty alcohol and a fatty acyl coenzyme A substrate. Previous in vitro enzymatic studies indicated that two different enzymes could be involved in the synthesis of the required fatty alcohol in Marinobacter aquaeolei VT8. In this study, we demonstrate through a series of gene deletions and transcriptional analysis that either enzyme is capable of fulfilling the role of providing the fatty alcohol required for wax ester biosynthesis in vivo, but evolution has clearly selected one of these, a previously characterized fatty aldehyde reductase, as the preferred enzyme to perform this reaction under typical wax ester-accumulating conditions. These results complement previous in vitro studies and provide the first glimpse into the role of each enzyme in vivo in the native organism.
INTRODUCTION
The global cycle of oil is of interest from the standpoints of both energy and the environment, as efforts by humankind to obtain this valuable resource can result in substantial releases of crude oil through incidents such as the Deepwater Horizon oil spill of 2010 in the Gulf of Mexico. We also note that crude oil from natural deposits is routinely released into aqueous environments, such as the oceans, by natural processes where geological reserves meet surface waters. These environments have allowed natural populations of organisms, such as marine bacteria, to evolve to utilize these supplies, rich in reduced carbon, for use as a biological fuel source. A primary focus related to oil-degrading marine bacteria is the oxidation of these oils to meet energy requirements of the living cell. Interestingly, for certain marine bacteria found to utilize and degrade oils, these bacteria are also capable of producing natural lipids that have economic values similar to those obtained from harvesting sperm whales prior to the late 20th century, even when the bacteria are grown on simple organic acids or carbohydrates. We selected the marine bacterium Marinobacter aquaeolei VT8, which was isolated from an oil well off the coast of Vietnam (1), as a model bacterial species to study metabolic processes in an oil-metabolizing and neutral lipid-accumulating bacteria. In addition to growing on long-chain hydrocarbons, M. aquaeolei VT8 also produces a natural hydrocarbon, the wax ester, when grown on simple citric acid cycle intermediates, such as succinate or citrate, as the sole carbon source (1–3), indicating that all of the precursors required for the biosynthesis of wax ester are indigenous to this strain.
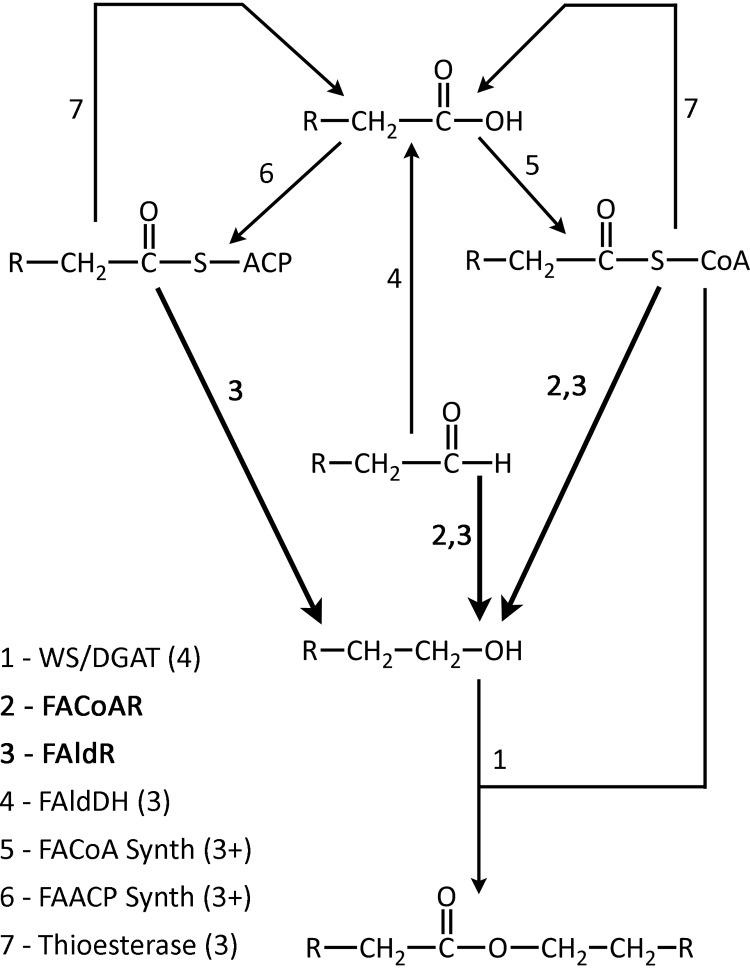
Biosynthesis of wax esters is accomplished by the combination of several different enzymes. The wax ester synthase/acyl-coenzyme A (CoA):diacylglycerol acyltransferase (WS/DGAT) enzyme catalyzes the reaction of a fatty acyl-CoA substrate with a fatty alcohol (Fig. 1). While the fatty acyl-CoA utilized by the WS/DGAT is proposed to come directly from the fatty acyl-CoA pool, the fatty alcohol is believed to be produced through the action of several reductase enzymes acting on activated fatty acids or fatty aldehydes. M. aquaeolei VT8 contains at least two enzymes that have been found to produce fatty alcohols from several different substrates in vitro, including fatty aldehydes, fatty acyl-CoAs, and fatty acyl-acyl carrier proteins (ACPs) (4–7). Additionally, both of the isolated enzymes from M. aquaeolei VT8 have significantly higher activities than those reported for other enzymes when tested in in vitro assays versus the enzyme isolated from Acinetobacter (4, 6–8). Thus, it was of interest to determine which of the two enzymes found in M. aquaeolei VT8 is responsible for the production of the fatty alcohol in this species under natural wax ester production conditions. To determine the roles of these different enzymes, efforts were undertaken to delete each of the genes by using homologous recombination, and also to track the transcription of the genes coding these enzymes during the wax ester accumulation stage of growth.
Fig 1.
Wax ester pathway. Shown are the proteins that comprise the current pathway for wax ester biosynthesis in lipid-accumulating bacteria such as M. aquaeolei VT8. Various proteins of the pathway are numbered, including the wax synthase (1), fatty acyl-CoA reductase (2), fatty aldehyde reductase (3), fatty aldehyde dehydrogenase (4), fatty acyl-CoA synthetase (5), fatty acyl-ACP synthetase (6), and thioesterase (7). Values shown following specific enzymes in parentheses indicate the number of known or putative homologs found in M. aquaeolei VT8. Enzymes shown in bold and with darker arrows are those featured in this study.
MATERIALS AND METHODS
Strains and reagents.
Marinobacter aquaeolei VT8 was obtained from the American Type Culture Collection (ATCC) and cultured aerobically on Miller lysogeny broth (LB) at 30°C. Escherichia coli WM3064 (9) was grown on LB supplemented with 20 μg/ml diaminopimelic acid (DAP) at 37°C. For wax ester production, M. aquaeolei VT8 was grown on a minimal medium with either citrate or succinate as a carbon source at a concentration of 7 g per liter (2). When appropriate, the medium was supplemented with kanamycin at 50 μg/ml. A list of reference names for genes and gene products is provided in Table 1 for quick reference. All reagents were obtained from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) unless otherwise specified.
Table 1.
Selected proteins for mRNA transcription analysis
| Gene product (gene)a | Protein (or ribosomal) productb | KEGG pathway (or putative pathway)c | Protein/nucleotide accession no. |
|---|---|---|---|
| FACoAR (acrB) | Fatty acyl-CoA reductase | Wax ester biosynthesis | YP_959769 |
| FAldR (farA) | Fatty aldehyde reductased | Wax ester biosynthesis | YP_959486 |
| FAldDH (aldF) | Fatty aldehyde dehydrogenase | Wax ester biosynthesis | YP_960668 |
| Medium ADH (adhM) | Medium alcohol dehydrogenase | Unknown | YP_958650 |
| 16S rRNAe | 16S rRNA | Ribosome | NR_027551 |
| Recombinase (recA) | Recombinase A | Homologous recombination | YP_959349 |
Provided as a simple reference for the gene products (or genes) shown in the figures.
The full name of the gene product, based on the NCBI reference number or previous annotations.
Potential pathway in which the gene product is involved, from the KEGG or the pathway shown in Fig. 1.
While annotated here as fatty aldehyde reductase, we also note that this gene product has been reported to have fatty acyl-CoA reductase and fatty acyl-ACP reductase activities by others (4, 5).
Three copies of the 16S rRNA gene are found in the genome.
Conjugation of M. aquaeolei VT8.
The M. aquaeolei VT8 conjugation procedure was derived from methods for the conjugation of Psychrobacter arcticus 273-4 and Marinobacter adhaerens (10, 11). Briefly, cultures of the donor cells, E. coli WM3064, containing the specific plasmid and recipient cells, M. aquaeolei VT8, were grown separately on LB plates, then mixed at a ratio of 1:3 donor to recipient cells, spotted onto an LB plate containing DAP, and then incubated at 30°C for approximately 24 h. Cells were collected from two spots, washed with LB broth, resuspended in 100 μl LB, and spread onto LB plates devoid of DAP but containing kanamycin for selection. These plates were then incubated at 30°C for 2 to 4 days, at which time colonies were selected and streaked several times to fresh plates prior to PCR verification of deletions by using primers flanking the regions of DNA that were manipulated.
Single-gene deletion experiments with M. aquaeolei VT8.
Single-gene deletions were accomplished by constructing a plasmid vector containing the mobilization element from the pBBR1MCS-2 vector incorporated into a pUC19 derivative vector, pBB053. The regions flanking the genes of interest were amplified by PCR and cloned into a separate pUC19 derivative vector (pBB053 or pBBTET3) with a different antibiotic marker and then shuffled to the deletion vector with the mobilization element. Finally, the kanamycin resistance cassette from pBBR1MCS-2 was placed between the two flanking regions to replace the target gene upon double homologous recombination. Specific details of the construction of these vectors are outlined in Table 2, and a list of the primers used to construct these vectors is shown in Table 3. A map of plasmid pPCRWEK29 is shown in Fig. 2. Plasmids pPCRWEK29 and pPCRWEK33 were transformed into E. coli strain WM3064 and used to conjugate M. aquaeolei VT8 as described in the previous section.
Table 2.
Key parent plasmids and their relevant derivatives used for the construction of Marinobacter aquaeolei VT8 manipulated strains
| Plasmida | Relevant gene cloned or plasmid manipulation(s) | Vector | Reference and/or source |
|---|---|---|---|
| pBBR1MCS-2 | Plasmid containing mobilization element | 14 | |
| pSMV3 | Plasmid containing sacB gene | 9 | |
| pBB053 | NdeI site removed from pUC19 by silent mutation | pUC19 | This study |
| pBB114 | Replaced pUC19 Amp resistance with Kan resistance cassette from pUC4K, then removed NsiI and HindIII sites from cassette by silent mutations | pUC19 | pUC4K (22) |
| pBBTET3 | Replaced pUC19 Amp resistance with Tet resistance cassette from pRK415 | pUC19 | pRK415 (23) |
| pPCRKAN4 | Cloned Kan cassette from pBBR1MCS-2 into pBBTET3 | pBBTET3 | This study |
| pPCRMOB4 | Moved mobilization element from pBBR1MCS-2 into pUC19 | pUC19 | This study |
| pPCRSACB6 | sacB gene from pSMV3 cloned into pBB053, and then EcoRI, HindIII, XbaI, and KpnI sites were removed by site-specific mutagenesis with silent mutations | pBB053 | This study |
| pPCRSACB7 | Moved sacB gene cassette from pPCRSACB6 to pBBTET3 | pBBTET3 | This study |
| pPCRWEK4 | Derivative of pBB053 for gene insertions | pBB053 | This study |
| pPCRWEK5 | Moved mobilization element from pPCRMOB4 into pPCRWEK4 | pBB053 | This study |
| pPCRWEK12 | Cloned acrB and flanking regions from M. aquaeolei VT8 genome with primers BBP1477 and BBP1478 into pBBTET3 EcoRI and XbaI sites | pBBTET3 | This study |
| pPCRWEK14 | Performed PCR with primers to remove acrB from pPCRWEK12, leaving flanking regions and adding BamHI site | pBBTET3 | This study |
| pPCRWEK20 | Cloned farA and flanking regions from M. aquaeolei VT8 genome with primers BBP1522 and BBP1523 into pBBTET3 EcoRI and XbaI sites | pBBTET3 | This study |
| pPCRWEK26 | Moved acrB flanking segments fragment from pPCRWEK14 into pPCRWEK5 | pBB053 | This study |
| pPCRWEK27 | Performed PCR with primers to remove farA from pPCRWEK20, leaving flanking regions and adding BamHI site | pBBTET3 | This study |
| pPCRWEK29b,c | Moved Kan cassette from pPCRKAN4 into BamHI-cut pPCRWEK26 | pBB053 | This study |
| pPCRWEK32 | Moved farA flanking segments fragment from pPCRWEK27 into pPCRWEK5 | pBB053 | This study |
| pPCRWEK33c | Moved Kan cassette from pPCRKAN4 into BamHI-cut pPCRWEK32 | pBB053 | This study |
| pPCRWEK48 | Moved sacB cassette from pPCRSACB7 into KpnI- and HindIII-cut pPCRWEK32 | pBB053 | This study |
| pPCRWEK50b,c | Moved Kan cassette cut with BamHI from pPCRKAN4 into BglII-cut pPCRWEK48 | pBB053 | This study |
The sequences of all plasmids used in this study are available upon request.
Plasmid map is also provided in Fig. 2.
Plasmids shown in bold are completed vectors used to transform M. aquaeolei VT8.
Table 3.
Primers used in this study
| Primer designation | Primer sequencea | Purpose |
|---|---|---|
| BBP1477 | 5′-GACATCTA GACTGGATCT TGTCTTCCCG GGAACCAC-3′ | acrB gene and flanking region cloning |
| BBP1478 | 5′-GACAGAAT TCTGGATTTC ACCGGCATCG ATCC-3′ | acrB gene and flanking region cloning |
| BBP1479 | 5′-GACAGGAT CCATATGTAC TCCATTCTGC CTGTTGTGTT TTTG-3′ | acrB gene deletion |
| BBP1480 | 5′-GACAGGAT CCGATATACT GGTAATCGTC GTTATAAACC AAG-3′ | acrB gene deletion |
| BBP1522 | 5′-GNNNGAAT TCGATCGCGC CAGTCTTGCT CGTCATTTG-3′ | farA gene and flanking region cloning |
| BBP1523 | 5′-GNNNTCTA GAAGCTTCGA AGCGTTCAGG ACACCGTCCT CGAAC-3′ | farA gene and flanking region cloning |
| BBP1524 | 5′-GNNNGGAT CCCTTCTCCG GGGCAGGAAA GCGTTTCTG-3′ | farA gene deletion |
| BBP1525 | 5′-GNNNGGAT CCGATAGAAC TCCTTCTCTG AGATCACTAA TGCCG-3′ | farA gene deletion |
| BBP1558 | 5′-CGAGATGC TGAACGTTCA TGTTGGC-3′ | farA deletion confirmation |
| BBP1559 | 5′-CACAGAGT GGATCGCACC AATACG-3′ | farA deletion confirmation |
| BBP1548 | 5′-GTATTCGC CTGCCTCCGG GTACTTC-3′ | acrB deletion confirmation |
| BBP1549 | 5′-CACACGCG AAAGACAAGA AGGAAGC-3′ | acrB deletion confirmation |
| BBP1678 | 5′-GTTCCGTT CCGCATCTAC CG-3′ | acrB qPCR |
| BBP1679 | 5′-CCAGTGCA TCGACCACGA AA-3′ | acrB qPCR |
| BBP1409 | 5′-CCGTCTTC GCGAGGCCGA TT-3′ | farA qPCR |
| BBP1410 | 5′-TGATGGCC AGCGCCTTGT CG-3′ | farA qPCR |
| BBP1403 | 5′-TTTCCGCT GCTGATGGCC GC-3′ | aldF qPCR |
| BBP1404 | 5′-CGCTTGCT GGTCGCCAAA GC-3′ | aldF qPCR |
| BBP1352 | 5′-TCCTGCCG TATCCACCGG CT-3′ | recA qPCR |
| BBP1353 | 5′-AAACCGGG TCCAGAGCGT GC-3′ | recA qPCR |
| BBP1126 | 5′-GCACGCTC TGGACCCGGT TT-3′ | recA qPCR |
| BBP1127 | 5′-CCACGTGG CTGTCGCCCA TT-3′ | recA qPCR |
| BBP1413 | 5′-TTGTTGGC CGGGTTACCG CC-3′ | adhM qPCR |
| BBP1414 | 5′-TAGCCGCC GTGAGTGACC GA-3′ | adhM qPCR |
| BBP1128 | 5′-CCGGCTAA CTCCGTGCCA GC-3′ | 16S rRNA qPCR |
| BBP1129 | 5′-ACGCATTT CACCGCTACA CAGG-3′ | 16S rRNA qPCR |
Specific restriction enzyme sites added to primers are underlined for clarity.
Fig 2.
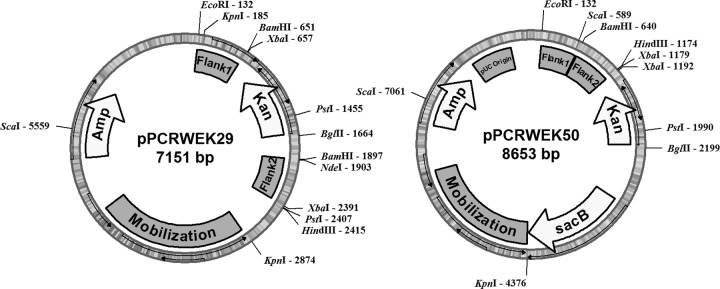
Key plasmids for gene deletion studies. Shown are representations of two of the final plasmid constructs utilized for the gene deletion studies. Plasmid pPCRWEK29 was used to perform double homologous recombination by replacing the gene of interest (acrB in this construct) with the antibiotic marker for kanamycin. Plasmid pPCRWEK50 was used to perform a single homologous recombination, which was selected by using the kanamycin marker. A counterselection following a second recombination event, based on the toxicity of the sacB gene product, resulted in markerless deletion of the farA gene. Images produced using the program pDRAW32 (Acaclone Software).
Single gene deletion with markerless counterselection.
Single gene deletions with markerless selection were constructed as described above for double homologous recombinations, with the following exceptions. For counterselections, the sacB gene and promoter from the pSMV3 plasmid (9) was cloned into a separate plasmid, and multiple restriction sites were removed by site-specific mutagenesis to optimize the gene construct for future use. A vector was then constructed containing the sacB gene, the mobilization element described above, and the kanamycin cassette described above, except that all fragments were inserted into the vector outside the segment containing the flanking region fragments. This new plasmid was called pPCRWEK50, and a map of the plasmid is shown in Fig. 2. Following the conjugation protocol described above, single homologous recombination was used to integrate the entire plasmid into the genome. Once isolated, a counterselection protocol was used by growing M. aquaeolei VT8 in the presence of sucrose. While this procedure has been used successfully in other strains, selection of the markerless gene deletion following a second recombination event was not very efficient and took several transfers in sucrose-containing medium before a successful gene deletion was obtained by screening multiple colonies grown on LB plates and then identifying those which no longer grew on LB plates supplemented with kanamycin.
Wax ester production in gene deletion strains.
Once gene deletion strains were confirmed, cultures were grown in a shaker flask in wax ester-producing medium, harvested, lyophilized, and extracted for wax ester analysis as described previously using a minimal medium with citrate as the carbon source (2). Each gene deletion strain was grown as three independent cultures and harvested along with a control of the wild-type strain. Cultures were harvested approximately 24 h after nitrate was depleted from the culture.
M. aquaeolei VT8 batch culture experiments for quantitative PCR (qPCR) and wax ester analysis.
M. aquaeolei VT8 wild type was first isolated as a single colony on an LB plate. Batch culture experiments were performed in a nitrogen-limited defined medium containing the following per liter: 50 g NaCl, 7 g sodium citrate, 5 g MgSO4 · 7H2O, 500 mg K2HPO4, 200 mg CaCl2 · 2H2O, 15 mg FeSO4 · 7H2O, and 640 mg NaNO3, adjusted to pH 7.3 with NaOH and HCl. A loop full of cells (∼50 μl total volume) was scraped from an LB plate containing a fresh lawn of cells and transferred to a Celstir flask (Wheaton, Millville, NJ) containing 5 liters of the nitrogen-limited medium and 100 μl of polypropylene glycol to minimize foaming during the culture. Aeration was provided by including a custom aeration bar with three pinholes, with filtered air (0.2 μm) provided by a simple aquarium pump. This represented the initial time of the culture experiment, and samples were taken at various time points based on nitrate consumption and cell density. At each time point, a series of samples were drawn, centrifuged, and flash-frozen for RNA isolation, and a separate sample was taken for isolation of cells for quantification of the wax ester fraction. The pH of the culture was adjusted by adding HCl following sampling to maintain the pH below 7.8. Wax esters at each time point were analyzed and quantified versus an external standard as described previously (2), and the lipid quantification and dry cell mass obtained from each sampling period were used to categorize the samples as the culture transitioned through three different phases of growth: exponential-growth stage with low wax esters, the wax ester production and accumulation stage, and the wax ester catabolism stage.
RNA isolation and RT-qPCR analysis.
RNA was isolated by resuspending frozen cells in 1 ml of TRIzol reagent (Invitrogen, Grand Island, NY), and then samples were vortexed for several minutes until fully dissolved. Following this, 200 μl of chloroform was added, vortexed, and then centrifuged at 12,000 × g for 2 min. The upper phase was removed and further purified using the Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA). RNA was eluted, then treated following the manufacturer's directions using the RNase-free DNase kit (Qiagen, Hilden, Germany) in a total volume of 100 μl for 10 min at room temperature, and then suspended in 300 μl of TRIzol and again isolated using the Direct-zol miniprep kit. The isolated RNA quantity was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA), and then 1 μg of total RNA was immediately converted to cDNA by using the Improm-II reverse transcriptase (RT) kit and random primers (Promega, Madison, WI). Once completed, cDNA was frozen and stored at −20°C. Samples for qPCR were prepared using the SYBR green master mix (Roche, Basel, Switzerland) in a total volume of 400 μl containing 100 ng of cDNA. Samples were prepared in 96-well plates with the addition of specific primer pairs and were analyzed following a standard qPCR protocol on a LightCycler 480 II instrument (Roche, Basel, Switzerland). Primers were designed using primer BLAST with a target PCR product size of approximately 200 bp.
All qPCR experiments were performed using cDNA generated from 1.0 μg of isolated RNA based on spectrophotometric quantification. Conditions for qPCR were as follows: an initial melting cycle of 95°C for 10 min, followed by the PCR conditions of 95°C for 10 s, 58°C for 10 s, and 72°C for 15 s, repeated 40 times. Data analysis was performed by using the crossing-point (Cp) calculation (LightCycler 480 software, release 1.5.0 SP3; Roche, Basel, Switzerland) and included the reference gene recombinase A (12) in addition to 16S rRNA as reference samples. Data analysis was done by calculating the ΔCp value between each data point and the final time point in the batch culture. Controls were performed for each of the gene targets by comparison of obtained Cp values over a range, including a 32-fold decrease in total cDNA using a serial dilution strategy with 6 sample points to confirm a linear relationship based on the exponential function. PCR products were further analyzed by agarose gel electrophoresis to confirm the correct sizes of the products.
RESULTS AND DISCUSSION
The marine bacterium Marinobacter aquaeolei VT8 produces wax esters under nutrient-limited conditions when grown in the presence of simple carbon sources (such as acetate, citrate, or succinate) in a minimal medium (2). Previous studies in our laboratory have characterized several key enzymes that could participate in the wax ester biosynthetic pathway (2, 6, 7). One feature associated with wax ester production in M. aquaeolei VT8 is redundancy of several of the enzymes involved in this pathway (Fig. 1). This includes multiple homologs for the wax ester synthase enzyme (2) and two alternative enzymes that have been found to reduce more-oxidized pathway intermediates (such as fatty aldehydes or activated fatty acids) to fatty alcohols (2, 4, 6, 7). The reasons why M. aquaeolei VT8 has enzyme redundancy within this pathway are unclear, as are the roles that different enzymes play in vivo during wax ester production. This feature of enzyme redundancy differentiates M. aquaeolei VT8 from other model wax ester-accumulating organisms, such as Acinetobacter calcoaceticus, that are reported to have only a single enzyme for each of these roles (2, 4, 6–8, 13).
The goal of these experiments was to determine the roles, under wax ester-accumulating conditions, of the two different enzymes reported to reduce fatty acid-derived precursors in the wax ester biosynthetic pathway to fatty alcohols in M. aquaeolei VT8 (enzymes 2 and 3 in the pathway shown in Fig. 1). Both of these enzymes have been shown to yield fatty alcohols from fatty aldehydes or activated fatty acids by using NADPH as a reductant during in vitro studies with the purified enzymes (4, 6, 7). The genes coding these enzymes do not appear to be part of an operon and are separated from one another in the genome by approximately 320 kb. Both genes are also significantly distanced from the known wax ester synthase genes (3). To address the question of what role these enzymes play and the function of the enzymes in vivo, two complementary approaches were taken as part of this work. The first consisted of single gene deletions followed by wax ester production analysis, and the second approach included a gene expression profile for these genes during batch culture in wax ester-accumulating medium for the wild-type strain.
Single gene deletions for acrB and farA.
To produce single gene deletions, we selected an approach utilizing double homologous recombination and conjugation strategies. A similar approach to what we have taken was recently reported for an alternative strain of Marinobacter (11). We found that pUC19-derived plasmids did not replicate in M. aquaeolei VT8 and could thus serve as suicide vectors for genome integration studies if they contained the proper mobilization element (14), and we found the kanamycin cassette from pBBR1MCS-2 (14) to be an ideal selection marker with M. aquaeolei VT8. This strategy was successful in isolating strain ΔacrB::kan, containing a single gene deletion of the gene coding for FACoAR (NCBI accession number YP_959769 for the fatty acyl-CoA reductase gene product, or Maqu_2507) and separately strain ΔfarA::kan, containing a single gene deletion of the gene coding for FAldR (NCBI accession number YP_959486 for the fatty aldehyde reductase gene product, or Maqu_2220). We will utilize our previous naming scheme of fatty aldehyde reductase (FAldR) for the latter gene product for clarity, although we acknowledge others have reported additional activities with fatty acyl-CoA and fatty acyl-ACP substrates for this gene product (4, 5).
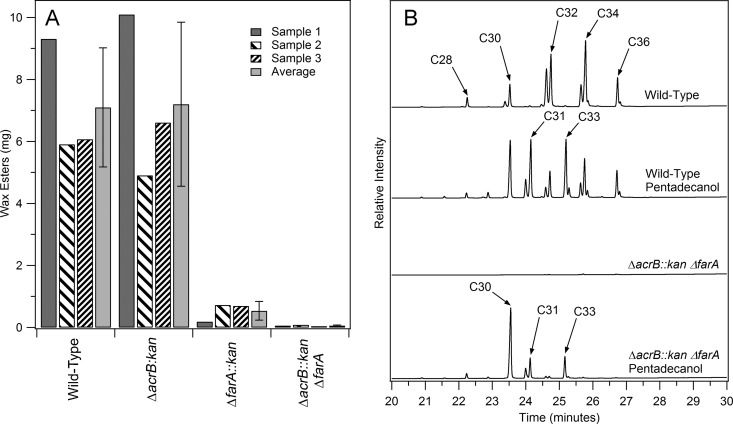
Once isolated, each of the individual deletion strains, ΔacrB::kan and ΔfarA::kan, and the wild-type M. aquaeolei VT8 were grown under wax ester-accumulating conditions using citrate as the primary carbon source (2). Figure 3A shows the results of the wax ester analysis from three independent cultures of these strains. Due to differences in peak lipid production (see the discussion related to Fig. 4, below), there was some degree of variance seen between results, although differences were clear for the specific strains. The wild-type and ΔacrB::kan strains yielded similar quantities of wax esters (Fig. 3A), while the ΔfarA::kan strain resulted in a significant decrease in the amounts of wax esters found. Neither single-gene deletion strain resulted in a wax ester deletion phenotype, indicating that both enzymes are capable of fulfilling this role in the wax ester biosynthesis pathway in the absence of the other gene.
Fig 3.
Gene deletion and wax ester production rescue studies. (A) Quantities of wax esters obtained from replicate cultures of wild-type M. aquaeolei VT8 cells under wax ester-accumulating conditions versus the two single-gene deletion strains and the double deletion strain. Two hundred milligrams of dried cell mass was extracted in each case. Error bars represent the standard deviations of the three samples. (B) Gas chromatograms illustrating the results of studies with wild-type cells and the ΔacrB::kan ΔfarA double deletion strain grown under normal wax accumulation conditions and under the same conditions with added extraneous pentadecanol, resulting in the accumulation of odd-numbered wax esters (for the wild type) and the rescue of wax ester production based on only these odd-numbered alcohols (for the double deletion).
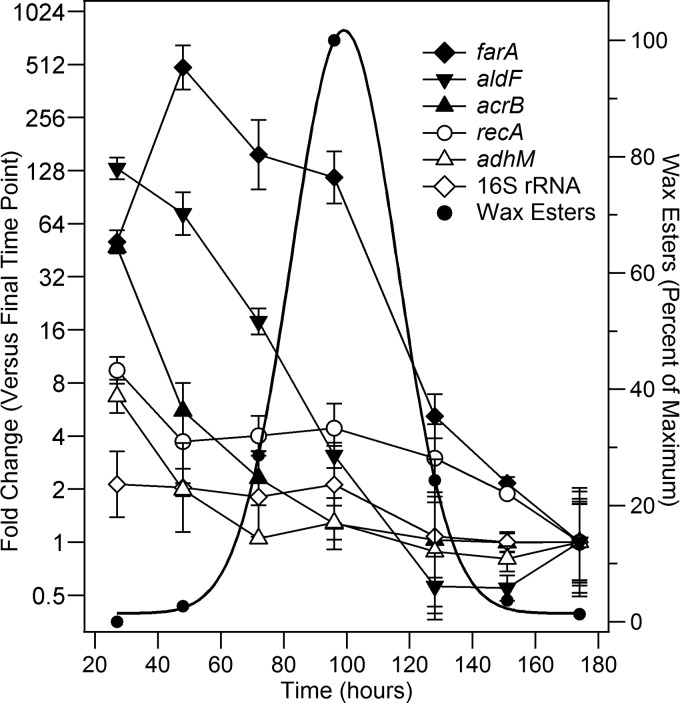
Fig 4.
Gene transcription during batch culture of M. aquaeolei VT8. Shown are results obtained from an RT-qPCR analysis for a single batch culture of wild-type M. aquaeolei VT8 cells grown under wax ester-accumulating conditions over the course of several days. Transcriptional levels (the fold change in mRNA levels) were compared against the results obtained for the last time point (normalized to 1-fold; left y axis) and are plotted on a log scale for simplicity. Results obtained from the wax ester analysis for this culture were fit to a simple Gaussian curve for clarity and are shown on the same graph (right y axis). Where shown, statistics represent the averages and standard deviations for three replicates.
Double gene deletion of both acrB and farA.
To probe and confirm the specific roles of both enzymes, a further effort was initiated to construct a double deletion strain. Here, the first deletion was accomplished by using a single homologous recombination event with a selectable marker for counterselection. We chose to utilize the sacB gene, which results in a sucrose-sensitive phenotype in certain bacteria (15–17). Following isolation and confirmation of the single homologous recombination based on antibiotic selection, M. aquaeolei VT8 colonies were grown in liquid culture containing sucrose in a minimal medium and enriched by several subsequent transfers to fresh medium before plating and screening for loss of the antibiotic marker. Strains containing the markerless deletion were confirmed by colony PCR. The second gene deletion was then isolated using the double homologous selection method described above for single gene deletions. The toxicity of sacB in M. aquaeolei VT8 was not as potent as was found for controls tested in Escherichia coli and thus is best characterized as a screening protocol under the current conditions. We suspect this may be related to poor sugar uptake by M. aquaeolei VT8, which does not grow well on simple sugars. However, by utilizing this procedure, a double deletion strain, ΔacrB::kan ΔfarA, was obtained. Characterization of wax ester production in the ΔacrBv::kan ΔfarA strain (Fig. 3A) revealed only a minimal background of wax esters. This confirms that either enzyme is capable of supporting the wax biosynthetic pathway independent of one another, though the ΔfarA gene product seems to play a greater role in M. aquaeolei VT8 under typical wax ester-accumulating conditions based on these single-gene deletion studies. It also supports the proposal that these are the two primary genes capable of producing fatty alcohols, as only trace amounts of wax esters were found in the absence of both genes.
Wax ester production phenotype rescue through addition of extraneous alcohols.
To confirm that loss of the wax ester production phenotype in these deletion strains is based on the lack of this enzymatic step in the pathway (and not a secondary effect related to poor culture health, for example), we utilized a strategy used previously to add foreign fatty alcohols (specifically, the odd-carbon-number pentadecanol, which results in unique wax ester products) in an attempt to rescue the wax ester production phenotype in this double deletion strain (2). This results in the production of odd-numbered wax esters when provided extraneously to wild-type cells (2), as shown in Fig. 3B. This strategy relies on the fact that waxes found in M. aquaeolei VT8 under the culture conditions utilized here are derived primarily from C16 and C18 fatty acids (2). Thus, addition of pentadecanol to a strain lacking only the enzyme(s) involved in the reduction of fatty acids to fatty alcohols should result in wax esters containing even-numbered fatty acids and only pentadecanol as the alcohol. This result is confirmed in Fig. 3B (bottom). Two peaks were found at approximately 24 min that corresponded to pentadecanol and C16 fatty acid (either C16:1 or C16) derived wax esters, and one additional peak was found just after 25 min that corresponded to pentadecanol and C18 fatty acid (predominantly C18:1) derived wax esters. An additional large peak corresponding to a C30 wax ester resulted from pentadecanol oxidation within the cell through alternative directions in the pathway (Fig. 1; see also reference 18), resulting in significant amounts of pentadecanol and C15 fatty acid-derived wax ester accumulating in the cell as well. This result was further confirmed by treatment of the wax esters from this sample with methanol and acid as described previously (2), followed by characterization of individual components by gas chromatography and mass spectrometry (GC/MS). This analysis found four primary fatty acids: C15, C16:1, C16, and C18:1, at a ratio of approximately 3.5:0.5:1:1, which agrees well with the wax ester profile of the chromatogram shown in Fig. 3B (bottom). Importantly, predominant wax esters, such as C32, derived from C16 fatty acids and fatty alcohols (at approximately 24.5 min) were not prominent in the double deletion strain, and C16 and C18 fatty alcohols were not found during GC/MS analysis in the double deletion strain, while they were present in a wild-type control sample. This confirmed that the wax ester biosynthesis pathway can be reconstituted in vivo in the double deletion strain by adding extraneous alcohol (pentadecanol).
Transcriptional analysis of acrB and farA during wax ester accumulation.
In addition to the gene deletion studies described above, it was also of interest to investigate the changes in gene transcriptional levels for these two fatty alcohol-producing enzymes during a typical batch culture in wild-type M. aquaeolei VT8. To pursue these studies, an approach was taken to grow M. aquaeolei VT8 as a batch culture using a medium recipe routinely utilized in our laboratory to induce lipid accumulation. The key feature of this defined medium is that cells exhaust the source of nitrogen prior to reaching maximum cell density for the specific culture conditions and enter into a nitrogen-limited state that results in wax ester accumulation. This is believed to occur because the carbon source needed for energy and cellular building blocks remains plentiful but the nitrogen required for DNA and protein synthesis is not available for further replication (19). The larger batch culture was selected here so that a thorough sampling of the culture could be made through various stages of the wax ester accumulation and declination process. The sampling strategy adopted included multiple samples for total RNA isolation and the harvest of cells for further drying and wax ester quantification. Lipids were extracted using a previously described protocol that isolates primarily wax esters from dried M. aquaeolei VT8 cells, which can be analyzed directly using a gas chromatography method with flame ionization detection (GC/FID) to measure specific classes of lipids (2). This method is preferred over indirect methods, such as gravimetric approaches, as the specific compound of interest (the wax ester) is separated and specifically quantified using external standards, while polar lipids, such as cell membrane components that would contribute to total lipids in certain gravimetric methods, are excluded from the measurement by this approach.
Figure 3B (upper trace) shows a typical GC/FID chromatogram obtained from extraction of dried M. aquaeolei VT8 cells, while Fig. 4 shows the typical wax ester accumulation cycle found with M. aquaeolei VT8 when cells are grown as a batch culture. Cells grow exponentially with sufficient amounts of nitrogen to support replication until approximately 40 h, when all the available nitrogen in the medium has been consumed. Wax ester accumulation begins shortly after that, reaching a maximum quantity at about 100 h (generally about 10% of dry cell mass, as was found for the experiment shown). Following this, the culture enters a final stage in which wax esters in the cell begin to decline to very low levels after about 150 h. The rise and decrease in levels of wax esters in the cell during culture was reproducible (n > 5 independent experiments) and was responsible for the high sample-to-sample variance for wax concentrations shown in Fig. 3A, which was the result of a single-time-point analysis versus analysis at multiple time points, as was done in the batch culture here. The wax ester analysis shown in Fig. 4 was arbitrarily fit to a Gaussian curve for the sake of visualization only and is not meant to indicate that the wax ester accumulation cycle behaves precisely in this specific manner (though this works well for most of the batch cultures analyzed in our laboratory). The changes in transcriptional levels (reported as the fold change) of the genes coding for the fatty aldehyde reductase (farA) and fatty acyl-CoA reductase (acrB) are plotted along with the wax ester analysis in Fig. 4. Additionally, several reference genes coding for recombinase (recA), a medium alcohol dehydrogenase (adhM), and a fatty aldehyde dehydrogenase (aldF) and the 16S rRNA are included for comparison (12, 18, 20). Two of the reference samples (adhM and recA) showed only slight changes in transcription levels during this experiment, and 16S rRNA levels were also consistent. aldF was included, as it may be involved in the pathway necessary to oxidize extraneously added pentadecanol to pentadecanoic acid to produce the C30 wax ester found in the phenotype rescue experiments (Fig. 3B) (2, 18). Transcriptional levels of aldF were 100-fold higher during exponential growth but dropped during wax ester accumulation and remained low during the stage of wax ester depletion. Transcription of acrB was also elevated during exponential growth but rapidly declined during the initial phase of wax ester accumulation, while only farA showed elevated transcriptional levels during the wax ester accumulation phase of the culture (between 40 and 100 h), dropping only after wax esters had reached their peak concentration during the culture. Relative levels of farA, acrB, and aldF were all within 1-fold (Cp value within one cycle) of one another at the final time point, though they were approximately 8-fold (Cp values within two to three cycles) lower than in the adhM and recA reference samples.
Agreement between RT-qPCR and gene deletion experiments.
The results from transcriptional analysis of mRNA for the fatty aldehyde reductase and fatty acyl-CoA reductase correlate well with what was found in the gene deletion studies. The fatty aldehyde reductase transcription appears to be upregulated during lipid accumulation. The fatty acyl-CoA reductase transcriptional levels were elevated primarily during exponential growth under the conditions utilized here but dropped substantially once the cell entered into the wax ester accumulation stage. Thus, the deletion of the gene encoding the fatty acyl-CoA reductase (ΔacrB::kan) had very little effect on levels of wax esters. However, deletion of the gene coding for the fatty aldehyde reductase (ΔfarA::kan) dramatically decreased the levels of wax esters that accumulated but did not completely delete the wax ester accumulation phenotype.
The findings from these studies, which looked at features of these enzymes in vivo in the indigenous organism, contrast with what has been found previously for these fatty alcohol-producing enzymes through in vitro experiments. Studies with isolated enzymes indicate that the fatty acyl-CoA reductase is the more active of the two enzymes following purification (6, 7). Homologs to the fatty acyl-CoA reductase are also more prevalent in other model wax ester-accumulating species (Acinetobacter, Psychrobacter, and Rhodococcus), while the fatty aldehyde reductase is found less frequently (7). This was a key motivator to pursue these studies, as M. aquaeolei VT8 provides an ideal test case to analyze which fatty alcohol-yielding enzyme evolution has “selected” to optimize wax ester production when both options exist within the repertoire of enzymes in the cell. As mentioned previously, additional activities toward fatty acyl-CoA and fatty acyl-ACP have been reported for the fatty aldehyde reductase from M. aquaeolei VT8 since the initial characterization of this enzyme (4, 5). Additionally, others have recently demonstrated that the fatty aldehyde reductase (Maqu_2220, or NCBI accession number YP_959486) resulted in improved fatty alcohol yields when heterologously expressed in E. coli versus several other fatty acyl-CoA reductases (21).
Summary of findings.
These results demonstrate that while M. aquaeolei VT8 has incorporated redundancy within the wax ester production pathway to yield the fatty alcohols required for wax ester production, evolution has selected one specific branch of this pathway as the clear preferred route when producing wax esters from simple organic acids, such as succinate or citrate.
ACKNOWLEDGMENTS
This work is supported by a grant from the National Science Foundation to B.M.B. (award number 0968781) and the Microbial Engineering Program through the Biotechnology Institute at the University of Minnesota for partial support of E.M.L. Further support was provided through generous startup funds through the University of Minnesota.
We thank Erin Ray, Robert Willis, and Zeyuan Wu, who assisted in preliminary studies related to this work. We are grateful to Jim Hu for helpful suggestions in the selection of acceptable gene nomenclature.
Footnotes
Published ahead of print 6 September 2013
REFERENCES
- 1.Huu NB, Denner EBM, Ha DTC, Wanner G, Stan-Lotter H. 1999. Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated from a Vietnamese oil-producing well. Int. J. Syst. Bacteriol. 49:367–375 [DOI] [PubMed] [Google Scholar]
- 2.Barney BM, Wahlen BD, Garner E, Wei JS, Seefeldt LC. 2012. Differences in substrate specificities of five bacterial wax ester synthases. Appl. Environ. Microbiol. 78:5734–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer E, Webb EA, Nelson WC, Heidelberg JF, Ivanova N, Pati A, Edwards KJ. 2011. Genomic potential of Marinobacter aquaeolei, a biogeochemical “opportunitroph.” Appl. Environ. Microbiol. 77:2763–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofvander P, Doan TTP, Hamberg M. 2011. A prokaryotic acyl-CoA reductase performing reduction of fatty acyl-CoA to fatty alcohol. FEBS Lett. 585:3538–3543 [DOI] [PubMed] [Google Scholar]
- 5.McDaniel R, Behrouzian B, Clark L, Hattendorf D, Valle F. February 2013. Production of fatty alcohols with fatty alcohol forming acyl-CoA reductases (FAR). US patent 20,130,040,352
- 6.Wahlen BD, Oswald WS, Seefeldt LC, Barney BM. 2009. Purification, characterization, and potential bacterial wax production role of an NADPH-dependent fatty aldehyde reductase from Marinobacter aquaeolei VT8. Appl. Environ. Microbiol. 75:2758–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willis RM, Wahlen BD, Seefeldt LC, Barney BM. 2011. Characterization of a fatty acyl-CoA reductase from Marinobacter aquaeolei VT8: a bacterial enzyme catalyzing the reduction of fatty acyl-CoA to fatty alcohol. Biochemistry 50:10550–10558 [DOI] [PubMed] [Google Scholar]
- 8.Reiser S, Somerville C. 1997. Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J. Bacteriol. 179:2969–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 100:10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakermans C, Sloup RE, Zarka DG, Tiedje JM, Thomashow MF. 2009. Development and use of genetic system to identify genes required for efficient low-temperature growth of Psychrobacter arcticus 273-4. Extremophiles 13:21–30 [DOI] [PubMed] [Google Scholar]
- 11.Sonnenschein EC, Gärdes A, Seebah S, Torres-Monroy I, Grossart HP, Ullrich MS. 2011. Development of a genetic system for Marinobacter adhaerens HP15 involved in marine aggregate formation by interacting with diatom cells. J. Microbiol. Methods 87:176–183 [DOI] [PubMed] [Google Scholar]
- 12.Takle GW, Toth IK, Brurberg MB. 2007. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol. 7:50. 10.1186/1471-2229-7-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Röttig A, Steinbüchel A. 2013. Acyltransferases in bacteria. Microbiol. Mol. Biol. Rev. 77:277–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 15.Blomfield IC, Vaughn V, Rest RF, Eisenstein BI. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive Psc101 replicon. Mol. Microbiol. 5:1447–1457 [DOI] [PubMed] [Google Scholar]
- 16.Pelicic V, Reyrat JM, Gicquel B. 1996. Generation of unmarked directed mutations in Mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919–925 [DOI] [PubMed] [Google Scholar]
- 17.van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197–202 [DOI] [PubMed] [Google Scholar]
- 18.Ishige T, Tani A, Sakai Y, Kato N. 2000. Long-chain aldehyde dehydrogenase that participates in n-alkane utilization and wax ester synthesis in Acinetobacter sp. strain M-1. Appl. Environ. Microbiol. 66:3481–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stöveken T, von Landenberg P, Steinbüchel A. 2005. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol. Microbiol. 55:750–763 [DOI] [PubMed] [Google Scholar]
- 20.Wei J, Timler JG, Knutson CM, Barney BM. 2013. Branched-chain 2-keto acid decarboxylases derived from Psychrobacter. FEMS Microbiol. Lett. 346:105–112 [DOI] [PubMed] [Google Scholar]
- 21.Liu A, Tan X, Yao L, Lu X. 2013. Fatty alcohol production in engineered E. coli expressing Marinobacter fatty acyl-CoA reductases. Appl. Microbiol. Biotechnol. 97:7061–7071 [DOI] [PubMed] [Google Scholar]
- 22.Taylor LA, Rose RE. 1988. A correction in the nucleotide sequence of the Tn9O3 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mather MW, McReynolds LM, Yu CA. 1995. An enhanced broad-host-range vector for Gram-negative bacteria: avoiding tetracycline phototoxicity during the growth of photosynthetic bacteria. Gene 156:85–88 [DOI] [PubMed] [Google Scholar]