Abstract
Sacoglossans are characterized by the ability to sequester functional chloroplasts from their algal diet through a process called kleptoplasty, enabling them to photosynthesize. The bacterial diversity associated with sacoglossans is not well understood. In this study, we coupled traditional cultivation-based methods with 454 pyrosequencing to examine the bacterial communities of the chemically defended Hawaiian sacoglossan Elysia rufescens and its secreted mucus. E. rufescens contains a defense molecule, kahalalide F, that is possibly of bacterial origin and is of interest because of its antifungal and anticancer properties. Our results showed that there is a diverse bacterial assemblage associated with E. rufescens and its mucus, with secreted mucus harboring higher bacterial richness than entire-E. rufescens samples. The most-abundant bacterial groups affiliated with E. rufescens and its mucus are Mycoplasma spp. and Vibrio spp., respectively. Our analyses revealed that the Vibrio spp. that were highly represented in the cultivable assemblage were also abundant in the culture-independent community. Epifluorescence microscopy and matrix-assisted laser desorption–ionization mass spectrometry (MALDI-MS) were utilized to detect the chemical defense molecule kahalalide F on a longitudinal section of the sacoglossan.
INTRODUCTION
Mollusca is the largest marine phylum, with approximately 23% of all known named marine invertebrates (1), yet there are few studies assessing bacterial symbiosis associated with members of this phylum. The term symbiosis is used to describe the close and often long-term association of two or more organisms, as originally defined by de Bary (2). Symbiosis is used here in this broad sense as by Taylor et al. (3). Microbial symbionts can provide nutrients to the host, assist in structural development, cause disease, protect the host from infectious microbes, or initiate chemical responses to the environment (4–7), among other roles. In addition to impacting the hosts, marine microbial symbionts can also be valuable resources for drug discovery. In some cases, the symbionts may be the true producers of compounds found in invertebrates (8–10). These bioactive products often possess anticancer and antimicrobial properties (11–13).
Most molluscan-associated bacterial community studies highlight the classes Bivalva and, less commonly, Gastropoda and, in many cases, focus on mollusks living in extreme environments. Sulfur-oxidizing and methanotrophic symbionts located in the ctenidia, the comblike respiratory gills of mollusks, are now recognized as a widespread nutritional strategy established in five separate families of marine bivalves that occupy habitats from intertidal zones to hadal depths (14–17). Hydrothermal vent gastropods from the same family in two separate locations harbor sulfur-oxidizing epsilon- and gammaproteobacteria (18).
Many mollusks live in several seawater habitats where bacteria are extremely abundant, achieving densities of up to 106 cells per ml of seawater (19). Mollusks do not have known acquired immunological memory (20, 21). These factors thus suggest that mollusks may have developed defense strategies to protect themselves from exposure to a high density of microorganisms, including potential pathogens. The use of secondary metabolites is well documented in the phylum Mollusca as part of their communication systems (22–24), predatory behavior (25, 26), and defensive secretions (27–30). The abundance, diversity, and chemical potential of mollusks make this phylum of interest for studies of microbial symbiosis.
About 90% of molluscan biological and chemical diversity is found in the class Gastropoda (1), with opisthobranchs being one of the most diverse groups of gastropods. Opisthobranchs reside in a wide range of marine habitats (31), and they possess biological features related to foraging or defensive strategies that are unique or rare in the animal kingdom (32). These strategies are common in the sacoglossans (a suborder of the opisthobranchs), also known as sea slugs. Sacoglossan means “sap sucking,” and these mollusks possess the ability to incorporate and use intact chloroplasts from their algal diet (33) by a process called kleptoplasty. Sacoglossans are the only metazoans known to photosynthesize, and the range of photosynthetic ability varies among sea slugs (34). The herbivorous sacoglossan sea slug Elysia chlorotica retains functional plastids in the cells lining the digestive tract for several months in the absence of the algal prey and continues to photosynthesize (33, 35). One hypothesis proposed to explain how plastids continue to function within the slug cells in the absence of the algae is horizontal gene transfer (HGT) of essential photosynthetic genes from the algal diet to the sea slug genome. Pierce et al. analyzed the transcriptome of E. chlorotica and reported the presence of at least 101 chloroplast-encoded gene sequences and 111 algal transcripts, 52 of which were nuclear genes (36). However, the mechanisms for long-term maintenance of photosynthesis are still unresolved, as recent genomic data from the E. chlorotica germ line (egg DNA) and from its algal prey, Vaucheria litorea, failed to provide evidence for alga-derived HGT into the germ line of the sea slug (37).
Many sea slugs are able to synthesize chemical precursors or incorporate secondary metabolites from their algal diet in a process called kleptochemistry (38). These sea slugs feed on specific algae that possess chemical compounds with antimicrobial, antifouling, and feeding-deterrent abilities (39, 40). These compounds or their modified forms can be detected in the slugs, specifically in the mucous secretion, where the compounds are thought to function in defense (41). Recently, 16S rRNA-based metagenomics of the bacterial communities associated with the sacoglossan E. chlorotica, its algal prey V. litorea from their native environments, and E. chlorotica after being starved of algal prey and bred in the laboratory revealed diverse bacterial profiles that varied between populations and among all conditions except for the laboratory-bred samples (42). These results provide an example of dense bacterial communities associated with a sacoglossan. However, from our current knowledge, there are no bioactive compounds that have been established as being produced by the sacoglossan or bacterial partners in this association.
The sacoglossan Elysia rufescens and the alga Bryopsis species form another sacoglossan-alga association, and in this case, the sacoglossan is chemically protected against fish predators by the deterrent properties of kahalalide F (KF) (43). KF is a promising anticancer compound that has been extracted from both Elysia rufescens and Bryopsis species (44). The bioactive compounds associated with E. rufescens are well investigated (45–47), and preliminary data revealed that two strains of Vibrio species isolated from E. rufescens produce the compound KF, with production confirmed by liquid chromatography-mass spectrometry (LCMS) and nuclear magnetic resonance (NMR) (48). However, recent attempts to isolate KF from cultured Vibrio strains have given inconsistent results, and more studies are needed to unequivocally determine the organism(s) responsible for the production of KF. In this study, we seek to better understand the bacteria associated with E. rufescens. Since sacoglossans are known to secrete bioactive compounds in their mucus (41), the bacterial communities associated with E. rufescens mucus, as well as with E. rufescens itself, were investigated.
We used a comprehensive approach by coupling traditional cultivation with cloning and deep pyrosequencing of 16S rRNA gene amplicons to characterize and compare the bacterial communities associated with E. rufescens and its secreted mucus.
MATERIALS AND METHODS
Sample collection and processing.
E. rufescens sea slugs were collected by snorkel at Black Point Bay, Honolulu, HI (21°15′N, 157°47′W), in March 2010. Sixteen sacoglossan individuals were placed in separate plastic collection bags filled with the surrounding seawater and transported to a laboratory for processing within 1 h of collection. Samples were rinsed 3 times with sterile 1× phosphate-buffered saline (PBS) to remove transiently and loosely attached bacteria. Five E. rufescens individuals were transferred to a sterile 50-ml conical tube (Corning, Tewksbury, MA) and removed after secreting copious amounts of mucus. The secreted mucus left after the removal of the sea slugs was collected and pooled for culture-based and molecular analyses. This process was repeated for a second set of five E. rufescens individuals, and the secreted mucus from these individuals was treated as a separate sample. One milliliter of the two separate secreted-mucus samples was used for bacterial cultivation, and the remaining mucus was immediately stored at −80°C and later lyophilized (Labconco, Kansas City, MO) for molecular analysis. Three separate individuals of E. rufescens were used for bacterial cultivation, and an additional set of three individuals were immediately stored at −80°C and later lyophilized (Labconco, Kansas City, MO) for molecular analysis. Water samples were collected near the sea slugs in sterile 20-liter containers, and approximately 5 liters was filtered through three 0.22-μm-pore-size Sterivex filters (Millipore, Billerica, MA) for each water sample. The Sterivex filters were immediately stored at −20°C for isolation of nucleic acids.
Bacterial cultivation and identification.
E. rufescens was homogenized in 9 ml of sterile 1× PBS using a mortar and a pestle. The homogenate was used to create a 10-fold dilution series of which 100-μl aliquots were plated on marine agar 2216 (Becton, Dickinson, Sparks, MD). These plates were incubated for 5 days at 30°C, and morphotypes were randomly picked for identification. The same procedure was used for the secreted mucus from E. rufescens. DNA was extracted from the bacterial isolates obtained from E. rufescens and secreted mucus as described by Montalvo et al. (49). Briefly, DNA from cultured representatives was extracted using the Mo Bio UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. Bacterial 16S rRNA gene fragments were PCR amplified with eubacterial primers 27F and 1492R (50). Reactions were run in a PTC-200 cycling system (MJ Research, Waltham, MA) using the following cycling parameters: 300 s of denaturation at 94°C, followed by 25 cycles of 30 s at 92°C (denaturing), 120 s at 46°C (annealing), and 90 s at 72°C (elongation), with a final extension at 72°C for 300 s. The PCR products were sequenced on an ABI 3130 XL genetic analyzer (Applied Biosystems, Foster City, CA), using the 27F primer. Sequences were classified with the RDP classifier (http://rdp.cme.msu.edu/). The closest relatives for each sequence were obtained from the GenBank database using the BLASTn tool (http://blast.ncbi.nlm.nih.gov/).
Total genomic DNA extraction.
Total genomic DNA from lyophilized E. rufescens and its mucus was extracted using the Mo Bio PowerBiofilm DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions, with the following modifications: a vortex adaptor (Scientific Industries, Inc., Bohemia, NY) was used for cell lysis, and doubling solution BF3 to increase amplification efficiency. DNA was extracted from the Sterivex filters obtained from seawater samples using the protocol described by Somerville et al. (51).
Bacterial 16S rRNA gene clone library construction and identification.
Bacterial 16S rRNA gene clone libraries were constructed from lyophilized E. rufescens and secreted mucus from E. rufescens. Bacterial 16S rRNA gene fragments were PCR amplified with universal primers 27F and 1492R (50), and clone libraries were constructed as described by Montalvo et al. (49). PCR products were gel visualized and purified by ethanol precipitation. Purified PCR products were ligated into pCR-XL-TOPO vector and transformed into One Shot TOP10 chemically competent Escherichia coli cells using the TOPO XL PCR cloning kit (Invitrogen-Life Technologies, Carlsbad, CA). Clones were sequenced as described above.
DGGE of bacterial communities.
Denaturing gradient gel electrophoresis (DGGE) was performed on PCR-amplified genomic DNA from E. rufescens, secreted mucus from E. rufescens, and surrounding-seawater samples using a 195-bp region corresponding to positions 341 and 534 in the 16S rRNA gene of E. coli using P2 and P3 primers as described by Muyzer et al. (52). DGGE was performed by using the DCode system (Bio-Rad, Hercules, CA) on an 8% (wt/vol) polyacrylamide gel with a denaturing gradient of 40 to 70% in 1× Tris-acetate-EDTA (TAE). Electrophoresis was performed for 16 h at 60 V at 60°C. The gel was stained with SYBR green in a 1× TAE staining bath and visualized with a Typhoon 9410 image system (Amersham Biosciences, Piscataway, NJ). DGGE banding patterns were compared using BioNumerics 7.0 software (Applied Maths NV, Sint-Martens-Latem, Belgium). An unweighted pair group with arithmetic mean (UPGMA) dendrogram based on the Dice similarity coefficient was generated to show the similarities in banding patterns across samples.
Pyrosequencing of barcoded 16S rRNA gene amplicons.
Total genomic DNA was extracted as described for the construction of clone libraries. Hypervariable regions V1 to V3 of the 16S rRNA gene fragments were amplified with primers 27F and 534R. DNA amplification of the 16S rRNA genes was performed using high-fidelity Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) and 20 to 50 ng of template DNA in a total reaction mixture volume of 25 μl, following the Platinum Taq product protocol. Reactions were run in a PTC-200 cycling system (MJ Research, Waltham, MA) using the following cycling parameters: 180 s of denaturation at 94°C, followed by 30 cycles of 30 s at 94°C (denaturing), 30 s at 46°C (annealing), and 45 s at 72°C (elongation), with a final extension at 72°C for 300 s. Negative controls were included for each amplification and barcoded primer pair, including amplification without template DNA. The presence of amplicons was confirmed by gel electrophoresis on a 2% agarose gel and staining with ethidium bromide. PCR products were quantified using a Quant-iT PicoGreen double-stranded DNA (dsDNA) assay. Equimolar amounts (50 ng) of the PCR amplicons were mixed in a single tube. Amplification primers and reaction buffer were removed using the AMPure kit (Agencourt, Beverly, MA), and purified amplicon mixtures sequenced by Roche/454 FLX pyrosequencing using 454 Life Sciences primer A by the Institute for Genome Sciences, University of Maryland School of Medicine, using protocols recommended by the manufacturer. All 16S rRNA gene amplicons were sequenced as part of the same pool in the same sequencing reaction.
Sequence processing and estimation of bacterial diversity.
16S rRNA gene sequences were processed using a combination of QIIME (53) and mothur (54). Pyrosequences were binned using sample-specific barcodes and trimmed by removal of barcodes and primer sequences using default parameters in QIIME (http://qiime.org/scripts/split_libraries.html). Chimeras were removed with UCHIME (55). Pyrosequences were rigorously filtered as described by Huse et al. (56), using mothur. Sequences were removed from the analysis if they were <400 bp and contained ambiguous characters. To ensure that the V1-to-V3 region was targeted, all sequences (including isolates and clones) were cut to 500 bp for the analysis. Following quality filtration, sequences were clustered at 3% with the average neighbor-joining method. The Yue and Clayton theta similarity coefficient was used to compare bacterial community structure (54, 57). Phylogenetic analyses were performed with the ARB software package (58), and phylogenetic trees were constructed using the neighbor-joining algorithm (59). Bootstrap values were generated using PHYLIP with 1,000-replicate data sets.
Epifluorescence imaging and MALDI-MS analysis.
Matrix-assisted laser desorption–ionization mass spectrometry (MALDI-MS) imaging of a longitudinal 14-μm-thick section of an individual E. rufescens was performed as described by Simmons et al. (60). The epifluorescence image obtained at 590 nm was overlaid by the MALDI image at m/z 1,500 to obtain differential localization of the known compound KF.
16S rRNA gene sequence accession numbers.
The bacterial 16S rRNA gene sequences obtained in this study have been deposited on the public MG-RAST server (http://metagenomics.anl.gov) under MG-RAST accession numbers 4534702.3 to 4534710.3. All sequences can be downloaded by accessing the Elysia rufescens Associated Bacteria Study using the MG-RAST Project link (http://metagenomics.anl.gov/linkin.cgi?project=5791).
RESULTS
Bacterial community analysis and methods.
Bacterial communities were characterized with the following four different methods, which provided consistent results: (i) DGGE, (ii) short-fragment 16S rRNA gene amplicon pyrosequencing, (iii) cloning by partial-length 16S rRNA gene amplicon Sanger sequencing, and (iv) cultivation and characterization of isolates by partial-length 16S rRNA gene Sanger sequencing. All 16S rRNA gene assignments were made at the species level (distance = 0.03).
The bacterial community structure associated with three individuals of E. rufescens (ER1 to ER3), two samples of its mucus (M1 and M2), and surrounding water samples (W1 to W3) were initially compared using DGGE coupled with BioNumerics clustering analysis (see Fig. S1A and B in the supplemental material). Although DGGE has limited sensitivity for detection of rare community members, cluster analysis showed that bacterial communities in replicate samples of E. rufescens, mucus, and water were consistent, while the communities in the E. rufescens individual and mucus were more similar to each other than to those from water samples. The DGGE analysis indicated major differences in the bacterial communities associated with E. rufescens and the surrounding seawater.
A total of 29,063 16S rRNA gene sequences from three E. rufescens individuals and two mucus samples were then analyzed (Table 1). Data sets from these three individuals of E. rufescens contained 10,405 (ER1), 9,108 (ER2), and 1,230 (ER3) 16S rRNA gene pyrosequences and were together assigned to 1,225 operational taxonomic units (OTUs) after the removal of 6,110 chloroplast-derived sequences (Table 1; see also Table S1 in the supplemental material). Data sets from the two mucus samples from E. rufescens contained 6,533 (M1) and 1,539 (M2) 16S rRNA gene pyrosequences and were together assigned to 1,873 OTUs after the removal of 5 chloroplast-derived sequences (Table 1; see also Table S1). The rarefaction curves did not reach a plateau for any sample (see Fig. S2 in the supplemental material), indicating that further sampling is needed to determine the full diversity of the bacterial communities associated with E. rufescens and its secreted mucus at the species level.
Table 1.
Number of 16S rRNA gene sequences analyzed and number of OTUs observed from E. rufescens and its mucus at 3% clusteringa
| Source of genetic material | Total no. of sequences | No. of chloroplast-derived sequences removed | No. of OTUs |
|---|---|---|---|
| Elysia rufescens | |||
| Isolates | 35 | — | 6 |
| Clones | 96 | 80 | 6 |
| Pyrosequences | 20,743 | 6,110 | 1,225 |
| Mucus | |||
| Isolates | 30 | — | 7 |
| Clones | 87 | — | 15 |
| Pyrosequences | 8,072 | 5 | 1,873 |
Distance of 0.03.
For the characterization of samples by partial-length 16S rRNA gene amplification, 96 clones containing 16S rRNA genes were analyzed from E. rufescens, of which only 16 were not chloroplast derived and were assigned to 6 OTUs, while 87 clones were analyzed from the secreted mucus, which did not contain any chloroplast-derived reads and were assigned to 15 OTUs (Table 1).
Cultivable isolates from E. rufescens and mucus samples were randomly picked and identified by partial-length sequencing. These isolates were assigned to 6 OTUs for E. rufescens and 7 OTUs for secreted mucus (Table 1).
Abundant bacterial groups.
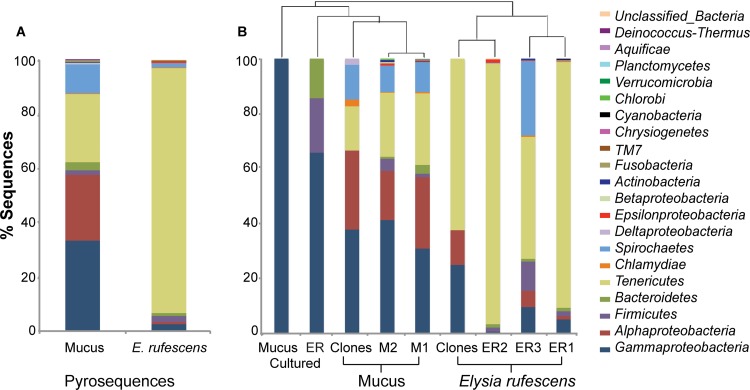
16S rRNA gene sequences affiliated with the phylum Tenericutes were abundant in the bacterial communities of both E. rufescens and its mucus (90% and 25% of the sequences, respectively) (Fig. 1A). The three individuals of E. rufescens varied greatly with respect to the percentages of sequences assigned to different bacterial phyla, but Tenericutes was the most-abundant phylum associated with all three E. rufescens individuals, including the clones (Fig. 1B). Gammaproteobacteria was the most-abundant bacterial group associated with the mucus pyrosequences (33%) and clones (38%) (Fig. 1A and B). Alphaproteobacteria (24%) and Spirochaetes (11%) were also abundant in the mucus of E. rufescens. The secreted-mucus samples were similar with respect to the percentages of sequences assigned to bacterial taxa, and Gammaproteobacteria was the major bacterial group in both replicates. All isolates cultured from secreted mucus were assigned to Gammaproteobacteria, and they represented over 60% of the isolates cultured from E. rufescens (Fig. 1B).
Fig 1.
Percentages of total sequences representing various bacterial phyla. Combined pyrosequences from secreted mucus from E. rufescens and entire E. rufescens (A) and from all samples (B). OTUs were defined by 3% sequence difference from the nearest neighbor. Groups are shown in the order in which they are listed.
Classification of abundant bacterial groups Tenericutes and Gammaproteobacteria.
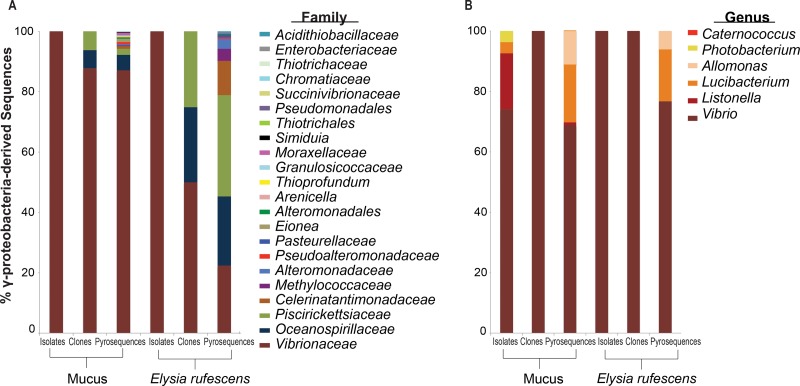
All of the Tenericutes-derived sequences were assigned to one genus, Mycoplasma. At least 50% or more of the Gammaproteobacteria-derived sequences belong to the family Vibrionaceae for all samples and methodologies except the pyrosequences of E. rufescens (Fig. 2A). However, Vibrionaceae was still abundant in the pyrosequences of E. rufescens, comprising 22% of the Gammaproteobacteria-derived sequences (Fig. 2A). Piscirickettsiaceae and Oceanospirillaceae were also abundant in the pyrosequences of E. rufescens (34% and 23%, respectively), as well as in the clones, but were rare in the mucus pyrosequences and clones. At the genus level, over 65% of the Gammaproteobacteria-derived sequences belonged to Vibrio spp. for all samples and methods (Fig. 2B).
Fig 2.
Percentages of total Gammaproteobacteria-derived sequences from secreted mucus from E. rufescens and entire E. rufescens. Gammaproteobacteria-derived sequences are classified at the family (A) and genus (B) level. OTUs were defined by 3% sequence difference from the nearest neighbor. Groups are shown in the order in which they are listed.
Shared OTUs.
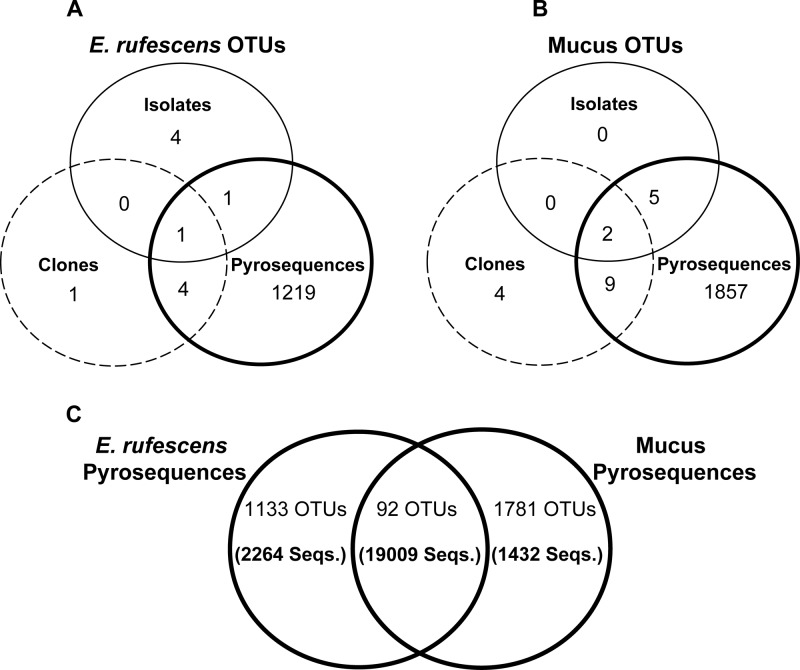
Gammaproteobacteria (Vibrio spp.) were the only OTUs shared across all samples and method types. There were a total of two OTUs shared by the isolates and pyrosequences of E. rufescens and one OTU shared between the isolates and clones of E. rufescens, all classified as Vibrio spp. The clones and pyrosequences of E. rufescens shared 5 OTUs belonging to Gammaproteobacteria (4 OTUs) and Mycoplasma (1 OTU) (Fig. 3A; see also Table S2 in the supplemental material). E. rufescens samples shared one OTU that was found in the cultured isolates, clones, and pyrosequences and was identified as ER-OTU-1, Vibrio sp. (Fig. 4). This OTU was 100% identical to Vibrio sp. strain UST061013-043 (GenBank accession number EF587982).
Fig 3.
Shared OTUs from entire E. rufescens (A), mucus secreted from E. rufescens (B), and pyrosequences from E. rufescens and its mucus (C). The numbers of shared pyrosequences are given in parentheses. OTUs were defined by 3% sequence difference from the nearest neighbor.
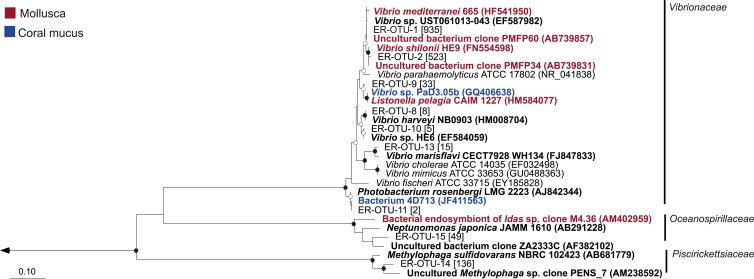
Fig 4.
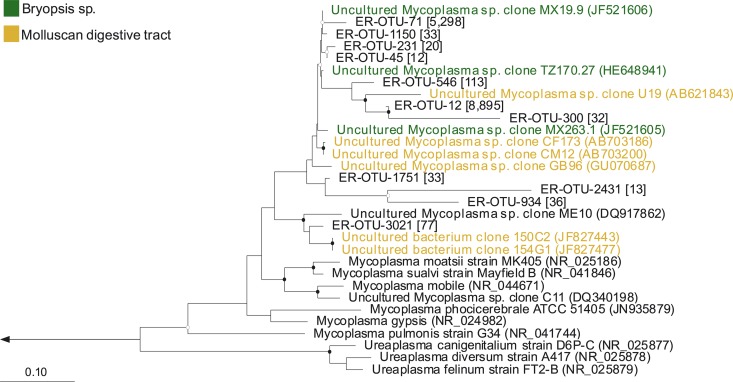
Neighbor-joining tree of Gammaproteobacteria-derived shared OTUs. The number of sequences in each OTU is listed in brackets, and the nearest neighbors in boldface. The nearest neighbors highlighted in red were isolated from mollusks, while the nearest neighbors highlighted in blue were isolated from the mucus of a coral. Bootstrap values (neighbor-joining method, 1,000 replicates) are indicated by closed circles (values >90%) and open circles (values >65%). The arrow goes to an outgroup, Streptomyces albolongus (AB184425). The scale bar represents 10% sequence divergence.
The secreted-mucus isolates and pyrosequences shared 7 OTUs, and the isolates and clones shared 2 OTUs; all were classified as Gammaproteobacteria, and 7 of the 9 OTUs were Vibrio spp. (Fig. 3B; see also Table S2 in the supplemental material). The clones and pyrosequences of the mucus shared 11 OTUs, including Gammaproteobacteria (5 OTUs), Alphaproteobacteria (2 OTUs), Spirochaetes (2 OTUs), Chlamydia (1 OTU), and Tenericutes (1 OTU). Secreted-mucus samples shared 2 OTUs found in the cultured isolates, clones, and pyrosequences that were identified as ER-OTU-1 and ER-OTU-2 (Fig. 4). The closest relatives in GenBank of ER-OTU-2 are an uncultured bacterium designated clone PMFP34 (GenBank accession number AB739831), isolated from the mollusk Pinctada margaritifera, and Vibrio shilonii HE9 (GenBank accession number FN554598), isolated from the gut of the mollusk Haliotis diversicolor. Both of the closest relatives share 99% identity to ER-OTU-2.
The pyrosequences of E. rufescens and its mucus shared 92 OTUs, including 27 OTUs assigned to Mycoplasma and 20 OTUs assigned to Gammaproteobacteria (Fig. 3C; see also Table S2 in the supplemental material). These 92 shared OTUs contain over 19,000 sequences and represent nearly 84% of the total pyrosequences associated with E. rufescens and its mucus.
Phylogeny of shared Gammaproteobacteria-derived OTUs.
Nine Gammaproteobacteria-derived OTUs were shared across multiple methodologies and samples, and six of those OTUs were closely related to Vibrio spp. (Fig. 4). The most dominant OTUs of this family include ER-OTU-1 (935 sequences), found in all samples, and ER-OTU-2 (523 sequences), found in all samples except the E. rufescens clones. These two OTUs represented 58% and 32% of the total sequences assigned to Vibrio spp. ER-OTU-9, ER-OTU-8, and ER-OTU-11 were exclusively found associated with the mucus samples, and two of the three OTUs' closest relatives were isolated from the mucus of a coral or from mollusks (Fig. 4). The closest relative to ER-OTU-13 based on BLAST analysis is Vibrio marisflavi, and this OTU was found in the clone and pyrosequence analyses of the E. rufescens bacterial community as well as by the pyrosequence analysis of the mucus-associated bacterial community. There was one Gammaproteobacteria-derived shared OTU found in the family Oceanospirillaceae, ER-OTU-15 (49 sequences), and the sequences from this OTU represented 22% of the total sequences in this family. This was a novel shared OTU, with 92% identity to an uncultured bacterial clone and 89% identity to the cultured representative Neptunomonas japonica. One Gammaproteobacteria-derived shared OTU, ER-OTU-14 (136 sequences), belonged to the family Piscirickettsiaceae, and the sequences affiliated with this OTU represented 67% of the total sequences in the family. This was also a novel OTU, with 95% identity to an uncultured Methylophaga sp. clone and 91% identity to the closest cultured relative, Methylophaga sulfidovorans.
Phylogeny of dominant Mycoplasma-derived OTUs.
Eleven OTUs dominated (>10 sequences) the abundant bacterial group Tenericutes, and all OTUs were Mycoplasma spp. Mycoplasma-derived OTUs clustered between common shared relatives, uncultured Mycoplasma sp. clones isolated from Bryopsis sp., and uncultured Mycoplasma sp. clones isolated from the digestive tract of mollusks (Fig. 5). The most-dominant shared OTUs, ER-OTU-71 (5,298 sequences) and ER-OTU-12 (8,895 sequences), together represented 93% of the total sequences assigned to Mycoplasma and were found in both the cloned and pyrosequenced samples of E. rufescens and its mucus. ER-OTU-71 was 97% identical to the uncultured Mycoplasma sp. clone MX19.9 (GenBank accession number JF521606), isolated from Bryopsis sp., with only 92% coverage, while ER-OTU-12 was 90% identical to the uncultured Mycoplasma sp. clone U19 (GenBank accession number AB621843), isolated from the abalone Haliotis gigantea. Mycoplasma-derived OTUs clustered with uncultured Mycoplasma sp. clones, were distantly related to well-known Mycoplasma type strains, and did not group with the only other Mycoplasmatacea genus, Ureaplasma.
Fig 5.
Neighbor-joining tree of the dominant Mycoplasma-derived OTUs. The number of sequences in each OTU is listed in brackets, and the nearest neighbors are in boldface. The nearest neighbors highlighted in green were isolated from Bryopsis species, while the nearest neighbors highlighted in yellow were isolated from the digestive tract of a mollusk. Bootstrap values (neighbor-joining method, 1,000 replicates) are indicated by closed circles (values >90%) and open circles (values >65%). The arrow goes to an outgroup, Nitrosomonas europaea (AB070982). The scale bar represents 10% sequence divergence.
Epifluorescence and MALDI-MS imaging.
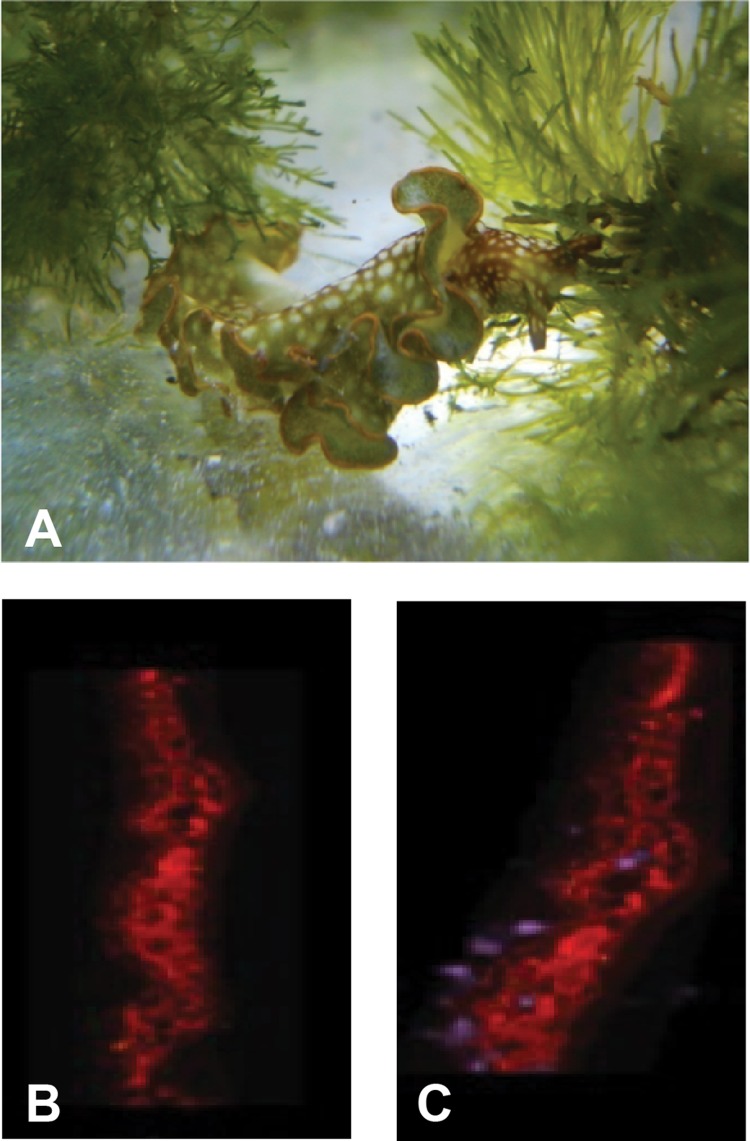
Imaging techniques confirmed the presence of KF in the sea slug, E. rufescens. The sea slug and its algal-diet Bryopsis species form a close association (Fig. 6A). Epifluorescence microscopy revealed the autofluorescence of chloroplasts associated with the sacoglossan (Fig. 6B). Autofluorescence imaging overlaid by MALDI-MS imaging revealed the presence of the KF compound in the outer region of an E. rufescens individual (Fig. 6C). It was not possible to distinguish whether KF was present in the outer tissue of the E. rufescens or in the mucus surrounding the mollusk.
Fig 6.
(A) E. rufescens feeding on algal-diet Bryopsis species in the Institute of Marine and Environmental Technology (IMET) Aquaculture Research Center (ARC). (B and C) Autofluorescence of Elysia rufescens 14-μm longitudinal thin section at 590 nm. (C) Autofluorescence is overlaid by MALDI image, m/z 1,500 (purple), indicating presence of compound with mass spectrum consistent with kahalalide F.
DISCUSSION
Our study provided insights into the bacterial communities associated with E. rufescens and its secreted mucus by using a variety of methods, which consistently uncovered the presence of the same abundant bacterial groups. The results presented here revealed that, although E. rufescens and its secreted mucus are hosts to many of the same bacterial groups, the bacterial communities are not identical. Two bacterial groups, Tenericutes (Mycoplasma spp.) and Gammaproteobacteria (mainly Vibrio spp.), were the most abundant in the bacterial communities associated with entire E. rufescens and secreted mucus from E. rufescens, respectively. Vibrio spp. were the most-abundant cultured bacteria and were also abundant in the pyrosequences. Notably, the phylogenetic analysis revealed that, in most cases, the closest relatives of the shared OTUs assigned to Vibrio spp. were isolated from other mollusks or the mucus layer of invertebrates, whereas the closest relatives of Mycoplasma-derived OTUs were uncultured clones obtained from the algal-diet Bryopsis species of E. rufescens or the gut of other mollusks that are known to forage on algae (Fig. 4 and 5).
Bacteria belonging to the genus Vibrio are well recognized for their symbiotic relationships, pathogenicity, and the production of bioactive secondary metabolites (61–64). Opisthobranchs are known to secrete metabolites in their mucus as a defense mechanism (41). The abundance and concentration of vibrios in the mucus combined with their well-documented metabolic flexibility makes it tempting to speculate that they are candidates to be the producers of the defense chemical KF found in E. rufescens. The MALDI-MS imaging of KF does reveal that the compound is present in the outer region of the E. rufescens individual, conceivably in the mucus. However, it is important to note that we cannot be certain that the KF is in the mucus and there is no direct connection between the presence of KF in the outer region of the mollusk and the dominance of vibrios in the mucus revealed in this study. Also, no metabolic functions were determined for the vibrios.
Mycoplasma spp. are a diverse group of bacteria and are known for pathogenicity in a wide range of mammalian hosts (65). Recent studies reveal that they are members of microbial communities associated with isopods, fish, and abalone, where they may provide nutritional benefits to the host (66–68). In the bacterial community analysis of E. rufescens and its mucus, all sequences assigned to Tenericutes belong to the genus Mycoplasma. However, no Mycoplasma organism was cultured, and it is not known if the sequenced Mycoplasma comes from the algal-diet Bryopsis species or from E. rufescens. The closest relatives of the dominant OTUs were uncultured Mycoplasma spp. clones obtained from Bryopsis spp. or the digestive tract of other mollusks, specifically, abalones of the genus Haliotis. A study performed on Haliotis species found that the animals could feed on a wide range of algae (69). The findings of Erasmus et al. (70) suggest that bacterial inhabitants of the digestive tract in Haliotis midae possess a polysaccharide-degrading ability to assist in breaking down algal diets. More specifically, the closest relative of the uncultured Mycoplasma sp. clone obtained from Bryopsis sp. in a study performed on the Mexican coast was the uncultured Mycoplasma sp. clone GB96 isolated from Haliotis diversicolor (71), and this is also one of the closest relatives of the dominant Mycoplasma-derived OTUs in this study (Fig. 5). The presence of Mycoplasma species in the digestive bacterial community of H. diversicolor has also been hypothesized to be algal food related (68, 71). It appears that the presence of Mycoplasma may be alga specific and might result from algal grazing. Consistent with previous studies, Mycoplasma could be a symbiont acquired through feeding on algal diets; however, this study did not examine any metabolic functions of the Mycoplasma.
Because the whole samples of E. rufescens that were processed for community analysis also contained mucus, higher richness was expected in these whole-E. rufescens samples than in secreted mucus alone. However, higher bacterial richness was found in secreted-mucus samples than in whole-E. rufescens samples (Table 1 and Fig. 1). One possibility for less bacterial richness in whole E. rufescens is the uneven distribution of bacterial groups sampled in the community. Almost all of the sequences represented (93%) were chloroplast derived or belonged to a single genus, Mycoplasma, whereas the mucus samples contained more rare phylotypes. This large proportion assigned to only two groups skews the overall bacterial richness and most likely is a result of the intimate association between E. rufescens and its diet, Bryopsis species. This symbiotic association enables E. rufescens to sequester intact plastids (33), hence the abundance of chloroplast-derived sequences. The alga-sacoglossan relationship may also allow for the exchange of specific bacterial groups, such as Mycoplasma sp., which as previously noted may result from algal grazing.
Recently, the first insights were provided into the characterization of the bacterial communities from two different Bryopsis spp. from the Mexican Pacific coast (71). 16S rRNA gene analysis revealed the presence of Mycoplasma sp. clones detected in several Bryopsis samples collected hundreds of kilometers apart. A follow-up study by the same group revealed that Bryopsis harbors rather stable endophytic bacteria, which showed little variability after a 1-year cultivation of the algal samples (72). Moreover, the same uncultured Mycoplasma sp. clone remained present over the entire algal cultivation period.
There were a total of 20,743 pyrosequences assessed from E. rufescens. Nearly 30% were chloroplast-derived (6,110 sequences) and were removed from the analysis. Of the remaining sequences, 90% were assigned to Mycoplasma species (13,207 sequences). There were only five chloroplast-derived sequences identified and removed from the secreted-mucus samples. Mycoplasma spp. were present in both the whole-E. rufescens and secreted-mucus pyrosequences. However, 87% of the total Mycoplasma sequences were from E. rufescens samples. The closest known relatives of the most-dominant OTUs, ER-OTU-71 (5,298 sequences) and ER-OTU-12 (8,895 sequences), were the same uncultured Mycoplasma spp. clones identified previously from Bryopsis spp. off the Mexican coast (71). Those OTUs represented 93% of total sequences assigned to the genus Mycoplasma. Further sequencing effort may allow for greater bacterial richness to be observed in entire E. rufescens compared with its secreted mucus. However, the 92 OTUs shared between the pyrosequences of E. rufescens and its secreted mucus captured the dominant OTUs of both communities and represent nearly 84% of the total pyrosequences (Fig. 3C). This demonstrates that, although secreted mucus appears to have higher bacterial richness, E. rufescens and its mucus share the same dominant bacterial groups.
The mucus samples not only had greater bacterial diversity than whole E. rufescens, but there also appear to be some rare specific Vibrio spp. that are only found in both the cultured and pyrosequenced communities associated with secreted-mucus samples. There is one classic example of how mucus secretion assists in the recruitment and selection of specific bacteria. In the bacterial symbiosis between the Hawaiian mollusk Euprymna scolopes and the bioluminescent symbiont Vibrio fischeri, host-derived mucus secretion is used to concentrate V. fischeri near sites of light organ colonization during initiation of the symbiosis (73). A further study revealed that mucus secretion and ciliated fields are used to recruit nonsymbiotic bacteria as well as V. fischeri over the first 24 to 48 h. However, once V. fischeri successfully colonizes, mucus secretion is suppressed, nonspecific bacteria are no longer recruited or present, and only specific strains of V. fischeri permanently colonize the light organ (74). In the case of E. rufescens, perhaps the mucus also selects for specific bacteria that provide functional roles, as illustrated in the example above. Indeed, copious mucus that bathes and protects sacoglossans is well known (35, 75, 76) and has been a challenge for many working with these organisms (77). However, the dynamics of the mucus layer are not yet known and may possess properties that can influence bacterial composition.
To understand the bacterial community associated with E. rufescens and its mucus, we coupled traditional cultivation with cloning and pyrosequencing. Our analysis revealed that the most-abundant bacteria captured by culturing, Vibrio spp., were also abundant in the culture-independent community, demonstrating the potential importance of Vibrio spp. as symbionts of this complex community. While 454 data provided the needed insight into Mycoplasma-associated bacteria, culturing provided a more-comprehensive view of the bacterial community associated with E. rufescens when coupled with next-generation sequencing.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Rappé and the Hawaii Institute of Marine Biology at Coconut Island for providing laboratory space in Hawaii. We thank Naomi Montalvo for bioinformatics advice and Jindong Zan and Leah Blasiak for comments on the manuscript.
J.D. thanks Tamara Hamilton, National Science Foundation, Louis Stokes Alliance for Minority Participation (LSAMP) Bridge to the Doctorate graduate fellowship for support. Rosemary Jagus and the National Oceanic Atmospheric Administration, Living Marine Resource Cooperative Science Center (LMRCSC) Graduate Fellowship are thanked for scholarship funding for J.D.
Footnotes
Published ahead of print 6 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01568-13.
REFERENCES
- 1.Benkendorff K. 2010. Molluscan biological and chemical diversity: secondary metabolites and medicinal resources produced by marine molluscs. Biol. Rev. Camb. Philos. Soc. 85:757–775 [DOI] [PubMed] [Google Scholar]
- 2.De Bary HA. 1879. Die erscheinung der symbiose. Privately printed, Strasbourg [Google Scholar]
- 3.Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmström C, Kjelleberg S. 1994. The effect of external biological factors on settlement of marine invertebrate and new antifouling technology. Biofouling 8:147–160 [Google Scholar]
- 5.Donachie SP, Zdanowski MK. 1998. Potential digestive function of bacteria in krill Euphausia superba stomach. Aquat. Microb. Ecol. 14:129–136 [Google Scholar]
- 6.Mearns-Spragg A, Bregu A, Boyd KG, Burgess JG. 1998. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett. Appl. Microbiol. 27:142–146 [DOI] [PubMed] [Google Scholar]
- 7.Hill RT. 2004. Microbes from marine sponges: a treasure trove of biodiversity for natural products discovery, p 177–190 In Bull AT. (ed), Microbial diversity and bioprospecting. ASM Press, Washington, DC [Google Scholar]
- 8.Schmidt EW, Nelson JT, Rasko Sudek DAS, Eisen JA, Haygood MG, Ravel J. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U. S. A. 102:7315–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. U. S. A. 101:16222–16227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. 2001. Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl. Environ. Microbiol. 67:4531–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proksch P, Edrada RA, Ebel R. 2002. Drugs from the sea—current status and microbiological implications. Appl. Microbiol. Biotechnol. 59:25–34 [DOI] [PubMed] [Google Scholar]
- 12.Piel J. 2004. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 21:519–538 [DOI] [PubMed] [Google Scholar]
- 13.Suarez-Jimenez GM, Burgos-Hernandez A, Ezquerra-Brauer JM. 2012. Bioactive peptides and depsipeptides with anticancer potential: sources from marine animals. Mar. Drugs 10:963–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher CR. 1990. Chemoautotrophic and methanotrophic symbiosis in marine invertebrates. Rev. Aquat. Sci. 2:399–436 [Google Scholar]
- 15.Cavanaugh CM, McKiness ZP, Newton ILG, Stewart FJ. 2006. Marine chemosynthetic symbiosis, p 475–507 In Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E. (ed), The prokaryotes, 3rd ed, vol 1 Springer, New York, NY [Google Scholar]
- 16.Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6:725–740 [DOI] [PubMed] [Google Scholar]
- 17.Taylor JD, Glover EA. 2010. Chemosymbiotic bivalves, p 107–135 In Kiel S. (ed), The vent and seep biota: aspects from microbes to ecosystems, vol. 33 Topics in Geobiology Springer, Dordrecht, Netherlands [Google Scholar]
- 18.Urakawa H, Dubilier N, Fujiwara Y, Cunningham DE, Kojima S, Stahl DA. 2005. Hydrothermal vent gastropods from the same family (Provannidae) harbour ε- and γ-proteobacterial endosymbionts. Environ. Microbiol. 7:750–754 [DOI] [PubMed] [Google Scholar]
- 19.Azam F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694–696 [Google Scholar]
- 20.Sminia T, Van der Knaap WPW. 1986. Immunorecognition in invertebrates with special reference to mollusks, p 112–124 In Brehelin M. (ed), Immunity in invertebrates. Springer-Verlag, Berlin, Germany [Google Scholar]
- 21.Hooper C, Day R, Slocombe R, Handlinger J, Benkendorff K. 2007. Stress and immune responses in abalone: limitations in current knowledge and investigative methods based on other models. Fish Shellfish Immunol. 22:363–379 [DOI] [PubMed] [Google Scholar]
- 22.Cimino G, Passeggio A, Sodano G, Spinella A, Villani G. 1991. Alarm pheromones from the Mediterranean opisthobranch Haminoea navicula. Experientia 47:61–63 [Google Scholar]
- 23.Zatylny C, Gagnon J, Boucaud-Camou E, Henry J. 2000. ILME: a waterbourne pheromonal peptide released by the eggs of Sepia officinalis. Biochem. Biophys. Res. Commun. 275:217–222 [DOI] [PubMed] [Google Scholar]
- 24.Cummins SF, Nichols AE, Schein CH, Nagle GT. 2006. Newly identified water-borne protein pheromones interact with attractin to stimulate mate attraction in Aplysia. Peptides 27:597–606 [DOI] [PubMed] [Google Scholar]
- 25.Craig AG. 2000. The characterization of conotoxins. J. Toxicol. Toxin Rev. 19:53–93 [Google Scholar]
- 26.Kanda A, Iwakoshi-Ukena E, Takuwa-Kuroda K, Minakata H. 2003. Isolation and characterization of novel tachykinins from the posterior salivary gland of the common octopus Octupus vulgaris. Peptides 24:35–43 [DOI] [PubMed] [Google Scholar]
- 27.Pawlik JR, Albizati KF, Faulkner DJ. 1986. Evidence of a defensive role for limatulone, a novel triterpene from the interdial limpet Collisella limatula. Mar. Ecol. Prog. Ser. 30:251–260 [Google Scholar]
- 28.Marin A, Alvarez LA, Cimino G, Spinella A. 1991. Chemical defense in cephalaspidean gastropods: origin, anatomical location and ecological roles. J. Molluscan Stud. 65:121–131 [Google Scholar]
- 29.Kelley WP, Wolters AM, Sack JT, Jockusch RA, Jurchen JC, Williams ER, Sweedler JV, Gilly WF. 2003. Characterization of a novel gastropod toxin (6-bromo-2-mercaptotryptamine) that inhibits shaker K channel activity. J. Biol. Chem. 278:34934–34942 [DOI] [PubMed] [Google Scholar]
- 30.Derby CD, Kicklighter CE, Johnson PM, Zang X. 2007. Chemical composition of inks of diverse marine molluscs suggests convergent chemical defenses. J. Chem. Ecol. 33:1105–1113 [DOI] [PubMed] [Google Scholar]
- 31.Carmona L, Malaquias MAE, Gosliner TM, Pola M, Cervera JL. 2011. Amphi-atlantic distributions and cryptic species in sacoglossan sea slugs. J. Molluscan Stud. 77:401–412 [Google Scholar]
- 32.Wägele H. 2004. Potential key characters in opisthobranchia (Gastropoda, Mollusca) enhancing adaptive radiation. Org. Div. Evol. 4:175–188 [Google Scholar]
- 33.Rumpho ME, Summer EJ, Green BJ, Fox TC, Manhart JR. 2001. Mollusc/algal chloroplast symbiosis: how can isolated chloroplasts continue to function for months in the cytosol of a sea slug in the absence of an algal nucleus? Zoology 104:303–312 [DOI] [PubMed] [Google Scholar]
- 34.Pelletreau KN, Bhattacharya D, Price DC, Worful JM, Moustafa A, Rumpho ME. 2011. Sea slug kleptoplasty and plastid maintenance in a metazoan. Plant Physiol. 155:1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rumpho ME, Summer EJ, Manhart JR. 2000. Solar-powered sea slugs. Mollusc/algal chloroplast symbiosis. Plant Physiol. 123:29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierce SK, Fang X, Schwartz JA, Jiang X, Zhao W, Curthis NE, Kocot KM, Yang B, Wang J. 2012. Transcriptomic evidence for the expression of horizontally transferred algal nuclear genes in the photosynthetic sea slug, Elysia chlorotica. Mol. Biol. Evol. 29:1545–1556 [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharya D, Pelletreau KN, Price DC, Sarver KE, Rumpho ME. 2013. Genome analysis of Elysia chlorotica egg DNA provides no evidence for horizontal gene transfer into the germ line of this kleptoplastic mollusc. Mol. Biol. Evol. 30:1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avila C. 1995. Natural products of opisthobranch molluscs: a biological review. Oceanogr. Mar. Biol. Annu. Rev. 33:487–559 [Google Scholar]
- 39.Smyrniotopoulos V, Abatis D, Tziveleka LA, Tsitsimpikou C, Roussis V, Loukis A, Vagias C. 2003. Acetylene sesquiterpenoid esters from the green alga Caulerpa prolifera. J. Nat. Prod. 66:21–24 [DOI] [PubMed] [Google Scholar]
- 40.Dobretsov S, Dahms HU, Harder T, Qian PY. 2006. Allelochemical defense against epibiosis in the macroalga Caulerpa racemosa var. turbinata. Mar. Ecol. Prog. Ser. 318:165–175 [Google Scholar]
- 41.Marin A, Ros J. 2004. Chemical defenses in sacoglossan opisthobranchs: taxonomic trends and evolutive implications. Sci. Mar. 68:227–241 [Google Scholar]
- 42.Devine SP, Pelletreau KN, Rumpho ME. 2012. 16S rDNA-based metagenomic analysis of bacterial diversity associated with two populations of the kleptoplastic sea slug Elysia chlorotica and its algal prey Vaucheria litorea. Biol. Bull. 223:138–154 [DOI] [PubMed] [Google Scholar]
- 43.Becerro MA, Goetz G, Paul VJ, Scheuer PJ. 2001. Chemical defenses of the sacoglossan mollusk Elysia rufescens and its host alga Bryopsis sp. J. Chem. Ecol. 27:2287–2299 [DOI] [PubMed] [Google Scholar]
- 44.Hamann MT, Scheuer PJ. 1993. Kahalalide F: a bioactive depsipeptide from the sacoglossan mollusk Elysia rufescens and the green alga Bryopsis sp. J. Am. Chem. Soc. 115:5825–5826 [Google Scholar]
- 45.Hamann MT, Otto CS, Scheuer PJ. 1996. Kahalalides: bioactive peptides from a marine mollusk Elysia rufescens and its algal diet Bryopsis sp. J. Org. Chem. 61:6594–6600 [DOI] [PubMed] [Google Scholar]
- 46.Hamann MT. 2004. Technology evaluation: Kahalalide F, PharmaMar. Curr. Opin. Mol. Ther. 6:657–665 [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Hamann MT. 2011. Chemistry and biology of kahalalides. Chem. Rev. 111:3208–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill RT, Enticknap J, Rao KV, Hamann MT. August 2007. Kahalalide-producing bacteria. US patent 20070196901
- 49.Montalvo NF, Mohamed NM, Entichnap JJ, Hill RT. 2005. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 87:29–36 [DOI] [PubMed] [Google Scholar]
- 50.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY [Google Scholar]
- 51.Somerville CC, Knight IT, Straube WL, Colwell RR. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muyzer G, DeWaal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Commun. Stat. Theory Methods 34:2123–2131 [Google Scholar]
- 58.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 60.Simmons TL, Coates RC, Clark BR, Engene N, Gonzalez D, Esquenazi E, Dorrestein PC, Gerwick WH. 2008. Biosynthetic origin of natural products isolated from marine microorganisms-invertebrate assemblages. Proc. Natl. Acad. Sci. U. S. A. 105:4587–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruby EG. 1996. Lessons from a cooperative, bacterial-animal association. The Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591–624 [DOI] [PubMed] [Google Scholar]
- 62.Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholera. Microbiol. Mol. Biol. Rev. 62:1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wietz M, Mansson M, Gotfredsen CH, Larsen TO, Gram L. 2010. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar. Drugs 8:2946–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mansson M, Gram L, Larsen TO. 2011. Production of bioactive secondary metabolites by marine Vibrionaceae. Mar. Drugs 9:1440–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whithear K. 2001. Diseases due to mycoplasmas, p 413–422 In Williams ES, Barker IK. (ed), Infectious diseases of wild mammals. Iowa University Press, Ames, IA [Google Scholar]
- 66.Fraune S, Zimmer M. 2008. Host-specificity of environmentally transmitted Mycoplasma-like isopod symbionts. Environ. Microbiol. 10:2497–2504 [DOI] [PubMed] [Google Scholar]
- 67.Bano N, DeRae SA, Bennett W, Vasquez L, Hollibaugh JT. 2007. Dominance of Mycoplasma in the guts of the Long-Jawed Mudsucker, Gillichthys mirabilis, from five California salt marshes. Environ. Microbiol. 9:2636–2641 [DOI] [PubMed] [Google Scholar]
- 68.Huang ZB, Guo F, Zhao J, Li WD, Ke CH. 2010. Molecular analysis of the intestinal bacterial flora in cage-cultured adult small abalone, Haliotis diversicolor. Aquac. Res. 41:760–769 [Google Scholar]
- 69.Naidoo K, Maneveldt G, Ruck K, Bolton J. 2006. A comparison of various seaweed-based diets and formulated feed growth rate of abalone in a land-based aquaculture system. J. Appl. Phycol. 18:437–443 [Google Scholar]
- 70.Erasmus JH, Cook PA, Coyne VE. 1997. The role of bacteria in the digestion of seaweed by the abalone Haliotis midae. Aquaculture 155:377–386 [Google Scholar]
- 71.Hollants J, Leroux O, Leliaert F, Decleyre H, De Clerck O, Willems A. 2011. Who is in there? Exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta). PLoS One 6:e26458. 10.1371/journal.pone.0026458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hollants J, Decleyre H, Leliaert F, De Clerck O, Willems A. 2011. Life without cell membrane: challenging the specificity of bacterial endophytes within Bryopsis (Bryopsidales, Chlorophyta). BMC Microbiol. 11:255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. U. S. A. 97:10231–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68:5113–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trench RK. 1975. Of leaves that crawl: functional chloroplast in animal cells. Symp. Soc. Exp. Biol. 29:229–265 [PubMed] [Google Scholar]
- 76.Paul VJ, Van Alstyne KL. 1988. Use of ingested algal diterpenoid by Elysia halimedae Macnae (Opisthobranchia: Ascoglossa) as antipredator defenses. J. Exp. Mar. Biol. Ecol. 119:15–29 [Google Scholar]
- 77.Rumpho ME, Mujer CV, Andrews DL, Manhart JR, Pierce SK. 1994. Extraction of DNA from mucilaginous tissues of a sea slug (Elysia chlorotica). Biotech. 17:1097–1101 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.