Abstract
Reductive dehalogenases are the key enzymes involved in the anaerobic respiration of organohalides such as the widespread groundwater pollutant tetrachloroethene. The increasing number of available bacterial genomes and metagenomes gives access to hundreds of new putative reductive dehalogenase genes that display a high level of sequence diversity and for which substrate prediction remains very challenging. In this study, we present the development of a functional genotyping method targeting the diverse reductive dehalogenases present in Sulfurospirillum spp., which allowed us to unambiguously identify a new reductive dehalogenase from our tetrachloroethene-dechlorinating SL2 bacterial consortia. The new enzyme, named PceATCE, shows 92% sequence identity with the well-characterized PceA enzyme of Sulfurospirillum multivorans, but in contrast to the latter, it is restricted to tetrachloroethene as a substrate. Its apparent higher dechlorinating activity with tetrachloroethene likely allowed its selection and maintenance in the bacterial consortia among other enzymes showing broader substrate ranges. The sequence-substrate relationships within tetrachloroethene reductive dehalogenases are also discussed.
INTRODUCTION
Widespread use of chlorinated solvents in cleaning and metal-degreasing operations over the last century has resulted in extensive environmental contamination by chlorinated ethenes. Tetrachloroethene (PCE) is currently one of the main contaminants of aquifers. Different remediation strategies have been developed, including bioremediation, which uses microorganisms for the degradation of pollutants (1). Several genera of strictly anaerobic bacteria, including Desulfitobacterium (2), Dehalococcoides (3), Dehalobacter (4), and Sulfurospirillum (5), are able to conserve energy via the reductive dehalogenation of chloroethenes by a respiratory metabolism (6–9) recently coined organohalide respiration (OHR). The enzyme class involved in OHR for the reduction of halogenated compounds is known as the reductive dehalogenase (RdhA) family (10, 11). Most RdhA enzymes have been isolated from OHR bacteria (OHRB) as 48- to 65-kDa monomers consisting of a single polypeptide chain and containing one corrinoid cofactor and two iron-sulfur clusters. The majority of currently known OHRB carry multiple rdhA genes, i.e., 1 to 36 genes, depending on the strain (12). Several studies have demonstrated that the presence of multiple nonidentical rdhA genes is a typical feature of OHRB (13–15). This suggests that the substrate range of the OHRB may be far greater than previously believed (16). However, substrate specificity has been determined for only about 15 enzymes within the several hundreds of putative rdhA sequences reported in databases (12). Moreover, transcriptomic studies on strains displaying multiple rdhA genes often failed to clearly identify which reductive dehalogenase was responsible for the dechlorination of specific substrates (12, 17–20).
The genus Sulfurospirillum comprises microaerophilic or facultative anaerobic sulfur-reducing bacteria belonging to the Gram-negative Epsilonproteobacteria. So far, nine species of Sulfurospirillum have been isolated, and all display a versatile respiratory metabolism (see references 21 and 22 for comparative tables). Sulfurospirillum species of particular environmental interest are Sulfurospirillum multivorans (5) and Sulfurospirillum halorespirans (23), as they were isolated based on their property of using PCE as an electron acceptor. Over the years, S. multivorans became a model OHRB. Indeed, the key enzyme, the PCE reductive dehalogenase (PceA) catalyzing the dechlorination of PCE to cis-dichloroethene (cis-DCE), and its corresponding gene were the first to be identified (24, 25). A new corrinoid, norpseudovitamin B12, has been isolated as the cofactor of S. multivorans PceA (26). Two types of rdhA genes were obtained at least partially from S. multivorans and S. halorespirans. In addition to PceA-encoding genes (GenBank accession no. AF022812 and AY013367), which can be considered Sulfurospirillum rdhA type 1 genes (Sul-rdhA1), fragments of a second type (Sul-rdhA2) have been identified in S. multivorans (rdhA; GenBank accession no. AJ539530) (27) and S. halorespirans (rdpA; GenBank accession no. AY013368). More recently, the full sequence of a type 2 rdhA (GenBank accession no. AB537449) (28) was reported in a metagenomic study revealing an overall protein sequence identity of 63% with S. multivorans PceA.
In an earlier study, we obtained a bacterial consortium (named SL2) from a fixed-bed bioreactor sludge treating groundwater that was pumped to the surface at a PCE-contaminated site (29). The SL2 consortium was considered a model of an OHR consortium degrading PCE completely to ethene and were shown to contain members of the genera Sulfurospirillum and Dehalococcoides (29, 30). Sulfurospirillum spp. were demonstrated to catalyze the first two steps of dechlorination (from PCE to cis-DCE), whereas members of the genus Dehalococcoides were involved in the final steps (from cis-DCE to ethene) (30), with the most striking feature being a clear stepwise dechlorination process. The succession of different enzymes and/or bacterial populations along the dechlorination pattern was then proposed but could not be demonstrated clearly.
A new consortium, SL2-PCEb, was selected from SL2 by serial dilutions and frequent culture transfers when trichloroethene (TCE) dechlorination was almost complete (30). This consortium had lost the ability to dechlorinate cis-DCE and was shown to contain only Sulfurospirillum spp. as OHRB. It was still characterized by a strong transient TCE accumulation, further suggesting that at least two Sulfurospirillum populations were maintained in SL2-PCEb. Initial identification and detection of rdhA genes by single-strand conformation polymorphism (SSCP) analysis revealed the presence of several gene fragments showing strong similarity to type 1 and type 2 rdhA genes of Sulfurospirillum isolates and suggested the involvement of the former enzymes in the stepwise dechlorination of PCE to cis-DCE (30). However, neither analysis of 16S rRNA genes nor that of the internal transcribed spacer region allowed a clear distinction of multiple Sulfurospirillum populations.
In the present work, after identifying the complete sequences of three rdhAB operons present in the SL2 consortia, and similar to an earlier study on Dehalococcoides (31), we developed a dedicated terminal restriction fragment length polymorphism (T-RFLP) method to detect the diverse population of Sulfurospirillum rdhA genes and to confirm their involvement in the stepwise dechlorination of PCE. Additional consortia subcultivated from SL2-PCEb allowed the unambiguous identification of a new PCE reductive dehalogenase, PceATCE, with reduced substrate specificity.
MATERIALS AND METHODS
Chemicals.
All chemicals were analytical grade and were used without purification. Gases (N2, CO2, and H2) were purchased from SLGas (Sauerstoffwerk Lenzburg, Switzerland).
Bacterial strains and plasmids.
In this work, several consortia were investigated which were previously selected from the SL2 consortium, which was established from a fixed-bed bioreactor treating groundwater pumped to the surface from a PCE-contaminated site (29, 30). Both pure isolates Sulfurospirillum multivorans DSM 12446 and S. halorespirans DSM 13726 were obtained from the DSMZ culture collection (DSMZ, Braunschweig, Germany). Escherichia coli DH5α was used as a host for molecular work. Competent E. coli cells were prepared using the standard CaCl2 method (32). This strain was cultivated at 37°C in lysogeny broth (LB) liquid medium and on plates containing 100 μg/ml of ampicillin when needed. The vector pGEM-T Easy (Promega, Madison, WI) was used for direct cloning of PCR products. The complete list of bacterial strains and plasmids used in this study is given in Table S1 in the supplemental material.
The cultivation of SL2 consortia and of pure Sulfurospirillum cultures was carried out in serum bottles of 100, 500, or 1,000 ml (VWR International AG, Merck, Dietikon, Switzerland) containing 50, 200, 300, or 600 ml of an anaerobic medium that has been described previously (30). PCE or TCE dissolved in hexadecane was added as a terminal electron acceptor, depending on the experiment. Two or three replicate cultures were performed systematically to confirm the trends in the observed physiological behavior and the data obtained by molecular analyses.
DNA and RNA work.
Fifty- to 100-ml SL2 cultures were centrifuged at 3,300 × g for 10 min at 4°C. For DNA extraction, cell pellets were washed in 1 ml of 50 mM Tris-HCl buffer (pH 8) and centrifuged in microcentrifuge tubes at 8,800 × g for 5 min. Biomass pellets were flash-frozen in liquid N2 and stored at −80°C. DNA extraction was performed with a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany), including the pretreatment for lysis of Gram-positive bacteria. The genomic DNA was quantified with a NanoDrop spectrophotometer (ND-1000; Thermo Fisher Scientific, Wohlen, Switzerland).
Sampling for RNA extraction was done from a volume of 50 to 200 ml of SL2 culture by centrifugation at 3,300 × g for 2 min at 12°C. The pellet was quickly resuspended in 1 ml of LifeGuard solution (MoBio, Carlsbad, CA), flash-frozen in liquid N2, and stored at −80°C. An optimized TRIzol-based RNA extraction protocol was used for total RNA extraction as previously described (33). Reverse transcription (RT) was performed as follows. RNA (0.2 μg) was mixed with 0.5 μg of random hexamers (Microsynth GmbH, Balgach, Switzerland) in a total volume of 10 μl, incubated for 5 min at 70°C, and cooled on ice for 3 min. From this mixture, 5 μl was mixed with 4 μl of 5× RT buffer, 1 μl of 10 mM (each) deoxynucleoside triphosphates (dNTPs), 2.4 μl of 25 mM MgCl2, 6.1 μl of RNase-free water, 0.5 μl of RNasin (Promega), and 1 μl of ImProm-II reverse transcriptase (Promega). Control reactions were done with 2.5 μl of the RNA-hexamer mixture in a similar reaction mixture, but omitting the RT enzyme. The reaction mixture was incubated for 5 min at 25°C and then for 90 min at 42°C, followed by enzyme denaturation for 15 min at 70°C.
Standard PCR for T-RFLP analysis or cloning was performed under the following conditions. Each 50-μl PCR mixture contained 5 μl of Taq DNA polymerase 10× buffer, 6 μl of 2.5 mM (each) dNTPs, 2.5 μl of 10 μM (each) primers (see Table S2 in the supplemental material), 0.4 μl of 25 mM MgCl2, 0.25 μl of Taq DNA polymerase (Peqlab Biotechnologie, Erlangen, Germany), and 5 μl of DNA template. The DNA was amplified in a T3 thermocycler (Biometra, Biolabo Scientific Instruments, Châtel-St-Denis, Switzerland), using the following program: 5 min of initial denaturation at 94°C followed by 30 cycles of 1 min of denaturation at 94°C, 1 min of primer annealing at 53°C (rdhA genes) or 56°C (16S rRNA genes), and 1 to 4 min of elongation at 72°C, depending on the target size. A final extension step of 10 min at 72°C was included. For cloning and sequencing of PCR products, the PCR products were directly cloned into pGEM-T Easy (Promega) and then selected using colony PCR with T7 and SP6 standard primers. The detailed cloning and sequencing procedure has been described previously (34). Quantitative PCR (qPCR) targeting Sulfurospirillum rdhA genes specifically was performed to validate the data obtained by T-RFLP analysis (see the supplemental material for details).
For T-RFLP analysis (see below), 16S rRNA genes and Sulfurospirillum rdhA genes were amplified by a PCR using the 6-carboxyfluorescein (FAM)-labeled forward primers 8f-FAM and Sul-rdhA-f-FAM with the reverse primers 518-r and Sul-rdhA-r, respectively (see Table S2 in the supplemental material). To 40 μl of PCR product, 128 μl of ethanol and 32 μl of water were added. After 20 min of incubation at room temperature in the dark, the samples were centrifuged at 16,000 × g for 20 min. The supernatant was removed, 250 μl of 70% ethanol was added, and the samples were centrifuged again for 10 min. The DNA pellet was then air dried for 5 to 10 min and resuspended in 15 μl of sterile double-distilled water (ddH2O). The DNA was resuspended by 5 min of incubation at 55°C. Concentrated PCR products (5 μl) were digested for 2 h in 10-μl reaction mixtures containing 1 μl of 10× restriction enzyme buffer and 0.5 μl of restriction enzyme (HaeIII at 37°C for 16S rRNA genes and TaqI at 65°C for rdhA genes). An aliquot of 1 μl of the digested PCR product was mixed with 8.5 μl of formamide and 0.5 μl of internal standard (GeneScanTM-600LIZ; ABI Applied Biosystems, Rotkreuz, Switzerland). The mixture was heated at 95°C for 2 min, cooled on ice for 3 min, and loaded into an ABI Prism 3130XL capillary sequencer equipped with a 50-cm array. The electropherograms were obtained with ABI Prism GeneScan software. The results were analyzed with Treeflap T-RFLP software (developed by C. Walsh; available on request) as described previously (35).
Protein work.
For assays of reductive dehalogenase activity, cell extracts were obtained from 600-ml SL2 cultures. The biomass was harvested anaerobically by centrifugation at 10,000 × g and 12°C for 20 min. Cells were resuspended in 50 mM Tris-HCl (pH 7.5) and washed 3 times. Cells were broken by repetitive sonication as previously described (36). The activity in crude extracts was determined anaerobically following a previously described protocol (37) for measuring spectrophotometrically the oxidation of reduced methyl viologen (MV) (extinction coefficient [ε578] = 9,700 M−1 cm−1) with PCE or TCE as the electron acceptor (see reference 38 for details).
For the detection of Sulfurospirillum reductive dehalogenases in SL2 crude extracts, Western blot analysis was performed as described previously (36). Anti-PceA serum raised against the reductive dehalogenase of S. multivorans (kindly provided by G. Diekert, University of Jena, Germany) was used at a 1:50,000 dilution.
Analytical methods.
The dechlorination activity of the cultures was followed by measuring the concentration of chloride in the culture medium by silver ion titration with a Chlor-o-counter (Flohr Instrument, Nieuwegein, Netherlands) (30). Chloroethenes were analyzed by gas chromatography as previously described (39). Protein quantification in crude extracts was performed with a Pierce bicinchoninic acid (BCA) protein assay kit according to the manufacturer's instructions. A standard curve using BSA was established to calculate the concentrations of proteins.
Nucleotide sequence accession numbers.
All sequences obtained in this study were deposited in GenBank under the following accession numbers: 16S rRNA gene, KF533073; SL2-pceABDCE, KF533074; SL2-pceABTCE, KF533075; and SL2-rdhAB2, KF533076.
RESULTS
Our previous study of the PCE-dechlorinating SL2 consortia showed that the SL2-PCEb culture was able to dechlorinate PCE to cis-DCE in a stepwise fashion and was characterized by low bacterial diversity, as it harbored mainly members of the genus Sulfurospirillum as OHRB (30). In the present work, two additional consortia showing distinct dechlorination patterns were obtained from SL2-PCEb. The first consortium, SL2-PCEc, was obtained after 10 successive rapid transfers of the SL2-PCEb culture when PCE was dechlorinated mainly to TCE, while the second one, SL2-TCE, was selected by cultivation directly on TCE during 15 successive transfers. The SL2-PCEc consortium had completely lost the ability to dechlorinate TCE, while SL2-TCE kept the initial dechlorination potential (PCE to cis-DCE). The two new SL2 consortia, together with SL2-PCEb, were investigated at the physiological and molecular levels in order to distinguish Sulfurospirillum subpopulations and to identify the reductive dehalogenases involved in both dechlorination steps.
Identification of rdh operons in the SL2 consortia.
A few partial sequences of the reductive dehalogenase (rdhA) genes were already identified from SL2 consortia (30), matching with both types of RdhA found in Sulfurospirillum spp. In order to identify the complete rdhAB sequences present in SL2 consortia, two primer pairs were designed to target the flanking regions of both type 1 (such as the S. multivorans pceAB operon; GenBank accession no. AF022812) and type 2 (S. multivorans rdhAB2; obtained from T. Schubert prior to publication) rdhAB sequences. Three rdhAB operons were amplified from SL2 DNA samples and cloned into plasmids pT1P-DS, pT1P-TS, and pT2RS. Sequence analysis of three clones of each plasmid revealed the following encoded proteins: PceADCE (type 1 RdhA identified from SL2-TCE and named according to its dechlorination of PCE to cis-DCE), PceATCE (type 1 RdhA from SL2-PCEc, with dechlorination of PCE to TCE only), and RdhA2 (type 2 RdhA present in both consortia, with unknown dechlorination potential). All three RdhA sequences and their cognate RdhB protein sequences were aligned and compared with those of RdhA (Fig. 1) and RdhB (see Fig. S1 in the supplemental material) of both pure Sulfurospirillum strains and environmental samples.
Fig 1.
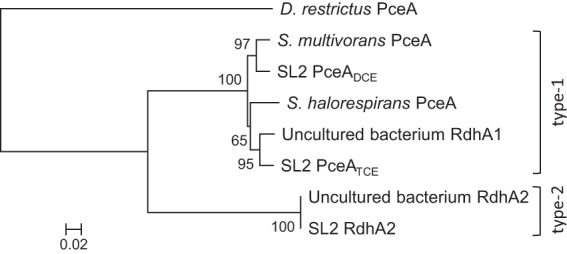
Sequence likelihood analysis showing the relationships of newly identified RdhA proteins from SL2 consortia to both Sulfurospirillum RdhA types. The neighbor-joining method of ClustalX was used to build the tree, including 100× bootstrap values. All sequences used in the alignment had similar lengths. The following sequences were used: S. multivorans PceA (GenBank accession no. AAC60788), S. halorespirans PceA (GenBank accession no. AAG46194), RdhA1 from an uncultured bacterium (GenBank accession no. BAJ09319), and RdhA2 from an uncultured bacterium (GenBank accession no. BAJ09321). The tree was rooted with PceA of Dehalobacter restrictus (GenBank accession no. CAD28790). The scale bar represents 2% sequence divergence.
Sequence homology values of SL2 RdhA proteins compared with available RdhA sequences for Sulfurospirillum clearly illustrate the evolutionary distance separating type 1 and type 2 RdhA proteins (Table 1). While type 2 sequences are very similar to each other, they share only around 60% sequence identity with type 1 sequences. Interestingly, sequence conservation within type 1 PceA sequences ranges from 92% to 97% identity, suggesting some divergent evolution from a common ancestor. Figure S2 in the supplemental material depicts the sequence alignment of both type 1 PceA identified in SL2 consortia and the characterized PceA proteins from S. multivorans and S. halorespirans.
Table 1.
Sequence identity comparison of Sulfurospirillum and SL2 RdhA proteins
| Proteina | % identity with: |
|||||
|---|---|---|---|---|---|---|
| Sul-RdhA2 | Smu-PceA | Sha-PceA | SL2-RdhA2 | SL2-PceADCE | SL2-PceATCE | |
| SL2-PceATCE | 61.9 | 92.1 | 92.6 | 61.9 | 93.3 | 100.0 |
| SL2-PceADCE | 63.4 | 96.6 | 91.1 | 63.4 | 100.0 | |
| SL2-RdhA2 | 100.0 | 63.0 | 61.6 | 100.0 | ||
| Sha-PceA | 61.6 | 91.7 | 100.0 | |||
| Smu-PceA | 63.0 | 100.0 | ||||
| Sul-RdhA2 | 100.0 | |||||
Newly identified PceB sequences accompanying the PceADCE and PceATCE proteins were identical to each other and to those of PceB proteins of pure Sulfurospirillum isolates. RdhB2 belonging to SL2-RdhA2 was identical to the uncharacterized RdhB protein of S. multivorans (T. Schubert, personal communication) (see Fig. S1 in the supplemental material).
Development of a T-RFLP method dedicated to Sulfurospirillum rdhA genes.
As neither classical T-RFLP analysis nor sequencing of Sulfurospirillum 16S rRNA genes nor analysis of the 16S-23S rRNA internal transcribed spacer allowed for discrimination of the different Sulfurospirillum populations present in the SL2-PCEb consortium (30) or in the SL2-PCEc and SL2-TCE consortia (data not shown), we designed a dedicated T-RFLP method to specifically detect the Sulfurospirillum rdhA gene types and subtypes. A single primer set (Sul-rdhA-f-FAM and Sul-rdhA-r) was used to amplify a fragment of all rdhA genes, covering the second insertion/deletion (indel) region (Fig. 2). Subsequent digestion with the TaqI restriction enzyme allowed us to produce three terminal restriction fragments (T-RFs): 272 bp for pceATCE, 284 bp for pceADCE, and 388 bp (undigested product) for rdhA2. In order to validate the data obtained with this method, qPCR was used to detect each of the three rdhA genes present in SL2 consortia and to calculate their relative abundances along the dechlorination profiles of the three consortia (Table 2). Although both experimental approaches gave slightly different abundances for rdhA genes, the trends observed in the T-RFLP data were confirmed by qPCR.
Fig 2.
Sequence alignment of Sul-rdhA gene fragments for rdhA-specific T-RFLP analysis. Sequences were aligned with ClustalX. The Sul-rdhA primers used in the dedicated T-RFLP analysis match the sequences of all 3 rdhA genes present in Sulfurospirillum spp. and are indicated above the alignment. Positions indicated by dashes represent a 12-nucleotide deletion in the sequence of pceATCE. Positions indicated by dots are identical to the sequence of pceATCE. A box indicates the position of the TaqI restriction site.
Table 2.
Detection and quantification of rdhA genes during PCE dechlorination in SL2 consortiaa
| Consortium | Time (days) | Concn of chloride formed (mM) | Relative abundance determined by: |
|||||
|---|---|---|---|---|---|---|---|---|
| T-RFLP analysis |
qPCR |
|||||||
| pceATCE | pceADCE | rdhA2 | pceATCE | pceADCE | rdhA2 | |||
| SL2-PCEc | 2 | 4.4 ± 0.0 | 57 ± 2 | ND | 44 ± 2 | 41 ± 2 | ND | 59 ± 2 |
| 3 | 8.2 ± 0.3 | 54 ± 0 | ND | 46 ± 0 | 43 ± 1 | ND | 57 ± 1 | |
| 4 | 9.5 ± 0.5 | 53 ± 1 | ND | 48 ± 1 | 36 ± 1 | ND | 64 ± 1 | |
| SL2-TCE | 1 | 0.9 ± 0.0 | ND | 51 ± 3 | 49 ± 3 | ND | 36 ± 2 | 64 ± 2 |
| 2 | 8.6 ± 0.2 | ND | 56 ± 3 | 44 ± 3 | ND | 43 ± 2 | 57 ± 2 | |
| 3 | 9.9 ± 0.3 | ND | 57 ± 1 | 44 ± 1 | ND | 45 ± 4 | 55 ± 4 | |
| 4 | 11.5 ± 0.4 | ND | 58 ± 2 | 43 ± 2 | ND | 44 ± 1 | 56 ± 1 | |
| SL2-PCEb | 3 | 4.0 ± 0.0 | 35 ± 3 | 17 ± 5 | 48 ± 2 | 35 ± 3 | 11 ± 2 | 54 ± 6 |
| 4 | 6.2 ± 0.0 | 39 ± 3 | 12 ± 4 | 49 ± 2 | 35 ± 6 | 7 ± 1 | 58 ± 4 | |
| 5 | 8.7 ± 0.3 | 40 ± 2 | 11 ± 2 | 50 ± 4 | 32 ± 4 | 7 ± 0 | 61 ± 5 | |
| 6 | 10.8 ± 0.4 | 35 ± 1 | 18 ± 1 | 48 ± 1 | 27 ± 2 | 15 ± 2 | 58 ± 2 | |
| 7 | 13.2 ± 0.2 | 29 ± 2 | 27 ± 1 | 44 ± 2 | 22 ± 3 | 27 ± 2 | 51 ± 5 | |
| 8 | 16.0 ± 0.0 | 26 ± 0 | 30 ± 1 | 44 ± 1 | 16 ± 1 | 31 ± 4 | 52 ± 8 | |
| 9 | 18.4 ± 0.0 | 20 ± 3 | 37 ± 6 | 43 ± 8 | 11 ± 1 | 31 ± 6 | 58 ± 5 | |
| 10 | 19.3 ± 0.9 | 22 ± 4 | 35 ± 3 | 44 ± 2 | 10 ± 1 | 35 ± 0 | 55 ± 8 | |
Data are means ± standard deviations. ND, not detected.
The dechlorination pattern of SL2-PCEb was characterized by a strong initial accumulation of TCE upon PCE dechlorination, with only a little formation of cis-DCE. Only when the PCE concentration was low were TCE dechlorination and formation of cis-DCE started (30). This stepwise dechlorination pattern suggested either a succession of different populations during degradation or a regulated expression of genes involved in each step. The rdhA-specific T-RFLP method described above and the emergence of two consortia either restricted or dedicated to a single dechlorination step allowed us to test this hypothesis.
Detection of rdhA genes in SL2-PCEc and SL2-TCE consortia.
The rdhA-specific T-RFLP analysis showed that pceATCE and rdhA2 were detected in SL2-PCEc, while pceADCE and rdhA2 were found in SL2-TCE. This observation strongly highlighted the restricted dechlorination substrate range of PceATCE. No significant gene fluctuation was observed during the single dechlorination steps, as all samples showed between 40% and 60% relative abundances of their respective pceA and rdhA2 genes, suggesting that each Sulfurospirillum population contains one copy of its respective pceA gene and one copy of rdhA2 in its genome (Table 2).
Interplay of both pceA genes in the SL2-PCEb consortium during the stepwise dechlorination of PCE to cis-DCE.
The rdhA-specific T-RFLP analysis was further used to evaluate the relative abundances of Sulfurospirillum rdhA genes, on the DNA level, along the stepwise dechlorination of PCE to cis-DCE occurring in the SL2-PCEb culture (Table 2 and Fig. 3). All three genes (pceATCE, pceADCE, and rdhA2) were detected in each sample. While rdhA2 was predominant and remained constant, at around 50%, confirming that this gene is present in both Sulfurospirillum populations present in SL2-PCEb, a clear gradual change occurred between pceATCE and pceADCE along the stepwise dechlorination of PCE to TCE and cis-DCE, an observation that is in complete agreement with the molecular analysis of the SL2-PCEc and SL2-TCE consortia.
Fig 3.
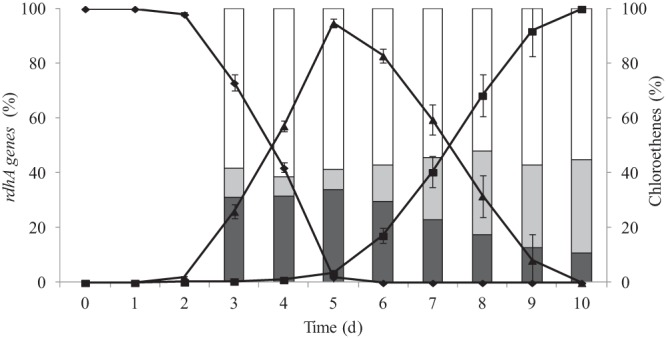
Transformation of chloroethenes and interplay of rdhA genes in the SL2-PCEb culture along the dechlorination of PCE to cis-DCE. Chloroethenes are represented by diamonds for PCE, triangles for TCE, and squares for DCE. Dark gray bars represent the relative abundances of pceATCE, light gray bars represent those of pceADCE, and white bars represent those of rdhA2. The figure was obtained by averaging the data obtained from two independent batch cultures.
Furthermore, in order to assess whether all three rdhA genes were transcribed during dechlorination, an SL2-PCEb culture was analyzed on the RNA level by using the combination of reverse transcription and T-RFLP analysis. This analysis revealed that pceATCE and pceADCE were transcribed, while rdhA2 was not (see Fig. S3 in the supplemental material).
PceATCE seems to be 5-fold more active than PceADCE with PCE as substrate.
The crude extracts of the SL2-PCEc and SL2-TCE consortia, expressing the PceATCE and PceADCE reductive dehalogenases, respectively, were tested for RdhA activity with PCE or TCE as the substrate. The crude extract of SL2-PCEc (PceATCE) showed a 5-fold higher RdhA activity with PCE than that of SL2-TCE (PceADCE) (Table 3). The SL2-PCEc crude extract displayed no activity at all with TCE, confirming its reduced dechlorination range. The SL2-TCE crude extract, however, kept the ability to reduce both PCE and TCE, even though the culture was cultivated directly on TCE.
Table 3.
Reductive dehalogenase enzymatic activities measured in crude extracts of SL2 consortia
| Culture | RdhAa | RdhA sp act in crude extracts (nkat/mg of total protein) |
|
|---|---|---|---|
| With PCE | With TCE | ||
| SL2-PCEc | PceATCE | 37.6 ± 0.4 | 0.0 |
| SL2-TCE | PceADCE | 7.5 ± 1.0 | 6.0 ± 0.5 |
| S. multivorans | PceA | 7.3b | NDc |
The corresponding gene was detected in the genomic DNA of the indicated culture.
Taken from reference 38.
ND, not determined.
The levels of PceA expression in both SL2 consortia were shown to be similar (see Fig. S4 in the supplemental material), strongly suggesting that the PceATCE enzyme has a higher specific activity with PCE than that of PceADCE.
DISCUSSION
Neither T-RFLP analysis nor sequencing of 16S rRNA genes allowed discrimination of Sulfurospirillum populations in SL2 consortia. A survey of gene databases (including the genomes of Sulfurospirillum deleyianum and Sulfurospirillum barnesii) confirmed that fingerprinting methods targeting rRNA sequences are not suited to distinguishing Sulfurospirillum spp. from each other (see Table S3 in the supplemental material). Moreover, OHR is not a conserved feature in the genus Sulfurospirillum, arguing for the need of a fingerprinting method directly targeting the key elements for OHR, i.e., the reductive dehalogenases, in this genus. Here we developed a simple T-RFLP method dedicated to detecting and distinguishing the three rdhA genes previously identified in Sulfurospirillum-containing SL2 consortia (30). Another functional T-RFLP study has already been reported for Dehalococcoides rdhA genes, which required 13 different primer pairs to cover the larger number of genes and their higher level of sequence diversity (31).
Using our dedicated T-RFLP method, the dynamic behavior of rdhA genes was analyzed in the SL2-PCEb consortium, which catalyzes the dechlorination of PCE to cis-DCE in a stepwise manner. This analysis revealed that at least two functionally distinct Sulfurospirillum populations were present and successively selected during the two dechlorination steps, explaining the peculiar transient TCE accumulation. Indeed, the latter feature has not been observed for pure Sulfurospirillum isolates (23, 40), in which PceA has been shown to catalyze the dechlorination of PCE and TCE to cis-DCE. While the hypothesis of population dynamics was clearly demonstrated, the data presented here do not per se exclude a possible regulation of the different rdhA genes at the transcriptional level. However, the experimental setup used here did not fulfill the conditions applied in the study by John et al. that reported the peculiar long-term regulation of the pceA gene in S. multivorans (41). The emergence of the new SL2-PCEc consortium, displaying a dechlorination substrate range restricted to PCE, further allowed the unambiguous identification of the key player in the PCE-to-TCE dechlorination, namely, an enzyme we called PceATCE. The gene encoding the second PCE-dechlorinating enzyme identified in SL2-PCEb, PceADCE, was enriched in the SL2-TCE consortium upon cultivation on TCE. The chlorinated substrate specificities of SL2-PCEc and SL2-TCE were tested in crude extracts, and they confirmed the restricted substrate range of PceATCE and further revealed that PceADCE was able to dechlorinate both PCE and TCE. Despite the high level of sequence identity of their respective PceA enzymes, the 5-fold higher PCE dechlorination activity of SL2-PCEc crude extract than that of SL2-TCE crude extract represents, to the best of our knowledge, a new situation in OHR bacterial consortia and explains why the Sulfurospirillum sp. population harboring the pceATCE gene was able to maintain itself in SL2-PCEb over long periods and repeated culture transfers. It also explains why TCE transiently accumulated in the dechlorination pattern of the SL2-PCEb consortium. The enrichment of specific dechlorinating populations by varying the added chlorinated compound has also been reported for the ANAS consortium, carrying several Dehalococcoides spp., where the focus has been given to the interplay of the tceA and vcrA gene homologs along the dechlorination of TCE to cis-DCE and vinyl chloride (VC) to ethene (19).
The identification of the PceATCE enzyme in the SL2 consortia, which displays dechlorinating activity toward PCE only, is of great interest for investigating the sequence-substrate relationships of Sulfurospirillum reductive dehalogenases. Indeed, in comparing the sequence of PceATCE to those of the other three Sulfurospirillum PceA enzymes, only 16 amino acid positions (3.2% of the total number of residues), which are rather scattered throughout the enzyme sequence, are specific to the enzyme with a restricted substrate range, suggesting that these residues participate in defining the substrate binding pocket of Sulfurospirillum reductive dehalogenases (see Fig. S2 in the supplemental material). Moreover, 42 additional amino acid positions (including 8 indel positions) vary among all four protein sequences and could also be involved in the substrate range definition. Seven unique residues are conserved in both the SL2-PceATCE and -PceADCE sequences but not in the other two enzymes isolated from S. multivorans and S. halorespirans, possibly reflecting the evolution of both SL2 enzymes from a common ancestor, which might have diverged toward their respective substrate specificities. The very high sequence identity of all Sulfurospirillum RdhB proteins clearly confirms the common origin of all rdh gene clusters in Sulfurospirillum spp. and suggests that the residues participating in the proposed interaction between the A and B components are fully conserved.
The divergence toward specific substrates of highly similar reductive dehalogenase sequences has already been highlighted for the Dehalobacter restrictus PceA (Dre-PceA) and Desulfitobacterium dichloroeliminans DcaA (Ddi-DcaA) enzymes (34, 42, 43). There are only 62 amino acid positions (11% of the total sequence length) that are responsible for the drastic change in substrate specificity of these enzymes (PCE versus 1,2-dichloroethane, respectively). In comparing sequence pairs of early-diverging proteins (Dre-PceA and Ddi-DcaA versus S. multivorans PceA and SL2-PceATCE), both pairs of sequences share only 11 common positions with substituted amino acids among the 62 and 40 substitutions present in each pair, respectively, indicating that the substrate binding pocket in at least these reductive dehalogenases is not likely to be defined by discrete amino acid positions but rather by two variable regions, as previously proposed by Marzorati et al. (43) (see Fig. S5 in the supplemental material for details).
Recent analysis of a curated set of reductive dehalogenases revealed that sequence similarity and substrate specificity are generally not correlated, making functional prediction from sequence information difficult (12). In conclusion, the results obtained in the present study further emphasize this observation by showing that minute changes in the sequences of highly similar enzymes are responsible for differences in their substrate range. Moreover, the identification of a new PCE reductive dehalogenase restricted to PCE as the substrate, along with the indication that it displays a 5-fold higher dechlorination activity, suggests that recent evolution likely promoted new catalytic properties that help individual OHRB populations to compete for the same substrate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cindy Kunze for technical assistance and Noam Shani and Nevenka Adler for fruitful discussions.
We thank the Swiss National Science Foundation for financial support (grant 31003A_138114).
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02312-13.
REFERENCES
- 1.Christ JA, Ramsburg CA, Abriola LM, Pennell KD, Löffler FE. 2005. Coupling aggressive mass removal with microbial reductive dechlorination for remediation of DNAPL source zones: a review and assessment. Environ. Health Perspect. 113:465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villemur R, Lanthier M, Beaudet R, Lepine F. 2006. The Desulfitobacterium genus. FEMS Microbiol. Rev. 30:706–733 [DOI] [PubMed] [Google Scholar]
- 3.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Muller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63:625–635 [DOI] [PubMed] [Google Scholar]
- 4.Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJ. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313–321 [DOI] [PubMed] [Google Scholar]
- 5.Scholz-Muramatsu H, Neumann A, Messmer M, Moore E, Diekert G. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163:48–56 [Google Scholar]
- 6.Mohn WW, Kennedy KJ. 1992. Reductive dehalogenation of chlorophenols by Desulfomonile tiedjei DCB-1. Appl. Environ. Microbiol. 58:1367–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher W, Holliger C. 1996. The proton/electron ratio of the menaquinone-dependent electron transport from dihydrogen to tetrachloroethene in “Dehalobacter restrictus.”. J. Bacteriol. 178:2328–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smidt H, de Vos WM. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43–73 [DOI] [PubMed] [Google Scholar]
- 9.van de Pas BA, Jansen S, Dijkema C, Schraa G, de Vos WM, Stams AJ. 2001. Energy yield of respiration on chloroaromatic compounds in Desulfitobacterium dehalogenans. Appl. Environ. Microbiol. 67:3958–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futagami T, Goto M, Furukawa K. 2008. Biochemical and genetic bases of dehalorespiration. Chem. Rec. 8:1–12 [DOI] [PubMed] [Google Scholar]
- 11.Holliger C, Regeard C, Diekert G. 2003. Dehalogenation by anaerobic bacteria, p 115–157 In Häggblom MM, Bossert IB. (ed), Dehalogenation: microbial processes and environmental applications. Kluwer Academic, Boston, MA [Google Scholar]
- 12.Hug LA, Maphosa F, Leys D, Löffler FE, Smidt H, Edwards EA, Adrian L. 2013. Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120322. 10.1098/rstb.2012.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holscher T, Krajmalnik-Brown R, Ritalahti KM, Von Wintzingerode F, Gorisch H, Löffler FE, Adrian L. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritalahti KM, Amos BK, Sung Y, Wu Q, Koenigsberg SS, Löffler FE. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rupakula A, Kruse T, Boeren S, Holliger C, Smidt H, Maillard J. 2013. The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120325. 10.1098/rstb.2012.0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maphosa F, Lieten SH, Dinkla I, Stams AJ, Smidt H, Fennell DE. 2012. Ecogenomics of microbial communities in bioremediation of chlorinated contaminated sites. Front. Microbiol. 3:351. 10.3389/fmicb.2012.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens S, Azizian MF, McMurdie PJ, Sabalowsky A, Dolan ME, Semprini L, Spormann AM. 2008. Monitoring abundance and expression of “Dehalococcoides” species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Appl. Environ. Microbiol. 74:5695–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Futamata H, Kaiya S, Sugawara M, Hiraishi A. 2009. Phylogenetic and transcriptional analyses of a tetrachloroethene-dechlorinating “Dehalococcoides” enrichment culture TUT2264 and its reductive dehalogenase genes. Microbes Environ. 24:330–337 [DOI] [PubMed] [Google Scholar]
- 19.Holmes VF, He J, Lee PK, Alvarez-Cohen L. 2006. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl. Environ. Microbiol. 72:5877–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahm BG, Morris RM, Richardson RE. 2006. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl. Environ. Microbiol. 72:5486–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodama Y, Ha LT, Watanabe K. 2007. Sulfurospirillum cavolei sp. nov., a facultatively anaerobic sulfur-reducing bacterium isolated from an underground crude oil storage cavity. Int. J. Syst. Evol. Microbiol. 57:827–831 [DOI] [PubMed] [Google Scholar]
- 22.Sorokin DY, Tourova TP, Muyzer G. 2013. Isolation and characterization of two novel alkalitolerant sulfidogens from a Thiopaq bioreactor, Desulfonatronum alkalitolerans sp. nov. and Sulfurospirillum alkalitolerans sp. nov. Extremophiles 17:535–543 [DOI] [PubMed] [Google Scholar]
- 23.Luijten MLGC, de Weert J, Smidt H, Boschker HTS, de Vos WM, Schraa G, Stams AJM. 2003. Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int. J. Syst. Evol. Microbiol. 53:787–793 [DOI] [PubMed] [Google Scholar]
- 24.Neumann A, Wohlfarth G, Diekert G. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515–16519 [DOI] [PubMed] [Google Scholar]
- 25.Neumann A, Wohlfarth G, Diekert G. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kräutler B, Fieber W, Ostermann S, Fasching M, Ongania KH, Gruber K, Kratky C, Mikl C, Siebert A, Diekert G. 2003. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo-B12, a new type of a natural corrinoid. Helv. Chim. Acta 86:3698–3716 [Google Scholar]
- 27.Regeard C, Maillard J, Holliger C. 2004. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J. Microbiol. Methods 56:107–118 [DOI] [PubMed] [Google Scholar]
- 28.Kimoto H, Suye S, Makishima H, Arai J, Yamaguchi S, Fujii Y, Yoshioka T, Taketo A. 2010. Cloning of a novel dehalogenase from environmental DNA. Biosci. Biotechnol. Biochem. 74:1290–1292 [DOI] [PubMed] [Google Scholar]
- 29.Rouzeau-Szynalski K, Maillard J, Holliger C. 2011. Frequent concomitant presence of Desulfitobacterium spp. and “Dehalococcoides” spp. in chloroethene-dechlorinating microbial communities. Appl. Microbiol. Biotechnol. 90:361–368 [DOI] [PubMed] [Google Scholar]
- 30.Maillard J, Charnay MP, Regeard C, Rohrbach-Brandt E, Rouzeau-Szynalski K, Rossi P, Holliger C. 2011. Reductive dechlorination of tetrachloroethene by a stepwise catalysis of different organohalide respiring bacteria and reductive dehalogenases. Biodegradation 22:949–960 [DOI] [PubMed] [Google Scholar]
- 31.Wagner A, Adrian L, Kleinsteuber S, Andreesen JR, Lechner U. 2009. Transcription analysis of genes encoding homologues of reductive dehalogenases in “Dehalococcoides” sp. strain CBDB1 by using terminal restriction fragment length polymorphism and quantitative PCR. Appl. Environ. Microbiol. 75:1876–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 33.Prat L, Maillard J, Rohrbach-Brandt E, Holliger C. 2012. An unusual tandem-domain rhodanese harbouring two active sites identified in Desulfitobacterium hafniense. FEBS J. 279:2754–2767 [DOI] [PubMed] [Google Scholar]
- 34.Duret A, Holliger C, Maillard J. 2012. The physiological opportunism of Desulfitobacterium hafniense strain TCE1 towards organohalide respiration with tetrachloroethene. Appl. Environ. Microbiol. 78:6121–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossi P, Gillet F, Rohrbach E, Diaby N, Holliger C. 2009. Statistical assessment of variability of terminal restriction fragment length polymorphism analysis applied to complex microbial communities. Appl. Environ. Microbiol. 75:7268–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maillard J, Genevaux P, Holliger C. 2011. Redundancy and specificity of multiple trigger factor chaperones in Desulfitobacteria. Microbiology 157:2410–2421 [DOI] [PubMed] [Google Scholar]
- 37.Palmer T, Berks BC, Sargent F. 2010. Analysis of Tat targeting function and twin-arginine signal peptide activity in Escherichia coli. Methods Mol. Biol. 619:191–216 [DOI] [PubMed] [Google Scholar]
- 38.Neumann A, Wohlfarth G, Diekert G. 1995. Properties of tetrachloroethene and trichloroethene dehalogenase of Dehalospirillum multivorans. Arch. Microbiol. 163:276–281 [Google Scholar]
- 39.Maillard J, Schumacher W, Vazquez F, Regeard C, Hagen WR, Holliger C. 2003. Characterization of the corrinoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl. Environ. Microbiol. 69:4628–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann A, Scholz-Muramatsu H, Diekert G. 1994. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch. Microbiol. 162:295–301 [DOI] [PubMed] [Google Scholar]
- 41.John M, Rubick R, Schmitz RP, Rakoczy J, Schubert T, Diekert G. 2009. Retentive memory of bacteria: long-term regulation of dehalorespiration in Sulfurospirillum multivorans. J. Bacteriol. 191:1650–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marzorati M, Balloi A, de Ferra F, Corallo L, Carpani G, Wittebolle L, Verstraete W, Daffonchio D. 2010. Bacterial diversity and reductive dehalogenase redundancy in a 1,2-dichloroethane-degrading bacterial consortium enriched from a contaminated aquifer. Microb. Cell Fact. 9:12. 10.1186/1475-2859-9-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marzorati M, de Ferra F, Van Raemdonck H, Borin S, Allifranchini E, Carpani G, Serbolisca L, Verstraete W, Boon N, Daffonchio D. 2007. A novel reductive dehalogenase, identified in a contaminated groundwater enrichment culture and in Desulfitobacterium dichloroeliminans strain DCA1, is linked to dehalogenation of 1,2-dichloroethane. Appl. Environ. Microbiol. 73:2990–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



