Abstract
Intraerythrocytic development of the human malaria parasite Plasmodium falciparum appears as a continuous flow through growth and proliferation. To develop a greater understanding of the critical regulatory events, we utilized piggyBac insertional mutagenesis to randomly disrupt genes. Screening a collection of piggyBac mutants for slow growth, we isolated the attenuated parasite C9, which carried a single insertion disrupting the open reading frame (ORF) of PF3D7_1305500. This gene encodes a protein structurally similar to a mitogen-activated protein kinase (MAPK) phosphatase, except for two notable characteristics that alter the signature motif of the dual-specificity phosphatase domain, suggesting that it may be a low-activity phosphatase or pseudophosphatase. C9 parasites demonstrated a significantly lower growth rate with delayed entry into the S/M phase of the cell cycle, which follows the stage of maximum PF3D7_1305500 expression in intact parasites. Genetic complementation with the full-length PF3D7_1305500 rescued the wild-type phenotype of C9, validating the importance of the putative protein phosphatase PF3D7_1305500 as a regulator of pre-S-phase cell cycle progression in P. falciparum.
INTRODUCTION
Malaria caused by Plasmodium falciparum infection is the major type of severe malaria and results in hundreds of thousands of deaths each year and hundreds of millions of clinical illnesses (1, 2). Within the blood of infected individuals, asexual forms of the intraerythrocytic parasite grow rapidly through successive cycles of growth and proliferation. In each asexual generation, active entry into erythrocytes is followed by a growth phase culminating in dynamic release of erythrocyte-invading merozoites. The Plasmodium mitotic cycle is not fully understood and differs from the well-defined models established in yeast and mammalian cells (3). Observing the exact mitotic transitions during the cycle has been difficult due to variations in processes such as chromatid segregation, nuclear division, and spindle formation (4). However, this pattern of development has been observed in other members of the Apicomplexa, such as Toxoplasma gondii and Eimeria tenella, demonstrating the importance and conservation of this process across the phylum (5, 6).
In eukaryotic systems, phosphorylation cascades are critical to cellular development and depend on the coordinated activity of kinases, which are in turn modulated by the activity of phosphatases. There is growing evidence that kinases are critical regulators of cell growth and development in Plasmodium species (7–9). Plasmodium kinases, for example, have been found to be involved with the initial invasion of host cells in addition to egress and differentiation (6, 10). Additional studies have identified kinases in phosphorylation cascades of the gametocyte-ookinete-oocyst transition in the mosquito midgut (11–17). In contrast, few studies have described the phosphatases involved in these processes, which would understandably function with kinases to coregulate cell cycle progression at key checkpoints (18–22). This dearth of information about phosphatases may be due to a smaller relative number of identifiable phosphatases than of kinases in the Plasmodium genome (20, 23). However, it is not uncommon for phosphatases to be fewer due to their nonspecific mechanism of targeting phosphorylated substrates (24).
The protein tyrosine phosphatase (PTP) superfamily is defined by a conserved CX5R motif located in a phosphate-binding pocket. Dual-specificity phosphatases (DUSPs) are a subset of this superfamily that includes the mitogen-activated protein kinase (MAPK) phosphatases (MKPs). MKPs are frequently involved in regulation of cell cycle progression, growth, and proliferation (25). One class of MKP is characterized by the presence of a noncatalytic N-terminal rhodanese (RHD)-like domain utilized for substrate recognition upstream from a catalytic DUSP domain (26–28). In P. falciparum, the PF3D7_1305500 gene is the only one encoding a product with these characteristics (20). In this study, we hypothesize that PF3D7_1305500 is an atypical MAPK phosphatase of P. falciparum expressed during intraerythrocytic development.
MATERIALS AND METHODS
In vitro parasite culture conditions.
P. falciparum NF54 and mutant C9 clones were cultured according to standard methods at 37°C (5% O2 and 5% CO2, nitrogen balanced) in 5% hematocrit (O+ blood) and RPMI 1640 medium with 0.5% Albumax II, 0.25% sodium bicarbonate, and 0.01 mg/ml gentamicin (29). The C9 mutant parasite line was created by random insertional mutagenesis using the piggyBac transposon pXL-BACII-HDGH. The location of the insertion in the PF3D7_1305500 open reading frame (ORF) was confirmed by thermal asymmetric interlaced (TAIL) sequence analysis.
Determination of merozoite number per schizont.
Parasite cultures were double synchronized by standard methods using 5% sorbitol (29). Merozoites were counted in 300 singly infected segmented schizonts in Giemsa-stained thin smears from NF54 and C9 cultured parasites 40 h postsynchronization to determine the average number of merozoites produced per schizont.
RNA extraction and analysis by qRT-PCR and RT-PCR.
NF54 RNA was collected from 6 intraerythrocytic developmental stages (early rings, late rings, early trophozoites, late trophozoites, early schizonts, and late schizonts [ER, LR, ET, LT, ES, and LS, respectively]) followed by saponin lysis, suspended in TRIzol reagent (Life Technologies), RNA purified, and treated with DNase I. Purity was confirmed by PCR carried out without the addition of reverse transcriptase (RT). PF3D7_1305500 was amplified using primers 5′-TCGATTTTGAGGAGCTGAA-3′ and 5′-GGGTAAAACATCCTTTTTGTT-3′ with the SuperScript III Platinum SYBR Green One-Step quantitative RT (qRT)-PCR kit (Life Technologies) following the manufacturer's protocol. The relative expression of PF3D7_1305500 was then normalized against actin (PFL2215w). For RT-PCR analysis, 100 ng DNAse I-treated total RNA was amplified using primers 5′-CACCATGGAATATAAAAGCATCGATTTTG-3′ and 5′-GTTTATGTAATTATTTATTACTATAAATGGTC-3′ and analyzed by horizontal agarose gel electrophoresis. As a control, 18 RNA (PF3D7_0112300) was amplified for each sample.
Plasmid constructs and genetic complementation.
The plasmid was developed with the full-length PF3D7_1305500 ORF and its native 700-bp 5′ untranslated region (UTR) using primers 5′-CACCCTACCCCTGTATTATTTCCTACCCTC-3′ and 5′-GTTTATGTAATTATTTATTACTATAAATGGTC-3′ along with a C-terminal hemagglutinin (HA) tag and a 3′UTR calmodulin (CAM) termination sequence. The transfection plasmid carried a blasticidin S deaminase (BSD) drug selection cassette under the control of the 5′UTR of the gene encoding histidine-rich protein (HRP3), and the 3′ UTR of the gene encoding histidine-rich protein-2 (HRP2) was used for drug selection. The vector included two inverted terminal repeat regions (ITR1 and ITR2), a feature included to promote the integration of a stable ORF as done in previous transgenic expression systems (30). Schizonts were isolated from a 20-ml culture with 3 to 5% parasitemia using a VarioMACS Separator (Miltenyi Biotec) and counted with a hemacytometer. Fresh 50% hematocrit blood was washed, combined with Cytomix (29) in a 1:1 (vol/vol) ratio, aliquoted into chilled 2-mm cuvettes, and electroporated using a Gene Pulser Xcell CE (Bio-Rad) to load erythrocytes (RBCs) with transposon and helper plasmids purified using methods described previously (31). Positive transfected clones were selected using blasticidin, diluted, transferred to 96-well culture plates, and maintained for 17 days to select individual clones.
Plasmodium falciparum growth assay and cell cycle determination.
Growth assays for cell cycle determination were performed by maintaining tightly synchronized cultures of P. falciparum NF54 and C9 clones at 0.5 to 2% parasitemia for 168 h. Parasite cultures were started at the same parasitemia, and time point cultures were collected every 2 h and then fixed in 0.05% glutaraldehyde after removal of culture medium. Parasitemia was estimated using flow cytometry as described previously (32) by staining parasites with ethidium bromide and analyzed using an Accuri C6 flow cytometry system (BD Accuri). A total of 100,000 cells were counted for each sample, and the data were analyzed using CFlow Plus software (BD Accuri). The cell cycle was determined by comparing the relative abundance of each developmental stage at each time point according to methods developed previously (33). The relative fold change was determined by calculating the fold increase in parasitemia between time zero and the endpoint. Each sample was then plotted as a percentage relative to NF54.
Whole-genome sequencing.
To create a reference genome for the parental line, NF54 genomic DNA (gDNA) was converted into Illumina sequencing libraries using both PCR-free (34) and Kappa HiFi methods (35) in order to minimize the bias against AT-rich DNA that is introduced by PCR enzymes in standard Illumina sequencing methodologies. NF54 libraries were sequenced with 76-bp and 250-bp paired end reads on an Illumina HiSeq and MiSeq (ENA accession numbers ERS038926 and ERS184445, respectively). The sequence data were assembled into an NF54 reference genome using ICORN (36), which iteratively mapped, compared, and corrected the reads against the P. falciparum 3D7 reference genome from GeneDB (37). ICORN ran 7 iterations and corrected 526 1-bp substitutions and >600 small insertion and deletions. Annotation was transferred onto this NF54 genome from the 3D7 reference using RATT (38).
An Illumina library was generated from PF3D7_1305500 mutant (C9) mutant strain gDNA using the Kappa HiFi amplification methodology and sequenced with 76-bp paired end reads on an Illumina HiSeq (ENA accession ERS038913). C9 and NF54 76 bp reads were mapped against the NF54 genome using SMALT (http://www.sanger.ac.uk/resources/software/smalt/; parameters: -r, 0; -i, 800; -y, 0.9; index was done with -k of 13 and -s of 3); 90.2% and 89.7% of reads mapped to the NF54 genome, with an average coverage of 54× and 57×, respectively, for the NF54 and C9 data. To identify locations where C9 differed from the parental NF54 genome, we merged the two BAM files using individual read group and run GATK (39) to realign the reads and call variants using the Plasmodium falciparum settings. Variations were called when the quality score was higher than 60 and when both samples had 10 or more reads mapping to the potential variant locus.
Illumina sequence data were also used to confirm the location of the piggyBac insertion site in the C9 genome. The sequence of the piggyBac transposon was added to the NF54 genome sequence, and then the C9 reads were mapped against this combined genome with SMALT using the -x parameter, which maps each read pair independently. We then searched for mate pairs in which one read mapped to the piggyBac sequence and the other within the reference genome.
Multiple alignments and phylogenetic analysis.
The sequence of PF3D7_1305500 and orthologous Plasmodium sequences were retrieved from PlasmoDB v9.2 (www.plasmodb.org). Outlier species were identified through BLAST searches with the DUSP domain using NCBI BLASTP. Sequences with the greatest homology to the PF3D7_1305500 ORF were retrieved and used to build the multiple alignments using ClustalW (40, 41). The evolutionary history was inferred using a phylogenetic tree created using the neighbor-joining method with 1,000 bootstrap replicates MEGA5 (42–44).
RESULTS
Identification of an attenuated growth mutant in P. falciparum.
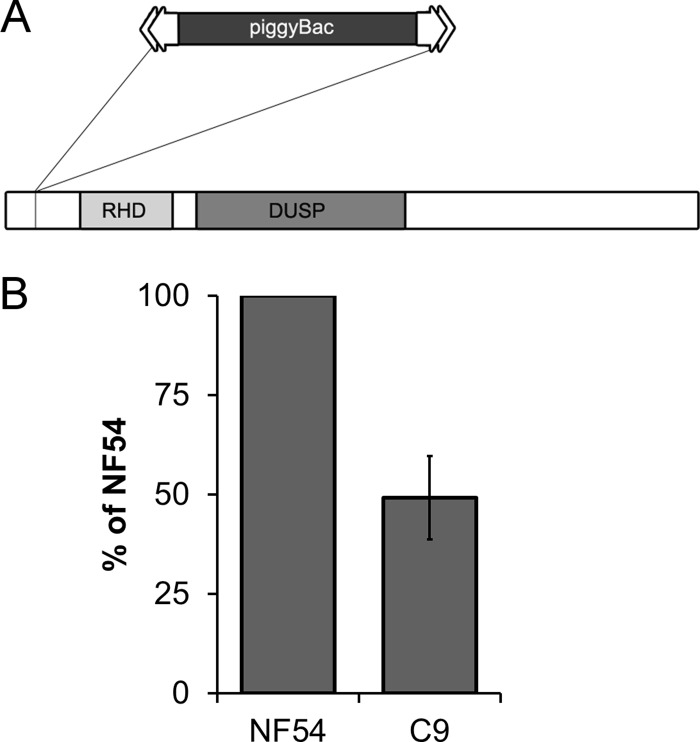
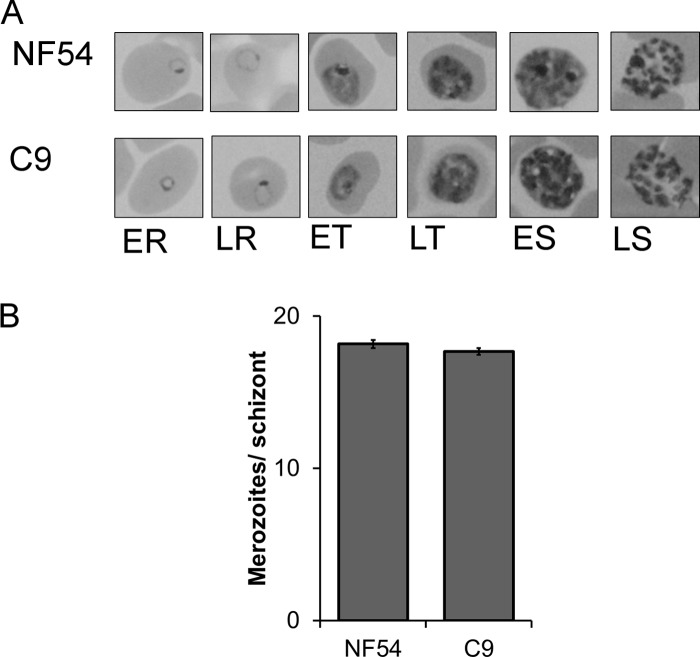
A collection of unique mutant clones was created from a laboratory line of P. falciparum NF54 using random insertional mutagenesis with a piggyBac transposon (31). The C9 parasite line carried one copy of a piggyBac transposon (pXL-BACII-HGDH) inserted at the TTAA nucleotides 199 to 202 downstream of the start codon at the 5′ end of the single ORF of PF3D7_1305500 (Fig. 1A) (see the supplemental material). Intraerythrocytic growth for clone C9 was analyzed and determined to be severely attenuated with a net growth rate consistently ∼50% of the NF54 parent (Fig. 1B). The growth-attenuating mutation in C9, though serious, is not fatal and allows the parasites to develop in a seemingly normal pattern in culture. During development, intraerythrocytic stages did not demonstrate any obvious differences in morphological characteristics (Fig. 2A), and the mean numbers of merozoites produced did not vary significantly from those of the NF54 parent (Fig. 2B). It was apparent that each cycle of the mutant parasite required a longer development time, but otherwise, the organism appeared morphologically similar in Giemsa-stained thin blood smears. While the longer cell cycle length can account for some of the lower growth rate, the merozoites also appear to have a lower invasion rate.
Fig 1.
The growth phenotype of the C9 mutant parasite is due to disruption of PF3D7_1305500. (A) Schematic of PF3D7_1305500 disrupted by a single insertion of the piggyBac transposon. Tandem RHD and DUSP domains are characteristic of MKPs. (B) Calculation of fold change reveals that the knockout of PF3D7_1305500 resulted in a reduced fold change of ∼50% relative to NF54.
Fig 2.
Morphological analysis of the C9 mutant compared to NF54. (A) Comparison of Giemsa-stained thin blood smears of the wild-type parent NF54 and C9 did not reveal an obvious difference in major developmental stages. The hours postinvasion indicated in the micrographs are as follows: ER, 8 h; LR, 16 h; ET, 24 h; LT, 32 h; ES, 40 h; LS, 48 h. (B) Average merozoite counts in segmented schizonts of NF54 and C9 were not statistically different.
Defining characteristics of PF3D7_1305500 MKP.
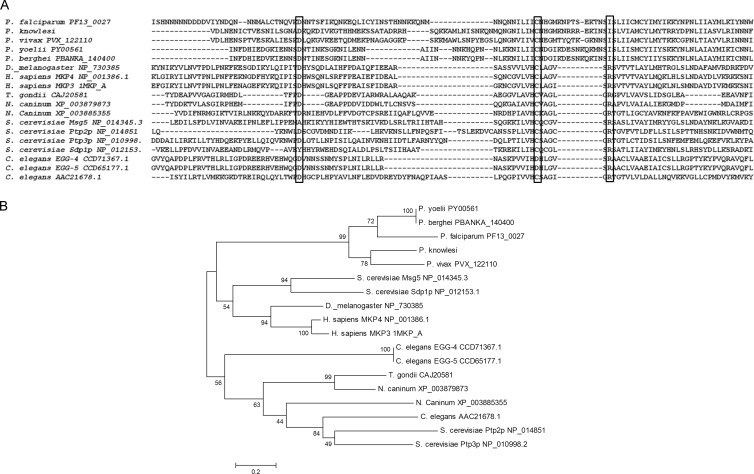
The protein encoded by PF3D7_1305500 was determined to have two key structural features, an RHD domain followed by a DUSP-like domain. Bioinformatics analysis defined this tandem arrangement as characteristic of certain MKPs conserved within humans and other model organisms such as fruit fly and yeast (45–48). It is for this reason that we refer to the PF3D7_1305500 product as P. falciparum MKP1. The conserved signature motif of CX5R, which is typically definitive of DUSP domains in MKPs, is only partially conserved in PF3D7_1305500 (Fig. 3A). The putative binding pocket of MKP1 was identified through BLAST searches and multiple sequence alignments with MKPs that have three-dimensional (3-D) crystal structures (Plasmodb version 9.2; NCBI GenBank Flat files release 193.0). Within this binding pocket, the amino acid residue cysteine-383 (C383) along with another residue of the catalytic triad aspartate-345 (D345) are conserved and align with conserved cysteine and aspartic acid of the other identified DUSPs. However, the third conserved catalytic residue, arginine, aligns with isoleucine-398 (I398) in PF3D7_1305500. During dephosphorylation, the conserved arginine is critical for dephosphorylation activity, since active DUSP domains often depend on arginine to maintain the transition state with the phosphorylated substrate (28). Absence of arginine is expected to drastically reduce the catalytic activity of the DUSP; therefore, this is an important departure from the consensus motif defined for catalytically active DUSP domain orthologs and would be expected to reduce phosphatase activity (28, 49).
Fig 3.
Multiple alignment and phylogenetic analysis of PF3D7_1305500. (A) Alignment of PF3D7_1305500 with its Plasmodium orthologs and outlier species showing the conservation of catalytic residues (boxes). Cysteine and aspartic acid align with all homologs. Isoleucine aligns with the position of the conserved arginine and is conserved in all species of Plasmodium. A string of residues (bracket) are inserted into the signature motif and are conserved among the Plasmodium orthologs. (B) Phylogenetic analysis using the neighbor-joining method with 1,000 bootstrap replicates shows grouping of the Plasmodium sequences independent of the other species, suggesting an early divergence in the evolutionary lineage.
Prior mutagenesis studies and analyses of catalytic domains in the DUSPs of model organisms suggest that unique characteristics, such as the ones identified in PF3D7_1305500, may be characteristic of a pseudophosphatase or a low-activity phosphatase (50–52). Additionally, there is an insertion of nine residues disrupting the spacing within the CX5R signature motif. Though it cannot be determined if this insertion changes the three-dimensional structure within the putative binding pocket, this unique stretch of residues is conserved in each of the Plasmodium orthologs. Conservation of these unique characteristics in Plasmodium species supports the formation of an individual clade (Fig. 3B).
PF3D7_1305500 regulates transition from pre-S phase to S/M phase.
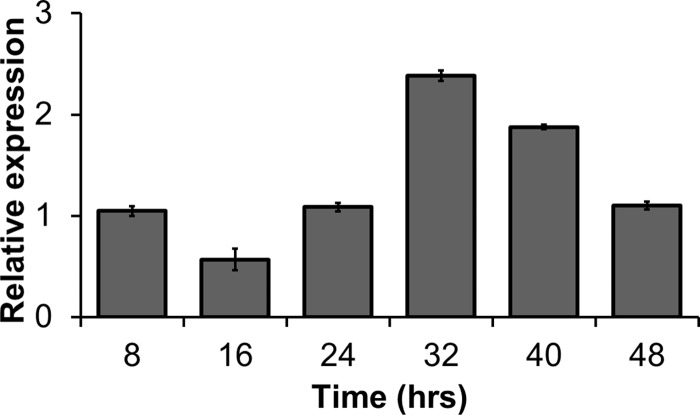
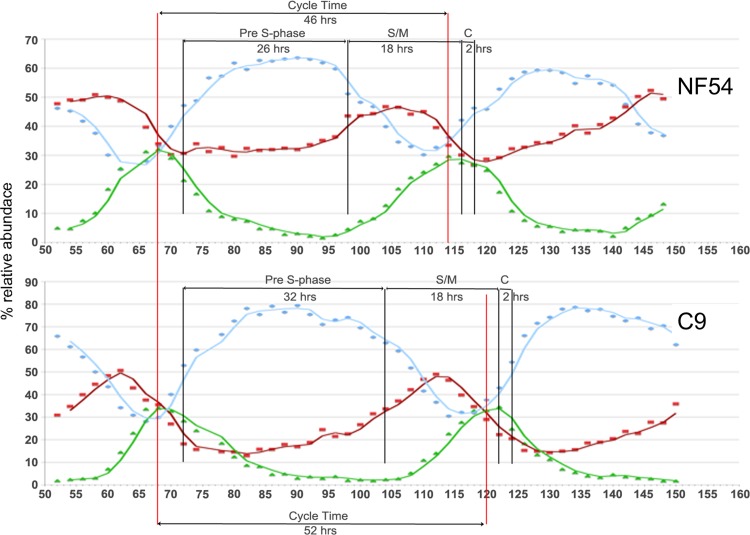
In wild-type parasites, the highest relative abundance of PF3D7_1305500 transcripts is at the end of the pre-S development phase (i.e., late trophozoite) during intraerythrocytic development (53). This expression profile coincides with the stage when the C9 mutant cycle deviates from the wild-type cell cycle (Fig. 4). Utilizing the detailed time course experimental protocol developed previously (33), the timing for NF54 was determined to be 46 h compared to 52 h in the C9 MKP-null mutant. The difference resulted entirely from a prolonged pre-S trophozoite stage causing late entry into the S/M schizont phase. The duration of schizont development (S/M − C) was similar in C9 and NF54, making pre-S phase the only abnormal growth phase of the intraerythrocytic cycle (Fig. 5).
Fig 4.
Transcription profile of PF3D7_1305500. Analysis by qRT-PCR showed that expression of PF3D7_1305500 has its highest expression relative to actin during the late trophozoite stage 32 h postinvasion. This stage corresponds to late pre-S development, during which the cell cycle of null mutants deviates from the wild-type development pattern. The time points 8, 16, 24, 32, 40, and 48 h postinvasion correspond to ER, LR, ET, LT, ES, and LS, respectively.
Fig 5.
The cell cycle of the C9 MKP-null mutant is altered. Cell cycle analysis reveals a prolonged pre-S phase in mutant parasite lines. Late entry into the S/M phase leads to an overall longer cycle time producing a slow-growing phenotype. The blue, red, and green graph lines represent the relative abundance of rings, trophozoites, and schizonts, respectively. The pre-S phase in NF54 is 26 h andis followed by a 16-h S/M phase and 2-h cytokinesis (C). In C9, the cycle time is increased by 6 h due to the longer 32-hour pre-S phase. Late entry into the S/M phase coincides with the timing for peak expression of PF3D7_1305500, suggesting that the deficiency in the mutant cycle can be correlated to the gene expression pattern.
Phenotype rescue of wild-type growth by genetic complementation.
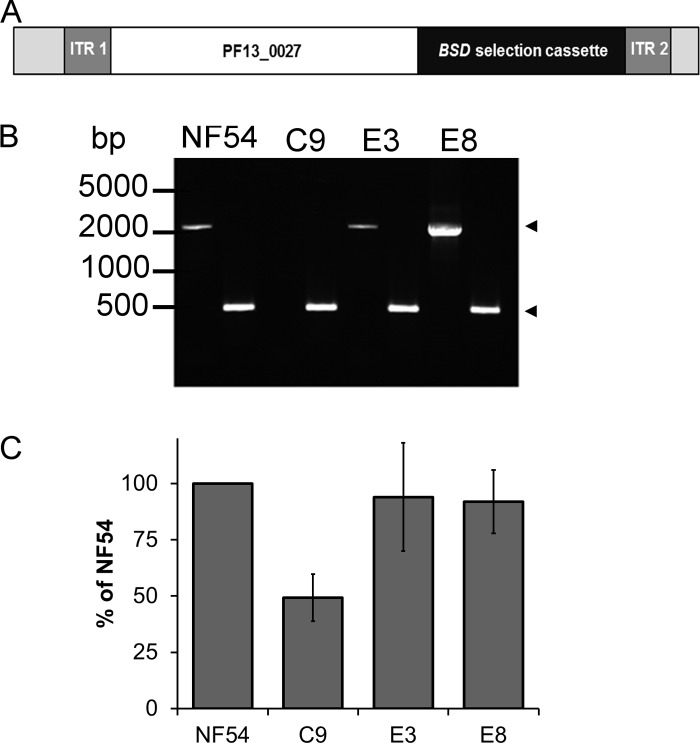
Attenuated growth of the C9 MKP-null mutant remained stable over multiple subsequent generations, suggesting that survival was not due to phenotype reversion. This is consistent with general experience using the piggyBac system as it is now extensively used in a number of organisms, and the transposable elements remain integrated in the genome in the absence of transposase (31, 54–58). However, the extended maintenance of P. falciparum intraerythrocytic cultures required for transfection and the selection process in the experimental studies increases the possibility for secondary mutations to alter the cell cycle or cause growth attenuation. Therefore, to validate that the phenotype was due to disruption of PF3D7_1305500, we genetically complemented the C9 mutant with a full-length copy of PF3D7_1305500, including its putative promoter region (Fig. 6A). The 700-bp 5′ intergenic region between PF3D7_1305500 and the upstream gene MAL13P1.28 was added to the ORF to ensure that the native promoter was included. Using the BSD resistance marker on the complement vector, we were able to select for two independent cloned lines, E3 and E8. RT-PCR analysis of both E3 and E8 revealed that the complemented parasite lines transcribed PF3D7_1305500, in contrast to what occurred with C9, which did not have detectable transcripts (Fig. 6B). By Southern blotting hybridization analysis, it was also determined that the complemented parasite lines maintained the full-length copy of PF3D7_1305500 as stable episomes (data not shown). This finding was also confirmed by whole-genome sequencing of the mutant and complemented parasite lines (Table 1). Forty-four positions met the criteria as potential differences between the C9 and parent strains, comprising 40 single-nucleotide polymorphisms (SNPs) and 4 indels. Of these potential variants, only 11 occurred within open reading frames. Closer examination of these 11 revealed that 9 fall into low-complexity regions, repetitive sequence and homopolymer tracts, and hence clear calling of variants is not possible. Only 2 SNPs were confirmed in coding sequences outside such low-complexity regions, one in PF3D7_0704000 and one in PF3D7_0730300. PF3D7_0704000 is a conserved protein of unknown function, and C9 contains a nonsynonymous mutation (D to Y) at position 4201 of the predicted protein sequence. PF3D7_0730300 is a member of the AP2 family of transcription factors that is known to play a role in liver stage parasite development, but with no known role in blood stage parasites. C9 contains a nonsynonymous mutation (G to E) at position 3047 of the predicted protein, which is near the C terminus of the protein and well outside the AP2 DNA binding domains. Using the Illumina sequence data to confirm the location of the piggyBac insertion site confirmed only a single piggyBac insertion site in the C9 genome, at position 271000 of chromosome 13.
Fig 6.
Gene complementation of the C9 mutant rescues the wild-type phenotype. (A) The plasmid construct used to complement the attenuated C9 parasite line was developed using the full-length PF3D7_1305500 ORF inserted adjacent to a BSD drug selection cassette. (B) RT-PCR analysis detected PF3D7_1305500 transcripts in NF54 and the complemented parasites (E3 and E8), but not in the C9 MKP-null mutant (upper arrow). As a positive control, 18S RNA was also included for each sample (lower arrow). (C) The relative fold changes of complemented parasites (E3 and E8) were statistically the same as those of wild-type NF54, demonstrating successful rescue of the phenotype.
Table 1.
Whole-genome sequencing data for NF54, C9, and the complemented parasite linesa
| Clone | Chromosome | Position | Presence of piggyBac insertion | Gene | Episomal complementation | No. of unique SNPs | SNP genes |
|---|---|---|---|---|---|---|---|
| NF54 | No | None | No | ||||
| C9 | 13 | 271000 | Yes | PF3D7_1305500 | No | 2 | PF3D7_0704000, PF3D7_0730300 |
| E3 | 13 | 271000 | Yes | PF3D7_1305500 | Yes | 2 | PF3D7_0704000, PF3D7_0730300 |
| E8 | 13 | 271000 | Yes | PF3D7_1305500 | Yes | 2 | PF3D7_0704000, PF3D7_0730300 |
A single SNP in the putative clathrin coat assembly protein AP180 was present in all samples. This experiment also validated that the complementation of C9 was episomal.
Complementation of the C9 mutant with the full-length copy of PF3D7_1305500 rescued the phenotype of both E3 and E8 as evident by their return to normal growth (Fig. 6C). Significance of the phenotypic rescue was determined by Kruskal-Wallis comparison (P = 0.0038) with Dunn's multiple-comparison test to determine significance between each sample.
DISCUSSION
Cell cycle progression in P. falciparum and completion of intraerythrocytic development is highly dependent on a precise pattern of metabolic events. Disruption of any of the numerous biochemical pathways and processes is anticipated to have detrimental effects on the efficiency of this process. In our study, we discovered that normal cell cycle development was delayed by disruption of PF3D7_1305500, indicating that this atypical phosphatase is a regulator of the P. falciparum cell cycle. The delayed transition from the pre-S trophozoite to the S/M schizont suggests this transition phase during the parasite's intraerythrocytic growth is a cell cycle checkpoint. Rescuing the phenotype in the null mutant through genetic complementation validated the notion that the attenuated phenotype was due to disruption of PF3D7_1305500.
Considering the attenuated phenotype along with the homology found between PF3D7_1305500 and the other well-characterized MKPs, it can be suggested that this putative atypical phosphatase might fulfill a similar function regulating MAPK in Plasmodium. MKPs of similar structure are often involved in signaling pathways, which is a likely function of PF3D7_1305500 (20, 59). Investigations in yeast demonstrate that MKPs are critical components of various signal transduction pathways that regulate transcription and maturation, which can also have an influence on the cell cycle (45, 46). Disruption of such functions in P. falciparum could produce the phenotype observed in C9. This domain structure is not evident in any other gene in the P. falciparum genome, but single-copy orthologs are evident in all of the other Plasmodium species with completed genomes, suggesting that its function is conserved among all malaria parasites. The presence of an RHD domain upstream of the DUSP is consistent with a secondary regulatory function that aids in substrate recognition and activity of MKPs (25, 27, 60). Conservation of this domain in PF3D7_1305500 as determined by bioinformatics analysis implicates an MAPK-like function for this atypical phosphatase.
Imprecise identification of PF3D7_1305500 as a putative protein phosphatase was likely due to the limited conservation of the conserved CX5R signature motif. Each catalytic residue is critical to optimal function of a phosphatase, and the modifications suggest that this DUSP may not be highly catalytic or possibly a pseudophosphatase. Noncatalytic pseudophosphatases typically maintain structural homology with active phosphatases, allowing them to trap phosphoproteins, thereby regulating cellular functions without dephosphorylation activity (51, 52). One such DUSP homology domain in pseudophosphatases is referred to as a serine/threonine/tyrosine interacting (STYX) domain, which has an endogenous substitution of one, or more, of the catalytic residues (51, 61, 62). Reports of pseudophosphatase activity in Caenorhabditis elegans demonstrate this function as an important role fulfilled by Egg-4 and Egg-5 in controlling oocyte-to-zygote transition (63). It has also been proposed that physical access of native phosphatases is blocked by these pseudophosphatases, which exert a “dominant negative” function, thereby protecting substrates from dephosphorylation (52). This is not a surprising regulatory mechanism, considering that the DUSP binding pockets generally lack substrate specificity. Interestingly, members of the Apicomplexa possess a unique group of pseudophosphatases with long N-terminal domains and EF-hand motifs termed “EFPPs” (64). However, a grouping of the variety of STYX domain pseudophosphatases, which have a substitution of the cysteine residue in the CX5R motif, has never been characterized in Plasmodium. The PF3D7_1305500 product does not have the EF-hand motif or Ca2+ binding sites typically associated with the EFPP grouping; however, MKP1 is missing the conserved arginine that would make it a unique classification of putative protozoan pseudophosphatase.
The pressing need for new antimalarial drugs and identification of new targets is critical due to emerging resistance to frontline drugs and the lack of diverse chemotherapeutic targets (65, 66). Furthermore, there have not been any new classes of antimalarial drugs introduced into clinical practice since 1996 (67–69). As a result, the preferred methods for use of antimalarial drugs have been combination therapies due to the foreseeable challenges associated with monotherapy or highly mutable drug targets (66, 70). Kinases and other regulators of phosphorylation pathways of malaria parasites represent potential high-value targets for future antimalarial drugs. However, the complex processes of phosphorylation cascades in Plasmodium are poorly understood and limit our ability to identify the highest-value targets. Our discovery of PF3D7_1305500 as an important regulator of the cell cycle helps elucidate the trophozoite-to-schizont transition stage as a potentially vulnerable step of the developmental cycle and will help create new avenues into understanding Plasmodium biology. The phenotype associated with C9 highlights this pathway and the regulated processes as potential targets. With further delineation of the function of the PF3D7_1305500 and identification of its interacting partners, additional knowledge arising from its study will aid future drug discovery.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the valuable contributions of support from Mandy Sanders and Matthew Berriman from the Wellcome Trust Sanger Institute for assistance in the whole-genome sequencing.
This study was supported by funds from the U.S. National Institutes of Health (R01AI033656 and R01AI094973 to J.H.A. and F31AI083053 to C.C.), European 7th framework EviMalaR (to T.D.O.), and The Wellcome Trust (to J.C.R.).
B.B., C.C., and J.H.A. designed the research. B.B., C.C., J.S., S.M., N.S., P.T., and A.P. performed the research. B.B., C.C., J.C.R., A.P., and J.H.A. analyzed the data; and B.B., C.C., and J.H.A. wrote the paper.
Footnotes
Published ahead of print 28 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00028-13.
REFERENCES
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434:214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO 2011. World Malaria Report 2011. World Health Organization, Geneva, Switzerland: http://www.who.int/malaria/world_malaria_report_2011/. [Google Scholar]
- 3.Arnot DE, Gull K. 1998. The Plasmodium cell-cycle: facts and questions. Ann. Trop. Med. Parasitol. 92:361–365 [DOI] [PubMed] [Google Scholar]
- 4.Arnot DE, Ronander E, Bengtsson DC. 2011. The progression of the intra-erythrocytic cell cycle of Plasmodium falciparum and the role of the centriolar plaques in asynchronous mitotic division during schizogony. Int. J. Parasitol. 41:71–80 [DOI] [PubMed] [Google Scholar]
- 5.Ng ST, Sanusi Jangi M, Shirley MW, Tomley FM, Wan KL. 2002. Comparative EST analyses provide insights into gene expression in two asexual developmental stages of Eimeria tenella. Exp. Parasitol. 101:168–173 [DOI] [PubMed] [Google Scholar]
- 6.Lim DC, Cooke BM, Doerig C, Saeij JP. 2012. Toxoplasma and Plasmodium protein kinases: roles in invasion and host cell remodelling. Int. J. Parasitol. 42:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucet IS, Tobin A, Drewry D, Wilks AF, Doerig C. 2012. Plasmodium kinases as targets for new-generation antimalarials. Future Med. Chem. 4:2295–2310 [DOI] [PubMed] [Google Scholar]
- 8.Ma J, Rahlfs S, Jortzik E, Schirmer RH, Przyborski JM, Becker K. 2012. Subcellular localization of adenylate kinases in Plasmodium falciparum. FEBS Lett. 586:3037–3043 [DOI] [PubMed] [Google Scholar]
- 9.Rached FB, Ndjembo-Ezougou C, Chandran S, Talabani H, Yera H, Dandavate V, Bourdoncle P, Meissner M, Tatu U, Langsley G. 2012. Construction of a Plasmodium falciparum Rab-interactome identifies CK1 and PKA as Rab-effector kinases in malaria parasites. Biol. Cell 104:34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, Bright AT, Westenberger S, Winzeler E, Blackman MJ, Baker DA, Wandless TJ, Duraisingh MT. 2010. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science 328:910–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorin-Semblat D, Schmitt S, Semblat JP, Sicard A, Reininger L, Goldring D, Patterson S, Quashie N, Chakrabarti D, Meijer L, Doerig C. 2011. Plasmodium falciparum NIMA-related kinase Pfnek-1: sex specificity and assessment of essentiality for the erythrocytic asexual cycle. Microbiology 157:2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reininger L, Garcia M, Tomlins A, Muller S, Doerig C. 2012. The Plasmodium falciparum, Nima-related kinase Pfnek-4: a marker for asexual parasites committed to sexual differentiation. Malaria J. 11:250. 10.1186/1475-2875-11-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian S, Brochet M, Collins MO, Schwach F, Jones ML, Goulding D, Rayner JC, Choudhary JS, Billker O. 2012. A Plasmodium calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs. Cell Host Microbe 12:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reininger L, Tewari R, Fennell C, Holland Z, Goldring D, Ranford-Cartwright L, Billker O, Doerig C. 2009. An essential role for the Plasmodium Nek-2 Nima-related protein kinase in the sexual development of malaria parasites. J. Biol. Chem. 284:20858–20868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tewari R, Straschil U, Bateman A, Bohme U, Cherevach I, Gong P, Pain A, Billker O. 2010. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe 8:377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alano P. 2007. Plasmodium falciparum gametocytes: still many secrets of a hidden life. Mol. Microbiol. 66:291–302 [DOI] [PubMed] [Google Scholar]
- 17.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, Brinkmann V. 2004. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117:503–514 [DOI] [PubMed] [Google Scholar]
- 18.Mamoun CB, Goldberg DE. 2001. Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor 1beta and inhibits its PKC-mediated nucleotide exchange activity in vitro. Mol. Microbiol. 39:973–981 [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama D, Saito-Ito A, Asao N, Tanabe K, Yamamoto M, Matsumura T. 1998. Modulation of the growth of Plasmodium falciparum in vitro by protein serine/threonine phosphatase inhibitors. Biochem. Biophys. Res. Commun. 247:18–23 [DOI] [PubMed] [Google Scholar]
- 20.Wilkes JM, Doerig C. 2008. The protein-phosphatome of the human malaria parasite Plasmodium falciparum. BMC Genomics 9:412. 10.1186/1471-2164-9-412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hills T, Srivastava A, Ayi K, Wernimont AK, Kain K, Waters AP, Hui R, Pizarro JC. 2011. Characterization of a new phosphatase from Plasmodium. Mol. Biochem. Parasitol. 179:69–79 [DOI] [PubMed] [Google Scholar]
- 22.Guttery DS, Poulin B, Ferguson DJ, Szoor B, Wickstead B, Carroll PL, Ramakrishnan C, Brady D, Patzewitz EM, Straschil U, Solyakov L, Green JL, Sinden RE, Tobin AB, Holder AA, Tewari R. 2012. A unique protein phosphatase with kelch-like domains (PPKL) in Plasmodium modulates ookinete differentiation, motility and invasion. PLoS Pathog. 8:e1002948. 10.1371/journal.ppat.1002948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung DW, Ponts N, Cervantes S, Le Roch KG. 2009. Post-translational modifications in Plasmodium: more than you think! Mol. Biochem. Parasitol. 168:123–134 [DOI] [PubMed] [Google Scholar]
- 24.Almo SC, Bonanno JB, Sauder JM, Emtage S, Dilorenzo TP, Malashkevich V, Wasserman SR, Swaminathan S, Eswaramoorthy S, Agarwal R, Kumaran D, Madegowda M, Ragumani S, Patskovsky Y, Alvarado J, Ramagopal UA, Faber-Barata J, Chance MR, Sali A, Fiser A, Zhang ZY, Lawrence DS, Burley SK. 2007. Structural genomics of protein phosphatases. J. Struct. Funct. Genomics 8:121–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondoh K, Nishida E. 2007. Regulation of MAP kinases by MAP kinase phosphatases. Biochim. Biophys. Acta 1773:1227–1237 [DOI] [PubMed] [Google Scholar]
- 26.Farooq A, Chaturvedi G, Mujtaba S, Plotnikova O, Zeng L, Dhalluin C, Ashton R, Zhou MM. 2001. Solution structure of ERK2 binding domain of MAPK phosphatase MKP-3: structural insights into MKP-3 activation by ERK2. Mol. Cell 7:387–399 [DOI] [PubMed] [Google Scholar]
- 27.Farooq A, Zhou MM. 2004. Structure and regulation of MAPK phosphatases. Cell. Signal. 16:769–779 [DOI] [PubMed] [Google Scholar]
- 28.Theodosiou A, Ashworth A. 2002. MAP kinase phosphatases. Genome Biol. 3:REVIEWS3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malaria Research and Reference Reagent Resource Center 2008. Methods in malaria research, vol 5.2 Malaria Research and Reference Reagent Resource Center, Paris, France [Google Scholar]
- 30.Balu B, Adams JH. 2006. Functional genomics of Plasmodium falciparum through transposon-mediated mutagenesis. Cell. Microbiol. 8:1529–1536 [DOI] [PubMed] [Google Scholar]
- 31.Balu B, Shoue DA, Fraser MJ, Jr, Adams JH. 2005. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proc. Natl. Acad. Sci. U. S. A. 102:16391–16396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balu B, Singh N, Maher SP, Adams JH. 2010. A genetic screen for attenuated growth identifies genes crucial for intraerythrocytic development of Plasmodium falciparum. PLoS One 5:e13282. 10.1371/journal.pone.0013282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo I, Oksman A, Vaupel B, Goldberg DE. 2009. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc. Natl. Acad. Sci. U. S. A. 106:1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. 2009. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6:291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyola SO, Otto TD, Gu Y, Maslen G, Manske M, Campino S, Turner DJ, Macinnis B, Kwiatkowski DP, Swerdlow HP, Quail MA. 2012. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics 13:1. 10.1186/1471-2164-13-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto TD, Sanders M, Berriman M, Newbold C. 2010. Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology. Bioinformatics 26:1704–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logan-Klumpler FJ, De Silva N, Boehme U, Rogers MB, Velarde G, McQuillan JA, Carver T, Aslett M, Olsen C, Subramanian S, Phan I, Farris C, Mitra S, Ramasamy G, Wang H, Tivey A, Jackson A, Houston R, Parkhill J, Holden M, Harb OS, Brunk BP, Myler PJ, Roos D, Carrington M, Smith DF, Hertz-Fowler C, Berriman M. 2012. GeneDB–an annotation database for pathogens. Nucleic Acids Res. 40:D98–D108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 39:e57. 10.1093/nar/gkq1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 41.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Peterson D, Tamura K. 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28:2685–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gustin MC, Albertyn J, Alexander M, Davenport K. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin H, Flandez M, Nombela C, Molina M. 2005. Protein phosphatases in MAPK signalling: we keep learning from yeast. Mol. Microbiol. 58:6–16 [DOI] [PubMed] [Google Scholar]
- 47.Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Jr, Dixon JE. 2007. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. U. S. A. 104:6596–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arino J. 2002. Novel protein phosphatases in yeast. Eur. J. Biochem. 269:1072–1077 [DOI] [PubMed] [Google Scholar]
- 49.Denu JM, Dixon JE. 1998. Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 2:633–641 [DOI] [PubMed] [Google Scholar]
- 50.Wishart MJ, Denu JM, Williams JA, Dixon JE. 1995. A single mutation converts a novel phosphotyrosine binding domain into a dual-specificity phosphatase. J. Biol. Chem. 270:26782–26785 [DOI] [PubMed] [Google Scholar]
- 51.Wishart MJ, Dixon JE. 1998. Gathering STYX: phosphatase-like form predicts functions for unique protein-interaction domains. Trends Biochem. Sci. 23:301–306 [DOI] [PubMed] [Google Scholar]
- 52.Tonks NK. 2009. Pseudophosphatases: grab and hold on. Cell 139:464–465 [DOI] [PubMed] [Google Scholar]
- 53.Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, Lemieux J, Barrell B, Pain A, Berriman M, Newbold C, Llinas M. 2010. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 76:12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraser MJ, Ciszczon T, Elick T, Bauser C. 1996. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 5:141–151 [DOI] [PubMed] [Google Scholar]
- 55.O'Brochta DA, Atkinson PW. 1996. Transposable elements and gene transformation in non-drosophilid insects. Insect Biochem. Mol. Biol. 26:739–753 [DOI] [PubMed] [Google Scholar]
- 56.Morales ME, Mann VH, Kines KJ, Gobert GN, Fraser MJ, Jr, Kalinna BH, Correnti JM, Pearce EJ, Brindley PJ. 2007. piggyBac transposon mediated transgenesis of the human blood fluke, Schistosoma mansoni. FASEB J. 21:3479–3489 [DOI] [PubMed] [Google Scholar]
- 57.Sethuraman N, Fraser MJ, Jr, Eggleston P, O'Brochta DA. 2007. Post-integration stability of piggyBac in Aedes aegypti. Insect Biochem. Mol. Biol. 37:941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fonager J, Franke-Fayard BM, Adams JH, Ramesar J, Klop O, Khan SM, Janse CJ, Waters AP. 2011. Development of the piggyBac transposable system for Plasmodium berghei and its application for random mutagenesis in malaria parasites. BMC Genomics 12:155. 10.1186/1471-2164-12-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozlov S, Waters NC, Chavchich M. 2010. Leveraging cell cycle analysis in anticancer drug discovery to identify novel plasmodial drug targets. Infect. Disord. Drug Targets 10:165–190 [DOI] [PubMed] [Google Scholar]
- 60.Andreeva AV, Kutuzov MA. 2008. Protozoan protein tyrosine phosphatases. Int. J. Parasitol. 38:1279–1295 [DOI] [PubMed] [Google Scholar]
- 61.Wishart MJ, Dixon JE. 2002. The archetype STYX/dead-phosphatase complexes with a spermatid mRNA-binding protein and is essential for normal sperm production. Proc. Natl. Acad. Sci. U. S. A. 99:2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hinton SD, Myers MP, Roggero VR, Allison LA, Tonks NK. 2010. The pseudophosphatase MK-STYX interacts with G3BP and decreases stress granule formation. Biochem. J. 427:349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng KC, Klancer R, Singson A, Seydoux G. 2009. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell 139:560–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kutuzov MA, Andreeva AV. 2008. Protein Ser/Thr phosphatases of parasitic protozoa. Mol. Biochem. Parasitol. 161:81–90 [DOI] [PubMed] [Google Scholar]
- 65.Hyde JE. 2005. Drug-resistant malaria. Trends Parasitol. 21:494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, von Seidlein L. 2010. Artemisinin resistance: current status and scenarios for containment. Nat. Rev. Microbiol. 8:272–280 [DOI] [PubMed] [Google Scholar]
- 67.Ekland EH, Fidock DA. 2008. In vitro evaluations of antimalarial drugs and their relevance to clinical outcomes. Int. J. Parasitol. 38:743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, Vanderwall DE, Green DV, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. 2010. Thousands of chemical starting points for antimalarial lead identification. Nature 465:305–310 [DOI] [PubMed] [Google Scholar]
- 69.Jensen K, Plichta D, Panagiotou G, Kouskoumvekaki I. 2012. Mapping the genome of Plasmodium falciparum on the drug-like chemical space reveals novel anti-malarial targets and potential drug leads. Mol. Biosyst. 8:1678–1685 [DOI] [PubMed] [Google Scholar]
- 70.Stepniewska K, Ashley E, Lee SJ, Anstey N, Barnes KI, Binh TQ, D'Alessandro U, Day NP, de Vries PJ, Dorsey G, Guthmann JP, Mayxay M, Newton PN, Olliaro P, Osorio L, Price RN, Rowland M, Smithuis F, Taylor WR, Nosten F, White NJ. 2010. In vivo parasitological measures of artemisinin susceptibility. J. Infect. Dis. 201:570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.