Abstract
Synaptobrevin, also called vesicle-associated membrane protein (VAMP), is a component of the plasma membrane N-methylmaleimide-sensitive factor attachment protein receptor (SNARE) complex, which plays a key role in intracellular membrane fusion. Previous studies have revealed that, similar to synaptobrevin in other organisms, the fission yeast synaptobrevin ortholog Syb1 associates with post-Golgi secretory vesicles and is essential for cytokinesis and cell elongation. Here, we report that Syb1 has a role in sporulation. After nitrogen starvation, green fluorescent protein (GFP)-Syb1 is found in intracellular dots. As meiosis proceeds, GFP-Syb1 accumulates around the nucleus and then localizes at the forespore membrane (FSM). We isolated a syb-S1 mutant, which exhibits a defect in sporulation. In syb1-S1 mutants, the FSM begins to form but fails to develop a normal morphology. Electron microscopy shows that an abnormal spore wall is often formed in syb1-S1 mutant spores. Although most syb1-S1 mutant spores are germinated, they are less tolerant to ethanol than wild-type spores. The syb1-S1 allele carries a missense mutation, resulting in replacement of a conserved cysteine residue adjacent to the transmembrane domain, which reduces the stability and abundance of the Syb1 protein. Taken together, these results indicate that Syb1 plays an important role in both FSM assembly and spore wall formation.
INTRODUCTION
Members of the soluble N-methylmaleimide-sensitive factor attachment protein receptor (SNARE) family contribute to transport specificity by regulating interactions between membrane vesicles and their appropriate target membranes (1). SNARE proteins exist as complementary sets of v-SNAREs, found on vesicle membranes, and t-SNAREs, found on target membranes. Recent classification, however, takes into account the structural features of SNARE proteins, subdividing them into R-SNAREs and Q-SNAREs (2).
There are approximately 40 SNAREs in an animal cell, and each associates with a particular organelle in the biosynthetic-secretory or endocytic pathway (3). A v-SNARE is a single polypeptide chain, whereas a t-SNARE complex is composed of two or three proteins. The v-SNAREs and t-SNAREs have characteristic helical domains, and when a v-SNARE interacts with a t-SNARE, the helical domains of one wrap around the helical domains of the other to form a stable four-helix bundle. The resulting trans-SNARE complex locks the two membranes together.
SNAREs have been well characterized in neurons, where they mediate the docking and fusion of synaptic vesicles at the nerve terminal's plasma membrane (PM) during the process of neurotransmitter release. The SNARE complex responsible for docking synaptic vesicles at the PM of nerve terminals consists of three proteins. The transmembrane proteins v-SNARE synaptobrevin (also called vesicle-associated membrane protein [VAMP]) and t-SNARE syntaxin each contribute one α-helix to the complex (4, 5), whereas the peripheral membrane protein t-SNARE SNAP-25 contributes two α-helices to the four-helix bundle.
The fission yeast Schizosaccharomyces pombe is widely used as a model system for eukaryotic cell biology. The components of the PM SNAREs are highly conserved in S. pombe. Psy1 and Sec9, which are fission yeast orthologs of mammalian syntaxin 1 and SNAP-25, respectively, localize to the PM and are essential for growth (6, 7). On the other hand, the synaptobrevin ortholog Syb1, which is also essential for growth, is observed as punctate structures in the cytoplasm and becomes enriched in the medial region and cell ends in a fashion dependent on the actin cytoskeleton (8). These observations suggest that the PM SNARE components of S. pombe cells function in a manner similar to those of mammalian cells.
In addition to their role in vegetative growth, Psy1 and Sec9 are also involved in sporulation. S. pombe cells initiate a sporulation program when challenged with nutrient starvation (9, 10). Spore formation requires the assembly of double-layered intracellular membranes, termed forespore membranes (FSMs). As the nucleus divides in meiosis II, the FSM expands and eventually encapsulates a haploid nucleus generated by two rounds of division, thereby producing the prespore, a membrane-bound precursor of the spore (11–13). Ultimately, the inner layer of the FSM becomes the spore PM. In the space between the inner and outer FSMs, spore wall materials are deposited to form layers of spore walls. Mature spores are then liberated from an ascus when the ascus walls are autolyzed.
Similar to other membranes, the FSM expands by membrane vesicle fusion (11, 12). Psy1 was originally identified by its ability to suppress the sporulation defect of the spo3 mutants when overexpressed. Psy1 localizes to the FSM during sporulation. A mutation in the psy1+ gene compromises expansion of the FSM (6). The sec9 mutant also shows a defect in FSM expansion. Furthermore, sec9+ genetically interacts with psy1+ (7). Thus, the PM t-SNARE proteins Psy1 and Sec9 are essential in sporulation. syb1+ is upregulated during sporulation (14), suggesting that Syb1 plays an important role in sporulation. However, it remains unclear how Syb1 is involved in this event.
The aim of this study was to examine the role of syb1+ in sporulation. Syb1 localization was dynamically changed under nitrogen starvation, and eventually the protein relocalized to the nascent FSM. Isolation and analysis of sporulation-deficient syb1 mutants revealed that Syb1 is important in both FSM expansion and spore maturation. Our analysis also supports the notion that spore wall materials are transported by secretory vesicles.
MATERIALS AND METHODS
Yeast strains, media, and plasmids.
The S. pombe strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Complete medium (YEA) and synthetic medium (MM+N) were used for growth, and malt extract medium (MEA) and synthetic sporulation media (SSA, SSL-N, and MM−N) were used for mating and sporulation (15, 16). Synchronous meiosis was induced by a temperature shift using strains carrying the pat1-114 allele as described previously (17).
Table 1.
Strains used in this study
| Strain (accession no.a) | Genotypeb | Source |
|---|---|---|
| TY1 (FY17356) | h90 ura4≪GFP-syb1 ura4-D18 | This study |
| TY5 (FY17360) | h90/h90 ade6-M210/ade6-M216 leu1-32/leu1-32 syb1::ura4+ ura4-D18/ura4-D18 | This study |
| TY58 (FY17418) | h90 ade6 leu1≪syb1-S1 syb1::ura4+ ura4-D18 | This study |
| TY128 (FY17488) | h90 ade6-M216 leu1-32 ura4≪GFP-syb1 | This study |
| TY149 (FY17513) | h−/h− pat1-114/pat1-114 leu1≪GFP-syb1/leu1≪GFP-syb1 ade6-M210/ade6-M216 | |
| leu1-32/leu1-32 ura4+/ura4-D18 | This study | |
| TY161 (FY17525) | h90 ade6 leu1≪syb1+ syb1::ura4+ ura4-D18 | This study |
| TY208 (FY17571) | h90 ade6≪GFP-psy1 leu1-32≪syb1-S1 syb1::ura4+ ura4-D18 | This study |
| TY209 (FY17572) | h90 leu1≪mCherry-psy1 ura4≪GFP-syb1 | This study |
| TY222 (FY17585) | h90 ade6≪GFP-psy1 leu1≪syb1-WT syb1::ura4+ ura4-D18 | This study |
| TY227 (FY17590) | h90 leu1≪GFP-syb1-S1 syb1::ura4+ ade6-M216 ura4-D18 | This study |
| TY229 (FY17592) | h90 leu1≪GFP-syb1 syb1::ura4+ ade6-M210 ura4-D18 | This study |
| TY234 (FY17601) | h90 ade6≪GFP-psy1 leu1≪syb1-S1 ura4≪mCherry-atb2 syb1::ura4+ | This study |
| TY235 (FY17602) | h90 ade6≪GFP-psy1 leu1≪syb1+ ura4≪mCherry-atb2 syb1::ura4+ | This study |
| TY245 (FY17612) | h90 leu1≪syb1-S1 syb1::ura4+ ura4-D18 | This study |
| TY246 (FY17613) | h90 leu1≪syb1+ syb1::ura4+ ura4-D18 | This study |
| MKW5 (FY7456) | h90 | 19 |
| KI101 | h90 leu1≪mCherry-syb1 ade6-M210 ura4-D18 + pKB282(rer1-GFP) | This study |
| JZ670 (FY7051) | h−/h− pat1-114/pat1-114 ade6-M210/ade6-M216 leu1-32/leu1-32 | Yamamoto |
Accession numbers are from the Yeast Genetic Resource Center of Japan supported by the National BioResource Project (YGRC/NBRP; http:/yeast.lab.nig.ac.jp/nig/).
x≪y means that gene y is integrated at gene x.
Table 2.
Plasmids used in this study
| Plasmid | Characteristics | Source |
|---|---|---|
| pAL-KS | ars1, LEU2-based multicopy shuttle vector | 43 |
| pAL(syb1) | pAL-KS, syb1+ | This study |
| pAL(sec9) | pAL-KS, sec9+ | This study |
| pAL(psy1) | pAL-KS, psy1+ | This study |
| pAL(mde10-GFP) | pAL-KS, mde10-GFP | 23 |
| pDblet | ars2004 × 2, ura4+-based vector | 44 |
| pKB282 | ade6+ in pDblet | 19 |
| pKB282(syb1) | pKB282, syb1+ | This study |
| pKB282(syb1-S1) | pKB282, syb1-S1 | This study |
| pKB282(rer1-GFP) | pKB282, rer1-GFP | 19 |
| pL-A | ars1, ade6+-based multicopy shuttle vector | 7 |
| pL-A(syb1) | pL-A, syb1+ | 7 |
| pL-A(sec9) | pL-A, sec9+ | 7 |
| pL-A(psy1) | pL-A, psy1+ | 7 |
| pBR(leu1) | leu1+ in pBR322 | 19 |
| pBR(leu1)(syb1) | pBR(leu1), syb1+ | This study |
| pBR(leu1)(syb1-S1) | pBR(leu1), syb1-S1 | This study |
| pBR(leu1)(GFP-syb1) | pBR(leu1), GFP-syb1 | This study |
| pBR(leu1)(GFP-syb1-S1) | pBR(leu1), GFP-syb1-S1 | This study |
Generation of anti-Sec9 antibody.
A glutathione S-transferase (GST)-tagged Sec9 protein produced in Escherichia coli was used to generate polyclonal antibodies. GST-Sec9 was obtained as follows. A 0.9-kb DNA fragment encoding the C-terminal half of Sec9 (amino acids [aa] 175 to 419) was amplified by PCR with two oligonucleotides, 5′-CACACGGATCCGGTACTGAAGGAGATGAATAT-3′ and 5′-AGAGAGCGGCCGCTTATCAATGGATATGGCGAAGTCG-3′ (underlined sequences indicate restriction enzyme sites, BamHI for the former and NotI for the latter). The amplified DNA was digested with BamHI and NotI and then inserted into the corresponding sites of the GST-tag expression vector pGEX-KG(NotI) (18) to make pGEX(sec9). This plasmid was introduced into E. coli BL21. The fusion protein was purified with glutathione Sepharose (GE Healthcare, Little Chalfont, United Kingdom) and used to immunize a rabbit. For affinity purification, rabbit sera were loaded onto an AffiGel-15 (Bio-Rad, Hercules, CA) column containing purified GST-tagged Sec9, and the antibody was eluted with 0.2 M glycine-HCl (pH 2.6). The eluates were immediately dialyzed against phosphate-buffered saline (pH 7.4) (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl) containing 30% glycerol and used in Western blot analysis.
Time-lapse analysis.
Time-lapse observation was performed as described previously (13). For the colocalization study, cells expressing only GFP-Syb1 or mCherry-Psy1 were also observed to rule out the possibility of channel bleed through. Digital images were processed with Image J and Adobe Photoshop CS5.
Isolation of syb1-S1.
PCR was used to introduce a random point mutation into the syb1+ gene as follows: 1 cycle at 95°C for 3 min, followed by 30 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 3 min with Ex Taq polymerase (TaKaRa-Bio, Otsu, Japan). The amplified fragment containing the promoter and terminator regions, in addition to the syb1+ open reading frame (ORF), was digested with SacI and NotI and cloned into pBR(leu1) (19). The resulting library was digested at the SnaBI site within the leu1+ gene and then integrated at the leu1 locus of a heterozygous diploid carrying a syb1::ura4+ allele (TY5). Colonies of transformants on SSA sporulation plates were treated with ethanol to kill contaminating vegetative cells and then spread again on SSA plus adenine sporulation plates, which were exposed to iodine vapor (16). Because wild-type spores accumulate amylose-like polysaccharides, they are stained dark brown by iodine. The iodine-negative (light brown) colonies were selected and inspected for zygotic ascus formation.
Nucleotide sequence accession numbers.
The S. pombe strains constructed in this study have been deposited with the Yeast Genetic Resource Center of Japan supported by the National BioResource Project (YGRC/NBRP; http://yeast.lab.nig.ac.jp/nig/) under the accession numbers shown in Table 1.
RESULTS
Expression of Syb1 during sporulation.
To address whether syb1+ is upregulated during sporulation, log-phase cells of a homothallic haploid strain (MKW5) were incubated in the sporulation medium MM−N, and syb1 mRNA abundance was monitored by Northern analysis. mRNA was detected in vegetative cells and was found to increase during sporulation (Fig. 1A). To determine exactly when the rise in syb1 mRNA level occurred, we used the pat1-114 temperature-sensitive strain, which enters meiosis in a highly synchronous manner when it is shifted to its restrictive temperature (17). The level of syb1 mRNA began to increase about 6 h after induction of meiosis and peaked at about 9 h when the cells were in meiosis II (see Fig. S1A and B in the supplemental material). Our previous study showed that the transcription of psy1+ and sec9+ is also upregulated during sporulation; however, the peak of syb1 mRNA was slightly later than that of psy1+ and sec9+ (see Fig. S1A in the supplemental material) (7), an observation that is consistent with a previous genome-wide transcriptional analysis (14).
Fig 1.
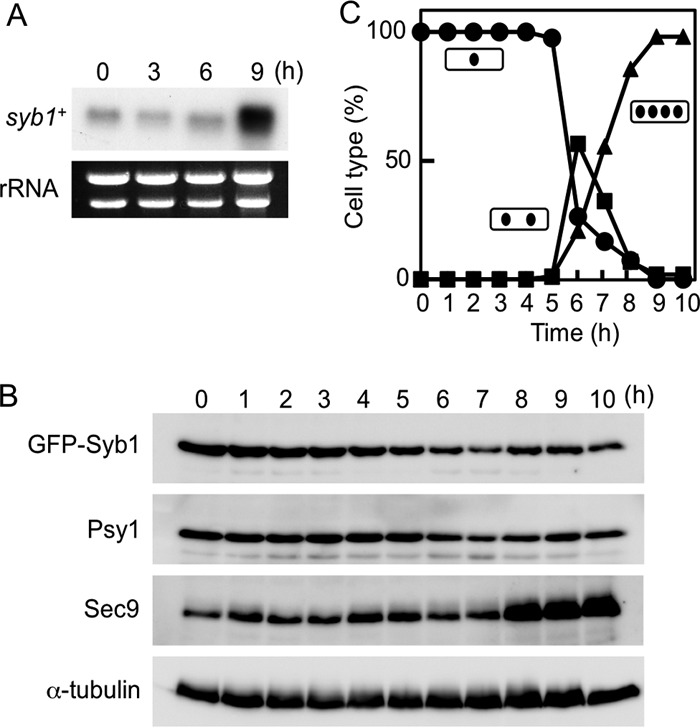
Expression of Syb1 during sporulation. (A) Transcription of the syb1+ gene. MKW5 (h90 wild-type) cells were precultured overnight in MM+N medium and then transferred to MM−N sporulation medium. Total RNA was analyzed by Northern blotting hybridization. The approximate quantity of RNA was checked by staining gels with ethidium bromide, which reveals rRNA. (B) Changes in the abundance of Syb1, Psy1, and Sec9 during sporulation. Cells expressing GFP-Syb1 (TY149) were allowed to proceed through synchronous meiosis. Aliquots were removed at hourly intervals, and the protein extract was subjected to Western blot analysis with mouse anti-GFP, anti-Psy1 (40), and anti-Sec9 antibodies. α-Tubulin was used as a loading control. (C) The meiotic nuclear division of cells from the analysis shown in panel B was monitored by counting the nuclei in each cell. Circles, mononucleate cells; squares, binucleate cells; triangles, tetranucleate cells.
To ascertain whether or not the levels of Syb1 protein paralleled those of syb1 mRNA, we next examined the abundance of GFP-Syb1 protein during sporulation. A TY149 strain carrying the pat1-114 mutation and expressing GFP-Syb1 was cultured at 34°C to induce synchronous meiosis. Unexpectedly, the abundance of GFP-Syb1 remained essentially constant after the induction of sporulation (Fig. 1B and C; see Fig. S2A in the supplemental material). Furthermore, Psy1 protein also showed a pattern similar to that of Syb1 protein. In contrast, Sec9 protein levels increased in a manner similar to that of sec9+ transcript levels (Fig. 1B and C). Interestingly, GFP-Syb1 was barely detectable in gradient-purified spores, whereas the syb1 mRNA was more abundant in spores than in vegetative growing cells (see Fig. S1C and S2B in the supplemental material).
Dual observation of Syb1 and Psy1.
A previous study has shown that Syb1 is localized on various sizes of vesicle-like structures in the cytoplasm and is enriched in the medial region and cell ends in cells in the vegetative growth phase (8, 20). However, the localization of Syb1 during sporulation is still unknown. We therefore studied the localization of GFP-Syb1 in meiotic and sporulating cells by fluorescence microcopy. In cells in meiotic prophase I, the GFP-Syb1 signal was dispersed in the cytoplasm as numerous dots (Fig. 2A). These dots of Syb1 did not overlap those of Rer1, a Golgi apparatus-associated protein (Fig. 2B). When cells proceeded to meiosis II, GFP-Syb1 first appeared as semicircle structures encircling dividing nuclei and was subsequently observed as ring-like structures encapsulating each nucleus (Fig. 2C). The meiosis-II spindle pole bodies were situated at the center of each semicircle (Fig. 2D). These data indicate that Syb1 localizes to the FSM during sporulation.
Fig 2.
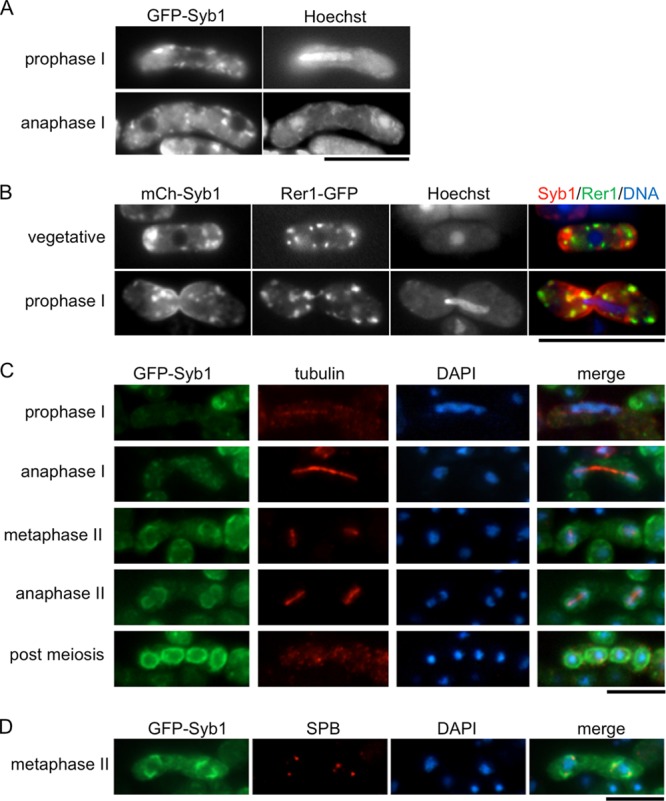
Localization of GFP-Syb1 during meiosis and sporulation. (A) A wild-type (TY128) strain expressing GFP-Syb1 was cultured on SSA at 28°C for 16 h. The chromatin region was stained with Hoechst 33342. (B) A wild-type strain (KI101) expressing mCherry-Syb1 and Rer1-GFP, a Golgi protein, was cultured on SSA at 28°C for 16 h. The chromatin region was stained with Hoechst 33342. (C and D) Cells expressing GFP-Syb1 (TY1) were cultured in SSL-N to induce sporulation. Cells fixed at different stages of meiosis were examined with 4′,6-diamidino-2-phenylindole (DAPI) and GFP as well as with anti-α-tubulin antibody TAT-1 as a loading control (C) or with anti-Sad1 antibody to highlight the spindle pole body (SPB) (D). Bars, 10 μm.
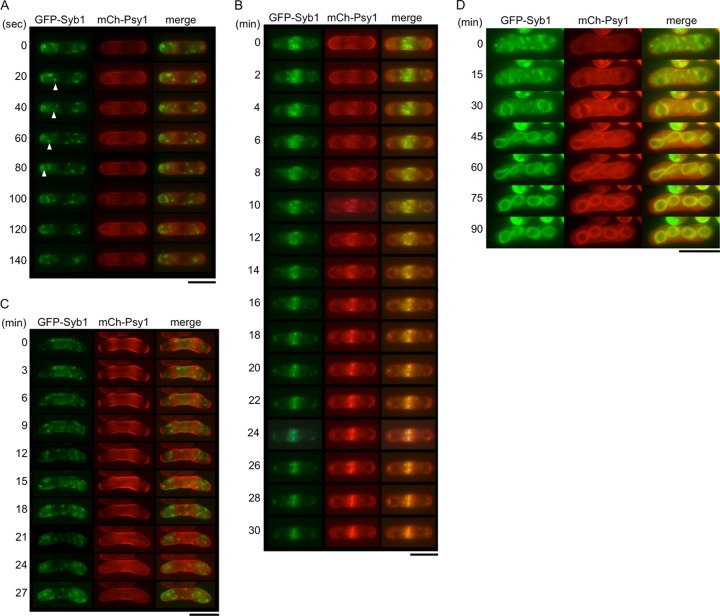
Previous studies have indicated that both Syb1 and Psy1 are involved in vesicle membrane fusion on the PM, but their locations differ considerably. As described above, Syb1 is located on vesicle-like structures in the cytoplasm and is enriched in the medial region and cell ends, whereas Psy1 is predominantly localized at the PM (6, 8, 20). To observe the location of these proteins in more detail, GFP-Syb1 was expressed together with mCherry-Psy1, and their expression was simultaneously traced by time-lapse fluorescence microscopy. During mitotic interphase, the fluorescence of GFP-Syb1 was detected as punctate structures in the cytoplasm and enriched in the medial region and cell ends, as expected (Fig. 3A; see Movie S1 in the supplemental material) (8, 20). Movement of the dots to the cell pole was often observed (Fig. 3A). In contrast, mCherry-Psy1 was essentially observed on the PM. In M-phase cells, dots of GFP-Syb1 accumulated at the center of the cell, where mCherry-Psy1 was subsequently observed (Fig. 3B). In dividing cells, GFP-Syb1 and mCherry-Psy1 colocalized on the nascent septum (Fig. 3B; see Movie S2 in the supplemental material). Thus, these results indicate the characteristic distribution of PM SNARE proteins during vegetative growth.
Fig 3.
Simultaneous observation of Syb1 and Psy1. GFP-Syb1 and mCherry-Psy1 expressed in wild-type strain TY209 were observed in cells during mitotic interphase (A), mitosis (B), early meiosis (C), and sporulation (D). Bars, 10 μm.
We also examined the behavior of Syb1 and Psy1 during sexual differentiation. In early meiotic cells, GFP-Syb1 was observed as dots. Unlike in vegetative growth, these dots were dispersed homogenously throughout the cytoplasm. On the other hand, mCherry-Psy1 localized on the PM in a fashion similar to that of cells in vegetative growth (Fig. 3C; see Movie S3 in the supplemental material). In cells at meiosis I, the dots of GFP-Syb1 accumulated around the nucleus, whereas the majority of mCherry-Psy1 persisted on the PM. mCherry-Psy1 was then gradually internalized from the PM, and observed as dots (Fig. 3D, 0 to 15 min; see Movie S4 in the supplemental material). During FSM formation, GFP-Syb1 and mCherry-Psy1 were colocalized on the nascent forespore membrane (Fig. 3D, 30 to 90 min; see Movie S4 in the supplemental material).
Isolation of sporulation-deficient syb1 mutants.
To determine how Syb1 is involved in sporulation, we attempted to isolate sporulation-deficient mutants of syb1 by using random PCR mutagenesis (see Materials and Methods). Approximately 52,000 colonies of transformants on the synthetic sporulation medium were exposed to iodine vapor. Iodine-negative (light brown) colonies were selected and inspected for zygotic ascus formation. Consequently, eight colonies, designated syb1-S1 to syb1-S8, exhibited a defect in sporulation (Fig. 4A). We next determined the sites mutated in these mutants. Nucleotide sequencing demonstrated that these mutants all had the same nucleotide change (C to T) that results in replacement of cysteine 98 with arginine (Fig. 4B). This amino acid residue is adjacent to the C-terminal transmembrane domain (Fig. 4B). The corresponding cysteine residue in the budding yeast synaptobrevin ortholog Snc1 and mammalian synaptobrevin 2 is known to be palmitoylated (21, 22). Although syb1-S1 cells formed asci, the spores were apparently smaller than wild-type spores (Fig. 4A and D). Small spores were also observed at lower temperatures, and the phenotype was not enhanced at higher temperatures (see Tables S1 and S2 in the supplemental material). In addition to causing a defect in ascospore formation, the syb1-S1 mutation compromised vegetative growth. As shown in Fig. 4C, the syb1-S1 mutant grew well at 25°C but exhibited a severe growth defect at 37°C. Thus, the syb1-S1 mutation confers temperature sensitivity for growth.
Fig 4.
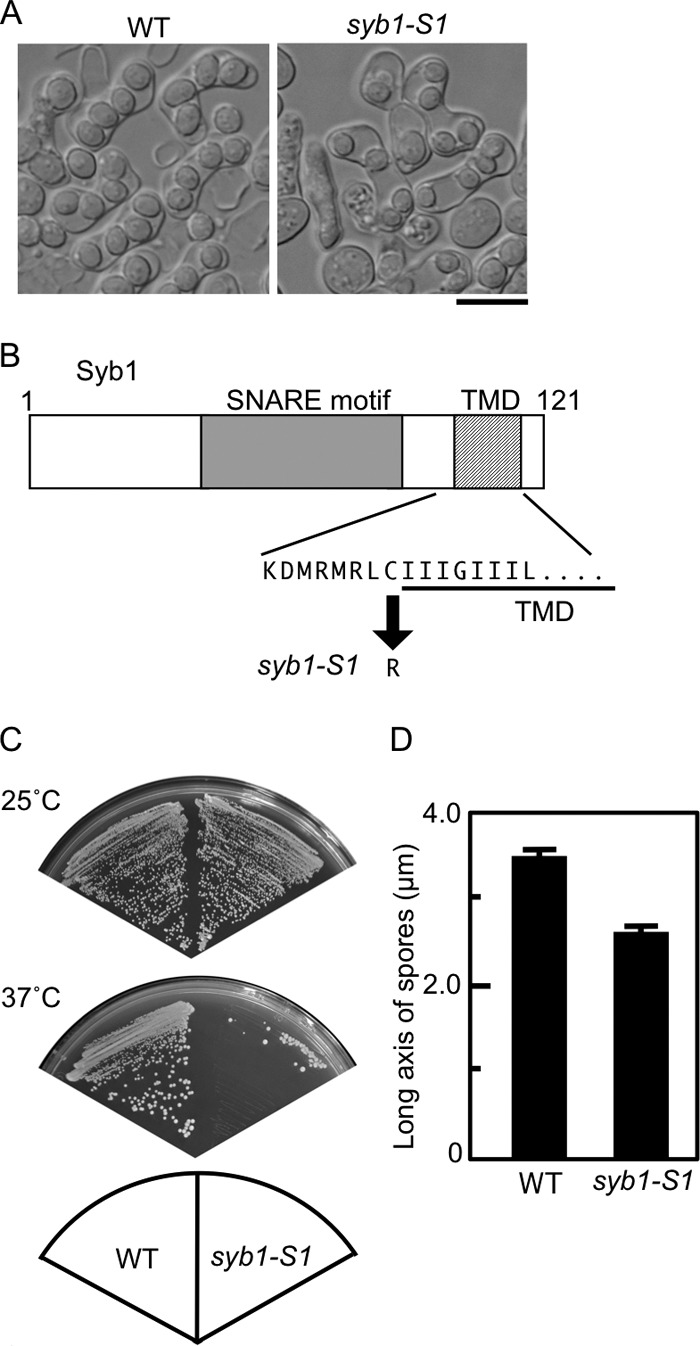
Phenotypes of the syb1-S1 mutant. (A) Wild-type (TY161) and syb1-S1 (TY58) cells were sporulated on SSA medium at 28°C for 2 days. Bar, 10 μm. (B) The syb1-S1 mutant allele carries a single nucleotide change (from C to T), which results in the replacement of cysteine 98 with arginine. TMD, transmembrane domain. (C) Wild-type (TY161) and syb1-S1 (TY58) cells were streaked on complete medium (YEA) and incubated at 25°C or 37°C for 3 days. (D) Sizes of wild-type (TY161) and syb1-S1 (TY58) spores.
The syb1-s1 mutant is defective in FSM formation and spore maturation.
To investigate the morphology of the abnormal syb1-S1 ascospores in greater detail, we observed them by thin-section electron microscopy (EM). Consistent with the data obtained by optical microscopy, the syb1-S1 ascospores were small compared with wild-type ascospores (Fig. 5A). In addition, many of the spore walls of syb1-S1 cells were apparently thicker than those of wild-type cells but were constructed of seemingly sparsely distributed material on the basis of their lower electron density. Moreover, abnormal membranous structures were often observed in the spore wall of syb1-S1 cells (Fig. 5A). Mde10 is a predicted metalloprotease that is essential for formation of the spore surface architecture (23). Because Mde10-GFP is localized at the peripheral region of spores at the end of meiosis, it is used as a marker to examine the spore surface structure. As shown in Fig. 5B, the Mde10-GFP signal was observed on most of the wild-type spore surface, whereas it was observed on only one-third of syb1-S1 spores. These data indicate that spore maturation is defective in syb1-S1 mutants.
Fig 5.
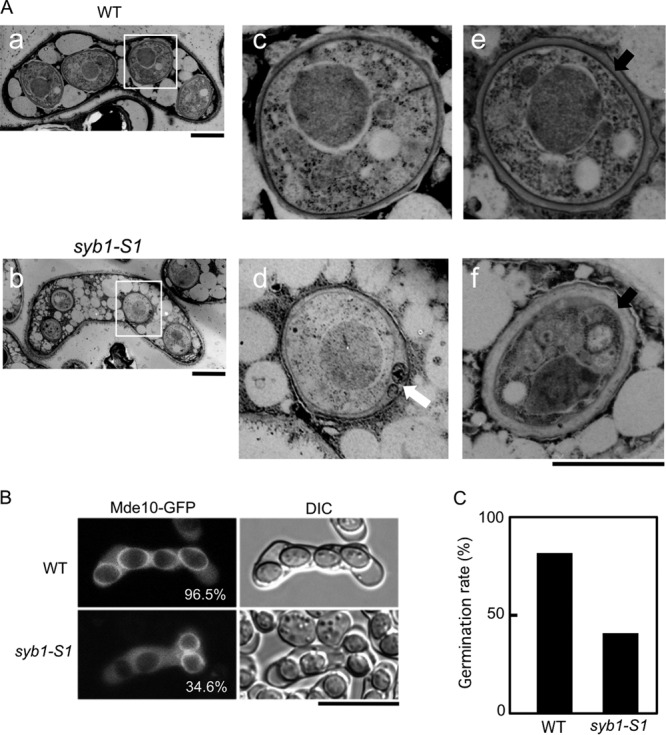
Immature spores in the syb1-S1 mutant. (A) Wild-type (TY161) and syb1-S1 (TY58) cells were sporulated on MEA medium for 3 days at 28°C and observed by electron microscopy. a, c, and e, wild-type spores; b, d, and f, syb1-S1 spores. Images in panels c and d are magnified images of the boxed regions in panels a and b, respectively. White and black arrows indicate the abnormal membranous structure and the spore wall, respectively. Bars, 2 μm (c to f). (B) Localization of Mde10-GFP. Wild-type (TY161) and syb1-S1 (TY58) strains expressing Mde10-GFP were sporulated in MM−N medium. The percentage of spores with an Mde10-GFP signal is also indicated. (C) Loss of ethanol resistance in syb1-S1 spores. Wild-type (TY246) and syb1-S1 (TY245) strains were cultured in MM−N for 10 days. Spores isolated by Urografin density gradient centrifugation (42) were treated with 30% ethanol at 28°C for 30 min and spread on YEA plates to score viability.
Next, we examined whether spore germination is affected by the syb1-S1 mutation. Single spores isolated by micromanipulation from homothallic syb1-S1 cells were allowed to germinate on growth medium. The frequency of colony formation was almost the same for wild-type spores (96.4%) and syb1-S1 spores (94.1%). S. pombe spores are relatively resistant to enzymatic digestion, heat, and some organic solvents. This resistance is thought to be dependent on the spore walls. Because the spore wall of syb1-S1 cells was abnormal, we considered that syb1-S1 spores might have lost this resistance. Examination of the sensitivity of syb1-S1 spores to Glusulase and heat shock (55°C) revealed no abnormalities (data not shown). We next tested the tolerance of the syb1-S1 spores to ethanol. The spores were suspended in 30% ethanol and then spread on YEA plates to score viability. The resistance of syb1-S1 spores to ethanol was about one-half of that of wild-type spores (Fig. 5C). Thus, the syb1-S1 mutant is defective in spore maturation.
To examine in detail why the syb1-S1 mutant forms small spores, the assembly of FSMs in the syb1-S1 mutant was analyzed by using GFP-Psy1. In wild-type cells, most haploid nuclei produced by meiotic second divisions were encapsulated by the FSM (Fig. 6). In syb1-S1 mutant cells, FSMs seemed to initiate normally at both poles of the meiosis II spindles, but their extension was blocked (Fig. 6). This result indicates that the small spore size was caused by defective expansion of the FSM. Furthermore, Syb1 is required for proper construction of the FSM in addition to spore maturation.
Fig 6.
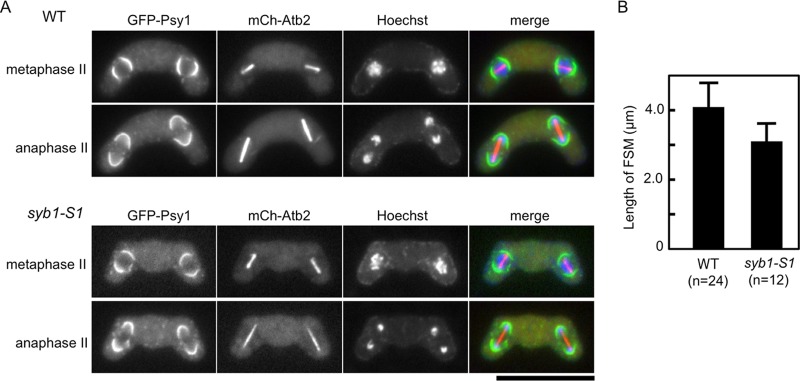
(A) Aberrant assembly of the FSM in syb1-S1 cells. Wild-type (TY235) and syb1-S1 (TY234) strains expressing GFP-Psy1 and mCherry-tagged Atb2 (α-tubulin) were sporulated on MEA medium at 28°C for 1 day. Chromosomal DNA was stained with Hoechst 33342 and analyzed by fluorescence microscopy. Bar, 10 μm. (B) Development of the FSM in syb1-S1 cells. The contour length of FSMs in wild-type (TY235) and syb1-S1 (TY234) cells at late meiosis II was measured by Image J software.
To test whether the Syb1 mutant protein (Syb1S1) is adequately localized at the FSM, GFP-Syb1S1 was expressed in wild-type cells. As shown in Fig. S3 in the supplemental material, the localization of GFP-Syb1S1 resembled that of wild-type GFP-Syb1 throughout vegetative growth and sporulation. Therefore, it is unlikely that the phenotypes of the syb1-S1 mutant are simply due to mislocalization of Syb1. We next examined the levels of Syb1S1 protein by Western analysis. The amount of GFP-Syb1S1 was markedly decreased compared to that of GFP-Syb1 (Fig. 7A), suggesting that the phenotypes of syb1-S1 mutants might be caused by instability of the Syb1S1 protein. In support of this notion, overexpression of syb1-S1 complemented the temperature sensitivity and the sporulation defect of the syb1-S1 mutant (Fig. 7B and C). Because the decrease in Syb1 was observed in both vegetative cells (even at the permissive temperature) and sporulating cells, these data also suggest that the sporulation process requires greater amounts of Syb1 protein than does vegetative growth.
Fig 7.
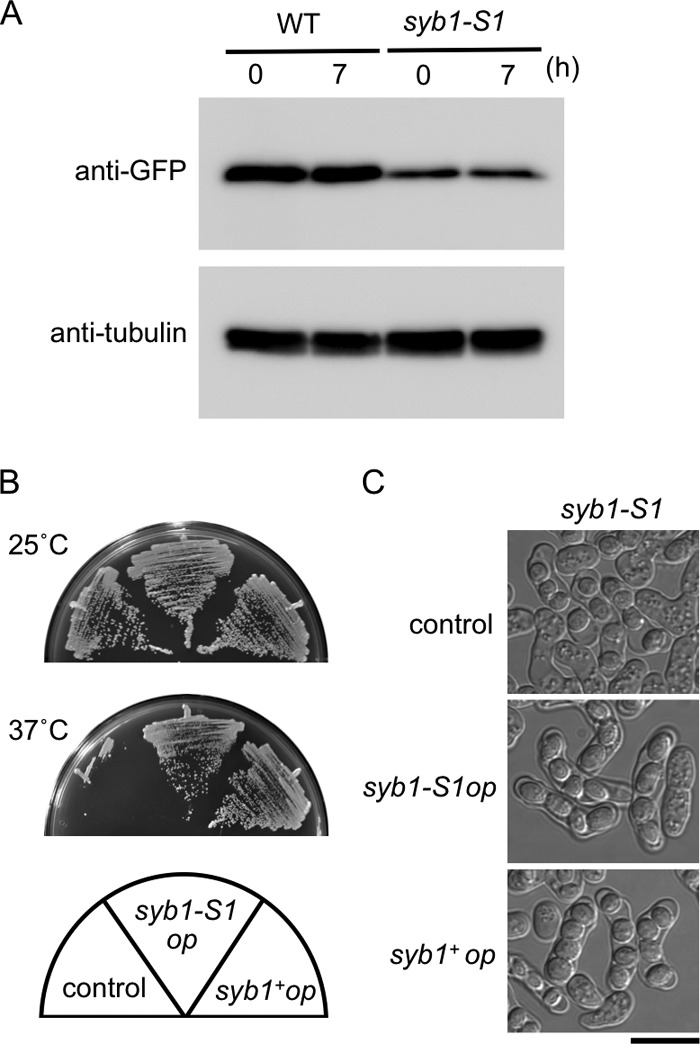
Expression of Syb1 in the syb1-S1 mutant. (A) Wild-type (TN229) and syb1-S1 (TN227) cells precultured in MM+N were incubated in MM−N at 28°C for 7 h. Protein extracts were subjected to Western blot analysis with anti-GFP antibody, as well as with anti-α-tubulin antibody as a loading control. (B and C) Overexpression of syb1-S1 complemented the temperature sensitivity (B) and the sporulation defect (C) of the syb1-S1 mutant. Strain TY58 (syb1-S1 mutant) carrying pKB282 (control), pKB282(syb1-S1), or pKB282(syb1) was incubated on YEA medium at 25 or 37°C for 3 days (B) or on SSA medium at 28°C for 1 day (C).
Genetic interaction of Syb1 with Psy1 and Sec9.
In higher eukaryotes, synaptobrevin forms a complex with syntaxin-1 and SNAP-25, and this interaction is important for specific membrane fusion. To examine whether overproduction of Psy1 and/or Sec9 suppresses the syb1-S1 mutation, a multicopy plasmid harboring either psy1+ or sec9+ was introduced into syb1-S1 strains. Overexpression of each gene was tested and found to restore the size of spores in the syb1-S1 mutant (Fig. 8A). Sec9 but not Psy1 suppressed the temperature sensitivity of syb1-S1 cells (Fig. 8B). These data indicate that syb1+ interacts genetically with the t-SNARE components psy1+ and sec9+.
Fig 8.
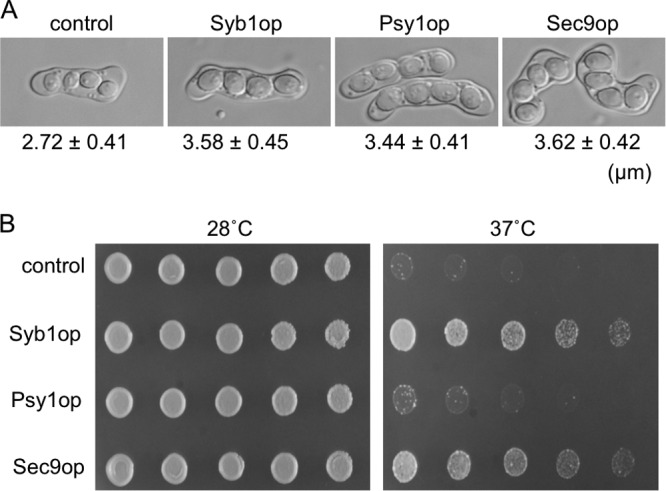
Effect of overexpression of psy1+ and sec9+ in the syb1-S1 mutant. (A) The syb1-S1 strain (TY58) was transformed with a multicopy plasmid (pL-A) carrying syb1+, psy1+, or sec9+. Transformants were sporulated in MM−N medium. The size of the spores is also indicated. (B) Transformants were cultured on YEA medium and incubated for 2 days at 28°C or 37°C.
DISCUSSION
Expression of the PM SNARE proteins during sporulation.
In this study, we sought to understand the role of the fission yeast synaptobrevin ortholog Syb1 in sporulation and showed that Syb1 is essential for proper expansion of the developing FSM and spore wall formation. Although syb1 mRNA was highly upregulated during sporulation (about 10-fold) (14) (see Fig. S1 in the supplemental material), the protein level of Syb1 was almost constant. A few possibilities might explain the difference between syb1 mRNA and GFP-Syb1 protein. First, GFP-Syb1 might be more stable than the native protein. However, a Syb1-GFP fusion protein with the tag at the C terminus was not functional; therefore, we could not confirm this possibility. Because psy1 mRNA and Psy1 showed a profile similar to that of syb1 mRNA and Syb1 (Fig. 1), it is unlikely that the difference is caused by the stability of the fusion protein. Second, Syb1 translation might be delayed during sporulation. Contrary to this possibility, however, the protein level was not enhanced even at 14 h after induction of sporulation (see Fig S2A in the supplemental material). Furthermore, we isolated spores and examined the amount of syb1 mRNA by reverse transcription quantitative PCR (RT-qPCR) and GFP-Syb1 protein by Western analysis. Interestingly, GFP-Syb1 protein was barely detectable, although syb1 mRNA was abundant in spores (see Fig. S1C and S2B in the supplemental material). Thus, these data indicate that Syb1 protein is degraded after sporulation is complete and suggest the possibility that the induced syb1 RNA is stored for germination. Indeed, most syb1Δ spores did not germinate (data not shown), supporting this possibility.
In contrast to Syb1 and Psy1, the levels of Sec9 protein increased during sporulation in a manner similar to that of the sec9+ transcript. Thus, it is possible that turnover of Syb1 and Psy1 proteins might be facilitated during sporulation. In Saccharomyces cerevisiae, the orthologs of synaptobrevin, syntaxin 1, and SNAP-25 are encoded by 3 respective pairs of genes: SNC1 and SNC2, SSO1 and SSO2, and ScSEC9 and SPO20. These genes are highly conserved and well characterized (24–27). It has been shown that the level of S. cerevisiae SNARE complexes that can form in vivo is limited by the availability of ScSec9 (28). On the other hand, during formation of the prospore membrane, which corresponds to the FSM of S. pombe, vesicle fusion requires Spo20 but not ScSec9 (29–31). Thus, in S. cerevisiae the SNAP-25 orthologs seem to fine-tune the function of the PM SNAREs. Similarly, S. pombe might control FSM formation by the upregulation of Sec9.
Behavior of the PM SNARE proteins.
Dual observation of Syb1 and Psy1 was used to address the dynamic aspects of the PM SNARE proteins. Syb1 and Psy1 showed a different localization profile in vegetative interphase cells, consistent with previous observations (6, 8, 20). In dividing cells, the location of Syb1 and Psy1 completely overlapped on the nascent septum. The transport of Syb1 to the cell tip is known to be mainly dependent on the actin cytoskeleton (8). The distribution of F-actin during sporulation is well documented. F-actin dots seem to be randomly scattered in prophase I and become concentrated around the nucleus at the early stage of meiosis II (32, 33). This behavior of F-actin is similar to that of Syb1, suggesting that, in sporulation, Syb1 localization is highly dependent on the actin cytoskeleton.
Our live-cell analysis also raised some interesting possibilities for membrane dynamics during FSM formation. First, the localization of distinct intracellular membranes containing Syb1 and Psy1 prior to FSM assembly suggests that the initial events in FSM formation may involve heterotypic fusion of different classes of vesicle, rather than homotypic fusion as has been generally presumed (34–36). Second, the steady-state localization of Syb1 shifts from predominantly endocytic/vesicular membranes in vegetative cells to predominantly the target membrane (FSM) in sporulating cells, suggesting that the kinetics of recycling of the v-SNARE from its target membrane are altered during FSM growth.
Sporulation-defective phenotype of the syb1-S1 mutant.
The syb-S1 mutant exhibited temperature sensitivity for growth and was expressed at lower levels than the wild type, suggesting that it is a hypomorphic allele. It is possible that the threshold activity level of Syb1 required for sporulation is higher than that required for vegetative growth. We previously reported that the spo14-B221 mutant shows a sporulation-specific defect; however, the level of Spo14 protein in spo14-B221 is low during both vegetative growth and sporulation (19). Other sporulation mutants such as spo20-KC104, sec9-10, and psy1-S1 also show a sporulation-defective phenotype at temperatures permissive for proliferation (6, 7, 37), supporting the above possibility. The syb1-S1 mutant carried a mutation in a conserved cysteine residue. The corresponding cysteine residue of S. cerevisiae Snc1p and mammalian synaptobrevin 2 is known to be palmitoylated (21, 22). Replacement of the cysteine residue of Snc1p by serine, which blocks Snc1p palmitoylation, affects the stability of Snc1 protein. However, this mutation does not affect the general growth characteristics of budding yeast and does not interfere with the secretory processes (21). Thus, the importance of the conserved cysteine may differ between the two yeasts.
Role of Syb1 in sporulation.
In S. cerevisiae sporulation, Sncp relocalizes to the prospore membrane, which corresponds to the fission yeast FSM, and interacts with Sso1p and the sporulation-specific SNAP-25 ortholog Spo20p (27). During sporulation, Snc1p is transported to the prospore membrane via a retrograde pathway from the PM (38). These results imply that Sncp has an important function in sporulation. However, the phenotype of snc mutants in sporulation has been described only briefly (39). To our knowledge, ours is the first study to document the phenotype of synaptobrevin mutants in sporulation. syb1-S1 cells formed small spores due to a defect in expansion of the FSM. To form the layers of the spore wall, spore wall materials are deposited in the space between the inner and outer membranes of the FSM (11, 12). As a result, it has been suggested that spore wall materials are transported by secretory vesicles. Because the sporulation-deficient sec9-10 and psy1-S1 mutant cells form no spores, possibly due to severe defects in FSM formation, we could not examine whether the PM SNAREs are indeed required for spore wall formation (7, 40). Compared with sec9-10 and psy1-S1, syb1-S1 exhibited a milder phenotype, and most of the syb1-S1 spores germinated. The spore wall of syb1-S1 was thicker than that of the wild type and often contained abnormal membrane-like structures. The phenotype of syb1-S1 seemed reminiscent of the spores with abnormal morphology observed in the meu10Δ mutant. meu10+ encodes a glycosylphosphatidylinositol (GPI)-anchored protein, which localizes to the spore wall. In the meu10Δ mutant, the spore walls are thick and 1,3-β-glucan is distributed abnormally throughout the spore wall, rather than being concentrated on the inner part of the spore wall as in the wild type (41). These data indicate that spore wall proteins may not be transported properly in syb1-S1 cells. Indeed, another spore wall protein, Mde10, was often mislocalized in syb1-S1 mutants (Fig. 5B). Thus, these results support the notion that spore wall components are transported by secretory vesicles.
The present study, together with our previous work (6, 7, 40), shows that all of the fission yeast PM SNARE components play a critical role in sporulation. Unlike S. cerevisiae, S. pombe has a sole gene encoding each of the PM SNARE components. Interestingly, Psy1 is dynamically internalized during sporulation and relocalizes to the nascent FSM, which is essential for sporulation. Thus, it is important to address how the meiotic signal regulates the function of PM SNARE proteins, which will aid in elucidation of the molecular mechanisms underlying gametogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kayoko Tanaka, Masayuki Yamamoto, and Roger Tsien for strains and plasmids and Osamu Niwa and Keith Gull for antibodies.
This study was partly supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. K.I. is a recipient of the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00061-13.
REFERENCES
- 1.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature 362:318–324 [DOI] [PubMed] [Google Scholar]
- 2.Fasshauer D, Sutton RB, Brunger AT, Jahn R. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U. S. A. 95:15781–15786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stow JL, Manderson AP, Murray RZ. 2006. SNAREing immunity: the role of SNAREs in the immune system. Nat. Rev. Immunol. 6:919–929 [DOI] [PubMed] [Google Scholar]
- 4.Baumert M, Maycox PR, Navone F, De Camilli P, Jahn R. 1989. Synaptobrevin: an integral membrane protein of 18,000 daltons present in small synaptic vesicles of rat brain. EMBO J. 8:379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trimble WS, Cowan DM, Scheller RH. 1988. VAMP-1: a synaptic vesicle-associated integral membrane protein. Proc. Natl. Acad. Sci. U. S. A. 85:4538–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, Nakamura-Kubo M, Hirata A, Shimoda C. 2001. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+ encoding syntaxin-like protein. Mol. Biol. Cell 12:3955–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura T, Kashiwazaki J, Shimoda C. 2005. A fission yeast SNAP-25 homologue, SpSec9, is essential for cytokinesis and sporulation. Cell Struct. Funct. 30:15–24 [DOI] [PubMed] [Google Scholar]
- 8.Edamatsu M, Toyoshima YY. 2003. Fission yeast synaptobrevin is involved in cytokinesis and cell elongation. Biochem. Biophys. Res. Commun. 301:641–645 [DOI] [PubMed] [Google Scholar]
- 9.Shimoda C, Nakamura T. 2003. Control of late meiosis and ascospore formation, p 311–327 Molecular biology of Schizosaccharomyces pombe. Springer, Berlin, Germany [Google Scholar]
- 10.Shimoda C. 2004. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 117:389–396 [DOI] [PubMed] [Google Scholar]
- 11.Yoo BY, Calleja GB, Johnson BF. 1973. Ultrastructural changes of the fission yeast (Schizosaccharomyces pombe) during ascospore formation. Arch. Mikrobiol. 91:1–10 [DOI] [PubMed] [Google Scholar]
- 12.Hirata A, Tanaka K. 1982. Nuclear behavior during conjugation and meiosis in the fission yeast Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 28:263–274 [Google Scholar]
- 13.Nakamura T, Asakawa H, Nakase Y, Kashiwazaki J, Hiraoka Y, Shimoda C. 2008. Live observation of forespore membrane formation in fission yeast. Mol. Biol. Cell 19:3544–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mata J, Lyne R, Burns G, Bahler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143–147 [DOI] [PubMed] [Google Scholar]
- 15.Egel R, Egel-Mitani M. 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88:127–134 [DOI] [PubMed] [Google Scholar]
- 16.Gutz H, Heslot H, Leupold U, Loprieno N. 1974. Schizosaccharomyces pombe, p 395–446 In King RC. (ed), Handbook of genetics 1. Plenum Press, New York, NY [Google Scholar]
- 17.Iino Y, Hiramine Y, Yamamoto M. 1995. The role of cdc2 and other genes in meiosis in Schizosaccharomyces pombe. Genetics 140:1235–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan KL, Dixon JE. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262–267 [DOI] [PubMed] [Google Scholar]
- 19.Nakamura-Kubo M, Nakamura T, Hirata A, Shimoda C. 2003. The fission yeast spo14+ gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol. Biol. Cell 14:1109–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kita A, Sugiura R, Shoji H, He Y, Deng L, Lu Y, Sio SO, Takegawa K, Sakaue M, Shuntoh H, Kuno T. 2004. Loss of Apm1, the micro1 subunit of the clathrin-associated adaptor-protein-1 complex, causes distinct phenotypes and synthetic lethality with calcineurin deletion in fission yeast. Mol. Biol. Cell 15:2920–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couve A, Protopopov V, Gerst JE. 1995. Yeast synaptobrevin homologs are modified posttranslationally by the addition of palmitate. Proc. Natl. Acad. Sci. U. S. A. 92:5987–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veit M, Becher A, Ahnert-Hilger G. 2000. Synaptobrevin 2 is palmitoylated in synaptic vesicles prepared from adult, but not from embryonic brain. Mol. Cell. Neurosci. 15:408–416 [DOI] [PubMed] [Google Scholar]
- 23.Nakamura T, Abe H, Hirata A, Shimoda C. 2004. ADAM family protein Mde10 is essential for development of spore envelopes in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3:27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Protopopov V, Govindan B, Novick P, Gerst JE. 1993. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74:855–861 [DOI] [PubMed] [Google Scholar]
- 25.Aalto MK, Ronne H, Keranen S. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12:4095–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. 1994. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell 79:245–258 [DOI] [PubMed] [Google Scholar]
- 27.Neiman AM, Katz L, Brennwald PJ. 2000. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155:1643–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehman K, Rossi G, Adamo JE, Brennwald P. 1999. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J. Cell Biol. 146:125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neiman AM. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jantti J, Aalto MK, Oyen M, Sundqvist L, Keranen S, Ronne H. 2002. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 115:409–420 [DOI] [PubMed] [Google Scholar]
- 31.Yang HJ, Nakanishi H, Liu S, McNew JA, Neiman AM. 2008. Binding interactions control SNARE specificity in vivo. J. Cell Biol. 183:1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen J, Nielsen O, Egel R, Hagan IM. 1998. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell Sci. 111:867–876 [DOI] [PubMed] [Google Scholar]
- 33.Kashiwazaki J, Yamasaki Y, Itadani A, Teraguchi E, Maeda Y, Shimoda C, Nakamura T. 2011. Endocytosis is essential for dynamic translocation of a syntaxin 1 orthologue during fission yeast meiosis. Mol. Biol. Cell 22:3658–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidow LS, Goetsch L, Byers B. 1980. Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics 94:581–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knop M, Strasser K. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19:3657–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bajgier BK, Malzone M, Nickas M, Neiman AM. 2001. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell 12:1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakase Y, Nakamura T, Hirata A, Routt SM, Skinner HB, Bankaitis VA, Shimoda C. 2001. The Schizosaccharomyces pombe spo20+ gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in forespore membrane formation. Mol. Biol. Cell 12:901–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morishita M, Mendonsa R, Wright J, Engebrecht J. 2007. Snc1p v-SNARE transport to the prospore membrane during yeast sporulation is dependent on endosomal retrieval pathways. Traffic 8:1231–1245 [DOI] [PubMed] [Google Scholar]
- 39.David D, Sundarababu S, Gerst JE. 1998. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J. Cell Biol. 143:1167–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda Y, Kashiwazaki J, Shimoda C, Nakamura T. 2009. The Schizosaccharomyces pombe syntaxin 1 homolog, Psy1, is essential in the development of the forespore membrane. Biosci. Biotechnol. Biochem. 73:339–345 [DOI] [PubMed] [Google Scholar]
- 41.Tougan T, Chiba Y, Kakihara Y, Hirata A, Nojima H. 2002. Meu10 is required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 7:217–231 [DOI] [PubMed] [Google Scholar]
- 42.Nishi K, Shimoda C, Hayashibe M. 1978. Germination and outgrowth of Schizosaccharomyces pombe ascospores isolated by Urografin density gradient centrifugation. Can. J. Microbiol. 24:893–897 [DOI] [PubMed] [Google Scholar]
- 43.Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Biol. Cell 20:3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brun C, Dubey DD, Huberman JA. 1995. pDblet, a stable autonomously replicating shuttle vector for Schizosaccharomyces pombe. Gene 164:173–177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.