Abstract
Many fungi respond to light and regulate fungal development and behavior. A blue light-activated complex has been identified in Neurospora crassa as the product of the wc-1 and wc-2 genes. Orthologs of WC-1 and WC-2 have hitherto been found only in filamentous fungi and not in yeast, with the exception of the basidiomycete pathogenic yeast Cryptococcus. Here, we report that the fission yeast Schizosaccharomyces japonicus responds to blue light depending on Wcs1 and Wcs2, orthologs of components of the WC complex. Surprisingly, those of ascomycete S. japonicus are more closely related to those of the basidiomycete. S. japonicus reversibly changes from yeast to hyphae in response to environmental stresses. After incubation at 30°C, a colony of yeast was formed, and then hyphal cells extended from the periphery of the colony. When light cycles were applied, distinct dark- and bright-colored hyphal cell stripes were formed because the growing hyphal cells had synchronously activated cytokinesis. In addition, temperature cycles of 30°C for 12 h and 35°C for 12 h or of 25°C for 12 h and 30°C for 12 h during incubation in the dark induced a response in the hyphal cells similar to that of light. The stripe formation of the temperature cycles was independent of the wcs genes. Both light and temperature, which are daily external cues, have the same effect on growing hyphal cells. A dual sensing mechanism of external cues allows organisms to adapt to daily changes of environmental alteration.
INTRODUCTION
Many organisms on Earth have adapted themselves to the alternating day/night environment, particularly with regard to light and temperature cycles. Responses to light have been thoroughly studied in unicellular organisms. Among eukaryotes, many fungi respond to light and regulate fungal development and behavior, and the wavelengths they respond to are mostly blue light (1). A blue light-activated photoreceptor has been identified in Neurospora crassa as the product of the wc-1 gene (2). The wc-2 gene is also essential for photoresponse. These gene products associate with each other and make a transcription factor activated by blue light via a flavin adenine dinucleotide (FAD) chromophore. WC-1 has an LOV (light, oxygen, or voltage) domain with a flavin binding site (3, 4) and two PAS (PER-ARNT-SIM) domains, and WC-2 has one PAS domain. The PAS domain is a protein structural motif that is often seen in a variety of signal sensor molecules and is involved in protein-protein interactions (5). Orthologs of WC-1 and WC-2 have hitherto been characterized only in filamentous fungi and not in yeast, with the exception of the basidiomycete pathogenic yeast Cryptococcus (6, 7).
N. crassa WC-1 and WC-2 are subject to light-dependent phosphorylation, and transient phosphorylation of WC-1 is crucial for desensitization of the photoreceptor (8). Importantly, the function of the WC complex is not only as a blue light sensor but also as a regulator of expression of FRQ (frequency) (3). FRQ is one of the components for the circadian clock oscillator (9). The circadian clock oscillator depends on regulatory interactions between the WC complex and FRQ (10). Thus, the WC complex is incorporated into the circadian feedback loop (10) and is a crucial component for the circadian rhythm that causes periodic formation of conidia in N. crassa (11). Once the circadian response of each cell of N. crassa is synchronized by the daily light/dark cycle (L/D), the daily development of conidiation is visible as a band pattern that is formed by altering zones of mycelium and sporulating mycelium. After synchronization by the light/dark cycle, this band pattern progresses during dark/dark cycles (D/D) or light/light cycles (L/L). In addition, the circadian rhythm is entrained by temperature changes (12).
It has recently been reported that a circadian rhythm of metabolic activity is shown in Saccharomyces cerevisiae after entrainment of temperature cycles, although the molecular basis of sensing temperature remains unknown (13). Here, we found orthologs of WC-1 and WC-2 in the genome of the fission yeast Schizosaccharomyces japonicus, which possesses characteristics common to filamentous fungi and whose cells change from yeast growth to hyphal growth in response to environmental stress, including nutrient loss or DNA damage (14, 15). If the DNA-damaging reagent camptothecin (CPT) is included in nutrient-rich medium, hyphal cells will develop. We discovered that hyphal cell growth is a response to light and temperature and that the former is dependent on the WC-1 and WC-2 orthologs.
MATERIALS AND METHODS
Media.
Schizosaccharomyces japonicus was cultivated as previously described (16). YE medium (yeast extract, 5 g; glucose, 30 g/liter) was used as a rich medium. To induce growth of nutrient-dependent hyphae, malt agar (malt extract, 30 g), EMM2 (17), and morphology agar (Difco Yeast Morphology agar; Becton, Dickinson and Company, USA) were used. If necessary, a final concentration of 2% agar was added to make solid medium. For marker selection in YE medium, 40 μg/ml of Geneticin was used. To induce hyphal cells on agar plates, 2 μl of overnight-cultured yeast cells was spotted at the center and incubated at 30°C for at least a week. Burkholder's synthetic medium (18) was modified in this study as follows: nicotinic acid (10.0 mg/liter), citric acid (1.0 mg/liter), potassium chloride (0.1 mg/liter), and sodium sulfate (40.0 mg/liter) were supplemented instead of riboflavin, pyridoxine, thiamine, and niacin. The optical density at 660 nm (OD660) of cultures was automatically recorded every 15 min with a TVS062CA biophotorecorder (Advantec Co. Ltd., Japan).
Strains.
Strains used in this study are summarized in Table 1. Transformation of plasmids into yeast cells was performed by electroporation (16). The S. japonicus orthologs of wc-1 and wc-2 genes were identified by searching the database available at the Broad Institute (http://www.broadinstitute.org//annotation/genome/schizosaccharomyces_group/MultiHome.html) (19). These S. japonicus genes were wcs1, SJAG_02860, and wcs2, SJAG_05242.
Table 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| NIG2017 | h+ mat-2017 | 16 |
| NIG2028 | h− mat-P2028 | 16 |
| NIG5097 | h− mat-P2028 hht3-GFP::Kan | 16 |
| NIG3001 | h+ mat-2017 wcs1::natMX6 | This study |
| NIG3002 | h+ mat-2017 wcs2::kanMX6 | This study |
| NIG3003 | h+ mat-2017 wcs1::natMX6 wcs2::kanMX6 | This study |
| NIG3004 | h− mat-P2028 wcs1::natMX6 | This study |
| NIG3005 | h− mat-P2028 wcs2::kanMX6 | This study |
| NIG3006 | h− mat-P2028 wcs1::natMX6 wcs2::kanMX6 | This study |
Photography.
Colonies with hyphal zones were photographed with illuminating light coming through a plate from the bottom. Images were taken by a Nikon D3s camera with AF-S Micro Nikkor 60-mm f/2.8G ED or a Nikon D5200 camera with AF-S DX Micro Nikkor 85-mm f/3.5G ED VR. Interval images were carried out by shutter release mode under red light-emitting diode (LED) light (Flat Beam Aurora; GEX Corporation, Osaka, Japan). Images were processed by iPhoto imaging software (Apple Inc., USA). A fluorescence stereomicroscope (SMZ1500; Nikon, Japan) was used for hyphal cells with green fluorescent proteins (GFP). Distance between septa was measured by using MacSCOPE image-processing software (Mitani Co., Ltd., Japan).
Incubation under light and temperature cycles.
Under light conditions, plates were placed in a stainless steel sterilizing box (240 mm by 240 mm by 200 mm) under an LED lamp. The intensity of the LED lamp was 50 lx on the surface of a plate. The temperature of the incubator was 30.6 ± 0.2°C when the LED lamp was on and 30.3 ± 0.2°C when the LED lamp was off. The time of lighting was controlled by a programmable timer. The hyphal response by temperature cycles was induced using incubators MIR-153 (Panasonic Corporation, Japan) and FMU-0541 and FMU-1331 (Fukushima Industries Corp., Japan).
Deletion of wcs genes.
To create a wcs1 deletion mutants (NIG3001), the 5′ untranslated region (UTR) and 3′ UTR of wcs1 were amplified from NIG2017 genomic DNA using the following primers: 5′ UTR, AACAGGATCCACGGTCATTTCACGCTTCC and AGTTTAATTAAACGGATGTTGGAGAATAGC, and 3′ UTR, ACAGTTTAAACTAAGTGGTCTGCCTGATGC and ATTGTGAATTCCGTGTGATGAGATGCGGAG. To create wcs2 deletion mutants (NIG3001), the following primers were used: 5′ UTR, ACCCAGGATCCACTGTATCCCAAAG and GGCATATTAATTAACCACACCTTGAGG, and 3′ UTR, CTGCGTTTAAACGTCGATCAGTG and GCAACAGAATTCTTAGCTGTTCTTAGC. The gene disruption mutants for each of these S. japonicus genes were constructed as described previously (14).
Cloning of wcs genes.
DNA fragments containing the wcs1 or wcs2 gene with its putative promoter and terminator regions were amplified from S. japonicus genomic DNA (NIG2017) using the following primers: for wcs1, TAGGCTGCGGCCGCATTGCATGCAGTTCCATTTC and CGTAAAGCGGCCGCATACACTTCGTAGCATG, and for wcs2, CAGACAGCGGCCGCTCAAGTGATCTTACCC and GCTCTTGCGGCCGCAAACTGTAATCTTGC. Both PCR DNA fragments were digested with NotI restriction enzyme. These DNA fragments were inserted into the NotI site of a vector plasmid, pSJK11 (20).
Phylogenetic tree and alignment of amino acid sequences.
Homologues of Neurospora crassa WC-1 (accession no. X94300) or WC-2 (accession no. Y09119) were searched for in genomic databases of fungi by using the Basic Local Alignment Search Tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi). Homologues of N. crassa WC-1 with E values lower than 3E−25 were further analyzed by the neighbor-joining algorithm, ClustalW (http://clustalw.ddbj.nig.ac.jp/index.php?lang=ja). A phylogenetic tree of the homologues of Wcs1 was drawn by using FigTree software (http://tree.bio.ed.ac.uk/software/figtree/). Bootstrap values in this analysis were performed based on 1,000 replications by using ClustalW. The homologues of Wcs1 were aligned by using clustalX (http://www.clustal.org/clustal2/).
RESULTS
Photoresponse of growing hyphae.
When a colony of yeast cells is formed, hyphal cells extend from the periphery of the colony. Eventually, hyphae cover a large area of an agar plate called the “hyphal zone.” We noticed that when culture dishes were kept in an incubator with a window, concentric rings were occasionally formed within the hyphal zone. We assumed that the elongating hyphae responded to daily alterations of light coming through the window. When regular cycles of light illumination (12-h light and 12-h dark [12-h L/12-h D]) were applied throughout the hyphal growth phase under experimentally controlled conditions, distinct dark- and bright-colored stripes formed on the agar plate (Fig. 1A). In contrast, those stripes were not formed in hyphal cells when the incubation was under constant light (LL) or darkness (DD). Thus, hyphal cells of the fission yeast made concentric circles of dark- and bright-colored stripes surrounding the colony according to the number of light cycles.
Fig 1.
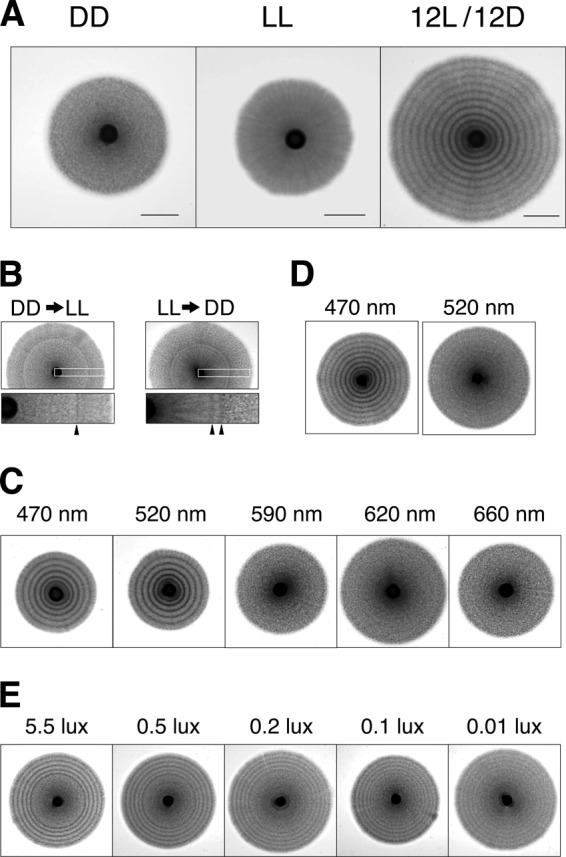
Photoresponse of hyphal cells. (A) Yeast cells (NIG2017) were spotted on the center of EMM2 agar plates and incubated at 30°C for longer than a week with constant dark (DD), constant light (LL), and the light cycle of 12-h light and 12-h dark (12L/12D). Bars, 1 cm. (B) An agar plate was incubated at 30°C until forming a hyphal zone during DD and then transferred into growth conditions at LL (DD→LL). Conversely, another was transferred from growth conditions under LL to DD (LL→DD). The portion of the bands shown in the white boxes is enlarged, and arrowheads indicate the dark-colored bands in each image. (C) Light from various LED lamps was tested for its ability to induce the photoresponse of hyphal cells. The power of the lamps was 50 lx at 10 cm of distance, and the light cycles were 12-h L/12-h D. (D) The power of the lamps was 2.5 lx at 10 cm. (E) Yeast cells were spotted on the center of EMM2 agar plates and incubated at 30°C for a week under constant dark and then under the light cycle of 12-h light and 12-h dark (12L/12D). The power of the LED lamp (470 nm) is indicated above each image.
The growth pattern of hyphal cells varied with the nutrient conditions of the agar plates. Concentric circles were clearly made on morphology agar and malt agar plates by elongated hyphal cells, given the proper light cycle, although sections were made within the hyphal zone (see Fig. S1 in the supplemental material). In conclusion, hyphal cells of S. japonicus have the ability to respond to light regardless of nutrient conditions for hyphal induction.
Effects of light and dark on growing hyphae.
Incubation of hyphal cells in continuous dark or light completely inhibited the formation of the hyphal stripes (Fig. 1A). This result suggests that the photoresponse is dependent either on periodic changes or on a single change of light or dark. We tested the effect of a single stimulus of light on hyphal growth. First, a yeast colony on an agar plate was incubated for several days in the dark to form a well-growing hyphal zone, and then light was applied. The plate was further incubated in the light for several days. A single transition from the darkness to the light induced a single dark-colored hyphal stripe band (Fig. 1B). Thus, a single stimulus of light after continuous darkness is sufficient for induction of the photoresponse of hyphal cells.
Next, we carried out an experiment with transition from continuous light to continuous darkness. Interestingly, darkness after continuous light also induced the hyphal photoresponse (Fig. 1B). In this case, two dark-colored bands appeared after the removal of light. We determined how long it took until the dark-colored stripes began to form after the transition. Several agar plates were incubated in continuous light until the hyphal zone was well formed, and then they were incubated in the dark. The first band appeared 10 h later, and the second stripe appeared 20 h later (see Fig. S2 in the supplemental material). Interestingly, we found that both light and darkness induced the photoresponse. These results suggested that the alteration of light intensity triggers the photoresponse. Incubation for many hours in the light is necessary for the induction of two dark-colored bands by darkness (see below).
Hyphal photoresponse by blue light.
It is known that many filamentous fungi have a photoresponse ability that is activated by a specific wavelength of light (1). We tested which wavelength activated the photoresponse of hyphal cells of S. japonicus. We used five different colors of light-emitting diode (LED) lamps, blue (470 nm), green (520 nm), orange (590 nm), red (620 nm), and deep red (660 nm), to illuminate hyphal cells. The blue and green lamps were highly effective in inducing the photoresponse, but the orange, red, and deep red lamps were not (Fig. 1C). Furthermore, the blue light was the most effective, as it could induce the formation of hyphal stripes at a lower power than could the green light (Fig. 1D). At least 0.1 lx of intensity of the blue lamp is sufficient for distinct formation of hyphal stripes (Fig. 1E). These results indicate that blue light contains the most effective wavelength to induce hyphal ring formation.
Hyphal photoresponse by blue light sensors.
Our results suggested that S. japonicus hyphae might have a blue light receptor for the photoresponse. In filamentous fungi, the photoreceptor system for blue light sensing is widespread, and genes encoding orthologs of photoreceptor proteins, WC-1 and WC-2, are well conserved in their genome. In the genome of S. japonicus, we found orthologs of WC-1 and WC-2 with low similarity of amino acid sequences (Fig. 2; see also Fig. S3 and S4 in the supplemental material) and referred to them as Wcs1 and Wcs2, respectively. Orthologs of cryptochrome and of photolyase, which are related to photoreactivation, are not found in the genome of S. japonicus. To confirm the involvement of these orthologs in blue light sensing, deletion mutations in these genes were constructed. Both deletion mutants were unable to form any stripes in response to blue light (Fig. 3A). Furthermore, a single stimulus of light or dark also had no effect on the photoresponse to hyphal cells (Fig. 3B). Complementation tests with cloned wcs1 and wcs2 genes showed that formation of the stripes by the blue light is recovered in each deletion mutant (Fig. 3C). Thus, Wcs1 and Wcs2 were certainly responsible for blue light sensing in S. japonicus, suggesting that they form the blue light receptor.
Fig 2.
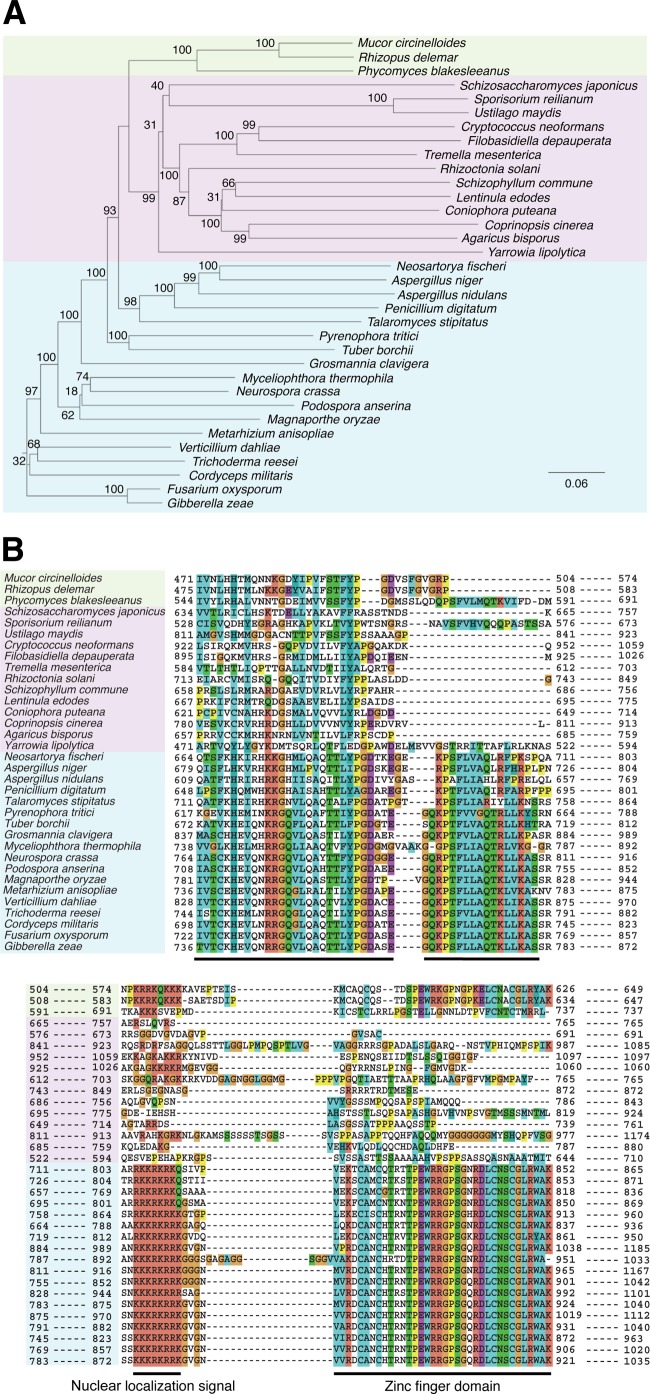
Phylogenetic tree and alignment of amino acid sequences of representative members (34 species) in the Wcs1 protein family. A phylogenetic tree (A) is shown for three phyla of fungi: Ascomycota (blue), Basidiomycota (pink), and Zygomycota (green). Bootstrap values are indicated as percentages. The scale bar indicates an evolutionary distance of 0.06 amino acid substitutions per position. Phylogenetic trees of all members of WC families are shown as Fig. S3 and S4 in the supplemental material. The color scheme in partial alignment of amino acid sequences is used as the default scheme of ClustalX (B). The orange indicates G as glycine. The yellow indicates P as proline. The green indicates polar amino acid residues STQN when those residues were conserved more than 50% or hydrophobic residues ACFHILMVWYP when they were conserved more than 60%. The blue indicates hydrophobic residues ACFHILMVWYP when those residues were conserved more than 60%. The cyan indicates H as histidine and Y as tyrosine when each residue was conserved more than 85% or hydrophobic residues ACFILMVWP when they were conserved more than 60%. The magenta indicates negatively charged residues DE when those residues were conserved more than 50%. The blood orange color indicates positively charged residues KR when those residues were conserved more than 60%. Black bars indicate the amino acid sequences that are well conserved in Ascomycota.
Fig 3.
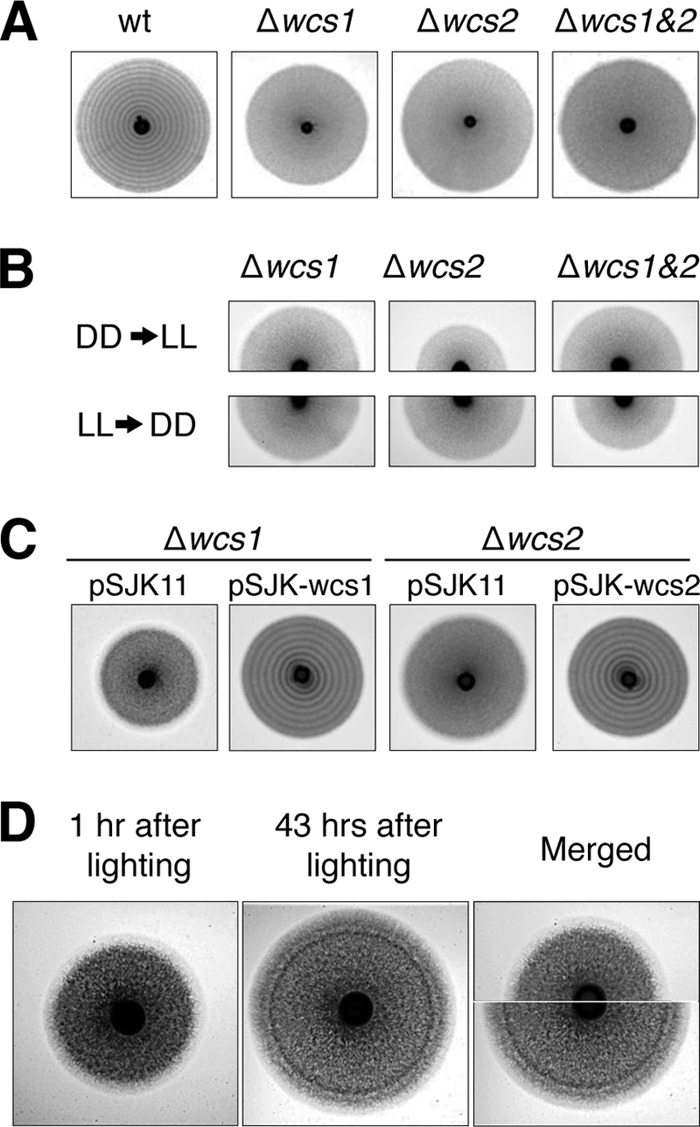
The effect of wcs genes on stripe formation. (A) Wild-type (wt) yeast cells (NIG2017) and deletion mutants for wcs genes (NIG3001, NIG3002, and NIG3003) were spotted on EMM2 agar plates and incubated at 30°C under 12-h L/12-h D light cycles. (B) EMM2 agar plates with deletion mutants of wcs genes (NIG3001, NIG3002, and NIG3003) were incubated at 30°C until forming a hyphal zone during DD and then were transferred into growth conditions at LL (DD→LL, top). Conversely, another was transferred from growth conditions under LL to DD (LL→DD, bottom). (C) The complementation tests of the deletion mutants of the wcs genes were performed by using the plasmid harboring wcs1 (pSJK-wcs1) or wcs2 (pSJK-wcs2). The deletion mutants (NIG3001 and NIG3002) harboring these plasmids or the vector plasmid pSJK11 were incubated at 30°C under 12-h L/12-h D light cycles. (D) An EMM2 agar plate with yeast strain NIG2017 was incubated at 30°C in the dark until forming a hyphal zone and then illuminated by a blue LED lamp for 1 h. It was further incubated under constant red light (620 nm). After stimulation by blue light, images were taken at 1-h intervals. Images shown are at 1 h and 43 h. Upper and lower halves of the images were combined.
To test whether a circadian rhythm was involved in the stripe formation of hyphal growth, a free-running experiment was carried out. The rhythm of the stripe formation was entrained to 12-h L/12-h D cycles for several days and then changed to DD or LL. The patterning of the dark- and bright-colored stripes ceased formation after the change (see Fig. S5 in the supplemental material). In fact, other genes related to the circadian rhythm of N. crassa (frq, frh, and vvd) are not found in the S. japonicus genome.
The growing front of the hyphae is photosensitive.
Hyphal cells show apical growth, which means the filamentous cells elongate at only one of the cell tips. When hyphal cells radially spread from a yeast colony, distal daughter cells of the hyphae continue to divide after cell growth while proximal daughter cells tend to suspend cell growth. As a result, hyphal cells that are located at the front of the hyphal zone are always actively growing. This suggests that cells at the forefront of the hyphal zone are likely to respond to light and then form a dark-stripe zone. To test whether cells at the forefront of the hyphal zone become dark colored after light stimuli, we carried out time-lapse imaging of the photoresponse. Hyphal cells on a plate were grown in the dark and then irradiated by a blue LED lamp for 1 h, and subsequently the cells were incubated under a red LED lamp that does not trigger the photoresponse. We were able to take images because of the red LED lamp during further incubation. Distinct dark-colored stripes were visible in the hyphal cell zone 43 h later (Fig. 3D). We compared the photoimages of the hyphal zone 1 h after blue light exposure with that 43 h after exposure (Fig. 3D). The position of the dark-colored stripes that appeared on the plate 43 h after blue light exposure corresponded to the forefront of the hyphal zone on the plate 1 h after exposure where there were apical growing hyphal cells. We determined how long it took until the dark stripes began to form after exposing the hyphal cells to blue light. A series of time-lapse images indicated that incubation for 20 h in light caused darkness at the edge of the hyphal zone (see Fig. S6 in the supplemental material). These results indicated that light stimulates only hyphal cells at the forefront, and although distal daughter cells continue to elongate, proximal daughter cells remain at their position and synchronously activate cell division 20 h after light stimulus.
Active cytokinesis in the dark zone of the stripes.
What is the nature of the dark-colored stripes of the hyphae? We directly observed hyphal cells under a fluorescence stereomicroscope. A strain of S. japonicus that constitutively expressed histone H3 fused with green fluorescent protein (GFP) was used so that nuclei were detectable in hyphal cells. Zones of the brightly colored stripes contained long filamentous hyphae that were transparent tubular cells without septa (Fig. 4A). In contrast, septated hyphae, in which septation occurred at a higher frequency, were enriched in the dark zones. Because many septations by cytokinesis had diminished the transparency of the hyphae, the zones had become dark colored. In addition, we confirmed that hypha septation was also accompanied by nuclear division (Fig. 4A). Fluorescent nuclei were distributed in each septated hypha- and yeast-like cell. Statistical analyses of distance between septa in hyphal cells showed that the length of cells in the bright-colored segment is greater than that in the dark-colored segment, indicating that hyphal cells undergo high-frequency division in the dark-colored segment (Fig. 4B and C). These results indicated that growing hyphal cells are actively undergoing cytokinesis and nuclear division. We conclude that the cell cycle of hyphal cells is synchronously stimulated by light.
Fig 4.
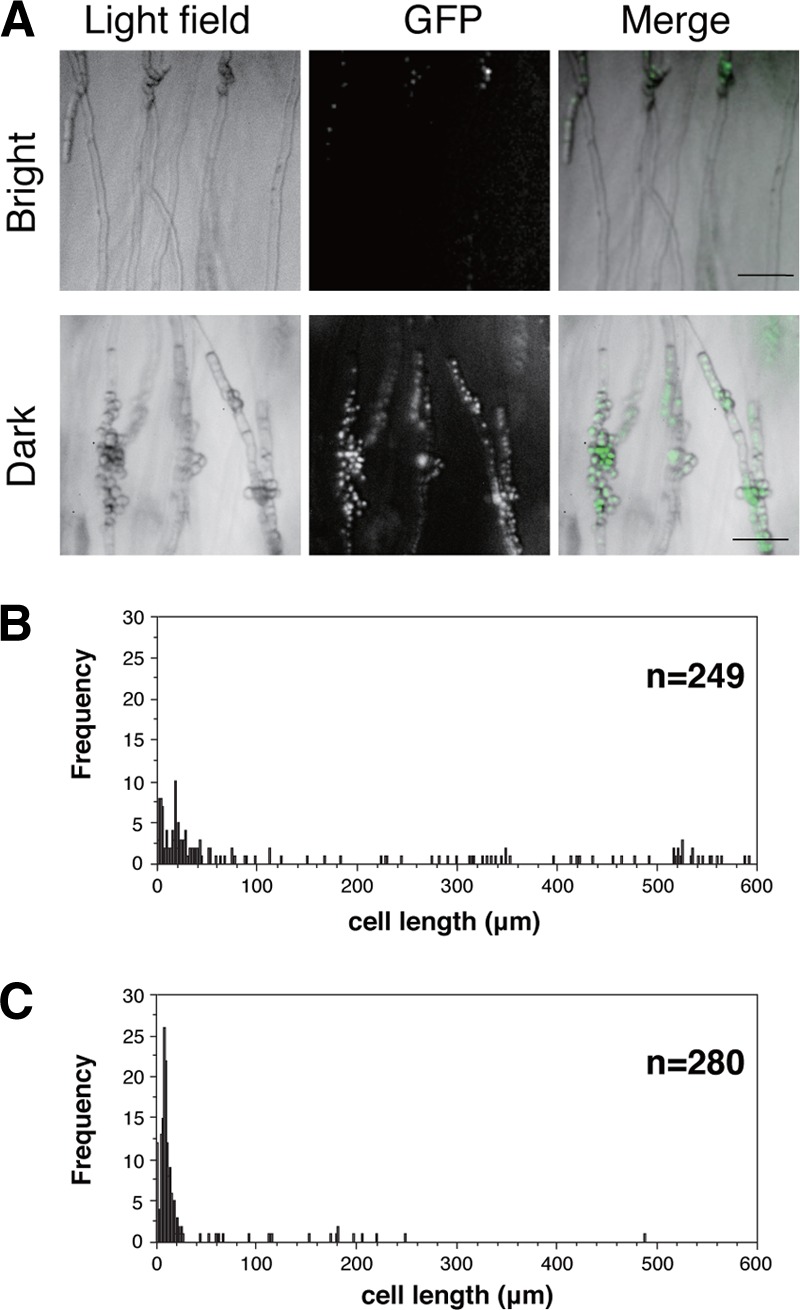
Hyphal cells in dark- and bright-colored stripes. (A) Images of hyphal cells taken by a fluorescence stereomicroscope. Hyphal cells are shown in a bright-colored stripe (upper row) and a dark-colored stripe (lower row). A yeast strain (NIG5097) with histone H3 proteins labeled by GFP was used to detect nuclei. Bar, 0.1 mm. (B and C) Distributions of cell length in the bright-colored stripe (B) and dark-colored zone (C). The average length of cells in the bright-colored segment is 142 ± 189 μm (mean ± standard deviation [SD]; n = 249), and the average length of cells in the dark-colored segment is 24 ± 51 μm (mean ± SD; n = 280).
Patterning of hyphal stripes in response to various ratios of L/D.
In the natural environment of the Earth, the light cycle is a constant 24 h of daily rhythms. However, the day-to-night ratio varies according to the seasons and latitude. It seemed that the photoresponse of S. japonicus adapts to the lighting cycles in its natural habitat. We tested the effect of varied ratios of light and dark (L/D) in a 24-h light cycle on the formation of dark- and bright-colored stripes. At least 1 min of blue light illumination was effective in causing the photoresponse in a 24-h light cycle (Fig. 5). Given a prolonged lighting time, the pattern of the dark- and bright-colored stripes changed. At first, the dark-colored area of the stripes was narrowed as the period of the dark was shortened. Simultaneously, the pattern of the stripes was clear and enhanced under 12-h L/12-h D. Interestingly, when 14-h L/10-h D was applied, an additional dark-colored stripe was induced between the usual dark stripes (Fig. 5). The additional dark-colored stripes were faint and thinner than the usual dark-colored stripes. They also appeared under 16-h L/8-h D, 18-h L/6-h D, and 21-h L/3-h D. Finally, the stripes of 21-h L/3-h D were weakened, and further prolonged illumination inhibited the formation of stripes in the hyphal zone. This result indicates that at least 3 h of darkness is necessary for the formation of stripes during the light cycle.
Fig 5.
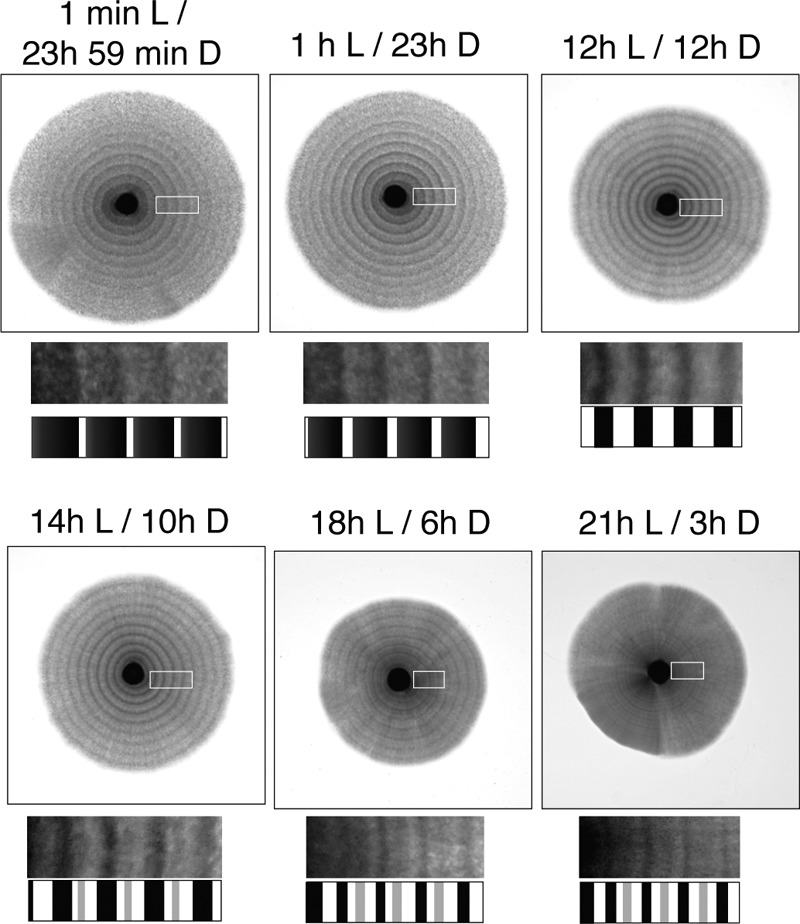
Photoresponse to various ratios of L/D. Yeast cells (NIG2017) were spotted on the center of agar plates and incubated at 30°C for 2 weeks under various lighting times and 24-h light cycles. Blue light was applied according to each light cycle as indicated above each image. A portion of the stripes shown in the white boxes is enlarged, and the patterns of the stripes are illustrated below each image.
Hyphal photoresponse by temperature cycles.
We tested whether other environmental cues could cause the formation of the hyphal stripes. Alternation in temperature can be a daily external cue. We incubated plates with hyphal cells in an incubator with temperature cycles of 30°C for 12 h and 35°C for 12 h. After incubation for more than 10 days, patterning with the dark- and bright-colored stripes was observed regardless of incubation in the dark (Fig. 6A). As seen in the photoresponse, hyphal cells in the dark-colored stripes were septated, and the cells in the bright-colored stripes were transparent and filamentous (see Fig. S7 in the supplemental material). It is obvious that alteration of temperature induced a similar response in the hyphal cells to light.
Fig 6.
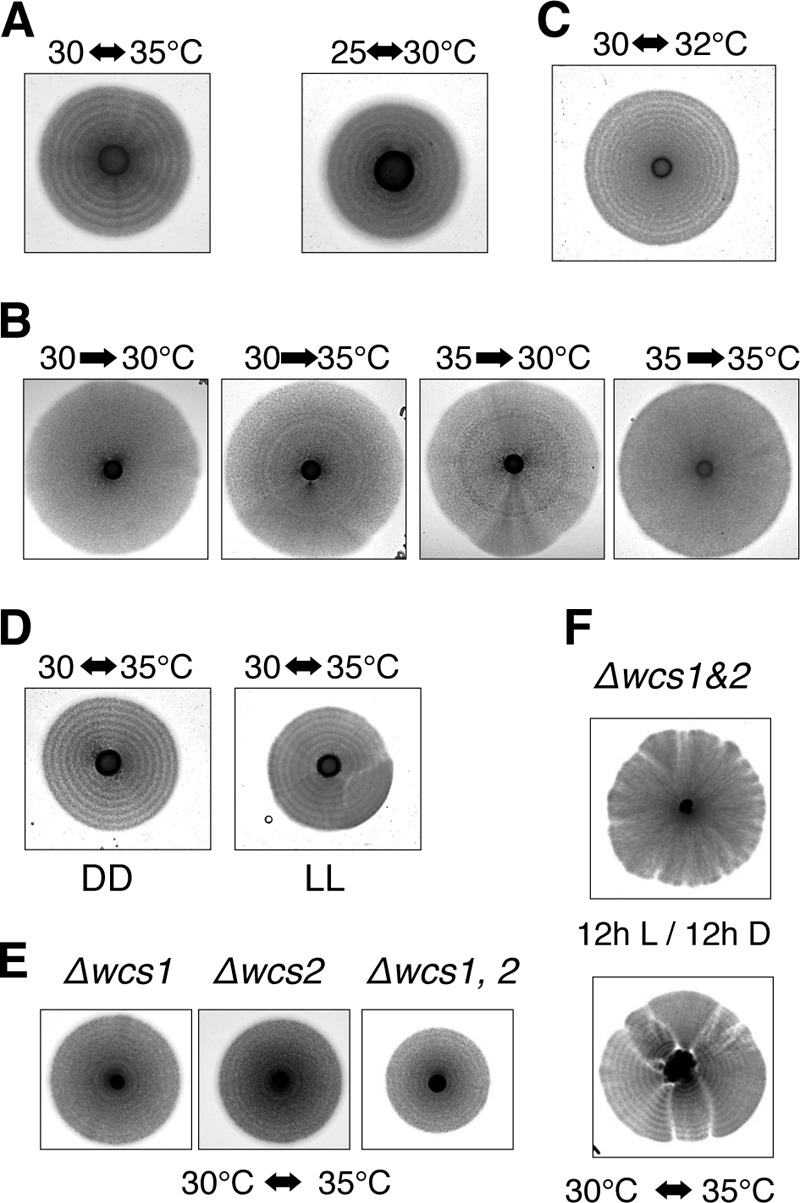
Effect of temperature shift on hyphal cells. (A) Yeast cells (NIG2017) were spotted on the center of EMM2 agar plates and incubated under DD for more than a week: temperature cycles were 12 h at 30°C and 12 h at 35°C or 12 h at 25°C and 12 h at 30°C. (B) EMM2 agar plates were incubated at 30°C until forming hyphal zones under DD and then were transferred into growth condition at 35°C. Conversely, others were transferred from growth conditions under 35°C to 30°C. (C) Yeast cells (NIG1056) were spotted on the center of YE agar plates and incubated under DD for more than a week: temperature cycles were 12 h at 30°C and 12 h at 32°C. (D) An EMM2 agar plate was incubated under constant blue light with temperature cycles of 12 h at 30°C and 12 h at 35°C. The power of the lamps was 50 lx at a 10-cm distance. (E) Yeast cells of deletion mutants of wcs genes were spotted on EMM2 agar plates and incubated at 30°C with temperature cycles of 12 h at 30°C and 12 h at 35°C. (F) The double deletion mutant of wcs1 and wcs2 genes was spotted on morphology agar plates and incubated at 30°C under a 12-h L/12-h D light cycle (top) with temperature cycles of 12 h at 30°C and 12 h at 35°C (bottom).
Temperature cycles of 25°C for 12 h and 30°C for 12 h also caused hyphal cells to form stripes (Fig. 6A). The stripes caused by alteration in temperature were induced by both a shift up, from 30°C to 35°C, and a shift down, from 35°C to 30°C (Fig. 6B). We previously isolated a spontaneous mutant that constitutively induces hyphal cells regardless of the nutrient conditions. We found that hyphal cells of the mutant strain were able to form stripes with temperature cycles of 30°C for 12 h and 32°C for 12 h (Fig. 6C). Only a 2°C alteration in external temperature was required to affect the cell division cycle of hyphal cells, suggesting that this heat effect is different from heat shock or cold shock that causes structural changes in proteins and macromolecules. Thus, the response to alterations in temperature was quite sensitive in the hyphal cells.
The effect of temperature cycles was tested either in constant dark or in constant light. Constant blue light had no obvious effect on the temperature cycles (Fig. 6D). The stripes caused by the temperature cycles formed in all wcs gene mutants (Fig. 6E). Although the contrast between the bright- and dark-colored stripes was weakened on EMM2 agar plates, the response to alteration in temperature was retained in mutant cells as seen by the formation of temperature-dependent hyphal rings on morphology agar plates (Fig. 6F). These results suggest that each signaling pathway for light and temperature independently functions to induce a synchronous cell division cycle in hyphal cells.
Wcs proteins in yeast cells.
It was previously reported that light enhances sexual flocculation in S. japonicus under conditions of vegetative growth (21). Sexual flocculation is made prior to sporulation when light is adequate (Fig. 7A). This sexual flocculation was inhibited in the dark (Fig. 7B). In addition, the sexual flocculation was completely inhibited in the wcs1 or wcs2 deletion mutant or double mutants, even though the culture was illuminated (Fig. 7C). As a result, zygotes could not develop in the mutants. On the other hand, when the nitrogen source was eliminated from the medium to induce sporulation, even in the mutants, they were able to induce sexual flocculation and succeed in sporulation regardless of the dark. Thus, although neither wcs1 nor wcs2 was essential for vegetative growth and sporulation per se, given the light-dependent physiology of this yeast, sporulation and vegetative growth were affected via the function of the wcs genes under certain nutrient conditions.
Fig 7.
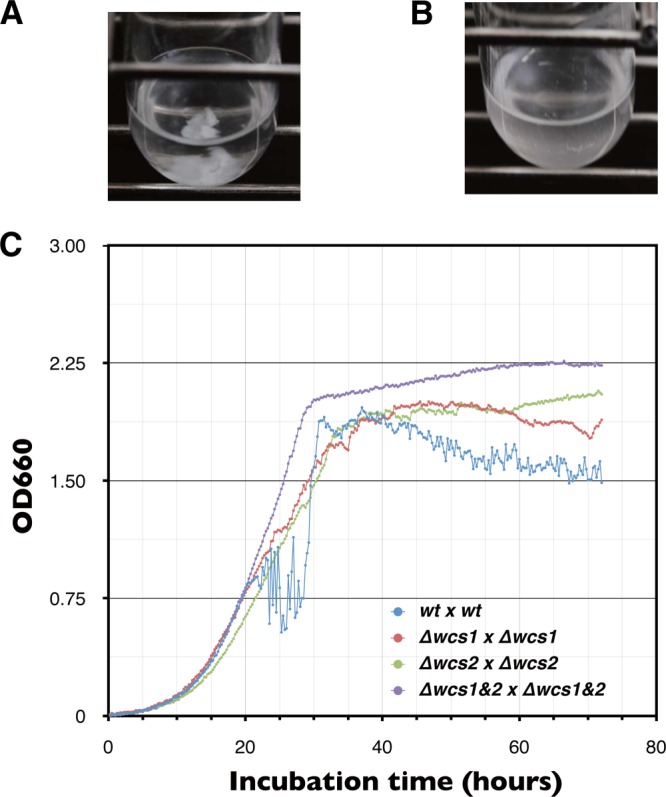
Effect of wcs genes on sexual flocculation. S. japonicus cells can sporulate in a nutrient-rich medium, Burkholder synthetic medium. (A) Sexual flocculation enhances precipitation of cells at the bottom of gently shaking test tubes in which wild type (wt) (h+) and wt (h−) are mixed. (B) Sexual flocculation is completely inhibited in test tubes incubated in the dark. Heterothallic strains of S. japonicus cells were independently cultivated in modified Burkholder synthetic medium overnight. Cells were washed once with fresh modified Burkholder synthetic medium and then mixed in test tubes according to each combination of mating types as follows: wt (h+) × wt (h−), Δwcs1 (h+) × Δwcs1 (h−), Δwcs2 (h+) × Δwcs2 (h−), Δwcs1&2 (h+) × Δwcs1&2 (h−). (C) The optical density of test tubes was automatically monitored during conjugation. Optical density of mating cultures was remarkably reduced because of precipitation of cells by sexual flocculation. After completion of spore formation, cells separated from one another and then optical density increased. The OD660 of mixing cultures was automatically recorded every 15 min.
DISCUSSION
Many organisms adjust the timing of cell division or differentiation to the night in order to escape photodamage. From our experiments applying various ratios of L/D, the light cycles tended to activate cytokinesis during the dark period. This might be beneficial to cells, as they can avoid light-induced damage during cell division. However, it seems that adjustment of the cell cycle to darkness is not perfect. In the case of S. japonicus, cellular responses to changes in either light or temperature lead the growing hyphal cells to the same consequence: synchronous activation of the cell division cycle. Both the temperature cycles and the light cycles might independently adjust the cellular activity to an ideal time of day in the natural environment. Given that aborting the light or temperature cycles immediately interrupted the hyphal stripe formation, each is a one-time-only response to daily alternating environmental stimuli, and circadian rhythmic responses are not involved. Without a circadian rhythm, this organism can still make periodic adaptations to environmental stresses by using a dual sensing mechanism for temperature and light. In other words, S. japonicus cells adapt their physiology and behavior directly to environmental circumstances rather than by anticipation of daily changes by a circadian clock.
S. japonicus has quite a high sensitivity to light and temperature, and it might be that even moonlight affects the hyphal cells, because the intensity of moonlight may reach 0.7 lx (22). Circadian clocks can sense even a 2°C difference in temperature (23–25). Curiously, the present sensing mechanism can respond to both negative and positive stimuli from light and temperature. In general, the WC complex, which includes FAD to catch photons, is activated by light. Loss of the wcs1 and wcs2 genes caused loss of sensitivity to light stimuli both from DD to LL and from LL to DD. The WC complex of S. japonicus is activated somehow not only by light but also by its absence.
By providing light for only 1 min after continuous dark, two stimuli are generated: exposure to light and its removal. In general, light is a major stimulus for organisms. An adequate lighting period is required for the effect of darkness, and this lighting period is longer than 14 h for S. japonicus. We inferred this requirement from the appearance of faint stripes due to stimulation by darkness after continuous illumination for longer than 14 h (Fig. 8). Dark-colored stripes caused by the effect of light emerged 20 h after illumination. Two dark-colored stripes emerged after removal of light: the first stripe emerged 10 h after darkness, and the second stripe emerged at 20 h. In the case of more than 14 h of light, the dark-colored stripes caused by light and the first stripes caused by darkness merged, and then the second stripes were seen as additional faint bands.
Fig 8.
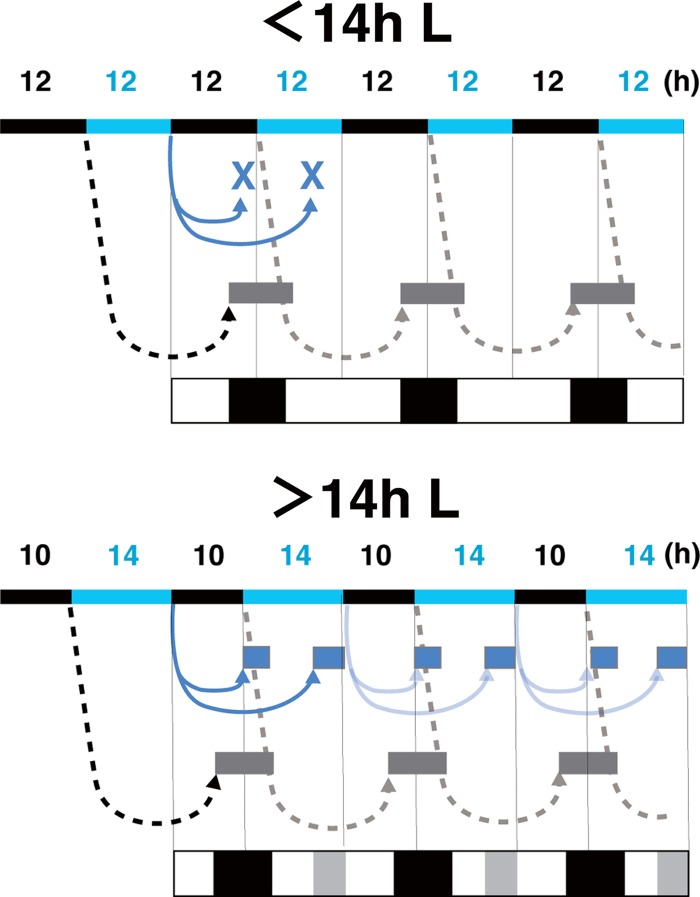
Diagrams of the photoresponse caused by light and darkness. The light cycles are indicated by the top bars (black, dark; blue, light). The effect of light (black dotted lines) induced synchronous cell division 20 h later (gray boxes). When the lighting time was longer than 14 h, the photoresponse by darkness (blue line) induced synchronous cell division 10 and 20 h later, respectively (blue boxes). The patterns of the dark- and bright-colored stripes are illustrated at the bottom, and additional dark-colored stripes are shown as gray.
Activation of the cell division cycle in hyphal cells is synchronously induced with a constant time delay after stimulation with light or temperature and then synchronously reduced so that regular patterning of hyphal cells is formed. This temporal regulation suggests that the response of hyphal cells is controlled by a timer, like an hourglass. This means that once triggered, an event will be performed. An hourglass timer requires resetting to be repeated. In fact, a 22-h L/2-h D light cycle does not result in formation of hyphal stripes. More than 3 h of darkness for reactivation of the photoresponse might be necessary to reset the putative hourglass timer.
A relatively broad range of the ratio of light to dark affects the photoresponse of hyphal cells. When lighting is more than 14 h, an additional effect is observed when darkness is applied. The photoresponse of S. japonicus has probably been adapted to its particular natural environment. While the natural habitat of S. pombe is in the tropics, S. japonicus is endemic to temperate regions such as Michigan, USA, and Fukuoka, Japan, where periods of daylight are more than 14 h before and after the summer solstice (26, 27).
There is a homologue of WC-1 in the basidiomycete yeast Cryptococcus neoformans (6). In addition, we recently found homologues of Neurospora crassa WC-1 in two species of the Ascomycota yeasts, S. japonicus and Yarrowia lipolytica. For genome databases of fungi that have been analyzed, S. japonicus and Y. lipolytica are the only ascomycete yeasts that retain proteins of the WC family. The WC-1 orthologs of these two species lack the conserved zinc finger domains, the nuclear localization signal sequences, and an unknown functional domain found in Ascomycota (Fig. 2B). Interestingly, their WC orthologs are more closely related to those of the Basidiomycota rather than to those of the Ascomycota. In S. japonicus, not only do hyphal cells show a periodic response to daily alterations, but also yeast cells show light-dependent flocculation during sporulation. Further analyses of gene expression will reveal whether yeast respond to light and temperature during vegetative growth.
Except for the WC orthologs, genes involved in circadian rhythms are not found in the genome of S. japonicus. This observation is consistent with the loss of a free-running rhythm after disruption of the light or temperature cycle. During adaptation to its particular environment, this yeast had apparently lost those genes and only the photoactivated proteins remained. Alternatively, these clock components arose later in a small branch of the Ascomycota. In any case, the acquirement of photoactivated proteins was a sufficient adaptation for S. japonicus. Moreover, the temperature cycle-dependent response provides this fission yeast with an additional strategy for hyphal cells to adapt to environmental conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank N. Ishihara for technical assistance and H. Iwasaki for discussions of our results.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to H.N. and an NIG Postdoctoral Fellowship to S.O.
Footnotes
Published ahead of print 19 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00109-13.
REFERENCES
- 1.Corrochano LM, Avalos J. 2010. Light sensing, p 417–441 In Borkovich KA, Ebbole DJ. (ed), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC [Google Scholar]
- 2.Linden H, Ballario P, Macino G. 1997. Blue light regulation in Neurospora crassa. Fungal Genet. Biol. 22:141–150 [DOI] [PubMed] [Google Scholar]
- 3.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. 2002. White collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297:815–819 [DOI] [PubMed] [Google Scholar]
- 4.He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y. 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297:840–843 [DOI] [PubMed] [Google Scholar]
- 5.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Idnurm A, Heitman J. 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3:e95. 10.1371/journal.pbio.0030095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu YK, Sun KH, Shen WC. 2005. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol. Microbiol. 56:480–491 [DOI] [PubMed] [Google Scholar]
- 8.Schwerdtfeger C, Linden H. 2001. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol. Microbiol. 39:1080–1087 [DOI] [PubMed] [Google Scholar]
- 9.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. 1994. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263:1578–1584 [DOI] [PubMed] [Google Scholar]
- 10.Wijnen H, Young MW. 2006. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40:409–448 [DOI] [PubMed] [Google Scholar]
- 11.Kaldi K, Gonzalez BH, Brunner M. 2006. Transcriptional regulation of the Neurospora circadian clock gene wc-1 affects the phase of circadian output. EMBO Rep. 7:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Merrow M, Loros JJ, Dunlap JC. 1998. How temperature changes reset a circadian oscillator. Science 281:825–829 [DOI] [PubMed] [Google Scholar]
- 13.Eelderink-Chen Z, Mazzotta G, Sturre M, Bosman J, Roenneberg T, Merrow M. 2010. A circadian clock in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 107:2043–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuya K, Niki H. 2010. The DNA damage checkpoint regulates a transition between yeast and hyphal growth in Schizosaccharomyces japonicus. Mol. Cell. Biol. 30:2909–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sipiczki M, Takeo K, Yamaguchi M, Yoshida S, Miklos I. 1998. Environmentally controlled dimorphic cycle in a fission yeast. Microbiology 144:1319–1330 [DOI] [PubMed] [Google Scholar]
- 16.Furuya K, Niki H. 2009. Isolation of heterothallic haploid and auxotrophic mutants of Schizosaccharomyces japonicus. Yeast 26:221–233 [DOI] [PubMed] [Google Scholar]
- 17.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 18.Burkholder PR. 1943. Vitamin deficiencies in yeast. Am. J. Bot. 30:206–211 [Google Scholar]
- 19.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Muller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C. 2011. Comparative functional genomics of the fission yeasts. Science 332:930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki K, Nakajima R, Furuya K, Niki H. 2010. Novel episomal vectors and a highly efficient transformation procedure for the fission yeast Schizosaccharomyces japonicus. Yeast 27:1049–1060 [DOI] [PubMed] [Google Scholar]
- 21.Itoh S, Takahashi S, Tsuboi M, Shimoda C, Hayashibe M. 1976. Effect of light on sexual flocculation in Schizosaccharomyces japonicus. Plant Cell Physiol. 17:1355–1358 [Google Scholar]
- 22.Bunning E, Moser I. 1969. Interference of moonlight with the photoperiodic measurement of time by plants, and their adaptive reaction. Proc. Natl. Acad. Sci. U. S. A. 62:1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis CD, Sargent ML. 1979. Effects of temperature perturbations on circadian conidiation in Neurospora. Plant Physiol. 64:1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Underwood H, Calaban M. 1987. Pineal melatonin rhythms in the lizard Anolis carolinensis: II. Photoreceptive inputs. J. Biol. Rhythms 2:195–206 [DOI] [PubMed] [Google Scholar]
- 25.Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. 1993. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J. Biol. Rhythms 8:67–94 [DOI] [PubMed] [Google Scholar]
- 26.Wickerham LJ, Duprat E. 1945. A remarkable fission yeast, Schizosaccharomyces versatilis nov. sp. J. Bacteriol. 50:597–607 [DOI] [PubMed] [Google Scholar]
- 27.Yukawa M, Maki T. 1931. Schizosaccharomyces japonicus. nov. spec. Bul. Sci. Fak. Terkult. Kjusu Imp. Univ. Fukuoka Japan 4:218–226 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.