Abstract
Cks1 was originally identified based on genetic interactions with CDC28, the gene that encodes Cdk1 in the budding yeast Saccharomyces cerevisiae. Subsequent work has shown that Cks1 binds Cdc28 and modulates its activity against certain substrates. However, the Cks1/Cdc28 complex also has a role in transcriptional chromatin remodeling not related to kinase activity. In order to elucidate protein networks associated with Cks1 transcriptional functions, proteomic analysis was performed on immunoaffinity-purified Cks1, identifying a physical interaction with the Paf1 complex. Specifically, we found that the Paf1 complex component Rtf1 interacts directly with Cks1 and that this interaction is essential for efficient recruitment of Cks1 to chromatin in the context of GAL1 gene induction. We further found that Cks1 in this capacity serves as an adaptor allowing Rtf1 to recruit 19S proteasome particles, shown to be required for efficient RNA production from some rapidly inducible genes such as GAL1.
INTRODUCTION
Cks1 (cyclin-dependent kinase subunit 1), first identified in yeasts based on genetic and physical interaction with Cdc28 and Cdc2, the budding and fission yeast orthologs of Cdk1, has been shown to be involved in cell cycle regulation (1, 2). Loss of Cks1 function impedes cell cycle progression due to inadequate expression of Cdc20, a positive regulator of mitosis (3). Recruitment of Cks1 to the promoter of CDC20 is correlated with the periodic expression of CDC20 mRNA, suggesting a direct role in CDC20 transcription (3). The observation that Cks1 interacts with the proteasome (4) implies that the transcriptional function of Cks1 may involve the proteasome. Indeed, both Cks1 and the proteasome bind to the promoter of CDC20 coordinately with its expression (3). Proteolysis is the primary function carried out by the 26S proteasome, composed of 19S regulatory and 20S proteolytic core particles (5), and degradation of RNA polymerase (Pol) II and some transcription factors has been shown to be an integral part of the transcription cycle (6, 7). However, nonproteolytic roles of the proteasome are also central to the transcription process (8, 9). In particular, the 19S regulatory particle acts as a distinct unit on chromatin during transcription regulation, independent of protein degradation functions (10). It has been suggested that the complete 26S proteasome is recruited to chromatin initially, but whether the proteolytic or nonproteolytic functions will be executed is context dependent (11).
The 19S proteasome affects many aspects of transcription, including (i) modulation of association between transcription factors and chromatin (12–14), (ii) regulation of interaction among transcription factors (15, 16), (iii) modification of histones (15, 17–21), and (iv) enhancement of transcription efficiency (22, 23). The interaction between Cks1 and the proteasome led us to investigate the contribution of Cks1 to aspects of transcription that require the function of the 19S proteasome. To this end, we chose the GAL1 gene, encoding galactokinase, as the readout system for study of Cks1 function because of its robust inducibility, a signature shared by most 19S proteasome-targeted genes (24), and because induction can be easily manipulated. We have previously shown that Cks1 is required for recruitment of the 19S proteasome to the GAL1 locus (25), leading to induction-specific nucleosome eviction required for efficient transcriptional elongation (26) and possibly other functions. Presumably the ATPase activities associated with the 19S proteasome provide energy required for this aspect of chromatin remodeling. However, how Cks1 is recruited to genes and executes regulatory roles during transcription remains to be elucidated. Therefore, to gain more insight into Cks1-mediated transcriptional functions, we have carried out analyses to identify Cks1-interacting proteins.
In the present study, a proteomic analysis of Cks1 immunoprecipitates identified components of the Paf1 complex (RNA polymerase II-associated factor 1). The Paf1 complex, consisting of Cdc73, Ctr9, Leo1, Paf1, and Rtf1, is thought to be a platform for assembling factors that facilitate RNA polymerase II movement along chromatin during transcriptional elongation, although each component has specific roles during these processes (27). Recent findings indicate that the Paf1 complex is also involved in Pol I transcription and RNA processing and export (28). Notably, the Paf1 complex is required for H2B ubiquitylation (29, 30), and the ubiquitylation of histone H2B is required for the recruitment of the 19S proteasome to GAL genes (17). Nevertheless, biochemical evidence linking the Paf1 complex to the 19S proteasome is lacking. Here we report that Cks1 serves as a mediator for recruitment of the 19S proteasome to the Paf1 complex, thereby promoting transcriptional elongation by providing energy for nucleosome eviction.
MATERIALS AND METHODS
Yeast strains.
All Saccharomyces cerevisiae strains were derived from 15Daub (31). Genotypes were generated by standard yeast genetic methods (32) and are listed in Table 1.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| 15D a | a bar1 ura3 ade1 his2 leu2-3 trp1 | 31 |
| 15D α | α bar1 ura3 ade1 his2 leu2-3 trp1 | 31 |
| Δcks1 | 15D α cks1Δ::KANR | 25 |
| Cks1-Flag | 15D a CKS1-Flag | 25 |
| Cks1-Flag, Rtf1-HA | 15D a CKS1-Flag RTF1-HA | This study |
| Cks1-Flag, Rtf1-Myc | 15D a CKS1-Flag RTF1-Myc | This study |
| Δcdc73 | 15D α cdc73Δ::HYGR | This study |
| Δctr9 | 15D α ctr9Δ::HYGR | This study |
| Δleo1 | 15D α leo1Δ::HYGR | This study |
| Δpaf1 | 15D α paf1Δ::HYGR | This study |
| Δrtf1 | 15D α rtf1Δ::HYGR | This study |
| Cks1-Flag, Δcdc73 | 15D a CKS1-Flag cdc73Δ::HYGR | This study |
| Cks1-Flag, Δctr9 | 15D a CKS1-Flag ctr9Δ::HYGR | This study |
| Cks1-Flag, Δleo1 | 15D a CKS1-Flag leo1Δ::HYGR | This study |
| Cks1-Flag, Δpaf1 | 15D a CKS1-Flag paf1Δ::HYGR | This study |
| Cks1-Flag, Δrtf1 | 15D a CKS1-Flag rtf1Δ::HYGR | This study |
| Rtf1-Flag | 15D α RTF1-Flag | This study |
| Rtf1-Flag, Δcks1 | 15D α RTF1-Flag cks1Δ::KANR | This study |
| Rtf1-HA | 15D a RTF1-HA | This study |
| Rpt1-Flag | 15D a RPT1-Flag | This study |
| Rpt1-Flag, Rtf1-HA | 15D a RPT1-Flag RTF1-HA | This study |
| Rpt1-Flag, Δrtf1 | 15D a RPT1-Flag rtf1Δ::HYGR | This study |
Yeast growth.
Regular growth medium for our yeast stains was yeast extract-peptone (YEP) with 2% dextrose (YEPD). For GAL1 induction experiments, yeast cells were grown in YEP medium with 2% raffinose, galactose (Gal) with a final concentration of 2% was added, and cells were incubated for 45 min. Mating pheromone arrest synchrony experiments were performed as described previously (33).
Antibodies.
The following antibodies were obtained from commercial sources: anti-Flag (Sigma), anti-glutathione S-transferase (anti-GST; Millipore), antihemagglutinin (anti-HA; Roche), anti-His (Millipore), anti-Myc (Sigma), and anti-Rpt1 (Abcam). Anti-Cdc28 antibodies were described previously (34).
Western blotting and coimmunoprecipitation.
Western blotting and coimmunoprecipitations were carried out as previously described (4).
Purification of proteins for mass spectrometry analysis.
Flag-Cks1 and untagged strains were grown to an optical density (OD) of 2.0, typically in 1 liter of YEPD medium. Cells were harvested and washed once with ice-cold water. The cell pellet was drop frozen in liquid nitrogen and ground to a fine powder using a Retsch grinder chilled with liquid nitrogen. The ground powder was collected in a 50-ml screw-cap tube and thawed in one pellet volume of 50 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, and 5 mM MgCl2 (buffer A). The thawed cell lysate was centrifuged for 20 min at 17,000 rpm, and the pellet was discarded. A 3-ml aliquot of the supernatant was mixed with 100 μl of anti-Flag M2 agarose beads (Sigma, St. Louis, MO) for 2 h on a rocker at 4°C. The beads were then collected, transferred to 2-ml microcentrifuge tubes, and washed with 50 volumes total of buffer A plus 0.2% Triton X-100. Specifically bound proteins were eluted for 3 h at 4°C with three bead volumes of elution buffer containing 25 mM Tris (pH 7.5), 150 mM NaCl, 15% glycerol, 5 mM MgCl2, and 100 μg/ml of Flag peptide. Eluates from both Flag-Cks1 and untagged strains were prepared for analysis by mass spectrometry.
Mass spectrometry analysis.
Mass spectrometry analysis was performed as previously described (35, 36). Proteins were considered candidate Cks1-interacting proteins only if they were identified in Cks1 affinity purifications and not in control purifications from untagged wild-type yeast strains.
ChIP.
Chromatin immunoprecipitation (ChIP) samples were prepared as previously described (37). Primer locations and sequences for GAL1 have been described previously (26).
RNA purification and RT-PCR.
Total RNA was purified using the RNeasy kit (Qiagen) per the manufacturer's instructions. Real-time PCR (RT-PCR) operations and primer sequences were described previously (26). Data analysis was performed using MJ Opticon Monitor analysis software, version 3.0.
Protein purification.
Escherichia coli BL21 cells expressing GST or GST fusion proteins were grown to an OD of 0.5 at 600 nm. The induction conditions were 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 12 h at 27°C. Cells were centrifuged and washed once with phosphate-buffered saline (PBS). They were then resuspended in 1/20 culture volume of buffer containing 50 mM Tris-HCl (pH 7.8), 500 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and then lysed by sonication on ice. Lysates were precleared at 35,000 × g for 15 min and then incubated with glutathione-Sepharose beads (Pharmacia Biotech) at 4°C. The GST fusion protein-bound beads were washed five times with 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM DTT, and 0.5 mM PMSF. For 6×His-tagged proteins, cells were lysed in lysis buffer (PBS, 1% Triton X-100, 5 mM imidizole [pH 8.0], 0.1% PMSF), followed by sonication on ice. Lysates were precleared at 35,000 × g for 15 min and then incubated with nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) at 4°C and washed according to the manufacturer's instructions. All binding assays were performed at 4°C for 4 h. Samples were resolved by 4 to 20% SDS-PAGE. Western blots were developed using antibodies as indicated below.
Reproducibility and significance.
All experiments were carried out at least two times. Error bars, where shown, correspond to standard deviations (SD). P values were determined using Student's t test.
RESULTS
Cks1 interacts with the Paf1 complex.
To identify Cks1-interacting proteins, immunoprecipitates of Cks1 were subjected to high-resolution mass spectrometric analysis (multidimensional protein identification technology [MuDPIT]) (35, 36). Specifically, proteins bound to Flag-Cks1 immunoprecipitated using anti-Flag resin were compared to proteins bound to the resin when incubated with extract from a nontagged strain. Among Flag-Cks1-specific interactions detected (Table 2), components of the Paf1 transcriptional elongation complex were of particular interest from a transcriptional perspective. To confirm the interaction, Cks1 was tagged with the Flag epitope and one component of the Paf1 complex, Rtf1, was tagged with the HA (Fig. 1A) or Myc (Fig. 1B) epitope. To maintain endogenous expression levels, both proteins were tagged at their carboxyl termini by insertion of sequences into the respective chromosomal loci. Immunoprecipitation of Cks1 and Rtf1 was performed, and in each case, the heterologous protein was detected by Western blotting, revealing an interaction and thus confirming the proteomic analysis. In addition, Cdc28 (Cdk1), a binding partner of Cks1, was detected in all immune complexes. Note that whereas Cks1 immune complexes were highly enriched for Rtf1, a relatively smaller proportion of Rtf1 immune complexes contained Cks1 and Cdc28. This suggests that a significant fraction of the Cks1 pool in the cell is associated with Rtf1, whereas a much smaller fraction of the Rtf1 pool is bound to Cks1. It has been reported that Rtf1 binds nonspecifically to anti-Flag resin (38, 39), possibly producing an artifactual result for the Flag-Cks1 pulldown component of the experiment. However, when extract containing HA-Rtf1 but not Flag-Cks1 was incubated with anti-Flag resin, HA-Rtf1 was not detected in the eluate, whereas it was detected in parallel anti-HA immunoprecipitates (see Fig. S1 in the supplemental material). Therefore, under the experimental conditions employed in the current study, Rtf1 does not bind to anti-Flag resin at a level that is detectable.
Table 2.
Cks1-interacting proteins
| Protein | Function | No. of peptides | No. of spectra | % sequence coverage |
|---|---|---|---|---|
| Cdc28 | CDK | 7 | 11 | 20.8 |
| Clb1 | B-type cyclin | 4 | 5 | 12.7 |
| Clb2 | B-type cyclin | 3 | 5 | 10.8 |
| Clb3 | B-type cyclin | 4 | 4 | 15.9 |
| Clb4 | B-type cyclin | 1 | 1 | 2.4 |
| Clb5 | B-type cyclin | 5 | 8 | 12.4 |
| Cln1 | G1 cyclin | 1 | 1 | 1.5 |
| Cln2 | G1 cyclin | 2 | 2 | 5.5 |
| Sic1 | CDK inhibitor | 10 | 16 | 27.5 |
| Hhf1,2 | Histone H4 | 3 | 3 | 34 |
| Hht1,2 | Histone H3 | 1 | 2 | 23.5 |
| Paf1 | Paf1 complex | 3 | 3 | 9.9 |
| Rtf1 | Paf1 complex | 2 | 2 | 4.7 |
| Cdc73 | Paf1 complex | 2 | 5 | 7.1 |
| Ctr9 | Paf1 complex | 6 | 6 | 9.6 |
Fig 1.
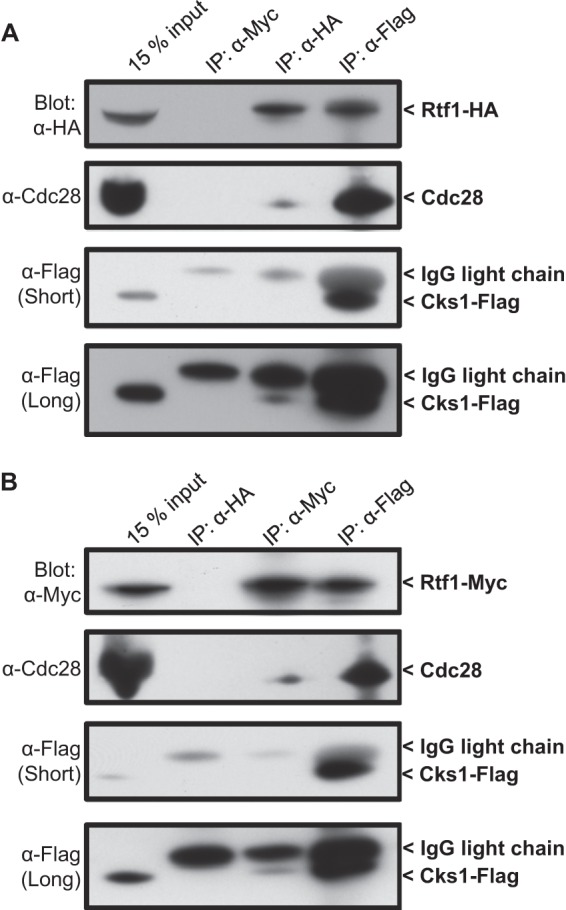
Rtf1 binds Cks1/Cdc28 in vivo. (A) Cells harboring Flag-tagged Cks1 and HA-tagged Rtf1 were lysed and immunoprecipitated (IP) using anti-Myc (as a negative control), anti-HA, or anti-Flag antibodies. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting using the indicated antibodies. The middle image shows a short exposure, and the bottom blot shows a long exposure. (B) Cells harboring Flag-tagged Cks1 and Myc-tagged Rtf1 were lysed and immunoprecipitated using anti-HA (as a negative control), anti-Myc, or anti-Flag antibodies. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting using the indicated antibodies. The middle blot shows a short exposure, and the bottom blot shows a long exposure. Note that Cks1-Flag runs immediately below the IgG light chain in immunoprecipitates.
GAL1 induction is defective in an rtf1 deletion mutant.
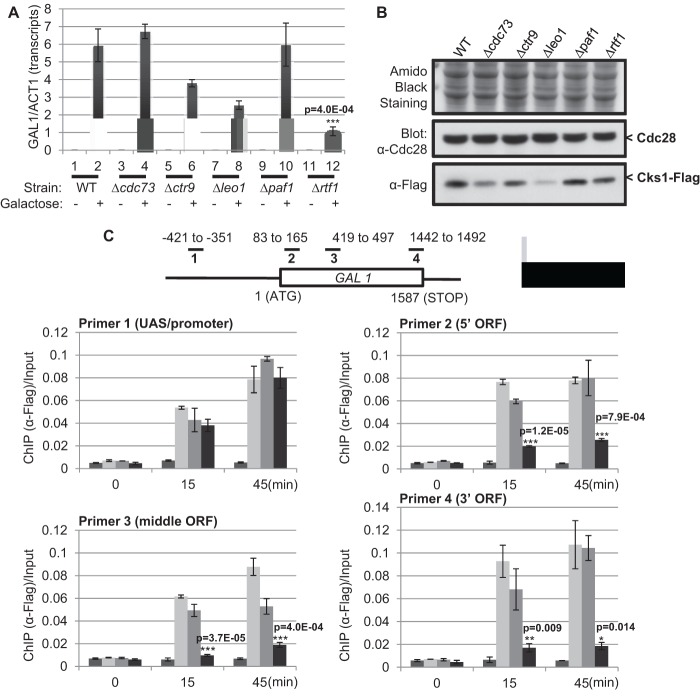
We have previously shown that Cks1 regulates transcription of GAL1 (25). Because of the physical interaction between Cks1 and Rtf1, we hypothesized that Cks1 is involved in GAL1 transcription through interaction with the Paf1 complex. Thus, mutants with deletions of each component in the Paf1 complex were generated and induction of GAL1 was measured 45 min after addition of galactose by quantitative real-time PCR analysis. Previously, it has been shown that deletion of some genes encoding Paf1 complex subunits confers defective galactose-dependent transcription at some loci (40, 41). However, a complete analysis of all component genes has not been reported, and strain background differences appear to affect Paf1 complex mutant phenotypes (our unpublished observations), justifying this partial repetition of previous work. As shown in Fig. 2A, the induction of GAL1 is attenuated in several of the mutants, but most severely in the rtf1 deletion mutant. To exclude the trivial possibility that the defect in GAL1 transcription in the Paf1 complex mutants is due to altered expression of Cks1, endogenous Cks1 was Flag tagged in the various deletion mutant strains and Western blotting was carried out to compare the levels of Cks1 protein expression. Whereas the leo1 and cdc73 mutants indeed showed reduced expression of Cks1, there was no detectable alteration of Cks1 expression in the rtf1 mutant (Fig. 2B). Therefore, the defect in GAL1 induction in the rtf1 mutant is not due to the impairment of Cks1 expression but most likely is attributable to a direct contribution of Rtf1 to GAL1 transcription.
Fig 2.
Rtf1 is required for effective recruitment of Cks1 to the GAL1 ORF. (A) Each component of the Paf1 complex was deleted separately. Wild-type (WT) and mutant cells were cultured in raffinose (−) for 2 h, and 2% galactose (+) was added for 45 min. GAL1 transcripts were quantified and normalized to actin (ACT1) mRNA. (B) Expression level of Flag-tagged Cks1 in WT and various Paf1 complex deletion mutant strains. Samples were analyzed by SDS-PAGE and Western blotting. Amido black staining (top) and Cdc28 (middle) served as loading controls. (C) Recruitment of Flag-tagged Cks1 to the GAL1 ORF in the absence of Paf1 or Rtf1. Upon galactose induction for 15 or 45 min, chromatin immunoprecipitation using anti-Flag antibodies was performed in WT, Flag-tagged Cks1, and paf1 and rtf1 deletion mutant strains. Cks1-associated chromatin fragments were isolated, amplified using the indicated primers, and normalized to the amount of input DNA prior to immunoprecipitation. All error bars represent SD. Statistical analysis was carried out comparing WT and Δrtf1 in panels A and C. P values were determined using Student's t test.
Rtf1 is required for recruitment of Cks1 onto the GAL1 ORF.
Because failure to express Cks1 was ruled out as the mechanism accounting for the deficiency of GAL1 induction in the rtf1 mutant, we investigated whether recruitment of Cks1 to the promoter or the open reading frame (ORF) regions of the GAL1 gene was defective. Note that recruitment of Cks1 to the GAL1 gene has been shown to be essential for efficient transcriptional induction (25). Therefore, ChIP was carried out using anti-Flag antibodies to determine the occupancy of Flag-Cks1 on GAL1 chromatin during a 45-min time course. Four primer sets designed to amplify the upstream activating sequence (UAS)/promoter, 5′ ORF region, middle ORF region, or 3′ ORF region of the GAL1 gene are shown schematically in Fig. 2C (upper portion). In contrast to wild-type cells and the paf1 deletion mutant, the rtf1 mutant showed highly compromised Cks1 binding along the entire GAL1 ORF (Fig. 2C). Thus, a failure to efficiently recruit Cks1 to the GAL1 ORF could account for the defect in GAL1 induction observed in the rtf1 mutant.
The failure to accumulate GAL1 mRNA observed in the rtf1 mutant could be due to a defect in transcription initiation or in a downstream function, such as elongation, 3′ end processing, or mRNA transport. We therefore carried out an RNA polymerase II ChIP in parallel with the Flag-Cks1 ChIP described above (see Fig. S2 in the supplemental material). Clearly, there is no defect in RNA polymerase II occupancy as a function of GAL1 induction in the rtf1 mutant, ruling out a defect in transcription initiation.
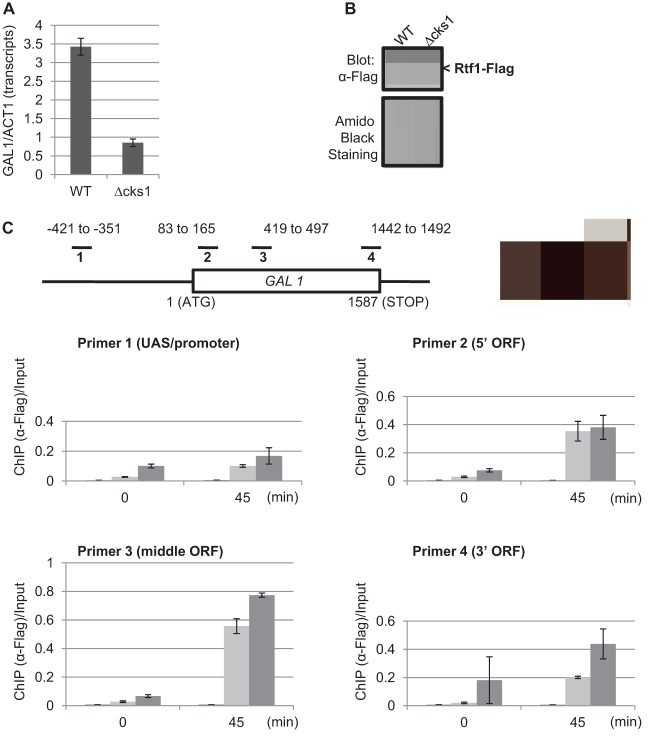
To determine if Cks1 and Rtf1 are required mutually for loading onto chromatin during transcription, ChIP analysis was conducted to compare the recruitment of Rtf1 to the GAL1 gene in the cks1 deletion mutant versus the wild-type strain. As previously shown (25), GAL1 expression is reduced in the absence of Cks1 (Fig. 3A). However, levels of Rtf1 (Fig. 3B) and binding of Rtf1 to the GAL1 gene are comparable in the csk1 mutant and wild-type strain (Fig. 3C). Taken together, these results indicate that whereas binding of Cks1 to the GAL1 ORF in the context of transcriptional induction is dependent on Rtf1, binding of Rtf1 is independent of Cks1.
Fig 3.
Cks1 is not required for the recruitment of Rtf1 to the GAL1 ORF. (A) Wild-type or cks1 deletion mutant cells were cultured in raffinose for 2 h, and 2% galactose was added for 45 min. GAL1 transcripts were quantified and normalized to actin (ACT1) mRNA. (B) Expression level of Flag-tagged Rtf1 in wild-type and cks1 deletion mutant strains. Samples were analyzed by SDS-PAGE and Western blotting. Amido black staining (bottom) was used as a loading control. (C) Recruitment of Flag-tagged Rtf1 to the GAL1 ORF in the absence of Cks1. Before and after galactose induction for 45 min, chromatin immunoprecipitation using anti-Flag antibodies was performed on wild-type untagged (control) wild-type Rtf1-Flag and cks1 Rtf1-Flag strains. Rtf1-associated chromatin fragments were isolated, amplified using the indicated primers, and normalized to the amount of input DNA prior to immunoprecipitation. Error bars represent SD.
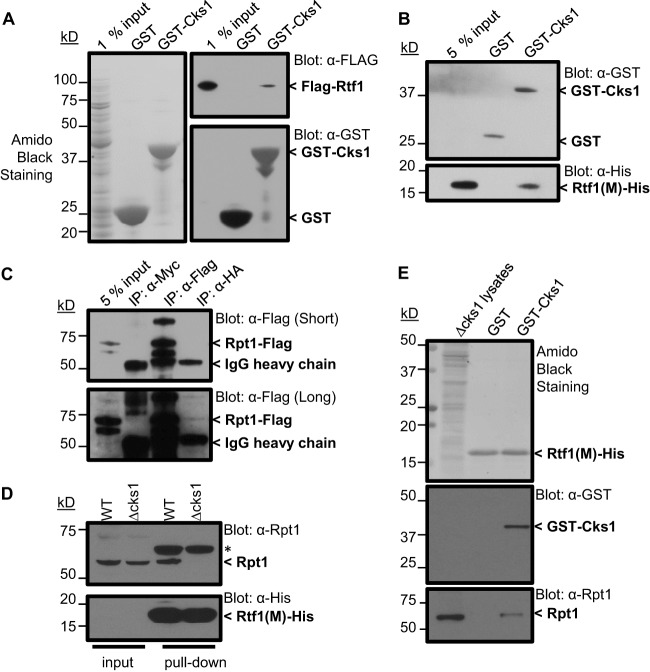
Interaction between Cks1 and Rtf is direct.
The binding detected between Cks1 and Rtf1 in Fig. 1 could be direct or mediated by other proteins. In order to distinguish between these mechanisms, Cks1 (GST tagged) and Rtf1 (Flag tagged) were expressed in E. coli. GST alone or GST-tagged Cks1 was immobilized on glutathione agarose beads and incubated with extracts containing Flag-Rtf1. The beads were washed, and eluted proteins were analyzed by SDS-PAGE and Western blotting. Rtf1 was captured on beads containing GST-Cks1 but not those containing GST (Fig. 4A), indicating a direct interaction. Through a series of domain mapping experiments analogous to the experiment described above, except that fragments of Rtf1 were tested for ability to bind Cks1 (see Fig. S3 in the supplemental material), the Cks1 binding domain of Rtf11 was identified as residing between residues 234 and 373 (Fig. 4B). In this case, the central fragment of Rtf1 was tagged with the 6×His epitope rather than the Flag epitope used to mark the full-length protein in Fig. 4A. This is a region shown genetically to be essential for a number of the transcriptional functions attributed to Rtf1 (42).
Fig 4.
Cks1 serves as an adaptor for recruiting the 19S proteasome to Rtf1. (A) Rtf1 binds directly to Cks1. GST or GST-Cks1 immobilized on glutathione resin was incubated with crude extract from E. coli expressing Flag-tagged Rtf1 (full length). Rtf1, GST, and GST-Cks1 were detected by Western blotting using the indicated antibodies. (B) A central domain of Rtf1 is essential for binding Cks1. Immobilized GST or GST-Cks1 was incubated with His-tagged Rtf1 central domain (234 to 373 amino acids) purified from E. coli extracts by Ni-NTA chromatography. Rtf1, GST, and GST-Cks1 were detected by Western blotting using the indicated antibodies. (C) Rtf1 associates with the 19S proteasome in vivo. Cells expressing Flag-tagged Rpt1 and HA-tagged Rtf1 were lysed and immunoprecipitated using anti-Myc (negative control), anti-HA, or anti-Flag antibodies. Samples were analyzed by SDS-PAGE and Western blotting. (D) The association between Rpt1 and the 19S proteasome requires Cks1. Lysates from wild-type (WT) or Δcks1 cells were incubated His-tagged Rtf1 central domain (234 to 373 amino acids) immobilized on Ni-NTA resin. Eluted proteins were analyzed by SDS-PAGE and Western blotting for the indicated proteins. (E) Cks1 is necessary and sufficient for binding of the 19S proteasome to Rtf1. Lysates from Δcks1 cells were incubated with His-tagged Rtf1 central domain (234 to 373 amino acids) immobilized on Ni-NTA resin, which was then incubated with GST or GST-Cks1. Bound Rpt1 was detected by SDS-PAGE, followed by Western blotting. Approximate molecular masses are indicated to the left of each blot.
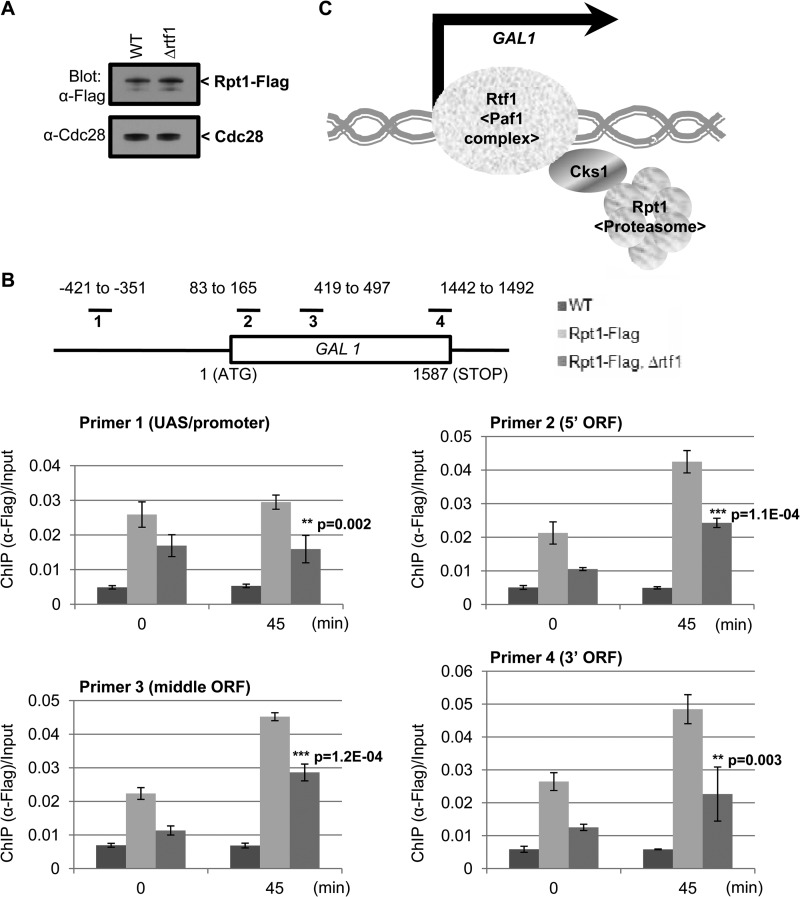
Cks1 is essential for recruiting the 19S proteasome particle to Rtf1.
Since Rtf1 is required for efficient recruitment of Cks1 to GAL1 (Fig. 2C and 3C) and Cks1 is required for recruitment of the 19S proteasome particle (25), we speculated that Cks1 might serve as an adaptor to allow binding the 19S proteasome particle to Rtf1. To test this idea, we first carried out an immunoprecipitation experiment to determine whether Rtf1 and the 19S proteasome interact in vivo. Rtf1 was tagged using the HA epitope, and Rpt1, an ATPase component of the 19S proteasome base, was tagged using the Flag epitope. When lysates were immunoprecipitated using anti-Myc (negative control), anti-Flag, and anti-HA antibodies (Fig. 4C) a strong band at the position of Rpt1 was detected in the lane corresponding to the anti-Flag immunoprecipitate, as expected (upper and lower portions). In the lane corresponding to the HA-Rtf1 pulldown, a weak band at the mobility corresponding to Rpt1 is detected over background. This minimal signal is not surprising, since only a very small fraction of 19S proteasome particles in the cell is likely to be involved in transcription rather than proteolysis, the primary function of proteasomes. It was not possible to determine whether Rtf1 was coimmunoprecipitated with Flag-Rpt1 because of the high background contributed by anti-Flag IgG at the relevant position on the blot. To examine whether Cks1 is essential for the interaction between Rtf1 and the 19S proteasome, we devised a modified pulldown experiment. The Cks1-interacting region of Rtf1, residues 234 to 373, containing a 6×His tag produced in E. coli, was immobilized on Ni-NTA beads, which were then incubated with wild-type yeast extract or extract from a cks1Δ strain. When proteins were eluted and subjected to SDS-PAGE and Western blotting, Rpt1 from wild-type but not cks1Δ extract was captured on the Rtf1 beads (Fig. 4D). Therefore, the 19S proteasome particle cannot bind directly to Rtf1 but can bind in the presence of Cks1. To confirm that Cks1 is necessary and sufficient to mediate this interaction, we performed an in vitro reconstitution experiment (Fig. 4E). Again, the Cks1-interacting region of Rtf1 was preloaded onto Ni-NTA beads, after which the beads were incubated with GST alone or GST-tagged Cks1. Lysates from a cks1Δ strain were then incubated with the beads supplemented with GST-Cks1 or GST alone. Capture of 19S proteasome particles was again determined by Western blotting using anti-Rpt1 antibody. 19S particles were only captured by beads that had been preincubated with GST-Cks1 (Fig. 4E), confirming that Cks1 can serve as a mediator for recruiting 19S particles to Rtf1. If this relationship between Rtf1, Cks1, and the 19S proteasome particle exists in vivo, then Rtf1 should contribute to recruitment of the 19S proteasome to chromatin. We therefore used ChIP analysis to compare recruitment of the 19S particle to the GAL1 ORF in wild-type versus rtf1 deletion strains. There was a significant decrease in Rpt1 binding to the GAL1 ORF in the rtf1 mutant after 45 min of induction (Fig. 5B), similar to the reduction observed for Cks1 binding (Fig. 2C). Deletion of rtf1 did not affect the level of Rpt1 (Fig. 5A), thereby excluding changes in Rpt1 expression as an explanation for reduced recruitment. These data are consistent with Cks1 serving as an adaptor that facilitates Rtf1-mediated recruitment of the 19S proteasome particle to actively transcribed chromatin in order to carry out transcriptional functions (Fig. 5C).
Fig 5.
Rtf1 is required for efficient recruitment of the 19S proteasome to the GAL1 ORF. (A) Expression level of Flag-tagged Rpt1 in wild-type (WT) and rtf1 deletion mutant strains. Samples were analyzed by SDS-PAGE and Western blotting. Cdc28 (Cdk1) was used as a loading control. (B) Recruitment of Flag-tagged Rpt1 to the GAL1 ORF in the absence of Rtf1. Before and after galactose induction for 45 min, chromatin immunoprecipitation using anti-Flag antibodies was performed in wild-type untagged protein (control), wild-type Rpt1-Flag, and rtf1 Rpt1-Flag. Rpt1-associated chromatin fragments were isolated, amplified using the indicated primers, and normalized to the amount of input DNA prior to immunoprecipitation. All error bars represent SD. Statistical analysis was carried out comparing WT and Δrtf1. P values determined using Student's t test. (C) Model for Cks1-mediated transcription. We propose that Cks1 functions as an adaptor for recruiting the 19S proteasome to Rtf1 in order to promote efficient transcription elongation.
DISCUSSION
Transcriptional elongation is a highly regulated process involving both positive and negative factors (43). In this context, the Paf1 complex serves as a multifunctional platform for recruiting factors that alter chromatin dynamics (28). Nevertheless, the molecular mechanisms downstream of the Paf1 complex have not been completely elucidated. Our proteomic analysis of Cks1-interacting proteins identified 4 out of 5 components of the Paf1 complex (Table 2), implying a potential role for Cks1 in transcriptional functions of the Paf1 complex. In the current study, we demonstrated that Cks1 serves as an adaptor that allows Rtf1, a component of the Paf1 complex, to recruit the 19S proteasome particle to target genes, necessary for efficient transcriptional elongation and transcript processing (22, 23). Although we have not directly determined the specific role of Rtf1 in this context at the GAL1 locus in our strain background, the fact that GAL1 message accumulation was impaired in the rtf1Δ mutant, but occupancy of RNA polymerase II on the GAL1 ORF was not, argues against a role in transcriptional activation or elongation and favors a role in mRNA processing. On the other hand, the facts that both Cks1 and the 19S proteasome particle have a role in nucleosome eviction (26) and that Rtf1 is required for efficient Cks1 and 19S proteasome particle recruitment are consistent with a role in transcriptional elongation. More direct experiments will be required to distinguish between these functions.
Rtf1-dependent recruitment of Cks1 and the 19S proteasome to the GAL1 gene during transcriptional induction.
The Paf1 complex was identified based on its physical association with RNA polymerase II (44). Using the GAL1 gene as a model system, it was previously shown that deletion of genes encoding Ctr9 or Paf1, components of the Paf1 complex, led to reduced expression by impairing nucleosome eviction as well as decreasing association of RNA Pol II (40). Yet the molecular mechanism whereby the Paf1 complex promotes nucleosome eviction has remained poorly understood. We have previously shown that Cks1 and the 19S proteasome particle are required for induction-dependent nucleosome eviction at the GAL1 gene (26). In the present study, we demonstrate that Cks1 promotes Rtf1-dependent recruitment of the 19S proteasome particle to the GAL1 gene, possibly providing an explanation for Paf1 complex-mediated nucleosome eviction. Interestingly, the recruitment of Cks1 was affected within the ORF region but not the UAS region of the GAL1 gene in the rtf1 deletion mutant (Fig. 2C). However, 19S proteasome particle loading was defective in both the UAS and ORF regions (Fig. 5B). This suggests that different mechanisms of proteasome recruitment might be operative with the UAS versus ORF regions. Indeed, it has been shown that recruitment of the 19S proteasome particle to the UAS region of the GAL1 gene for transcriptional activation functions is dependent on the transcription factor Gal4 and the chromatin-remodeling complex SAGA (16). Therefore, it appears that Cks1 is only important for recruitment of the 19S proteasome particle for elongation functions of Rtf1. This is also consistent with previous reports suggesting that the 19S proteasome particle has distinct functions in transactivation and elongation (22, 23).
General applicability of the mechanisms proposed in this study.
Cks1 has been shown to regulate expression of two inducible genes, CDC20 (3) and PHO5 (26). Other inducible cell cycle regulated genes (45, 46) and PHO5 (47) have also been reported to be regulated by the Paf1 complex. These data suggest that Rtf1-Cks1-19S proteasome particle axis comes into play particularly when rapid gene induction requires energy for nucleosome eviction. Presumably the ATPases of the 19S particle are mobilized for this function. Interestingly, Rtf1 seemed to be the most critical member of the Paf1 complex in GAL1 expression in our genetic background (Fig. 2A). This is consistent with a recent study in which a fragment of Rtf1 could support H2B ubiquitylation even in the absence of other Paf1 complex members (48). In addition, Rtf1 is essential for the association of the Paf1 complex with chromatin and RNA Pol II in yeast (49). Therefore, Rtf1 appears to be central to the Paf1 complex and in some cases can carry out functions independently. Indeed, we observed no defect in GAL1 induction in the paf1 deletion mutant but a strong defect in the rtf1 deletion mutant. It should be pointed out, though, that in another study, the paf1 mutant was defective in GAL1 induction (40), most likely attributable to differences in genetic background.
Possible parallels between Cks protein transcriptional functions in yeast and mammalian cells and possible roles in cancer.
We discovered novel physical and functional interactions between Cks1 and a component of the Paf1 complex in yeast. Several observations suggest that a similar functional relationship may exist in mammals. First, Cks1 (50) and the Paf1 complex (28) share significant structural similarity with their counterparts in mammals. Second, Cks paralogs, Cks1 and Cks2, are required for efficient expression of CDK1, CCNA2, and CCNB1 (encoding Cdk1, cyclin A2, and cyclin B1) (51, 52). A similar relationship exists between the Paf1 complex and expression of the same three genes (53). Moreover, in some ductal breast carcinomas, the expression of Cdk1, cyclin B1, and cyclin A2, as well as Cks proteins, is coordinately upregulated (54). However, although Cks protein overexpression is a characteristic of many forms of cancer (55–73) and Cks1 deletion has protective effects in some mouse cancer models and cell line studies (74–76), it is not clear that Cks-mediated oncogenicity is related to transcriptional functions. As Cks1 is a component of the SCFSkp2 ubiquitin ligase, it has been suggested that oncogenic functions of Cks1 might be mediated by affecting the stability of the Cdk inhibitor p27Kip1, an SCFSkp2 target (77). Yet disruption of Skp2 has only a modest effect on Myc-mediated lymphomagenesis in a mouse model, whereas deletion of Cks1 significantly attenuates the disease (78). On the other hand, it is more likely that oncogenic functions of Cks proteins are linked to override of cell cycle checkpoints and oncogene-induced stress barriers associated with Cks protein overexpression (76). A thorough understanding of the Cks1-proteasome-Paf1 interaction in humans, should it exist, will allow the determination of whether Cks protein-mediated transcription functions contribute to oncogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Scripps Cell Cycle Group for discussion.
This research was supported by NIH grants GM038328 and CA074224.
Footnotes
Published ahead of print 3 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00151-13.
REFERENCES
- 1.Hadwiger JA, Wittenberg C, Mendenhall MD, Reed SI. 1989. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol. Cell. Biol. 9:2034–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayles J, Beach D, Durkacz B, Nurse P. 1986. The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol. Gen. Genet. 202:291–293 [DOI] [PubMed] [Google Scholar]
- 3.Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. 2003. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423:1009–1013 [DOI] [PubMed] [Google Scholar]
- 4.Kaiser P, Moncollin V, Clarke DJ, Watson MH, Bertolaet BL, Reed SI, Bailly E. 1999. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 13:1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer RT, Baker TA. 2011. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80:587–612 [DOI] [PubMed] [Google Scholar]
- 6.Lonard DM, O'Malley BW. 2009. Emerging roles of the ubiquitin proteasome system in nuclear hormone receptor signaling. Prog. Mol. Biol. Transl. Sci. 87:117–135 [DOI] [PubMed] [Google Scholar]
- 7.Wilson MD, Harreman M, Svejstrup JQ. 2013. Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim. Biophys. Acta 1829:151–157 [DOI] [PubMed] [Google Scholar]
- 8.Bhat KP, Greer SF. 2011. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. Biophys. Acta 1809:150–155 [DOI] [PubMed] [Google Scholar]
- 9.Geng F, Wenzel S, Tansey WP. 2012. Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 81:177–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296:548–550 [DOI] [PubMed] [Google Scholar]
- 11.Geng F, Tansey WP. 2012. Similar temporal and spatial recruitment of native 19S and 20S proteasome subunits to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U. S. A. 109:6060–6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archer CT, Delahodde A, Gonzalez F, Johnston SA, Kodadek T. 2008. Activation domain-dependent monoubiquitylation of Gal4 protein is essential for promoter binding in vivo. J. Biol. Chem. 283:12614–12623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostendorff HP, Peirano RI, Peters MA, Schluter A, Bossenz M, Scheffner M, Bach I. 2002. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature 416:99–103 [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Sun L, Liang J, Yu W, Zhang Y, Wang Y, Chen Y, Li R, Sun X, Shang Y. 2006. The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 25:4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123:423–436 [DOI] [PubMed] [Google Scholar]
- 16.Malik S, Shukla A, Sen P, Bhaumik SR. 2009. The 19 S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 284:35714–35724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezhkova E, Tansey WP. 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell 13:435–442 [DOI] [PubMed] [Google Scholar]
- 18.Koues OI, Dudley RK, Mehta NT, Greer SF. 2009. The 19S proteasome positively regulates histone methylation at cytokine inducible genes. Biochim. Biophys. Acta 1789:691–701 [DOI] [PubMed] [Google Scholar]
- 19.Koues OI, Dudley RK, Truax AD, Gerhardt D, Bhat KP, McNeal S, Greer SF. 2008. Regulation of acetylation at the major histocompatibility complex class II proximal promoter by the 19S proteasomal ATPase Sug1. Mol. Cell. Biol. 28:5837–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koues OI, Mehta NT, Truax AD, Dudley RK, Brooks JK, Greer SF. 2010. Roles for common MLL/COMPASS subunits and the 19S proteasome in regulating CIITA pIV and MHC class II gene expression and promoter methylation. Epigenetics Chromatin 3:5. 10.1186/1756-8935-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truax AD, Koues OI, Mentel MK, Greer SF. 2010. The 19S ATPase S6a (S6′/TBP1) regulates the transcription initiation of class II transactivator. J. Mol. Biol. 395:254–269 [DOI] [PubMed] [Google Scholar]
- 22.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7:981–991 [DOI] [PubMed] [Google Scholar]
- 23.Ferdous A, Kodadek T, Johnston SA. 2002. A nonproteolytic function of the 19S regulatory subunit of the 26S proteasome is required for efficient activated transcription by human RNA polymerase II. Biochemistry 41:12798–12805 [DOI] [PubMed] [Google Scholar]
- 24.Ottosen S, Herrera FJ, Triezenberg SJ. 2002. Transcription. Proteasome parts at gene promoters. Science 296:479–481 [DOI] [PubMed] [Google Scholar]
- 25.Yu VP, Baskerville C, Grunenfelder B, Reed SI. 2005. A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol. Cell 17:145–151 [DOI] [PubMed] [Google Scholar]
- 26.Chaves S, Baskerville C, Yu V, Reed SI. 2010. Cks1, Cdk1, and the 19S proteasome collaborate to regulate gene induction-dependent nucleosome eviction in yeast. Mol. Cell. Biol. 30:5284–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaehning JA. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799:379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomson BN, Arndt KM. 2013. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta 1829:116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng HH, Dole S, Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625–33628 [DOI] [PubMed] [Google Scholar]
- 30.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739–34742 [DOI] [PubMed] [Google Scholar]
- 31.Reed SI, Hadwiger JA, Lorincz AT. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. U. S. A. 82:4055–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 33.de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J, III, Russell P, Wittenberg C. 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23:483–496 [DOI] [PubMed] [Google Scholar]
- 34.Mendenhall MD, Jones CA, Reed SI. 1987. Dual regulation of the yeast CDC28-p40 protein kinase complex: cell cycle, pheromone, and nutrient limitation effects. Cell 50:927–935 [DOI] [PubMed] [Google Scholar]
- 35.Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. 2008. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 22:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. 2009. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bruin RA, Kalashnikova TI, Aslanian A, Wohlschlegel J, Chahwan C, Yates JR, III, Russell P, Wittenberg C. 2008. DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc. Natl. Acad. Sci. U. S. A. 105:11230–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ezhkova E, Tansey WP. 2006. Chromatin immunoprecipitation to study protein-DNA interactions in budding yeast. Methods Mol. Biol. 313:225–244 [DOI] [PubMed] [Google Scholar]
- 39.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183 [DOI] [PubMed] [Google Scholar]
- 40.Marton HA, Desiderio S. 2008. The Paf1 complex promotes displacement of histones upon rapid induction of transcription by RNA polymerase II. BMC Mol. Biol. 9:4. 10.1186/1471-2199-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan CD, Holland MJ, Winston F. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280:913–922 [DOI] [PubMed] [Google Scholar]
- 42.Warner MH, Roinick KL, Arndt KM. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27:6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu. Rev. Biochem. 81:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16:669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268:272–285 [DOI] [PubMed] [Google Scholar]
- 46.Porter SE, Washburn TM, Chang M, Jaehning JA. 2002. The yeast pafl-rNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot. Cell 1:830–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvin CD, Kladde MP. 2004. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J. Biol. Chem. 279:33057–33062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM. 2012. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc. Natl. Acad. Sci. U. S. A. 109:10837–10842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447–456 [DOI] [PubMed] [Google Scholar]
- 50.Richardson HE, Stueland CS, Thomas J, Russell P, Reed SI. 1990. Human cDNAs encoding homologs of the small p34Cdc28/Cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 4:1332–1344 [DOI] [PubMed] [Google Scholar]
- 51.Martinsson-Ahlzén HS, Liberal V, Grunenfelder B, Chaves SR, Spruck CH, Reed SI. 2008. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol. Cell. Biol. 28:5698–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westbrook L, Manuvakhova M, Kern FG, Estes NR, II, Ramanathan HN, Thottassery JV. 2007. Cks1 regulates cdk1 expression: a novel role during mitotic entry in breast cancer cells. Cancer Res. 67:11393–11401 [DOI] [PubMed] [Google Scholar]
- 53.Moniaux N, Nemos C, Deb S, Zhu B, Dornreiter I, Hollingsworth MA, Batra SK. 2009. The human RNA polymerase II-associated factor 1 (hPaf1): a new regulator of cell-cycle progression. PLoS One 4:e7077. 10.1371/journal.pone.0007077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. 2006. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9:121–132 [DOI] [PubMed] [Google Scholar]
- 55.Chow LS, Lam CW, Chan SY, Tsao SW, To KF, Tong SF, Hung WK, Dammann R, Huang DP, Lo KW. 2006. Identification of RASSF1A modulated genes in nasopharyngeal carcinoma. Oncogene 25:310–316 [DOI] [PubMed] [Google Scholar]
- 56.de Vos S, Krug U, Hofmann WK, Pinkus GS, Swerdlow SH, Wachsman W, Grogan TM, Said JW, Koeffler HP. 2003. Cell cycle alterations in the blastoid variant of mantle cell lymphoma (MCL-BV) as detected by gene expression profiling of mantle cell lymphoma (MCL) and MCL-BV. Diagn. Mol. Pathol. 12:35–43 [DOI] [PubMed] [Google Scholar]
- 57.de Wit NJ, Rijntjes J, Diepstra JH, van Kuppevelt TH, Weidle UH, Ruiter DJ, van Muijen GN. 2005. Analysis of differential gene expression in human melanocytic tumour lesions by custom made oligonucleotide arrays. Br. J. Cancer 92:2249–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inui N, Kitagawa K, Miwa S, Hattori T, Chida K, Nakamura H, Kitagawa M. 2003. High expression of Cks1 in human non-small cell lung carcinomas. Biochem. Biophys. Res. Commun. 303:978–984 [DOI] [PubMed] [Google Scholar]
- 59.Kawakami K, Enokida H, Tachiwada T, Gotanda T, Tsuneyoshi K, Kubo H, Nishiyama K, Takiguchi M, Nakagawa M, Seki N. 2006. Identification of differentially expressed genes in human bladder cancer through genome-wide gene expression profiling. Oncol. Rep. 16:521–531 [PubMed] [Google Scholar]
- 60.Kitajima S, Kudo Y, Ogawa I, Bashir T, Kitagawa M, Miyauchi M, Pagano M, Takata T. 2004. Role of Cks1 overexpression in oral squamous cell carcinomas: cooperation with Skp2 in promoting p27 degradation. Am. J. Pathol. 165:2147–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M, Lin YM, Hasegawa S, Shimokawa T, Murata K, Kameyama M, Ishikawa O, Katagiri T, Tsunoda T, Nakamura Y, Furukawa Y. 2004. Genes associated with liver metastasis of colon cancer, identified by genome-wide cDNA microarray. Int. J. Oncol. 24:305–312 [PubMed] [Google Scholar]
- 62.Masuda TA, Inoue H, Nishida K, Sonoda H, Yoshikawa Y, Kakeji Y, Utsunomiya T, Mori M. 2003. Cyclin-dependent kinase 1 gene expression is associated with poor prognosis in gastric carcinoma. Clin. Cancer Res. 9:5693–5698 [PubMed] [Google Scholar]
- 63.Ouellet V, Guyot MC, Le Page C, Filali-Mouhim A, Lussier C, Tonin PN, Provencher DM, Mes-Masson AM. 2006. Tissue array analysis of expression microarray candidates identifies markers associated with tumor grade and outcome in serous epithelial ovarian cancer. Int. J. Cancer 119:599–607 [DOI] [PubMed] [Google Scholar]
- 64.Ouellet V, Provencher DM, Maugard CM, Le Page C, Ren F, Lussier C, Novak J, Ge B, Hudson TJ, Tonin PN, Mes-Masson AM. 2005. Discrimination between serous low malignant potential and invasive epithelial ovarian tumors using molecular profiling. Oncogene 24:4672–4687 [DOI] [PubMed] [Google Scholar]
- 65.Shapira M, Ben-Izhak O, Bishara B, Futerman B, Minkov I, Krausz MM, Pagano M, Hershko DD. 2004. Alterations in the expression of the cell cycle regulatory protein cyclin kinase subunit 1 in colorectal carcinoma. Cancer 100:1615–1621 [DOI] [PubMed] [Google Scholar]
- 66.Slotky M, Shapira M, Ben-Izhak O, Linn S, Futerman B, Tsalic M, Hershko DD. 2005. The expression of the ubiquitin ligase subunit Cks1 in human breast cancer. Breast Cancer Res. 7:R737–R744. 10.1186/bcr1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, Febbo PG, Balk SP. 2006. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 66:2815–2825 [DOI] [PubMed] [Google Scholar]
- 68.Urbanowicz-Kachnowicz I, Baghdassarian N, Nakache C, Gracia D, Mekki Y, Bryon PA, Ffrench M. 1999. ckshs expression is linked to cell proliferation in normal and malignant human lymphoid cells. Int. J. Cancer 82:98–104 [DOI] [PubMed] [Google Scholar]
- 69.Wong YF, Cheung TH, Tsao GS, Lo KW, Yim SF, Wang VW, Heung MM, Chan SC, Chan LK, Ho TW, Wong KW, Li C, Guo Y, Chung TK, Smith DI. 2006. Genome-wide gene expression profiling of cervical cancer in Hong Kong women by oligonucleotide microarray. Int. J. Cancer 118:2461–2469 [DOI] [PubMed] [Google Scholar]
- 70.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536 [DOI] [PubMed] [Google Scholar]
- 71.Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. 2005. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer 103:1336–1346 [DOI] [PubMed] [Google Scholar]
- 72.Kawakami K, Enokida H, Tachiwada T, Nishiyama K, Seki N, Nakagawa M. 2007. Increased SKP2 and CKS1 gene expression contributes to the progression of human urothelial carcinoma. J. Urol. 178:301–307 [DOI] [PubMed] [Google Scholar]
- 73.Lan Y, Zhang Y, Wang J, Lin C, Ittmann MM, Wang F. 2008. Aberrant expression of Cks1 and Cks2 contributes to prostate tumorigenesis by promoting proliferation and inhibiting programmed cell death. Int. J. Cancer 123:543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keller UB, Old JB, Dorsey FC, Nilsson JA, Nilsson L, MacLean KH, Chung L, Yang C, Spruck C, Boyd K, Reed SI, Cleveland JL. 2007. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. EMBO J. 26:2562–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee EK, Kim DG, Kim JS, Yoon Y. 2011. Cell-cycle regulator Cks1 promotes hepatocellular carcinoma by supporting NF-kappaB-dependent expression of interleukin-8. Cancer Res. 71:6827–6835 [DOI] [PubMed] [Google Scholar]
- 76.Liberal V, Martinsson-Ahlzen HS, Liberal J, Spruck CH, Widschwendter M, McGowan CH, Reed SI. 2012. Cyclin-dependent kinase subunit (Cks) 1 or Cks2 overexpression overrides the DNA damage response barrier triggered by activated oncoproteins. Proc. Natl. Acad. Sci. U. S. A. 109:2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harper JW. 2001. Protein destruction: adapting roles for Cks proteins. Curr. Biol. 11:R431–R435. 10.1016/S0960-9822(01)00253-6 [DOI] [PubMed] [Google Scholar]
- 78.Old JB, Kratzat S, Hoellein A, Graf S, Nilsson JA, Nilsson L, Nakayama KI, Peschel C, Cleveland JL, Keller UB. 2010. Skp2 directs Myc-mediated suppression of p27Kip1 yet has modest effects on Myc-driven lymphomagenesis. Mol. Cancer Res. 8:353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.