Abstract
The pathogenicity of enterohemorrhagic Escherichia coli (EHEC) strains depends on the production of Shiga toxins that are encoded on lambdoid prophages. Effective production of these toxins requires prophage induction and subsequent phage replication. Previous reports indicated that lytic development of Shiga toxin-converting bacteriophages is inhibited in amino acid-starved bacteria. However, those studies demonstrated that inhibition of both phage-derived plasmid replication and production of progeny virions occurred during the stringent as well as the relaxed response to amino acid starvation, i.e., in the presence as well as the absence of high levels of ppGpp, an alarmone of the stringent response. Therefore, we asked whether ppGpp influences DNA replication and lytic development of Shiga toxin-converting bacteriophages. Lytic development of 5 such bacteriophages was tested in an E. coli wild-type strain and an isogenic mutant that does not produce ppGpp (ppGpp0). In the absence of ppGpp, production of progeny phages was significantly (in the range of an order of magnitude) more efficient than in wild-type cells. Such effects were observed in infected bacteria as well as after prophage induction. All tested bacteriophages formed considerably larger plaques on lawns formed by ppGpp0 bacteria than on those formed by wild-type E. coli. The efficiency of synthesis of phage DNA and relative amount of lambdoid plasmid DNA were increased in cells devoid of ppGpp relative to bacteria containing a basal level of this nucleotide. We conclude that ppGpp negatively influences the lytic development of Shiga toxin-converting bacteriophages and that phage DNA replication efficiency is limited by the stringent control alarmone.
INTRODUCTION
While the majority of Escherichia coli strains are not pathogenic, some have evolved the ability to cause human disease. One example is Shiga toxin-producing E. coli (STEC) and particularly a subset of strains classified as enterohemorrhagic E. coli (EHEC). The pathogenicity of these strains, including the most extensively studied serotype, O157:H7, depends on the production of Shiga toxins (for a review, see reference 1). Infection of humans by EHEC usually causes hemorrhagic colitis and may result in hemolytic-uremic syndrome (HUS) (2). A recent EHEC outbreak, which occurred in Germany in 2011, resulted in over 4,000 severe infections and about 50 deaths, confirming again that Shiga toxin-producing E. coli is a very dangerous pathogen (3, 4).
Genes coding for Shiga toxins (stx genes) are not of bacterial chromosome origin. These genes are inserted into genomes of lambdoid bacteriophages (viruses related to members of the λ family) that can lysogenize E. coli strains and are called Shiga toxin-converting bacteriophages (5, 6). In all STEC strains analyzed to date, the stx genes are under the control of the late phage promoter, called pR′ (7, 8). In all lambdoid phages, expression of almost all genes (except cI) is strongly inhibited in the prophage state, due to activity of the cI repressor (for reviews, see references 9 and 10). This repressor is responsible for inactivation of the major phage early promoters, pL and pR, which are necessary for expression of genes involved in lytic development. One of these genes, Q, encodes an antitermination protein that is indispensable for effective transcription from the pR′ promoter (10). The pR′-derived transcripts encode proteins required for host cell lysis, as well as structural proteins which are required for capsid formation; in Shiga toxin-converting bacteriophages, stx genes are located between pR′ and the “lysis genes” (for reviews, see references 6 and 10). Thus, in bacteria lysogenic for Shiga toxin-converting bacteriophages, expression of the stx genes is blocked, and production of Shiga toxins must be preceded by prophage induction (11, 12). Moreover, after prophage induction, effective expression of stx genes by the host cell depends on efficient phage DNA replication and resultant multiple copies of these genes (6).
Studies on conditions that might inhibit development of Shiga toxin-converting bacteriophages indicated that low rates of bacterial growth slowed down production of progeny phage significantly (13), and amino acid starvation blocked this process almost completely (14). In bacteria, depletion of amino acids causes a specific metabolic response called the stringent response (for a review, see reference 15). This response is mediated by a specific nucleotide, ppGpp, which is then produced over 100 times more efficiently than in unstarved cells (15). In E. coli, this nucleotide directly interacts with RNA polymerase and significantly modulates its transcriptional properties, so that most promoters are downregulated while some are activated. Although dramatic changes in transcription of many genes are observed during the stringent response due to binding of ppGpp to RNA polymerase (15, 16), it is important to note that ppGpp may also directly interact with other proteins (17). ppGpp can be synthesized in E. coli cells by the products of the relA or spoT genes. The RelA protein is a ribosome-bound ppGpp synthetase that is activated upon binding of uncharged tRNA to the ribosome during translation. SpoT is a bifunctional enzyme, acting not only as an alternative, ribosome-independent ppGpp synthetase but also as an efficient hydrolase responsible for rapid degradation of this nucleotide (15). Mutants with mutations in the relA gene are unable to produce ppGpp in large amounts upon amino acid starvation, and they are called “relaxed” (accordingly, the response of relA strains to amino acid starvation is called the relaxed response) due to their phenotype of unrestricted synthesis of rRNA and tRNA in the event of lack of amino acids, which leads to energetic exhaustion of the cell. relA spoT double mutants, which are devoid of both ppGpp synthetases, contain no detectable levels of this alarmone under any conditions, and thus they are designated ppGpp0 (15).
A role for ppGpp in the control of the pathogenicity of various bacteria has been demonstrated (for a review, see reference 15 and references therein). This control is based on ppGpp-mediated modulation of transcription of genes coding for factors involved in virulence. Changes in transcription from bacteriophage λ promoters during the stringent and relaxed responses were also described, and it was reported that λ DNA replication and the “lysis-versus-lysogenization” decision may be influenced by elevated ppGpp levels (18, 19; reviewed in reference 20). In fact, various effects of high ppGpp concentrations on particular λ promoters were observed, with pR, pE, and pI being examples of negatively regulated promoters, paQ being an example of a positively regulated promoter, and pL being a neutral promoter (neither repressed nor stimulated by ppGpp) (19, 21).
Inhibition of the development of Shiga toxin-converting bacteriophages in slowly growing and starved E. coli hosts was speculated to be due to inefficient phage DNA replication (14). However, similar effects were observed during amino acid starvation of both wild-type cells and relA mutants. Thus, the data were inconclusive as to the role of ppGpp in mitigating phage replication and progeny production, because impairment of protein synthesis due to depletion of amino acids might potentially block these processes irrespective of the stringent response. Note that although a basal level of ppGpp is present in unstarved relA mutants due to SpoT-mediated synthesis of this nucleotide, in amino acid-starved relA strains (i.e., during the relaxed response), such an alternative (SpoT-dependent) production of ppGpp is inefficient, and its intracellular level decreases rather than increases (22; for a review, see reference 15). Therefore, it was not possible to determine whether ppGpp is the negative regulator of Shiga toxin-converting phage development, as the observed effects could be caused by either ppGpp action or amino acid deprivation or both (14).
Replication regions of several Shiga toxin-converting bacteriophages were found to be similar to that of phage λ, and plasmids derived from these phages, analogous to λ plasmids, have been constructed (23). Nevertheless, perhaps surprisingly, some regulatory mechanisms vary between λ and Shiga toxin-converting phages (23, 24). In contrast to the case for λ plasmids, revealing the classical stringent control (i.e., strong inhibition in amino acid-starved wild-type hosts but not in relaxed mutants), replication of plasmids derived from Shiga toxin-converting bacteriophages was negatively influenced by depletion of amino acids under conditions of both stringent and relaxed responses (24). These effects resembled the influence of starvation on development of the bacteriophages; however, again no conclusion could be drawn about a potential role for ppGpp in mediating the negative regulation of DNA replication.
In the light of the above uncertainty, we aimed to investigate the effects of ppGpp deficiency on prophage induction and DNA replication of Shiga toxin-derived bacteriophages. We assumed that the use of ppGpp0 hosts might allow us to learn about effects of ppGpp on prophage induction and phage DNA replication. While wild-type E. coli strains contain a low basal level of ppGpp, even if unstarved (25, 26), the relA spoT double mutants are fully devoid of this nucleotide (22). This is true for E. coli cells growing in both minimal medium (22) and nutrient broth, like LB (19). In fact, it was reported that when cultured in LB, wild-type cells contained a lower basal level of ppGpp (4 pmol/unit of optical density at 600 nm [OD600]) than in a minimal medium (6 pmol/OD600 unit) (19). Nevertheless, both these values were considerable, in contrast to a total lack of detectable ppGpp in the relA spoT double mutant bacteria under any tested conditions (19, 22). Therefore, such hosts were employed in our experiments, in comparison to wild-type ones.
MATERIALS AND METHODS
Escherichia coli strains, bacteriophages, and plasmids.
The E. coli strains, bacteriophages, and plasmids used in this work are presented in Table 1. The relA spoT strains were checked for the ppGpp0 phenotype by assessing their growth on minimal medium and production of ppGpp (bacteria devoid of ppGpp are auxotrophic for several amino acids), as described previously (22); such tests were performed during every set of experiments. The strains were tested for growth on minimal agar medium both before experiments and after overnight growth of chosen colonies, simultaneously with the main experiment. The intention was to pick only true ppGpp0 mutants (not second-site suppressor mutants, which were considered pseudorevertants) for experiments and to ignore the results if a suppressor mutant(s) appeared during cultivation of the chosen clones. When testing bacteria taken out of the stock, between 10 and 25% of colonies appeared to be pseudorevertants, as they could grow on minimal agar plates (data not shown). On the other hand, when true ppGpp0 clones were picked for experiments, no pseudorevertants were detected in the tests performed simultaneously with experiments, so it was not necessary to discard any results.
Table 1.
Escherichia coli strains, bacteriophages, and plasmids
| Escherichia coli strain, bacteriophage, or plasmid | Genotype or description | Reference, construction |
|---|---|---|
| E. coli strains | ||
| MG1655 | F− λ− ilvG rfb-50 rph | 37 |
| ppGpp0 | As MG1655 but relA256 spoT212 | 38 |
| CF1652 | As MG1655 but relA251::kan | 22 |
| MC1061(ϕ24B) | MC1061 genetic background, bearing the ϕ24B prophage | 39 |
| SCUC34 | 933W Δstx2::catGFP lysogen | 40 |
| SCUC143 | P22 Δstx2::catGFP lysogen | 40 |
| SCUC140 | P27Δstx2::catGFP lysogen | 40 |
| SCUC161 | P32 Δstx2::catGFP lysogen | 40 |
| MG1655(ϕ24B) | MG1655 lysogenic with ϕ24B Δstx2::cat | This work, by lysogenization |
| MG1655(933W) | MG1655 lysogenic with 933W Δstx2::catGFP | This work, by lysogenization |
| MG1655 (P22) | MG1655 lysogenic with P22 Δstx2::catGFP | This work, by lysogenization |
| MG1655(P27) | MG1655 lysogenic with P27 Δstx2::catGFP | This work, by lysogenization |
| MG1655(P32) | MG1655 lysogenic with P32 Δstx2::catGFP | This work, by lysogenization |
| ppGpp0(ϕ24B) | ppGpp0 lysogenic with ϕ24B Δstx2::cat | This work, by lysogenization |
| ppGpp0(933W) | ppGpp0 lysogenic with 933W Δstx2::catGFP | This work, by lysogenization |
| ppGpp0(P22) | ppGpp0 lysogenic with P22 Δstx2::catGFP | This work, by lysogenization |
| ppGpp0(P27) | ppGpp0 lysogenic with P27 Δstx2::catGFP | This work, by lysogenization |
| ppGpp0(P32) | ppGpp0 lysogenic with P32 Δstx2::catGFP | This work, by lysogenization |
| CF1652(ϕ24B) | CF1652 lysogenic with ϕ24B Δstx2::cat | This work, by lysogenization |
| CF1652(933W) | CF1652 lysogenic with 933W Δstx2::catGFP | This work, by lysogenization |
| CF1652(P22) | CF1652 lysogenic with P22 Δstx2::catGFP | This work, by lysogenization |
| CF1652(P27) | CF1652 lysogenic with P27 Δstx2::catGFP | This work, by lysogenization |
| CF1652(P32) | CF1652 lysogenic with P32 Δstx2::catGFP | This work, by lysogenization |
| Phages | ||
| ϕ24B | ϕ24B Δstx2::cat | 39 |
| 933W | 933W Δstx2::catGFP | 40 |
| P22 | P22 Δstx2::catGFP | 40 |
| P27 | P27Δstx2::catGFP | 40 |
| P32 | P32 Δstx2::catGFP | 40 |
| Plasmids | ||
| p933Wcmr | Plasmid derived from phage 933W | 23 |
| p22cmr | Plasmid derived from phage P22 | 23 |
| p27cmr | Plasmid derived from phage P27 | 23 |
| p32cmr | Plasmid derived from phage P32 | 23 |
| pCB104 | Plasmid derived from phage λ | 41 |
To construct bacteria lysogenic with a particular bacteriophage (obtained after induction of the corresponding prophage from the source host, as described in the next subsection), the cell culture (grown in LB at 37°C with shaking at 180 rpm) was centrifuged (2,000 × g for 5 min at room temperature) and the pellet was suspended in 1/10 volume of TMC buffer (10 mM Tris-HCl, 10 mM MgSO4, 10 mM CaCl2, pH 7.2). Following incubation of such a suspension with phage lysate (multiplicity of infection [MOI] = 10) for 50 min at 37°C, the mixture was spread on LB plates containing 20 μg ml−1 chloramphenicol. The plates were incubated overnight at 37°C, and bacterial colonies were tested for the presence of prophages by PCR-mediated detection of phage-specific sequences included in chromosomal DNA. (The following primers were used in these tests: catR, 5′-GCA TTC TGC CGA CAT GGA AG; catF 5′-CAC TGG ATA TAC CAC CGT TG; gfpR, 5′-GGT CTG CTA ATT GAA CGC TTC C; and gfpF, 5′-CAT GGC CAA CAC TTG TCA CTA C. The length of the specific reaction product was 375 bp.)
Media and growth conditions.
Cells were grown in LB medium supplemented with 10 mM CaCl2, 10 mM MgSO4, and 20 μg ml−1 chloramphenicol (if necessary). The same broth, supplemented with 1.5% bacteriological agar, was used as a bottom agar for pouring plates. Top agar consisted of 1% tryptone, 0.5% NaCl, and 0.7% bacteriological agar. Cultures were grown, and phage infections were carried out, at 37°C under aerobic conditions, unless specified otherwise. Starvation for serine was induced by addition of serine hydroxamate (to final concentration of 0.5 mg ml−1) to bacterial cultures grown in LB medium.
To prepare phage lysates, cell cultures were grown overnight in shake flasks with LB broth. Following 1,000-fold dilution in fresh LB, the cultivation was continued to an A600 of 0.1. Mitomycin C was added to final concentration of 1 μg ml−1, and the incubation was continued for 8 to 10 h. The cultures were then treated with chloroform (several drops per flask) and centrifuged (2000 × g for 10 min at 4°C) to remove bacterial debris. The obtained lysates were then filtered (0.22-μm-pore-size filters; Millipore, USA).
Growth of bacterial liquid cultures was monitored by spectrophotometry, with measurement of absorbance at a wavelength of 600 nm (A600).
Phage titration.
The phage titration procedure was performed according to the standard double agar overlay method, using plastic petri dishes (90 mm; Alchem, Poland) filled with 25 ml of the bottom LB agar (1.5% agar) and 3 ml of the top agar (0.7% agar) mixed with 300 μl of the indicator cell culture (A600 = 2.5 to 3.0). Sublethal supplementation of the bottom agar with chloramphenicol (2.5 μg ml−1) was used, as described previously (27), to obtain visible plaques formed on the bacterial lawn. The plates were incubated at 37°C overnight, and the PFU were counted.
Plaque size determination.
Following overnight incubation of plates, as described in the preceding paragraph, pictures were taken using the digital scanner HP Scanjet G4050 and assigned software. Diameters of plaques were measured manually, by using Adobe Photoshop CS3 software to provide a scale. The results of each experiment were based on measurement of 150 plaques, randomly selected from 5 different agar plates.
Prophage induction.
Induction of prophages was provoked in lysogenic bacteria, growing at 37°C in LB medium at an A600 of 0.1, by addition of mitomycin C to a final concentration of 1 μg ml−1, as previously reported (13). Following induction, cultures were incubated for the indicated times, and 1-ml samples were withdrawn at indicated times. A drop of chloroform was added to each sample, and after 10 s of vigorous shaking and centrifugation in a microcentrifuge for 1 min, the phage lysate was titrated on E. coli MG1655 host cells. The relative phage titer, shown in PFU per ml, was calculated by subtracting the mean value in the control experiment (without an inducer, which allows estimation of effects of spontaneous prophage induction) from the mean value determined in the main experiment.
Efficiency of lysogenization.
To estimate the efficiency of lysogenization, the procedure used to construct lysogenic strains (see “Escherichia coli strains, bacteriophages, and plasmids” above) was used, but the following modifications were introduced: (i) different MOIs (1, 5, or 10) were used, (ii) mixtures of bacteria and phages were incubated in TMC buffer for 30 min, (iii) one half of the mixture described above was spread on LB agar plates and the second half on LB agar plates containing 20 μg ml−1 chloramphenicol, and (iv) efficiency of lysogenization was calculated as a fraction of lysogens (all PCR-tested chloramphenicol-resistant colonies were lysogens) among all bacterial cells (determined on the basis of number of colonies appearing on LB agar plates with no antibiotic).
One-step growth experiments.
Intracellular phage lytic development was investigated in one-step growth experiments in phage-infected bacteria, as described previously (28), but removal of unadsorbed virions was performed by centrifugation (2,000 × g for 5 min at 4°C) and washing the bacterial pellet with TM buffer (10 mM Tris-HCl, 10 mM MgSO4, pH 7.2) rather than by the use of specific antibodies. The number of infected bacteria was determined as follows. Culture samples were withdrawn at times between 0 and 10 min after infection, 0.1 ml of each serial dilution of such samples was mixed with 0.3 ml of an overnight culture of E. coli strain MG1655, and then 3 ml of the top agar, prewarmed to 45°C, was added, mixed, and poured onto an LB plate. Plates were incubated overnight at 37°C, and the number of plaques was determined (infected cells were “infection centers” [IC] since they were sources of viruses, which after one lytic developmental cycle could be released from host cells and after infection of neighboring cells and subsequent lytic cycles could form plaques on a bacterial lawn). Samples withdrawn at later times (10 min and later) were treated with an equal volume of chloroform and titrated to determine the number of PFU. Burst size was calculated as a ratio of phage titer to the titer of infection centers. In these experiments, burst size values lower than 1 represent unadsorbed and unwashed phages rather than effects of intracellular virus development, and thus they are not taken into consideration in real burst size analyses.
Estimation of relative phage and plasmid DNA amounts.
Bacteriophage DNA was isolated and separated as described previously (28). Plasmid DNA was isolated from bacterial cells by using GenElute Plasmid Miniprep (Sigma-Aldrich). DNA was quantified by staining with Quant-iT PicoGreen double-stranded DNA (dsDNA) (Invitrogen), according to the manufacturer's instructions. Concentrations of phage DNA (in ng ml−1) were calculated relative to λ DNA standards of known concentrations stained under exactly the same conditions as for tested DNAs. For analogous determination of plasmid DNA concentration, λ plasmid (pCB104) DNA was used as a standard.
Efficiency of transformation.
The calcium chloride method of transformation improved by Hanahan (29) was used.
Microscopic analyses.
Samples of bacterial cultures (1 ml) were withdrawn for fluorescence microscopy studies. Cell membranes were visualized by staining with the fluorescent dye Synaptored/FM4-64 (Sigma-Aldrich) at a final concentration of 5 μg ml−1 for 10 min. DNA was visualized by staining with DAPI (4,6-diamidino-2-phenylindole) at 1 μg ml−1 for 10 min. Samples were immobilized on 1-mm 1.5% agarose pads dissolved in LB medium and visualized using a Leica DMI4000B microscope fitted with a DFC365FX camera (Leica). The following Leica filter sets were used: N2.1 (for FM4-64), green fluorescent protein (GFP), and A4 (for DAPI). Images were collected and processed using LAS AF 3.1 software (Leica).
Statistical analyses.
The significance of differences between mean values of two measured parameters was assessed by the t test. Differences were considered significant when the P value was <0.05.
RESULTS
Shiga toxin-converting bacteriophages form large plaques in the absence of ppGpp.
Many Shiga toxin-converting bacteriophages form small or very small (hardly visible) plaques on wild-type E. coli hosts (27). We confirmed this for the phages used in our work that are derivatives of ϕ24B, 933W, P22, P27, and P32 (Fig. 1A). However, we found that these phages form considerably larger plaques when titrated on the E. coli ppGpp0 strain (Fig. 1A).
Fig 1.
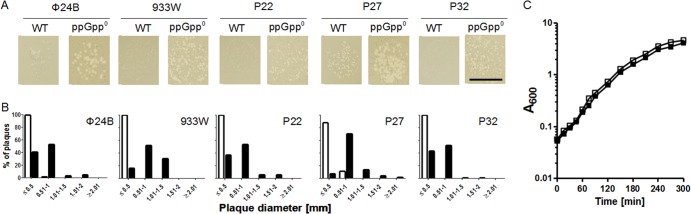
Effects of ppGpp deficiency on Shiga toxin-converting phage plaque morphology. Plaques formed by bacteriophages ϕ24B, 933W, P22, P27, and P32 on lawns of E. coli strain MG1655 (wild type [WT]) and its relA spoT derivative (ppGpp0), grown in LB, were analyzed. (A) Pictures of plaques were taken using an HP Scanjet G4050 digital scanner. The bar represents 1 cm. (B) Measured sizes of plaques on lawns of E. coli strain MG1655 (WT) (open bars) and its relA spoT derivative (ppGpp0) (closed bars). One hundred fifty randomly chosen plaques were measured for each phage titrated on each host, and the percentage of plaques included in each size range was determined. (C) Growth curves of E. coli MG1655 (WT) (open symbols) and its relA spoT derivative (ppGpp0) (closed symbols) cultured in LB medium at 37°C. Mean values from 3 experiments are shown (standard deviations [SD] were below 10%, and thus error bars would be smaller than sizes of symbols, especially in the logarithmic scale; therefore, they are not presented).
The sizes of plaques formed by the tested phages on wild-type and ppGpp0 bacteria were evaluated quantitatively. As shown in Fig. 1B, diameters of the plaques appearing on the double mutant (relA spoT) host were significantly larger than those observed on the wild-type strain. The growth rates of wild-type and ppGpp0 hosts were similar, determined as 30 and 32 min, respectively (Fig. 1C); therefore, the differences in plaque sizes could not result from any putative effects of relA and spoT alleles on the kinetics of host cell growth under the conditions employed (LB medium, 37°C). These results suggest that lytic development of Shiga toxin-converting phages could be more efficient in the absence of ppGpp than in cells bearing low, basal levels of this nucleotide. Therefore, we proceeded to study the phages' lytic development.
Lytic development of Shiga toxin-converting bacteriophages after infection or prophage induction in the presence and absence of ppGpp.
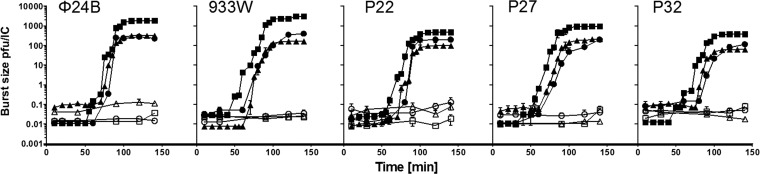
To test the effects of ppGpp on phage development, we monitored formation of progeny viruses in E. coli wild-type, relA, and ppGpp0 hosts after either infection or prophage induction, with and without amino acid starvation.
Infection of E. coli cells by lambdoid phages at a low multiplicity of infection (MOI) results in almost exclusive lytic development instead of lysogenization (for a review, see reference 29). Induction of amino acid starvation in wild-type E. coli cells provokes the stringent response, which is characterized by rapid production of ppGpp, while relA mutants, which are unable to produce ppGpp during such starvation, show the relaxed response (15). Starvation for serine (induced by addition of serine hydroxamate) resulted in a strong inhibition of development of all tested bacteriophages, irrespective of the ability of their hosts to produce ppGpp (Fig. 2, open symbols). Despite this clear result, these experiments could not allow us to determine whether ppGpp influences development of the tested phages, as inhibition of phage progeny formation might result from amino acid deprivation and deficiency in synthesis of structural proteins of virions, irrespective of ppGpp action. Therefore, we decided to test phage lytic development in unstarved cells, either producing basal levels of ppGpp (wild-type cells) or devoid of this nucleotide (ppGpp0 hosts). Under such conditions, in one-step growth experiments, we observed significantly higher burst sizes of all tested phages in the ppGpp0 host than in its wild-type counterpart (average differences at times after 100 min were 6.1-, 17.8-, 4.8-, 5.9-, and 6.1-fold for ϕ24B, 933W, P22, P27, and P32, respectively) (Fig. 2, closed symbols). Statistical analysis (t test) indicated significant differences (P < 0.05) between unstarved wild-type and ppGpp0 hosts and between relA and ppGpp0 hosts but not between wild-type and relA hosts for every phage at every time point after 100 min (but not at earlier times). For phage P22, the effects of the relA mutation might seem to be intermediate relative to those for wild-type and ppGpp0 hosts; however, the result of the statistical analysis was the same as for other phages (note that a logarithmic scale was used on the y axis, and thus visual assessment of these results must be careful). A slightly shorter latent period was also evident in the absence of ppGpp, particularly in phages 933W, P27, and P32 (Fig. 2).
Fig 2.
ppGpp influences lytic development of Shiga toxin-converting phages after infection. Lytic development of bacteriophages ϕ24B, 933W, P22, P27, and P32 after infection (at time zero) of E. coli strain MG1655 (triangles) and its relA (circles) and relA spoT (ppGpp0) (squares) derivatives, either unstarved (closed symbols) or starved for serine due to addition of serine hydroxamate to the culture up to 0.5 mg ml−1 at time zero (open symbols), was studied. Diagrams show mean results from 3 experiments, with error bars indicating SD (note that in many cases the SD were smaller than sizes of symbols). The results are presented as PFU per infection center (IC). Statistical analysis (t test) was performed for results from each time point and indicated significant differences (P < 0.05) between unstarved wild-type and ppGpp0 hosts and between relA and ppGpp0 hosts but not between wild-type and relA, hosts for every phage at every time point after 100 min (but not at earlier times).
We also assessed the effects of a lack of ppGpp on lysogenization of host cells by the employed phages (Table 2). Although in some cases (indicated in Table 2), statistically significant differences between the lysogenization efficiencies of wild-type and ppGpp0 hosts were found, under various MOI conditions these differences were often occurring in opposite directions. Therefore, we have calculated mean fold change values (cumulative for all MOI conditions in each phage) and tested the significance of differences (relative to the control) of this parameter. No statistical significance was found for any phage; therefore, we concluded that a lack of ppGpp has no drastic effects on the efficiency of lysogenization by the tested Shiga toxin-converting bacteriophages. Therefore, we used E. coli wild-type and ppGpp0 cells bearing the relevant prophages and tested lytic development of phages after prophage induction.
Table 2.
Efficiency of lysogenization of E. coli MG1655 (wild-type) and ppGpp0 hosts with Shiga toxin-converting bacteriophages
| Phage | Efficiency of lysogenizationa with the indicated host and MOI |
|||||
|---|---|---|---|---|---|---|
| Wild type (MG1655) |
ppGpp0 |
|||||
| 1 | 5 | 10 | 1 | 5 | 10 | |
| ϕ24B | 0.38 ± 0.02 | 0.47 ± 0.02 | 0.66 ± 0.02 | 0.40 ± 0.03 | 0.51 ± 0.01* | 0.65 ± 0.08 |
| 933W | 0.40 ± 0.01 | 0.56 ± 0.01 | 0.78 ± 0.08 | 0.28 ± 0.02* | 0.51 ± 0.04* | 0.59 ± 0.02* |
| P22 | 0.25 ± 0.02 | 0.27 ± 0.01 | 0.62 ± 0.02 | 0.28 ± 0.03 | 0.47 ± 0.01* | 0.60 ± 0.05 |
| P27 | 0.32 ± 0.03 | 0.48 ± 0.02 | 0.64 ± 0.04 | 0.29 ± 0.01 | 0.46 ± 0.03 | 0.58 ± 0.01* |
| P32 | 0.28 ± 0.03 | 0.52 ± 0.02 | 0.69 ± 0.03 | 0.31 ± 0.10 | 0.39 ± 0.03* | 0.66 ± 0.06 |
A value of 1 would correspond to 100% lysogenization efficiency, and a value of 0 would correspond to no lysogens found. The presented values reflect these theoretical values. The numbers are mean values from 3 experiments ± SD. Asterisks (marked only for the ppGpp0 host) indicate statistical significance of differences (P < 0.05, assessed by t test) between results obtained for wild-type and ppGpp0 hosts under the same conditions (phage and MOI). Differences in the mean fold change values (ppGpp0 host versus wild type), cumulative for all MOI conditions in each phage, did not reach statistical significance in the t test (P values for ϕ24B, 933W, P22, P27, and P32 were 0.184, 0.094, 0.300, 0.184, and 0.635, respectively).
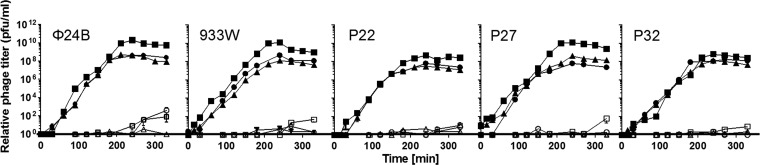
Similar to results of experiments with phage infection (Fig. 2), a significantly increased phage titer in cultures of cells devoid of ppGpp, relative to the wild-type host, was observed after mitomycin C-provoked induction of Shiga toxin-converting prophages (average differences at times after 200 min were 32.3, 66.1, 13.5, 24.4, and 6.2-fold for ϕ24B, 933W, P22, P27, and P32, respectively) (Fig. 3). Statistical analysis (t test) indicated significant differences (P < 0.05) between unstarved wild-type and ppGpp0 hosts and between relA and ppGpp0 hosts but not between wild-type and relA hosts for every phage at every time point after 200 min. Interestingly, the smallest differences between hosts strains were observed for phages P22 (like in the infection experiments) and P32. Specific reasons for such phenotypes remain to be elucidated. Amino acid starvation resulted in the dramatic impairment of formation of virions in all tested hosts (wild-type, relA, and relA spoT) (Fig. 3). However, it is not possible to distinguish whether this effect was due to ppGpp action (in strains producing this nucleotide) or to halted protein synthesis in cells devoid of amino acids.
Fig 3.
ppGpp influences lytic development of Shiga toxin-converting phages after prophage induction. Lytic development of bacteriophages ϕ24B, 933W, P22, P27, and P32, after prophage induction with mitomycin C (added up to 1 μg ml−1) at time zero of lysogenic E. coli strain MG1655 (triangles) and its relA (circles) and relA spoT (ppGpp0) (squares) derivatives, either unstarved (closed symbols) or starved for serine due to addition of serine hydroxamate to the culture up to 0.5 mg ml−1 at time zero (open symbols), was assessed. Diagrams show mean results from 3 experiments, with error bars indicating SD (note that in many cases the SD were smaller than sizes of symbols). Statistical analysis (t test) was performed for results from each time point and indicated significant differences (P < 0.05) between unstarved wild-type and ppGpp0 hosts and between relA and ppGpp0 hosts but not between wild-type and relA hosts for every phage at every time point after 200 min.
In summary, despite some differences of effects of the ppGpp0 host phenotype on burst sizes of various phages, the results presented in Fig. 2 and 3 strongly suggested that intracellular development of Shiga toxin-converting bacteriophages is negatively regulated by ppGpp, irrespective of the kind of initiation of the phage lytic developmental mode, by either infection or prophage induction.
Phage DNA replication in wild-type and ppGpp0 hosts.
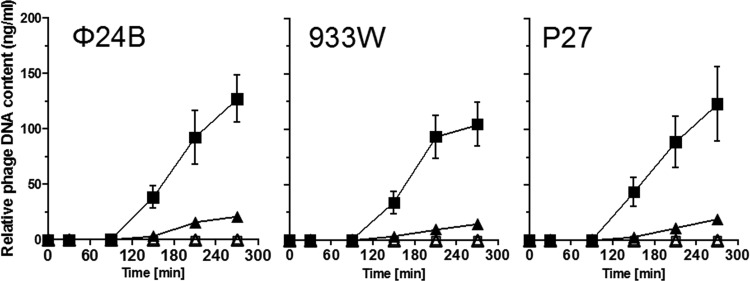
To test effects of the absence of ppGpp on DNA replication of Shiga toxin-converting bacteriophages, we estimated the levels of phage DNA in wild-type and ppGpp0 E. coli cells at different times after induction of prophages. Three phages (ϕ24B, 933W, and P27) were selected for these experiments (phages P22 and P32 were omitted since their replication regions are highly similar to that of phage P27 [23]). In all cases, a significantly higher increase in relative phage DNA amount was observed in the relA spoT double mutant than in the wild-type strain (Fig. 4). This suggests that replication of DNA of Shiga toxin-converting bacteriophages is negatively regulated by ppGpp.
Fig 4.
Effects of ppGpp on Shiga toxin-converting phage DNA amount after prophage induction. Amounts of DNA of phages ϕ24B, 933W, and P27, in E. coli strain MG1655 (triangles) and its relA spoT derivative (ppGpp0) (squares) either without prophage induction (open symbols) or after induction with mitomycin C (1 μg ml−1) at time zero (closed symbols), were measured. The results are mean values from 3 experiments, with error bars representing SD.
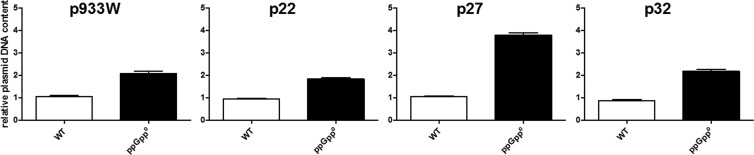
The results of experiments with phage DNA replication (Fig. 4) were corroborated by studies on plasmids derived from Shiga toxin-converting bacteriophages. Although the efficiencies of transformation of ppGpp0 and wild-type hosts by these plasmids were similar (Table 3), relative plasmid DNA levels were significantly higher in cells devoid of ppGpp than in the wild-type counterparts for all tested replicons (Fig. 5).
Table 3.
Efficiency of transformation of E. coli wild-type and ppGpp0 strains with lambdoid plasmids
| Plasmid | Efficiency of transformation (transformants per 1 μg of plasmid DNA)a |
|
|---|---|---|
| Wild type (MG1655) | ppGpp0 | |
| p933Wcmr | (1.5 ± 0.06) × 104 | (1.7 ± 0.04) × 104 |
| p22cmr | (0.9 ± 0.05) × 104 | (0.8 ± 0.07) × 104 |
| p27cmr | (2.3 ± 0.14) × 104 | (2.1 ± 0.03) × 104 |
| p32cmr | (1.2 ± 0.11) × 104 | (1.1 ± 0.08) × 104 |
Mean values from 3 experiments ± SD are presented. No statistical significance (P > 0.05 in t test) in differences between results obtained for all tested pairs (wild type versus ppGpp0) was found.
Fig 5.
Effects of ppGpp on DNA amounts of plasmids derived from Shiga toxin-converting bacteriophages. Relative levels of DNA of plasmids p933Wcmr (p993W), p22cmr (p22), p27cmr (p27), and p32cmr (p32) (derived from phages 933W, P22, P27, and P32, respectively) in an exponentially growing E. coli MG1655 host (WT) (open bars) and its relA spoT derivative (ppGpp0) (closed bars) were estimated. The DNA level of each plasmid determined in the wild-type host was considered to be 1, and other values reflect this value for each particular plasmid. The results are mean values from 3 experiments, with error bars representing SD.
ppGpp inhibits expression of stx genes.
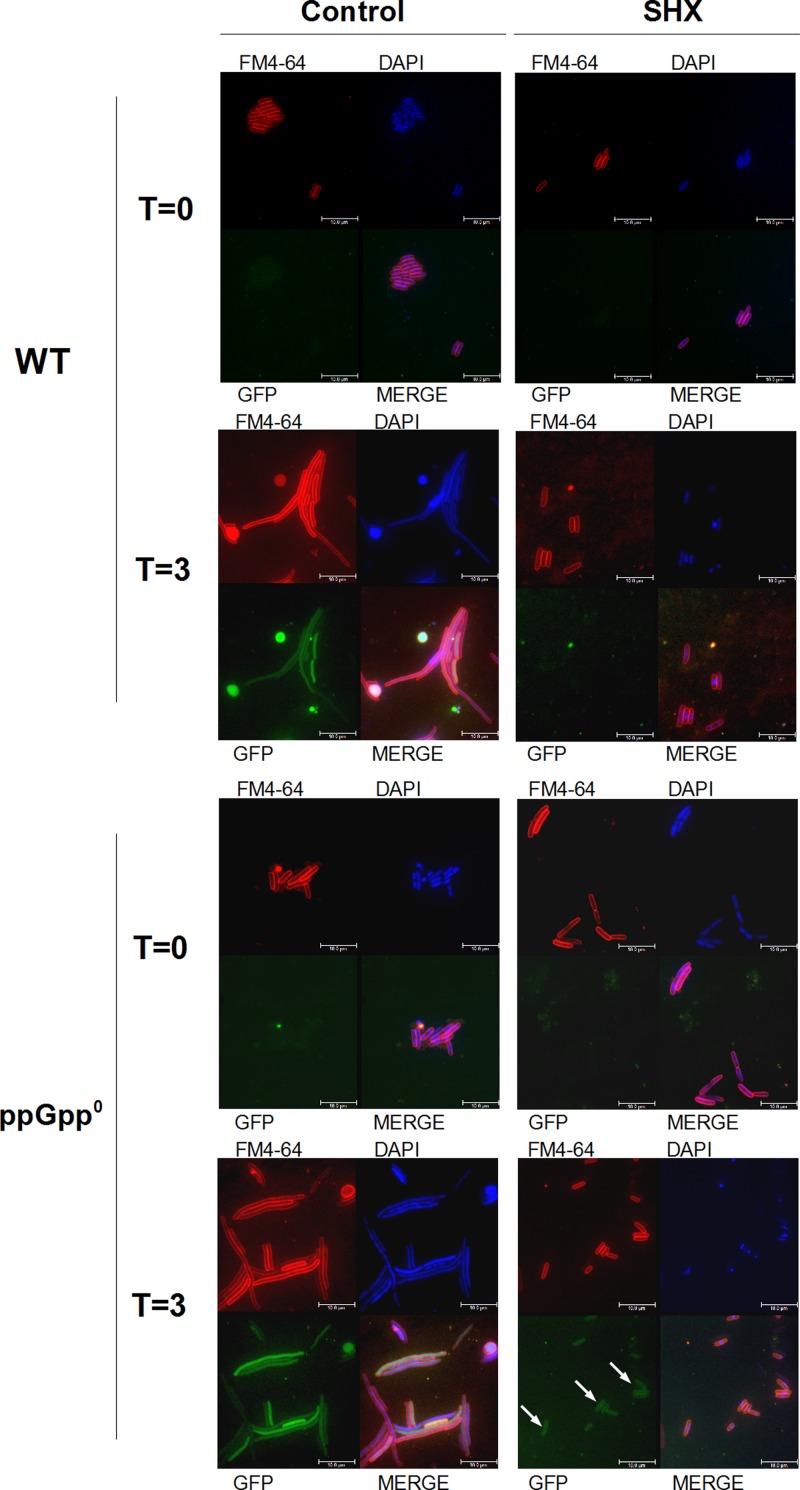
Since efficient phage DNA replication was postulated to be required for effective expression of stx genes upon Shiga toxin-converting prophage induction (6), we used a derivative of the 933W phage bearing a gene coding for GFP instead of stx and estimated its expression under various conditions. Mitomycin C-mediated prophage induction in the wild-type host caused filamentation of cells, a phenotype characteristic for intracellular phage lytic development, and effective expression of GFP (Fig. 6). Simultaneous treatment with serine hydroxamate, causing serine starvation and induction of the stringent response due to ppGpp production, halted both cell filamentation and GFP synthesis (Fig. 6).
Fig 6.
ppGpp influences stx gene expression after prophage induction. Effects of prophage induction and amino acid starvation on cells of wild-type (WT) (MG1655) and ppGpp0 hosts lysogenic with phage 933W Δstx2::catGFP were assessed. Prophage induction was caused by addition of mitomycin C up to 1 μg ml−1 to the bacterial culture at time zero (T = 0). Serine starvation was caused by addition of serine hydroxamate (SHX) up to 0.5 mg ml−1 at time zero (T = 0). Cell membranes were stained with Synaptored/FM4-64 (FM4-64), and DNA was stained with DAPI. GFP fluorescence corresponds to the strength of transcription and translation signals for expression of stx genes in native phages. Samples of cultures were withdrawn at time (T = 0) and 3 h (T = 3) after addition of mitomycin C (all panels) and serine hydroxamate (right panels only). Arrows indicate production of GFP, which corresponds to activity of signals for expression of stx genes. Bars represent 10 μm.
The ppGpp0 hosts treated with both mitomycin C and serine hydroxamate did not filament but produced significant amounts of GFP (Fig. 6). These results strongly suggest that completion of phage lytic development is inhibited by both ppGpp and amino acid deprivation, but expression of stx genes is blocked efficiently by ppGpp and may proceed in the absence of this nucleotide even under conditions of amino acid deficiency.
DISCUSSION
The stringent control is a bacterial response to amino acid starvation and some other environmental stresses, and ppGpp is the major alarmone of this response (15). It was demonstrated previously that replication of plasmids derived from Shiga toxin-converting bacteriophages is inhibited in amino acid-starved bacteria (24). However, this inhibition, similar to impairment of production of progeny virions, occurred during the stringent as well as the relaxed response to amino acid starvation, i.e., in the presence as well as in the absence of high levels of ppGpp (14). Therefore, the use of E. coli strains completely devoid of this nucleotide has been considered by us a straightforward way to answer the question whether ppGpp influences these processes. In this study, we have demonstrated that in the absence of ppGpp (in the relA spoT double mutant), both production of progeny virions and replication of phage DNA are significantly more efficient than in the wild-type strain, which bears basal levels of this alarmone. Therefore, we conclude that ppGpp negatively regulates replication of genomes of Shiga toxin-converting bacteriophages, and we suggest that this impairment may be responsible for inhibition of phage development in amino acid-starved host cells. Moreover, ppGpp appears to inhibit expression of stx genes from genomes of these phages, possibly by impairment of phage DNA replication (and a resultant low copy number of stx) or repression of the pR′ promoter (though such strong effects as those presented in Fig. 6 are unlikely to be solely due to the presence/absence of basal ppGpp amounts in cells) or both.
We suggest that the most plausible mechanism of ppGpp-mediated impairment of DNA replication of Shiga toxin-converting bacteriophages may be inhibition of the activity of the pR promoter by this alarmone. In fact, negative effects of ppGpp on pR-initiated transcription were demonstrated previously for two such phages, ST2-8624 and 933W (24). Transcription from this promoter provides mRNA for proteins necessary during early stages of the phage lytic development, and in phage λ such a transcription is in addition required to activate initiation of phage DNA replication (20). An alternative hypothesis that inhibition of replication of Shiga toxin-converting bacteriophages by ppGpp results from impaired priming reactions seems to be less likely. In fact, recent studies indicated that apart from influencing transcription of genes, ppGpp also inhibits activity of DnaG primases from Bacillus subtilis (30) and E. coli (31). However, although this inhibition negatively regulates DNA replication in B. subtilis cells (30), severe ppGpp-mediated impairment of the elongation stage of E. coli replicative DNA synthesis could be observed only in vitro and not in vivo (32, 33). This was proposed to stem from interactions of ppGpp with E. coli RNA polymerase but not with the homologous protein from B. subtilis (discussed in reference 33). Briefly, DnaG may be outcompeted for ppGpp binding by RNA polymerase in E. coli cells but not in B. subtilis, and thus the inhibitory effect of this nucleotide on the primase activity can be masked in the former bacterium (33).
Interestingly, λ replicons respond differently to ppGpp than replicons derived from Shiga toxin-converting phages, despite a very high level of similarity between the DNA sequences of their replication origin regions, as identified previously (23). Namely, replication of λ plasmids is strongly inhibited during the stringent response, but it proceeds in amino acid-starved relA mutants due to the activity of the replication complex which is inherited after each replication round by one of two daughter DNA molecules (18). In contrast, replication of plasmids derived from Shiga toxin-converting phages was inhibited during both the stringent and relaxed responses (24). The stable fraction of the O protein (replication initiator), which reflects the presence of the heritable replication complex (reviewed in reference 34), exists at the same level in λ and other phages mentioned above (24), which strongly suggests that it is not responsible for the observed differences.
It is intriguing that in the absence of ppGpp in cells, production of progeny virions of all tested Shiga toxin-converting phages was considerably more efficient than in wild-type bacteria (this report), while no significant differences in phage burst size and kinetics of formation of progeny phages were observed for phage λ between wild-type and ppGpp0 hosts, as demonstrated previously (19). It is worth mentioning that the presence of the Ser282Gly substitution in the O protein of bacteriophage λ resulted in the ability for replication of λ plasmids during both the relaxed and stringent responses, while replication of all tested plasmids derived from Shiga toxin-converting bacteriophages was strongly inhibited in amino acid-starved relA+ and relA cells irrespective of the presence of Ser or Gly residue at the position 282 (24). All these results suggest that unlike the case for λ, replicons derived from Shiga toxin-converting phages require efficient protein synthesis for initiation of new rounds of DNA replication and that replication of these phages is considerably more sensitive to ppGpp than replication of λ. The molecular reason for these differences is not known; however, it may be important that while there are 4 iteron (the O protein-binding) sequences in the replication region of the phage λ genome, the number of such sequences in tested Shiga toxin-converting phages is 6 (23). It is tempting to speculate that these differences provide a basis for the mechanisms of various responses of these replicons to amino acid starvation and to ppGpp. Such a proposal is supported by following facts: (i) the number of iterons must influence the efficiency of binding of the O protein to the origin region, (ii) the pR promoter activity is negatively regulated by ppGpp in λ (21) and in Shiga toxin-converting bacteriophages (24), (iii) transcription starting from pR is responsible for activation of the lambdoid origin and is plausibly coupled with the rearrangement of the replication complex (34), and (iv) the O protein interacts directly with RNA polymerase (35), a target for ppGpp in E. coli (36).
ACKNOWLEDGMENTS
We thank Katarzyna Potrykus for discussions.
This work was supported by the National Science Center (Poland) (project grant no. N N301 192439 to A.W. and project grant no. 2011/02/A/NZ1/00009 to G.W.) and was operated within the Foundation for Polish Science Ventures Programme cofinanced by the EU European Regional Development Fund (project grant no. VENTURES/2012-9/7 to D.N.).
Footnotes
Published ahead of print 30 August 2013
REFERENCES
- 1.Hunt JM. 2010. Shiga toxin-producing Escherichia coli (STEC). Clin. Lab. Med. 30:21–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85(Suppl 13):E45–E62 [DOI] [PubMed] [Google Scholar]
- 3.Bloch SA, Felczykowska A, Nejman-Faleńczyk B. 2012. Escherichia coli O104:H4 outbreak—have we learnt a lesson from it? Acta Biochim. Pol. 59:483–488 [PubMed] [Google Scholar]
- 4.Karch H, Denamur E, Dobrindt U, Finlay BB, Hengge R, Johannes L, Ron EZ, Tønjum T, Sansonetti PJ, Vicente M. 2012. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol. Med. 4:841–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allison HE. 2007. Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2:165–174 [DOI] [PubMed] [Google Scholar]
- 6.Łoś JM, Łoś M, Węgrzyn G. 2011. Bacteriophages carrying Shiga toxin genes: genomic variations, detection and potential treatment of pathogenic bacteria. Future Microbiol. 6:909–924 [DOI] [PubMed] [Google Scholar]
- 7.Wagner PL, Livny J, Neely MN, Acheson DW, Friedman DI, Waldor MK. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957–970 [DOI] [PubMed] [Google Scholar]
- 8.Wagner PL, Neely MN, Zhang X, Acheson DW, Waldor MK, Friedman DI. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ptashne M. 2004. A genetic switch: phage lambda revisited, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 10.Węgrzyn G, Węgrzyn A. 2005. Genetic switches during bacteriophage lambda development. Prog. Nucleic Acid Res. Mol. Biol. 79:1–48 [DOI] [PubMed] [Google Scholar]
- 11.Herold S, Karch H, Schmidt H. 2004. Shiga toxin-encoding bacteriophages—genomes in motion. Int. J. Med. Microbiol. 294:115–121 [DOI] [PubMed] [Google Scholar]
- 12.Waldor MK, Friedman DI. 2005. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8:459–465 [DOI] [PubMed] [Google Scholar]
- 13.Łoś JM, Łoś M, Węgrzyn G, Węgrzyn A. 2009. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb. Pathog. 47:289–298 [DOI] [PubMed] [Google Scholar]
- 14.Nejman-Faleńczyk B, Golec P, Maciąg M, Wegrzyn A, Węgrzyn G. 2012. Inhibition of development of Shiga toxin-converting bacteriophages by either treatment with citrate or amino acid starvation. Foodborne Pathog. Dis. 9:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 16.Chatterji D, Fujita N, Ishihama A. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 15:279–287 [DOI] [PubMed] [Google Scholar]
- 17.Kanjee U, Ogata K, Houry WA. 2012. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85:1029–1043 [DOI] [PubMed] [Google Scholar]
- 18.Szalewska-Pałasz A, Węgrzyn A, Herman A, Węgrzyn G. 1994. The mechanism of the stringent control of λ plasmid DNA replication. EMBO J. 13:5779–5785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slomińska M, Neubauer P, Wegrzyn G. 1999. Regulation of bacteriophage λ development by guanosine 5′-diphosphate-3′-diphosphate. Virology 262:431–441 [DOI] [PubMed] [Google Scholar]
- 20.Węgrzyn G, Licznerska K, Węgrzyn A. 2012. Phage λ—new insights into regulatory circuits. Adv. Virus Res. 82:155–178 [DOI] [PubMed] [Google Scholar]
- 21.Wróbel B, Murphy H, Cashel M, Wegrzyn G. 1998. Guanosine tetraphosphate (ppGpp)-mediated inhibition of the activity of the bacteriophage lambda pR promoter in Escherichia coli. Mol. Gen. Genet. 257:490–495 [DOI] [PubMed] [Google Scholar]
- 22.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990 [PubMed] [Google Scholar]
- 23.Nejman B, Łoś JM, Łoś M, Węgrzyn G, Węgrzyn A. 2009. Plasmids derived from lambdoid bacteriophages as models for studying replication of mobile genetic elements responsible for the production of Shiga toxins by pathogenic Escherichia coli strains. J. Mol. Microbiol. Biotechnol. 17:211–220 [DOI] [PubMed] [Google Scholar]
- 24.Nejman B, Nadratowska-Wesołowska B, Szalewska-Pałasz A, Węgrzyn A, Węgrzyn G. 2011. Replication of plasmids derived from Shiga toxin-converting bacteriophages in starved Escherichia coli. Microbiology 157:220–233 [DOI] [PubMed] [Google Scholar]
- 25.Sokawa Y, Sokawa J, Kaziro Y. 1975. Regulation of stable RNA synthesis and ppGpp levels in growing cells of Escherichia coli. Cell 5:69–74 [DOI] [PubMed] [Google Scholar]
- 26.Sarubbi E, Rudd KE, Cashel M. 1988. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol. Gen. Genet. 213:214–222 [DOI] [PubMed] [Google Scholar]
- 27.Łoś JM, Golec P, Węgrzyn G, Węgrzyn A, Łoś M. 2008. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl. Environ. Microbiol. 74:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wegrzyn G, Wegrzyn A, Konieczny I, Bielawski K, Konopa G, Obuchowski M, Helinski DR, Taylor K. 1995. Involvement of the host initiator function dnaA in the replication of coliphage lambda. Genetics 139:1469–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 30.Wang JD, Sanders GM, Grossman AD. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maciąg M, Kochanowska M, Łyżeń R, Węgrzyn G, Szalewska-Pałasz A. 2010. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid 63:61–67 [DOI] [PubMed] [Google Scholar]
- 32.DeNapoli J, Tehranchi AK, Wang JD. 2013. Dose-dependent reduction of replication elongation rate by (p)ppGpp in Escherichia coli and Bacillus subtilis. Mol. Microbiol. 88:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maciąg-Dorszyńska M, Szalewska-Pałasz A, Węgrzyn G. 2013. Different effects of ppGpp on Escherichia coli DNA replication in vivo and in vitro. FEBS Open Biol. 3:161–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wegrzyn A, Wegrzyn G. 2001. Inheritance of the replication complex: a unique or common phenomenon in the control of DNA replication? Arch. Microbiol. 175:86–93 [DOI] [PubMed] [Google Scholar]
- 35.Szambowska A, Pierechod M, Wegrzyn G, Glinkowska M. 2011. Coupling of transcription and replication machineries in λ DNA replication initiation: evidence for direct interaction of Escherichia coli RNA polymerase and the λO protein. Nucleic Acids Res. 39:168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toulokhonov II, Shulgina I, Hernandez VJ. 2001. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta′-subunit. J. Biol. Chem. 276:1220–1225 [DOI] [PubMed] [Google Scholar]
- 37.Jensen KF. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnusson LU, Gummesson B, Joksimović P, Farewell A, Nyström T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 189:5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allison HE, Sergeant MJ, James CE, Saunders JR, Smith DL, Sharp RJ, Marks TS, McCarthy AJ. 2003. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 71:3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gamage SD, Patton AK, Hanson JF, Weiss AA. 2004. Diversity and host range of Shiga toxin-encoding phage. Infect. Immun. 72:7131–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyd AC, Sherratt DJ. 1995. The pCLIP plasmids: versatile cloning vectors based on the bacteriophage λ origin of replication. Gene 154:57–62 [DOI] [PubMed] [Google Scholar]