Abstract
The spirochete Leptospira interrogans causes a systemic infection that provokes a febrile illness. The putative lipoproteins LigA and LigB promote adhesion of Leptospira to host proteins, interfere with coagulation, and capture complement regulators. In this study, we demonstrate that the expression level of the LigA and LigB proteins was substantially higher when L. interrogans proliferated at 37°C instead of the standard culture temperature of 30°C. The RNA comprising the 175-nucleotide 5′ untranslated region (UTR) and first six lig codons, whose sequence is identical in ligA and ligB, is predicted to fold into two distinct stem-loop structures separated by a single-stranded region. The ribosome-binding site is partially sequestered in double-stranded RNA within the second structure. Toeprint analysis revealed that in vitro formation of a 30S-tRNAfMet-mRNA ternary complex was inhibited unless a 5′ deletion mutation disrupted the second stem-loop structure. To determine whether the lig sequence could mediate temperature-regulated gene expression in vivo, the 5′ UTR and the first six codons were inserted between the Escherichia coli l-arabinose promoter and bgaB (β-galactosidase from Bacillus stearothermophilus) to create a translational fusion. The lig fragment successfully conferred thermoregulation upon the β-galactosidase reporter in E. coli. The second stem-loop structure was sufficient to confer thermoregulation on the reporter, while sequences further upstream in the 5′ UTR slightly diminished expression at each temperature tested. Finally, the expression level of β-galactosidase was significantly higher when point mutations predicted to disrupt base pairs in the second structure were introduced into the stem. Compensatory mutations that maintained base pairing of the stem without restoring the wild-type sequence reinstated the inhibitory effect of the 5′ UTR on expression. These results indicate that ligA and ligB expression is limited by double-stranded RNA that occludes the ribosome-binding site. At elevated temperatures, the ribosome-binding site is exposed to promote translation initiation.
INTRODUCTION
Leptospirosis is a globally endemic zoonosis caused by Leptospira interrogans and other pathogenic members of the genus Leptospira. The leptospiral life cycle involves a host stage in the renal tubules of mammalian reservoir hosts. Urinary shedding of the spirochetes into the external environment is followed by acquisition of infection by new host animals. Humans exposed to water or soil contaminated by L. interrogans are at risk of being infected by the spirochete through skin abrasions or mucous membranes. Leptospirosis patients often experience a sudden onset of fever as the spirochetes disseminate in the bloodstream and settle in the internal organs of the patient (1, 2).
L. interrogans encounters diverse environmental signals at different stages of its life cycle. Studies of individual genes and whole transcriptomes and proteomes have revealed a number of L. interrogans genes that are differentially expressed when the spirochete is incubated under conditions likely to be encountered during host infection (3–11). Levels of specific outer membrane proteins also differ during acute infection of the liver (12) and in leptospires exiting the host in urine (13). The mechanisms by which these changes occur have not been explored for any L. interrogans gene in part because the genetic tools available for manipulation of L. interrogans are limited (14).
The ligA and ligB genes encode probable lipoproteins that serve as multifunctional ligands that bind a number of host proteins. LigA and LigB may aid in the colonization of L. interrogans by binding with high affinity to components of the extracellular matrix (15–19). The Lig proteins may also facilitate dissemination by interacting with proteins that control hemostasis (19–21). Furthermore, ectopic expression of LigB in the nonpathogen Leptospira biflexa improved survival of the spirochete in human serum (22). Both LigA and LigB may promote complement resistance by capturing several complement regulators, including factor H and C4-binding protein (23). LigB also binds with high affinity to the complement proteins C3b and C4b, targets of the activities of factor H and C4-binding protein (22).
The expression pattern of the lig genes is consistent with a role of their gene products during infection of the mammalian host. ligB mRNA is upregulated in the bloodstream of infected rodent models, and Lig protein is detected in the kidneys of infected hamsters (24, 25). L. interrogans and the closely related species L. kirschneri are typically cultivated under low-osmolarity conditions; exposure of the organisms to higher levels of osmolarity, such as those found in the host, causes the expression level of ligA and ligB to increase markedly (26, 27).
Many microbial pathogens sense temperature to gauge successful entry into a warm-blooded mammalian host. Expression of virulence genes is triggered at 37°C by a variety of molecular mechanisms (28). For example, expression of Yersinia pseudotuberculosis lcrF, Listeria monocytogenes prfA, and Borrelia burgdorferi rpoS, which encode key regulators of virulence gene transcription, are inhibited at low temperatures by intramolecular base pairs within the mRNA that sequester the ribosome-binding site (RBS) (29–31). At mammalian host temperatures, the lcrF and prfA RBSs unfold due to their intrinsic thermolability. As the RBSs within these RNA thermometers unfold, translational efficiency increases, and/or rates of mRNA degradation are shifted to enhance production of the gene product. On the other hand, formation of the inhibitory RNA structure in the B. burgdorferi rpoS transcript appears to be blocked by trans-acting factors during growth at mammalian host temperatures at low bacterial densities (31–33).
In addition to temperature fluctuations, bacteria also sense shifts in external osmolarity. Exposure of Escherichia coli to increased osmolarity is accompanied by shrinkage of the cytoplasm and changes in the concentrations of cytoplasmic ions (34). The structure of RNA can be influenced by ionic concentrations. For example, the accessibility of the RBS of the E. coli rpoS transcript is sensitive to magnesium concentration changes in vitro, indicating that conformational changes in the mRNA leader may facilitate osmotic regulation of rpoS (35). Additionally, the melting point of the Salmonella fourU thermometer is altered by shifts in magnesium concentrations within a physiological range (36).
In this study, we demonstrate that the expression of ligA and ligB is upregulated by increasing the culture temperature from 30°C to 37°C. The (identical) mRNA leader sequences of the ligA and ligB genes are predicted to form an extended secondary structure that sequesters the RBS. Using translational fusions, we demonstrate that an extended mRNA leader sequence overlapping the ligA and ligB start codons imparts pronounced temperature regulation, but minimal osmotic regulation, on a β-galactosidase reporter in E. coli. We present genetic evidence that base pairing at the ribosome-binding site must be overcome to allow ribosomes to initiate translation of the lig mRNA.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Leptospira interrogans serovar Copenhageni strain Fiocruz L1-130 was maintained in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium supplemented with 1% heat-inactivated rabbit serum at 30°C. EMJH medium was assembled with Probumin vaccine-grade bovine serum albumin (BSA) (Millipore) and included 100 μg/ml 5-fluorouracil (Sigma). Rabbit serum (Rockland Immunochemical, Gilbertsville, PA) was heat inactivated at 56°C for 30 min prior to its addition to EMJH medium.
For the lig expression experiments, actively growing L. interrogans cells were diluted to a cell density of 4 × 106 cells/ml in 25 ml fresh EMJH medium supplemented with 1% rabbit serum in 125-ml Erlenmeyer flasks and incubated in a rotary shaker at 30°C or 37°C until a density of 1 × 108 to 2 × 108 cells/ml was achieved. A third 25-ml culture containing an additional 50 mM sodium chloride was incubated in parallel with the other cultures.
Plasmids.
Restriction enzymes were purchased from New England BioLabs (Ipswich, MA). Oligonucleotides were synthesized by Life Technologies (Grand Island, NY). PCRs were performed with Phusion DNA polymerase (Thermo Scientific, Asheville, NC) with genomic DNA from L. interrogans strain Fiocruz L1-130 as the template. Plasmid pBAD2-bgaB was obtained from Birgit Klinkert (37).
The 5′ deletion mutations with endpoints at positions −175, −64, and −42 relative to the start codon were generated with forward primers ligL(Nh)-15F, ligL(Nh)-13F, and ligL(Nh)-7F, respectively (oligonucleotide sequences are provided in Tables 1 and 2). The reverse primer was ligL(Ec)-6R. NheI and EcoRI restriction sites were introduced near the 5′ ends of the forward and reverse primers, respectively. The amplicons were digested with NheI and EcoRI and inserted into pBAD2-bgaB, which was digested with the same restriction enzymes, to create plasmids pRAT668 (position −175), pRAT669 (position −64), and pRAT662 (position −42). Primers ligL(Nh)-8F and ligL(Ec)-8R were annealed to generate the position −21 5′ deletion endpoint with NheI and EcoRI sticky ends and inserted into pBAD2-bgaB to create pRAT663.
Table 1.
Oligonucleotides for plasmid constructions
| Oligonucleotide | Nucleotide sequencea | Positionb |
|---|---|---|
| ligL(Nh)-15F | 5′-ATCTgctagcAGAGAACTTCTATAAAATTGATTTTTAATT-3′ | −175 |
| ligL(Nh)-13F | 5′-GTAAGGgctagcAATAAATAAACACAGCAAATACACA-3′ | −64 |
| ligL(Nh)-7F | 5′-ACACAGgctagcACAAAATGAAAACTTCAAATAAAACA-3′ | −42 |
| ligL(Ec)-6R | 5′-AAATCGgaattcACAAAATATTTTCTTCATAAACACTCA-3′ | 18 |
| ligL(Nh)-8F | 5′-ctagcAAACAATTAGAGTGAGTGTTTATGAAGAAAATATTTTGTg-3′ | −21 |
| ligL(Nh)-8R | 5′-aattcACAAAATATTTTCTTCATAAACACTCACTCTAATTGTTTg-3′ | 18 |
| ligL(Nh)-21F | 5′-ACACAGgctagcACAAAATGAAAACTTCAAATAAATGA-3′ | −42 |
| ligL(Ec)-18R | 5′-AAATCGgaattcACAAAATATTTTCTTCATAATGACTCA-3′ | 18 |
| sph2(Nh)-24F | 5′-ctagcATACAGAATGGAGATAGAGAACGATGATAAACAAAATAACAg-3′ | −23 |
| sph2(Ec)-24R | 5′-aattcTGTTATTTTGTTTATCATCGTTCTCTATCTCCATTCTGTATg-3′ | 18 |
| lipL41(Nh)-9F | 5′-ctagcATTCCCCAAAATCAAACAGGATTGGTGTTACTTTTCATGAGAAAATTATCTTCTg-3′ | −36 |
| lipL41(Ec)-9R | 5′-aattcAGAAGATAATTTTCTCATGAAAAGTAACACCAATCCTGTTTGATTTTGGGGAATg-3′ | 18 |
Restriction sites and sticky ends are in lowercase type. Start codons are italicized.
The position of the nucleotide underlined in the sequence is relative to the first nucleotide in the start codon.
Table 2.
Oligonucleotides for site-directed mutagenesis
| Oligonucleotide | Nucleotide sequencea | Template |
|---|---|---|
| ligL-12F | 5′-CAAATAAATGAATTAGAGTGAGTGTTTatgAAGAAAATA-3′ | pRAT670 |
| ligL-12R | 5′-CTCTAATTCATTTATTTGAAGTTTTCATTTTGTGTA-3′ | |
| ligL-14F | 5′-GAGTGAGTCATTatgAAGAAAATATTTTGTATTTCGA-3′ | pRAT670 |
| ligL-14R | 5′-CTTcatAATGACTCACTCTAATTGTTTTATTTGAAGTT-3′ | |
| ligL-19F | 5′-ATAAATAATGACAGCAAATACACAAAATGAAAAC-3′ | pRAT671 |
| ligL-19R | 5′-TTTGCTGTCATTATTTATTGAGAGCCTTTACAAAATT-3′ |
Nucleotide changes are underlined. The start codon is shown in lowercase type.
The BamHI-EcoRI fragment containing lig sequences from positions −175 to +18 was transferred from pRAT668 to pUC19 to generate pRAT670, which served as a template for PCR-based oligonucleotide-directed mutagenesis. The AC-to-TG mutation at positions −19 and −18 of lig in pRAT671 was generated with oligonucleotides ligL-12F and ligL-12R by using the Geneart site-directed mutagenesis System from Life Technologies. Similarly, the −4,−3/GT-to-CA mutation in pRAT672 was generated with ligL-14F and ligL-14R. All other oligonucleotide-directed mutagenesis reactions were performed as described previously by Zheng et al. (38), using Phusion DNA polymerase for 20 cycles of PCR. Oligonucleotides ligL-19F and ligL-19R were used to combine all four nucleotide changes in pRAT673 from pRAT671 as the template. Following mutagenesis, the BamHI-EcoRI lig fragments were transferred back into pBAD2-bgaB to generate plasmids pRAT683, pRAT684, and pRAT685. To construct the bgaB fusions with the point mutations combined with the 5′ deletion endpoint at position −43, pRAT671, pRAT672, and pRAT673 were used as templates for PCR with primer pairs ligL(Nh)-21F/ligL(Ec)-6R, ligL(Nh)-7F/ligL(Ec)-18R, and ligL(Nh)-21F/ligL(Ec)-18R, respectively. The amplicons were digested with NheI and EcoRI and inserted into pBAD2-bgaB to generate pRAT691, pRAT692, and pRAT693, respectively. To construct the lipL41′-′bgaB fusion plasmid, the oligonucleotides lipL41(Nh)-9F and lipL41(Ec)-9R were annealed together and inserted into pBAD2-bgaB to create pRAT667. The sequences of all inserted fragments were verified by DNA sequencing (Laragen, Culver City, CA).
Immunoblots.
The optical density of each L. interrogans culture was measured at 420 nm with an Ultrospec 2000 spectrophotometer (Amersham Biosciences). Two milliliters of the culture was collected by centrifugation at 9,000 × g in an Eppendorf 5424 microcentrifuge. The bacteria were washed once with ice-cold phosphate-buffered saline (PBS)–5 mM MgCl2 and resuspended in 1× NuPAGE LDS loading buffer (Invitrogen) containing 1× reducing agent (Invitrogen). The volume for resuspension was determined by multiplying the optical density at 420 nm (OD420) of the culture by 500 μl. Samples were boiled for 5 min, and 10 μl was loaded onto a 10% SDS-acrylamide gel (Lonza, Walkersville, MD) for electrophoresis at 150 V. The material in the gel was transferred onto an Immobilon-FL polyvinylidene fluoride (PVDF) membrane (Millipore) in methanol-free Western blot transfer buffer (Thermo Scientific) at 25 V for 90 min. The membrane was blocked overnight at 4°C in a blocking solution consisting of 5% (wt/vol) nonfat powdered milk (MP Biomedicals, Santa Ana, CA) dissolved in PBS-Tween 20 (Thermo Scientific). The membrane was probed with a 1:2,000 dilution of Lig antiserum (39) and a 1:10,000 dilution of LipL41 antiserum (40) for 60 min, washed three times with PBS-Tween 20, and then probed with a 1:5,000 dilution of a donkey anti-rabbit IgG-horseradish peroxidase conjugate (Amersham). The blot was developed with Pierce ECL 2 Western blot detection reagents (Thermo Scientific), covered with plastic wrap, and placed in contact with Amersham Hyperfilm to visualize the bands by chemiluminescence. Band intensities were measured by scanning the membrane with a Storm 840 instrument (Amersham Biosciences) in fluorescence mode and quantitated with ImageQuant TL software (Amersham Biosciences).
RNA purification.
L. interrogans cultures were quickly chilled in a dry-ice–ethanol bath, and leptospires were collected by centrifugation in a prechilled Sorvall SA-600 rotor at 8,000 rpm at 4°C for 15 min. The culture supernatant was removed, and RNA was extracted with TRIzol (Invitrogen). The RNA was then treated with 10 units of Turbo DNase (Ambion) in a final volume of 100 μl for 2 h at 37°C and purified by phenol-chloroform extraction, chloroform extraction, and ethanol precipitation. The RNA concentration was measured with a Nanovue spectrophotometer (GE Healthcare). The integrity of the RNA was checked by electrophoresis into a 1.4% agarose gel (Lonza).
5′-end mapping.
RNA was extracted from L. interrogans cells cultivated in a 1:1 mixture of EMJH medium and modified Eagle's minimal essential medium (MEM) (American Type Culture Collection, Manassas, VA). Two micrograms of L. interrogans RNA was subjected to 5′ rapid amplification of cDNA ends (RACE) with the 5′/3′ RACE kit from Roche, according to the manufacturer's instructions. Separate reverse transcription reactions were conducted with primers ligA-32R (5′-AGAAGCGATTCCTCTGACTCTGTTA-3′) and ligB-48R (5′-AGAAGCGATTCCTTTGACTCTGTTG-3′). After poly(A) tailing of the 3′ end of the cDNA, the cDNA was subjected to PCR with an oligo(dT) anchor primer (5′-GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV-3′, where V is A, C, or G; Roche) and lig-specific primer ligAB-22R (5′-GATCCCTGATGCCAACCCT-3′) with HotStarTaq (Qiagen) in an MJ Research PTC-200 thermal cycler (Bio-Rad). To generate the 5′ RACE amplicon, the PCR mixture was subjected to an initial denaturation step at 95°C for 15 min; a 35-cycle amplification of 94°C for 20 s, 54°C for 30 s, and 72°C for 40 s; and a final extension step at 72°C for 2 min.
Real-time PCR.
cDNA was generated from 1 μg of total L. interrogans RNA with iScript reverse transcription supermix according to the manufacturer's instructions (Bio-Rad). cDNA from 10 ng RNA was subjected to real-time PCR with iQ5 SYBR green PCR mix (Bio-Rad) with primer pairs ligA-29F/ligA-30R (5′-TCCTGAGCGAAGGTCTTACACTACAA-3′/5′-TCTCCAAATGCAAGAGCCGTTAC-3′), ligB-51F/ligB-52R (5′-AGGTAACTCCGGCACAATTGATTT-3′/5′-GTCACGTCTTGTACGGAATGATCC-3′), and lipL41-7F/lipL41-8R (5′-TCGGAAATCTGATTGGAGCGGAAGCA-3′/5′-AGAAGCGGCGAAACCTGCCACT-3′). Real-time PCR was performed with the iQ5 real-time PCR detection system (Bio-Rad). Standard curves were generated for each primer pair with cDNA synthesized from 50 ng, 10 ng, 2 ng, 0.4 ng, and 0.08 ng RNA. The PCR efficiencies for the primer pairs were between 90% and 110%. The parameters for PCR were as follows: an initial denaturation step at 95°C for 15 min; amplification at 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s 40 times; and a final extension step at 72°C for 2 min.
Toeprint assays.
The 30S ribosomal subunit was purified from E. coli as described previously (41). lig RNA transcripts were synthesized with T7 RNA polymerase from PCR products by using procedures described previously (35). The template for the PCR was plasmid pRAT659, which harbored a lig′-′bglB fusion. The forward primers contained the T7 promoter (lowercase type in sequences) linked to the 5′ untranslated region (UTR) of lig at position −175 (5′-taatacgactcactataggAGAGAACTTCTATAAAATTG-3′; −64, 5′-taatacgactcactataggCAATAAATAAACACAGCAAATACAC-3′; −43, 5′-taatacgactcactataggCACAAAATGAAAACTTC-3′; −21, 5′-taatacgactcactataggAAACAATTAGAGTGAGTG-3′), and the reverse primer was the bgaB-specific oligonucleotide 5′-CATCTTCATACCAAATTTC-3′. Toeprint assays were performed with 300 nM the E. coli 30S ribosomal subunit as described previously (31), except for the following changes: the reaction mixture was incubated for 25 min at 37°C, and the primer extension reaction was allowed to proceed with avian myeloblastosis virus (AMV) reverse transcriptase (RT) (Promega) and the primer 5′-ATACCAAATTTCCTCTGGCC-3′ for 15 min at 42°C. The quenched reaction mixtures were analyzed by electrophoresis in a 10% polyacrylamide gel in 1× Tris-borate-EDTA.
β-Galactosidase assay.
Fusion plasmids were transformed into E. coli NEB-5α (New England BioLabs), and transformants were selected on LB agar plates containing 100 μg/ml ampicillin at 37°C. Colonies from each plate were inoculated into 2 ml of LB with 100 μg/ml ampicillin in culture tubes in triplicate and incubated with shaking overnight at 30°C. The next morning, 50 μl of each culture was transferred into three culture tubes containing 4 ml of LB with 100 μg/ml ampicillin and 0.02% l-arabinose. The tubes were placed into shaking incubators at 30°C, 37°C, and 42°C until the cultures reached an OD600 of 0.8 to 1.0. β-Galactosidase activities were measured as described previously (42), except that reaction mixtures were incubated at 55°C.
For growth of the transformants at different culture osmolarities, three colonies of each transformant were incubated overnight in 2 ml of LB with 100 μg/ml ampicillin at 37°C. Twenty microliters of each culture was then transferred into 2 ml LB that was assembled without sodium chloride. After overnight incubation at 37°C, 50 μl of each culture was placed into 4 ml of LB lacking sodium chloride (80 mosmol/liter), LB with 110 mM sodium chloride (300 mosmol/liter), and LB with 360 mM sodium chloride (800 mosmol/liter). Cultures were grown to late log phase (OD600 of 0.9 to 1.0), and β-galactosidase activities were measured.
Statistics.
Statistical analysis of the β-galactosidase assay data was conducted with GraphPad Prism, version 5.04 (GraphPad, La Jolla, CA). The data were analyzed by one-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test.
RESULTS
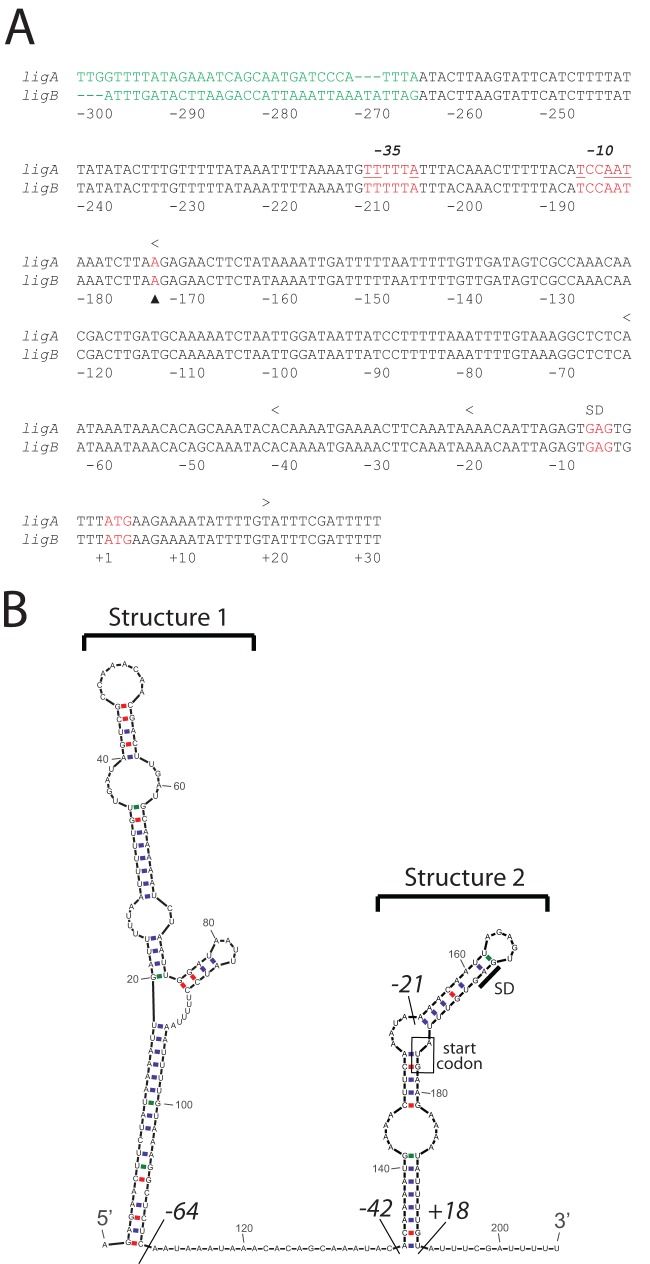
In the course of our ongoing study of lig gene regulation by osmolarity, we mapped the 5′ ends of the ligA and ligB transcripts by 5′ RACE. In the Fiocruz L1-130 strain of L. interrogans, the ligA and ligB sequences are identical beginning 266 nucleotides upstream of the start codon (Fig. 1A) and extending 75 nucleotides into the lig coding region. ligA- and ligB-specific primers were used for the reverse transcriptase step. The major 5′ end for both transcripts was located 175 nucleotides upstream of the start codon (Fig. 1A). Inspection of the sequence preceding the 5′ end revealed potential E. coli-type −10 and −35 sequences separated by 17 bp (Fig. 1A). The lengthy 5′ untranslated region (UTR) and the first 10 codons of lig were analyzed by the secondary structure prediction program MFOLD (43). The RNA is predicted to fold into two stem-loop structures separated by a 22-nucleotide single-stranded segment (Fig. 1B). Structure 2 includes the first six codons of the lig transcripts. Nucleotides within the Shine-Dalgarno sequence and start codon are involved in base pairing within structure 2 (Fig. 1B). Because translation initiation is typically blocked when the ribosome-binding site is sequestered within folded RNA (44, 45), the 5′ UTR of lig may thermally regulate translation of the ligA and ligB transcripts by unzipping at higher temperatures to provide ribosomes access to the ribosome-binding site (RBS). In contrast, the Shine-Dalgarno sequence and start codon of lipL41, the control gene for our study, are present in separate stem-loop structures (data not shown). These predicted structures were calculated by MFOLD to be much less stable thermodynamically (−3.2 and −3.6 kcal/mol for the Shine-Dalgarno sequence and start codon, respectively) than structure 2 of lig (−9.7 kcal/mol).
Fig 1.
Sequence and predicted RNA structure of the 5′ end of ligA and ligB. (A) The coding and upstream sequences of ligA and ligB were aligned with LALIGN (72). Only the 300 nucleotides upstream of the start codon and the first 10 lig codons of the alignment are shown. Nucleotides identical in ligA and ligB are shown in black and red. The triangle marks the major 5′ end of the lig transcripts. Nucleotides matching the −10 and −35 promoter consensus sequences are underlined. Endpoints of the 5′ deletion mutations are indicated by “<.” The 3′ boundary of the lig sequences fused to bgaB is indicated by “>.” The Shine-Dalgarno sequence (SD) was predicted from its complementarity to the 3′ end of the 16S rRNA of L. interrogans. (B) The secondary structure of first 205 nucleotides of the lig transcript was predicted with MFOLD, version 4.4 (43). Endpoints of 5′ deletion mutations at positions −64, −43, and −21, relative to the first nucleotide in the start codon, are marked. The lig sequence was fused at position +18 to bgaB.
Because many virulence genes in pathogens are regulated by temperature (28), we examined the expression of ligA and ligB from cultures of L. interrogans that were cultivated at 30°C and 37°C. Both cultures attained a density of 2 × 108 to 3 × 108 cells/ml after 4 days and at least five generations of growth. As a positive control for induction of lig expression, a third EMJH culture containing an additional 50 mM sodium chloride was cultivated at 30°C. A concentration of 50 mM is sufficient to upregulate ligA and ligB expression without inhibiting bacterial growth (27).
Figure 2 shows that small amounts of the Lig proteins were expressed when L. interrogans was grown at 30°C. In contrast, large amounts of LigA and LigB were produced when the leptospires were incubated at 37°C. For quantitation, band intensities of LigA and LigB were normalized to that of LipL41, which is stably expressed across a broad range of growth conditions. Over three experiments, LigA protein levels were a mean of 20-fold higher in L. interrogans cells grown at 37°C than at 30°C, and LigB levels were 14-fold higher. For comparison, LigA and LigB protein levels were 31- and 23-fold higher, respectively, when the culture medium was supplemented with 50 mM sodium chloride and the culture was grown at 30°C. lig transcript levels were measured from the same cultures by quantitative real-time PCR and normalized to lipL41 transcript levels. lig mRNA levels were higher in the culture grown at 37°C as well: 6.6-fold for ligA and 5.1-fold for ligB. In the cultures containing the extra sodium chloride, ligA mRNA levels were 18-fold higher, and ligB mRNA levels were 13-fold higher.
Fig 2.
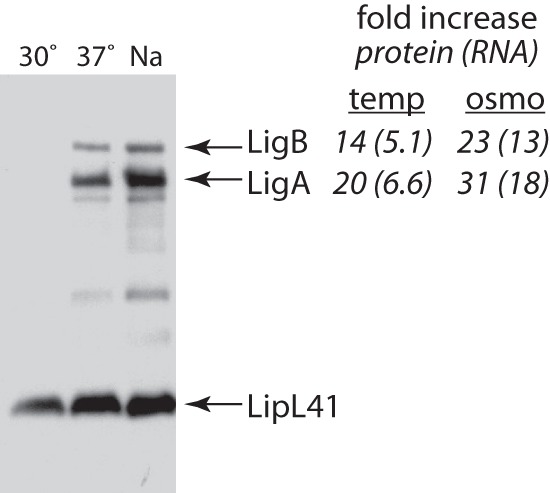
Immunoblot analysis of ligA and ligB expression. Cultures of L. interrogans were incubated at 30°C in EMJH medium, at 37°C in EMJH medium, or at 30°C in EMJH medium supplemented with 50 mM sodium chloride (Na). Immunoblots of the cell lysates were probed with Lig and LipL41 antisera. A representative immunoblot from three experiments is shown. The fold differences of LigA and LigB protein levels at a growth temperature of 37°C relative to those at 30°C (“temperature”) and in EMJH medium supplemented with 50 mM sodium chloride relative to those in unsupplemented EMJH medium (“osmo”) are shown in italics. Lig protein band intensities were normalized to that of LipL41 prior to calculation of the fold increase. Similarly, CT (threshold cycle) values for ligA and ligB transcripts were normalized to that for the lipL41 transcript; relative increases in transcript levels are shown in parentheses.
To determine whether the 5′ lig sequences containing the predicted RNA stem-loop structures were involved in temperature regulation, DNA segments consisting of the 5′ UTR plus the first six codons of lig and various 5′ deletion constructs were cloned into pBAD2-bgaB, a plasmid designed to monitor posttranscriptional temperature-dependent gene expression in E. coli (37). pBAD2-bgaB and its predecessor pBAD-bgaB have been used to examine the function of a number of prokaryotic RNA thermometers (37). In this plasmid, the inserted gene fragment is fused in frame to the bgaB reporter gene lacking its start codon. The bgaB gene encodes a thermostable β-galactosidase enzyme found in Bacillus stearothermophilus, thereby allowing expression studies at culture temperatures above 37°C. The promoter of the E. coli arabinose operon, PBAD, is located immediately upstream of the inserted fragment and drives transcription of the gene fusion. The plasmid also carries the araC gene, which encodes the positive regulator of PBAD. As previously noted, RNA structural changes that are important for temperature regulation often function both in their natural context and when fused to bgaB in the model organism E. coli (37). A fragment from the 5′ end of L. interrogans lipL41 was also cloned into pBAD2-bgaB to serve as a negative control for temperature-dependent regulation.
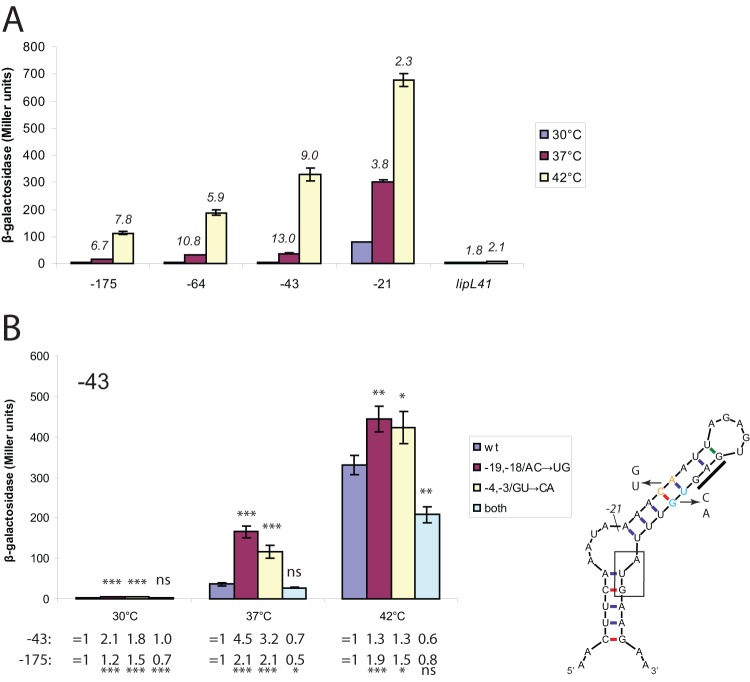
The plasmids were transformed into E. coli. The transformants were grown to late exponential phase in the presence of arabinose, and β-galactosidase levels were measured. Expression levels of the full-length position −175 lig fusion were low at 30°C and nearly 7-fold higher in cultures grown at 37°C (Fig. 3A), qualitatively similar to the differences in expression observed for LigA and LigB in L. interrogans (Fig. 2). In contrast, temperature had a <2-fold effect on expression from the lipL41 fusion (Fig. 3A). Expression of the lig fusion was an additional 7.8-fold higher in the 42°C cultures, while the increase in the expression level of the lipL41 fusion was only 2.1-fold. These results demonstrate that the 5′ UTR and the first six codons of lig contain the elements necessary to confer thermoregulation on a reporter gene.
Fig 3.
Thermoregulation of lig′-′bgaB fusion expression. (A) 5′ deletion analysis of the lig 5′ UTR. E. coli cells harboring various lig′-′bgaB fusions were incubated at 30°C, 37°C, and 42°C. lipL41′-′bgaB fusion expression level values were normalized to that of the lig′-′bgaB fusion at 30°C. Ratios of expression levels at 37°C to those at 30°C and of expression levels at 42°C to those at 37°C are displayed in italics. (B) Effect of point mutations predicted to disrupt base pairing within the lig ribosome-binding site. Point mutations were introduced into the lig leader (cyan, positions −4 and −3 relative to the A of the start codon; orange, positions −19 and −18) of the −43 and −175 fusions. The bar graph shows the results for the −43 fusion. Expression levels relative to those for the wild-type (wt) fusion for each temperature are shown below the graph. Error bars indicate standard deviations from the means (n = 3). ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ns, not significant (by one-way ANOVA, with Dunnett's multiple-comparison test with wild-type base-paired lig as the control).
Toeprint assays were performed to determine whether the lig 5′ UTR contained sequence elements that inhibited ribosome binding in vitro. lig transcripts were transcribed from a T7 promoter in vitro and purified. Additional lig transcripts with 5′ deletion endpoints at positions −64, −43, and −21 were also synthesized (Fig. 1B). The four transcripts were mixed with the 30S ribosomal subunit and tRNAfMet from E. coli, and primer extension was performed to detect the toeprint of the ternary 30S-tRNAfMet-mRNA complex (Fig. 4). The −175 (full-length), −64, and −43 lig transcripts generated weak toeprint signals. In contrast, the −21 transcript generated a strong signal at position +15 (relative to the A in the start codon), indicating that sequences upstream of position −21 harbored an inhibitory element that blocked formation of the ternary complex. The 30S subunit alone failed to generate a toeprint signal on any lig transcript. These results provide direct evidence for inhibition of ribosome binding to the lig RBS by an inhibitory element within the 5′ UTR.
Fig 4.
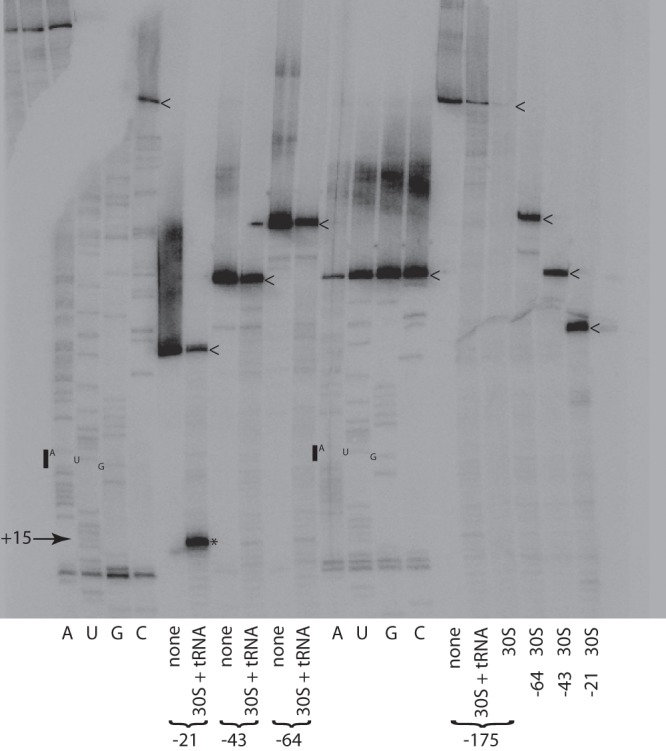
Toeprint analysis of lig transcripts. The complete binding reaction mixtures (“30S + tRNA”) were comprised of lig transcript, the E. coli 30S ribosomal subunit, and initiator tRNAfMet. Additional reaction mixtures (“30S”) lacked tRNAfMet. Control reactions (“none”) contained only lig transcript. The toeprint at position +15 is marked with an asterisk. The start codon is denoted by a bar next to the sequencing ladder. The 5′ ends of the lig transcripts are marked with arrowheads.
To assess the activity of the negative regulatory element in vivo, the 5′ deletions used for toeprint analysis were examined with pBAD2-bgaB in E. coli at 30°C, 37°C, and 42°C (Fig. 3A). For all constructs, expression of the fusion increased as temperature increased. At all three growth temperatures, maximum β-galactosidase expression was observed with the −21 lig fusion, which removed structure 1 and most of the left stem of structure 2, while expression of the full-length −175 fusion was minimal, confirming the presence of an inhibitory element. In addition, the −43 deletion mutant, which retained the predicted structure that sequestered the RBS, was sufficient for temperature regulation. The −21 mutation exhibited the lowest level of temperature induction, suggesting that the structure of the stem-loop is important for the temperature regulation of the ligA and ligB genes. This outcome was a consequence of much higher levels of fusion expression at 30°C and 37°C when the left stem of structure 2 between positions −43 and −21 was removed. In addition, the slightly lower β-galactosidase expression level from the full-length −175 fusion compared with the −43 fusion at all three temperatures indicates the presence of an additional inhibitory element or elements upstream of structure 2 (Fig. 3A).
To determine whether base pairing within the RBS was involved in inhibiting expression of the fusion, point mutations predicted to disrupt two base pairs between the initiation codon and Shine-Dalgarno sequence were introduced into the left strand of the −43 construct, which contains enough lig 5′ UTR sequence to confer temperature regulation upon bgaB (Fig. 3). The mutations caused a significant increase in fusion expression levels at each temperature (Fig. 3B). Mutations in the right stem introduced between the Shine-Dalgarno sequence and the start codon led to a similar increase in fusion expression levels. The combination of the two sets of mutations, which restored the two base pairs, restored repression of fusion expression to wild-type levels or lower. These results suggest that base pairing within the RBS is necessary to fully inhibit gene expression from lig. When the mutations were examined in the full-length −175 fusion, similar results were obtained, although expression levels in general were lower (Fig. 3B).
Since the concentration of cytoplasmic ionic solutes changes during shifts in external osmolarity (34), we hypothesized that changes in medium osmolarity would also affect expression of the lig fusion in E. coli. The osmolality of LB assembled without sodium chloride has been measured to be 0.08 osmol/kg (46). We added sodium chloride to obtain LB with calculated osmolarities of 300 mosmol/liter and 800 mosmol/liter. When E. coli cells harboring the full-length lig′-′bgaL fusion were incubated in the three LB culture media, the differences in β-galactosidase expression levels at the different osmolarities were <50% (Fig. 5), much weaker than the effect of temperature on lig fusion expression (Fig. 3A). Moreover, expression of the lig fusion did not vary relative to expression from the lipL41 fusion (Fig. 5).
Fig 5.
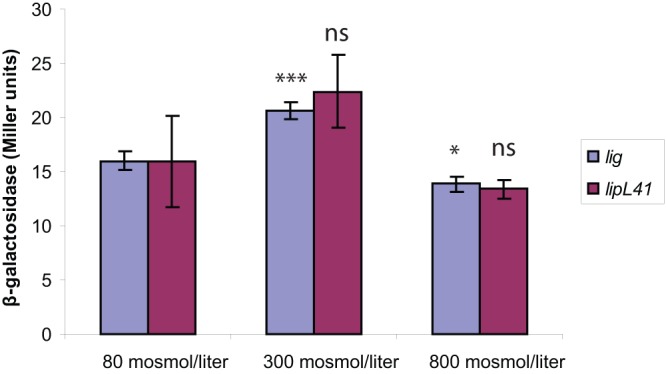
Osmotic effects on lig′-′bgaB fusion expression. E. coli cells harboring fusion plasmids were grown at 37°C in LB adjusted to various osmolarities with sodium chloride. Expression values of the lipL41′-′bgaB fusion were normalized to that of the lig′-′bgaB fusion at 80 mosmol/liter. Error bars indicate standard deviations from the means (n = 3). ∗, P < 0.05; ∗∗∗, P < 0.001; ns, not significant (by one-way ANOVA, with Dunnett's multiple-comparison test with 80 mosmol/liter as a control).
DISCUSSION
The structure of mRNA sequences can have large effects on translational efficiency and mRNA turnover in prokaryotes (47, 48). In this study, we have determined that the 5′ leader of the lig transcript is important in setting the expression level of the coding region that follows. We found that the length and sequence of the 5′-untranslated leader region influence the expression and degree of temperature regulation of a reporter gene in E. coli; deletion analysis of the 5′ UTR revealed the presence of an RNA-based negative regulatory element that repressed translation initiation of lig in vitro (Fig. 4) and expression in vivo (Fig. 3A). Although there is a modest temperature dependence of the expression of all sequences tested, the lig sequences comprising structure 2 alone are sufficient to confer significant temperature regulation on the bgaB reporter (Fig. 3A). This minimal lig regulatory element is predicted to fold into a stem-loop structure that sequesters the RBS within double-stranded RNA (structure 2 in Fig. 1B). The ratio of the expression level at high temperatures to that at low temperatures is reduced significantly in the −21 construct (Fig. 3A), which lacks most of the upstream sequences required to sequester the RBS. The small amount of repression exhibited by the −21 fusion may be due to the short stem-loop structure that remains (Fig. 1B). Our genetic evidence indicates that full repression of lig′-′bgaB fusion expression in vivo required base pairing within the RBS in addition to a low culture temperature (Fig. 3B). Other constructs that retain these base pairs (positions −175, −64, and −43) exhibited weak translation initiation in vitro and substantial temperature induction in vivo, whereas the 5′ deletion mutant that disrupts these base pairs (position −21) displayed the opposite behavior, suggesting that translational efficiency is correlated with the presence of base-paired RNA in the RBS. Mutations that reduce the stability of intramolecular base pairing that sequesters the RBS result in increased translational efficiency and protein expression in many systems (29, 30, 44, 45, 49, 50).
Based on our results, we propose that the 5′ UTR is in part responsible for temperature-dependent regulation of ligA and ligB expression in L. interrogans. The regulation mediated by the lig 5′ UTR was observed in translational fusions that are expressed in E. coli. If trans-acting factors are necessary for lig thermoregulation, they must be present in both L. interrogans and E. coli. At low cell densities, temperature-dependent induction of the rpoS gene in the spirochete Borrelia burgdorferi requires a trans-acting noncoding RNA unrelated in sequence to the one that controls rpoS translation in E. coli and the RNA-binding protein Hfq, which is produced by both bacteria (31–33). Alternatively, the lig sequence by itself may be sufficient for temperature-dependent regulation by functioning as a thermolabile RNA thermometer. Most prokaryotic RNA thermometers described to date control expression of proteins involved in the heat and cold shock responses and λ bacteriophage development (51). Exceptions include the Yersinia pseudotuberculosis lcrF gene and the Listeria monocytogenes prfA gene, which encode transcriptional regulators of virulence genes (29, 30).
Both transcript and protein levels can be influenced by translational efficiency. The density of ribosomes translating the mRNA can profoundly affect the chemical half-life of the transcript by shielding RNase target sites (52). Translational inhibition of small heat shock genes by the ROSE (repression of heat shock gene expression) thermometer is accompanied by large reductions in mRNA levels (53, 54). Although growth temperature affected ligA and ligB transcript levels as well as protein levels in L. interrogans (Fig. 2), we demonstrated posttranscriptional temperature-dependent induction of gene expression by investigating regulation using a heterologous promoter driving the transcription of the lig sequence.
Sequences upstream of structure 2 reduced lig fusion expression, especially at mammalian host temperatures (Fig. 3A). Although the mechanisms by which these sequences diminish lig expression are unknown, there are precedents for additional helices upstream of the one harboring the RBS influencing regulation. Like the lig 5′ UTR, ROSE sequences are predicted to harbor additional stem-loop structures upstream of the thermometer (50). Mutational analysis of the stem present upstream of the thermometer structure within the Bradyrhizobium japonicum ROSE1 sequence indicates that base pairing is necessary for the thermometer to exert full repression of expression at 30°C (50). The first two hairpins within the ROSE element of the E. coli ibpA gene are stable at all temperatures from 20°C to 50°C in vitro, whereas the structure containing the RBS becomes accessible to single-strand-specific ribonucleases at temperatures of 35°C and higher (55). The function of structures upstream of the thermometer may be to assist in the folding of the thermometer. Another function proposed for sequences upstream of the thermometer is to harbor targets for ribonucleases involved in decay of the transcript (56).
The limitation imposed on lig translation at 30°C by base pairing within the RBS raises the question of how ribosomes access the RBS to generate the Lig proteins when L. interrogans is incubated at higher culture osmolarities (Fig. 2) (26). The increased cytoplasmic concentration of ions that accompanies growth at higher external osmolarity (34) may alter the lig RNA structure to expose the RBS. However, the poor response of lig fusion expression in E. coli over a 10-fold range in external osmolarity (Fig. 5) fails to account for the >20-fold increase in Lig protein levels observed in L. interrogans when the culture osmolarity was increased <3-fold (Fig. 2). These observations suggest that a trans-acting factor aids in invasion of the base-paired lig RBS by ribosomes when L. interrogans is incubated at higher osmolarities at 30°C. This model assumes that L. interrogans responds to shifts in osmolarity like E. coli. As the major cationic osmolyte in bacteria, potassium has a major role in maintaining turgor. During osmotic shock, turgor is restored in E. coli by a rapid influx of potassium through the Trk and Kdp transporters (57). The L. interrogans genome encodes homologs of the Kdp and Trk-like Ktr potassium transporters (58, 59). Disruption of genes encoding Trk, Ktr, and Kdp transporters attenuates growth of diverse bacteria during osmotic stress (60–64).
Although shifts in L. interrogans gene expression in response to environmental changes have been described previously, to our knowledge, cis-acting sites responsible for differential gene expression have yet to be identified experimentally. Data from ongoing projects to obtain whole-genome sequences from a variety of Leptospira strains will aid in the analysis of candidate cis- and trans-acting factors that underlie the genetic regulatory circuitry controlling expression of leptospiral genes (65–70). One outcome of these efforts is the computational identification of potential riboswitches, including those bound by cobalamin and thiamine pyrophosphate (68, 71). Here we have presented experimental evidence for an RNA-based cis-acting element that controls gene expression in L. interrogans in response to temperature changes. Although we could not test mutations directly in L. interrogans because of the difficulty in genetic manipulation of the spirochete, we believe that our mutational analyses involving toeprinting in vitro and a β-galactosidase reporter system in E. coli provide strong evidence for an RNA stem-loop structure operating as a posttranscriptional inhibitory element.
ACKNOWLEDGMENTS
We are grateful to Birgit Klinkert for providing plasmid pBAD2-bgaB. We thank Justin Ching for providing technical support.
This study was supported by a Veterans Affairs Merit Review award (to J.M.) and Public Health Service grant AI-034431 from the National Institute of Allergy and Infectious Diseases (to D.A.H.). P.J.S. was supported by National Center for Research Resources grant 5P20RR016463 and National Institute of General Medical Sciences grant 8 P20 GM103423 from the National Institutes of Health.
The results and conclusions presented are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies.
Footnotes
Published ahead of print 6 September 2013
REFERENCES
- 1.Farr RW. 1995. Leptospirosis. Clin. Infect. Dis. 21:1–8 [DOI] [PubMed] [Google Scholar]
- 2.Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsunaga J, Lo M, Bulach DM, Zuerner RL, Adler B, Haake DA. 2007. Response of Leptospira interrogans to physiologic osmolarity: relevance in signaling the environment-to-host transition. Infect. Immun. 75:2864–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo M, Bulach DM, Powell DR, Haake DA, Matsunaga J, Paustian ML, Zuerner RL, Adler B. 2006. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 74:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo M, Murray GL, Khoo CA, Haake DA, Zuerner RL, Adler B. 2010. Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect. Immun. 78:4850–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patarakul K, Lo M, Adler B. 2010. Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol. 10:31. 10.1186/1471-2180-10-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue F, Dong H, Wu J, Wu Z, Hu W, Sun A, Troxell B, Yang XF, Yan J. 2010. Transcriptional responses of Leptospira interrogans to host innate immunity: significant changes in metabolism, oxygen tolerance, and outer membrane. PLoS Negl. Trop. Dis. 4:e857. 10.1371/journal.pntd.0000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin JH, Sheng YY, Zhang ZM, Shi YZ, He P, Hu BY, Yang Y, Liu SG, Zhao GP, Guo XK. 2006. Genome-wide transcriptional analysis of temperature shift in L. interrogans serovar Lai strain 56601. BMC Microbiol. 6:51. 10.1186/1471-2180-6-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen PA, Cordwell SJ, Bulach DM, Haake DA, Adler B. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 70:2311–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo M, Cordwell SJ, Bulach DM, Adler B. 2009. Comparative transcriptional and translational analysis of leptospiral outer membrane protein expression in response to temperature. PLoS Negl. Trop. Dis. 3:e560. 10.1371/journal.pntd.0000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt A, Beck M, Malmstrom J, Lam H, Claassen M, Campbell D, Aebersold R. 2011. Absolute quantification of microbial proteomes at different states by directed mass spectrometry. Mol. Syst. Biol. 7:510. 10.1038/msb.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nally JE, Whitelegge JP, Bassilian S, Blanco DR, Lovett MA. 2007. Characterization of the outer membrane proteome of Leptospira interrogans expressed during acute lethal infection. Infect. Immun. 75:766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monahan AM, Callanan JJ, Nally JE. 2008. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect. Immun. 76:4952–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choy HA, Kelley MM, Chen TL, Moller AK, Matsunaga J, Haake DA. 2007. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 75:2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin YP, McDonough SP, Sharma Y, Chang YF. 2010. The terminal immunoglobulin-like repeats of LigA and LigB of Leptospira enhance their binding to gelatin binding domain of fibronectin and host cells. PLoS One 5:e11301. 10.1371/journal.pone.0011301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin YP, Greenwood A, Nicholson LK, Sharma Y, McDonough SP, Chang YF. 2009. Fibronectin binds to and induces conformational change in a disordered region of leptospiral immunoglobulin-like protein B. J. Biol. Chem. 284:23547–23557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin YP, Lee DW, McDonough SP, Nicholson LK, Sharma Y, Chang YF. 2009. Repeated domains of Leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. J. Biol. Chem. 284:19380–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choy HA, Kelley MM, Croda J, Matsunaga J, Babbitt JT, Ko AI, Picardeau M, Haake DA. 2011. The multifunctional LigB adhesin binds homeostatic proteins with potential roles in cutaneous infection by pathogenic Leptospira interrogans. PLoS One 6:e16879. 10.1371/journal.pone.0016879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin YP, McDonough SP, Sharma Y, Chang YF. 2011. Leptospira immunoglobulin-like protein B (LigB) binding to the C-terminal fibrinogen alphaC domain inhibits fibrin clot formation, platelet adhesion and aggregation. Mol. Microbiol. 79:1063–1076 [DOI] [PubMed] [Google Scholar]
- 21.Figueira CP, Croda J, Choy HA, Haake DA, Reis MG, Ko AI, Picardeau M. 2011. Heterologous expression of pathogen-specific genes ligA and ligB in the saprophyte Leptospira biflexa confers enhanced adhesion to cultured cells and fibronectin. BMC Microbiol. 11:129. 10.1186/1471-2180-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choy HA. 2012. Multiple activities of LigB potentiate virulence of Leptospira interrogans: inhibition of alternative and classical pathways of complement. PLoS One 7:e41566. 10.1371/journal.pone.0041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castiblanco-Valencia MM, Fraga TR, Silva LB, Monaris D, Abreu PA, Strobel S, Jozsi M, Isaac L, Barbosa AS. 2012. Leptospiral immunoglobulin-like proteins interact with human complement regulators factor H, FHL-1, FHR-1, and C4BP. J. Infect. Dis. 205:995–1004 [DOI] [PubMed] [Google Scholar]
- 24.Palaniappan RU, Chang YF, Jusuf SS, Artiushin S, Timoney JF, McDonough SP, Barr SC, Divers TJ, Simpson KW, McDonough PL, Mohammed HO. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70:5924–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui M, Soupe ME, Becam J, Goarant C. 2012. Differential in vivo gene expression of major Leptospira proteins in resistant or susceptible animal models. Appl. Environ. Microbiol. 78:6372–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsunaga J, Medeiros MA, Sanchez Y, Werneid KF, Ko AI. 2007. Osmotic regulation of expression of two extracellular matrix-binding proteins and a haemolysin of Leptospira interrogans: differential effects on LigA and Sph2 extracellular release. Microbiology 153:3390–3398 [DOI] [PubMed] [Google Scholar]
- 27.Matsunaga J, Sanchez Y, Xu X, Haake DA. 2005. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 73:70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro RS, Cowen LE. 2012. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. mBio 3(5):e00238–12. 10.1128/mBio.00238-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, Dersch P. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. 10.1371/journal.ppat.1002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551–561 [DOI] [PubMed] [Google Scholar]
- 31.Archambault L, Linscott J, Swerdlow N, Boyland K, Riley E, Schlax P. 2013. Translational efficiency of rpoS mRNA from Borrelia burgdorferi: effects of the length and sequence of the mRNA leader region. Biochem. Biophys. Res. Commun. 433:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lybecker MC, Samuels DS. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 64:1075–1089 [DOI] [PubMed] [Google Scholar]
- 33.Lybecker MC, Abel CA, Feig AL, Samuels DS. 2010. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 78:622–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cayley S, Lewis BA, Guttman HJ, Record MT., Jr 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 222:281–300 [DOI] [PubMed] [Google Scholar]
- 35.Worhunsky DJ, Godek K, Litsch S, Schlax PJ. 2003. Interactions of the non-coding RNA DsrA and RpoS mRNA with the 30 S ribosomal subunit. J. Biol. Chem. 278:15815–15824 [DOI] [PubMed] [Google Scholar]
- 36.Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. 2011. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res. 39:8258–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klinkert B, Cimdins A, Gaubig LC, Rossmanith J, Aschke-Sonnenborn U, Narberhaus F. 2012. Thermogenetic tools to monitor temperature-dependent gene expression in bacteria. J. Biotechnol. 160:55–63 [DOI] [PubMed] [Google Scholar]
- 38.Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. 10.1093/nar/gnh110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, Bolin CA, Reis MG, Riley LW, Haake DA, Ko AI. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang ES, Summers TA, Haake DA. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64:2322–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spedding G, Draper DE. 1993. Allosteric mechanism for translational repression in the Escherichia coli alpha operon. Proc. Natl. Acad. Sci. U. S. A. 90:4399–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 43.Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Smit MH, van Duin J. 1994. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol. 244:144–150 [DOI] [PubMed] [Google Scholar]
- 45.de Smit MH, van Duin J. 1990. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U. S. A. 87:7668–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilizota T, Shaevitz JW. 2012. Fast, multiphase volume adaptation to hyperosmotic shock by Escherichia coli. PLoS One 7:e35205. 10.1371/journal.pone.0035205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Regnier P, Arraiano CM. 2000. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays 22:235–244 [DOI] [PubMed] [Google Scholar]
- 48.Marzi S, Fechter P, Chevalier C, Romby P, Geissmann T. 2008. RNA switches regulate initiation of translation in bacteria. Biol. Chem. 389:585–598 [DOI] [PubMed] [Google Scholar]
- 49.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. 2007. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65:413–424 [DOI] [PubMed] [Google Scholar]
- 50.Nocker A, Hausherr T, Balsiger S, Krstulovic NP, Hennecke H, Narberhaus F. 2001. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 29:4800–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10:255–265 [DOI] [PubMed] [Google Scholar]
- 52.Kaberdin VR, Blasi U. 2006. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 30:967–979 [DOI] [PubMed] [Google Scholar]
- 53.Nocker A, Krstulovic NP, Perret X, Narberhaus F. 2001. ROSE elements occur in disparate rhizobia and are functionally interchangeable between species. Arch. Microbiol. 176:44–51 [DOI] [PubMed] [Google Scholar]
- 54.Gaubig LC, Waldminghaus T, Narberhaus F. 2011. Multiple layers of control govern expression of the Escherichia coli ibpAB heat-shock operon. Microbiology 157:66–76 [DOI] [PubMed] [Google Scholar]
- 55.Waldminghaus T, Gaubig LC, Klinkert B, Narberhaus F. 2009. The Escherichia coli ibpA thermometer is comprised of stable and unstable structural elements. RNA Biol. 6:455–463 [DOI] [PubMed] [Google Scholar]
- 56.Narberhaus F, Waldminghaus T, Chowdhury S. 2006. RNA thermometers. FEMS Microbiol. Rev. 30:3–16 [DOI] [PubMed] [Google Scholar]
- 57.Rhoads DB, Epstein W. 1978. Cation transport in Escherichia coli. IX. Regulation of K transport. J. Gen. Physiol. 72:283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsunaga J, Coutinho ML. 2012. Positive regulation of Leptospira interrogans kdp expression by KdpE as demonstrated with a novel beta-galactosidase reporter in Leptospira biflexa. Appl. Environ. Microbiol. 78:5699–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanelt I, Tholema N, Kroning N, Vor der Bruggen M, Wunnicke D, Bakker EP. 2011. KtrB, a member of the superfamily of K+ transporters. Eur. J. Cell Biol. 90:696–704 [DOI] [PubMed] [Google Scholar]
- 60.Holtmann G, Bakker EP, Uozumi N, Bremer E. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185:1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A. 2010. Identification of trkH, encoding a potassium uptake protein required for Francisella tularensis systemic dissemination in mice. PLoS One 5:e8966. 10.1371/journal.pone.0008966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez-Ferreras A, Munoz S, Olivares J, Soto MJ, Sanjuan J. 2009. Role of potassium uptake systems in Sinorhizobium meliloti osmoadaptation and symbiotic performance. J. Bacteriol. 191:2133–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gowrishankar J. 1985. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J. Bacteriol. 164:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuda N, Kobayashi H, Katoh H, Ogawa T, Futatsugi L, Nakamura T, Bakker EP, Uozumi N. 2004. Na+-dependent K+ uptake Ktr system from the cyanobacterium Synechocystis sp. PCC 6803 and its role in the early phases of cell adaptation to hyperosmotic shock. J. Biol. Chem. 279:54952–54962 [DOI] [PubMed] [Google Scholar]
- 65.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, Creno S, Kuczek ES, Bommezzadri S, Davis JC, McGrath A, Johnson MJ, Boursaux-Eude C, Seemann T, Rouy Z, Coppel RL, Rood JI, Lajus A, Davies JK, Medigue C, Adler B. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. 10.1371/journal.pone.0001607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, Davis J, Johnson M, Kuczek E, Alt DP, Peterson-Burch B, Coppel RL, Rood JI, Davies JK, Adler B. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chou LF, Chen YT, Lu CW, Ko YC, Tang CY, Pan MJ, Tian YC, Chiu CH, Hung CC, Yang CW. 2012. Sequence of Leptospira santarosai serovar Shermani genome and prediction of virulence-associated genes. Gene 511:364–370 [DOI] [PubMed] [Google Scholar]
- 68.Ricaldi JN, Fouts DE, Selengut JD, Harkins DM, Patra KP, Moreno A, Lehmann JS, Purushe J, Sanka R, Torres M, Webster NJ, Vinetz JM, Matthias MA. 2012. Whole genome analysis of Leptospira licerasiae provides insight into leptospiral evolution and pathogenicity. PLoS Negl. Trop. Dis. 6:e1853. 10.1371/journal.pntd.0001853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 70.Nascimento AL, Verjovski-Almeida S, Van Sluys MA, Monteiro-Vitorello CB, Camargo LE, Digiampietri LA, Harstkeerl RA, Ho PL, Marques MV, Oliveira MC, Setubal JC, Haake DA, Martins EA. 2004. Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 37:459–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrick JE, Breaker RR. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8:R239. 10.1186/gb-2007-8-11-r239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang XQ, Miller W. 1991. A time efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12:337–357 [Google Scholar]