Abstract
Regulation of gene expression by small noncoding RNAs (sRNAs) plays a critical role in bacterial response to physiological stresses. NrrF, a trans-acting sRNA in Neisseria meningitidis and Neisseria gonorrhoeae, has been shown in the meningococcus to control indirectly, in response to iron (Fe) availability, the transcription of genes encoding subunits of succinate dehydrogenase, a Fe-requiring enzyme. Given that in other organisms, sRNAs target multiple mRNAs to control gene expression, we used a global approach to examine the role of NrrF in controlling gonococcal transcription. Three strains, including N. gonorrhoeae FA1090, an nrrF deletion mutant, and a complemented derivative, were examined using a custom CombiMatrix microarray to assess the role of this sRNA in controlling gene expression in response to Fe availability. In the absence of NrrF, the mRNA half-lives for 12 genes under Fe-depleted growth conditions were longer than those in FA1090. The 12 genes controlled by NrrF encoded proteins with biological functions including energy metabolism, oxidative stress, antibiotic resistance, and amino acid synthesis, as well as hypothetical proteins and a regulatory protein whose functions are not fully understood.
INTRODUCTION
Limiting access to the essential element iron (Fe) by sequestration on host proteins or intracellular reservoirs (“nutritional immunity”) is a first line of defense used by the mammalian host against a variety of pathogens (1). Conversely, bacterial pathogenesis is dependent on the acquisition of Fe from the host, and invading microbes have evolved a variety of mechanisms to obtain this essential nutrient (2). To survive in a Fe-deficient environment, such as that found in host tissues, pathogens globally sense Fe availability and respond by controlling Fe acquisition and storage genes (3) to maintain intracellular Fe concentrations within an appropriate physiological range (4). The ferric uptake (Fur) protein is responsible for sensing Fe availability and acts as a central controller of the iron response regulon in many bacteria (5). During growth under Fe-rich conditions, Fur binds as a dimer to conserved Fur box sequences in the promoters of Fe-regulated genes, using ferrous Fe (Fe2+) as a corepressor to block transcription at these promoters. As intracellular Fe stores are depleted, the Fur-Fe2+ complex dissociates from the promoter, allowing transcription (6). In this instance, Fur functions as a classical repressor protein and directly regulates the expression of genes involved in Fe acquisition and storage, as well as that of genes found in several cellular pathways important for a variety of functions, including stress and oxidative responses, and other functions essential for pathogenesis (7–10). Fur also has the capacity to activate transcription (11–15; reviewed in reference 16). A recent study by Yu and Genco (16) investigated the Fur-mediated activation in Neisseria gonorrhoeae F62 of 37 genes we previously reported to be activated in FA1090 in response to high levels of Fe (15). Using electrophoretic mobility shift assays (EMSAs) and DNase footprinting, Yu and Genco showed that Fur can operate as an activator protein through both direct and indirect mechanisms, emphasizing the complex mode of action of this global regulatory protein.
Bacterial small RNAs (sRNAs) are a ubiquitous group of regulators that impact many facets of bacterial physiology (for a recent review, see reference 17). sRNAs that target multiple pathways integrate divergent cellular functions and enable a rapid response to environmental stress (18). In Escherichia coli, several trans-acting sRNAs were initially reported to be expressed at increased levels under stress conditions such as Fe or glucose starvation and anaerobic or oxidative stress (19). These sRNAs exert their effects at the posttranscriptional level either by affecting protein activity or, more commonly, through limited complementary antisense base pairing with target mRNAs, typically promoting mRNA degradation by RNases; alternatively, mRNA translation may be increased in some instances (20). In E. coli, indirect control of transcription by Fur appears to be due in part to Fur-mediated repression of a small regulatory RNA, RyhB (21).
A similar arrangement between Fur and at least one sRNA occurs in Neisseria species (22, 23). The trans-acting, Fe-regulated sRNA NrrF (unrelated to RyhB) was initially identified in Neisseria meningitidis and N. gonorrhoeae by using a bioinformatics approach (22). We confirmed the identity of the NrrF transcript in the intergenic space between NGO2002 and NGO2003 in N. gonorrhoeae FA1090 (24). In N. meningitidis, NrrF is derepressed under low-iron conditions, facilitating the regulation of succinate dehydrogenase subunits sdhA and sdhC through the binding of NrrF to limited complementary regions of the sdh transcript, thus promoting increased turnover of the sdh message (22, 23). In the present study, we employed a global analysis to assess the impact of NrrF expression on the transcriptome of N. gonorrhoeae FA1090. Under low-Fe conditions, NrrF appears to control the level of sdh mRNA, as observed in N. meningitidis, as well as transcript levels for gonococcal genes involved in amino acid synthesis or oxidative stress and for a gene encoding a regulatory protein. We report that NrrF also affects mRNA levels for MtrF, a tightly regulated accessory protein of the mtrCDE antibiotic efflux pump. In most but not all instances, gonococcal NrrF appears to directly control the expression of these genes by affecting mRNA stability, a mechanism similar to the action of RyhB in E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All strains were routinely grown on GCB agar (Becton Dickinson) supplemented with 2% IsoVitaleX (Becton Dickinson) from frozen stock cultures under a 5% CO2 atmosphere at 37°C. Fe-deplete medium (CDM-0) and Fe-replete medium [CDM-10, supplemented with 10 μM Fe(NO3)3] were prepared as reported previously (25). Organisms were inoculated to an optical density at 600 nm (OD600) of 0.100 from overnight growth on GCB agar plates into a 500-ml side-arm flask containing 125 ml of CDM-0. Cultures were grown to an OD600 of 0.200 to starve the cells of Fe. Half of the culture for each strain was transferred to a separate 500-ml flask; 62.5 ml of CDM-0 was then added to both flasks; and Fe(NO3)3 was added to one of the flasks (CDM-10). The cultures were then allowed to grow to stationary phase in a 37°C shaking incubator (225 rpm), and cells were collected at designated time points for RNA isolation (13, 15). See Fig. S1 in the supplemental material for growth curve comparisons.
Construction of the NrrF deletion mutant and mutant complement.
An nrrF deletion mutant (designated LJ001) was constructed as follows (for construction details and primers, see Fig. S2 in the supplemental material). Genomic FA1090 DNA was used as a template with Pfu Turbo Hotstart DNA polymerase (Stratagene) and PCR primers 2002NrrF_534 and 2002NrrF_1041 to amplify a region of DNA 3′ to nrrF. A second amplicon, containing a kanamycin cassette, was amplified from pUC4K as the template by using primers Kan1014_and Kan_433 (see Fig. S2). A third amplicon, representing a region 5′ to nrrF, was generated from the genomic FA1090 template using primers Prom2004_124 and Prom2004_606. After agarose gel purification, the first and second amplicons were joined by overlap extension PCR (26) using the following steps: 15 PCR cycles without any primer were run using the first and second amplicons as templates; then the two fragments were joined by PCR (35 cycles) with end primers 2002NrrF_534 and Kan_433. By the same overlap extension approach, the DNA fragment containing the first and second amplicons was joined to the third amplicon with end primers 2002NrrF_534 and Prom2004_606. In order to block transcription initiated downstream of the Fur box, a transcription termination signal was introduced between the kanamycin cassette and the Fur box upstream of NrrF by adding oligonucleotides to the 5′ ends of primers Kan_433 and Prom2004_124 (see Fig. S2). The final joined DNA fragment was transformed into N. gonorrhoeae strain FA1090. The NrrF deletion mutant was identified from transformants on GC agar containing 100 μg/ml kanamycin, followed by sequence confirmation on an ABI 3730 DNA analyzer. Deletion of NrrF was confirmed by quantitative real-time PCR (qRT-PCR) (data not shown).
Similarly, the complemented mutant strain (designated LJ002) was constructed as detailed in Fig. S3 in the supplemental material. A wild-type copy of NrrF was inserted between NGO1029 and NGO1030, taking advantage of a bidirectional termination signal between the two open reading frames (ORFs). The following DNA fragments were joined by overlapping PCR using the primers and templates listed in Fig. S3 in the supplemental material: a short fragment of NGO1029 (N. gonorrhoeae FA1090), the NrrF gene with the Fur box, a chloramphenicol resistance cassette (27), and a short fragment of the termination signal of NGO1030 (N. gonorrhoeae FA1090). The gel-purified joined DNA fragment was transformed into the nrrF mutant. The complemented mutant LJ002 was selected out of transformants on GC agar containing 0.6 μg/ml chloramphenicol and was sequenced to confirm the structure of the complementing construct. Expression of NrrF under Fe-limited conditions by the complemented strain was also confirmed by qRT-PCR (data not shown). In growth assays, the wild-type, mutant, and complemented mutant strains were indistinguishable, suggesting that no obvious phenotypic deficits were incurred by the nrrF deletion mutation (see Fig. S1 in the supplemental material).
RNA isolation.
All N. gonorrhoeae strains were grown under Fe-depleted (Fe−) and Fe-replete (Fe+) conditions, as described above. Total RNA was isolated from cells collected at the 3-h time point for microarray studies; this time was chosen because we had previously determined this to be the peak of NrrF expression under low-Fe conditions (24). Three separate biological replicates for both growth conditions (Fe− and Fe+) were carried out at the 3-h time point for each gonococcal strain. RNA was isolated using a hot-phenol method described previously (13, 15) with the following changes: RNAs were digested with DNase (1 U RQ1 RNase-free DNase to 1 μg RNA [Promega]), followed by RNA cleanup according to the TRI Reagent protocol (Molecular Research Center, Inc.). This was done instead of using the Qiagen RNeasy columns to ensure that RNA of <200 bp was collected. Quantitation was done using the NanoDrop ND-100 spectrophotometer instrument (Thermo Scientific), and RNA integrity was assessed with an Agilent RNA 6000 Nano Kit on the Agilent 2100 bioanalyzer (28).
N. gonorrhoeae microarray design.
The present studies were carried out using a custom 12K microarray designed by CombiMatrix Corporation (Mukilteo, WA). There were 4 to 5 probes (lengths, 34 to 35 nucleotides) for each ORF. The probes represented 1,916 genes out of 2,069 identified in the NCBI annotation of the N. gonorrhoeae FA1090 genome, in addition to sense and antisense probes covering intergenic areas. Controls on the array included no-oligonucleotide and negative-control bacterial probes, factory probes, and three Arabidopsis thaliana genes as positive controls. Experimental and control probe sets were distributed randomly across the microarray.
cDNA labeling and array hybridization.
cDNA was generated from 10 μg of DNase-treated total RNA using an Applied Biosystems High-Capacity cDNA archive kit. A. thaliana mRNA positive controls (SpotReport Cab, RCA, and RBCL mRNAs) were spiked into each reverse transcription reaction mixture at a final concentration of 1 ng each (Stratagene). After synthesis, cDNA was labeled using the Mirus Label IT μArray biotin labeling kit. The QIAquick PCR purification kit (Qiagen) was used both to purify the cDNA after the removal of the RNA template and before labeling and to purify the biotinylated cDNA probes. A 3-μg aliquot of biotin-labeled cDNA was hybridized to the custom 12K N. gonorrhoeae expression array, according to the CombiMatrix hybridization protocol (28). Arrays were hybridized at 50°C for 14 h before electrochemical detection using an ElectraSense reader (CombiMatrix).
Microarray analysis.
Data files generated by the ElectraSense Reader software were imported into GeneSpring GX 12.5 for analysis after background subtraction of the “no-oligonucleotide” control probes. The microarray data were normalized per chip by dividing each measurement by the 50th percentile of all measurements in that sample and were normalized per gene by dividing the measurement for each gene by the median of its measurement across all samples. The three biological replicates for each growth condition were averaged together, and the normalized data were used for further analysis. Unless otherwise specified, the expression of an individual gene was reported as the ratio of the expression level in an isolate grown under low-Fe conditions to the expression level of the same gene in the same isolate grown under high-Fe conditions. Genes exhibiting a ≥1.5-fold-change in expression with a P value of ≤0.05 were considered differentially expressed.
N. gonorrhoeae qRT-PCR.
Primers for qRT-PCR are shown in Table S1 in the supplemental material. Real-time PCR was carried out as we described previously (15). Briefly, cDNA was generated from DNase-treated total RNA using the Applied Biosystems High-Capacity cDNA kit, including a negative-control reaction mixture (without reverse transcriptase) for each RNA sample. Primers were designed using Primer Express software, version 1.5 (Applied Biosystems), and primer efficiencies were measured on all primer pairs to confirm equal amplification rates (user bulletin no. 2; Applied Biosystems). qRT-PCR amplification was performed on an ABI Prism 7500 system (Applied Biosystems) using Power SYBR green master mix (Applied Biosystems). The relative mRNA concentration was calculated by the comparative threshold cycle (2−ΔΔCT) method with porA as the endogenous reference. Each mRNA was assayed in duplicate.
mRNA half-life measurements.
FA1090 and the nrrF mutant were grown for 3 h under Fe− conditions as described above. Cells collected at 3 h represented time zero without rifampin. Rifampin (final concentration, 200 μg/ml) was then added to each culture to stop transcription (see Fig. S4 in the supplemental material), and cells were harvested for RNA isolation at 0, 2, 4, 8, 12, and 20 min. Bulk DNase-treated RNA was purified and was reverse transcribed as described above, for use in estimating mRNA levels by qRT-PCR. The primers used for measurement of the half-life of mRNA are listed in Table S1 in the supplemental material, and qRT-PCR was performed as described above. Relative gene expression was quantified using the standard-curve method with porA as the endogenous reference. Gene expression values for each time point, normalized to the endogenous reference, were log transformed and were imported into GraphPad Prism, version 5. The mRNA half-lives were estimated (average of 3 experiments) using nonlinear regression analysis with one-phase dissociation.
EMSAs.
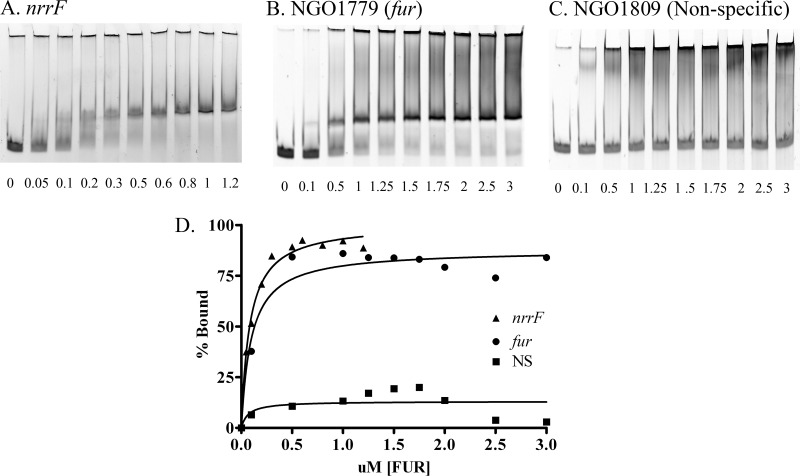
The cloned gonococcal Fur protein was expressed and purified, and electrophoretic mobility shift assays (EMSAs) were carried out, as we have described previously (15). Briefly, EMSAs were performed using the Invitrogen EMSA kit (catalog no. E-33075; Life Technologies). The DNA fragments used were PCR amplified (see Table S1 in the supplemental material for primer sequences) and were purified with a QIAprep kit (Qiagen). EMSA reactions were carried out by incubation of a 25 nM concentration of each respective DNA target with increasing concentrations of gonococcal Fur protein. For the NrrF DNA fragment, Fur concentrations ranged from 0 to 1.2 μM, while for NGO1779 (fur) and NGO1809 (nonspecific DNA control), Fur concentrations ranged from 0 to 3 μM protein. These mixtures were incubated for 20 min at room temperature with the respective DNA fragment in EMSA binding buffer and were loaded onto a 4% nondenaturing polyacrylamide gel. Products of the EMSA reactions were stained with SYBR green for 20 min and were visualized using the Kodak Gel Logic 1500 imaging system (Carestream Molecular Imaging). Band intensities were determined with ImageJ imaging software (http://rsb.info.nih.gov/ij/), and analysis was carried out using GraphPad Prism. The KD (equilibrium dissociation constant) for the binding of Fur to the DNA sequence was defined as the concentration of Fur needed for a 50% shift of the target DNA. Experiments were repeated twice.
Microarray data accession number.
Microarray data are deposited at Gene Expression Omnibus (GEO) (http://www.ncbi.nih.gov/geo) under GEO accession number GSE48526.
RESULTS AND DISCUSSION
Complementation and transcriptional profile of the N. gonorrhoeae nrrF mutant.
We examined the relative levels of nrrF expression in FA1090 and in the complemented mutant to confirm that the complemented strain expressed appropriate levels of the transcript. Fe−/Fe+ ratios, as determined by qRT-PCR and estimation of Fe-repressed NrrF expression at 3 h for FA1090 and the complemented mutant, were 59.9 ± 10.1 and 50.2 ± 18.9 (means ± standard errors of the means [SEM]), respectively, indicating that the complementation construct restored NrrF expression to wild-type levels and that expression was appropriately Fe regulated. The absence of nrrF transcription in nrrF mutant cells was confirmed by qRT-PCR (data not shown). In agreement with Fe repression of nrrF and the location of a Fur box in the NrrF promoter (24), a DNA fragment containing the NrrF promoter bound gonococcal Fur with high affinity (KD, 82 nM) as measured by EMSA (Fig. 1).
Fig 1.
The NrrF promoter binds with high affinity to the gonococcal Fur protein. EMSA reactions were carried out using 25 nM each respective DNA target with increasing concentrations (μM) of gonococcal Fur protein (given below each lane). Promoter controls included NGO1779 (fur) and the nonspecific DNA fragment NGO1809 (NS), which does not contain a Fur box. (A, B, and C) Representative EMSA gels of nrrF, fur, and nonspecific NGO1809, respectively. (D) The amounts of DNA in bound and unbound bands were measured and were plotted to estimate the percentage of binding. The KD was defined as the concentration of Fur required for a 50% shift of the target DNA and is representative of two separate experiments.
The Fe-regulated global transcription profile of FA1090 in these experiments was similar to previous microarray data (see Table S2 in the supplemental material) (13, 15), as were global transcript levels in the complemented strain in response to Fe starvation (see Table S2). Of the 349 genes whose expression in FA1090 was identified as being differentially regulated, 124 were upregulated and 225 were downregulated in response to Fe availability (see Table S2). Additionally, the growth of wild-type, mutant, and complemented strains under iron-depleted and iron-replete conditions was essentially the same (see Fig. S1 in the supplemental material). Transcriptional analysis of the nrrF mutant strain showed that 107 genes were upregulated and 338 genes were downregulated when iron was limiting (see Table S3 in the supplemental material). When we sorted the differentially expressed genes according to COGs (Clusters of Orthologous Groups) categories, hypothetical proteins accounted for the majority, followed by phage-associated proteins, proteins involved in cellular processes, and transport proteins (see Fig. S5 in the supplemental material); the distribution of genes in these COGs categories is comparable to the distribution of Fe-regulated genes in FA1090 (present study; also reference 15).
Effects of NrrF on the sdhCDAB operon.
The sdhC and sdhA loci have been reported to be regulated by NrrF in N. meningitidis (22, 23). Thus, it was not surprising to find an increase in the half-life of sdhA (NGO0921) in the mutant (Table 1); however, we detected no differences in half-life for the mRNA corresponding to the first gene in the operon, sdhC (Table 1). We observed a slight increase in the half-life of sdhD and a significant increase in the half-life of sdhB (Table 1). The succinate dehydrogenase enzyme complex, comprising four subunits, functions in the Krebs cycle as well as the electron transport chain. The SdhC and SdhD (SdhC/D) subunits form the membrane-bound portion of the complex, which interacts with ubiquinone and anchors the catalytic domain, while the cytoplasmic portion of the complex includes SdhA (a flavoprotein) and SdhB, a protein containing three iron-sulfur clusters (29, 30). Our data suggest that NrrF appears to preferentially affect the turnover of the 3′ portion of the mRNA. Given the fact that SdhA/B can exhibit succinate dehydrogenase activity in the absence of SdhC/D, NrrF appears to target sdhAB for degradation as a means of both regulating enzymatic activity and lowering the Fe requirement of the organism. In E. coli, RyhB has been proposed to bind between the first and second genes (sdhCD) in the succinate dehydrogenase operon (31). Similarly, meningococcal NrrF binds to the region in the mRNA that overlaps the sdhDA junction (23), consistent with our findings of increased message degradation of sdhA and the downstream gene sdhB (Table 1). Since N. gonorrhoeae has an incomplete tricarboxylic acid (TCA) cycle (32), interruption of Sdh activity by this combined mechanism would uncouple the TCA cycle from oxidative phosphorylation, reducing the necessity for Fe-containing enzymes in a variety of systems. If the regulation of the sdh transcript were entirely under the control of NrrF, then the repression by NrrF during Fe-limited growth should have been alleviated in the mutant. In fact, all genes in the sdh operon remained downregulated in the mutant under Fe− conditions: expression levels were roughly half of those seen in FA1090 (Table 1). This suggests the presence of a second, NrrF-independent regulator, possibly another Fe-regulated sRNA, controlling sdh operon expression levels.
Table 1.
NrrF-mediated regulation of the Sdh operon
| Locus tag | Annotation | Fold change in expression (Fe−/Fe+)a |
Half-life (min)b |
||
|---|---|---|---|---|---|
| FA1090 | nrrF mutant | FA1090 | nrrF mutant | ||
| NGO0923 | sdhC | −9.4 ± 2.6 | −3.8 ± 1.1 | 1.2 ± 0.2 | 1.5 ± 0.2 |
| NGO0922 | sdhD | −10.2 ± 2.3 | −3.3 ± 0.9 | 1.4 ± 0.2 | 2.1 ± 0.4 |
| NGO0921 | sdhA | −7.1 ± 2.1 | −3.9 ± 0.5 | 1.3 ± 02 | 2.7 ± 0.5c |
| NGO0920 | sdhB | −6.7 ± 2.6 | −3.0 ± 0.9 | 1.3 ± 0.2 | 2.5 ± 0.6c |
qRT-PCR data from the 3-h microarray samples. Values reported are means for three experiments ± SEM. For fold change ratios of <1, the reciprocal was taken and a minus sign added.
Half-lives were measured in Fe− medium. Values reported for each gene are means of at least two measurements ± SEM.
P, <0.05 for comparison of the nrrF mutant to FA1090.
Identification of additional genes potentially regulated by NrrF.
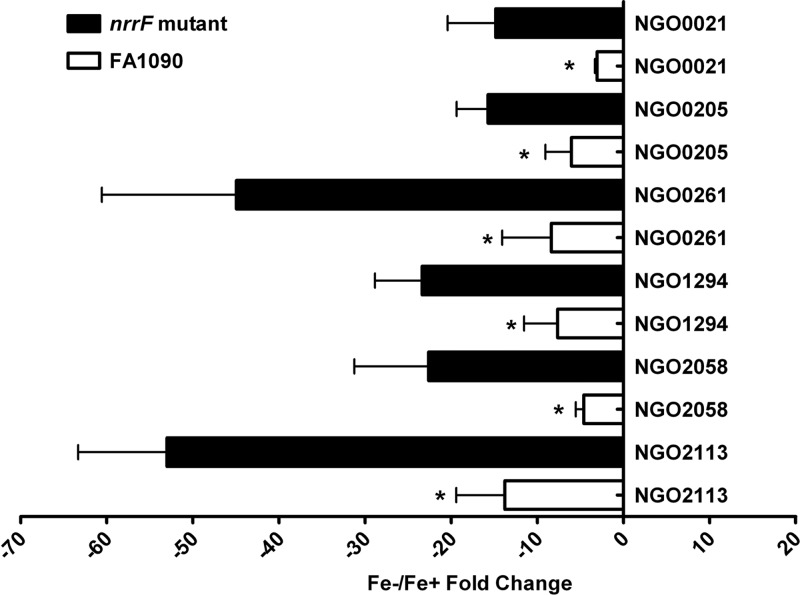
In general, NrrF appears to counteract the effect of transcriptional repression mediated by Fur, essentially buffering the repressive effect of Fur on the transcriptome. Comparison of genes whose Fe−/Fe+ expression ratios were significantly changed revealed a subset of genes that appeared to be dysregulated in the nrrF mutant, in that the Fe−/Fe+ expression ratio differed 2-fold or more from the expression observed in FA1090. Compared to that in wild-type FA1090, the repression of several downregulated genes was increased in the NrrF mutant (Fig. 2; see also Table S2 in the supplemental material), including that of genes encoding two hypothetical proteins (NGO2058 and NGO2113). This gene set includes the gene encoding TdfF (NGO0021), a putative TonB-dependent transporter (33) present only in pathogenic Neisseria species (34). A gonococcal strain with a mutation in tdfF, encoding this uncharacterized iron transporter, was unable to survive in human cervical epithelial cell growth assays, and addition of iron rescued the defect, implicating TdfF in intracellular iron transport under conditions similar to those seen in the host (35). Modulation of Fur-mediated repression of the tdfF transcript by the action of NrrF could be necessary to ensure that modest amounts of this protein would be expressed to prime the organism for intracellular growth even if extracellular bacteria encounter an environment high in available iron, such as that during menses. Lipoprotein transport may also be affected; while the level of repression of the lipoprotein carrier NGO0205 (lolA) was approximately 10-fold in FA1090, it was >40-fold in the NrrF mutant (Fig. 2). LolA acts as a chaperone to ferry lipoproteins transported into the periplasmic space to LolB for incorporation into the outer membrane (36). Lipoproteins are important in bacterial cellular functions such as signal transduction, antibiotic resistance, adhesion to tissues, and establishment of host adaptive immune responses (37). We detected no transcriptional changes for the lolCDE loci, involved in transporting the lipoproteins across the inner membrane, in either the mutant or the wild-type strain (see Table S2). NrrF may modulate the lipoprotein content of the outer membrane, but if so, this effect may not be straightforward, and additional factors may be involved. Compared to that of FA1090, a putative lipoprotein (NGO1321) exhibited enhanced (2-fold) expression in the NrrF mutant (see Table S2); this would run counter to the reduced expression of LolA.
Fig 2.
Global effect of NrrF. The level of repression of gene expression in the nrrF mutant was increased over that in FA1090. Each bar represents the mean fold change for 3 separate microarray experiments; error bars, SEM. Asterisks indicate a P value of ≤0.05 for comparison of the mean fold change in the expression of a gene in the nrrF mutant with that of the same gene in FA1090.
Control of mRNA turnover.
If NrrF controls gene expression by affecting the rate of mRNA turnover, then apparent Fe “induction” of a transcript could be due to an increase in the half-life of this transcript under Fe+ conditions, when NrrF is repressed by Fe. Accordingly, potential NrrF-regulated candidates were identified as transcripts whose expression in FA1090 was significantly decreased 1.5-fold or more under Fe− conditions from that under Fe+ conditions, while no significant fold change was observed in the mutant under the same conditions (see Table S2 in the supplemental material). Candidates were further examined using quantitative real-time PCR to validate the microarray results (Table 2). To examine whether our results may be due to an effect of nrrF on mRNA turnover, we measured the half-lives of transcripts for these genes after rifampin was added to stop transcription. In each instance, we found that the nrrF mutation significantly increased the mRNA half-life, suggesting that NrrF acts to control transcript levels by increasing the rate of mRNA turnover (Table 2). NrrF appears to affect the transcript levels of genes encoding proteins involved in a variety of distinct processes, including energy metabolism (see above), biosynthesis of amino acids, response to oxidative stress and DNA damage, and regulation of gene expression, as well as a complex regulatory circuit controlling at least one antibiotic efflux pump. Although further work will be needed to fully understand the role of NrrF in the iron regulon and infection, we can combine the observations from Fig. 2 and Table 2 to propose reasonable hypotheses to describe the effects NrrF may have on Fe demands, metabolism, and the overall stress response of the organism.
Table 2.
Candidate NrrF-regulated genes
| Locus tag | Annotation | Fold change in expression (Fe−/Fe+)a |
Half-life (min)b |
||
|---|---|---|---|---|---|
| FA1090 | nrrF mutant | FA1090 | nrrF mutant | ||
| NGO0224 | ntpA (pyrophosphohydrolase) | −1.9 ± 0.1 | NC | 0.4 ± 0.1 | 1.7 ± 0.2 |
| NGO0588 | Hypothetical protein | −1.9 ± 0.4 | NC | 1.6 ± 0.2 | 3.1 ± 0.5 |
| NGO0797 | XRE transcriptional regulator | −2.0 ± 0.7 | NC | 0.7 ± 0.1 | 2.1 ± 0.4 |
| NGO0802 | Hypothetical protein | −2.0 ± 0.6 | NC | 1.0 ± 0.1 | 3.1 ± 0.1 |
| NGO1024 | Hypothetical protein | −3.1 ± 0.7 | NC | 0.7 ± 0.1 | 1.8 ± 0.3 |
| NGO1368 | mtrF (efflux pump component) | −2.1 ± 0.5 | NC | 1.3 ± 0.1 | 2.3 ± 0.3 |
| NGO1446 | Hypothetical protein, YadA-like | −3.2 ± 1.0 | NC | 0.6 ± 0.1 | 1.7 ± 0.5 |
| NGO1561 | exoIII (exoDNase III) | −3.0 ± 0.9 | NC | 0.9 ± 0.3 | 2.3 ± 0.3 |
| NGO1861 | Hypothetical protein | −2.6 ± 0.8 | 3.3 ± 1.2 | 1.6 ± 0.2 | 3.1 ± 0.5 |
| NGO2075 | thrB (homoserine kinase) | −4.6 ± 1.1 | NC | 1.7 ± 0.3 | 3.3 ± 0.6 |
qRT-PCR data from the 3-h microarray samples. Values reported are means for three experiments ± SEM. For fold change ratios of <1, the reciprocal was taken and a minus sign added. NC (no change) indicates a fold change of <1.5.
Half-lives were measured in Fe− medium. Values reported for each gene are means of at least two measurements ± SEM. For all genes listed, the differences in half-life between the nrrF mutant and FA1090 were significant (P, <0.05).
DNA metabolism and oxidative stress.
Two observations suggest that NrrF may aid in fine-tuning the oxidative health of the cell. We noted an increase in the half-life of NGO0224 (ntpA) mRNA in the mutant (Table 2). NGO0224 encodes a protein in the Nudix hydrolase superfamily of pyrophosphohydrolases, which typically targets intact and oxidatively damaged nucleoside triphosphates, dinucleotide polyphosphates, nucleotide sugars, and dinucleotide enzymes, removing potentially mutagenic nucleotide metabolites (38, 39). Nudix hydrolases have also been implicated in the regulation of metabolism (40). The transcript for gonococcal exoDNase (NGO1561) had an increased half-life in the nrrF mutant (Table 2). In N. meningitidis, the exoIII homolog NMB0399, encoding a 3′ phosphodiesterase, neisserial exonuclease (NExo), protects the cell from oxidative stress and is necessary for virulence in a rat model of infection (41). DNA repair by several mechanisms, including base excision repair (42), is essential during the oxidative burst from phagocytic cells in the host or oxidative stress due to the accumulation of potentially cytotoxic mutations through replication during pathogenesis. However, under low-Fe conditions, oxidative stress on the organism is likely to be reduced, suggesting a reduced need for NExo activity. In this instance, NrrF may act to adjust the NExo transcript levels accordingly.
Amino acid biosynthesis.
Included in the subset of dysregulated genes (Fig. 2) was NGO0261 [N-(5′-phosphoribosyl)anthranilate isomerase], the second committed step in tryptophan biosynthesis. The half-life of NGO2075 (thrB) mRNA was increased in the NrrF mutant compared to wild-type FA1090. NGO2075 (thrB) encodes a homoserine kinase involved in threonine biosynthesis and has been well characterized in E. coli (43) (Table 2). Reduction of thrB mRNA levels by NrrF could impact the accumulation of branched-chain amino acids and glycine (44). Thus, NrrF could also stimulate cysteine and methionine biosynthesis (45). Finally, we observed that the ∼10-fold repression of NGO1294 (an Lrp [leucine-responsive regulatory protein] transcriptional regulator) in FA1090 was increased to approximately 45-fold repression in the NrrF mutant (Fig. 2). The Lrp family of global regulators consists of small DNA-binding proteins involved in the synthesis and degradation of amino acids (46). By monitoring leucine as an indicator of protein availability (leucine is the most abundant amino acid in proteins), E. coli Lrp facilitates the transition between growth under nutrient-rich and nutrient-poor conditions (47, 48). Assuming that gonococcal Lrp acts similarly, these data may suggest that NrrF modulation of Fur-mediated repression of Lrp acts to buffer the Lrp-mediated response to nutrient-poor conditions. However, considering the pleiotropic nature of Lrp regulation, the result of this action by NrrF is not clear.
Hypothetical proteins.
NGO0797 is a member of the helix-turn-helix (HTH) xenobiotic response element (XRE) family of transcription factors (49). These global regulatory proteins have two alpha-helices, with the second helix containing the DNA recognition motif (50). The half-life of NGO0797 mRNA was approximately 3-fold longer in the NrrF mutant than in the wild type (Table 2). This suggests that NrrF participates in adjusting the transcript levels for this protein in response to Fe availability. While we do not currently know the targets of this regulatory protein, these data indicate that NrrF alters the control that this protein exerts over downstream gene expression and thus extends the effect of the Fur protein deeper into the gonococcal transcriptome.
The half-lives of the mRNAs for five additional hypothetical proteins were increased in the mutant strain (Table 2). Of these, two warrant mention. NGO0588 is 140 amino acids long, with a predicted signal peptide cleavage site between amino acids 22 and 23 (51), and is included in pfam13827 (52), a family of proteins of unknown function but containing six well-conserved cysteines. It may be that NGO0588 expression is regulated by NrrF due to the effect of this protein on cellular Fe demand, since cysteine residues are often used as coordination sites for Fe3+ ions (53). NGO1446 includes a conserved domain belonging to the pfam03895 family, representing the C-terminal 120 amino acids of an outer membrane adhesion protein, YadA, present in enteropathogenic Yersinia species. The YadA-like domain in NGO1446 is located midway in the protein sequence. YadA is essential for the invasion of epithelial cells and for increased resistance to the adaptive immune response; later in infection, it is responsible for adhesion to collagen, initiating a sterile inflammation of the joints (54). The general topology for this protein involves the oligomerization of three YadA proteins to form a lollipop structure with the head at the N termini, an intervening stalk, and the C-terminal end harboring the outer membrane anchor. As noted above, these data suggest that NrrF may modulate the expression of genes important for interactions with the host.
Antibiotic efflux.
The clearest picture that we have of the regulatory relationships in which NrrF is modulating transcript levels is presented by mtrF (NGO1368) and NGO1024. We observed that the half-lives for these mRNAs were significantly increased in the nrrF mutant (Table 2), suggesting that NrrF facilitates the turnover of these mRNAs. Recent data for N. gonorrhoeae indicate a role for NGO1024 in unsaturated fatty acid biosynthesis (55). MtrF is a putative inner membrane protein involved in high-level resistance to hydrophobic agents mediated by the mtrCDE efflux pump (56). MtrF has been proposed either to shuttle antimicrobials to the pump for export or to help stabilize one or more components of the efflux pump (56, 57). MtrF and NGO1024 appear to be linked in a regulatory sense, in that the transcripts for these proteins are controlled directly or indirectly by at least four regulatory elements, including NrrF, Fur, and two additional transcriptional repressors, MtrR and MpeR (57, 58). The relationships between these regulatory elements are summarized in Fig. 3. Under high-Fe conditions (Fig. 3A), Fur directly represses both MpeR (58) and NrrF (24). This relieves the MpeR-mediated repression of mtrF, which is nevertheless still transcribed at a low level, because this locus is repressed by MtrR under high-Fe conditions; MtrR repression is itself relieved indirectly by the Fur-mediated repression of MpeR. MtrR also holds the transcription of NGO1024 at low levels, although it appears that this locus may be transcribed at a somewhat higher level than mtrF, even under MtrR repression (57). NrrF in this instance is efficiently repressed by Fur, and its effects on the mtrF and NGO1024 transcripts appear negligible. MtrR also directly represses the mtrCDE efflux pump and rpoH (sigma 32) (57) (not shown in Fig. 3). Our data do not suggest a role for NrrF in controlling these other loci.
Fig 3.
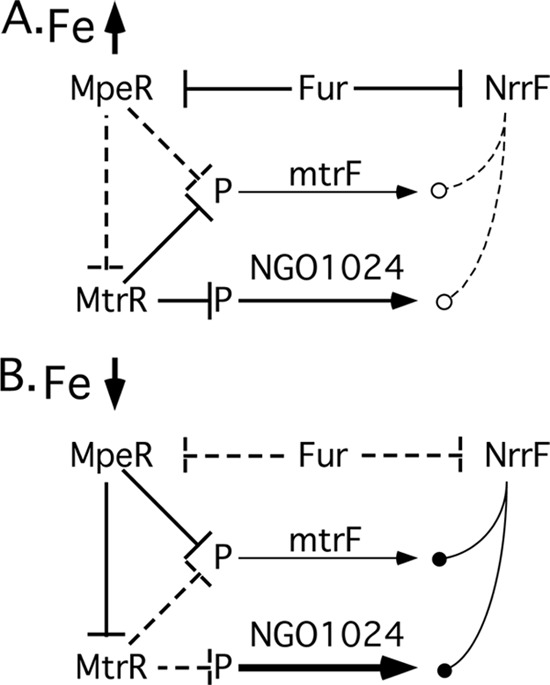
Proposed model of MtrF regulation, including NrrF. (A) Under Fe-replete conditions, MpeR and NrrF transcription is repressed by Fur, relieving MpeR-mediated repression of mtrF and nominal effects of NrrF on either mtrF or NGO1024 transcripts. (B) Under low-Fe conditions, transcription of MpeR and NrrF is derepressed, allowing MpeR to suppress MtrR and MtrF transcription, and allowing increased message turnover of mtrF and NGO1024 by NrrF. Solid perpendicular lines indicate repression of the transcript, and dashed perpendicular lines indicate derepression. Solid lines with filled circles represent enhanced message degradation by NrrF; conversely, diminution of the effects of NrrF are represented by dashed lines with open circles. P, promoter region.
In contrast, under low-Fe conditions (Fig. 3B), Fur repression of both MpeR and NrrF is relieved. This results in the production of active MpeR repressor, which inhibits MtrR transcription, thereby relieving the repression of NGO1024 transcription for higher-level expression. MpeR also replaces MtrR at the mtrF promoter, continuing to control mtrF transcription at a low level. NrrF transcription is derepressed, and the expression of this sRNA acts to significantly increase the turnover of both the mtrF and NGO1024 transcripts. This has the effect of further lowering the level of mtrF transcript in the cell, suggesting that overexpression of MtrF may have deleterious effects on the cell and that mtrF is therefore rigorously regulated at both the transcriptional (MtrR and MpeR) and posttranslational (NrrF) levels. Similarly, NrrF expression reduces NGO1024 transcript levels, buffering the cell against overexpression of NGO1024 under low-Fe conditions due to the relief of MtrR repression by MpeR. Coordinating the expression of one component of an efflux pump for hydrophobic compounds (mtrF) with the expression of at least one locus that may participate in synthesizing these hydrophobic compounds (NGO1024) makes sense from a biological viewpoint. Assuming that NGO1024 is involved in unsaturated fatty acid biosynthesis, the role of NrrF here may be to modulate the synthesis of these hydrophobic compounds. Overproduction of these fatty acids may be deleterious to the cell under low-iron conditions, where the activity of the mtr efflux pump may be unchanged and perhaps would be unable to deal with the increased load of intracellular hydrophobic compounds produced were NGO1024 expression to be unrestricted.
In summary, we deleted NrrF in FA1090 and globally assessed the role of NrrF under iron-limited conditions by using microarray analysis and monitoring mRNA turnover for selected genes. NrrF appears to act as an accessory regulator that modulates overall transcriptional control by posttranscriptionally adjusting mRNA levels in response to Fe availability and by buffering Fur repression.
Supplementary Material
ACKNOWLEDGMENTS
This project and its publication were supported by the National Center for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health through grant 8P20GM103447.
Footnotes
Published ahead of print 13 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00839-13.
REFERENCES
- 1.Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron—a metal at the host-pathogen interface. Cell. Microbiol. 12:1691–1702 [DOI] [PubMed] [Google Scholar]
- 2.Rohde K, Dyer D. 2003. Mechanisms of iron acquisition by the human pathogens Neisseria meningitidis and Neisseria gonorrhoeae. Front. Biosci. 8:d1186–d1218 [DOI] [PubMed] [Google Scholar]
- 3.Andrews S, Robinson A, Rodriquez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237 [DOI] [PubMed] [Google Scholar]
- 4.Cartron M, Maddocks S, Gillingham P, Craven CJ, Andrews S. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157 [DOI] [PubMed] [Google Scholar]
- 5.Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect. Immun. 77:2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escolar L, Pérez-Martín J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal S, Sebastian S, Szmigielski B, Rice PA, Genco CA. 2008. Expression of the gonococcal global regulatory protein Fur and genes encompassing the Fur and iron regulon during in vitro and in vivo infection in women. J. Bacteriol. 190:3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies BW, Bogard RW, Mekalanos JJ. 2011. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc. Natl. Acad. Sci. U. S. A. 108:12467–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao H, Zhou D, Li Y, Guo Z, Han Y, Song Y, Zhai J, Du Z, Wang X, Lu J, Yang R. 2008. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190:3063–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK. 2010. Transcriptomic response of Listeria monocytogenes to iron limitation and fur mutation. Appl. Environ. Microbiol. 76:406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delany I, Grifantini R, Bartolini E, Rappuoli R, Scarlato V. 2006. Effect of Neisseria meningitidis Fur mutations on global control of gene transcription. J. Bacteriol. 188:2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delany I, Rappuoli R, Scarlato V. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081–1090 [DOI] [PubMed] [Google Scholar]
- 13.Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi A, Rappuoli R, Grandi G, Genco C. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U. S. A. 100:9542–9547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson L, Ducey T, Day M, Zaitshik J, Orvis J, Dyer DW. 2010. Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J. Bacteriol. 192:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu C, Genco CA. 2012. Fur-mediated global regulatory circuits in pathogenic Neisseria species. J. Bacteriol. 194:6372–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards GR, Vanderpool CK. 2011. Molecular call and response: the physiology of bacterial small RNAs. Biochim. Biophys. Acta 1809:525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43:880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3(12):a003798. 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papenfort K, Vogel J. 2010. Regulatory RNA in bacterial pathogens. Cell Host Microbe 8:116–127 [DOI] [PubMed] [Google Scholar]
- 21.Massé E, Salvail H, Desnoyers G, Arguin M. 2007. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10:140–145 [DOI] [PubMed] [Google Scholar]
- 22.Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. 2007. A novel Fur- and iron-regulated small RNA, NrrF, is required for indirect Fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189:3686–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metruccio MME, Fantappiè L, Serruto D, Muzzi A, Roncarati D, Donati C, Scarlato V, Delaney I. 2009. The Hfq-dependent small noncoding RNA NrrF directly mediates Fur-dependent positive regulation of succinate dehydrogenase in Neisseria meningitidis. J. Bacteriol. 191:1330–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ducey T, Jackson L, Orvis J, Dyer D. 2009. Transcript analysis of nrrF, a Fur repressed sRNA of Neisseria gonorrhoeae. Microb. Pathog. 46:166–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyer D, West E, Sparling P. 1987. Effects of serum carrier proteins on the growth of pathogenic neisseriae with heme-bound iron. Infect. Immun. 55:2171–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932 [DOI] [PubMed] [Google Scholar]
- 27.Mehr IJ, Seifert HS. 1997. Random shuttle mutagenesis: gonococcal mutants deficient in pilin antigenic variation. Mol. Microbiol. 23:1121–1131 [DOI] [PubMed] [Google Scholar]
- 28.Jackson L, Dyer D. 2012. Protocol for gene expression profiling using DNA microarrays in Neisseria gonorrhoeae. Methods Mol. Biol. 903:343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNeil MB, Fineran PC. 2013. Prokaryotic assembly factors for the attachment of flavin to complex II. Biochim. Biophys. Acta 1827:637–647 [DOI] [PubMed] [Google Scholar]
- 30.Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. 2003. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299:700–704 [DOI] [PubMed] [Google Scholar]
- 31.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holten E. 1976. Radiospirometric studies in genus Neisseria. 2. The catabolism of glutamate and fumarate. Acta Pathol. Microbiol. Scand. B 84:1–8 [PubMed] [Google Scholar]
- 33.Cornelissen CN, Hollander A. 2011. TonB-dependent transporters expressed by Neisseria gonorrhoeae. Front. Microbiol. 2:117. 10.3389/fmicb.2011.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marri PR, Paniscus M, Weyand NJ, Rendón MA, Calton CM, Hernandez D, Higashi D, Scodergren E, Weinstock G, Rounsley S, So M. 2010. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One 5:e11835. 10.1371/journal.pone.0011835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagen TA, Cornelissen CN. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol. Microbiol. 62:1144–1157 [DOI] [PubMed] [Google Scholar]
- 36.Liechti G, Goldberg J. 2012. Outer membrane biogenesis in Escherichia coli, Neisseria meningitidis, and Helicobacter pylori: paradigm deviations in H. pylori. Front. Cell. Infect. Microbiol. 2:29. 10.3389/fcimb.2012.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J. 279:4247–4268 [DOI] [PubMed] [Google Scholar]
- 38.McLennan A. 2006. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63:123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLennan A. 2013. Substrate ambiguity among the nudix hydrolases: biologically significant, evolutionary remnant, or both? Cell. Mol. Life Sci. 70:373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galperin MY, Moroz OV, Wilson KS, Murzin AG. 2006. House cleaning, a part of good housekeeping. Mol. Microbiol. 59:5–19 [DOI] [PubMed] [Google Scholar]
- 41.Carpenter EP, Corbett A, Thomson H, Adacha J, Jensen K, Bergeron J, Kasampalidis I, Exley R, Winterbotham M, Tang C, Baldwin G, Freemont P. 2007. AP endonuclease paralogues with distinct activities in DNA repair and bacterial pathogenesis. EMBO J. 26:1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidsen T, Tuven HK, Bjørås M, Rødland EA, Tønjum T. 2007. Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J. Bacteriol. 189:5728–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chassagnole C, Raïs B, Quentin E, Fell DA, Mazat JP. 2001. An integrated study of threonine-pathway enzyme kinetics in Escherichia coli. Biochem. J. 356:415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fondi M, Brilli M, Emiliani G, Paffetti D, Fani R. 2007. On the origin and evolution of biosynthetic pathways: integrating microarray data with structure and organization of the Common Pathway genes. BMC Bioinformatics 8(Suppl 1):S12. 10.1186/1471-2105-8-S1-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brinkman AB, Ettema TJG, De Vos WM, Van Der Oost J. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287–294 [DOI] [PubMed] [Google Scholar]
- 47.Calvo JM, Matthews RG. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landgraf JR, Boxer JA, Calvo JM. 1999. Escherichia coli Lrp (leucine-responsive regulatory protein) does not directly regulate expression of the leu operon promoter. J. Bacteriol. 181:6547–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aravind L, Anantharamna V, Balaji S, Babu M, Iyer L. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231–262 [DOI] [PubMed] [Google Scholar]
- 50.Wintjens R, Rooman M. 1996. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262:294–313 [DOI] [PubMed] [Google Scholar]
- 51.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 52.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger L, Sonnhammer E, Eddy S, Bateman A, Finn D. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon M, Bachovchin D, Mowen K, Baker D, Cravatt B. 2010. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468:790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isabella VM, Clark VL. 2011. Identification of a conserved protein involved in anaerobic unsaturated fatty acid synthesis in Neisseria gonorrhoeae: implications for facultative and obligate anaerobes that lack FabA. Mol. Microbiol. 82:489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Folster JP, Shafer WM. 2005. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J. Bacteriol. 187:3713–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Folster JP, Johnson PJT, Jackson L, Dhulipali V, Dyer DW, Shafer WM. 2009. MtrR modulates rpoH expression and levels of antimicrobial resistance in Neisseria gonorrhoeae. J. Bacteriol. 191:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mercante A, Jackson L, Johnson P, Stringer V, Dyer D, Shafer WM. 2012. MpeR regulates the mtr efflux locus in Neisseria gonorrhoeae and modulates antimicrobial resistance by an iron-responsive mechanism. Antimicrob. Agents Chemother. 56:1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.