Abstract
The gene transfer agent of Rhodobacter capsulatus (RcGTA) is a bacteriophage-like genetic element with the sole known function of horizontal gene transfer. Homologues of RcGTA genes are present in many members of the alphaproteobacteria and may serve an important role in microbial evolution. Transcription of RcGTA genes is induced as cultures enter the stationary phase; however, little is known about cis-active sequences. In this work, we identify the promoter of the first gene in the RcGTA structural gene cluster. Additionally, gene transduction frequency depends on the growth medium, and the reason for this is not known. We report that millimolar concentrations of phosphate posttranslationally inhibit the lysis-dependent release of RcGTA from cells in both a complex medium and a defined medium. Furthermore, we found that cell lysis requires the genes rcc00555 and rcc00556, which were expressed and studied in Escherichia coli to determine their predicted functions as an endolysin and holin, respectively. Production of RcGTA is regulated by host systems, including a putative histidine kinase, CckA, and we found that CckA is required for maximal expression of rcc00555 and for maturation of RcGTA to yield gene transduction-functional particles.
INTRODUCTION
Gene transfer agents (GTAs) are bacteriophage-like particles that have no known function other than the transfer of host-derived DNA segments from a donor bacterium to recipient cells. In recent years, GTA-transducing particles or genes encoding putative GTAs have been discovered in a wide variety of bacterial species, indicating that GTA-mediated horizontal gene transfer is widespread and diverse in nature (1). Furthermore, it has been suggested that the rate at which gene transfer occurs under natural conditions is extremely high and may rival or exceed the rate at which other horizontal gene transfer mechanisms occur (2). The GTA produced by Rhodobacter capsulatus (RcGTA) was the first GTA to be discovered (3), and most members of the family Rhodobacteraceae harbor a gene cluster homologous to the RcGTA structural genes (4). These RcGTA-like genes appear to have been propagated by vertical transmission in the alphaproteobacterial lineage from a single ancestor. Recently, several other alphaproteobacteria have been reported to produce functional RcGTA-like particles: Roseovarius nubinhibens, Nitratireductor sp. strain 44B9s, and Ruegeria pomeroyi (2, 5, 6). In addition, species of Bartonella and Brachyspira produce functional GTA particles (BaGTA and VSH-1, respectively) that are not homologous to RcGTA (7–10). The genetic conservation and demonstrated functionality of RcGTA-like particles indicate that they serve a beneficial role for the host population, because deletion bias eliminates genes that serve no benefit to the host (11, 12), and this view was supported by an analysis of RcGTA codon substitution rates (1). Similarly, Guy et al. argued that the BaGTA produced by Bartonella has been maintained to facilitate adaptive evolution between the bacterium and its host (8).
Morphologically, RcGTA resembles a small, tailed bacteriophage (phage); however, RcGTA particles do not form plaques on a bacterial lawn, and the ∼4 kb of packaged DNA (13) is insufficient to encode the RcGTA gene cluster (∼15 kb). RcGTA packages essentially random genomic DNA segments from the host, as has been found for other GTAs, such as VSH-1 (Brachyspira hyodysenteriae) (13, 14).
RcGTA production is regulated by several host proteins, including the quorum-sensing proteins GtaI and GtaR (15), and proteins homologous to the sensor kinase CckA and response regulator CtrA proteins that control the cell cycle in Caulobacter crescentus (16, 17). Disruption of ctrA in R. capsulatus severely diminishes RcGTA mRNA levels and transduction frequencies, indicating that CtrA is required for transcription of the gene cluster (18). RcGTA transduction is similarly reduced by disruption of cckA; however, the RcGTA capsid protein is still produced, and so it appears that CckA is not needed for transcription of the RcGTA gene cluster (16, 19). Mercer et al. (16) recently reported that the CckA protein is needed for release of the RcGTA capsid protein from cells, but it remained unclear how release is blocked and whether the capsid protein that accumulates in ΔcckA mutant cells comprises functional RcGTA particles. Two recent reports indicated that RcGTA is produced by a subset of the bacterial population and that the fraction of RcGTA gene-expressing cells in the total cell population is increased in an RcGTA overproducer strain, DE442 (14, 20). Additionally, maximal gene transduction is dependent on the growth phase of recipient cultures, largely due to quorum-sensing regulation of the production of a capsule receptor needed for binding of RcGTA (21). The production of RcGTA is induced as cultures enter the stationary phase and particles are released through cell lysis (19, 20), which requires the endolysin encoded by rcc00555 (14).
Shortly after the discovery of RcGTA, a difference in transduction frequency was observed between different growth media: transduction frequencies were very low when cells were cultured in the defined medium RCV, while they were elevated in the complex medium YPS (13, 22). We have previously reported that the level of cell lysis of an RcGTA overproducer strain is similarly reduced in RCV compared to that in YPS (20). The reason for this difference between the growth media was never determined.
In this study, we analyzed the RcGTA gene cluster promoter region, and in this report, we describe two processes involved in the release of RcGTA particles from R. capsulatus cells. We report that millimolar concentrations of inorganic phosphate inhibit the release of RcGTA from cells and that high transduction frequencies are obtained in an RCV-derived defined medium containing reduced concentrations of phosphate. This effect did not require a homologue of PhoB or the depletion of inorganic phosphate from the medium. We provide direct evidence that the endolysin (rcc00555) and holin (rcc00556) are required for cell lysis of R. capsulatus. Expression of rcc00555 in Escherichia coli made cells susceptible to lysis, and expression of rcc00556 halted growth and permeabilized cells to propidium iodide. We also discovered that the putative histidine kinase CckA regulates the expression of the endolysin encoded by rcc00555 in a phosphate concentration-independent pathway. Furthermore, CckA was found to be required for maximal intracellular production of mature, fully functional RcGTA particles. The RcGTA promoter region was investigated, and we propose that the RcGTA open reading frame (ORF) g1 (rcc01682) transcriptional start site is located approximately 215 bp 5′ of the coding region.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains are described in Table 1. RcGTA production and cell lysis experiments were carried out on the genome-sequenced strain SB1003 (23) and the RcGTA overproducer DE442. RcGTA promoter mapping experiments were carried out on strain Y262 (13). For GTA-mediated transduction bioassays, wild-type (WT) rifampin-sensitive strain B10 was used as the recipient (24). For all experiments, cultures were inoculated to ∼20 Klett units (optical density at 660 nm [OD660], approximately 0.15; 7.2 × 107 CFU/ml), grown photoheterotrophically in 16.5-ml capped glass tubes at 30°C with illumination, and harvested after 36 h (early stationary phase), unless otherwise specified. The R. capsulatus culture growth media were modifications of either the complex yeast extract/peptone medium YPS (25) or the defined medium RCV (26). The modified YPS (YPSm) medium contained a reduced concentration of MgSO4 and CaCl2 (0.5 mM each), pH 6.8; the modified RCV (RCVm) medium lacked KPO4 (the only phosphate source in RCV), contained 14.7 mM K+ (by neutralizing the malic acid carbon source with KOH), and was buffered with 20 mM 3-morpholinopropane-1-sulfonic acid (MOPS), adjusted to pH 6.8 with HCl. Cultures were supplemented with salts adjusted to pH 6.8 with HCl or NaOH to an initial concentration, as described below, and supplemented with kanamycin sulfate (10 μg/ml), gentamicin sulfate (3 μg/ml), or tetracycline HCl (0.5 μg/ml), as appropriate. Culture turbidity was monitored using a Klett-Summerson photometer (filter 66). For resuspension experiments, strains were cultured in RCVm containing 10 mM KPO4, and cells were harvested by centrifugation and resuspended in RCVm defined medium or 20 mM Tris-HCl buffer (pH 6.8) to an OD660 of 5.0 or 0.5.
Table 1.
Bacterial strains and plasmids
| Strain | Reference or source | Description |
|---|---|---|
| Strains | ||
| SB1003 | 54 | R. capsulatus WT, Rifr |
| B10 | 24 | R. capsulatus WT, Rifs |
| DE442 | ?a | R. capsulatus RcGTA overproducer, Rifr |
| Y262 | 13 | R. capsulatus RcGTA overproducer, Rifr |
| DH5α | 27 | E. coli, general cloning strain |
| S17-1 λ pir | 28 | E. coli pir, plasmid conjugation |
| TEC5 | 29 | E. coli, plasmid conjugation |
| BL21(DE3) | Invitrogen | E. coli, protein overexpression |
| SB555 | 14 | R. capsulatus SB1003 Δrcc00555 Rifr Kanr |
| DE555 | This work | R. capsulatus DE442 Δrcc00555 Rifr Kanr |
| SB1003 cckA | 16 | R. capsulatus SB1003 ΔcckA Rifr Kanr |
| DEKOcckA | This work | R. capsulatus DE442 ΔcckA Rifr Kanr |
| Plasmids | ||
| pUC19 | Invitrogen | Cloning vector, Ampr |
| TOPO TA | Invitrogen | Cloning vector, Ampr |
| pZJD29A | J. Jiang and C. E. Bauer, unpublished data | Suicide plasmid, sacB Gmr |
| pRK415 | 31 | Broad-host-range vector, Tetr |
| pRCckA | This work | CckA complementation plasmid, Tetr |
| pR555 | 14 | rcc00555 complementation plasmid, Tetr |
| pIND4 | 55 | Inducible expression plasmid, Kanr |
| pIND4Gm | This work | Inducible expression plasmid, Gmr |
| pI556 | This work | rcc00556 complementation plasmid, inducible, Kanr |
| pI556Gm | This work | rcc00556 complementation plasmid, inducible, Gmr |
| pXCA601 | 32 | Reporter plasmid, lacZ Tetr |
| pXCA-555 | This work | rcc00555 promoter::lacZ fusion plasmid, Tetr |
| pET28a(+) | Novagen | E. coli T7 expression vector, inducible, Kanr |
| pET-555C | This work | rcc00555 expression plasmid, C-terminal 6-His tag, inducible, Kanr |
| p601-g65 | 15 | ORF g1 promoter::lacZ fusion plasmid, Tetr |
| p601-g15 | This work | Truncated ORF g1 promoter::lacZ fusion plasmid, no start codon, Tetr |
| p601-g64 | This work | Truncated ORF g1 promoter::lacZ fusion plasmid lacking the 5′-most 5′ end and the −10/−35 sequence, Tetr |
| p601-g61 | This work | Truncated ORF g1 promoter::lacZ fusion plasmid lacking both 5′ ends and the −10/−35 sequence, Tetr |
The strain is of uncertain provenance but is a crtD mutant probably derived from Y262 (B. Marrs, personal communication).
For construction of R. capsulatus chromosomal deletion mutants, E. coli strain DH5α (27) was used for general cloning work. E. coli strains S17-1 (28) containing λ pir and TEC5 (29) were used for conjugation of plasmids into R. capsulatus. For overexpression of rcc00555, E. coli BL21(DE3) was used, and cells were induced at an OD600 of 0.5 by addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). E. coli strains were cultured in LB medium (27) supplemented with either ampicillin (100 μg/ml), tetracycline HCl (10 μg/ml), kanamycin sulfate (50 μg/ml), or gentamicin sulfate (10 μg/ml), as appropriate.
Construction of targeted mutants and trans-complementation.
The plasmids used in this study are described in Table 1, and the PCR primer sequences are presented in Table 2. The R. capsulatus strain DE442-derived ΔcckA mutant was created by amplifying the genomic segment of SB1003 ΔcckA (16) containing the truncated cckA (interrupted by a kanamycin resistance cassette), using primers CckA-F and CckA-R. The resultant ∼1.8-kb amplicon was cloned into the plasmid pUC19 SmaI site. Replacement of the native DE442 cckA gene was performed by transduction, as described by Aklujkar et al. (30). DE555 (DE442 Δrcc00555) was created by RcGTA-mediated transduction of a KIXX-disrupted (kanamycin resistance) knockout fragment as described by Hynes et al. (14).
Table 2.
Primer sequences
| Name | Sequence | Purpose or description of sequence |
|---|---|---|
| CckA-F | TAAGTAGTCGACGATCTGGTGCTGGT | Amplification of cckA::KIXX fragment |
| CckA-R | ACAATGGTCGACACGCTTTCGCACAG | |
| CckAComp-F | ATATTCTAGAGGTGCTGGTCGATGCGCCCT | cckA complementation plasmid pRCckA; XbaI (italic) and HindIII (bold) sites are indicated |
| CckaComp-R | ATATAAGCTTCTGCAGCCCGAGACCGAGGC | |
| GTA2.6 | GAACCGGATCCATCGCCAGGG | Plasmids p601-g64 and p601-g61; the BamHI (underlined) site is indicated |
| GTA4 | CGCCTGCAGCAACCCTGAATATAGC | Plasmid p601-g64; the PstI (bold) site is indicated |
| GTA1 | ACGCTTCAAGCTGCAGATAAGGCATG | Plasmid p601-g61; the PstI (bold) site is indicated |
| GTA5 | GATGCGGCTGCAGACCGATCC | Plasmid p601-g15; the PstI (bold) site is indicated |
| GTA2.1 | CTCCAGCGGATCCACCGGAGG | Plasmid p601-g15; the BamHI (underlined) site is indicated |
| 555-F | ATATCTGCAGGCGTGCTGCCCGACCTCTTT | Plasmid pXCA-555; the PstI (bold) and BamHI (underlined) sites are indicated |
| 555-R | ATATGGATCCATCCGATCCCCCTTGGCTGAG | |
| GSP1 | CTTGTCTGCGAAGTTTTTCA | ORF g1 RACE |
| GSP3 | GCAATTTCCCGATAAAGCTCCTC | |
| 556-F | ATATCCATGGGGCTGATCGGGACGAT | Plasmid pI556 and pI556Gm; the NcoI (bold) and BamHI (underlined) sites are indicated |
| 556-R | ATATGGATCCTGCCTTTGCGGTGCCCGAAAA | |
| 555C-F | ATATCCATGGGATCTGTCTACGAGATTGC | Plasmid pET-555C; the NcoI (bold) and XhoI (underlined) sites are indicated |
| 555C-R | ATATCTCGAGGCCCCATGCCGCCACCCG | |
| aacC1-F | ATATCCCGGGTTGACATAAGCCTGTTCGGT | aacC1 gene conferring Gmr; the XmaI (bold) sites are indicated |
| aacC1-R | ATATCCCGGGCTTGAACGAATTGTTAGGTG |
The trans-complementing cckA-containing plasmid was constructed by amplifying DE442 genomic DNA with primers CckAComp-F and CckaComp-R. The resultant 2.8-kb fragment was cloned into the low-copy-number, broad-host-range plasmid pRK415 (31) by ligation to the introduced XbaI and HindIII sites to create pRCckA. Plasmid pRCckA was conjugated from E. coli S17-1 to DE442 ΔcckA using selection for tetracycline resistance.
Plasmid pIND4Gm was created by amplifying the aacC1 gene from plasmid pZJD29A using primers aacC1-F and aacC1-R. The resultant 0.78-kb fragment was ligated into the XmaI site of plasmid pIND4 to create plasmid pIND4Gm. The rcc00556 complementation/expression plasmids were constructed by amplifying DE442 genomic DNA with primers 556-F and 556-R. The resultant 0.67-kb fragment was cloned into pIND4 or pIND4Gm to create pI556 or pI556Gm, respectively, using the introduced NcoI and BamHI sites.
Construction of lacZ-fusion reporter and rcc00555 overexpression plasmid.
The RcGTA promoter reporter plasmid p601-g65 contains an in-frame fusion between the RcGTA ORF g1 ATG codon indicated codon 5 in Fig. 2A and the E. coli lacZ gene. The truncated promoter region plasmids p601-g64 and p601-g61 were constructed by amplifying R. capsulatus B10 chromosomal DNA using reverse primer GTA2.6 and forward primer GTA4 or GTA1 for p601-g64 and p601-g61, respectively. Similarly, plasmid p601-g15 was constructed using forward primer GTA5 and reverse primer GTA2.1. Plasmid pXCA-555, used to measure rcc00555 expression, was constructed using forward primer 555-F and reverse primer 555-R to amplify strain SB1003 DNA. The resultant PCR fragments were digested with PstI and BamHI and ligated into plasmid pXCA601 (32) to create an in-frame fusion to lacZ, as described for p601-g65 (15).
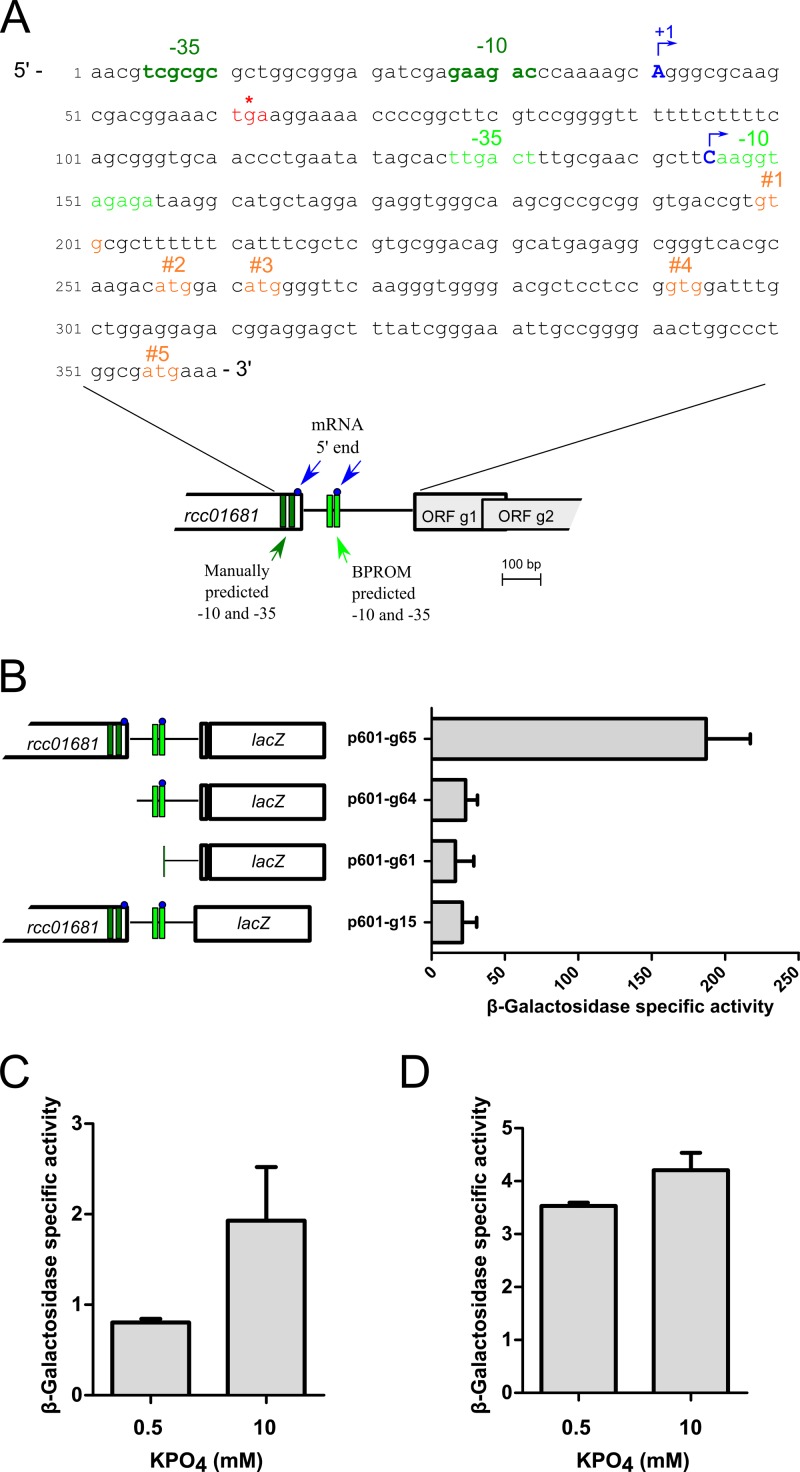
Fig 2.
Mapping of the RcGTA promoter and effect of phosphate concentration on RcGTA gene transcription. (A) Annotated sequence and schematic representation of the promoter region of ORF g1 of the RcGTA promoter region. Dark green and light green, −10 and −35 boxes manually annotated and identified by BPROM, respectively; blue, 5′ ends of mRNA identified using 5′ RACE; orange, putative translational start codons for ORF g1 from the literature; red asterisk, annotated stop codon of rcc01681. (B) Schematic representation of ORF g1::lacZ fusions constructs (left) and β-galactosidase activities of strain Y262 containing the indicated plasmid (right); plasmid names ending in -g61, -g64, and -g65 encode fusions to putative start codon 5, whereas plasmid p601-g15 encodes a fusion to putative start codon 4. (C) β-Galactosidase specific activities of WT strain SB1003 containing ORF g1::lacZ fusion plasmid p601-g65 grown in RCVm containing 0.5 mM or 10 mM KPO4 for 36 h. (D) β-Galactosidase specific activities of strain SB555 containing the ORF g1::lacZ fusion plasmid p601-g65 grown in RCVm containing 0.5 mM or 10 mM KPO4 for 24 h. Error bars represent the standard deviations of three biological replicates.
The rcc00555 overexpression plasmid pET-555C was created by amplifying SB1003 genomic DNA using primers 555C-F and 555C-R. The ∼0.63-kb amplicon was ligated between pET28a(+) NcoI/XhoI sites to create plasmid pET-555C. Plasmid pET-555C contained a mutation resulting in an amino acid substitution from phenylalanine to leucine at the C-terminal end (the catalytic site is predicted to be in the N terminus).
Purification and determination of the activity of the rcc00555 endolysin.
IPTG-induced cells were incubated at 30°C for 2 h (lysis assay) or 4 h (purification). For purification of the 6-His-tagged Rcc00555 protein (555C), cells were resuspended in 20 mM NaCl, 20 mM Tris, pH 8.0, 20 mM imidazole lysis buffer, passed through a French press, and purified using a nickel-nitrilotriacetic acid (Ni-NTA) column. Protein was eluted by addition of 450 mM imidazole to the buffer.
To determine lytic activity, induced cells were diluted to an OD600 of 0.5 in 1 ml of LB medium or LB medium containing 10 mM KPO4, pelleted at 16,000 relative centrifugal force (RCF) for 5 min, and resuspended in a 100-μl volume of distilled H2O (dH2O) or dH2O containing 10 mM KPO4, pH 6.8. Samples were incubated for 10 min and brought up to 1 ml, and the turbidity was determined. Zymography was performed as described previously (20), using peptidoglycan isolated from a stationary-phase SB1003 culture, and the reaction mixture was incubated at 30°C.
Western blotting of RcGTA capsid protein.
Production of RcGTA capsid protein was measured essentially as described by Fogg et al. (20), but in a 10% (vol/vol) separation polyacrylamide gel with commercially available RcGTA major capsid antiserum (AS08 365; Agrisera AB). Detection was performed as described by Haan and Behrmann (33). Samples from SB1003 and derived mutants were normalized relative to biomass on the basis of the OD660 of the cultures. Due to extensive cell lysis of DE442, samples of this strain and derived mutants were normalized relative to the total protein concentration after passage of cultures through a French press (Aminco), as described below.
GTA transduction assay.
GTA activity was determined essentially as described by Fogg et al. (20), except that RCV medium was added after 1 h and cells were harvested after an additional 3 h.
Cell pellet lysis for GTA transduction assay.
Ten milliliters of culture was centrifuged at 3,500 RCF for 10 min, and the supernatant was discarded. The cell pellet was resuspended in 1 ml of 20 mM Tris-HCl, pH 7.8, 50 mM EDTA, 250 mM sucrose, and 0.5 mg/ml lysozyme and subjected to four freeze-thaw cycles between dry ice and ∼40°C water bath. Nine milliliters of 20 mM Tris-HCl, pH 7.8, 0.5 mM MgCl2, and 0.1 mg/ml DNase I was added, and samples were incubated at room temperature for 10 min to reduce viscosity. Samples were cleared by centrifugation, and the supernatant was filtered through a 0.2-μm-pore-size sterile filter before being used in GTA transduction assays. The concentration of protein in the lysis buffer was subtracted from the values for the sample before normalization to the total amount of protein.
Visualization of spheroplast-like structures.
For low-speed centrifugation of DE442-derived strains, cultures were centrifuged at 16,000 RCF for 30 min. The resultant pigmented and semitransparent layers were separated from the dense cell pellet by careful resuspension using a micropipette. For ultracentrifugation, 10 ml of culture was initially pelleted by centrifugation at 3,500 RCF for 10 min, and the supernatant was transferred to a fresh tube. For SB1003-derived strains, the low-speed centrifugation step was repeated 3 times, after which 3 ml of the cleared supernatant was centrifuged at 37,000 RCF for 10 min. The resultant pellet was resuspended in 10 μl of 2.5 mM NaCl, 100 mM Tris-HCl, pH 7.8. Vesicles were visualized at ×100 magnification and photographed.
Lysozyme treatment was performed by adding 0.25 mg/ml lysozyme (Sigma) and 1.3 mM EDTA (pH 8) to resuspended cell pellets, followed by incubation for 30 min at 30°C.
Transcription start site mapping.
The transcription start site of RcGTA ORF g1 was determined using a 5′ rapid amplification of cDNA ends (RACE) kit (Invitrogen). RNA was isolated from approximately 5 × 108 cells using an RNeasy minikit (Qiagen) following the supplier's protocol, yielding ∼25 μg of RNA. Three to four micrograms of RNA was used for cDNA synthesis by the use of SuperScript II enzyme and primer GSP1, which binds downstream of ATG codon 5 (see Fig. 2A), according to the kit's protocol. A homopolymeric tail was added to cDNA using terminal deoxynucleotidyl transferase and dCTP and amplified using the nested primer GSP3 and the reverse primers supplied in the RACE kit. The resultant product was cloned into a TOPO TA vector (Invitrogen), and its DNA was sequenced to determine the RNA 5′ ends.
Malate dehydrogenase assays.
Malate dehydrogenase activity from strain DE442 and derived mutants was determined as previously described by Fogg et al. (20). For the WT strain SB1003, filtered culture supernatant was ultracentrifuged at 37,000 RCF, and the supernatant was concentrated 18-fold in a 50-kDa-cutoff ultrafiltration device (Amicon Ultracel). A 0.5-ml sample of concentrated supernatant was mixed with 0.2 mM NADH and 100 mM KPO4, pH 7.4, to a total volume of 980 μl. For resuspension experiments, cells were incubated at room temperature (RT) for 30 min, and the supernatant was obtained by centrifuging samples at 6,800 RCF for 5 min. Twenty to 30 μl (OD660 = 5.0) or 200 μl (OD660 = 0.5) was assayed for malate dehydrogenase activity. For experiments where protein synthesis was inhibited by gentamicin, cells were initially resuspended in RCVm containing 10 mM KPO4 for 15 min to allow inhibition and then resuspended in RCVm and incubated for 30 min before the supernatant was obtained for assay. Gentamicin sulfate (60 μg/ml) was present in both steps.
Native agarose gel electrophoresis of RcGTA particles.
Filtered culture supernatant or lysates of cells passed through a French press (see above) were separated in a 0.8% (wt/vol) slab agarose gel using 0.5× TBE (Tris-borate-EDTA) buffer, and DNA was visualized by ethidium bromide staining. Centrifugation of RcGTA particles was performed at 200,000 RCF for 15 min, and DNA was released from particles resuspended in sterile growth medium by incubating samples at 70°C for 10 min.
Propidium iodide staining.
E. coli cultures were centrifuged to pellet the cells, which were washed and resuspended in 10 mM MgSO4, pH 6.5. Propidium iodide was added to 0.2 mM, and the cells were incubated for 30 min at RT before visualization on slides coated with agarose. Fluorescence microscopy was performed using a mercury lamp (HBO, 50 W) for excitation and a Zeiss filter set 15.
Other enzyme assays.
Promoter activity was determined by measuring the β-galactosidase activity of lacZ fused to promoters, as described by Leung et al. (15). β-Galactosidase activity was normalized to the total amount of protein and calculated by U = {1,000 · [(ΔA420/Δtmin) · (Vreaction/Vlysate)]/(4.5 · Cprotein)}, where ΔA420 is the change in the absorbance at 420 nm, Vreaction is the volume of the reaction mixture, Δtmin is the change in time in minutes, Vlysate is the volume of the cell lysate, and Cprotein is the concentration (mg/ml) of total protein. The concentration of protein in cell lysates was measured in a Lowry protein assay (34).
Image processing.
Digital overlays of fluorescence on phase-contrast images and adjustments of brightness and contrast for Western blots, microscopy images, and zymograms were performed using the GNU image manipulation program (www.gimp.org).
RESULTS
High concentrations of phosphate inhibit RcGTA-mediated gene transduction.
Prior studies found that RcGTA-mediated gene transduction is greater in a yeast extract/peptone-based medium (YPS) than in the RCV defined medium (13, 22), but the cause of this effect was not investigated. One of the many differences between these two media is the concentration of phosphate. It was found that YPS complex medium contains ∼0.5 mM phosphate (data not shown), whereas the RCV defined medium contains almost 20-fold more phosphate (9.6 mM potassium phosphate). To evaluate the effect of the phosphate concentration on RcGTA production, the R. capsulatus WT strain SB1003 was grown in YPSm complex medium supplemented with 10 mM either KPO4, NaPO4, or the buffer MOPS, all adjusted to an initial pH of 6.8.
The presence of 10 mM KPO4 or NaPO4 in the YPSm complex medium resulted in decreased frequencies of gene transduction (to 37% and 27%, respectively, compared to the frequency for cultures grown in nonsupplemented YPSm medium (Fig. 1A). Increased transduction frequencies were observed in cultures grown in the YPSm medium supplemented with 10 mM MOPS buffer (Fig. 1A), which we attribute to the stabilization of RcGTA particles by maintenance of the culture pH relatively close to neutrality.
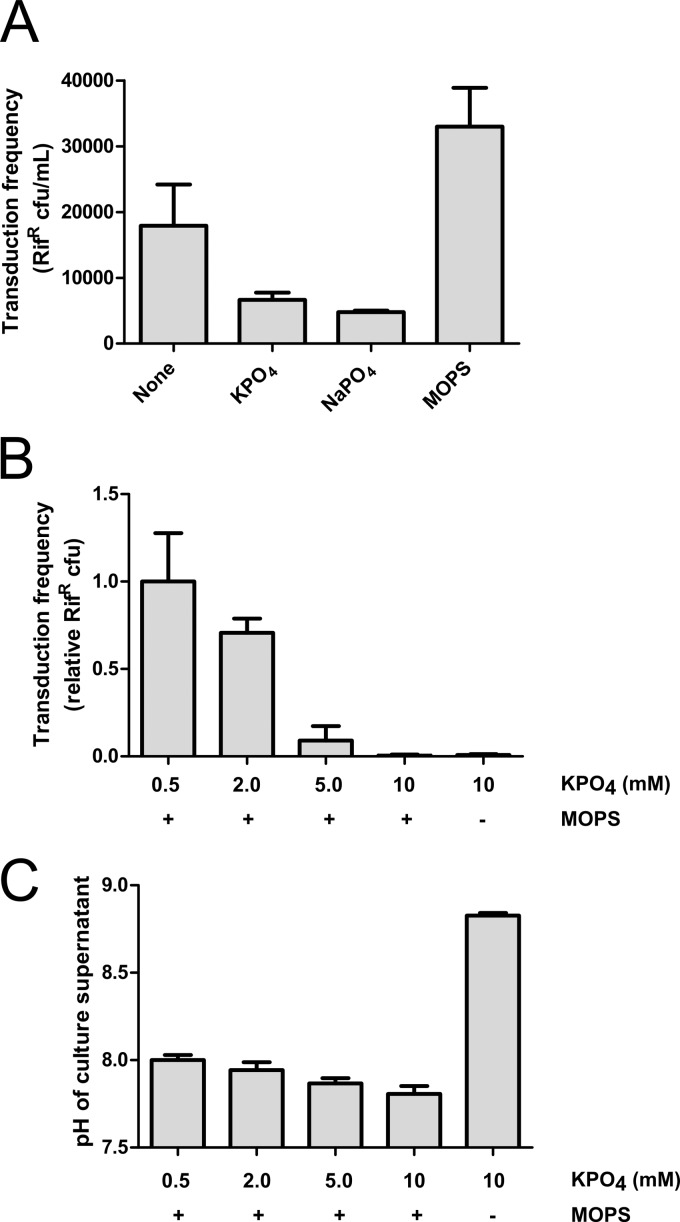
Fig 1.
High concentrations of phosphate inhibit gene transduction. The transduction frequencies of SB1003 culture supernatants are shown. (A) Transduction frequencies of cultures grown in YPSm medium (None) or YPSm supplemented with 10 mM KPO4, NaPO4, or MOPS; (B) transduction frequencies of cultures grown in RCVm medium containing various concentrations of KPO4 (0.5, 2.0, 5.0, and 10 mM); (C) pH of culture supernatant at harvest. Error bars represent the standard deviations of three biological replicates. Samples were normalized to the culture turbidity at 660 nm (A and B).
To study the influence of phosphate in a defined medium, we formulated RCVm, a phosphate-free defined medium containing 20 mM MOPS (pH 6.8) as a pH buffer, with RCV-like levels of K+ (14.7 mM). In cultures with elevated concentrations of phosphate in the RCVm medium, the transduction frequency decreased (to 0.4%) in the presence of 10 mM phosphate compared to that obtained with 0.5 mM phosphate (Fig. 1B). Omitting the MOPS buffer from a culture grown in the presence of 10 mM phosphate did not change the transduction frequency, and there was not a correlation between the culture pH at harvest (Fig. 1C) and the transduction frequency (Fig. 1B).
Analysis of the RcGTA gene cluster promoter region and effect of the phosphate concentration on RcGTA gene expression.
To study the expression of RcGTA genes in response to phosphate, we fused a lacZ reporter gene translationally in frame to the first gene of the RcGTA gene cluster, ORF g1, which is thought to encode the small subunit of the terminase complex (1). However, construction of an appropriate reporter plasmid required that the promoter region of the RcGTA gene cluster be characterized. In the current annotation of the R. capsulatus genome, the start codon of ORF g1 (rcc01682) is designated the first ATG (underlined) in the sequence 5′-ATG GAC ATG −3′ (labeled codon 2 in Fig. 2A), but inspection of the DNA sequence revealed four other in-frame ATG or GTG codons potentially encoding the start codon for ORF g1, and these are designated codons 1, 3, 4, and 5 in Fig. 2A.
Plasmid p601-g65, in which the lacZ gene is fused to codon 5 and which contains all four upstream potential start codons, produced maximal expression of 187 units of β-galactosidase activity (Fig. 2A and B). In contrast, the p601-g15 construct, in which the lacZ gene is fused in frame to the putative start codon 4 and which excludes codon 5, produced only 11% of the activity of p601-g65. This reduced activity level was not significantly different (P = 0.64) from that for the p601-g61 no-promoter control (9% of that of p601-g65). Therefore, we chose the region 3′ of codon 5 as the target for an oligonucleotide to prime reverse transcriptase extension to mRNA 5′ ends.
We used the RACE methodology for mRNA 5′ end mapping and identified two potential start sites for the RcGTA ORF g1 transcript (Fig. 2A). Analysis of the region upstream of ORF g1 using the bioinformatic tool BPROM (Softberry) indicated putative −10 and −35 sequences (light green in Fig. 2A). However, the location of these sequences did not correspond to either of the two RNA 5′ ends mapped. By manual inspection of the sequence upstream of the most distal mRNA end (designated +1 in Fig. 2A), we identified another set of putative −10 and −35 sequences (dark green in Fig. 2A). To determine which sequences in this 5′ region are required for maximal ORF g1 transcription, we used plasmids containing deletions within the ORF g1 5′ region of the in-frame ORF g1::lacZ reporter plasmid p601-g65 and measured the β-galactosidase activity in strains containing these plasmids.
Plasmid p601-g65 contains sequences extending to about 320 bp 5′ of the rcc001681 stop codon and appears to contain all cis-active sequences needed for normal expression of RcGTA genes (15, 18, 20). Plasmid p601-g64 lacks sequences at the distal 5′-most 5′ end (+1 in Fig. 2A) and the manually predicted −10 and −35 sequences but contains the proximal 5′ end and the BPROM-predicted −10 and −35 sites. Plasmid p601-g64 yielded only 23 units of β-galactosidase activity (∼12% of the p601-g65 activity), which was not significantly different (P = 0.47) from the activity yielded by the no-promoter control plasmid p601-g61 (Fig. 2B). Therefore, we suggest that the promoter of the RcGTA gene cluster is located within the 3′ coding region of rcc01681 with the −10 and −35 sites indicated in Fig. 2A. In summary, the maximal β-galactosidase activity of an ORF g1::lacZ translational fusion reporter required inclusion of codon 5 and sequences extending upstream to the proposed −10 and −35 sites.
Based on the information gained from the promoter and ORF g1 start codon analyses, we chose plasmid p601-g65 to evaluate RcGTA promoter activity in response to the phosphate concentration. Surprisingly, we found that the β-galactosidase activity of the SB1003 WT strain was 2.4-fold greater for cells cultured in 10 mM phosphate than those cultured in 0.5 mM phosphate (Fig. 2C). RcGTA production had previously been reported to be associated with cell lysis (20), and cell lysis of lacZ-expressing cells would reduce the intracellular β-galactosidase activity measured in these experiments. Therefore, we compared the difference in β-galactosidase activity for cells cultured in high and low phosphate using the rcc00555-knockout, RcGTA nonreleaser strain SB555 (14) containing plasmid p601-g65. The β-galactosidase activity was similar for cells cultured in the presence of 10 mM and 0.5 mM phosphate (1.2-fold difference; Fig. 2D). Therefore, the increased transduction frequency under low concentrations of phosphate appeared not to be due to increased transcription of the RcGTA gene cluster.
High concentrations of phosphate inhibit release of RcGTA from cells.
To determine whether the phosphate concentration inhibits the release of RcGTA, we investigated the intracellular (cell pellet) and extracellular (culture supernatant) amounts of the RcGTA capsid protein (encoded by ORF g5 [rcc01687]) in Western blots of cultures grown in the presence of different concentrations of phosphate.
Cellular and extracellular fractions of cultures grown in both YPSm and RCVm were probed with RcGTA capsid protein antiserum, which showed that the capsid protein was produced with and without the addition of phosphate (Fig. 3A and B). However, the release of the RcGTA capsid protein from cells decreased in response to the addition of phosphate to the culture medium, whereas other anions, cations, or MOPS had little or no effect. The amount of capsid protein present in the supernatant fraction of samples grown in RCVm increased with decreasing concentrations of phosphate, while the pellet fraction showed a converse trend, with larger amounts of capsid protein being detected in cells grown in medium containing higher phosphate concentrations than in medium containing lower concentrations (Fig. 3B). These data confirm that high concentrations of phosphate do not inhibit the production of RcGTA but, rather, inhibit the release of particles from cells grown in both YPSm complex and RCVm defined media.
Fig 3.
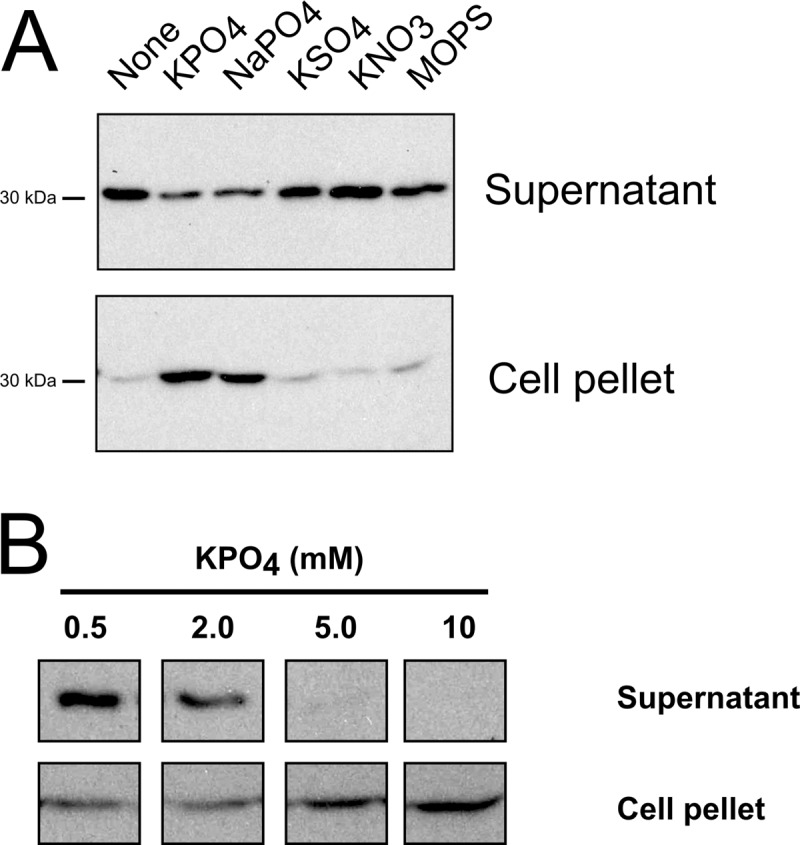
Release of RcGTA is inhibited by high phosphate concentrations. Western blots of culture supernatant and cell pellet fractions probed using RcGTA capsid protein antiserum are shown. (A) Western blots of cells grown in YPSm complex medium (None) and YPSm supplemented with 10 mM KPO4, NaPO4, KSO4, KNO3, or MOPS, as indicated; (B) Western blots of cells grown in RCVm defined medium containing the indicated concentrations of KPO4. Samples were normalized to the culture turbidity at 660 nm.
The release of RcGTA is independent of the Pho regulon.
We initially hypothesized that the phosphate effect involved PhoB, a two-component response regulator known to upregulate multiple regulons in response to phosphate starvation in many bacteria (35). However, little or no difference in RcGTA transduction frequency or RcGTA capsid production and release was observed in a ΔphoB mutant compared to the findings for the WT (see Fig. S1B and C in the supplemental material). Furthermore, depletion of inorganic phosphate was not required for RcGTA release (not shown).
CckA is required for RcGTA release and maximal production of mature RcGTA particles independently of the phosphate concentration.
A previous study found that the CckA sensor kinase protein is needed for maximal release of RcGTA from WT SB1003 cells (16), and so the possibility of phosphate signal transduction through CckA was investigated. To facilitate the study of RcGTA production and release, we exploited the RcGTA-overproducer phenotype of DE442, which has a higher proportion of cells in the population expressing RcGTA than does SB1003 (14, 20).
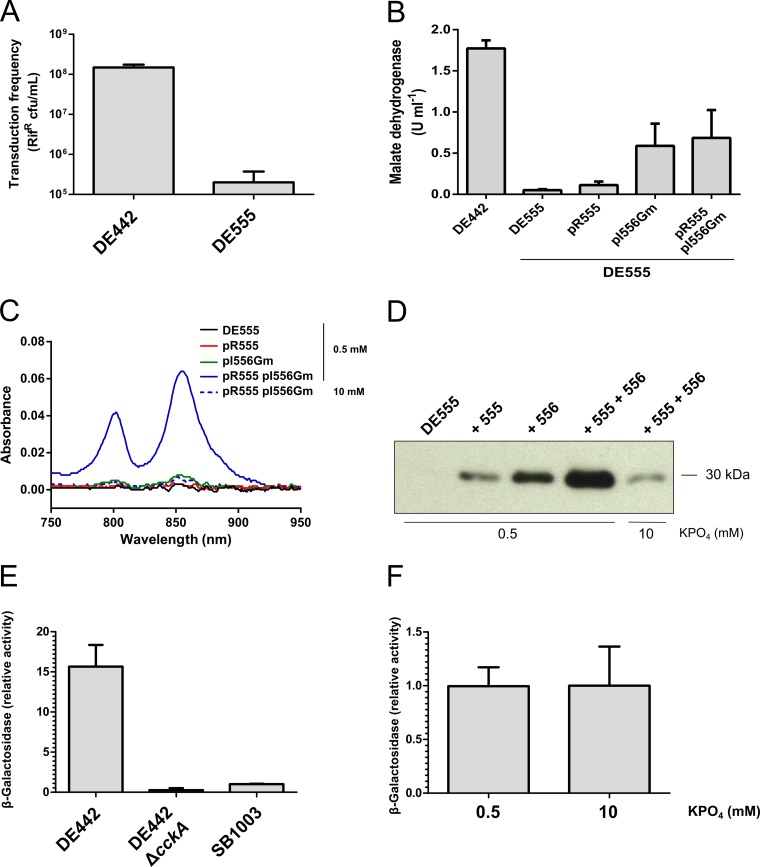
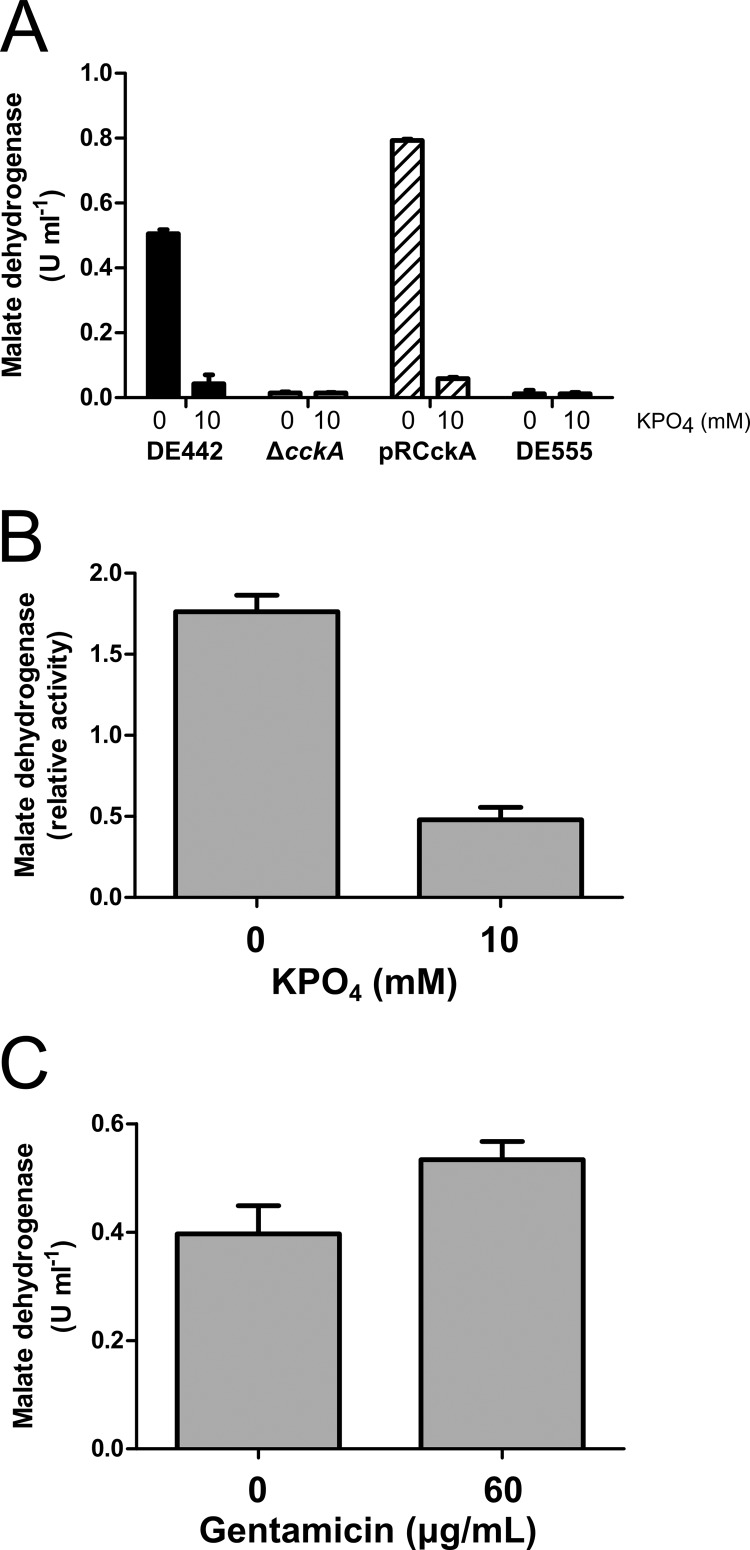
The patterns of transduction and RcGTA capsid production for DE442 cultured in 0.5 mM and 10 mM phosphate were comparable to those in WT strain SB1003 (compare Fig. 4A and B to Fig. 1B and 3B). To investigate whether CckA is involved in phosphate-dependent RcGTA release, we created a DE442 ΔcckA null mutant strain and an in trans complementation plasmid, pRCckA. No extracellular capsid was detected in DE442 ΔcckA cultures grown in the presence of either 0.5 or 10 mM phosphate, and transduction frequencies from the supernatant were reduced 4 to 6 orders of magnitude compared to those for DE442, whereas the intracellular capsid protein was present at high levels regardless of the phosphate concentration (Fig. 4A and B). Nevertheless, the transduction frequencies of the ΔcckA mutant cultures appeared to respond to the phosphate concentration, because the frequencies obtained with filtered supernatants of cultures grown in the presence of 10 mM phosphate were ∼3% of the frequencies obtained with cultures grown in the presence of 0.5 mM phosphate. Therefore, the CckA protein is required for the appreciable release of RcGTA from DE442, as was shown for WT strain SB1003 (16), and the phosphate concentration effect appears to be independent of CckA, because the increased phosphate concentration inhibited RcGTA release in ΔcckA mutant cultures.
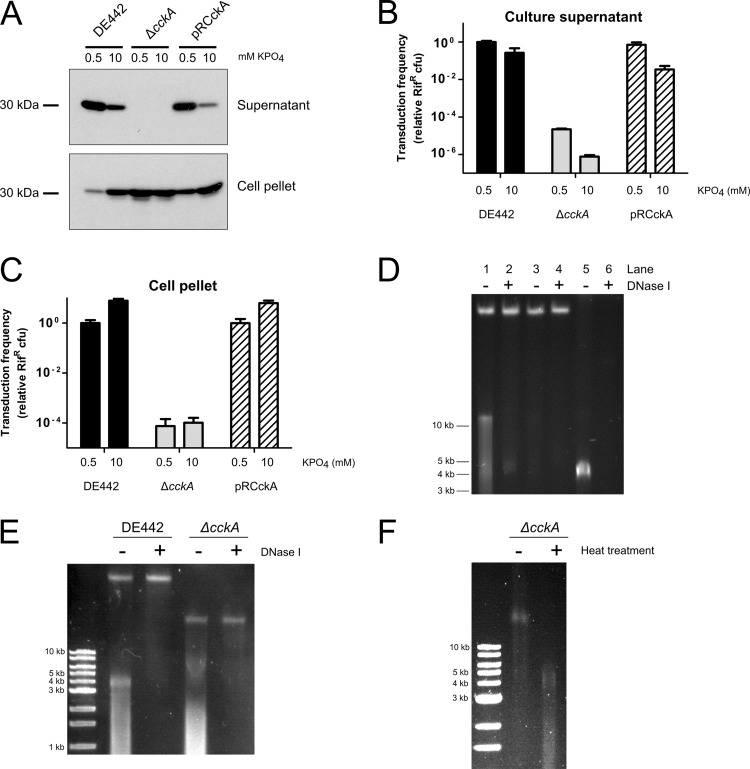
Fig 4.
Independent roles of the phosphate concentration and the CckA protein in production and release of mature RcGTA particles. (A) Western blots of strains DE442, DE442 ΔcckA, and DE442 ΔcckA complemented in trans by pRCckA. Culture supernatant and cell pellet fractions were probed using RcGTA capsid protein antiserum. The approximate migration of the 30-kDa marker is indicated. (B) Transduction frequencies of culture supernatants of DE442, DE442 ΔcckA, and DE442 ΔcckA complemented in trans by pRCckA. (C) Transduction frequencies of freeze-thaw-lysed cells of DE442, DE442 ΔcckA, and DE442 ΔcckA complemented in trans by pRCckA. (D) Native agarose gel of RcGTA. Lanes 1 and 2, filtered DE442 culture supernatant; lanes 3 and 4, resuspended pellet of supernatant after ultracentrifugation; lanes 5 and 6, heat-treated samples. (E) Native agarose gel comparing DE442 and DE442 ΔcckA lysates obtained by passage through a French press; cells were cultured in RCV. (F) Native agarose gel of DE442 ΔcckA lysate obtained by passage through a French press subjected to DNase I, followed by heat treatment. Cells were cultured in RCVm medium (A, B, C, E) or YPS (D). Samples were normalized to the total amount of protein in French press lysates of culture (A and B) or cell pellet (C). Error bars represent the standard deviations of three biological replicates.
Spheroplasts of DE442 parental strain and ΔcckA mutant cells were prepared to investigate whether the intracellular capsid protein represented functional RcGTA. The level of transduction using DE442 cell lysates was 7.8-fold higher in the presence of 10 mM than in the presence of 0.5 mM phosphate (Fig. 4C), similar to the pattern obtained with the intracellular capsid (Fig. 4A). In contrast, extremely low transduction frequencies were observed for equivalent lysates of ΔcckA cells, with transduction levels of 0.0075% or 0.010% of the level for parental strain DE442 obtained when ΔcckA cell lysates were grown in the presence of 0.5 or 10 mM phosphate, respectively. Complementation of the DE442 ΔcckA strain with plasmid pRCckA restored the production of functional, intracellular RcGTA particles and release of capsid to the culture medium (Fig. 4A to C). Therefore, the majority of the capsid protein present in ΔcckA cells is not associated with functional RcGTA particles, thereby indicating a role for the CckA protein in the intracellular assembly of transduction-capable particles.
The RcGTA capsid protein detected in Western blots (Fig. 4A) of ΔcckA cells was the 30-kDa proteolytically cleaved protein, indicating formation of a processed head structure analogous to that of prohead II or a later maturation stage of phage HK97 (36). To determine whether CckA is required for DNA packaging and/or maturation of the RcGTA particle, we analyzed culture supernatants and cell lysates by native agarose gel electrophoresis (37).
Filtered (pore size, 0.2 μm) culture supernatant from strain DE442 contained an apparently large-molecular-size (≫10 kb) band after ethidium bromide staining (Fig. 4D, lane 1). This band was protected from DNase I digestion (Fig. 4D, lane 2), in contrast to other DNA present (Fig. 4D, compare lanes 1 to 3), and was pelleted by ultracentrifugation. Heat treatment, used to release DNA from viral particles (38), resulted in an essentially complete conversion of the large-molecular-size band to an ∼4-kb band that was no longer protected from DNase I digestion (Fig. 4D, lanes 5 and 6). Similarly, gel excision of the large-molecular-size band, followed by guanidine thiocyanate protein denaturation and DNA purification by silica column chromatography, resulted in an ∼4-kb band (not shown). These results strongly indicate that the large-molecular-size band represents the ∼4-kb DNA fragments inside RcGTA particles (13).
Lysates of the DE442 ΔcckA mutant cells contained a DNase I-protected band that migrated at an apparent large molecular size; however, the migration rate was clearly faster than that of the band from DE442 lysates (Fig. 4E). Heat treatment of ΔcckA mutant lysates resulted in a conversion of this large-molecular-size band to species migrating to the ≤4-kb region of the gel (Fig. 4F). Therefore, the ΔcckA mutant packages RcGTA DNA; however, almost all the particles fail to undergo complete maturation to produce transduction-capable RcGTAs.
Inhibition of cell lysis by phosphate.
We previously showed that strain DE442 releases the cytoplasmic enzyme malate dehydrogenase and photosynthetic pigments when cultivated in the YPS complex medium, whereas this effect was mitigated in the RCV defined medium (20). Independently, Hynes et al. recently reported that rcc00555 encodes a putative endolysin which is required for release of RcGTA from the WT strain SB1003 (14).
The turbidity of DE442 cultures grown in RCVm defined medium containing 0.5, 2.0, or 10 mM phosphate was monitored over time to investigate the effects of phosphate on cell lysis (Fig. 5A). The turbidity of DE442 cultures containing 0.5 or 2.0 mM phosphate showed a marked decline after ∼14 h of growth, with the degree of decline being inversely proportional to the phosphate concentration (Fig. 5A). Furthermore, addition of 9.5 mM phosphate to a culture grown in the presence of 0.5 mM phosphate, after the onset of the decline in turbidity, rapidly halted the decline (Fig. 5B). No decline was observed for DE442 cultured in 10 mM phosphate or the WT strain SB1003 (Fig. 5A).
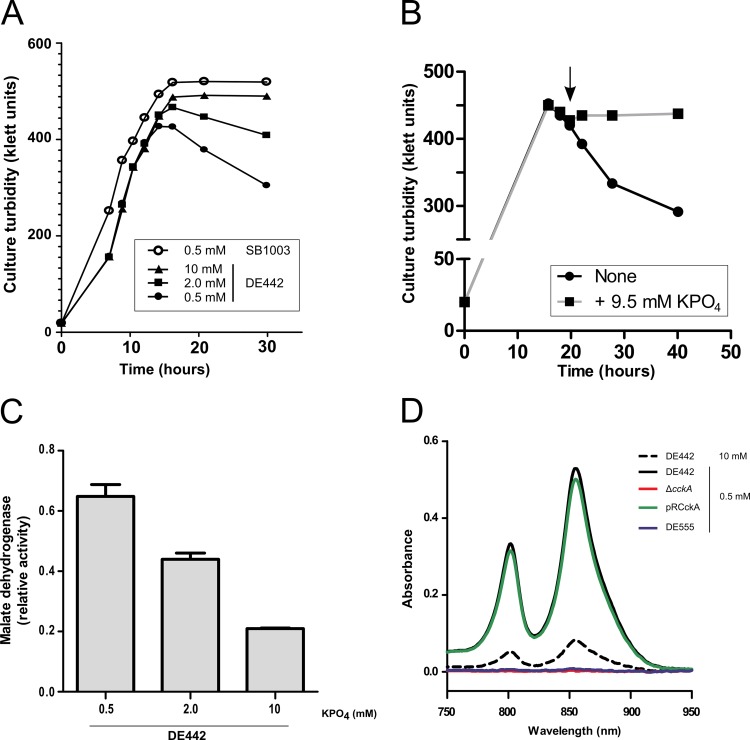
Fig 5.
A decrease in culture turbidity and the release of a cytoplasmic enzyme and intracellular pigments indicate modulation of cell lysis by phosphate. (A) Kinetics of changes in culture turbidity of the overproducer DE442 and WT SB1003 strains grown in the presence of several concentrations of phosphate; (B) kinetics of change in the culture turbidity of overproducer DE442 after addition of phosphate; (C) extracellular fraction of cytoplasmic enzyme malate dehydrogenase relative to the total (extracellular plus intracellular) specific activity in DE442 cultures grown in the presence of different concentrations of phosphate; (D) absorption spectra of cell-free culture supernatants of strains DE442, DE442 ΔcckA, DE442 ΔcckA complemented in trans with pRCckA, and DE555 in the region from 750 to 950 nm, showing transmembrane LH2 complex peaks at 802 and 855 nm. Samples were blanked against sterile RCVm medium. All samples were from cultures grown in RCVm containing 0.5 mM KPO4, unless otherwise indicated. Growth curves (A and B) show the average of two biological samples. Error bars represent the standard deviations of three biological replicates.
To confirm that DE442 cultures underwent phosphate-dependent cell lysis, we measured the release of the cytoplasmic enzyme malate dehydrogenase (Fig. 5C) and photosynthetic membrane pigments (Fig. 5D) to the culture supernatant. The specific activity of extracellular malate dehydrogenase increased with decreasing phosphate concentration and was 3-fold higher in DE442 cultures grown in the presence of 0.5 mM phosphate than in cultures grown in the presence of 10 mM phosphate (Fig. 5C). Similarly, higher levels of light-harvesting complex 2 pigments, which are components of intracellular membranes (39), were observed in cell-free supernatants from cells cultured in 0.5 mM phosphate than in those from cells cultured in 10 mM phosphate (Fig. 5D).
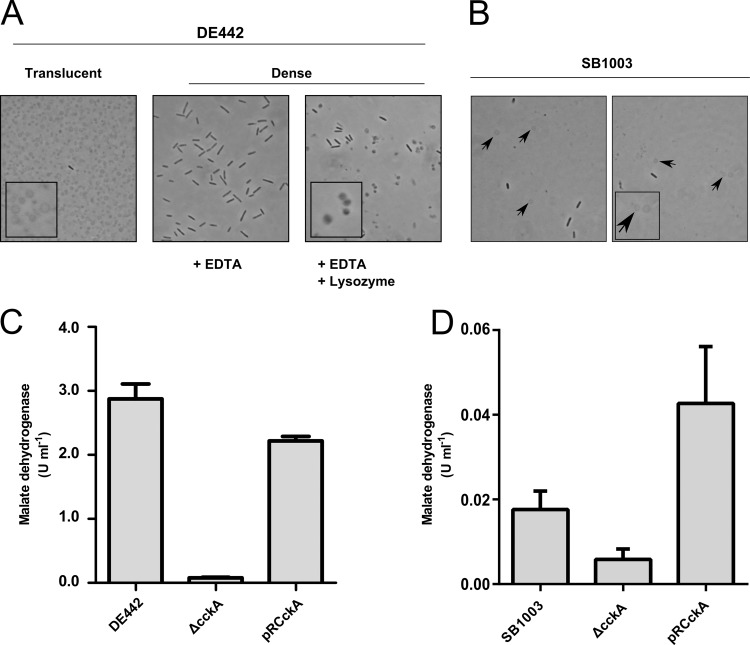
The cell pellet obtained from DE442 cultured in the presence of 0.5 mM phosphate, but not that from DE442 cultured in the presence of 10 mM phosphate, was found to be composed of two layers (see Fig. S2A in the supplemental material): (i) a lower dense pellet and (ii) an upper translucent layer that was separated from the dense pellet by gentle pipetting. The translucent layer primarily contained spheroplast-like vesicles of various diameters, whereas the lower dense pellet was almost exclusively composed of cells (Fig. 6A). To assess whether these spheroplast-like structures resulted from bacterial cells lacking the structural support of peptidoglycan (40), we treated the resuspended dense cell pellet with EDTA and lysozyme. This treatment resulted in structures similar to the vesicles observed in the translucent layer, whereas EDTA alone did not affect cell morphology (Fig. 6A).
Fig 6.
Production of spheroplast-like vesicles accompanies RcGTA release, and CckA is required for maximal cell lysis. (A) Phase-contrast microscopy of pellet layers after centrifugation of DE442 cells; an upper translucent layer and a lower dense layer, with and without lysozyme treatment, are indicated; (B) phase-contrast microscopy of a resuspended pellet of an ultracentrifuged supernatant from an SB1003 culture shows the presence of spheroplast-like structures (arrow). (Insets) Digital enlargements. Magnifications, ×100. (C) Malate dehydrogenase specific activities of culture supernatants of DE442, DE442 ΔcckA, and DE442 ΔcckA complemented in trans with pRCckA. (D) Activity of malate dehydrogenase in culture supernatants of SB1003, SB1003 ΔcckA, and SB1003 ΔcckA complemented in trans with pRCckA. All samples were from cultures grown in RCVm containing 0.5 mM KPO4, unless otherwise indicated. Error bars represent the standard deviations of three biological replicates.
The SB1003 WT strain produces much less RcGTA than DE442 (20). SB1003 grown in the presence of 0.5 mM phosphate showed no measurable decrease in turbidity (Fig. 5A), and initially, we did not observe spheroplast-like vesicles in such cultures. To improve the detection of spheroplast-like vesicles, SB1003 cells were pelleted by low-speed centrifugation, followed by ultracentrifugation of the supernatant to pellet vesicles possibly present at a low concentration. This yielded a pigmented pellet for SB1003 cultured in the presence of 0.5 mM phosphate, which was small in comparison with that for DE442 (see Fig. S2B in the supplemental material). Microscopy of resuspended pellets revealed many spheroplast-like structures in the overproducer strain DE442 pellet (not shown) and, to a lesser extent, the WT strain SB1003 pellet (Fig. 6B), indicating that SB1003 cultures produce a low level of spheroplast-like vesicles, apparently due to the low percentage of cell lysis.
In summary, cultures decreased in turbidity and cells released cytoplasmic malate dehydrogenase and photosynthetic pigments accompanied by the production of spheroplast-like vesicles when grown in the presence of low (0.5 mM) concentrations of phosphate, consistent with cell lysis (Fig. 5A to D and 6A). This cell lysis was inhibited in the presence of high (10 mM) concentrations of phosphate.
The CckA protein is required for maximal lysis of cells.
Because ΔcckA cells release little of the RcGTA capsid into the growth medium when cultured in 0.5 mM phosphate (Fig. 4A), we hypothesized that CckA is required for cell lysis. The malate dehydrogenase activity in culture supernatants of the DE442 ΔcckA mutant was 2.7% of the DE442 level, whereas the level for the pRCckA-complemented DE442 ΔcckA strain was restored to 77% of the DE442 level (Fig. 6C). Similarly, the release of photosynthetic pigments from DE442 ΔcckA was greatly diminished compared to that from WT DE442, and complementation by pRCckA restored this release (Fig. 5D). Additionally, the DE442 ΔcckA mutant cultured in the presence of 0.5 mM phosphate did not form a translucent layer after centrifugation, and this layer was restored by complementation with plasmid pRCckA (see Fig. S2A in the supplemental material). CckA was also required for maximal release of malate dehydrogenase from WT SB1003 cells (Fig. 6D). Additionally, the pellets obtained by ultracentrifugation of the culture supernatant from the SB1003 ΔcckA and DE442 ΔcckA mutant cultures were found to be much smaller than those of the parental strains (see Fig. S2B in the supplemental material), and few (DE442 ΔcckA) or no (SB1003 ΔcckA) spheroplast-like vesicles were observed after resuspension (not shown). Therefore, the CckA protein is required for maximal cell lysis and the consequent release of cytoplasmic proteins and RcGTA particles, with an equivalent role in both the overproducer DE442 and WT SB1003 strains.
Cell lysis of strain DE442 requires the endolysin-holin system encoded by rcc00555 and rcc00556, and rcc00555 expression is modulated by CckA.
To establish whether the predicted rcc00555 endolysin gene (14) is required for phosphate concentration-modulated cell lysis and gene transduction in the RcGTA overproducer strain DE442, we disrupted rcc00555 in the DE442 background. The frequency of RcGTA-mediated transduction from the resultant strain, DE555, was greatly impaired (Fig. 7A), extending the observation of Hynes et al. (14) from the WT strain SB1003 to the overproducer strain DE442. Furthermore, the cell-free malate dehydrogenase activity of strain DE555 was 3% of the parental DE442 level (Fig. 7B), and production of the translucent vesicle-containing layer and release of pigments were mitigated (Fig. 5D; see Fig. S2A in the supplemental material). Complementation of strain DE555 in trans with plasmid pR555 (14) did not restore the release of malate dehydrogenase or pigments to the culture supernatant (Fig. 7B and C), and because the open reading frames of rcc00555 and rcc00556 overlap, we hypothesized that the kanamycin resistance cartridge in the disrupted rcc00555 impaired transcription of the downstream holin (rcc00556). Complementation of rcc00555-disrupted strain DE555 with both pR555 and pI556Gm increased the release of pigments to the supernatant, and this release was dependent on the phosphate concentration (Fig. 7C). Similarly, the release of the RcGTA capsid protein to the culture supernatant was greatly increased for DE555 complemented with both pR555 and pI556Gm compared to that for DE555 or DE555 containing either of the plasmids singly (Fig. 7D). Furthermore, this release was diminished for cells cultured in 10 mM phosphate compared to that for cells cultured in 0.5 mM (Fig. 7D). Therefore, our complementation experiments show that both the endolysin and holin encoded by rcc00555 and rcc00556 are required for cell lysis under conditions with low phosphate concentrations and that these two genes appear to be cotranscribed, and so DE555 is, in effect, a rcc00555 rcc00556 double mutant.
Fig 7.
The rcc00555 endolysin is essential for maximal release of RcGTA by cell lysis and requires CckA for expression that is independent of the phosphate concentration. Comparisons of parental strain DE442, mutant DE555, and DE555 complemented in trans with plasmids pR555 and pI556Gm, either singly or together, are shown (A to D). (A) Transduction frequencies of cells cultured in RCVm containing 0.5 mM KPO4. (B) Extracellular levels of cytoplasmic enzyme malate dehydrogenase activity of cells cultured in RCVm containing 0.5 mM KPO4. (C) Absorption spectra of cell-free culture supernatants in the region from 750 nm to 950 nm, showing transmembrane LH2 complex peaks at 802 and 855 nm. Samples were blanked against sterile RCVm medium. (D) Western blots of culture supernatant fractions of DE555 and DE555 complemented with plasmid pR555 and/or pI556Gm probed using RcGTA capsid protein antiserum. The KPO4 concentration of the medium and the approximate migration of the 30-kDa marker are indicated. (E) Promoter activity of the rcc00555 endolysin measured by determination of the β-galactosidase activity in DE442, DE442 ΔcckA, and SB1003 strains containing reporter plasmid pXCA-555 cultured in RCV medium (9.6 mM KPO4). (F) rcc00555 promoter activity measured by determination of the β-galactosidase activity of DE555(pXCA-555) cultured in RCVm defined medium containing 0.5 mM or 10 mM KPO4. Error bars represent the standard deviations of a minimum of three biological replicates (A, B, F) or the ranges for two biological replicates (E).
To determine whether CckA or the phosphate concentration influences the expression of the rcc00555 endolysin, we constructed the reporter plasmid pXCA-555, containing 247 bp of 5′ sequences and the annotated start codon of rcc00555 fused in frame to a lacZ reporter gene. The β-galactosidase activity encoded by pXCA-555 was 62-fold greater in the parental strain DE442 than in the ΔcckA mutant, indicating that CckA is required for maximal expression of rcc00555 (Fig. 7E). Furthermore, the β-galactosidase activity of pXCA-555 was 15.7-fold higher in the overproducer DE442 than in the WT strain SB1003 (Fig. 7E), consistent with the increased degree of cell lysis observed in DE442 cultures compared to SB1003 cultures. Because the lysis of cells expressing functional rcc00555 would skew the cell-associated β-galactosidase activities, as observed for the RcGTA ORF g1::lacZ fusions (Fig. 2C and D), we used the nonlysing DE555 mutant to investigate whether the phosphate concentration modulates the expression of rcc00555. No difference in the average β-galactosidase activity was observed between cells cultured in the presence of 0.5 mM phosphate and those cultured in the presence of 10 mM phosphate (Fig. 7F), indicating that the phosphate concentration does not modulate the expression of rcc00555.
rcc00555 encodes an endolysin.
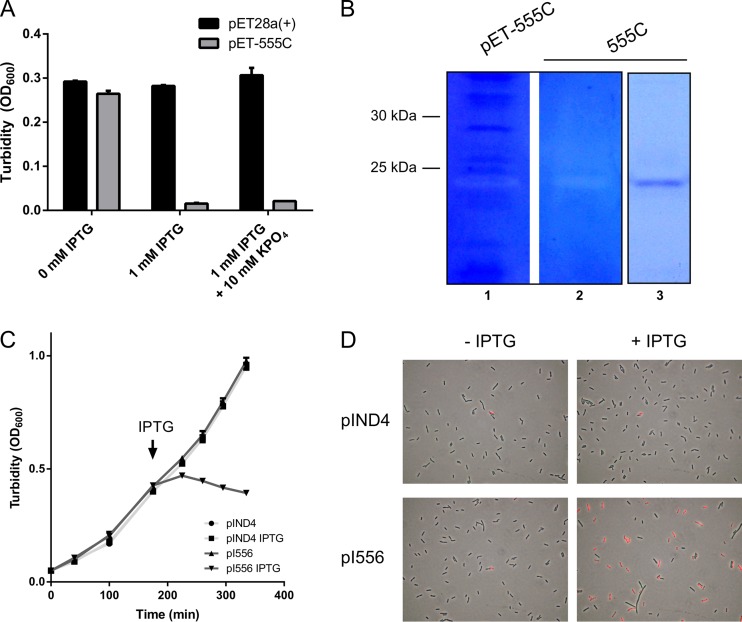
To verify that rcc00555 encodes an endolysin, we expressed the rcc00555 gene in E. coli. Cells induced to express the C-terminally 6-His-tagged rcc00555 gene product (555C; carried by plasmid pET-555C) were centrifuged and resuspended in 0.1 volume of water. The turbidity of resuspended cells induced to express rcc00555 was markedly less than that of both the empty vector control and noninduced cultures (Fig. 8A), consistent with rcc00555 encoding an endolysin. Phase-contrast microscopy revealed the production of spheroplasts and extensive lysis of cells expressing rcc00555 (not shown). We did not detect a marked phosphate-dependent inhibition of cell lysis for cells cultured and resuspended in 10 mM phosphate (Fig. 8A), and so phosphate does not appear to inhibit this lytic activity of rcc00555.
Fig 8.
Expression of the rcc00555 endolysin and rcc00556 holin in E. coli. The lytic and peptidoglycan-degrading activities of rcc00555 (A and B) and holin-like activity of rcc00556 (C and D) expressed in E. coli are shown. (A) Turbidity after resuspension in dH2O of induced E. coli. (B) Lane 1, Zymogram of E. coli culture lysate; lane 2, affinity-purified 555C protein; lane 3, SDS-PAGE of purified 555C protein. The approximate migration of the markers is indicated. (C) Growth curves of E. coli. Arrow, induction time point. (D) Propidium iodide staining of E. coli after induction of rcc00556 expression. Cells contained the inducible rcc00555 expression plasmids pET-555C (C-terminally 6-His tagged) or the empty vector control pET28a(+) (A and B) and the inducible rcc00556 expression plasmid pI556 or the empty vector control pIND4 (C and D). Cells were induced by addition of 1 mM IPTG. Error bars represent the ranges for two (A) or the standard deviations for three (C) biological replicates.
The 555C protein was purified using Ni-NTA chromatography, and the eluate yielded a band of the predicted size of 22.9 kDa (Fig. 8B, lane 3). Zymography was performed in an SDS-polyacrylamide gel that contained SB1003-derived peptidoglycan and that was stained with Coomassie blue. A clearing at the position corresponding to the 555C band against a blue background was observed for the cell lysate (Fig. 8B, lane 1) and purified 555C (Fig. 8B, lane 2). Therefore, the rcc00555 gene product has peptidoglycan-degrading activity, consistent with the lytic activity due to an endolysin.
rcc00556 encodes a holin.
To confirm that rcc00556 encodes a holin, we induced expression of rcc00556 from plasmid pI556 in E. coli. Expression rapidly halted growth, whereas no effect on growth was observed with the empty vector control (Fig. 8C). To confirm that the membrane potential of the cells was disrupted, we stained cells with propidium iodide. About 50% of the cells induced to express rcc00556 fluoresced when treated with propidium iodide, whereas very few fluorescent cells were observed for uninduced and empty vector control cells (Fig. 8D). Therefore, the rcc00556 gene product alone appears to be sufficient to depolarize bacterial cytoplasmic membranes by forming a hole, as has been found for bacteriophage holins (41).
Inhibition of cell lysis by phosphate does not require de novo protein synthesis.
To investigate whether the effect of the phosphate concentration on cell lysis requires active growth, DE442 cells cultured in 10 mM phosphate were collected and resuspended in RCVm medium in the presence or absence of 10 mM phosphate. Malate dehydrogenase was readily detected after 30 min incubation in the absence of phosphate, but to a lesser degree in the presence of phosphate, in a concentration-dependent manner, whereas other salts did not affect the release (Fig. 9A; see Fig. S3A in the supplemental material). Similarly, phosphate concentration-modulated release of malate dehydrogenase was observed using cells resuspended in a salt-free Tris-HCl buffer (Fig. 9B). Treatment with gentamicin, an inhibitor of protein synthesis (42), inhibited growth (see Fig. S3B in the supplemental material) but did not reduce the amount of malate dehydrogenase released compared to that for untreated cells (Fig. 9C). As found for actively growing cultures (Fig. 6C and 7B), very little malate dehydrogenase was released from resuspended ΔcckA and DE555 mutant cells (Fig. 9A).
Fig 9.
Inhibition of cell lysis by phosphate occurs in the absence of de novo protein synthesis. The malate dehydrogenase activity in cell-free supernatants is shown. (A) DE442, DE442 ΔcckA, DE442 ΔcckA(pRCckA), and DE555 cells resuspended in RCVm defined medium lacking phosphate (bars labeled 0) or containing 10 mM added phosphate (bars labeled 10); (B) DE442 resuspended in Tris-HCl buffer containing no (bars labeled 0) or 10 mM (bars labeled 10) added phosphate (C) DE442 treated with 0 or 60 μg/ml gentamicin and resuspended in RCVm defined medium lacking phosphate. Cells were resuspended to an OD660 of 5.0 (A and B) or 0.5 (C). For panel B, activity is expressed relative to the activity in RCVm lacking phosphate. For panel C, activity was multiplied by 10 for direct comparison with experiments using an OD660 of 5.0. Error bars represent the standard deviations of three biological replicates.
The malate dehydrogenase activity in the culture supernatant of the actively growing rcc00556-positive and rcc00555-negative strain DE555 containing plasmid pI556Gm was similar to that of this strain complemented by both pR555 and pI556Gm and ∼33% of the activity measured for WT DE442 (Fig. 7B). In contrast, the release of photosynthetic pigments required both the rcc00555 endolysin and rcc00556 holin, and this release was inhibited for cells cultured in 10 mM KPO4 (Fig. 7C). However, no effect of phosphate concentration on the release of malate dehydrogenase was observed after resuspension of rcc00556-positive and rcc00555-knockout strain DE555(pI556Gm) cells in RCVm containing no additional phosphate or 10 mM KPO4 (see Fig. S3C in the supplemental material). Therefore, malate dehydrogenase appears to be released from cells by a combination of diffusion through rcc00556 holin-mediated holes and rcc00555 endolysin-dependent cell lysis, whereas the release of intracytoplasmic membrane vesicles containing photosynthetic pigments requires endolysin-mediated cell lysis.
DISCUSSION
Identification of the transcriptional start site of RcGTA ORF g1.
Prior to this study, the RcGTA promoter was imprecisely defined as a sequence located in a region ∼600 bp upstream of the start codon of ORF g1 (18). We identified an RNA 5′ end and putative −10 and −35 sequences (Fig. 2A) which result in a long, presumably untranslated region 5′ of RcGTA ORF g1 which could potentially contain binding sites for regulatory proteins, such as GtaR and CtrA (15, 16, 18). Although Leung et al. identified a putative lux box in the RcGTA promoter region, the LuxR-type protein GtaR did not bind to this sequence under the conditions tested (15), and we have similarly been unable to detect binding to this region of the CtrA protein or CtrA protein treated with acetyl phosphate (not shown).
During construction of the reporter plasmid for RcGTA expression, we found that a fusion to codon 4 did not yield activity greater than that of a no-promoter control (Fig. 2B), which indicated that either codon 5 is the ORF g1 start codon or sequences between codons 4 and 5 are required for maximal expression of ORF g1, with translation initiating 5′ of codon 5. Multiple-sequence alignment of top BLASTP hits to the ORF g1 protein showed that several amino acids encoded between potential start codon 2 and codon 5 and some amino acids between codons 1 and 2 are conserved between R. capsulatus SB1003, Rhodobacter sphaeroides, Rhodobacter spp., Paracoccus sp. strain TRP, and Oceaniovalibus guishaninsula JLT2003 (see Fig. S4 in the supplemental material). This conservation indicates that the genuine start codon of the R. capsulatus ORF g1 is upstream of codon 5 in Fig. 2A.
More research is needed to fully investigate this region, and it is conceivable that there is more than one translational start codon, yielding a mixture of ORF g1 proteins with different N termini.
rcc00555 encodes an endolysin and rcc00556 encodes a holin which are regulated by CckA.
The release of RcGTA is accompanied by cell lysis (20), and Hynes et al. previously reported that rcc00555 and rcc00556 encode an endolysin and a holin, respectively (14). However, other than the absence of an extracellular RcGTA capsid and low transduction frequencies in an rcc00555 mutant, direct evidence of a role for rcc00555 and rcc00556 in cell lysis was not provided.
For the E. coli phage λ, the timing of cell lysis is regulated by the relative expression of holin and antiholin, both encoded by gene S (43). Functional lysozyme (encoded by gene R) accumulates in the cytoplasm during late gene expression and is released to the periplasm through cytoplasmic membrane lesions formed by the holin, apparently in response to a sudden drop in the proton motive force (44, 45). We present direct results indicating that rcc00555 encodes an endolysin and rcc00556 encodes a holin, both of which are required for release of RcGTA by cell lysis: rcc00556 expressed in E. coli rapidly halted growth and made the cells permeable to propidium iodide (Fig. 8C and D), and expression of rcc00556 in trans in the endolysin-disrupted strain DE555 released malate dehydrogenase (Fig. 7B), indicating formation of a hole large enough to release this ∼130- to 140-kDa cytoplasmic enzyme (46, 47). Expression of rcc00555 in E. coli resulted in cell lysis after resuspension in dH2O (Fig. 8A), and the purified Rcc0555 protein degraded SB1003-derived peptidoglycan (Fig. 8B), indicating that rcc00555 encodes an endolysin. Furthermore, disruption of the endolysin (and, evidently, the holin; see below) in R. capsulatus greatly inhibited cell lysis and the release of RcGTA (Fig. 5D, 7A to D, and 9A).
Because the rcc00555 and rcc00556 coding sequences overlap, we suggest that the rcc00555 endolysin and the rcc00556 holin are cotranscribed, consistent with the finding that complementation with both rcc00555 and rcc00556 was required to restore release of pigment-containing vesicles from rcc00555-disrupted strain DE555 (Fig. 7C). The expression of the endolysin (and, therefore, the holin) is highly upregulated in the overproducer DE442 compared to the level of regulation in WT strain SB1003 (Fig. 7E), confirming the microarray results of Hynes et al. (14) and supporting our previous (20) and current (Fig. 5A and 6C and D) findings that a greater proportion of DE442 cells than of SB1003 cells lyse. Furthermore, expression of rcc00555 was regulated by the putative histidine kinase CckA (Fig. 7E), which was also required for cell lysis (Fig. 5D, 6C and D, and 9A).
Millimolar concentrations of phosphate inhibit the lytic release of RcGTA.
Although the high transduction frequencies observed in R. capsulatus cultures grown in the complex medium YPS compared to those observed in cultures grown in the defined medium RCV (13, 22) have been exploited for decades to transduce alleles between strains of R. capsulatus (48), the reason for this difference was unknown. Similarly, we reported that release of RcGTA by cell lysis is greatly increased in YPS medium compared to that in the minimal medium RCV for an unknown reason (20). We found that the concentration of phosphate in the growth medium modulates RcGTA transduction (Fig. 1) by inhibition of the lytic release of RcGTA by high phosphate concentrations (Fig. 5 and 6A and B), and this release requires the endolysin/holin system (Fig. 7A to D). The effects of phosphate concentration on RcGTA release did not appear to be a stress response to low/limiting phosphate concentrations: there was no requirement for the response regulator PhoB, which regulated alkaline phosphatase activity in R. capsulatus (see Fig. S1 in the supplemental material); homologues of the phosphate-transporter system encoding pstSCAB; or a complete depletion of phosphate from the medium (not shown). Furthermore, there was a rapid inhibition of cell lysis by the addition of phosphate (Fig. 5B) and a rapid phosphate-dependent release of a cytoplasmic enzyme from cells not actively growing (Fig. 9A and B; see Fig. S3A in the supplemental material). Furthermore, the inhibitory effect of phosphate appears to be posttranslational (Fig. 9C), and no effect of phosphate concentration on expression of the rcc00555 endolysin was observed (Fig. 7F). Addition of 10 mM phosphate did not inhibit lysis of E. coli cells expressing recombinant endolysin (Fig. 8A), which indicates that phosphate does not inhibit the catalytic activity of the rcc00555 endolysin.
Several bacteriophages encode proteins in addition to the endolysin and holin required for efficient lysis of the host cell. The genes Rz and Rz1, located adjacent to the endolysin and holin genes in E. coli phage λ, encode the spanin complex, which performs the final step in the lysis pathway by disrupting the outer membrane (49). Interestingly, the spanin complex is required for cell lysis in a medium containing millimolar but not low concentrations of divalent cations (50). We have not been able to identify spanin complex genes in R. capsulatus, and it is possible that the inhibition of cell lysis by high concentrations of phosphate is due to stabilization of the cells, as opposed to a direct inhibitory action on a factor involved in the lysis pathway.
rcc00555 homologues are present in other GTA-producing organisms.
There is a growing interest in the study of GTAs from species other than R. capsulatus (2, 5, 6). Using BLASTP, we found that several other alphaproteobacteria, including Ruegeria pomeroyi DSS-3, Roseovarius nubinhibens ISM, and Rhodobacter sphaeroides 2.4.1, encode rcc00555 homologues (not shown) which could be involved in release of the particles. The millimolar concentration of phosphate required for inhibition of release of RcGTA from cells makes it unlikely that this is a mechanism for regulation of RcGTA release in most aquatic ecosystems, apart from highly eutrophic environments, such as the environment in Lake Erie (51). However, optimization of the phosphate concentration in the growth medium used to culture strains for production of RcGTA-like particles may be essential for the discovery of additional RcGTA-like particles. Similarly, attention to the possible inhibition of GTA release by high concentrations of phosphate is essential when comparing levels of production of RcGTA-like particles from strains cultured in different growth media (6). Therefore, our finding that the phosphate concentration modulates RcGTA release is highly relevant for the larger scientific community studying GTAs.
CckA is required for RcGTA maturation.
CckA was previously reported to be required for the release of RcGTA from the SB1003 WT strain, on the basis of the absence of the RcGTA capsid protein in the culture supernatant of a cckA mutant (16). In addition to regulating cell lysis (see above), we found that the DE442 ΔcckA mutant accumulated nonfunctional RcGTA particles inside the cells (Fig. 4A and C). This is in contrast to the functional RcGTA particles accumulating inside WT cells grown in 10 mM phosphate (Fig. 4A and C). The capsid proteins produced by the ΔcckA mutant (Fig. 4A) appeared to have undergone assembly and proteolytic cleavage (18, 52), similar to the maturation of phage HK97 capsid protein (36, 53), indicating formation of a structure similar to the HK97 prohead II-like structure or a later maturation step. Furthermore, this structure contained DNA but migrated more rapidly than the WT particle (Fig. 4E), indicating a structural difference between WT and ΔcckA mutant particles. We note that the sharpness and intensity of the large-molecular-size band observed from the ΔcckA mutant were relatively variable between experiments (compare the sharpness of the large-molecular-size band from the ΔcckA mutant in Fig. 4E and F). This variability could be due to a diversity of partially assembled particles. Therefore, we suggest that CckA is required for a maturation step after DNA packaging in RcGTA biogenesis, such that a cckA mutant yields intermediates on the pathway to mature, transduction-capable particles. Because the CckA protein appears to be a sensor kinase (16), we speculate that the function of CckA in RcGTA maturation is at the beginning of a signal transduction pathway to a response regulator that differentially regulates expression of one or several genes required for assembly of functional RcGTA. To our knowledge, this is the first example of a host-encoded sensor kinase being required for maturation of a GTA or phage particle.
Summary of results.
In this paper, we identified the RcGTA structural gene cluster promoter. We present other new data in support of an rcc00555 (endolysin)- and rcc00556 (holin)-dependent lytic release mechanism for RcGTA, and expression of rcc00555 and rcc00556 in E. coli demonstrated their functions as an endolysin and a holin, respectively. It was found that lytic release requires the sensor kinase homologue CckA, which regulates the expression of the rcc00555 endolysin. We also provide evidence that the CckA protein is needed for the maturation of functional RcGTA particles, perhaps by inducing the expression of one or more genes encoding factors needed for postprohead particle assembly.
We discovered a role for inorganic phosphate in modulating the release of RcGTA particles from cells, apparently by posttranslational inhibition of cell lysis. Our findings solve the decades-long mystery of the large difference in RcGTA-mediated transduction in the RCV defined medium and yeast extract/peptone-based complex medium (13, 22). We suggest that of the multiple differences between these media, the major effect on RcGTA-mediated transduction frequencies is due to the effect of the phosphate concentration on the lysis of cells to release RcGTA particles. Our finding that high frequencies of RcGTA transduction are obtained in a defined RCV-derived medium containing reduced phosphate concentrations allows a shift from use of the complex growth media currently used for most GTA transduction studies to facilitate incisive studies of the effects of specific nutrients on gene expression.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by Canadian Institutes of Health Research grant 93779 to J.T.B.
We thank A. Hynes and A. Lang for kindly contributing plasmids and strains for creating strain DE555 and C. Brimacombe for valuable comments.
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00669-13.
REFERENCES
- 1.Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10:472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH. 2010. High frequency of horizontal gene transfer in the oceans. Science 330:50. [DOI] [PubMed] [Google Scholar]
- 3.Marrs B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. U. S. A. 71:971–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang AS, Beatty JT. 2007. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 15:54–62 [DOI] [PubMed] [Google Scholar]
- 5.Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA. 2008. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl. Environ. Microbiol. 74:2933–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDaniel LD, Young EC, Ritchie KB, Paul JH. 2012. Environmental factors influencing gene transfer agent (GTA) mediated transduction in the subtropical ocean. PLoS One 7:e43506. 10.1371/journal.pone.0043506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berglund EC, Frank AC, Calteau A, Vinnere Pettersson O, Granberg F, Eriksson AS, Naslund K, Holmberg M, Lindroos H, Andersson SG. 2009. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet. 5:e1000546. 10.1371/journal.pgen.1000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy L, Nystedt B, Toft C, Zaremba-Niedzwiedzka K, Berglund EC, Granberg F, Naslund K, Eriksson AS, Andersson SG. 2013. A gene transfer agent and a dynamic repertoire of secretion systems hold the keys to the explosive radiation of the emerging pathogen Bartonella. PLoS Genet. 9:e1003393. 10.1371/journal.pgen.1003393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mason PE, Neilson GW, Dempsey CE, Barnes AC, Cruickshank JM. 2003. The hydration structure of guanidinium and thiocyanate ions: implications for protein stability in aqueous solution. Proc. Natl. Acad. Sci. U. S. A. 100:4557–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motro Y, La T, Bellgard MI, Dunn DS, Phillips ND, Hampson DJ. 2009. Identification of genes associated with prophage-like gene transfer agents in the pathogenic intestinal spirochaetes Brachyspira hyodysenteriae, Brachyspira pilosicoli and Brachyspira intermedia. Vet. Microbiol. 134:340–345 [DOI] [PubMed] [Google Scholar]
- 11.Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589–596 [DOI] [PubMed] [Google Scholar]
- 12.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yen HC, Hu NT, Marrs BL. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157–168 [DOI] [PubMed] [Google Scholar]
- 14.Hynes AP, Mercer RG, Watton DE, Buckley CB, Lang AS. 2012. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol. Microbiol. 85:314–325 [DOI] [PubMed] [Google Scholar]
- 15.Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. 2012. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol. Microbiol. 83:759–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. 2012. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol. Lett. 331:53–62 [DOI] [PubMed] [Google Scholar]
- 17.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93 [DOI] [PubMed] [Google Scholar]
- 18.Lang AS, Beatty JT. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. U. S. A. 97:859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florizone SM. 2006. Studies in the regulation of the gene transfer agent (GTA) of Rhodobacter capsulatus. M.Sc. thesis University of British Columbia, Vancouver, British Columbia, Canada [Google Scholar]
- 20.Fogg PC, Westbye AB, Beatty JT. 2012. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One 7:e43772. 10.1371/journal.pone.0043772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brimacombe CA, Stevens A, Jun D, Mercer R, Lang AS, Beatty JT. 2013. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol. Microbiol. 87:802–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solioz M. 1975. The gene transfer agent of Rhodopseudomonas capsulata. Ph.D. thesis St. Louis University, St. Louis, MO [Google Scholar]
- 23.Strnad H, Lapidus A, Paces J, Ulbrich P, Vlcek C, Paces V, Haselkorn R. 2010. Complete genome sequence of the photosynthetic purple nonsulfur bacterium Rhodobacter capsulatus SB 1003. J. Bacteriol. 192:3545–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver PF, Wall JD, Gest H. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105:207–216 [DOI] [PubMed] [Google Scholar]
- 25.Wall JD, Weaver PF, Gest H. 1975. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch. Microbiol. 105:217–224 [DOI] [PubMed] [Google Scholar]
- 26.Beatty JT, Gest H. 1981. Generation of succinyl-coenzyme A in photosynthetic bacteria. Arch. Microbiol. 129:335–340 [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic-engineering—transposon mutagenesis in gram-negative bacteria. Biotechnology (NY) 1:784–791 [Google Scholar]
- 29.Taylor DP, Cohen SN, Clark WG, Marrs BL. 1983. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J. Bacteriol. 154:580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aklujkar M, Harmer AL, Prince RC, Beatty JT. 2000. The orf162b sequence of Rhodobacter capsulatus encodes a protein required for optimal levels of photosynthetic pigment-protein complexes. J. Bacteriol. 182:5440–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197 [DOI] [PubMed] [Google Scholar]
- 32.Adams CW, Forrest ME, Cohen SN, Beatty JT. 1989. Structural and functional analysis of transcriptional control of the Rhodobacter capsulatus puf operon. J. Bacteriol. 171:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haan C, Behrmann I. 2007. A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J. Immunol. Methods 318:11–19 [DOI] [PubMed] [Google Scholar]
- 34.Peterson GL. 1983. Determination of total protein. Methods Enzymol. 91:95–119 [DOI] [PubMed] [Google Scholar]
- 35.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendrix RW, Johnson JE. 2012. Bacteriophage HK97 capsid assembly and maturation. Adv. Exp. Med. Biol. 726:351–363 [DOI] [PubMed] [Google Scholar]
- 37.Serwer P, Griess GA. 1999. Advances in the separation of bacteriophages and related particles. J. Chromatogr. B Biomed. Sci. Appl. 722:179–190 [DOI] [PubMed] [Google Scholar]
- 38.Duda RL, Ross PD, Cheng N, Firek BA, Hendrix RW, Conway JF, Steven AC. 2009. Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J. Mol. Biol. 391:471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feick R, van Grondelle R, Rijgersberg CP, Drews G. 1980. Fluorescence emission by wild-type- and mutant-strains of Rhodopseudomonas capsulata. Biochim. Biophys. Acta 593:241–253 [DOI] [PubMed] [Google Scholar]
- 40.Fischetti VA. 2005. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 13:491–496 [DOI] [PubMed] [Google Scholar]
- 41.Wang IN, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825 [DOI] [PubMed] [Google Scholar]
- 42.Sarre SG, Hahn FE. 1967. Mode of action of gentamicin. Trans. N. Y. Acad. Sci. 29:575–578 [DOI] [PubMed] [Google Scholar]
- 43.Young R, Wang I-N. 2006. Phage lysis. In Calendar R. (ed), The bacteriophages, 2nd ed. Oxford University Press, New York, NY [Google Scholar]
- 44.Doermann AH. 1952. The intracellular growth of bacteriophages. I. Liberation of intracellular bacteriophage T4 by premature lysis with another phage or with cyanide. J. Gen. Physiol. 35:645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grundling A, Manson MD, Young R. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. U. S. A. 98:9348–9352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tayeh MA, Madigan MT. 1987. Malate dehydrogenase in phototrophic purple bacteria: purification, molecular weight, and quaternary structure. J. Bacteriol. 169:4196–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang IN, Deaton J, Young R. 2003. Sizing the holin lesion with an endolysin–beta-galactosidase fusion. J. Bacteriol. 185:779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scolnik PA, Marrs BL. 1987. Genetic research with photosynthetic bacteria. Annu. Rev. Microbiol. 41:703–726 [DOI] [PubMed] [Google Scholar]
- 49.Berry J, Rajaure M, Pang T, Young R. 2012. The spanin complex is essential for lambda lysis. J. Bacteriol. 194:5667–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang N, Young R. 1999. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage lambda. Mol. Gen. Genet. 262:659–667 [DOI] [PubMed] [Google Scholar]
- 51.Howell T, Nakamoto L. 26 August 2009, posting date Detroit River-Western Lake Erie basin indicator project. U.S. Environmental Protection Agency, Washington, DC: http://www.epa.gov/med/grosseile_site/indicators/western-lake-phos.html [Google Scholar]
- 52.Spano AJ, Chen FS, Goodman BE, Sabat AE, Simon MN, Wall JS, Correia JJ, McIvor W, Newcomb WW, Brown JC, Schnur JM, Lebedev N. 2007. In vitro assembly of a prohead-like structure of the Rhodobacter capsulatus gene transfer agent. Virology 364:95–102 [DOI] [PubMed] [Google Scholar]
- 53.Aksyuk AA, Rossmann MG. 2011. Bacteriophage assembly. Viruses 3:172–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yen HC, Marrs B. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ind AC, Porter SL, Brown MT, Byles ED, de Beyer JA, Godfrey SA, Armitage JP. 2009. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl. Environ. Microbiol. 75:6613–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.