Abstract
Type IV secretion system (T4SS) substrates are recruited through a translocation signal that is poorly defined for conjugative relaxases. The relaxase TrwC of plasmid R388 is translocated by its cognate conjugative T4SS, and it can also be translocated by the VirB/D4 T4SS of Bartonella henselae, causing DNA transfer to human cells. In this work, we constructed a series of TrwC variants and assayed them for DNA transfer to bacteria and human cells to compare recruitment requirements by both T4SSs. Comparison with other reported relaxase translocation signals allowed us to determine two putative translocation sequence (TS) motifs, TS1 and TS2. Mutations affecting TS1 drastically affected conjugation frequencies, while mutations affecting either motif had only a mild effect on DNA transfer rates through the VirB/D4 T4SS of B. henselae. These results indicate that a single substrate can be recruited by two different T4SSs through different signals. The C terminus affected DNA transfer rates through both T4SSs tested, but no specific sequence requirement was detected. The addition of a Bartonella intracellular delivery (BID) domain, the translocation signal for the Bartonella VirB/D4 T4SS, increased DNA transfer up to 4% of infected human cells, providing an excellent tool for DNA delivery to specific cell types. We show that the R388 coupling protein TrwB is also required for this high-efficiency TrwC-BID translocation. Other elements apart from the coupling protein may also be involved in substrate recognition by T4SSs.
INTRODUCTION
Bacterial type IV secretion systems (T4SS) are versatile multiprotein channels involved in processes such as conjugative DNA transfer between bacteria or effector translocation to eukaryotic cells (1). Examples of these two types of T4SS are present in the conjugative plasmid R388, originally isolated from Escherichia coli (2), and the human intracellular pathogen Bartonella henselae. The conjugative machinery mediates horizontal gene transfer among bacteria and is composed of three functional modules (3): (i) the T4SS itself; (ii) a relaxosome constituted mainly by the relaxase (TrwC in R388), the origin of transfer (oriT), and sometimes an accessory protein and host factors (TrwA and IHF in R388); and (iii) a type IV coupling protein (T4CP) (TrwB in R388), which links the relaxosome and the T4SS. Although the conjugation process implies transfer of a relaxase-DNA nucleoprotein complex, some relaxases, such as VirD2 of Agrobacterium tumefaciens (4) or TrwC of R388 (5), can be translocated by their T4SS in the absence of DNA transfer (5). Others, such as TraI of R1, however, are not translocated under the same conditions (6). In addition to the relaxase, T4SS involved in DNA transfer may translocate other proteins, such as Sog (plasmid ColIb-P9 [7]) or VirE2 (A. tumefaciens, [4]). Other T4SS dedicated to protein translocation constitute bona fide virulence factors in diverse human-pathogenic bacteria, such as Legionella pneumophila, Brucella suis, Brucella abortus, Helicobacter pylori, or B. henselae (8).
In order to be recruited by their respective T4SS, substrates must carry a specific signal for their recognition (Fig. 1) called the translocation signal (TS). Relaxase TSs in conjugative T4SS have been localized to various internal positions using a Cre recombinase assay for translocation (CRAfT) (9, 10). In the case of the relaxase MobA of R1162, they were designated sig 1, located at positions 204 to 323, and sig 2, at 322 to 387 (9). In the relaxase TraI of R1 and F, they were named TSA for the signal at positions 530 to 816 and TSB at 1255 to 1564 (10). The three-dimensional (3D) structure of TSA has recently been reported (11). The initial study on the TS of TraI identified conserved residues in TSA and TSB defining the consensus sequence (G[E/D]R[L/M]R[V/F]T) embedded in a larger TS. The authors proposed an extension of their results to other relaxases based on conservation of the consensus within a RecD2-like part of the proteins (10). Using this sequence, a single TS can be predicted for R388 TrwC within residues 705 to 895.
Fig 1.
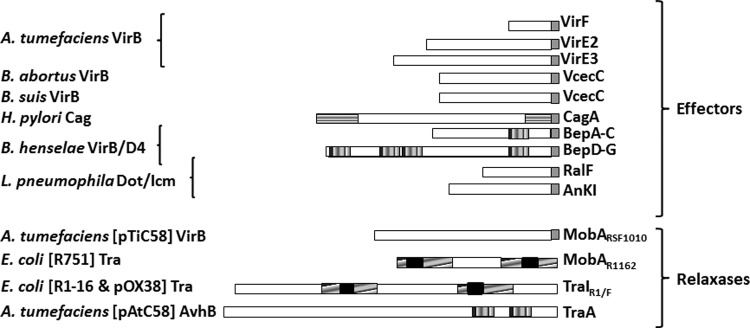
Known translocation signals in T4SS substrates. The T4SS is indicated on the left. Substrate features determined as relevant for secretion are indicated as follows. Gray boxes, C-terminal TSs, including a positively charged secretion motif (R-X7-R-X-R-X-R-X-Xn) (4, 29, 30); vertically hatched boxes, BID domains (13); horizontally hatched boxes, N-terminal and C-terminal regions of H. pylori CagA (12); diagonally hatched boxes, TS-containing regions in relaxases, as determined by CRAfT assays (9, 10); black boxes, consensus TraI/F sequences (G[E/D]R[L/M]R[V/F]T) (10).
In substrates of T4SS involved in effector translocation to eukaryotic cells, the C terminus of effector proteins is necessary, but not always sufficient, for translocation. The positive charge and the hydrophobicity profile of this C-terminal domain have been shown to be relevant characteristics of the TS, rather than a specific amino acid sequence (4). In some cases, additional intrinsic motifs and/or chaperones were also required for secretion (1). VirE2 and other effectors from A. tumefaciens, RalF and other effectors from L. pneumophila, VceC and VceA from Brucella, and CagA from H. pylori require intact C termini. CagA also requires the N terminus to be translocated (12), and all Bartonella effector proteins (BepA to -G) require a bipartite signal consisting of a positively charged C terminus plus at least one copy of the Bartonella intracellular delivery (BID) domain (13).
Several reports have demonstrated the possibility of switching substrates between different T4SS. Relaxase MobA of plasmid R1162 contains an internal bipartite TS for recruitment by the T4SS of conjugative plasmid R751 (9), while MobA from the virtually identical plasmid RSF1010 is recruited through its C-terminal 48 amino acids by A. tumefaciens VirB in the absence of DNA transfer (4). These data indicate that MobA may possess two different TS sites, one involved in recognition by a conjugative system (sig 1 and sig 2) and another (in the C-terminal domain) that interacts with the A. tumefaciens T4SS. The BID domain is naturally present in the conjugative relaxase TraA of plasmid pAtC58 from A. tumefaciens and mediates protein transfer through the VirB/D4 Bartonella T4SS into human endothelial cells (13). TrwC of plasmid R388 is translocated by its T4SS during conjugation (5) and is efficiently recognized by the B. henselae VirB/D4 T4SS (14). The relaxase Mob from the Bartonella cryptic plasmid pBGR1 can also be recognized by this T4SS with low efficiency, which is increased 100-fold by the addition of the secretion signal of BepD (15). In both cases, the translocated protein was linked to DNA, since the detection was based on the expression of a plasmid-encoded enhanced green fluorescent protein (eGFP) (14, 15). R388 and pBGR1 derivative plasmids were transferred from B. henselae to human vascular endothelial cells in a manner that depended on TrwC and the Bartonella VirB system (14, 15). Since no BID domain is apparent in TrwC, the nature of recognition of this protein by the Bartonella VirB/D4 T4SS remains unknown.
In this work, we characterize TrwC requirements for translocation through two different T4SS. We have constructed TrwC derivatives carrying different C termini, including a BID domain, to test plasmid mobilization to other bacteria through the R388 Trw T4SS and to human cells through the Bartonella VirB/D4 T4SS. The C terminus affects T4SS recruitment but does not display sequence specificity. We mapped the TrwC TS for recruitment by the R388 machinery to a short motif in region 796 to 802 showing homology to that of R1-TraI. A mutation affecting this motif almost abolished conjugation. Interestingly, the same mutation barely altered DNA transfer by the T4SS of B. henselae. These results indicate that the recruiting motif needed for conjugation in TrwC is different from that required by the Bartonella VirB/D4 T4SS. This work provides insight into how each T4SS recognizes its particular set of substrates, a main issue in understanding T4SS biology and a key question to accomplish heterologous secretion by substrate manipulation. In fact, we show that addition of a BID to TrwC significantly increases rates of DNA transfer to human cells.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains are listed in Table 1. E. coli strains were grown on Luria-Bertani broth supplemented with agar for solid culture. Strain D1210 was used in all cloning procedures. Bartonella sp. strains were grown on Columbia blood agar plates at 37°C in a 5% CO2 environment. L. pneumophila CECT 7109T was grown on buffered charcoal yeast extract (BCYE) plates (Oxoid) at 37°C in a 5% CO2 atmosphere. Brucella strains were grown on Brucella broth (BB) plates (10 g meat peptone, 10 g casein peptone, 2 g yeast extract, 1 g dextrose, 5 g NaCl, and 0.1 g NaHSO3 [Pronadisa] supplemented with 1.5% [wt/vol] agar) at 37°C in a 5% CO2 environment. Selective media included antibiotics at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 10 μg/ml; kanamycin (Km), 50 μg/ml; nalidixic acid (Nx), 20 μg/ml; streptomycin (Sm), 300 μg/ml (E. coli) or 100 μg/ml (Bartonella); gentamicin (Gm), 10 μg/ml; and tetracycline (Tc), 10 μg/ml.
Table 1.
Bacterial strains used in this work
| Strain | Genotype | Reference |
|---|---|---|
| B. henselae RSE247 | Smr spontaneus mutant from Houston-1 | 31 |
| B. suis 1330 | B. suis biotype 1; wild type | Laboratory strain |
| B. abortus 2308 | B. abortus wild type 2308 | Laboratory strain |
| E. coli D1210 | Smr recA hspR hsdM rpsI lac1 | 32 |
| E. coli DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 33 |
| L. pneumophila subsp. pneumophila CECT7109T | Serotype1; human lung source | Spanish type culture collection |
Plasmid constructs.
Bacterial plasmids are listed in Table 2 (published plasmids) and Table 3 (plasmids constructed for this work). An outline of each plasmid construction is shown in Table 3. Plasmids were constructed using standard techniques (16). Restriction enzymes, shrimp alkaline phosphatase, and T4 DNA ligase were purchased from Fermentas. Kapa high-fidelity polymerase was purchased from Kapa Biosystems. The DNA sequences of all cloned PCR segments were determined.
Table 2.
Published plasmids used in this work
| Plasmid | Antibiotic resistance | Descriptiona | Reference or source |
|---|---|---|---|
| CFP B | Apr | Cre fusion plasmid; cre from phage P1 cloned into the NheI and SalI site of pBR322 | 9 |
| pCIG1077 | Apr | pKK223-3::PABCtrwA-trwL | 34 |
| pCMS1 | Apr | trwC with XhoI site before stop codon | 14 |
| pCre-TrwC | Apr | R388 trwC encoding residues 2 to 966 in CFP B | 35 |
| pHP159 | Gmr | pBBR6::oriT trwABC::eGFP→ | 14 |
| pHP161 | Gmr | pBBR6::oriT trwABC::eGFP← | 14 |
| pHP178 | Gmr, Cmr | pHP161ΔtrwB::Cm | 14 |
| pHP181 | Gmr | pBBR6::oriT trwAB::eGFP | 14 |
| pLA24 | Gmr | pHP159::trwC::BID | 14 |
| pPG104 | Gmr | Source of bepD-BID sequence | 13 |
| pSU1395 | Apr, Kmr | pHG327::oriT trwABC with Tn5tac1 in trwC | 20 |
| pSU1443 | Kmr | pSU1425::Tn5tac1 in trwB | 20 |
| pSU1445 | Kmr | pSU1425::Tn5tac1 in trwC | 20 |
| pSU4058 | Apr | pHG327::trwL-trwD | 36 |
Arrows indicate the orientation of the eGFP cassette.
Table 3.
Plasmids constructed in this work
| Plasmid name | Descriptiona | Construction |
||
|---|---|---|---|---|
| Vectorb | Insert/templatec | Enzymes/oligonucleotides (5′–3′)d | ||
| pAA11 | trwC::ralF TS | pCMS1 | L. pneumophila CECT7109 genomic DNA | CAACTCGAGGACTGGCACTTAAGGAGGGC |
| CAACTCGAGTATCGATACACTATGAGACCGAATGATTT | ||||
| pAA12 | pHP159::trwC::ralF TS | pHP159 | pAA11 | SphI/ClaI |
| pAA14 | trwC*::BIDb | pSU1395 | pPG104 | CCAAGGATCCGCCCCTCTACGAAGGAGTTGGCCCA |
| CCAATGATCATATCGATTACATACCAAAGGCCATTCC | ||||
| pAA15 | pHP159::trwC::BID | pHP159 | pAA14 | SphI/ClaI |
| pAA16 | pHP159::trwC(TS2*) | pHP159 | pDEL39 | SphI/ClaI |
| pAA22 | pHP159::trwC(TS1*) | pHP159 | pDEL40 | SphI/ClaI |
| pAA23 | pHP159::trwC (TS1* + TS2*) | pHP159 | pDEL41 | SphI/ClaI |
| pAA26 | pLA24ΔtrwB::Cm | pLA24 | pHP178 | PmlI/SphI |
| pDEL39e | Cre-TrwC (TS2*) | pCre-TrwC | pSU1443 | TGGCTAACGGTGATCAAATGAAAGTTGTCGCGG (primer a) |
| CTCGGCATAGATTTCA (primer b) | ||||
| ClaI/SalI CCCGTCGACATCGATTTACCTTCCGGCCT (primer c) | ||||
| pDEL40e | Cre-TrwC(TS1*) | pCre-TrwC | pSU1443 | GGGCCGAGCTGGCCGTTGCTGGCGGCGCAATACGCATCACGCGAAAC (primer a) |
| CTCGGCATAGATTTCA (primer b) | ||||
| ClaI/SalI CCCGTCGACATCGATTTACCTTCCGGCCT (primer c) | ||||
| pDEL41e | Cre-TrwC (TS1* + TS2*) | pCre-TrwC | pDEL39 | GGGCCGAGCTGGCCGTTGCTGGCGGCGCAATACGCATCACGCGAAAC (primer a) |
| CTCGGCATAGATTTCA (primer b) | ||||
| ClaI/SalI CCCGTCGACATCGATTTACCTTCCGGCCT (primer c) | ||||
| pEF018 | Step 1: pCMS1::VceC from B. suis | pCMS1 | B. suis 1330 | CCAACTCGAGAACGTTCAGAGCGTCCAGAA |
| CCAACTCGAGTATCGATTATCAACTCGCCAAGCAGCTTT | ||||
| Step 2: pHP159::TrwC::VceC signal from B. suis | pHP159 | pCMS1::VceC | SphI/ClaI | |
| pEF019 | Step 1: pCMS1::VceC from B. abortus | pCMS1 | PCR on genomic DNA from B. abortus 2308 | CCAACTCGAGAACGTTCAGAGCGTCCAGAA |
| CCAACTCGAGTATCGATTACTAATTGCGGGTTTCTC | ||||
| CCTTG | ||||
| Step 2: pHP159::TrwC::VceC signal from B. abortus | pHP159 | pCMS1::VceC | SphI/ClaI | |
| pYG2 | TrwC* | pHP159 | pSU1395 | ACCAAAGCTTATAGCTCAGTCACATGGTATT |
| CCAAATCGATTAGTTGCTCCCTCTGGCGCT | ||||
TrwC*, TrwC derivative with 32 C-terminal residues replaced by LIDCLTAYHRRSGQRCCHCCRRRTGRYGRSIH; TS1*, TrwC variant carrying substitutions G796A, D797G, and T98G and an A insertion; TS2*, TrwC variant Q815R; TS1* + TS2*, double variant.
Vector, backbone plasmid.
Insert/template, plasmid from which the insert was obtained or that used as the template for PCR.
Enzymes/oligonucleotides, either the restriction enzymes used for cloning or the oligonucleotides used for PCR amplification of the desired fragment, with the restriction sites underlined.
The vector was constructed by the megaprimer method (37). Briefly, primers a and c were used to obtain the megaprimer containing mutations and a cloning site (ClaI/SalI). This PCR product (the megaprimer) and the forward-flanking primer b were used to generate the final PCR product, which was introduced into the pCre-TrwC plasmid by restriction with SphI and SalI. Mutations are in boldface.
Mating assays.
Standard E. coli quantitative mating assays were performed as described previously (17). Equal numbers of donor and recipient strains from overnight cultures were mixed and placed on Millipore filters on a prewarmed LB agar plate for 1 h at 37°C. Strains D1210 and DH5α were used as donors and recipients, as indicated. The results are shown as the frequency of transconjugants per donor and are the means of 5 independent experiments. For E. coli-Bartonella matings, plasmids were routinely introduced into Bartonella by conjugation from E. coli DH5α with a helper plasmid (pSU4058 or pCIG1077). Bartonella cells were previously grown on Columbia blood agar plates for 3 to 4 days with appropriate antibiotic selection; bacteria from one plate were collected and washed with 1 ml phosphate-buffered saline (PBS) and then pelleted and resuspended in 1 ml PBS. Donor E. coli strains were grown overnight on liquid LB broth with appropriate antibiotics; 200 μl of donor strains was resuspended in 1 ml of PBS, pelleted, and resuspended in 200 μl PBS, and 1 ml of the Bartonella suspension was mixed with 50 μl of the E. coli suspension. The mixture was pelleted, the supernatant was discarded, and the pellet was placed in a nitrocellulose filter that had been previously placed in a prewarmed Columbia blood agar plate. The mating plate was incubated at 37°C for 6 h. Transconjugant selection was performed on Columbia blood agar with appropriate antibiotics, and the cells were incubated for 6 to 9 days. Total transconjugant DNA was extracted with InstaGene Matrix (Bio-Rad) following the manufacturer's instructions. The transconjugants were then confirmed by multiplex PCR of Bartonella (trwE amplification with primers 5′ CCAGTCGACGGAGGAAAATAATGTTGACA3′ and 5′CCAGAATTCTCTTTTTGTATAGG3′) and of the plasmid (amplification of R388 oriT with primers 5′TTACTCTAGACTCATTTTATGC3′ and 5′TTACTCTAGATTGTAGTGGCAT3′).
Electroporation of B. henselae.
A plate of B. henselae grown for 2 to 3 days was harvested with a sterile cotton swab and resuspended in 950 μl of LB. The suspension was centrifuged at 4,000 rpm for 5 min at 4°C, and the pellet was resuspended in 950 μl of ice-cold 10% glycerol (3 times); 40 μl of these competent cells was transferred to a cooled tube, and 3 μl of DNA (300 ng/μl) was added. The mixture was incubated on ice for 15 min and transferred to a cooled Bio-Rad 0.2-cm cuvette for electroshock with a Bio-Rad Pulse controller II at 2.5 kV/cm, 25 μF, and 200 Ω. After electroporation, 1 ml of SB broth (RPMI 1640 plus l-glutamine, 42 mM HEPES, 1% sodium pyruvate, 5% heat-inactivated fetal calf serum, and 5% sheep blood lysate) was added, and the mixture was transferred to an Eppendorf tube for incubation for 3.5 h at 37°C under 5% CO2 conditions with slow shaking. The cells were then centrifuged at 4,000 rpm for 4 min at room temperature. The pellet was resuspended in 40 μl SB broth and plated on a Columbia agar plate supplemented with the appropriate antibiotics.
Western blots.
The amount of TrwC derivatives in the cells was estimated by Western blotting of total protein extracts, as described previously (18). E. coli D1210 cells containing the indicated plasmids were grown overnight, and the optical density at 600 nm (OD600) was measured. Cells were collected, centrifuged, resuspended in 1/10 volume of 2× SDS gel loading buffer (16), and frozen at −20°C. Samples were boiled for 5 min, and an adequate amount of each sample was applied to SDS-PAGE gels to load the same number of cells per well. After the run, the gels were transferred to nitrocellulose filters. Primary antibody (anti-TrwC [19]) and secondary antibody (peroxidase-conjugated anti-rabbit IgG; ICN) were used at 1:10,000 dilution. Detection was performed with a Supersignal kit (Pierce), and the bands were analyzed on a Bio-Rad ChemiDoc apparatus.
Cell culture, infection, and flow cytometry.
Infection of EA.hy926 cells and flow cytometry analysis were performed as described previously (14). The immortalized hybridoma EA.hy926 cells (ATCC CRL-2922), a fusion cell line of human umbilical vein endothelial cells (HUVEC) with a thioguanine-resistant clone of A549, were grown on Dulbecco's modified Eagle's medium (DMEM) plus Glutamax (Gibco) supplemented with 10% fetal bovine serum (Cambrex) at 37°C in 5% CO2. Once the cells were confluent, they were cultured in 6-well plates (80,000 cells per well) in 3 ml of DMEM. After 16 h, the DMEM was replaced with M199 medium (Gibco) with 10% fetal bovine serum. B. henselae strains were grown directly from stocks (stored at −80°C in 50% glycerol) for 3 to 4 days on Columbia blood agar plates. Later, the cultures were collected and washed with PBS. Bacteria were added to EA.hy926 cells in M199 at a multiplicity of infection (MOI) of 400. Infected cultures were incubated for 3 days in the absence of antibiotics. The cells were then washed with PBS, treated with trypsin, centrifuged, resuspended in 500 μl PBS, and analyzed in a Cytomics FC500 flow cytometer (Beckman Coulter) to quantify green fluorescent protein (GFP)-positive cells.
Computational analysis.
Determination of possible TrwC TS regions was carried out by BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) comparing the previously defined TSA and TSB of R1-TraI against the whole TrwC sequence. Multiple alignments of translocation signal amino acid sequences were performed with ClustalO (http://www.ebi.ac.uk/Tools/msa/-clustalw2/). Predictions of the secondary structure were performed by the PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/). The charge and hydrophobicity of the TrwC tails were calculated by ExPASy ProtParam (http://web.expasy.org/protparam/).
Statistical analysis.
Statistical analysis tests were performed using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA, USA). Student's t test was used to analyze the data referred to the positive control TrwC (pHP161). P values are indicated in the figures.
RESULTS
Construction of TrwC derivatives to test conjugation and DNA transfer to human cells.
The conjugation genes of plasmid R388 include those coding for the T4SS plus a region known as Dtr (for DNA transfer and replication). The Dtr region includes the oriT and the genes coding for the relaxase TrwC, the accessory protein TrwA, and the T4CP TrwB. In order to determine the requirements for TrwC recruitment by different T4SS types, we constructed a set of plasmids derived from pHP161 (14), which contains the R388 Dtr region (oriT plus trwABC). In addition, the plasmids contain a gfp cassette joined with eukaryotic expression signals to detect DNA transfer to eukaryotic cells. The constructs are outlined in Table 3 and described below.
To search for putative TrwC translocation motifs, we performed a BLAST alignment of TrwC with R1-TraI and found two regions that resembled the translocation motif, defined by Lang et al. (10), contained within the TrwC 705-to-895 fragment. We designated the conserved motifs TS1 (GDTIRIT at positions 796 to 802) and TS2 (GDRMKVV at positions 813 to 819). We also performed a ClustalO alignment with the TraI-TS region and with the relaxase TraI of plasmid pKM101 (TraIpKM101), which is more closely related to TrwC. TraIpKM101 showed the same putative translocation motifs as TrwC (Fig. 2A). In order to determine if any of these TrwC motifs are functionally relevant, point mutations were created. Since Lang et al. (10) reported an R-to-Q variant which significantly affected TSB translocation, a similar R815Q variant was constructed in TS2 and denoted TS2*. The R residue is absent in TS1, so we altered the most conserved GDT residues to AGGA, generating TrwC variant TS1*. A mutant form containing both substituted motifs, TS1* plus TS2*, was also constructed. To assess the effects on the overall protein structure, a secondary-structure prediction of the TrwC wild type and variants was performed (Fig. 2B) and also compared to the model, RecD2. Although TS2* was predicted to change the secondary structure at the N-terminal end of the TS1 motif, no significant changes were observed within the proposed TS sequence motifs.
Fig 2.
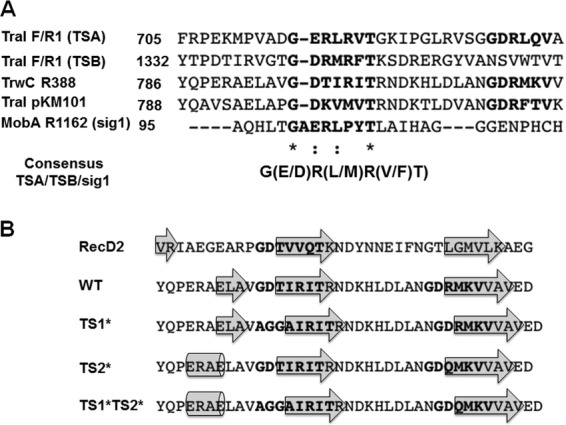
(A) ClustalO alignment of TSs determined for R1/F-TraI and R1162-MobA against the whole R388-TrwC and pKM101-TraI sequences. The asterisks indicate positions that have a single, fully conserved residue; the colons indicate conservation between groups of strongly similar properties. (B) The protein secondary-structure prediction was obtained using Phyre2 for RecD2; wild-type TrwC; and TrwC variants TS1*, TS2*, and TS1* plus TS2*. To simplify the results, the consensus secondary structures are represented by showing the amino acid sequences and the β-strands (arrows) or α-helices (cylinders).
We also constructed TrwC derivatives with different C termini, since the C termini of T4SS substrates have been shown to be essential for translocation in most T4SS involved in virulence (see the introduction). From a previous transposon insertion mutation in trwC (pSU1395 [20]), we knew that insertion at the position 32 amino acids (aa) from the C terminus did not affect the TrwC function in conjugation. Thus, we constructed a plasmid (pYG2) coding for TrwC but with the C-terminal 32 residues replaced with a different sequence (Table 3). This protein is referred to as TrwC*. The other constructs contained defined TSs for T4SS of diverse human intracellular pathogens fused to the C-terminal domain of TrwC: a RalF signal (25 aa) for L. pneumophila Dot/Icm T4SS, VceC signals for B. suis and B. abortus VirB T4SS (VceCBs and VceCBa; 107 and 115 residues, respectively), and the BID signal (183 aa) for the B. henselae VirB/D4 T4SS. The last was also fused at residue 934 of TrwC (TrwC*-BID).
These constructs were (i) introduced into donor E. coli cells carrying a helper plasmid to provide the R388 T4SS to test conjugative DNA transfer into recipient bacteria (referred to here as the conjugation frequency) and (ii) introduced into B. henselae to test DNA transfer through its VirB/D4 T4SS into the cytoplasm of infected human cells, as inferred from eGFP expression. The results obtained are compiled in Fig. 3 and 4 and are described below.
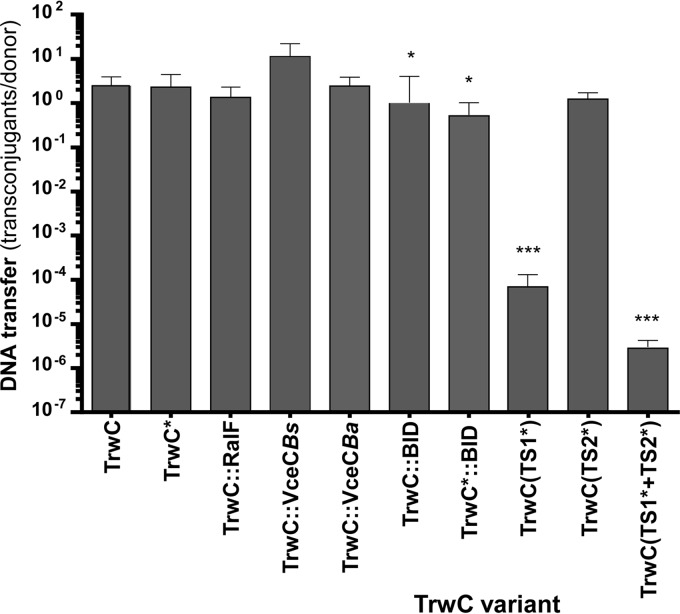
Fig 3.
Conjugative DNA transfer mediated by TrwC derivatives. Donor bacteria contained the plasmid coding for the Dtr region of R388 (oriT-trwABC) and a helper plasmid (pSU4058) to provide R388 T4SS. The TrwC variant in each construct is indicated at the bottom: TrwC*, a variant with different C-terminal 32 residues; double colons indicate domains fused to the C terminus; variants in TS motifs are indicated with asterisks (see the text for details). DNA transfer is expressed as the number of transconjugants per donor. The bars represent means of 5 independent experiments. The error bars indicate standard errors of the mean. Student's t test was used to analyze the data referred to the positive control, TrwC. *, P ≤ 0.01; **, P ≤ 0.001; ***, P ≤ 0.0001.
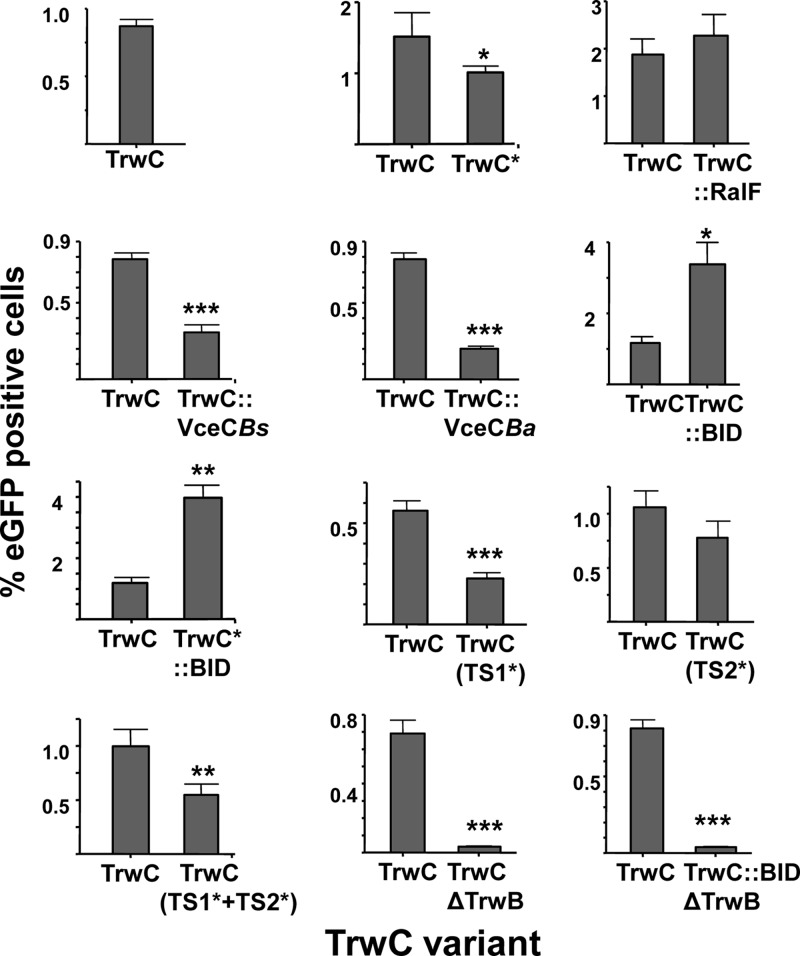
Fig 4.
Percentages of eGFP-positive EA.hy926 cells infected by B. henselae carrying the indicated TrwC variant compared with its own positive control. The TrwC derivative in each construct is indicated at the bottom, as in Fig. 3. The data shown come from the subset of experiments in which the particular construct was assayed in parallel with its positive control. The bars represent means from at least 3 independent experiments done in triplicate. The error bars indicate standard errors of the mean. Student's t test was used to analyze the data referred to the positive control, TrwC. *, P ≤ 0.01; **, P ≤ 0.001; ***, P ≤ 0.0001.
TrwC TS1 is relevant in conjugation but has a minor effect on DNA transfer by the B. henselae VirB/D4 T4SS.
Plasmids containing trwC wild-type or mutant alleles affecting the proposed translocation motifs were assayed for DNA transfer. The results in Fig. 3 and 4 show that the conjugation frequency of TrwC(TS1*) and the double variant in TS1* plus TS2* demonstrated a dramatic decrease compared to wild-type TrwC (more than 5-log-unit difference), while TrwC(TS2*) showed only a slight decrease. In contrast, DNA transfer from B. henselae to human vascular endothelial cells through the B. henselae VirB/D4 T4SS was only reduced to 50% of the wild-type efficiency by the TS1* variant. Thus, TrwC is recognized differently by its own T4SS and by the B. henselae VirB/D4 T4SS.
The C terminus of TrwC is relevant, though not essential, for translocation through both the R388 T4SS and the B. henselae VirB/D4 T4SS.
C termini have been shown to be relevant in the translocation of many T4SS substrates (Fig. 1), where charge and hydrophobicity, rather than specific amino acid sequences, were determined to be key features for TS (4). We evaluated the effects of different C termini in TrwC for translocation through the R388 T4SS during conjugation and through the Bartonella VirB/D4 T4SS into human cells. Western blots with anti-TrwC antibodies detecting proteins with the different C termini showed that all fusion proteins are present in the cell in amounts similar to that of wild-type TrwC, except for the two TrwC::BID fusions, which were present at approximately one-third the amount of wild-type TrwC (data not shown). The results in Fig. 3 show that conjugation frequencies are only slightly affected by the different C termini. TrwC*, whose C-terminal 32 residues are different, is efficiently translocated during conjugation. The addition of C termini shown to function as TSs in other T4SS effectors did not affect DNA transfer significantly, except for the addition of the BID domain, which decreased the DNA transfer rate. However, this could be attributed to the lower steady-state level of the BID fusions.
DNA transfer rates through the Bartonella VirB/D4 T4SS are shown in Fig. 4. To determine significant differences, each value was compared to that of the wild type in the same experiment. The results indicate a stronger effect of the different C termini on DNA transfer to human cells and, to a certain extent, show an effect opposite to that on conjugative DNA transfer (compare the data in Table 4). TrwC* was translocated at a significant rate, indicating that the C-terminal 32 residues of the relaxase TrwC per se are not essential. TrwC fusion to different C termini provoked a wide range of effects, from a significant decrease (TrwC::VceC) to a substantial increase (TrwC::BID) in DNA transfer rates. Significantly, addition of the BID signal tripled DNA transfer rates to human cells, even if the amount of TrwC-BID was smaller than that of wild-type TrwC. This result is easily explained, since the BID domain has been defined as the TS for substrates of the B. henselae VirB/D4 T4SS. In a previous study, we added the BID translocation signal to the C-terminal domain of TrwC in an attempt to improve DNA transfer through the VirB/D4 T4SS of B. henselae to human vascular endothelial cells, without success; however, this was probably due to the construct used in those assays, in which the TrwC-BID levels were severely reduced, as judged by Western blotting (14).
Table 4.
Summary of rates of DNA transfer to bacteria and human cells
| TrwC derivative | Description | C terminusa | Relative conjugation frequencyb | Relative DNA transfer to human cellsb |
|---|---|---|---|---|
| TrwC | Wild type (966 aa) | WT | 100 | 100 |
| TrwC* | 934 aa + 32-aa tail | +, F | 95 | 67 |
| TrwC-RalF | 966 aa + 25-aa RalF TS | = | 55 | 114 |
| TrwC-VceC | 966 aa + 107-aa B. suis VceC | FF | 457 | 39 |
| TrwC-VceC | 966 aa + 115-aa B. abortus VceC | ++, F | 99 | 25 |
| TrwC-BID | 966 aa + 183-aa BID | = | 21 | 278 |
| TrwC*-BID | 934 aa + 183-aa BID | = | 16 | 314 |
| TrwC-BID (no TrwB) | 966 aa + 183-aa BID (no trwB) | = | NAc | 5 |
| TrwC (TS2*) | 966 aa (TS2 R-Q) | WT | 51 | 73 |
| TrwC (TS1*) | 966 aa (TS1 GDT-AGGA) | WT | 3 × 10−3 | 41 |
| TrwC (TS1* + TS2*) | 966 aa (TS1 + TS2) | WT | 1 × 10−4 | 55 |
C terminus, relevant characteristics of the C-terminal 32 residues (extracted from data shown in Table 5). WT, wild type; =, similar to the wild type in terms of charge and hydrophobicity; + and ++, positive net charge >2-fold and 3-fold more, respectively, than the WT; F and FF, more hydrophobic than the WT (grand average of hydropathicity [GRAVY], an index that indicates the solubility of proteins, less than half- or one-third that of the WT, respectively).
Values are expressed as percentages relative to the positive-control mean value from the experiments in which the indicated construction was assayed, as shown in Fig. 3 and 4.
NA, not applicable; this plasmid (which lacks TrwB) was assayed for conjugation in the presence of pSU1445 (R388 trwC mutant), so the results are not comparable with the rest of the matings.
We analyzed the charges and hydrophobicities of the different C termini assayed, since they have been reported to be relevant features for TSs in several T4SS effectors (4). Table 5 shows the net positive charges and hydrophobicities of (i) the C-terminal 20 residues, defined as sufficient for T4SS recruitment in several T4SS effectors; (ii) the C-terminal 32 residues that correspond to the fragment replaced in TrwC*; and (iii) the C-terminal 183 residues, which together correspond to the size of the BID. The C termini of TrwC*, TrwC-VceCBs, and especially TrwC-VceCBa showed a net positive charge higher than that in wild-type TrwC; however, these three TrwC variants showed the lowest DNA transfer rates through the B. henselae VirB/D4 T4SS, so a positive charge at the C terminus was not a necessary feature for recruitment by the B. henselae VirB/D4 T4SS. It can also be observed that C termini with higher hydrophobicity also show lower levels of DNA transfer, supporting a possible influence on translocation. In any case, hydrophobicity does not seem to be a prominent feature of TSs; the BID domain, which is by far the best substrate for the B. henselae VirB/D4 T4SS, provides a C terminus more closely related to that of wild-type TrwC in terms of charge and hydrophobicity (Table 5).
Table 5.
Net charges and hydrophobicities of the C termini of TrwC derivatives
| Plasmid | TrwC description | C-terminal 183 aa |
C-terminal 32 aa |
C-terminal 20 aa |
|||
|---|---|---|---|---|---|---|---|
| Charge | GRAVYa | Charge | GRAVY | Charge | GRAVY | ||
| pHP161 | 966 aa (wild type) | +7 | −1.039 | +3 | −2.416 | +1 | −1.705 |
| pYG2 | 934 aa + 32-aa C-terminal tail | +10 | −0.738 | +7 | −0.891 | +6 | −1.320 |
| pAA12 | 966 aa + 25-aa C- terminal tail (RalF TS) | +8 | −1.068 | +2 | −1.278 | +1 | −1.530 |
| pEF18 | 966 aa + 107-aa C-terminal tail (VceC B. suis TS) | +18 | −0.868 | +5 | 0.056 | +5 | −0.810 |
| pEF19 | 966 aa + 115-aa C-terminal tail (VceC B. abortus TS) | +26 | −0.969 | +9 | −0.769 | +8 | −1.440 |
| pAA15 | 934 aa + 183-aa C-terminal tail (BID) | +5 | −1.141 | +2 | −2.013 | +2 | −1.740 |
| pLA24 | 966 aa + 183-aa C-terminal tail (BID) | +5 | −1.141 | +2 | −2.013 | +2 | −1.740 |
GRAVY (grand average of hydropathicity) is an index that indicates the solubility of proteins; positive is hydrophobic, and negative is hydrophilic.
Role of R388 T4CP TrwB in DNA transfer through the VirB/D4 T4SS.
We previously reported that in the absence of TrwB, the R388 T4CP, DNA transfer rates through the B. henselae VirB/D4 T4SS to human endothelial cells decreased by 1 log unit (14). This residual DNA transfer, which was TrwB independent, led us to speculate that the B. henselae T4CP, VirD4, could also be acting in substrate recruitment. In the presence of the BID domain, we would expect that the role of VirD4 would be more predominant. Accordingly, we constructed a plasmid coding for TrwC::BID but lacking trwB (pAA26). This plasmid was tested for TrwC function in conjugation by complementation of an R388 trwC mutant (pSU1445) to exclude a polar effect of trwB deletion in trwC. The plasmid displayed wild-type conjugation frequencies (not shown). In contrast, the DNA transfer rate through the B. henselae VirB/D4 T4SS decreased drastically and was comparable to that obtained with wild-type TrwC in the absence of TrwB (Fig. 4). These results argue that R388 TrwB may serve as the T4CP for the VirB/D4 T4SS under these conditions.
DISCUSSION
Type IV secretion systems translocate specific substrates, implying the recognition of a specific TS by each T4SS. Previous works have elucidated several TSs in substrates of both T4SS types involved in pathogenesis and in horizontal DNA transfer. While effectors translocated by T4SS into eukaryotic cells are mainly recruited through their C termini, bacterial relaxases show extended TS regions along the protein (see the introduction). TrwC, the relaxase of plasmid R388, is the protein substrate of the R388 T4SS, previously shown to also be secreted through the VirB/D4 T4SS of B. henselae (14). Thus, TrwC may be an excellent model system to ascertain molecular determinants for recognition by different T4SS. In this work, we analyzed the TrwC regions necessary for its translocation by its cognate conjugative T4SS, as well as through the VirB/D4 T4SS of B. henselae. We targeted two regions of TrwC: the C terminus and the proposed TrwC TS region based on its homology to TraI-TS. The different TrwC derivatives were assayed for DNA transfer to recipient bacteria (through the R388 T4SS) and human cells (through the T4SS of B. henselae). The results are compiled in Table 4 to facilitate their discussion.
Alignment of TrwC with TraI and MobA defined two possible motifs resembling the one described by Lang et al. as critical for protein translocation (10); while the first one (motif TS1) aligned with the defined consensus, the nearby sequence (motif TS2) also resembled this consensus and, moreover, included an Arg residue that, according to the previous work in TraI, was a key residue in TSB (10). We constructed point mutations affecting both putative TS motifs and analyzed their effects on DNA transfer. We found that TS1* exerted dramatic effects on conjugation frequency, while TS2* had no significant impact. Interestingly, when assayed for DNA transfer through the VirB/D4 T4SS of B. henselae, the same TS1* variant does not provoke such a drastic effect on DNA transfer rates, implying that TrwC(TS1*) can be efficiently translocated with DNA into recipient cells through another T4SS. This result implies that the most plausible explanation for the extreme conjugation phenotype observed for TS1* is a lack of recognition by its own T4SS. These results confirm that the TS1 motif presents key determinants for TrwC translocation through its own T4SS. Moreover, this result reveals a trwC point mutation with a drastic effect on relaxase function not observed previously.
In addition, we constructed TrwC derivatives fused to different C termini and assayed them for DNA transfer to both bacterial and human cells. We have demonstrated that the addition of different C-terminal domains to TrwC does not substantially affect conjugative DNA transfer through the R388 T4SS; on the contrary, the addition of different C termini affects DNA transfer through the B. henselae VirB/D4 T4SS more significantly. In turn, the TS variants affected recruitment by VirB/D4 to a much lesser extent than that by the R388 T4SS. In summary, we observed different recruitment requirements of different T4SS for a single T4SS substrate. These results are in agreement with previous reports regarding the TSs of conjugative relaxases and effectors of T4SS involved in virulence, particularly previous findings for relaxase MobA, in which different groups working with differently named plasmids (R1162 and RSF1010) identified different TSs for MobA recruitment by a conjugative T4SS or by the A. tumefaciens VirB T4SS (see the introduction).
The C terminus of TrwC is not required for recruitment by the B. henselae VirB/D4 T4SS, since TrwC* (with different C-terminal 32 aa) is efficiently recruited. It has been reported that C-terminal TS elements in different effector proteins are defined by the presence of positive charges and a hydrophilic profile (4). We analyzed these parameters in the TrwC variants used in this work (Table 5). Physical features of the C terminus other than charge and hydrophobicity must be relevant to substrate recognition, since we did not observe any correlation between the C-terminal charge or hydrophobicity values and apparent DNA transfer rates.
TrwC was previously shown to be recruited by the VirB/D4 T4SS of B. henselae; at the same time, DNA transfer was shown to be totally dependent on VirD4 and, to a lesser extent, on TrwB, raising questions about the role of each T4CP in this DNA transfer process (14). In agreement with a previous report (15), the addition of a BID domain increases the DNA transfer rate through the B. henselae VirB/D4 T4SS (Fig. 4). In the absence of TrwB, this TrwC::BID-mediated DNA transfer is as impaired as when wild-type TrwC is present (Fig. 4). This result suggests that TrwC::BID and TrwC use R388 TrwB similarly as a T4CP and that the increase in DNA transfer in the presence of BID is not due to preferential recruitment by the B. henselae T4CP VirD4, which would not be hindered by the absence of TrwB.
Since TrwB is involved in TrwC recruitment to both T4SS types while the effects of TS1* are so different in conjugation and in DNA transfer to eukaryotes, it seems that the TS1 motif may be interacting with another component rather than the T4CP TrwB. It has been suggested previously (9, 10) that other factors in addition to the T4CP must be involved in substrate recognition by T4SS. sig1 and sig2 on MobA probably possess elements for binding to the T4CP, to MobB, or to both (9). No cytoplasmic chaperones have been reported to interact with the Bartonella VirB/D4 T4SS, but there are several examples of chaperones implicated in substrate recognition and translocation through T4SS, such as the IcmS and IcmW protein complex of L. pneumophila Dot/Icm (21) or VirE1 of A. tumefaciens VirB/D4 (22). In addition to chaperones, molecules involved in presentation and spatial positioning of the nucleoprotein substrate have been reported for A. tumefaciens VirB/D4 as coming both from the plasmid and from the host (23, 24). VBP proteins, with homologues in multiple bacterial genera, were shown to recruit conjugative substrates to different transfer systems (24). Similar factors could be present in Bartonella, which would help in recruitment of the TrwC-DNA complex, and especially TrwC::BID.
It can be anticipated that the determination of the exact requirements for substrate recruitment by different T4SS may pave the way for the development of customized DNA and protein delivery machines. TrwC can be targeted to the nucleus, and it can integrate the commutatively transferred DNA into its target sequence present in the recipient cell genome (25, 26). These capacities of the system confer a great biotechnological potential (27). With the addition of the BID domain to TrwC, we have increased the number of eGFP-positive cells up to 4% of the infected cells. This result improves the potential of TrwC as a biotechnological tool to deliver long DNA molecules into specific human cells (28).
ACKNOWLEDGMENTS
A.A. and D.L. were supported by a JAE-Doc and a JAE-Predoc fellowship, respectively, from the Consejo Superior de Investigaciones Científicas (CSIC, Spain). This work was supported by grant BIO2010-11623-E from the Spanish Ministry of Science and Innovation to M.L.
We thank Yolanda González-Flores for the pYG2 construct, Coral González-Prieto for help with Bartonella transformation, Jorge Ripoll for help with the Western blots, and Lukas Matern for helpful comments on the manuscript.
Footnotes
Published ahead of print 30 August 2013
REFERENCES
- 1.Alvarez-Martínez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datta N, Hedges RW. 1972. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J. Gen. Microbiol. 72:349–355 [DOI] [PubMed] [Google Scholar]
- 3.Llosa M, de la Cruz F. 2005. Bacterial conjugation: a potential tool for genomic engineering. Res. Microbiol. 156:1–6 [DOI] [PubMed] [Google Scholar]
- 4.Vergunst AC, van Lier MC, den Dulk-Ras A, Grosse Stuve TA, Ouwehand A, Hooykaas PJ. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. U. S. A. 102:832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draper O, César CE, Machón C, de la Cruz F, Llosa M. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. U. S. A. 102:16385–16390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang S, Zechner EL. 2012. General requirements for protein secretion by the F-like conjugation system R1. Plasmid 67:128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkins BM, Thomas AT. 2000. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol. Microbiol. 38:650–657 [DOI] [PubMed] [Google Scholar]
- 8.Llosa M, Roy C, Dehio C. 2009. Bacterial type IV secretion systems in human disease. Mol. Microbiol. 73:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker C, Meyer RJ. 2007. The R1162 relaxase/primase contains two type IV transport signals that require the small plasmid protein MobB. Mol. Microbiol. 66:252–261 [DOI] [PubMed] [Google Scholar]
- 10.Lang S, Gruber K, Mihajlovic S, Arnold R, Gruber CJ, Steinlechner S, Jehl MA, Rattei T, Frohlich KU, Zechner EL. 2010. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol. Microbiol. 78:1539–1555 [DOI] [PubMed] [Google Scholar]
- 11.Redzej A, Ilangovan A, Lang S, Gruber CJ, Topf M, Zangger K, Zechner EL, Waksman G. 2013. Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Mol. Microbiol. 89:324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohlfeld S, Pattis I, Puls J, Plano GV, Haas R, Fischer W. 2006. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol. Microbiol. 59:1624–1637 [DOI] [PubMed] [Google Scholar]
- 13.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, Carena I, Dehio C. 2005. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. U. S. A. 102:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-González E, de Paz HD, Alperi A, Agúndez L, Faustmann M, Sangari FJ, Dehio C, Llosa M. 2011. Transfer of R388 derivatives by a pathogenesis-associated type IV secretion system into both bacteria and human cells. J. Bacteriol. 193:6257–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schröder G, Schülein R, Quebatte M, Dehio C. 2011. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc. Natl. Acad. Sci. U. S. A. 108:14643–14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17.Grandoso G, Avila P, Cayón A, Hernando MA, Llosa M, de la Cruz F. 2000. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J. Mol. Biol. 295:1163–1172 [DOI] [PubMed] [Google Scholar]
- 18.de Paz HD, Larrea D, Zunzunegui S, Dehio C, de la Cruz F, Llosa M. 2010. Functional dissection of the conjugative coupling protein TrwB. J. Bacteriol. 192:2655–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandoso G, Llosa M, Zabala JC, de la Cruz F. 1994. Purification and biochemical characterization of TrwC, the helicase involved in plasmid R388 conjugal DNA transfer. Eur. J. Biochem. 226:403–412 [DOI] [PubMed] [Google Scholar]
- 20.Llosa M, Bolland S, de la Cruz F. 1994. Genetic organization of the conjugal DNA processing region of the IncW plasmid R388. J. Mol. Biol. 235:448–464 [DOI] [PubMed] [Google Scholar]
- 21.Ninio S, Zuckman-Cholon DM, Cambronne ED, Roy CR. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55:912–926 [DOI] [PubMed] [Google Scholar]
- 22.Vergunst AC, Van Lier MC, Den Dulk-Ras A, Hooykaas PJ. 2003. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 133:978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 26:2540–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo M, Jin S, Sun D, Hew CL, Pan SQ. 2007. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc. Natl. Acad. Sci. U. S. A. 104:20019–20024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agúndez L, González-Prieto C, Machón C, Llosa M. 2012. Site-specific integration of foreign DNA into minimal bacterial and human target sequences mediated by a conjugative relaxase. PLoS One 7:e31047. 10.1371/journal.pone.0031047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agúndez L, Machón C, César CE, Rosa-Garrido M, Delgado MD, Llosa M. 2011. Nuclear targeting of a bacterial integrase that mediates site-specific recombination between bacterial and human target sequences. Appl. Environ. Microbiol. 77:201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Prieto C, Agundez L, Linden RM, Llosa M. 2013. HUH site-specific recombinases for targeted modification of the human genome. Trends Biotechnol. 31:305–312 [DOI] [PubMed] [Google Scholar]
- 28.Llosa M, Schroder G, Dehio C. 2012. New perspectives into bacterial DNA transfer to human cells. Trends Microbiol. 20:355–359 [DOI] [PubMed] [Google Scholar]
- 29.de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. 2008. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 70:1378–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U. S. A. 102:826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid MC, Schulein R, Dehio M, Denecker G, Carena I, Dehio C. 2004. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Mol. Microbiol. 52:81–92 [DOI] [PubMed] [Google Scholar]
- 32.Sadler JR, Tecklenburg M, Betz JL. 1980. Plasmids containing many tandem copies of a synthetic lactose operator. Gene 8:279–300 [DOI] [PubMed] [Google Scholar]
- 33.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87:4645–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.César CE, Llosa M. 2007. TrwC-mediated site-specific recombination is controlled by host factors altering local DNA topology. J. Bacteriol. 189:9037–9043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kienesberger S, Schober Trummler C, Fauster A, Lang S, Sprenger H, Gorkiewicz G, Zechner EL. 2011. Interbacterial macromolecular transfer by the Campylobacter fetus subsp. venerealis type IV secretion system. J. Bacteriol. 193:744–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolland S, Llosa M, Avila P, de la Cruz F. 1990. General organization of the conjugal transfer genes of the IncW plasmid R388 and interactions between R388 and IncN and IncP plasmids. J. Bacteriol. 172:5795–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar G, Sommer SS. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 8:404–407 [PubMed] [Google Scholar]