Abstract
d-Amino acids have been shown to play an increasingly diverse role in bacterial physiology, yet much remains to be learned about their synthesis and catabolism. Here we used the model soil- and rhizosphere-dwelling organism Pseudomonas putida KT2440 to elaborate on the genomics and enzymology of d-amino acid metabolism. P. putida KT2440 catabolized the d-stereoisomers of lysine, phenylalanine, arginine, alanine, and hydroxyproline as the sole carbon and nitrogen sources. With the exception of phenylalanine, each of these amino acids was racemized by P. putida KT2440 enzymes. Three amino acid racemases were identified from a genomic screen, and the enzymes were further characterized in vitro. The putative biosynthetic alanine racemase Alr showed broad substrate specificity, exhibiting measurable racemase activity with 9 of the 19 chiral amino acids. Among these amino acids, activity was the highest with lysine, and the kcat/Km values with l- and d-lysine were 3 orders of magnitude greater than the kcat/Km values with l- and d-alanine. Conversely, the putative catabolic alanine racemase DadX showed narrow substrate specificity, clearly preferring only the alanine stereoisomers as the substrates. However, DadX did show 6- and 9-fold higher kcat/Km values than Alr with l- and d-alanine, respectively. The annotated proline racemase ProR of P. putida KT2440 showed negligible activity with either stereoisomer of the 19 chiral amino acids but exhibited strong epimerization activity with hydroxyproline as the substrate. Comparative genomic analysis revealed differences among pseudomonads with respect to alanine racemase genes that may point to different roles for these genes among closely related species.
INTRODUCTION
Of the 20 canonical proteinogenic amino acids, 19 are chiral about their α carbon and therefore exist in one of two spatial arrangements, referred to as the l- and d-stereoisomers. Nature has effectively selected for l-amino acids to serve as the building blocks of ribosomally synthesized peptides and as important metabolic intermediaries in the cell. Their corresponding d-enantiomers are far less prevalent in most biological systems. Nonetheless, the d-enantiomer of each of the 19 amino acids is detected in biological systems (1–6), and in certain environments, d-amino acids are abundant. These include microbe-rich environments, such as topsoil (7), fermented foods (8), and the rumen (9). Where free d-amino acid content is measured in bacterial culture supernatants or ethanolic extracts of freeze-dried bacterial samples, the most abundant free d-amino acid is typically d-alanine, but high concentrations of d-aspartate, d-glutamate, d-leucine, and d-methionine are also noted in some species (1, 9). The d-stereoisomer of alanine comprises up to 65% of the free alanine in some samples (9), and millimolar d-alanine concentrations have been measured in culture supernatants (1). Consequently, when d-amino acids are detected in environmental samples, their presence is typically attributed to bacteria.
Because certain d-amino acids are essential components in bacterial peptidoglycan (e.g., d-alanine and d-glutamate), their abundance in bacterial culture is not surprising (10). Nonetheless, the d-amino acid distribution in culture does not necessarily match that expected simply for synthesis of peptidoglycan (1). Recent work has shown that complex physiological processes, such as biofilm formation, peptidoglycan remodeling, and sporulation, are influenced by the presence of certain d-amino acids (1, 6, 11). For example, the d-stereoisomers of leucine, methionine, tryptophan, and tyrosine have been shown to disassemble mature biofilms of Bacillus subtilis at concentrations as low as 10 nM (6). The corresponding l-enantiomer does not have the same effect. In this case, the bacterium is shown to synthesize the specific d-amino acid of note, but little is known about how this is accomplished. Further, bacterial catabolism of certain d-amino acids is noted and may be important for colonization of d-amino-acid-rich environments (12, 13).
Bacterial synthesis of d-amino acids proceeds via enzymatic racemization of the corresponding l-enantiomer. Amino acid racemases catalyze the interconversion of the l- and d-enantiomers using either a pyridoxal-5′-phosphate (PLP)-dependent or a PLP-independent mechanism. The PLP-dependent alanine racemase enzyme class has been studied extensively, owing to its potential as a target for antimicrobials (14). These enzymes are known to be necessary for d-alanine synthesis for peptidoglycan, as targeted disruption results in d-alanine auxotrophy (15). However, many bacteria encode more than one annotated alanine racemase in their genome. Catabolism of d-amino acids can also be initiated through enzymatic racemization to form the corresponding l-amino acid, although racemase-independent catabolic mechanisms also exist.
Pseudomonads are noted as models in ecological genomics (16, 17), pathogenesis (18, 19), and host-microbe interactions (20, 21). They may also be considered models in d-amino acid biology. Recent work in Pseudomonas aeruginosa PAO1 and Pseudomonas putida KT2440 has revealed unique mechanisms by which d-amino acids are catabolized (12, 13, 22). Here we build on this work by assessing the d- and l-amino acid catabolic capacity of P. putida KT2440, employing a functional screen to identify genes involved in d-amino acid metabolism, characterizing the enzymes involved in amino acid racemization, and identifying key differences in the genomics of d-amino acid metabolism between related pseudomonads.
MATERIALS AND METHODS
Growth studies using d- and l-amino acids as the sole source of carbon and nitrogen.
Sterile phosphate plus glucose (PG) medium (23) was prepared without a carbon or nitrogen source and was supplemented with a sterile amino acid stock to a final concentration of 25 mM. Three 3-ml growth tests of each amino acid enantiomer were conducted. An overnight culture of Pseudomonas putida KT2440, incubated in liquid PG medium containing the conventional carbon and nitrogen sources (0.5% glucose and 25 mM ammonium sulfate [23]), was washed with sterile water, and 10 μl was used to inoculate each tube. Growth controls consisted of uninoculated culture tubes. The culture tubes were maintained at 28°C with shaking, and the optical density at 600 nm (OD600) was measured after 24, 48, and 72 h (BioMate 3 spectrophotometer; Thermo Scientific). The respective growth control for each amino acid was used as a blank.
Genomic library construction and screening.
Genomic DNA from Pseudomonas putida KT2440 was isolated using a GenElute bacterial genomic DNA kit (Sigma-Aldrich) according to the instructions supplied with the kit. Genomic DNA was fragmented by hydrodynamic shearing (GeneMachines hydroshear; speed, 19; number of cycles, 20). It was subsequently end repaired (DNATerminator end-repair kit; Lucigen), and 8-kb to 15-kb fragments were gel extracted after performing agarose gel electrophoresis (Thermo Scientific GeneJET gel extraction kit). The purified DNA was ligated into a blunt-ended pUC19 vector using T4 DNA ligase (2 units; Thermo Scientific), and the entire volume of all reaction mixtures was transformed into electrocompetent Epi300 Escherichia coli cells (Epicentre). The library was prepared by incubating the recovery in LB medium with carbenicillin selection (100 μg ml−1) overnight at 37°C with shaking, harvesting the cells by centrifugation, and preparing a stock in 20% glycerol maintained at −80°C (24). DNA was isolated from the recombinant genomic library by use of a miniprep kit (GeneJET plasmid miniprep kit; Thermo Scientific) and used to construct four libraries in E. coli amino acid auxotrophic strains obtained from the Coli Genetic Stock Center (www.cgsc.biology.yale.edu; leucine, strain JW5807-2; lysine, strain JW2806-2; proline, strain JW0233-2; phenylalanine, strain JW2580-1). Auxotrophs were in-frame, single-gene knockouts from the Keio Collection (25).
Recombinant auxotrophic genomic libraries were screened for recovery of growth on minimal medium supplemented with d-proline (10 mM), d-lysine (1 mM), d-leucine (10 mM), or d-phenylalanine (10 μM) (Sigma-Aldrich) in lieu of the corresponding l-amino acid. Briefly, portions of the glycerol stock were grown in LB medium overnight at 37°C with shaking, after which the cultures were washed with sterile water and 50 μl of 1/100 dilutions was plated on minimal PG medium containing the respective d-amino acid. Serial dilutions were also plated on LB medium supplemented with carbenicillin to assess the number of CFU plated for each screen. Screen coverage was estimated using the total number of CFU screened and the average insert size from the genomic library. The screens were developed at 28°C. Positive clones, defined as those recombinant genomic clones that conferred growth of the auxotroph in the presence of the tested d-amino acid but not in its absence, were restreaked for isolation on PG medium supplemented with the d-amino acid of note, and cultures were started for plasmid DNA isolation (GeneJET plasmid miniprep kit; Thermo Scientific). Positive clones were retransformed into a fresh auxotrophic background and verified as described above. Additionally, open reading frames (ORFs) of interest from recovered clones (alanine racemase/DadX [PP5269; accession no. NP_747370], alanine racemase/Alr [PP3722; accession no. NP_745855], proline racemase [PP1258; accession no. NP_743418]) were cloned into pUC19 with primers dadX F (5′-ATATGGATCCTATGCGTCCCGCCCGCGCCCTGATC-3′; the BamHI sequence is underlined) and dadX R (5′-CCGCGAGCTCTCATTCGCCGATGTAGTCCCGTGGT-3′; the SacI sequence is underlined), primers alr F (5′-ATATGGATCCTATGCCCTTTCGCCGTACCCTTCTG-3′; the BamHI sequence is underlined) and alr R (5′-CCGCGAATTCTCAGTCGACGAGTATCTTCGGGTTG-3′; the EcoRI sequence is underlined]), and primers proR F (5′-CGCCGGATCCTATGAAACAGATTCACGTCATCGAC-3′; the BamHI sequence is underlined]) and proR R (5′-TAACGAATTCTCAGATGCCCCAGGCGAAAGGGTCT-3′; the EcoRI sequence is underlined), respectively. The constructs were transformed via electroporation into proline, leucine, or lysine auxotrophic cells to verify that the identified open reading frame itself confers growth on the d-amino acid substrate.
DNA was sequenced on an ABI 3730 instrument (Applied Biosystems) at the University of Kentucky Advanced Genetic Technology Center using a cycle sequencing kit (BigDye Terminator, v3.1, cycle sequencing kit; Applied Biosystems) to elucidate the insert DNA from each positive clone. The genomic region harboring the DNA insert from positive clones was identified using the bioinformatics resources provided by the Pseudomonas Genome Database (26).
Preparation of constructs and protein purification.
Open reading frames encoding the three putative amino acid racemases described above were cloned into pET28b using the same PCR primers. The constructs were employed for the production of an N-terminal histidine-tagged protein using E. coli Rosetta2(DE3) (EMD4Biosciences, EMD Millipore). For gene overexpression, an overnight culture of Rosetta2(DE3) cells containing the appropriate construct was used to inoculate 500 ml LB culture, which was grown at 37°C with shaking until the OD600 was approximately 0.6 to 0.7. The culture was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and shaken vigorously (270 rpm) for 3 h at 28°C. Induced cells were harvested by centrifugation and stored at −80°C.
For protein purification, the cells were resuspended in 2 ml buffer A2X [50 mM HEPES, pH 7.4, 200 mM NaCl, 1.95 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP), 10% glycerol] and lysed via sonication (10 20-s pulses at 60 V with 2-min breaks on ice), and the cell lysate was subjected to immobilized metal affinity column chromatography using a His-Select nickel affinity gel (Sigma-Aldrich). Following loading, the column was washed with buffer A (50 mM HEPES, pH 7.4, 100 mM NaCl, 0.97 mM TCEP, 5% glycerol) supplemented with 20 mM imidazole and then eluted with buffer A containing 250 mM imidazole. Fractions containing the purified protein were identified by SDS-PAGE and were concentrated and buffer exchanged into buffer A2X using centrifugal concentration (7 ml, 9,000-molecular-weight cutoff; Pierce concentrators; Thermo Scientific). Protein concentrations were determined via use of the protein assay dye reagent from Bio-Rad. Purified samples were analyzed by SDS-PAGE (Precise Tris-HEPES gels; Thermo Scientific) to assess apparent protein purity. The concentration of the purified protein was determined as described above, and 50-μl aliquots were snap-frozen in liquid nitrogen and stored at −80°C until use.
In vitro enzyme assays.
Amino acid racemization assays were performed in 2-ml Eppendorf tubes for 1 min at 37°C. The total volume for each assay was 200 μl and comprised the following at the indicated final concentrations: 50 mM HEPES (pH 7.4) and 20 μM PLP (PLP was added only for the characterization of the annotated alanine racemases and was not included for the proline racemase). Reactions were initiated by the addition of purified enzyme. For substrate specificity assays, the following were used at the indicated concentrations: alanine racemase/Alr, 1 μM; alanine racemase/DadX, 0.7 μM; and proline racemase, 1.9 μM. For kinetic assays, the following were used at the indicated concentrations: alanine racemase/Alr, 1 μM with alanine as the substrate and 1 nM with lysine as the substrate; alanine racemase/DadX, 0.7 μM with alanine as the substrate; proline racemase, 9.5 nM with cis-d-hydroxyproline (cis-d-HyPro) as the substrate and 1.9 μM with trans-l-hydroxyproline (trans-l-HyPro) as the substrate. To quench the enzyme reaction, 40 μl of 2 M HCl was added. The molar concentration of enzyme used in each reaction mixture was 10% or less of the initial substrate concentration, and we chose a reaction duration that allowed the conversion of 10% or less of the initial substrate to minimize the reverse reaction.
Marfey's reagent (1-fluoro-2,4-dinitrophenyl-5-l-alanine amide [FDAA]; Sigma-Aldrich) was used for the derivatization of amino acids and reverse-phase high-pressure liquid chromatography (HPLC; Dionex Ultimate 3000; Waters Nova-Pak C18 column [3.9 mm by 150 mm]) was employed to determine the concentration of each enantiomer (27). To avoid the hydrolysis of Marfey's reagent, 40 μl of 2 M NaOH was added to neutralize the quenched enzyme reaction prior to derivatization, followed by addition of 100 μl Marfey's reagent (0.5% solution in acetone) and 20 μl 1 M NaHCO3. The derivatization mixture was incubated at 40°C for 1 h, after which it was allowed to cool to room temperature and diluted 10-fold in 80% 0.05 M triethylamine-phosphate (TEAP) buffer, pH 3.0–20% acetonitrile (concentrated phosphoric acid was used to adjust the pH of the triethylamine solution to prepare the TEAP buffer). The resulting solution was passed through a 0.22-μm-pore-size filter (nonsterile hydrophobic polytetrafluoroethylene [PTFE] syringe filters; Tisch Scientific) and placed into an amber glass vial prior to HPLC analysis. A gradient method was used for HPLC separation of most amino acids, starting at 80% TEAP buffer (pH 3.0) and 20% acetonitrile and ramping to 70% buffer and 30% acetonitrile over 5 min, followed by a ramp to 50% buffer and 50% acetonitrile over 15 min (see Table S1 in the supplemental material for additional HPLC gradients). The injection volume was 10 μl, and the flow rate was 0.5 ml min−1. Products of the derivatization were detected at 340 nm. Authentic standards of each d- and l-amino acid were used to establish retention times, and a standard curve was generated from known amino acid concentrations in the reaction buffer in the absence of enzyme.
Phylogenetic analysis.
To determine any relationships between the enzymes identified in this study and other racemases, phylogenetic trees were constructed using software provided by Phylogeny.fr (28). The MUSCLE program was used for multiple-sequence alignment (no alignment curation; run mode, default; maximum number of iterations, 16), the PhyML program was used for tree construction (statistical test for branch support, aLERT, SH-like; substitution model, default), and the TreeDyn program was used for tree visualization (with default parameters). The Easyfig program (29) was used for comparison and visualization of the genomic loci. The FASTA sequences used in the tree construction were obtained from the GenBank database of the National Center for Biotechnology Information (NCBI) (30).
RESULTS
Growth profile: amino acids as the sole carbon and nitrogen source.
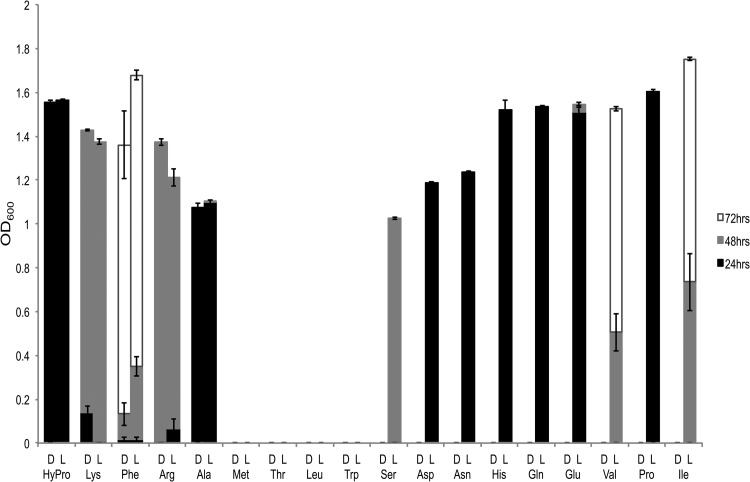
Other studies have described the catabolic versatility of P. putida KT2440, yet a comprehensive analysis of growth on d- and l-amino acids as the sole carbon and nitrogen source has not been reported (12, 31, 32). To assess the potential for racemization of d-amino acids, we performed a series of growth studies on P. putida KT2440 in which d- and l-amino acids were provided as the sole carbon and nitrogen source (Fig. 1). No growth was recorded on either stereoisomer of methionine, threonine, leucine, or tryptophan. In instances where only the l-stereoisomer was catabolized, aspartate, asparagine, histidine, glutamine, glutamate, and proline were the most rapidly catabolized. Rapid growth was observed with either epimer of 4-hydroxyproline, cis-d- and trans-l-HyPro, or either enantiomer of alanine. The lysine and arginine enantiomers were catabolized more slowly. Marginal growth was recorded on d- and l-phenylalanine after 24 h. However, after 72 h, cultures with these amino acids reached growth levels similar to those of cultures with the other amino acid enantiomer pairs that were more rapidly catabolized. While previous growth experiments have established that P. putida KT2440 catabolizes both d- and l-lysine (13, 33), P. putida KT2440 has not previously been shown to catabolize as the sole carbon and nitrogen sources d-phenylalanine, the epimers of HyPro (cis-d- and trans-l-HyPro), or d-arginine. Nonetheless, P. putida KT2440 can use both enantiomers of phenylalanine, arginine, and HyPro as the sole nitrogen source (31, 32).
Fig 1.
Ability of Pseudomonas putida KT2440 to use d- and l-amino acids as the sole source of carbon and nitrogen. The bar of each color represents the net increase in growth during the designated time period. The absence of a bar of a particular color signifies a lack of growth during the corresponding time period. Cultures were grown in PG liquid minimal medium. Average values are the result of three replicates.
Recovered genes with a potential role in amino acid racemization.
Screens were conducted in E. coli auxotrophs to identify mechanisms for racemization of lysine, proline, phenylalanine, and leucine. Lysine and phenylalanine auxotrophs were chosen due to the d-amino acid growth profiles of P. putida KT2440 (Fig. 1). The proline auxotroph was chosen due to the presence of an annotated proline racemase in the P. putida KT2440 genome. The leucine auxotroph was chosen for two reasons. First, no alanine auxotroph was available from the Coli Genetic Stock Center, leucine exhibits reasonable structural similarity to alanine, and a racemase that uses alanine as a substrate may also use leucine. Second, it is possible that P. putida KT2440 may also racemize amino acids that it cannot use as a sole carbon and nitrogen source. Figure 1 shows the four amino acids that are not catabolized as the sole carbon and nitrogen source by P. putida KT2440; leucine is a representative from this group. A screen on d-arginine was not conducted because of the high activity of the recovered Alr enzyme in conversion of d-arginine to l-arginine.
Screening commenced until coverage of the P. putida KT2440 genome of at least 500 times was achieved in each screen. DNA sequences recovered from the positive clones were compared with the P. putida KT2440 genome sequence using the sequences in the Pseudomonas genome database (26) to identify ORFs within each region. Because of the magnitude of the screen, identified genomic regions were typically recovered more than once, allowing us to estimate a minimal genomic region necessary for recovery of the phenotype. Multiple, independent genomic regions were recovered from the d-proline, d-lysine, and d-leucine screens, while only one region was found to rescue the phenotype in the d-phenylalanine screen. These regions (see Table S2 in the supplemental material) vary in size and in many cases include more than one gene that could be responsible for the observed phenotype. A genomic region containing the putative alanine racemase alr gene (PP3722) was recovered in both the d-lysine and d-leucine screens. The genomic region containing the putative alanine racemase dadX (PP5269) was recovered only in the d-leucine screen. The putative proline racemase (PP1258) was recovered only in the d-proline screen, and no putative racemase gene was identified from the sole genomic region that conferred growth in the d-phenylalanine screen.
Among genomic regions in which an annotated racemase was identified, a subclone containing the racemase gene by itself conferred the same phenotype as the insert from which it originated. We concluded that the racemase genes were responsible for the observed phenotype in these cases and did not further assess other genes from these regions. Metabolism of amino acids is known to involve enzymes such as dehydrogenases, oxidoreductases, and aminotransferases (13, 22, 34), and genes annotated as such were recovered in each of the four screens (see Table S2 in the supplemental material). The current study focuses on mechanisms for amino acid racemization. Further work beyond the scope of this study will be necessary to establish a role for these genes in d-amino acid metabolism in P. putida KT2440.
Substrate specificity of Pseudomonas putida KT2440 amino acid racemases.
Genes encoding putative racemase activity (proline racemase [PP1258], alanine racemase/Alr [PP3722], alanine racemase/DadX [PP5269]) were subcloned into pET28b. Each enzyme was purified to homogeneity and analyzed for its activity on both stereoisomers of each of the 19 chiral amino acids (Table 1). The protein product (Alr) of the gene annotated as alanine racemase (alr) demonstrated the broadest substrate specificity of the recovered racemases. Despite its annotation as a putative alanine racemase, substrates that conferred the highest activity (Table 1) were lysine (normalized to 100% for both d- and l-lysine) and arginine. The protein product (DadX) of the gene annotated as alanine racemase (dadX) demonstrated activity with both alanine stereoisomers, showed negligible activity with d-cysteine and l-serine, and exhibited no activity with the remaining amino acids. The putative proline racemase encoded by the proR gene (ProR) did not exhibit measureable racemase activity with any of the 19 chiral amino acid enantiomers. Nonetheless, the gene did rescue the phenotype of the E. coli proline auxotroph strain in the presence of d-proline, which is indicative of at least minimal racemization in vivo. Because previous work identified hydroxyproline epimerase activity in a cell extract of the related strain Pseudomonas putida KT2442 (31), we assessed the activity of the putative proline racemase with four epimers of HyPro. The different epimers arise from chirality about the α carbon and the γ carbon in the amino acid. Isomerization about the α carbon, typical of amino acid racemization, results in interconversion of the cis-d-HyPro and trans-l-HyPro epimers. The conversion of either l-HyPro epimer does not appear to be affected by the chirality about the γ carbon atom, described by the cis and trans notations (Table 1). However, cis-d-HyPro confers a much higher activity than trans-d-HyPro, suggesting a potential influence of γ-carbon chirality in the racemization of the d-HyPro epimers.
Table 1.
Normalized relative activity of racemases on d- and l-amino acids
| Purified enzyme |
d-Amino acid or HyProa |
l-Amino acida |
||
|---|---|---|---|---|
| Amino acid | Normalized % activity ± SD | Amino acid | Normalized % activity ± SD | |
| Alanine racemase (Alr) | Lys | 100.00 ± 0.00 | Lys | 100.00 ± 0.00 |
| Arg | 87.49 ± 1.64 | Arg | 10.79 ± 0.79 | |
| Met | 11.56 ± 0.20 | Met | 6.39 ± 0.22 | |
| Gln | 6.94 ± 0.09 | Gln | 3.07 ± 0.09 | |
| Ala | 3.10 ± 0.15 | Ser | 1.75 ± 0.09 | |
| Ser | 2.57 ± 0.09 | Ala | 1.18 ± 0.03 | |
| Leu | 1.37 ± 0.08 | Asn | 0.65 ± 0.08 | |
| His | 1.01 ± 0.03 | Leu | 0.57 ± 0.02 | |
| Asn | 0.96 ± 0.07 | His | 0.56 ± 0.02 | |
| Others | NDb | Others | ND | |
| Alanine racemase (DadX) | Ala | 100.00 ± 0.00 | Ala | 100.00 ± 0.00 |
| Cys | 8.35 ± 0.20 | Ser | 0.90 ± 0.04 | |
| Others | ND | Others | ND | |
| Proline racemase (ProR) | cis-d-HyPro | 100.00 ± 0.00 | ||
| trans-l-HyPro | 32.34 ± 0.82 | |||
| cis-l-HyPro | 32.23 ± 1.82 | |||
| trans-d-HyPro | 22.73 ± 0.62 | |||
| Others | ND | |||
Percent activity values were normalized for the difference in derivatization efficiency between each d- and l-enantiomer (52). The final assay concentration for each amino acid was 50 mM, except for Tyr (2.21 mM) and Asp (25 mM). Assays were performed in triplicate only for those amino acids that showed detectable activity. The others category comprises all remaining chiral proteinogenic amino acids.
ND, no activity detected.
Kinetic parameters of amino acid racemases.
The alanine racemase Alr demonstrated a Km in the μM range for d-lysine, while the Km for l-lysine was significantly higher (Table 2). However, the kcat values suggested a more rapid racemization in the direction of d-lysine formation. Nonetheless, the kcat/Km values indicated a 4-fold higher overall enzymatic efficiency in the d → l direction.
Table 2.
Kinetic parameters of racemases determined on select d- and l-amino acidsa
| Enzyme | Substrate |
Km (mM) |
kcat (s−1) |
kcat
Km−1 (M−1 s−1) |
|||
|---|---|---|---|---|---|---|---|
| d → l | l → d | d → l | l → d | d → l | l → d | ||
| Alanine racemase Alr | Lys | 0.36 ± 0.06 | 8.96 ± 1.52 | 274.5 ± 6.7 | 1,681.7 ± 161.7 | 7.63 × 105 | 1.88 × 105 |
| Ala | 15.71 ± 2.79 | 12.62 ± 0.74 | 8.83 ± 0.00 | 7.33 ± 0.00 | 5.62 × 102 | 5.81 × 102 | |
| Alanine racemase DadX | Ala | 7.73 ± 0.63 | 24.57 ± 2.64 | 37.47 ± 2.67 | 92.0 ± 11.6 | 4.85 × 103 | 3.74 × 103 |
| Proline racemase | HyPro (cis-d- and trans-l-) | 5.26 ± 1.12 | 15.04 ± 4.19 | 69.63 ± 8.60 | 7.74 ± 0.00 | 1.32 × 104 | 5.15 × 102 |
The substrates with the highest normalized percent activity were chosen for kinetic analysis, as were d-/l-Ala for Alr and trans-l-HyPro for proline racemase. Vmax and Km values were determined via nonlinear regression according to the Michaelis-Menten equation. kcat values were calculated by dividing Vmax by the enzyme concentration.
Considering that both Alr and DadX catalyze the racemization of alanine, we established the kinetic parameters of both enzymes with each alanine enantiomer (Table 2). For the conversion of d-alanine to l-alanine, the Km value of Alr was approximately twice that of DadX, while the kcat value for the DadX reaction was ∼4 times greater than that for the Alr reaction, contributing to an ∼9 times higher kcat/Km value for the DadX reaction for the conversion of d-alanine to l-alanine. In the direction of l-Ala to d-Ala, Alr showed an ∼2 times lower Km value and an ∼12 times lower kcat value, which contribute to an ∼6 times higher kcat/Km value for DadX.
Kinetic values for the isomerization of cis-d-HyPro and trans-l-HyPro (epimers about the α carbon) by ProR are shown in Table 2. The enzyme exhibited a lower Km value for cis-d-HyPro as well as a higher maximum velocity in the d → l direction, resulting in a kcat/Km value for the conversion of cis-d-HyPro to trans-l-HyPro 26 times greater than that in the opposite direction.
There is significant variability in reported enzyme parameters for racemases; this can likely be attributed to differences in reaction conditions. Owing to the reversible nature of the reaction, the lack of a depleting cosubstrate, and the reasonably similar catalytic efficiencies in either direction, it is necessary to limit the overall reaction time and minimize substrate turnover or the reverse reaction will confound the data. The assay conditions utilized in this work were optimized for such a reaction, with low (below 2 μM) enzyme concentrations with 1-min reaction times being used, which ensured that less than 10% of the total substrate from each reaction was turned over.
Bioinformatic analysis of P. putida KT2440 racemase genes.
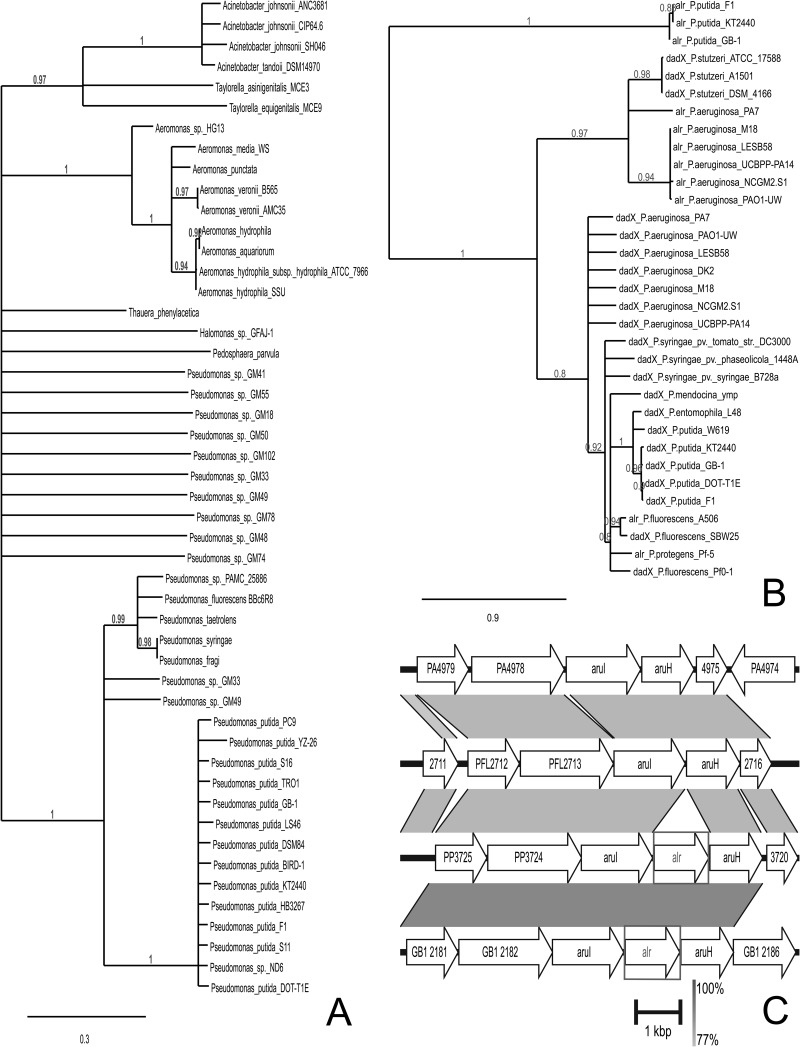
The genome of P. putida KT2440 possesses 894 paralogous gene domain families, which is the highest number known relative to other sequenced bacterial genomes (35). Only the genome of P. aeruginosa PAO1 has a number that approaches that number, with 809, while other genomes possess 50% of that number or less (17). Consequently, we assessed whether the genetic loci characterized in this study demonstrate identity and genomic synteny to those found in the genomes of other bacteria. We performed a BLASTp analysis using the deduced protein sequence of alanine racemase/Alr (Fig. 2A). The displayed branches of the constructed tree have high support values of ≥0.8, suggesting a confirmation of the tree topology. We found that the P. putida KT2440 Alr clusters only with similar proteins from P. putida strains and is quite different from similar proteins found in other Pseudomonas species and other bacteria (Fig. 2A).
Fig 2.
Phylogenetic analysis. (A) Tree of the 50 nonredundant protein sequences most identical to alanine racemase/Alr. (B) Tree of the annotated DadX and Alr from the sequenced genomes in the Pseudomonas Genome Database. Distance bars represent the expected proportion of substitutions per amino acid site. (C) Genomic synteny analysis on regions bordering the alr gene from Pseudomonas putida KT2440 (from the top, listed descending: P. aeruginosa PAO1, P. protegens Pf-5, P. putida KT2440, and P. putida GB-1). Gene annotations are based on the Pseudomonas Genome Database. The H bracket shows the actual DNA length. The gradient scale provides the percent identity for the genomic loci.
Because of differences in substrate specificity between the Alr and DadX enzymes, we considered the relationship among substrate specificity, gene class, and genomic neighborhood between the alr and dadX gene families. Figure 2B shows a phylogenetic tree of all annotated alr and dadX gene entries in the Pseudomonas genome database (accessed on 20 June 2013). While this is not a complete assessment of alanine racemase genes among pseudomonads, a clear delineation with strong branch support can be made between alr genes from P. putida entries and the remaining alr and dadX genes from these pseudomonads. Figure 2B also indicates strong conservation among pseudomonad dadX genes but significant divergence among the remaining alr genes.
Analysis of alr genomic synteny among pseudomonads revealed a strong conservation of the selected genetic loci between the two Pseudomonas putida strains KT2440 and GB-1 (Fig. 2C), as well as a similarly high synteny between P. aeruginosa and P. protegens Pf-5 (formerly P. fluorescens Pf-5). The notable difference is the lack of the alr gene in this region in the last two species. This difference was previously noted by Yang and Lu (36) when describing the l-arginine transaminase pathway (initiated by the aruH and aruI genes from Fig. 2C) in P. aeruginosa PAO1. The genomic synteny of the regions surrounding dadX and the gene for proline racemase (proR) was also explored (see Fig. S1A in the supplemental material). We observed high synteny for the regions bordering dadX among each of the tested pseudomonads (the same species used for the alr analysis), a finding similar to that reported by He et al. (37). Some conservation of the proR gene was noted among these species (see Fig. S1B in the supplemental material). Conservation of genes involved in HyPro metabolism between P. putida and P. aeruginosa was previously noted by Watanabe et al. (31).
DISCUSSION
In addition to elaborating on the extent of d-amino acid utilization by P. putida KT2440, we reasoned that growth profiles may give us information on which amino acids undergo racemization. For those amino acids in which both enantiomers are catabolized, the genomic screen should recover any mechanisms by which the enantiomers are directly interconverted. However, more than one mechanism for recovering growth using this screen exists. In addition to amino acid racemization, this includes deamination of the d-amino acid to produce its corresponding achiral α-keto acid, followed by stereospecific amination to produce the l-amino acid. The latter two-enzyme process is shown to be involved in the conversion of d-arginine to l-arginine via coupled catabolic and anabolic dehydrogenases encoded within the dauBAR operon in P. aeruginosa (22). A genomic library clone encoding an enzyme that catalyzes only the first step of this two-step process may recover growth due to the presence of native α-keto acid transamination activity (the second step of the above-described process) within the E. coli screening host. The loci recovered from these genomic screens indicate that both racemization and deamination clones were identified (see Table S2 in the supplemental material). Based on the annotated function of the recovered genes, we chose to further study the proline racemase (proR, PP1258) and the two alanine racemases (alr, PP3722; dadX, PP5269).
The proR gene product exhibited no in vitro racemization of proline; rather, it appeared to be a 4-hydroxyproline epimerase on the basis of its enzymatic activity. This activity is widespread among bacteria that colonize animals and plants (38, 39), owing to the abundance of trans-l-HyPro in both collagen and plant cell wall proteins (40, 41). In the catabolic pathway described by Gryder and Adams for a P. putida isolate (42), trans-l-HyPro is first isomerized to cis-d-HyPro by a (then unknown) 4-hydroxyproline epimerase. The product is then further oxidatively metabolized in several steps to produce α-ketoglutarate. We report here that the P. putida KT2440 proR gene encodes the enzyme necessary to initiate this catabolic pathway via epimerization and recommend that the current “proline racemase” designation be changed to “4-hydroxyproline epimerase.” Interestingly, the in vitro reaction appears to be much more efficient in the opposite direction, that is, in conversion of the d-HyPro epimer to the l-HyPro epimer (Table 2). Despite the lack of in vitro proline racemase activity, the enzyme must provide sufficient reactivity in vivo to account for the relatively small amount of l-proline necessary to recover growth of the proline auxotroph. Nonetheless, any proline racemase activity that it exhibits in P. putida KT2440 would be minimal, as it is not sufficient to allow growth on d-proline. Figure 1 shows that the bacterium grows readily on l-proline.
The P. putida KT2440 DadX enzyme exhibits tight substrate specificity, suggesting a single role in interconversion of alanine stereoisomers. This is consistent with the activity of those DadX orthologs that have been characterized (43, 44). Often referred to as a “catabolic racemase” (45), DadX enzymes are proposed to serve a role in the catabolism of l-alanine, whereby l-alanine is converted to d-alanine before enzymatic dehydrogenation to form pyruvate (15). The dadAX genomic locus, which also encodes the d-amino acid dehydrogenase (dadA) used in this catabolic process (37), exhibits significant synteny among closely related pseudomonads (see Fig. S1 in the supplemental material) (37). The P. putida KT2440 DadX enzyme shows overall similar kcat/Km values for both the l → d and d → l conversions (Table 2). Incidentally, the kcat/Km values obtained using l- and d-alanine as the substrates are ∼6- and 9-fold lower for the P. putida KT2440 Alr enzyme, respectively, largely stemming from lower kcat values with both l- and d-alanine. Nonetheless, in instances where both alr and dadX genes exist in the same organism, the Alr isozyme is considered the anabolic alanine racemase and is attributed to be the source for periplasmic d-alanine used in peptidoglycan synthesis (46). The P. putida KT2440 Alr exhibits minimal activity with alanine compared to other amino acids, however, raising questions about its role in d-alanine synthesis for peptidoglycan in P. putida KT2440. Indeed, the kcat/Km value with lysine is nearly 3 orders of magnitude greater than that with alanine in both reaction directions.
On the basis of the sequences available in GenBank, alr-encoded alanine racemases are more widespread than dadX-encoded alanine racemases. However, the presence of both annotated alanine racemases in one genome is not unusual, as this is seen in a large number of proteobacteria. Each of the 184 bacterial dadX alanine racemase gene entries in GenBank is from a member of the alpha-, beta-, or gammaproteobacteria, while the 1,088 bacterial alr alanine racemase entries are more widely distributed (accessed 13 June 2013). No uniform racemase gene nomenclature exists, however, and many genes are annotated as simply “alanine racemase.” A comparative analysis between the 10 P. aeruginosa genomes and 6 P. putida genomes currently available at the Pseudomonas Genome Database (26) (accessed 12 June 2013) revealed that each of these strains carries both the alr gene and the dadX gene. The dadAX locus is conserved in each of these strains, yet the alr locus differs according to species. With the exception of P. putida W619, each of the P. putida strains shares the same alr locus organization depicted for P. putida KT2440 in Fig. 2C. The location of the P. putida KT2440 alr gene between the aruI and aruH genes (Fig. 2C; first noted by Yang and Lu [36]) is noteworthy. The aruHI locus is shown to be necessary for the arginine transaminase (ATA) pathway for catabolism of l-arginine in P. aeruginosa PAO1 (36).
While both P. putida KT2440 and P. aeruginosa PAO1 catabolize d-arginine, they appear to use distinctly different mechanisms to initiate this process. In P. aeruginosa PAO1, catabolism of d-arginine is initiated by conversion to l-arginine via the coupled dehydrogenases DauA and DauB of the dauBAR operon (22). DauA and DauB are both required for P. aeruginosa PAO1 to grow on d-arginine as the sole carbon and nitrogen source, implying that d-arginine is metabolized only via conversion to l-arginine and that no other enzymatic arginine racemization mechanism exists in P. aeruginosa PAO1. Further, previous work demonstrates that d-lysine cannot complement an l-lysine auxotroph in P. aeruginosa PAO1 (47). Taken together, this indicates that no direct arginine or lysine racemization mechanism exists in P. aeruginosa PAO1. This indicates that the alr gene in P. aeruginosa PAO1 does not have the same substrate specificity as its P. putida KT2440 ortholog. Further, while the dauBAR locus is conserved among P. aeruginosa strains, it is not present in any of the P. putida strains from the Pseudomonas Genome Database.
On the basis of the observations made above, it appears to be plausible that the Alr enzyme in P. putida KT2440 serves a role analogous to that of DauA and DauB in P. aeruginosa PAO1 in conversion of d-arginine to l-arginine for further catabolism. In this scenario, the P. putida KT2440 alr gene is responsible for conversion of d-arginine into l-arginine, which is then catabolized via the ATA pathway by the products of the aruHI operon, in which the alr gene is incorporated (Fig. 2C).
Previous work on the catabolism of d- and l-lysine by P. putida KT2440 indicates that each proceeds through an independent pathway and that racemization of d-lysine to l-lysine prior to degradation is not required (13). While it is possible that the Alr enzyme is involved in the degradation of lysine, its potential for synthesis of d-lysine and any additional role that d-lysine may play should be further explored. Catabolic routes in pseudomonads for certain l-amino acids, such as alanine (15) and 4-hydroxyproline (42), involve initial conversion from the l-stereoisomer into the d-stereoisomer. This clearly prevents the amino acid from being incorporated into peptides and may minimize the changes in gene regulation that accompany increased pools of free amino acids in the cell (48, 49).
In environments where d-amino acids are abundant, their presence is typically attributed to the associated microbial communities (9, 50), yet outside of a few amino acids, nothing is known regarding their synthesis. Indeed, the d-stereoisomer of each of the 19 chiral amino acids has been identified in some capacity either from microbial cultures (51) or from microbe-enriched environments (1). The work presented here further establishes that the pseudomonads are good models for comparative genomic analysis of bacterial metabolism and points to differences in d-amino acid metabolism between two of the most well studied members of this clade. However, despite an increased awareness of the roles that d-amino acids play in bacterial ecophysiology, much remains to be discovered concerning the synthetic mechanisms and reasons for deployment of these compounds.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by grant 2011-67020-30195 from the USDA National Institute of Food and Agriculture.
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00761-13.
REFERENCES
- 1.Lam H, Oh D-C, Cava F, Takacs CN, Clardy J, De Pedro MA, Waldor MK. 2009. d-Amino acids govern stationary phase cell wall remodeling in bacteria. Science 325:1552–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishikawa M, Ogawa K. 2004. Occurrence of d-histidine residues in antimicrobial poly(arginyl-histidine), conferring resistance to enzymatic hydrolysis. FEMS Microbiol. Lett. 239:255–259 [DOI] [PubMed] [Google Scholar]
- 3.Bodanszky M, Stahl GL. 1974. The structure and synthesis of malformin A. Proc. Natl. Acad. Sci. U. S. A. 71:2791–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brückner H, Westhauser T. 2003. Chromatographic determination of l- and d-amino acids in plants. Amino Acids 24:43–55 [DOI] [PubMed] [Google Scholar]
- 5.Brückner H, Becker D, Lüpke M. 1993. Chirality of amino acids of microorganisms used in food biotechnology. Chirality 5:385–392 [DOI] [PubMed] [Google Scholar]
- 6.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. d-Amino acids trigger biofilm disassembly. Science 328:627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodowski S, Amelung W, Lobe I, Du Preez CC. 2004. Losses and biogeochemical cycling of soil organic nitrogen with prolonged arable cropping in the South African Highveld—evidence from d- and l-amino acids. Biogeochemistry 71:17–42 [Google Scholar]
- 8.Friedman M. 2010. Origin, microbiology, nutrition, and pharmacology of d-amino acids. Chem. Biodivers. 7:1491–1530 [DOI] [PubMed] [Google Scholar]
- 9.Schieber A, Brückner H, Ling JR. 1999. GC-MS analysis of diaminopimelic acid stereoisomers and amino acid enantiomers in rumen bacteria. Biomed. Chromatogr. 13:46–50 [DOI] [PubMed] [Google Scholar]
- 10.Vollmer W, Blanot D, De Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167 [DOI] [PubMed] [Google Scholar]
- 11.Siranosian KJ, Ireton K, Grossman AD. 1993. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J. Bacteriol. 175:6789–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Revelles O, Wittich R-M, Ramos JL. 2007. Identification of the initial steps in d-lysine catabolism in Pseudomonas putida. J. Bacteriol. 189:2787–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muramatsu H, Mihara H, Kakutani R, Yasuda M, Ueda M, Kurihara T, Esaki N. 2005. The putative malate/lactate dehydrogenase from Pseudomonas putida is an NADPH-dependent Δ1-piperideine-2-carboxylate/Δ1-pyrroline-2-carboxylate reductase involved in the catabolism of d-lysine and d-proline. J. Biol. Chem. 280:5329–5335 [DOI] [PubMed] [Google Scholar]
- 14.Conti P, Tamborini L, Pinto A, Blondel A, Minoprio P, Mozzarelli A, De Micheli C. 2011. Drug discovery targeting amino acid racemases. Chem. Rev. 111:6919–6946 [DOI] [PubMed] [Google Scholar]
- 15.Wasserman SA, Walsh CT, Botstein D. 1983. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 153:1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Monchy S, Taghavi S, Zhu W, Ramos J, Van der Lelie D. 2011. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 35:299–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martins dos Santos VAP, Heim S, Moore ERB, Strätz M, Timmis KN. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6:1264–1286 [DOI] [PubMed] [Google Scholar]
- 18.Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7:e1002132. 10.1371/journal.ppat.1002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collmer A, Badel JL, Charkowski AO, Deng WL, Fouts DE, Ramos AR, Rehm A, Anderson DM, Schneewind O, Van Dijk K, Alfano JR. 2000. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. U. S. A. 97:8770–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, Elbourne LDH, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, Van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS, Pierson EA, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. 10.1371/journal.pgen.1002784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matilla MA, Ramos JL, Bakker PA, Doornbos R, Badri DV, Vivanco JM, Ramos-González MI. 2010. Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ. Microbiol. Rep. 2:381–388 [DOI] [PubMed] [Google Scholar]
- 22.Li C, Lu C-D. 2009. Arginine racemization by coupled catabolic and anabolic dehydrogenases. Proc. Natl. Acad. Sci. U. S. A. 106:906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studier FW. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 24.Uchiyama T, Watanabe K. 2008. Substrate-induced gene expression (SIGEX) screening of metagenome libraries. Nat. Protoc. 3:1202–1212 [DOI] [PubMed] [Google Scholar]
- 25.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winsor GL, Lam DKW, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock REW, Brinkman FSL. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marfey P. 1984. Determination of d-amino acids. II. Use of bifunctional reagent 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 49:591–596 [Google Scholar]
- 28.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2011. GenBank. Nucleic Acids Res. 39:D32–D37. 10.1093/nar/gkq1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe S, Morimoto D, Fukumori F, Shinomiya H, Nishiwaki H, Kawano-Kawada M, Sasai Y, Tozawa Y, Watanabe Y. 2012. Identification and characterization of d-hydroxyproline dehydrogenase and Δ1-pyrroline-4-hydroxy-2-carboxylate deaminase involved in novel l-hydroxyproline metabolism of bacteria: metabolic convergent evolution. J. Biol. Chem. 287:32674–32688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina-Henares MA, De la Torre J, García-Salamanca A, Molina-Henares AJ, Herrera MC, Ramos JL, Duque E. 2010. Identification of conditionally essential genes for growth of Pseudomonas putida KT2440 on minimal medium through the screening of a genome-wide mutant library. Environ. Microbiol. 12:1468–1485 [DOI] [PubMed] [Google Scholar]
- 33.Revelles O, Espinosa-Urgel M, Fuhrer T, Sauer U, Ramos JL. 2005. Multiple and interconnected pathways for l-lysine catabolism in Pseudomonas putida KT2440. J. Bacteriol. 187:7500–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Lu C-D. 2007. Characterization of an arginine:pyruvate transaminase in arginine catabolism of Pseudomonas aeruginosa PAO1. J. Bacteriol. 189:3954–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Lu C-D. 2007. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J. Bacteriol. 189:3945–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He W, Li C, Lu C-D. 2011. Regulation and characterization of the dadRAX locus for d-amino acid catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 193:2107–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith EA, Macfarlane GT. 1997. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe 3:327–337 [DOI] [PubMed] [Google Scholar]
- 39.Maclean AM, White CE, Fowler JE, Finan TM. 2009. Identification of a hydroxyproline transport system in the legume endosymbiont Sinorhizobium meliloti. Mol. Plant Microb. Interact. 22:1116–1127 [DOI] [PubMed] [Google Scholar]
- 40.Cassab GI. 1998. Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:281–309 [DOI] [PubMed] [Google Scholar]
- 41.Bella J, Eaton M, Brodsky B, Berman H. 1994. Crystal and molecular structure of a collagen-like peptide at 1.9Å resolution. Science 266:75–81 [DOI] [PubMed] [Google Scholar]
- 42.Gryder R, Adams E. 1969. Inducible degradation of hydroxyproline in Pseudomonas putida: pathway regulation and hydroxyproline uptake. J. Bacteriol. 97:292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ju J, Yokoigawa K, Misono H, Ohnishi K. 2005. Cloning of alanine racemase genes from Pseudomonas fluorescens strains and oligomerization states of gene products expressed in Escherichia coli. J. Biosci. Bioeng. 100:409–417 [DOI] [PubMed] [Google Scholar]
- 44.Strych U, Huang H-C, Krause KL, Benedik MJ. 2000. Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr. Microbiol. 41:290–294 [DOI] [PubMed] [Google Scholar]
- 45.Wasserman SA, Daub E, Grisafi P, Botstein D, Walsh CT. 1984. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence, enzyme purification, and characterization. Biochemistry 23:5182–5187 [DOI] [PubMed] [Google Scholar]
- 46.Esaki N, Walsh CT. 1986. Biosynthetic alanine racemase of Salmonella typhimurium: purification and characterization of the enzyme encoded by the alr gene. Biochemistry 25:3261–3267 [DOI] [PubMed] [Google Scholar]
- 47.Li C, Yao X, Lu C-D. 2010. Regulation of the dauBAR operon and characterization of d-amino acid dehydrogenase DauA in arginine and lysine catabolism of Pseudomonas aeruginosa PAO1. Microbiology 156:60–71 [DOI] [PubMed] [Google Scholar]
- 48.Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. 2010. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J. Bacteriol. 192:6357–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno R, Martínez-Gomariz M, Yuste L, Gil C, Rojo F. 2009. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9:2910–2928 [DOI] [PubMed] [Google Scholar]
- 50.Hancock R. 1960. The amino acid composition of the protein and cell wall of Staphylococcus aureus. Biochim. Biophys. Acta 37:42–46 [DOI] [PubMed] [Google Scholar]
- 51.Cava F, De Pedro MA, Lam H, Davis BM, Waldor MK. 2011. Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J. 30:3442–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodlett DR, Abuaf PA, Savage PA, Kowalski KA, Mukherjee TK, Tolan JW, Corkum N, Goldstein G, Crowther JB. 1995. Peptide chiral purity determination: hydrolysis in deuterated acid, derivatization with Marfey's reagent and analysis using high performance liquid chromatography-electrospray ionization-mass spectrometry. J. Chromatogr. A 707:233–244 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.