Abstract
The acs operon of Gluconacetobacter is thought to encode AcsA, AcsB, AcsC, and AcsD proteins that constitute the cellulose synthase complex, required for the synthesis and secretion of crystalline cellulose microfibrils. A few other genes have been shown to be involved in this process, but their precise role is unclear. We report here the use of Tn5 transposon insertion mutagenesis to identify and characterize six non-cellulose-producing (Cel−) mutants of Gluconacetobacter hansenii ATCC 23769. The genes disrupted were acsA, acsC, ccpAx (encoding cellulose-complementing protein [the subscript “Ax” indicates genes from organisms formerly classified as Acetobacter xylinum]), dgc1 (encoding guanylate dicyclase), and crp-fnr (encoding a cyclic AMP receptor protein/fumarate nitrate reductase transcriptional regulator). Protein blot analysis revealed that (i) AcsB and AcsC were absent in the acsA mutant, (ii) the levels of AcsB and AcsC were significantly reduced in the ccpAx mutant, and (iii) the level of AcsD was not affected in any of the Cel− mutants. Promoter analysis showed that the acs operon does not include acsD, unlike the organization of the acs operon of several strains of closely related Gluconacetobacter xylinus. Complementation experiments confirmed that the gene disrupted in each Cel− mutant was responsible for the phenotype. Quantitative real-time PCR and protein blotting results suggest that the transcription of bglAx (encoding β-glucosidase and located immediately downstream from acsD) was strongly dependent on Crp/Fnr. A bglAx knockout mutant, generated via homologous recombination, produced only ∼16% of the wild-type cellulose level. Since the crp-fnr mutant did not produce any cellulose, Crp/Fnr may regulate the expression of other gene(s) involved in cellulose biosynthesis.
INTRODUCTION
Cellulose, the most abundant natural polymer, is produced not only by plants but also by diverse bacterial species, including soil and pathogenic bacteria (1). Cellulose synthesis and assembly is recently a prime interest because of its potential use in renewable energy production. In addition, cellulose is known for its extensive commercial usage in the paper and food industries. Gluconacetobacter hansenii and G. xylinus (both formerly named Acetobacter xylinum), Gram-negative bacteria, have long been used as a model system for studying the “simplest” cellulose synthase complex (CSC) required for cellulose biosynthesis (2–4), because they do not produce lignin or hemicellulose, hence providing an excellent platform for studying pure cellulose, and because they are able to synthesize and secrete large amounts of crystalline cellulose into growth media.
Several Gluconacetobacter genes have been shown to be involved in the synthesis and secretion of crystalline cellulose microfibrils. Four of them—acsA, acsB, acsC, and acsD—are thought to constitute an operon, named acs (acetobacter cellulose synthase) (3). Their protein products have been proposed to form the CSC that synthesizes cellulose using UDP-glucose as substrate and secrets the cellulose microfibrils through the outer membrane. Some G. xylinus strains (e.g., ATCC 53582) produce separate AcsA and AcsB proteins, whereas G. hansenii ATCC 23769 synthesizes AcsA and AcsB as a fusion protein AcsAB (5), which is processed into three polypeptides of molecular masses 34, 46, and 95 kDa (6). Although the precise cleavage sites are unknown, the 46-kDa polypeptide was named AcsAcat since it contains a conserved “D,D,D,QXXRW” motif found in cellulose synthases or cellulose synthase-like proteins (7, 8). The 34-kDa polypeptide was named AcsAreg since it contains the PilZ domain, the c-di-GMP binding domain in bacterial cellulose synthases (9, 10). The 95-kDa polypeptide corresponds to approximately AcsB, whose function in cellulose biosynthesis is as yet unknown.
Based on the predicted hydrophobicity and membrane localization, AcsC is hypothesized to form a pore-like structure for extrusion of the cellulose product. AcsD is the only Acs protein of Gluconacetobacter whose crystal structure has been determined (11); it is an octameric protein assembly localized to the periplasmic space (11, 12). It may function as a cellulose chaperone, directing the newly synthesized cellulose chains to AcsC in the outer membrane to prevent accumulation of rogue strands in the periplasm.
ccpAx (the subscripts “Ax” and “Ax” indicate proteins and genes from organisms formerly classified as Acetobacter xylinum) and cmcAx, located immediately upstream from the acs operon, are thought to be required for cellulose biosynthesis in Gluconacetobacter based on the following experimental results. ccpAx encodes a protein named cellulose-complementing protein, and cmcAx encodes a secreted carboxymethylcellulase (endo-β-1,4-glucanase), which has been shown to have cellulose-hydrolyzing activity in vitro (13). Standal et al. (14) showed that a non-cellulose-producing mutant of G. hansenii has an indigenous transposable element inserted in ccpAx. In G. hansenii, Sunagawa et al. (15) showed by fluorescence microscopy that CcpAx colocalizes with AcsD along one side of the cell and by a pulldown assay and isothermal titration calorimetry that CcpAx and AcsD interact with each other. These results suggest that CcpAx may be a component of the CSC; however, its precise role in cellulose biosynthesis is still unknown. In both G. hansenii and G. xylinus, CmcAx may play a role in the regulation of cellulose biosynthesis via its cellulose-hydrolyzing activity, since the addition of an anti-CmcAx antibody to the culture medium severely inhibited the formation of cellulose fiber (16) and since the addition of a small amount of CmcAx to the culture medium, or overexpression of the protein, enhanced the yield of cellulose production (13, 17).
bglAx is located immediately downstream from the acs operon. It is predicted to encode a β-glucosidase of 79 kDa (18, 19) and, based on amino acid sequence similarity, BglAx was classified with family 3 glycoside hydrolases (19). The enzyme from G. xylinus BPR2001 has been shown to have exo-1,4-β-glucosidase activity, cleaving from the nonreducing end of cellotriose and larger cello-oligosaccharides, and to possibly also have glucosyltransferase activity (20). However, the role of BglAx, if any, in cellulose biosynthesis, is still unknown.
Despite extensive study of cellulose biosynthesis in Gluconacetobacter, the CSC has not yet been purified, and thus its biochemical nature remains unknown. Nor is it known whether there are any additional proteins required for the synthesis of cellulose, the assembly of cellulose fibers to form crystalline cellulose microfibers, and the secretion of the microfibrils. To gain a comprehensive understanding of this process, we set out to systematically identify the genes involved by using Tn5 transposon mutagenesis to isolate mutants that either fail to synthesize/secrete cellulose into the culture medium or produce cellulose with altered properties (e.g., with reduced crystallinity). G. hansenii ATCC 23769 offers an attractive system for this effort, because its complete genome sequence has recently been determined (21), making gene identification relatively straightforward.
We report here the isolation and characterization of six non-cellulose-producing mutants generated by insertion of Tn5 transposon DNA into acsA, acsC, ccpAx, dgc1 (two different independent mutants were obtained), and crp-fnr (predicted to encode a Crp [cyclic AMP receptor protein]/Fnr [fumarate nitrate reductase] transcriptional regulator). Dgc1, a diguanylate cyclase, catalyzes the synthesis of cyclic dimeric GMP (c-di-GMP), a well-characterized activator of cellulose biosynthesis (9), but the involvement of a Crp/Fnr transcriptional regulator in cellulose biosynthesis in Gluconacetobacter was not previously known. We also report the findings (i) that acsD is expressed under its own promoter and not as part of the acs operon, as was previously found for several G. xylinus strains, (ii) that mutation in ccpAx results in a significant decrease in the levels of AcsB and AcsC, and (iii) that Crp/Fnr regulates the expression of bglAx and likely other gene(s) involved in cellulose biosynthesis.
MATERIALS AND METHODS
Isolation of a stable cellulose-producing clone of G. hansenii ATCC 23769.
A stock of G. hansenii ATCC 23769 that overproduces cellulose was obtained from Candace Haigler (22). Cells were grown on Schramm-Hestrin (SH) (23) agar plates at 30°C for 3 days. Both cellulose-producing colonies (small and rough appearance) and non-cellulose-producing colonies (large and smooth appearance) were obtained. A single cellulose-producing colony was selected and streaked onto a fresh SH agar plate, and the plate was incubated at 30°C for 2 to 3 days. Twenty single colonies were randomly selected and separately inoculated into 5 ml of SH medium in 17 × 100 mm culture tubes, and grown under static conditions at 30°C. One of the colonies found to produce cellulose in static culture was selected and streaked on a fresh SH agar plate, and 20 single colonies were selected, separately inoculated into 5 ml of SH medium in culture tubes (17 by 100 mm), and grown under static conditions at 30°C. This process was repeated until all of the colonies derived from the starting single colony showed identical cellulose-producing morphology on plates and all 20 colonies selected for growth in static culture produced cellulose. The clone used in the final assay was designated wild-type (WT) G. hansenii ATCC 23769 and used in all of the experiments described here. A similar procedure was used to select a stable non-cellulose-producing clone, designated WT (Cel−), from a single non-cellulose-producing colony grown from the original stock of G. hansenii ATCC 23769. For a colorimetric assay of cellulose-producing colonies on plates, G. hansenii cells were grown on SH agar plates containing 0.001% Pontamine Fast Scarlet 4B (Sigma-Aldrich, St. Louis, MO), and the stained cells were further observed by using an SZX12 microscope (Olympus, Japan) equipped with a SPOT Insight camera (Diagnostic Instruments, Sterling Heights, MI).
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in the present study are listed in Table S1 in the supplemental material. Wild-type G. hansenii ATCC 23769 (i.e., the stable cellulose-overproducing clone isolated from the original stock) and mutants generated from Tn5 transposon mutagenesis were grown under static and shaking conditions at 30°C in SH medium for 2 to 3 days. Escherichia coli strain DH5α and Stellar competent cells (Clontech, Mountain View, CA) were used for cloning and grown at 37°C in Luria-Bertani (LB) medium. Ampicillin (100 μg/ml), kanamycin (50 μg/ml), and tetracycline (20 μg/ml) were used to select E. coli transformants, and tetracycline (20 μg/ml) and spectinomycin (100 μg/ml) were used to select G. hansenii transformants. Plasmid EZ-Tn5 pMOD-3⟨R6Kγori/MCS⟩ (Epicentre Biotechnologies, Madison, WI) was used for constructing Tn5 transposon DNA. pGEM-T Easy vector (Promega, Madison, WI) was used for cloning and sequencing. A shuttle vector, pUCD2 (24), was used for making constructs for complementing Cel− mutants. pUC18 was used for generating targeted gene knockout mutants of G. hansenii via homologous recombination.
Transformation of G. hansenii.
Competent cells were prepared from wild-type G. hansenii ATCC 23769 and Cel− mutants for electroporation according to the procedure of Dower et al. (25) with some minor modifications. Briefly, G. hansenii cells were grown in 50 ml of SH medium with 0.02% (vol/vol) cellulase (Worthington Biochemical, Lakewood, NJ) at 30°C with shaking at 250 rpm to an optical density at 600 nm (OD600) of ∼0.8. The cells were washed twice with 1 mM HEPES (pH 7), and the pellet was resuspended in 15% (vol/vol) glycerol to yield an approximate concentration of 107 to 108 cells/ml. For transformation, 100 μl of freshly prepared competent cells were mixed with 0.2 μg of plasmid DNA, and the mixture was electroporated at 2.5 kV/cm using a MicroPulser (Bio-Rad Laboratories, Richmond, CA). After electroporation, the cells were transferred to 1 ml of SH medium containing 0.02% (vol/vol) cellulase, followed by incubation at 30°C with shaking at 200 rpm overnight. The cell suspension was diluted and plated on SH agar plates containing an appropriate antibiotic and then incubated at 30°C for 2 to 3 days.
Generation of Tn5 transposon construct and introduction of Tn5 transposon DNA into G. hansenii.
To make a Tn5 transposon construct containing a tetracycline resistance gene, the tetC gene in pUCD2 was amplified by PCR, using primers TetC F1 and TetC R1 (see Table S2 in the supplemental material), to generate a 1,340-bp fragment, which contained 56 bp of the sequence immediately upstream from the translation initiation codon, the entire coding sequence, and 97 bp of the 3′-untranslated region (3′-UTR). The cycling conditions were 98°C for 30 s for the initial denaturation, followed by 35 cycles of 98°C for 10 s, 57°C for 30 s, and 72°C for 45 s, and then 72°C for 10 min. The resulting fragment was ligated at the HindIII/EcoRI sites of EZ-Tn5 pMOD-3⟨R6Kγori/MCS⟩. The Tn5 transposon DNA was obtained by PCR using a pair of “ME (Mosaic End) plus 9 bp” primers (see Table S2 in the supplemental material). The cycling conditions were 94°C for 2 min for the initial denaturation, followed by 30 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 1.5 min, and then 72°C for 10 min. The transposon DNA was incubated with EZ-Tn5 transposase (Epicentre Biotechnologies) in the absence of Mg2+ so that the transposase formed a complex with the DNA fragment but did not cleave at its recognition sequence. The complex was electroporated into electrocompetent cells of wild-type G. hansenii, and the transformants were selected on SH agar plates with tetracycline. To identify mutants that did not synthesize cellulose or did not secrete cellulose into the medium, individual colonies were grown in shaking and static cultures in 5 ml of SH medium with tetracycline for 2 and 7 days, respectively.
Cloning and sequencing of genes disrupted by Tn5 transposon.
Genomic DNA was isolated from each Cel− mutant using a Wizard Genomic DNA purification kit (Promega). For each mutant, the site of the transposon insertion in the genome was identified by the DNA Walking SpeedUp Premix kit (Seegene, Inc., Seoul, Korea) using genomic DNA as a template and TS1 and TS2 (see Table S2 in the supplemental material) provided in the kit as primers. The cycling conditions for the first round of PCR were 94°C for 5 min for the initial denaturation, 42°C for 1 min and 72°C for 2 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 100 s, and then 72°C for 7 min. The cycling conditions for the second round of PCR were 94°C for 3 min for the initial denaturation, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 100 s, and then 72°C for 7 min. PCR fragments obtained were ligated to the pGEM-T Easy Vector. The ligation products were transformed into Stellar competent cells (Clontech) or DH5α competent cells (New England BioLabs, Beverly, MA), and the transformants were grown on ampicillin-containing LB agar plates at 37°C overnight. All plasmids were prepared using a NuceloSpin plasmid miniprep kit (Macherey-Nagel, Bethlehem, PA), and plasmid DNA was sequenced using an ABI 3730XL DNA Analyzer (Life Technologies, Carlsbad, CA) at Penn State's Genomics Core Facility. Sequences were analyzed with the Lasergene (DNAstar, Inc., Madison, WI) software package. Database searches were carried out using BLASTN and BLASTtx (blast.ncbi.nlm.nil.gov).
Promoter characterization.
The DNA fragments tested for promoter activity were obtained by PCR using gene-specific primers listed in Table S2 in the supplemental material and genomic DNA of wild-type G. hansenii as a template. The cycling conditions were 98°C for 30 s for initial denaturation, followed by 35 cycles of 98°C for 10 s, 57 to 60°C (depending on the primers used) for 30 s, and 72°C for 30 s, and then 72°C for 10 min. For the acs operon, a 221-bp fragment, amplified using the primers AcsA-PF and AcsA-PR, spanned the sequence between the translation termination codon of ccpAx and the translation initiation codon of acsA; for acsD, a 321-bp fragment, amplified using the primers AcsD-PF and AcsD-PR, spanned the sequence between the translation termination codon of acsC and the translation initiation codon of acsD; for the cmcAx-ccpAx operon, a 504-bp fragment, amplified using the primers CmcAx-PF and CmcAx-PR, contained the sequence immediately upstream from the translation initiation codon. A promoterless tetC gene (1,288 bp), containing the entire coding sequence and 97 bp of the 3′-UTR, was obtained by PCR using TetC pro-F and TetC pro-R as primers and pUCD2 as a template. An In-Fusion HD cloning kit (Clontech) was used to make all promoter constructs via the ligation of two DNA fragments, a promoter fragment and the 1,288-bp tetC fragment, to PvuI/SalI-digested pUCD2. All of the promoter constructs were transformed into E. coli and G. hansenii, and the promoter activity was assessed by the ability of the E. coli transformants to grow on LB/tetracycline agar plates and by the ability of the G. hansenii transformants to grow on SH/tetracycline agar plates. As a negative control, the 1,288-bp promoterless tetC fragment was ligated to the PvuI/SalI sites of pUCD2, and the construct was similarly transformed into E. coli and G. hansenii. This construct and all of the above-mentioned promoter constructs are listed in Table S3 in the supplemental material.
Complementation assay.
To make constructs for complementing acsA, dgc1 and crp-fnr mutants, PCRs were performed using genomic DNA of wild-type G. hansenii as a template and AcsA-PF/AcsC-CER, Dgc1-CEF/Dgc1-CER, and Crp-CEF/Crp-CER, respectively, as primers (see Table S2 in the supplemental material). All of the resulting DNA fragments were ligated to the SalI/PvuI sites of pUCD2 using the In-Fusion HD Cloning Kit. The construct for complementing the acsA mutant (mutant 5) contained an 8,551-bp fragment (the 221-bp promoter fragment for the acs operon and the entire coding sequences of acsAB and acsC, plus 93 bp of the 3′-UTR). The dgc1 construct for complementing mutant I-7 contained 108 bp of the 5′-UTR, 1,719 bp of the coding sequence, and 108 bp of the 3′-UTR. The crp-fnr construct for complementing mutant II-23 contained 136 bp of the 5′-UTR, 744 bp of the coding sequence, and 106 bp of the 3′-UTR. The acsC construct for complementing mutant 10 contained the 221-bp promoter fragment of the acs operon (amplified using the primers AcsA-PF and AcsA-PR2), and the coding sequence of acsC plus 93 bp of the 3′-UTR (amplified using the primers AcsC-CEF and AcsC-CER). The ccpAx construct for complementing mutant I-13, amplified using the primers CmcAx-PF and CcpAx-CER, contained the 504-bp promoter fragment for the cmcAx-ccpAx operon and the entire coding sequences of cmcAx and ccpAx, plus 111 bp of the 3′-UTR. The sequences of all of the above-mentioned primer pairs are listed in Table S2 in the supplemental material. All of the constructs were transformed into E. coli DH5α, and the transformants were selected on LB agar plates containing kanamycin. The plasmid DNA purified from the transformant of each gene was electroporated into the mutant in which the respective gene was disrupted by Tn5 transposon insertion. The transformed cells were grown in SH medium with cellulase (0.02%) at 30°C overnight, and the cells were resuspended and plated on SH agar plates containing spectinomycin.
Protein gel blot analysis.
Cells of wild-type, Cel− mutant, and complemented Cel− mutant strains were harvested from 100-ml shaking cultures at an OD600 of ∼0.7 and centrifuged at 5,000 × g for 15 min at 4°C. The cell pellets were suspended in 5 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 5% glycerol, 1% β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride) and lysed by sonication (30 s, 5 cycles for each cycle). Each cell lysate containing total protein was collected by centrifugation, and the protein concentration was estimated by the Bradford method (26). Protein gel blotting was performed according to standard procedures. Briefly, for each sample, total protein (40 μg) was boiled in sodium dodecyl sulfate (SDS) sample buffer and loaded into a lane of a 12% SDS-polyacrylamide gel. Electrophoresis was carried out at 200 V for ∼50 min. The proteins were transferred onto polyvinylidene difluoride membranes, followed by blotting with anti-AcsAcat, -AcsB, -AcsC, and -AcsD antibodies (6) or an anti-BglAx antibody. The details of these antibodies are listed in Table 1. Each polyclonal antibody was affinity purified by repeated binding and elution of antisera with nitrocellulose bound protein antigen (27) to yield a monospecific antibody. The blots were separately incubated with the anti-AcsAcat, -AcsB, -AcsC, -AcsD, and -BglAx antibodies (1:500 dilution) and then with the secondary antibody of anti-rabbit IgG conjugated with alkaline phosphatase (1:15,000 dilution; Sigma-Aldrich). The immunoreactive proteins were visualized with BCIP (5-bromo-4-chloro-3-indolylphosphate)/nitroblue tetrazolium (Amresco, Solon, OH). The molecular mass of each protein band was estimated from a molecular mass ladder (Fisher Scientific, Pittsburgh, PA).
Table 1.
Antibodies used in protein blot analysis
| Antibody | Targeta |
|---|---|
| Anti-AcsAcat | Heterologously expressed and purified truncated protein (aa 128 to 397) |
| Anti-AcsB | Heterologously expressed and purified truncated protein (aa 608 to 1510) |
| Anti-AcsC | Two synthetic peptides (RNRKHEVRVGVNLTYFGYK, aa 309 to 328; ARQDGFKALNAGRLSAA, aa 1146 to 1164) |
| Anti-AcsD | Heterologously expressed and purified AcsD |
| Anti-BglAx | Synthetic peptide (LEPRVLAHFDEKHDRRNV, aa 684 to 701) |
aa, amino acids.
Measurement of growth rates of Cel− mutants and cellulose production of complemented mutants.
The Cel− mutants and WT (Cel−) were grown in 100 ml of SH medium under static conditions for 15 days, with growth monitored daily by measuring the absorbance at 600 nm according to the procedure of Kawano et al. (5). After each Cel− mutant had been complemented with a functional copy of the gene disrupted by Tn5 transposon insertion, the complemented mutant was grown in 100 ml of SH medium under static conditions for 15 days. The cellulose pellicles were washed with 0.1 M NaOH at 80°C twice, for 1 h each time, with gentle stirring and then washed with distilled water until the pH decreased to 7. The pellicles were air dried, and the dry weight was measured. The wild type was similarly grown, and the amount of cellulose produced was similarly determined.
Quantitative real-time PCR (qRT-PCR).
Total RNA was isolated using TRIzol reagent (Invitrogen), according to the manufacturer's instructions, from 10-ml cultures of mutant II-23 (with crp-fnr disrupted), the wild type, and the wild type that had been transformed with a functional copy of crp-fnr (in pUC18). The quality of RNA was assessed by agarose gel electrophoresis after treatment with 1 U of DNase (Fisher Scientific). cDNA was synthesized from 2 μg of total RNA by SMARTScribe reverse transcriptase (Clontech) using random primers (Promega). qPCR was carried out with cDNA as a template and gene-specific primers (see Table S2 in the supplemental material). qPCR was performed with PerfecTA SYBR green FastMix ROX (Quanta Biosciences, Gaithersburg, MD) on a 7300 Sequence Detection System (Life Technologies), with three biological replicates for each gene tested. The cycling conditions were 95°C for 30 s for the initial denaturation, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 29 s, and then the melting-curve stage. Melting-curve analysis was used to confirm the absence of nonspecific amplification products. The threshold cycles (CT) were calculated by plotting normalized fluorescence in relation to the cycle number. Transcripts from the 16S rRNA gene were also detected, using the primers (see Table S2 in the supplemental material) described by Kawano et al. (28), and served as internal control to normalize expression of other genes. For each gene, the ratio of the amount of transcripts produced by the crp-fnr mutant, or by the crp-fnr overexpressing transformant, to that of the wild type was obtained from the ratio of the 2−ΔΔCT values.
Targeted gene knockout via homologous recombination.
A fragment of bglAx, bp 558 to 1129 of the coding sequence, was amplified using BglAx-F and BglAx-R as primers (see Table S2 in the supplemental material) and genomic DNA of the wild type as a template. The tetC gene in pUCD2 was amplified by using the primers TetC-HF and TetC-HR to yield an ∼1.3-kb fragment that contained the promoter region, entire coding sequence, and 93 bp of 3′-UTR. The cycling conditions were 98°C for 30 s for the initial denaturation, followed by 35 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 to 60 s, and then 72°C for 10 min. The two PCR fragments and BamHI/EcoRI-digested pUC18 were ligated using an In-Fusion HD cloning kit, and the resulting construct was introduced into the wild type via electroporation. The transformants were selected on SH/tetracycline agar plates. To confirm insertion of the tetC gene into the chromosomal bglAx, the genomic DNA of a transformant was amplified using as the primers Bgl-CF (see Table S2 in the supplemental material), which corresponded to bp 144 to 166 of the bglAx coding sequence, and TSP4 (see Table S2 in the supplemental material), which corresponded to bp 139 to 162 of the tetC coding sequence. The cycling conditions were 94°C for 5 min for the initial denaturation, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min, and then 72°C for 10 min. Cloning and sequencing of the PCR fragments were performed as described in “Cloning and sequencing of genes disrupted by Tn5 transposon” above.
RESULTS
Isolation of stable cellulose-producing (Cel+) and stable non-cellulose-producing (Cel−) clones of G. hansenii ATCC 23769.
It has long been known that cellulose-producing strains of Gluconacetobacter species when cultured in growth medium produce a mixture of cellulose-producing colonies and non-cellulose-producing colonies (23). This was initially thought to be due to high frequencies of mutation, but now it seems that the appearance of Cel− cells may be simply due to either impurity of the starting culture (containing both Cel+ and Cel− cells) or growth conditions (e.g., in static culture, cells that are not embedded in the pellicle on the surface of the medium generally do not produce cellulose) (4, 29). For our use of Tn5 transposon mutagenesis to identify mutants that fail to synthesize/secrete cellulose, it was imperative that we start with a stable cellulose-producing clone. Our original stock of G. hansenii ATCC 23769 was isolated as a cellulose-overproducing strain, but when cells were streaked onto SH agar plates and incubated at 30°C for 2 to 3 days, both small rough cellulose-overproducing colonies and large round non-cellulose-producing colonies were obtained. To examine whether our original stock contained both Cel+ and Cel− cells, we randomly selected one Cel+ colony and one Cel− colony and streaked out each colony onto separate SH agar plates. Twenty colonies were randomly picked from each plate and cultured in SH medium under both static and shaking conditions. All 20 colonies from the Cel+ clone produced cellulose under both conditions, whereas all 20 colonies from the Cel− clone did not. This process was repeated two more times, and both times all of the colonies derived from the Cel+ colony randomly picked from the previous round were Cel+, and all of the colonies derived from the Cel− colony were Cel− (see Fig. S1A in the supplemental material). One of those cellulose-producing colonies from the last round of plating was designated as wild-type (WT) G. hansenii and used for all subsequent experiments. Similarly, one of those non-cellulose-producing colonies from the last round of plating was designated WT (Cel−) and was used as a negative control for cellulose production and as a wild-type control when assessing growth rates of the Cel− mutants described below. On the SH agar plate containing 0.001% Pontamine Fast Scarlet 4B, all colonies derived from the Cel+ clone showed a red color, and all colonies derived from the Cel− clone showed a white color (Fig. S1B).
Isolation of non-cellulose-producing mutants.
We first examined four antibiotics—ampicillin (100 μg/ml), kanamycin (50 μg/ml), spectinomycin (100 μg/ml) and tetracycline (20 μg/ml)—to see whether they were appropriate for use as selectable markers for transformation of wild-type cells. At the concentrations used, wild-type cells were sensitive to tetracycline and spectinomycin but were completely resistant to ampicillin and kanamycin. We then tested the feasibility of using pUCD2, which contains a tetracycline resistance gene, tetC, as a shuttle vector between E. coli and G. hansenii. Electroporation of pUCD2 into competent cells of wild-type G. hansenii conferred on the transformants tetracycline resistance, verifying that this plasmid can autonomously replicate in G. hansenii and that the promoter of the tetC gene is active in G. hansenii. We thus used electroporation to transform competent cells of wild-type G. hansenii with a complex of Tn5 transposon DNA fragment (containing the tetC gene from pUCD2) and EZ-Tn5 transposase (see Fig. S2 in the supplemental material). A total of 769 individual colonies were examined under both static and shaking growth conditions, and the cellulose production of each colony was compared to that of the wild type. Wild-type cells produced cellulose as a round pellicle at the bottom of the culture under shaking conditions, and the culture medium remained clear. Under static conditions, the cellulose pellicle formed as a white layer at the top of the culture medium. Among the mutants screened, six did not show any cellulose in the medium after 7 days of growth under static conditions and after 2 days of growth under shaking conditions. The colonies of these Cel− mutants appeared smooth on SH agar plates and under a light microscope, confirming the lack of cellulose production (see Fig. S3 in the supplemental material).
Identification of genes disrupted by Tn5 transposon insertion in Cel− mutants.
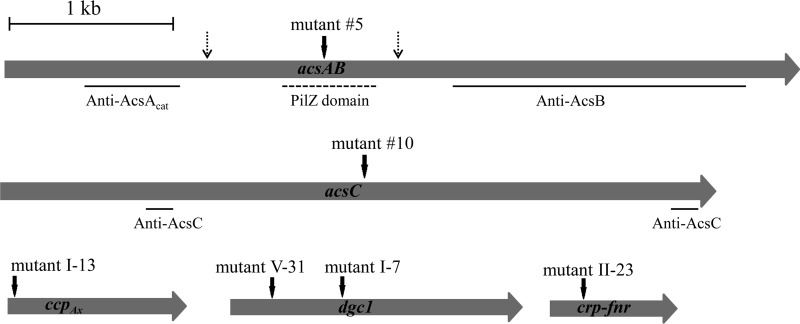
Genomic DNA was isolated from each of the six Cel− mutants, and a PCR-based genomic DNA walking method, as described in Materials and Methods, was used to amplify and clone DNA fragments containing part of the tetC gene and the immediate flanking regions of the gene disrupted. The nucleotide sequences of the cloned DNA fragments were determined and compared to the genome sequence of G. hansenii ATCC 23769 (GenBank accession number NZ_CM000920.1) (21) to identify the gene disrupted and the insertion site of the Tn5 transposon DNA in each mutant. The results are schematically shown in Fig. 1 and summarized in Table 2. Mutant 5 had the transposon inserted in the coding sequence for the PilZ domain (bp 1710 to 2058) of acsA (locus tag GXY_04277 of accession number NZ_CM000920.01). Mutant 10 had the transposon inserted in the coding sequence of acsC (GXY_04282). Mutant I-13 had the transposon inserted in the coding sequence for ccpAx (GXY_04267). Mutants I-7 and V-31 had the transposon inserted at different sites in the coding region of dgc1 (GXY_15547). (Mutant I-7 was used as the representative of dgc1 mutants in all subsequent experiments.) Mutant II-23 had the transposon inserted in the coding region of crp-fnr (GXY_00863).
Fig 1.
Schematics of genes inserted by Tn5 transposon DNA into six Cel− mutants. Dark vertical arrows indicate the sites of insertion. For acsAB, the regions encoding the truncated proteins used to raise anti-AcsAcat and anti-AcsB antibodies and the region encoding the PilZ domain are indicated. For acsC, the regions encoding two peptides used to raise anti-AcsC antibody are indicated. Broken vertical arrows indicate the putative cleavage sites of AcsAB as it is processed to AcsAcat, AcsAreg, and AcsB.
Table 2.
Genes disrupted by Tn5 transposon insertion in Cel− mutants
| Mutant | Disrupted gene | Protein encoded | Insertion sitea | Length (bp) of truncated protein/full length (no. of amino acids) |
|---|---|---|---|---|
| 5 | acsA | Cellulose synthase subunit A | 1893–1894 | 631/1,550 |
| 10 | acsC | Cellulose synthase subunit C | 1984–1985 | 661/1,303 |
| I-7 | dgc1 | Diguanylate cyclase | 656–657 | 218/572 |
| I-13 | ccpAx | Cellulose complementing protein | 40–41 | 13/353 |
| II-23 | crp-fnr | Transcriptional regulator, Crp/Fnr family protein | 187–188 | 62/157 |
| V-31 | dgc1 | Diguanylate cyclase | 242–243 | 80/572 |
That is, the base pair number counting from the translation initiation codon.
Analysis of production of Acs proteins in Cel− mutants.
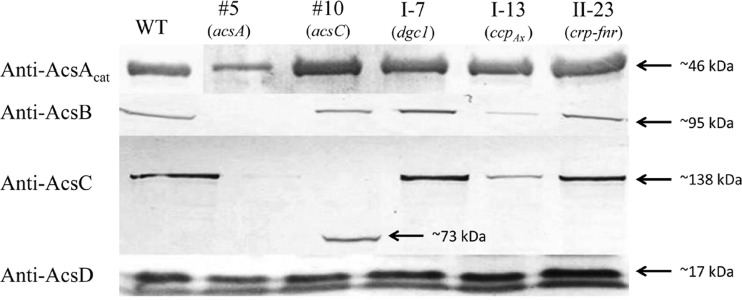
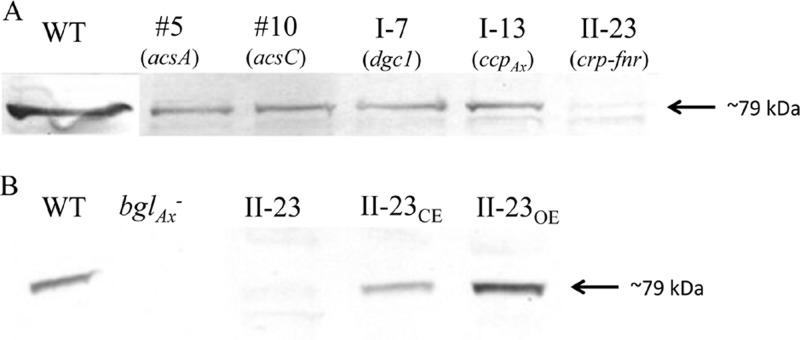
We next examined whether Tn5 transposon insertion into any of the Cel− mutants listed in Table 2 affected the production of other proteins involved in cellulose synthesis. Affinity-purified monospecific anti-AcsAcat, -AcsB, -AcsC, and -AcsD antibodies (Table 1) were used to detect production via protein blot analysis (Fig. 2). The anti-AcsAcat antibody detected an ∼46-kDa protein band in all of the Cel− mutants and the wild type. The detection of this protein band in mutant 5 is consistent with the finding that the Tn5 transposon DNA was inserted in the coding sequence for the PilZ domain of AcsA (Fig. 1) and is consistent with our previous findings that AcsAB is processed via a protease at a site upstream from the insertion site of Tn5 transposon (6). The anti-AcsB antibody detected an ∼95-kDa protein band in all but mutant 5. This is consistent with the Tn5 transposon insertion site being upstream from the coding region for AcsB (Fig. 1). The anti-AcsC antibody detected an ∼138-kDa protein in all but mutants 5 and 10. In mutant 10, the anti-AcsC antibody detected a protein band of ∼73 kDa, a size consistent with the molecular mass of the truncated AcsC predicted based on the Tn5 transposon insertion site (Fig. 1). Both the AcsB and the AcsC bands detected in mutant I-13 were much less intense than the corresponding bands in the wild type, mutant I-7, and mutant II-23. The anti-AcsD antibody cross-reacted with two protein bands, an ∼17-kDa band and a band with a slightly lower molecular mass. The ∼17-kDa band corresponded to AcsD, since this band, but not the other, was absent in an acsD knockout mutant of G. hansenii ATCC 23769 (Y. Deng and T.-h. Kao, unpublished results). The AcsD bands in all of the Cel− mutants and the wild type were of similar intensities.
Fig 2.
Protein blot analysis of Acs proteins in the wild type (WT) and five Cel− mutants. For each mutant, the gene disrupted is indicated in parentheses. Equal amounts (40 μg) of total proteins were used for all samples. The antibody used for each blot is shown to the left.
Assay of promoter activity.
Based on the organization of the acs operon of several G. xylinus strains studied, it was thought that this operon in G. hansenii ATCC 23769 also contained acsA, acsB, acsC, and acsD and, as such, these four genes would be expected to be transcribed from the same promoter into a polycistronic message. The finding that AcsC was not detected in mutant 5 (Fig. 2) could be explained by the polar effect caused by the insertion of a 1.7-kb Tn5 transposon DNA in the coding region of acsAB immediately upstream from acsC. However, the production of AcsD was not affected in mutant 5, and in mutant 10, disruption of acsC did not affect the production of AcsD (Fig. 2). These unexpected results, coupled with the presence of a 321-bp spacer between the translation termination codon of acsC and the translation initiation codon of acsD (see Fig. S4 in the supplemental material), raised the possibility that in G. hansenii ATCC 23769 acsD might not be part of the acs operon and might instead be expressed by its own promoter. To examine this possibility, we fused the 321-bp DNA fragment to the coding sequence of tetC in pUCD2 and introduced the construct into both E. coli and G. hansenii. The construct conferred on the G. hansenii transformants, but not on the E. coli transformants, tetracycline resistance (see Table S3 in the supplemental material), confirming the existence of a promoter in this fragment. Thus, in contrast to several G. xylinus strains, acsD in G. hansenii ATCC 23769 is expressed by its own promoter and is not part of the acs operon.
We further confirmed the existence of the promoter of the acs operon in the 221-bp sequence upstream from the translation initiation codon of acsA, since the construct containing this fragment conferred on the G. hansenii, but not the E. coli transformants, tetracycline resistance (see Table S3 in the supplemental material). We also examined the 504-bp sequence upstream from the translation initiation codon of cmcAx and found that, unlike the promoters of the acs operon and acsD, the promoter contained in this fragment was active in both E. coli and G. hansenii (see Table S3 in the supplemental material). Since there is a 4-bp overlap between the translation termination codon of cmcAx and the translation initiation codon of ccpAx, these two genes most likely form an operon and are expressed by the same promoter.
Complementation of each Cel− mutant by a functional copy of the gene disrupted by Tn5 transposon insertion.
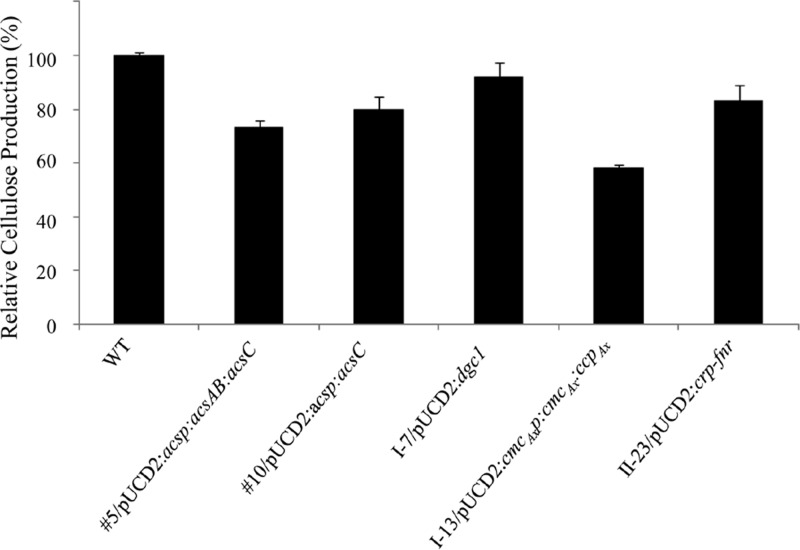
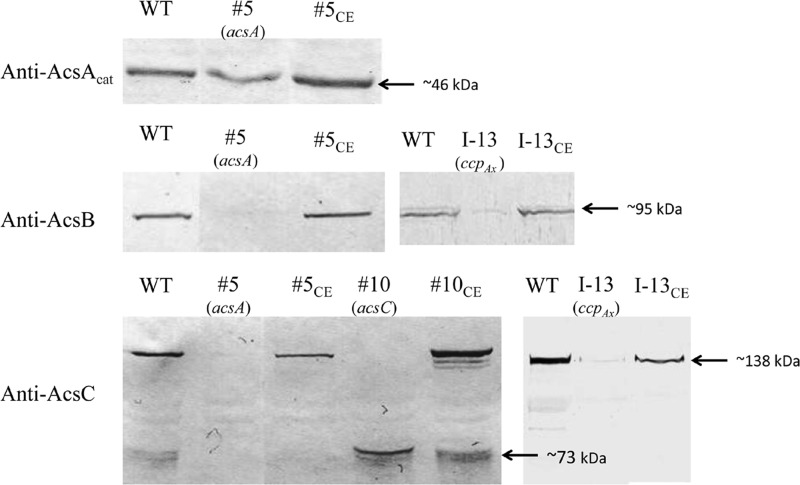
To verify that the phenotype of each Cel− mutant was indeed caused by Tn5 transposon insertion in the gene identified by genomic walking, we complemented each mutant with a functional copy of the gene carried by pUCD2. For mutant 5, since the production of both AcsAB and AcsC was affected (Fig. 2), we used PCR to amplify an 8.5-kb fragment from the genomic DNA of the wild type, which included the acs promoter and the entire coding sequences of acsAB and acsC, and ligated the fragment to pUCD2. The construct (Table 3) was introduced into mutant 5 via electroporation, and the transformants were selected on SH agar plates containing spectinomycin. Three independent colonies were chosen and grown in SH medium containing tetracycline and spectinomycin under both static and shaking conditions. All three colonies produced cellulose under both growth conditions. To determine the extent to which the cellulose-producing ability was restored in the complemented mutant, we inoculated each colony in 100 ml of SH medium containing both antibiotics, grew the cells under static conditions for 7 days, and processed and weighed the cellulose produced. The mean amount of cellulose produced by the three independent colonies was 72.9% ± 2.6% that of the wild type (Fig. 3). We then used protein blotting to examine the levels of AcsAcat, AcsB, and AcsC produced by one of the colonies, 5CE (Fig. 4). The levels of AcsB and AcsC were comparable to their respective wild-type levels, but AcsAcat was present at a higher level than in mutant 5, a finding consistent with the additional amount of AcsAcat produced from the introduced functional acsAB gene. Thus, the results from the complementation experiments confirmed that the Cel− phenotype of mutant 5 was caused by the insertion of the Tn5 transposon DNA in its acsA gene.
Table 3.
Constructs used in complementation experiments
| Construct | Promoter (bp)a | Coding sequence | Length of gene + 3′-UTR (bp) |
|---|---|---|---|
| pUCD2::acsp::acsAB::acsC | Promoter of acs operon (221) | acsAB::acsC | 8,237 + 93 |
| pUCD2::acsp::acsC | Promoter of acs operon (221) | acsC | 3,588 + 93 |
| pUCD2::cmcAxp::cmcAx::ccpAx | Promoter of cmcAx-ccpAx operon (504) | cmcAx::ccpAx | 2,087 + 111 |
| pUCD2::dgc1 | dgc1 promoter (108) | dgc1 | 1,719 + 108 |
| pUCD2::crp-fnr | crp-fnr promoter (136) | crp-fnr | 744 + 106 |
The base pair number in parentheses denotes the size of the fragment used in each construct and not the actual size of the promoter.
Fig 3.
Amount of cellulose produced by each complemented Cel− mutant relative to that of the wild type (WT). Each Cel− mutant was transformed with pUCD2 carrying a functional copy of the gene disrupted by Tn5 transposon. The promoter of the acs operon (acsp) was used to drive the expression acsC in mutant 10. For all of the other mutants, either the entire operon (mutants 5 and I-13) or the entire gene driven by its native promoter (mutants I-7 and II-23) was introduced. After growth in 100 ml of SH medium with spectinomycin (100 μg/ml) and tetracycline (20 μg/ml) under static conditions for 7 days, equal amounts of cells, based on the OD600, were used for determining the dry weights of cellulose present in the medium. Each bar represents the percentage of the mean of three biological replicates of a complemented mutant relative to that of the wild type. Error bars represent means ± the standard deviations (SD).
Fig 4.
Protein blot analysis of Acs proteins produced in the wild type (WT) and complemented Cel− mutants. The gene disrupted in each mutant is indicated in parentheses, and the antibody used for each blot is shown on the left. For each mutant, its complemented transformant is indicated by a subscript “CE”.
For mutant 10, as only the production of full-length AcsC was affected by the disruption of acsC, we used the promoter of the acs operon to express the coding sequence of acsC for complementation (Table 3). For mutant I-13, we used PCR to amplify the cmcAx-ccpAx operon for complementation, in case translation of the ccpAx transcript in the polycistronic mRNA might depend on the translation of the upstream cmcAx transcript. For mutant I-7, we used PCR to amplify the dgc1 gene for complementation. For mutant II-23, we used PCR to amplify the crp-fnr gene for complementation. All of these constructs, made in pUCD2 (Table 3), were introduced into the respective mutants, and the transformants obtained were analyzed as described above for complementation of mutant 5. All of the complemented Cel− mutants regained the ability to produce cellulose in the SH medium with spectinomycin (100 μg/ml) under both static and shaking conditions, albeit to different degrees (Fig. 3). For mutant 10, the mean of the amounts of cellulose produced by three independent colonies was 80.0% ± 4.7% that of the wild type. The protein blotting results of one of the colonies, 10CE, showed that, in addition to producing a truncated AcsC from the disrupted acsC as did mutant 10, it also produced full-length AcsC to a level comparable to that of the wild type (Fig. 4). For mutant I-7, the mean amount of cellulose produced by three independent colonies was 91.7% ± 5.4% that of the wild type. For mutant I-13, the mean amount of cellulose produced by three independent colonies was 58.0% ± 2.3% that of the wild type. Since mutant I-13 produced substantially lower amounts of AcsB and AcsC than did the wild type (Fig. 2), we examined the production of these two proteins in one of the colonies of the complemented mutant, I-13CE. The amounts of both proteins were comparable to the respective amounts in the wild type (Fig. 4). For mutant II-23, the mean amounts of cellulose produced by three independent colonies was 82.9% ± 6.2% that of the wild type.
Growth rates of Cel− mutants.
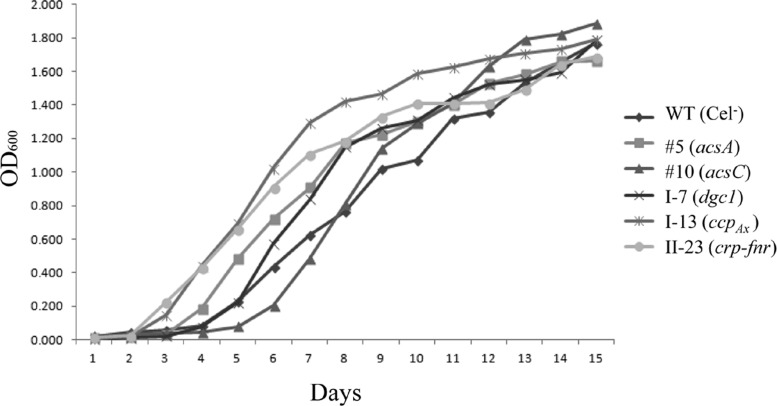
We examined the growth rates of the Cel− mutants over a period of 15 days to rule out the possibility that their lack of cellulose production was due to the negative effect on cell growth caused by the disruption of the genes. Under the static growth condition, cellulose-producing cells are embedded in the cellulose pellicle formed at the surface of the medium, making monitoring cell growth by optical density inaccurate. We thus used the stable non-cellulose-producing clone of ATCC 23769, WT (Cel−), as the wild-type control for growth rates of the Cel− mutants. Up to day 7, the growth rates of all except mutant 10 (with acsC disrupted) were slightly higher than that of WT (Cel−); however, at the end of day 15, all of the mutants and WT (Cel−) reached similar cell densities (Fig. 5).
Fig 5.
Growth curves of Cel− mutants and WT (Cel−), the stable non-cellulose-producing clone of the wild type. The gene disrupted in each mutant is indicated in parentheses. Cells were grown under static conditions in 100 ml of SH medium with tetracycline (20 μg/ml), and the OD600 values were taken for all of the samples at the same time on each day during a 15-day period.
Effect of crp-fnr mutation on transcription of genes involved in cellulose biosynthesis.
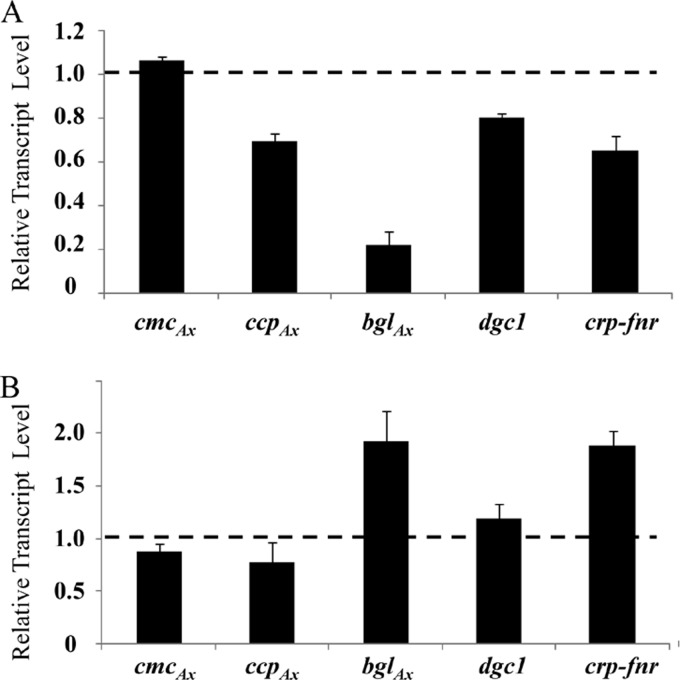
The crp-fnr gene disrupted in mutant II-23 is predicted to encode a transcription factor and, based on the protein blotting results shown in Fig. 2, this mutant produced wild-type levels of AcsAcat, AcsB, AcsC, and AcsD. Hence, transcription of the acs operon and acsD is not regulated by this transcription factor. To determine whether transcription of cmcAx, ccpAx, dgc1, and bglAx might be regulated by Crp/Fnr, we used qRT-PCR to determine their transcript levels in mutant II-23 and the wild type (Fig. 6A). The transcript levels of cmcAx, ccpAx, and dgc1 were all comparable to, or slightly lower than, their respective wild-type levels, whereas the transcript level of bglAx was only 16.1% ± 1.4% that of the wild type. To further assess whether any of these genes is under the control of Crp/Fnr, we introduced a functional crp-fnr gene in pUCD2 (Table 3) into wild-type G. hansenii to generate transformants that overexpressed crp-fnr and used qRT-PCR to determine the transcript levels of these genes. Analysis of one such transformant, II-23OE, showed that the transcript level of crp-fnr was approximately twice that of the wild type. Among the genes tested, the transcript level of bglAx was also approximately twice that of the wild type, whereas the transcript levels of cmcAx, ccpAx, and dgc1 were similar to their respective wild-type levels (Fig. 6B).
Fig 6.
Quantitative RT-PCR analysis of transcript levels of cmcAx, ccpAx, bglAx, dgc1, and crp-fnr in mutant II-23 (A) and in the wild type carrying a functional copy of crp-fnr in pUCD2 (B). The data are normalized to 16S RNA levels, and for each gene in panels A and B the data are presented as a level relative to that of the wild type, denoted by the dashed line at 1.0. Three independent biological replicates were used for each gene and, for each biological replicate, at least three qRT-PCRs were performed. Error bars represent means ± the SD.
Role of BglAx in cellulose biosynthesis.
To further confirm the role of Crp/Fnr in regulating the expression of bglAx, we first used an anti-BglAx antibody to examine the levels of BglAx in all of the Cel− mutants. As shown in Fig. 7A, an ∼79-kDa protein band, consistent with the predicted molecular mass of BglAx, was detected in the acsA, acsC, dgc1, and ccpAx mutants but was virtually undetectable in the crp-fnr mutant. We then examined the levels of BglAx in II-23CE and II-23OE (Fig. 7B). The intensity of the BglAx band in II-23CE was comparable to that of the wild type, whereas the intensity of this band in II-23OE, which produced twice the levels of crp-fnr transcript (Fig. 6B), was stronger than that of the wild type. Thus, restoring the production of Crp/Fnr in II-23CE led to normal production of BglAx, and overexpressing Crp/Fnr in II-23OE led to a higher-than-normal level of BglAx.
Fig 7.
Protein blot analysis of BglAx. (A) Wild type (WT) and five Cel− mutants. The gene disrupted in each mutant is indicated in parentheses. (B) Mutants II-23, II-23CE (complemented mutant II-23), II-23OE (WT transformed with a functional copy of crp-fnr in pUCD2), and bglAx− (a bglAx knockout mutant). An anti-BglAx antibody was used for both blots.
We next examined whether the Cel− phenotype of mutant II-23 might be at least partially due to the absence of BglAx. We generated a bglAx knockout mutant by introducing a construct in pUC18, containing bp 557 to 1129 of the bglAx coding sequence fused to tetC, into wild-type G. hansenii. The transformants were selected on SH agar plates containing tetracycline. Since pUC18 cannot autonomously replicate in G. hansenii, the tetracycline-resistant colonies obtained should have the entire plasmid construct integrated into the chromosomal DNA via homologous recombination between the sequence from bp 557 to 1129 in the construct and the same sequence in the chromosomal bglAx. To verify the insertion, we performed PCR on genomic DNA isolated from a transformant, using a primer corresponding to bp 144 to 166 of the bglAx coding sequence and a primer corresponding to bp 139 to 162 of the tetC coding sequence. Sequencing of the ∼1.1-kb PCR fragment confirmed that it contained the tetC gene inserted in the coding region of the chromosomal bglAx. Protein blotting analysis of this bglAx knockout transformant showed that it indeed did not produce any BglAx (Fig. 7B).
We then compared the amounts of cellulose produced by the bglAx mutant, II-23OE, and the wild type after 7 days of growth under static conditions. Three independent clones each were analyzed. As expected, the amount of cellulose produced by II-23OE was higher than that of the wild type (166.1% ± 7.6% that of the wild type). Unlike disruption of crp-fnr in mutant II-23, knocking out bglAx did not completely abolish cellulose production, since the cellulose produced by the bglAx mutant was 16.1% ± 1.4% the the wild-type level. This finding suggests that Crp/Fnr may regulate the expression of additional gene(s) involved in cellulose biosynthesis.
DISCUSSION
In this work, we have established the use of an EZ-Tn5 transposome/transposase complex as an effective means to generate random transposon insertions in the genome of G. hansenii. Screening of non-cellulose-producing (Cel−) mutants generated from a stable cellulose-overproducing clone is relatively easy when grown on agar plates (23, 30, 31). Since G. hansenii ATCC 23769 is sensitive to tetracycline (but not to ampicillin or kanamycin), we have included a tetracycline resistance gene in the Tn5 transposon DNA construct to allow the selection of colonies that have the Tn5 transposon inserted in their genomes.
From the screening of fewer than 800 colonies that are tetracycline resistant, we have already obtained six Cel− mutants, as well as ∼80 mutants that produce cellulose with altered morphologies (Deng and Kao, unpublished). Among the six Cel− mutants, four each have a different gene disrupted, and two have disruption at different sites of yet another different gene. Thus, based on the gene disrupted and the insertion site, all of these six Cel− mutants resulted from independent insertion of the Tn5 transposon DNA, suggesting that we have not yet saturated all possible genes involved in cellulose biosynthesis. Nonetheless, analysis of these mutants has already revealed a number of unexpected and interesting findings about cellulose biosynthesis.
Of the Cel− mutants characterized, we found that their lack of cellulose production is not due to compromised rates of growth. We used the stable non-cellulose-producing clone, WT (Cel−), instead of the stable cellulose-overproducing clone (WT) for comparison of cell growth under static conditions, as WT cells are embedded in the cellulose pellicle at the surface of the medium, making measurement of cell growth by OD600 inaccurate. Of these Cel− mutants, only mutant 10, with Tn5 insertion in the acsC gene, initially grew more slowly than WT (Cel−) (Fig. 5). Since this mutant still produced AcsAB (Fig. 2), it presumably was able to synthesize cellulose, but the cellulose produced likely remained in the periplasm due to lack of a functional AcsC for secretion. We had initially identified the Cel− mutants based on their inability to produce cellulose after 7 days of growth, thus we further examined the cellulose production of mutant 10 after 15 days of growth, when its cell density was comparable with that of the wild type. Still, no cellulose was observed in the medium.
Although our results confirm previous findings showing the importance of acsA and acsC in cellulose synthesis (3, 10), we have provided here new information on the genetic arrangement and the regulation of the genes involved in cellulose synthesis. For example, we found for the first time that acsD is not part of the acs operon. If it were, one would expect that Tn5 transposon insertion in acsA and acsC should affect the translation of acsD from the polycistronic message for AcsAB, AcsC, and AcsD due to a polar effect. The finding that the production of acsD is not affected in either mutant 5 or mutant 10 has led to confirmation by a promoter activity test that acsD is transcribed from its own promoter and is not part of the acs operon, as previously thought based on the analysis of the acs operon of several G. xylinus strains, e.g., ATCC 53582 and BPR2001 (32, 33). For these two strains, there is a single base overlap between the coding sequences of acsC and acsD, whereas for G. hansenii ATCC 23769, there is a 321-bp spacer (wherein the promoter of acsD was found to lie) between the translation termination codon of acsC and the translation initiation codon of acsD. Consistent with acsAB and acsC being organized in the acs operon of G. hansenii ATCC 23769, there is a 4-bp overlap between the termination codon of acsAB and the initiation codon of acsC. It is not clear what the physiological significance is, if any, between the different organizations of the acs genes in G. hansenii and G. xylinus.
It is interesting that the genome of G. hansenii ATCC 23769 has two additional acsAB-like genes (named acsAB1 and acsAB2) and acsC-like genes (named acsC1 and acsC2), but acsD is present in only one copy (21). The deduced amino acid sequences of acsAB1 (GXY_08864) and acsAB2 (GXY_14452) are 40 and 46% identical, respectively, to that of acsAB, and the deduced amino acid sequences of acsC1 (GXY_08869) and acsC2 (GXY_014472) are 28 and 30% identical, respectively, to that of acsC. Our finding that the absence of AcsAB and AcsC in mutant 5 results in the Cel− phenotype suggests that, despite the sequence homology, the two acsAB-like genes and two acsC-like genes are not involved in cellulose biosynthesis in vivo. Saxena et al. (34) also identified an additional acs-like operon in G. hansenii AY201 (a derivative of ATCC 23769), which contains genes homologous to acsA (named acsAII), acsB, and acsC, and showed that AcsAII has cellulose synthase activity in vitro but is not required for cellulose production when grown under laboratory conditions.
Our finding that the Tn5 insertion in ccpAx (mutant I-13) results in a complete loss of cellulose production is consistent with the finding by Standal et al. (14) that a non-cellulose-producing mutant of G. hansenii has an indigenous transposable element inserted in this gene. Recently, Sunagawa et al. (15) showed that, in G. hansenii, ccpAx interacts with AcsD and colocalizes with AcsD and proposed that CcpAx is a member of the CSC and responsible for its proper structure. If CcpAx is indeed a component of the CSC, its absence is expected to affect the assembly of a functional complex. We did not observe any reduction in the level of AcsD in the ccpAx mutant but instead found that the levels of two other presumed components of the CSC, AcsB and AcsC, are drastically reduced (Fig. 2). This finding provides an explanation for the Cel− phenotype of the ccpAx mutant; however, it is not clear why the absence of CcpAx affects the levels of AcsB and AcsC but not AcsD. It is also not known whether any additional protein might be affected by the absence of a functional CcpAx. It would be of interest to compare the proteomes of the wild type and the ccpAx mutant.
c-di-GMP is a second messenger in bacteria and is known to regulate biofilm formation, motility, and the production of extracellular polysaccharides (35). c-di-GMP binds to the PilZ domain in the C-terminal region of AcsA (10) and acts as a positive regulator of cellulose synthase in G. hansenii (9). Formation of c-di-GMP is catalyzed by diguanylate cyclase (Dgc), and degradation is catalyzed by phosphodiesterase A (PdeA) in conjunction with phosphodiesterase B (PdeB) (9). dgc and pdeA are located in the cdg (cyclic diguanylate) operon, and pdeA lies upstream from dgc (36). The genome of G. hansenii ATCC 23769 encodes two cdg operons (21). The deduced amino acid sequence of dgc2 (GXY_01661) of the second cdg operon is 59.9% identical to that of dgc1. However, disruption of dgc1 alone completely abolishes cellulose production, suggesting that the dgc2, despite its sequence homology with dgc1, cannot substitute for the function of dgc1 under the growth conditions used in our isolation of Cel− mutants. G. xylinus BPR2001 has three cdg operons and disrupting one of the dgc genes, named dgc1, has no effect on the cellulose yield (37). Consistent with this finding, the phylogenetic tree of the two dgc genes of ATCC 23769 and three dgc genes of BPR2001 shown in Fig. S5 in the supplemental material suggests that dgc1 of ATCC 23769 is more closely related to dgc2 of BPR2001.
Another notable finding of the present study is that disruption of crp-fnr (in mutant II-23), which is predicted to encode a transcriptional regulator, completely abolishes cellulose production. This suggests that at least one of the genes whose transcription is regulated by Crp/Fnr is required for the synthesis and secretion of cellulose microfibrils. Interestingly, a Crp/Fnr protein in Burkholderia cenocepacia (a pathogenic bacterium), named Bcam1349, is also involved in regulating biofilm formation (38). Bcam1349 functions as a c-di-GMP-responsive transcriptional regulator, as c-di-GMP greatly enhances its binding to the promoter of the cellulose synthase operon to activate the expression of the genes involved in cellulose biosynthesis. In this capacity, Bcam1349 also regulates the virulence of B. cenocepacia. To identify the target(s) of Crp/Fnr in G. hansenii, we first examined whether production of any of the Acs proteins in the crp-fnr mutant is affected. All of these Acs proteins were found to be present at wild-type levels (Fig. 2), suggesting that, unlike Bcam1349 of B. cenocepacia, Crp/Fnr in G. hansenii does not regulate expression of the cellulose synthase operon (the acs operon) or acsD. We next examined whether transcription of cmcAx, ccpAx, dgc1, and bglAx might be under the control of Crp/Fnr. cmcAx and ccpAx had previously been shown to be required for cellulose synthesis; the disruption of dgc1 is responsible for the Cel− phenotype of two of the mutants identified in this work, and bglAx is located immediately downstream from acsD. The results from qRT-PCR analysis of the transcript levels of bglAx in mutants II-23 and II-23OE and the wild type (Fig. 6) strongly suggest that the transcription of bglAx is regulated by Crp/Fnr. Moreover, the protein blotting results are consistent with the qRT-PCR results, i.e., mutant II-23 does not produce any detectable amount of BglAx, whereas mutant II-23OE that produces twice the wild-type level of the crp-fnr transcript produces a higher than wild-type level of BglAx (Fig. 7B).
Unlike the genes (acsA, acsC, ccpAx, dgc1, and crp-fnr) that were disrupted in the Cel− mutants identified here, bglAx, when knocked out, does not completely abolish the production of cellulose but instead results in the severe reduction (∼84%) of cellulose production. This is similar to the cmcAx knockout mutant of G. xylinus BPR2001 reported by Nakai et al. (39). CmcAx is a β-1,4-endoglucanase (endocellulase) and has cellulose-hydrolyzing activity in vitro (13). Since cellulose microfibrils produced by the cmcAx mutant are more twisted than those produced by the wild type, Nakai et al. (39) suggest that cellulose-hydrolyzing activity of CmcAx may be required to remove distorted glucan chains during the assembly of cellulose fibrils. However, an β-1,4-endoglucanase alone cannot completely hydrolyze a glucan chain to glucose. Exoglucanases are required to digest the oligosaccharide chains to cellobiose and cellotetrose, both of which are then hydrolyzed by β-glucosidases to glucose monomers. Moreover, it has been shown that cellobiose inhibits both endo- and exoglucanases (40). BglAx is predicted to be a β-glucosidase and, as such, it may be required, along with CmcAx and exoglucanases, to completely hydrolyze distorted glucan chains. This action may also be required to minimize product inhibition by cellobiose on CmcAx and the exoglucanases involved in the assembly of crystalline cellulose microfibrils. BglAx purified from cultured G. xylinus BPR2001 has exo-1,4-β-glucosidase activity toward cellotriose or larger cello-oligosaccharides, but only slight activity toward cellobiose, and may also have glucosyltransferase activity (20). It will be interesting to express BglAx of G. hansenii ATCC 23769 and examine its enzymatic activity, and to compare the levels of cello-oligosaccharides in the wild type and the bglAx knockout mutant.
Crp/Fnr of G. hansenii most likely regulates the expression of any additional genes that are involved in cellulose production, since the crp-fnr mutant does not produce any cellulose, whereas the bglAx knockout alone still produces a small amount of cellulose. It will be of interest to compare the transcriptomes of mutants II-23 and II-23OE and the wild type to determine whether there are any additional genes whose transcripts increase or decrease in response to the level of crp-fnr transcript.
In conclusion, from the characterization of five Cel− mutants, each having the Tn5 transposon inserted in a different gene, we found that (i) disruption of acsA in mutant 5 also affected the production of AcsB and AcsC but not the production of AcsD; (ii) disruption of acsC in mutant 10 resulted in the production of a truncated AcsC but did not affect the production of AcsAB or AcsD; (iii) disruption of dgc1 in mutant I-7 did not affect the production of any of the Acs proteins; (iv) disruption of ccpAx in mutant I-13 did not affect the production of AcsA or AcsD but resulted in substantial reduction in AcsB and AcsC levels; and (v) disruption of crp-fnr in mutant II-23 did not affect the production of any Acs protein or transcription of cmcAx, ccpAx, and dgc1 but resulted in a virtually undetectable level of BglAx.
Supplementary Material
ACKNOWLEDGMENTS
This material is based upon work supported as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under award DE-SC0001090.
We thank Candace Haigler for providing the cellulose-overproducing strain of G. hansenii ATCC 23769, Daniel Cosgrove for the use of an Olympus SZX12 microscope, Prashanti Iyer and Penglin Sun for valuable advice on protein blotting, and Prashanti Iyer and John McManus for help with purification of the antibodies used.
Footnotes
Published ahead of print 6 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00767-13.
REFERENCES
- 1.Römling U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205–212 [DOI] [PubMed] [Google Scholar]
- 2.Benziman M, Haigler CH, Brown RM, Jr, White AR, Cooper KM. 1980. Cellulose biogenesis: polymerization and crystallization are coupled processes in Acetobacter xylinum. Proc. Natl. Acad. Sci. U. S. A. 77:6678–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena IM, Kudlicka K, Okuda K, Brown RM., Jr 1994. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J. Bacteriol. 176:5735–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valla S, Ertesvåg H, Tonouchi N, Fjærvik E. 2009. Bacterial cellulose production: biosynthesis and applications, p 43–77 In Rehm BHA. (ed), Microbial production of biopolymers and polymer precursors: applications and perspectives. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 5.Kawano S, Tajima K, Uemori Y, Yamashta H, Erata T, Munekata M, Takai M. 2002. Cloning of cellulose synthesis related gene from Acetobacter xylinum ATCC 23769 and ATCC 53582: comparison of cellulose synthetic ability between strains. DNA Res. 9:149–156 [DOI] [PubMed] [Google Scholar]
- 6.Iyer PR, Liu YA, Deng Y, McManus JB, Kao TH, Tien M. 2013. Processing of cellulose synthase (AcsAB) from Gluconacetobacter hansenii ATCC 23769. Arch. Biochem. Biophys. 529:92–98 [DOI] [PubMed] [Google Scholar]
- 7.Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. 1996. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U. S. A. 93:12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxena IM, Brown RM, Jr, Dandekar T. 2001. Structure-function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry 57:1135–1148 [DOI] [PubMed] [Google Scholar]
- 9.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylate. Nature 325:279–281 [DOI] [PubMed] [Google Scholar]
- 10.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6 [DOI] [PubMed] [Google Scholar]
- 11.Hu SQ, Gao YG, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, Munekata M, Tanaka I, Yao M. 2010. Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc. Natl. Acad. Sci. U. S. A. 107:17957–17961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer PR, Catchmark JM, Brown NR, Tien M. 2011. Biochemical localization of a protein involved in synthesis of Gluconacetobacter hansenii cellulose synthesis. Cellulose 18:739–747 [Google Scholar]
- 13.Kawano S, Tajima K, Kono H, Erata T, Munekata M, Takai M. 2002. Effects of endogenous endo-β-1,4-glucanase on cellulose biosynthesis in Acetobacter xylinum ATCC 23769. J. Biosci. Bioeng. 94:275–281 [DOI] [PubMed] [Google Scholar]
- 14.Standal R, Iversen TG, Coucheron DH, Fjarvik E, Blatny JM, Valla S. 1994. A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. J. Bacteriol. 176:665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunagawa N, Fujiwara T, Yoda T, Kawano S, Satoh Y, Yao M, Tajima K, Dairi T. 2013. Cellulose complementing factor (Ccp) is a new member of the cellulose synthase complex (terminal complex) in Acetobacter xylinum. J. Biosci. Bioeng. 115:607–612 [DOI] [PubMed] [Google Scholar]
- 16.Koo HM, Song SH, Pyun YR, Kim YS. 1998. Evidence that a β-1,4-endoglucanase secreted by Acetobacter xylinum plays an essential role for the formation of cellulose fiber. Biosci. Biotechnol. Biochem. 62:2257–2259 [DOI] [PubMed] [Google Scholar]
- 17.Tonouchi N, Tahara N, Tsuchida T, Yoshinaga F, Beppu T, Horinouchi S. 1995. Addition of a small amount of endoglucanase enhances cellulose production by Acetobacter xylinum. Biosci. Biotechnol. Biochem. 59:805–808 [Google Scholar]
- 18.Tonouchi N, Tahara N, Kojima Y, Nakai T, Sakai F, Hayashi T, Tsuchida T, Yoshinaga F. 1997. A beta-glucosidase gene downstream of the cellulose synthase operon in cellulose-producing Acetobacter. Biosci. Biotechnol. Biochem. 61:1789–1790 [DOI] [PubMed] [Google Scholar]
- 19.Tajima K, Nakajima K, Yamashita H, Shiba T, Munekata M, Takai M. 2001. Cloning and sequencing of the beta-glucosidase gene from Acetobacter xylinum ATCC 23769. DNA Res. 8:263–269 [DOI] [PubMed] [Google Scholar]
- 20.Tahara N, Tonoucji N, Yano H, Yoshinaga F. 1998. Purification and characterization of exo-1,4-β-glucosidase from Acetobacter xylinum BPR2001. J. Ferment. Bioeng. 85:589–594 [Google Scholar]
- 21.Iyer PR, Geib SM, Catchmark JM, Kao TH, Tien M. 2010. Genome sequence of a cellulose-producing bacterium, Gluconacetobacter hansenii ATCC 23769. J. Bacteriol. 192:4256–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haigler CH. 1982. Alteration of cellulose assembly in Acetobacter xylinum by fluorescent brightening agents, direct dyes, and cellulose derivatives. Ph.D. dissertation University of North Carolina, Chapel Hill, NC [Google Scholar]
- 23.Hestrin S, Schramm M. 1954. Synthesis of cellulose by Acetobacter xylinum: preparation of freeze dried cells capable of polymerizing glucose to cellulose. Biochem. J. 58:345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall PE, Anderson SM, Johnston DM, Cannon RE. 1992. Transformation of Acetobacter xylinum with plasmid DNA by electroporation. Plasmid 28:194–200 [DOI] [PubMed] [Google Scholar]
- 25.Dower WJ, Miller JF, Ragsdale CW. 1988. High-efficiency transformation of Escherichia coli by high-voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 27.Robinson PA, Anderton BH, Loviny TLF. 1988. Nitrocellulose-bound antigen repeatedly used for the affinity purification of specific polyclonal antibodies for screening DNA expression libraries. J. Immunol. Methods 108:115–122 [DOI] [PubMed] [Google Scholar]
- 28.Kawano S, Tajima K, Kono H, Numata Y, Yamashita H, Satoh Y, Munekata M. 2008. Regulation of endoglucanase gene (cmcAx) expression in Acetobacter xylinum. J. Biosci. Bioeng. 106:88–94 [DOI] [PubMed] [Google Scholar]
- 29.Valla S, Kjosbakken J. 1982. Cellulose-negative mutants of Acetobacter xylinum. J. Gen. Microbiol. 128:1401–1408 [Google Scholar]
- 30.Steel R, Walker TK. 1957. A comparative study of cellulose-producing cultures and celluloseless mutants of certain Acetobacter spp. J. Gen. Microbiol. 17:445–452 [DOI] [PubMed] [Google Scholar]
- 31.Steel R, Walker TK. 1957. Celluloseless mutants of certain Acetobacter species. J. Gen. Microbiol. 17:12–18 [DOI] [PubMed] [Google Scholar]
- 32.Saxena IM, Lin FC, Brown RM., Jr 1991. Identification of a new gene in an operon for cellulose biosynthesis in Acetobacter xylinum. Plant Mol. Biol. 16:947–954 [DOI] [PubMed] [Google Scholar]
- 33.Nakai T, Moriya A, Tonouchi N, Tsuchida T, Yoshinaga F, Horinouchi S, Sone Y, Mori H, Sakai F, Hayashi T. 1998. Control of expression by the cellulose synthase (bcsA) promoter region from Acetobacter xylinum BPR 2001. Gene 213:93–100 [DOI] [PubMed] [Google Scholar]
- 34.Saxena IM, Borwn RM., Jr 1995. Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J. Bacteriol. 177:5276–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218–228 [DOI] [PubMed] [Google Scholar]
- 36.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bae SO, Sugano Y, Ohi K, Shoda M. 2004. Features of bacterial cellulose synthesis in a mutant generated by disruption of the diguanylate cyclase 1 gene of Acetobacter xylinum BPR 2001. Appl. Microbiol. Biotechnol. 65:315–322 [DOI] [PubMed] [Google Scholar]
- 38.Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. 2011. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82:327–341 [DOI] [PubMed] [Google Scholar]
- 39.Nakai T, Sugano Y, Shoda M, Sakakibara H, Oiwa K, Tuzi S, Imai T, Sugiyama J, Takeuchi M, Yamauchi D, Mineyuki Y. 2013. Formation of highly twisted ribbons in a carboxymethylcellulase gene-disrupted strain of a cellulose-producing bacterium. J. Bacteriol. 195:958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gusakov AV, Sinitsyn AP, Klyosov AA. 1985. Kinetics of the enzymatic hydrolysis of cellulose: 1. A mathematical model for a batch reactor process. Enzyme Microb. Technol. 7:346–352 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.