Abstract
Extraintestinal pathogenic Escherichia coli (ExPEC) is capable of colonizing outside of the intestinal tract and evolving into a systemic infection. Avian pathogenic E. coli (APEC) is a member of the ExPEC group and causes avian colibacillosis. Transfer-mRNA-small protein B (tmRNA-SmpB)-mediated trans-translation is a bacterial translational control system that directs the modification and degradation of proteins, the biosynthesis of which has stalled or has been interrupted, facilitating the rescue of ribosomes stalled at the 3′ ends of defective mRNAs that lack a stop codon. We found that disruption of one, or both, of the smpB or ssrA genes significantly decreased the virulence of the APEC strain E058, as assessed by chicken infection assays. Furthermore, the mutants were obviously attenuated in colonization and persistence assays. The results of quantitative real-time reverse transcription-PCR analysis indicated that the transcription levels of the transcriptional regulation gene rfaH and the virulence genes kpsM, chuA, and iss were significantly decreased compared to those of the wild-type strain. Macrophage infection assays showed that the mutant strains reduced the replication and/or survival ability in the macrophage HD11 cell line compared to that of the parent strain, E058. However, no significant differences were observed in ingestion by macrophages and in chicken serum resistance between the mutant and the wild-type strains. These data indicate that the tmRNA-SmpB system is important in the pathogenesis of APEC O2 strain E058.
INTRODUCTION
Escherichia coli is a diversified bacterial species and is probably one of the most widespread microbes on earth. E. coli, which is known to colonize the intestinal tract with no harmful effects or cause systemic infections in the host (1, 2), can be classified into three groups: nonpathogenic E. coli, intestinal pathogenic E. coli, and extraintestinal pathogenic E. coli (3). Avian pathogenic E. coli (APEC) strains cause one of the most significant extraintestinal infections (4), which take many forms and are collectively termed colibacillosis. Such strains are responsible for serious economic losses to the poultry industry (5).
E. coli has diverse serotypes, the most frequently observed in APEC being O1, O2, and O78 (6), although the order of prevalence varies in different countries and farms (7). APEC attach and colonize the respiratory tract and evolve into systemic infections. These generalized infections employ a variety of pathogenic mechanisms, which are categorized as adhesion, iron acquisition, hemolysis, and protection from bactericidal host factors, and toxin production (8, 9).
Quality control during protein synthesis is important for the maintenance of both the speed and fidelity of gene expression. Transfer-messenger RNA (tmRNA) is a small stable RNA molecule also known as SsrA or 10Sa RNA. In E. coli, the mature SsrA transcript consists of 363 nucleotides (10). The tmRNA acts as both a tRNA and an mRNA, which, in collaboration with small protein B (SmpB), plays a key role in protein quality control (11, 12). SmpB is a tmRNA-binding protein that is encoded immediately upstream of ssrA in E. coli (13). SsrA activity depends on four proteins: SmpB, elongation factor Tu (EF-Tu), elongation factor G (EF-G), and ribosomal protein S1 (12, 14–16).
The tmRNA-SmpB-mediated trans-translation is a bacterial translational control system that directs the modification and degradation of proteins, the biosynthesis of which has stalled or has been interrupted leading the rescue of ribosomes stalled at the 3′ ends of defective mRNAs lacking a stop codon, also known as “non-stop mRNAs” (11, 12, 17). smpB and ssrA genes have been found in almost all species in the bacterial kingdom examined to date (18–20).
E. coli has long been used as a living model for investigation of the tmRNA-SmpB system, and it has become increasingly clear that the rescue of ribosomes stalled at non-stop mRNAs occurs via three mechanisms. The classical tmRNA-SmpB system is considered to be the typical bacterial system; in addition, two additional backup systems that perform this function have been identified in E. coli (21–26).
Translational problems occur fairly frequently, and the tmRNA-SmpB system plays a key role in intracellular protein quality control; therefore, it is unsurprising that this system plays an important role in bacterial pathogenesis, although the underlying mechanism remains to be fully elucidated. Previous studies have shown that the SmpB-SsrA system plays a critical role in Salmonella pathogenesis through controlling the expression of virulence factors and improving the ability of this organism to survive within macrophages (27, 28). The deletion of ssrA significantly decreased the virulence of Salmonella enterica, as assessed in a mouse model (28). smpB-ssrA mutants of Yersinia pestis and Yersinia pseudotuberculosis exhibit abolished virulence compared to the wild-type (29, 30). Furthermore, the smpB-ssrA mutant of Y. pestis induced a strong antibody response in a mouse model and functions as a candidate live attenuated vaccine against pulmonary plague infection (30).
In the present study, we assessed the contribution of the tmRNA-SmpB system to the virulence of APEC O2 strain E058 by evaluating the pathogenicity of the single mutants E058ΔsmpB and E058ΔssrA, the double mutant E058ΔsmpBΔssrA, and their parental strain E058 both in vitro and in vivo. We demonstrated that mutant strains have significantly decreased levels of virulence. Consistent with these observations, we confirmed that the tmRNA-SmpB system affected the transcription of the antiterminator, RfaH, and several more important virulence factors in APEC.
MATERIALS AND METHODS
Bacterial strains, primers, and growth conditions.
The strains and plasmids used in the present study are listed in Table 1. The oligonucleotide primers are listed in Table S1 in the supplemental material. Bacteria were routinely cultured at 37 or 42°C in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl) or LB plates containing 1.5% agar. Ampicillin (60 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (30 μg/ml) were added to the media when appropriate.
Table 1.
Bacterial strains and plasmid constructions used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E058 | Wild-type avian E. coli serotype O2 | 70 |
| E058ΔsmpB | E058 ΔsmpB::kan | This study |
| E058ΔssrA | E058 ΔssrA::cat | This study |
| E058ΔsmpBΔssrA | E058 ΔsmpB ΔssrA | This study |
| ReE058ΔsmpBΔssrA | Complementation of E058ΔsmpBΔssrA | This study |
| DH5α | endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1Δ(lacIZYA-argF)U169 deoR [ϕ80dlacZΔ(lacZ)M15] | Invitrogen |
| Plasmids | ||
| pMD18-T Simple vector | TA cloning vector; Ampr | TaKaRa |
| pGEM-T Easy vector | TA cloning vector; Ampr | TaKaRa |
| pMD-T-smpB | smpB cloned into pMD18-T Simple vector | This study |
| pMD18-T-smpB::kan | Kanr gene inserted into pMD-T-smpB | This study |
| pMD18-T-ssrA | ssrA cloned into pMD18-T Simple vector | This study |
| pMD18-T-ssrA::cat | Cmr gene inserted into pMD-T-ssrA | This study |
| pGEM-T-Easy-BA | pGEM-T Easy carrying smpB and ssrA complete ORF and its native promoter | This study |
| pKD46 | Expresses λ Red recombinase | 31 |
| pUC4K | Kanr cassette | Invitrogen |
| pKD3 | Cmr cassette | 31 |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Nalr, nalidixic acid resistance; Kanr, kanamycin resistance.
Construction of deletion mutants.
The smpB and ssrA genes are located adjacent to each other in the genome of APEC strain E058 and separated by a sequence of 214 nucleotides. The ssrA gene nucleotide sequence exhibits complete homology with that of E. coli K-12 W3110 and E. coli K-12 MG1655. The smpB gene nucleotide sequence harbors 100% homology with that of APEC strain O1:K1:H7 (GenBank accession number NC_008563). Deletion of smpB and ssrA from the chromosome of APEC E058 was performed using allelic exchange based on the Lambda Red recombinase system (31, 32). A 146-bp fragment of smpB was substituted with the kanamycin resistance gene (kan) and half of the entire ssrA gene was replaced by the chloramphenicol (cat) cassette. Strain E058ΔsmpB was constructed as follows: the smpB fragment containing an introduced EcoRI site was amplified by PCR using the primers smpB-F and smpB-R (see Table S1 in the supplemental material) and cloned into the pMD18-T simple vector to form pMD-T-smpB. To insert the kan resistance cassette into the DNA segment, the reverse PCR product containing an introduced PstI site was amplified by PCR from pMD18-T-smpB using the primers smpB-R-F and smpB-R-R (see Table S1 in the supplemental material). To generate pMD18-T-smpB::kan, both the reverse PCR product and the pUC4K plasmid were digested with PstI and ligated. The same method was used to construct pMD18-T-ssrA and pMD18-T-ssrA::cat. The chloramphenicol (cat) resistance cassette was obtained from pKD3. The cat cassette was then introduced into the ssrA gene at the EcoRI and EcoRV sites. E058 was initially electroporated with pKD46 to express Red recombinase. We constructed the E058ΔsmpB::kan and E058ΔssrA::cat mutants and E058ΔsmpBΔssrA double mutants as described previously. All mutations were confirmed by DNA sequencing and reverse transcription-PCR (RT-PCR) analysis.
Complementation of E058ΔsmpBΔssrA.
To complement the mutant, we created a complementation plasmid, designated pGEM-T-Easy-BA. In this process, the entire smpB and ssrA genes and a 608-nucleotide region upstream of smpB, including its native putative promoter, was amplified and cloned into the pGEM-T Easy vector using the primers ReBA-F and ReBA-R (see Table S1 in the supplemental material). Incorporation of the DNA fragment into the plasmid was verified by PCR and DNA sequencing. The recombinant plasmid pGEM-T-Easy-BA was then purified and transformed into the mutant strain E058ΔsmpBΔssrA.
Chicken infection assays.
The virulence of APEC E058 and its mutants was assessed in vivo in a 1-day-old specific-pathogen-free (SPF) chickens (White Leghorn; Jinan SPAFAS Poultry Co., Ltd., Jinan, China) challenge model performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (approved by the State Council on 31 October 1988). Cultures of the wild-type strain and its mutants, as well as the complementation strain, were grown to logarithmic phase at 37°C. The bacteria were then collected, washed twice, and suspended in phosphate-buffered saline (PBS) prior to dilution to an appropriate optical density at 600 nm. Groups of 10 birds were challenged via the air sac with 0.1 ml of each culture suspension containing 107 CFU of bacteria. The number of bacteria contained in the inoculum was confirmed by culture of serial dilutions on LB agar plates. Five chickens were inoculated with PBS as a negative control. Survival among the infected chickens was monitored twice daily until 7 days postchallenge.
Bacterial colonization and persistence assays.
For colonization and persistence assays, a total of 15 5-week-old SPF chickens were challenged via the left thoracic air sac with the wild-type, mutant, and complementation strains. Each bird was inoculated with a bacterial suspension containing 108 CFU. Infected chickens were sacrificed at 24 h postinfection by CO2 asphyxiation. The cardiac blood, liver, spleen, lungs, and kidneys were individually and aseptically collected, weighed, and homogenized in 1 ml of PBS. Serial dilutions of homogenates were spread onto LB plates and incubated at 37°C for 18 h for CFU counting.
Bactericidal activity of SPF chicken serum assays.
Complement-sufficient SPF chicken serum was prepared and pooled. The bactericidal activity assay was performed in a 96-well plate. Cultures of the wild-type, mutant, and complementation strains were grown to log phase in LB medium at 37°C. Bacteria were then collected, washed twice, and suspended in PBS (pH 7.2). Each culture suspension (0.01 ml containing 106 CFU bacteria) was inoculated into the 96-well plate containing 190 μl of the 100% SPF chicken serum or heat-inactivated SPF chicken serum, and then incubated at 37°C for 1 h. Serial dilutions (1:10) of the contents of each well were plated onto LB agar plates and incubated for 24 h at 37°C for CFU counting. This assay was repeated at least three times on at least two separate occasions.
Macrophage ingestion and intracellular replication assays.
The avian macrophage cell line HD11 was cultured in Dulbecco modified Eagle medium (HyClone, USA) containing 4.0 mM l-glutamine and 4,500 mg of glucose/liter and supplemented with 10% fetal bovine serum (HyClone) in a humidified incubator with 5% CO2 at 41.5°C. HD11 cells were seeded into 24-well tissue culture plates (2 × 105 cells/well) 24 h prior to infection. Meanwhile, cultures of the wild-type, mutants, and complementation strains were grown to log phase in LB medium at 37°C. Bacteria were then collected, washed twice, and suspended in PBS prior to inoculation of HD11 cells at a multiplicity of infection of 100 (2 × 107 CFU/well). Plates were centrifuged for 10 min to facilitate interactions between the bacteria and macrophage cells. After centrifugation, the plates were incubated for 2 h to allow ingestion at 41.5°C in 5% CO2. To kill extracellular bacteria, HD11 cells were washed twice with PBS and then incubated for 1.5 h in the appropriate cell culture medium containing 100 μg of gentamicin/ml (time point, 0 h). The culture medium was collected, and the CFU represented the 0-h control. To liberate intracellular bacteria, monolayer cells were washed three times with PBS and lysed with 0.1% Triton X-100 in PBS. The lysates were collected in 1.5-ml tubes and vortexed. Serial dilutions of each sample were plated onto LB agar plates, and the resulting colonies were counted after 24 h of incubation at 37°C.
For determination of intracellular replication, HD-11 cells were washed three times with PBS, and fresh medium containing 10 μg of gentamicin/ml was added after incubation for 1.5 h in the appropriate cell culture medium containing 100 μg of gentamicin/ml. Samples were taken for the enumeration of intracellular bacteria and determination of intracellular replication at 2, 6, 12, and 24 h postinfection. Intracellular growth was expressed as the change (n-fold) in the bacterial number at a given time point relative to the internalized bacteria at 0 h postinfection.
Quantitative real-time PCR.
The wild-type E058 and E058ΔsmpBΔssrA strains were grown to log phase in LB medium at 37°C, collected, washed twice, and suspended in PBS. Total RNA was isolated using an RNAiso Plus kit (TaKaRa, Dalian, China) according to the manufacturer's recommendations, and genomic DNA was removed with gDNA Eraser at 42°C for 2 min. Then, 1 μg of RNA was used to synthesize cDNA using the PrimeScript RT reagent kit (TaKaRa) according to the manufacturer's instructions. Quantitative analysis of cDNAs was performed with a LightCycler instrument using SYBR green I DNA binding dye (Roche Applied Sciences, USA) to detect PCR products. The PCR mixture was prepared using a Fast Start Essential DNA Green Master kit (Roche) according to the manufacturer's instructions. Virulence genes, including the regulatory gene rfaH (33), the siderophore receptor gene iroN (34), the capsule gene kpsM (35), the ferric aerobactin uptake gene iutA (36), the hemin uptake gene chuA (37), and the serum survival gene iss (38) were investigated in both the wild-type and isogenic E058ΔsmpBΔssrA mutant strains by quantitative real-time PCR. Nonvirulence genes were amplified using gene-specific primers (see Table S1 in the supplemental material). These included mreD that encodes MreD, which is required to maintain the shape of rod-shaped bacteria (39), murC that encodes MurC as an ATP-dependent ligase involved in the biosynthesis of peptidoglycan (40), and ftsK, which encodes FtsK as an important cell division protein (41). The parameters for the amplification were as follows: initial preincubation at 95°C for 10 min, followed by 37 cycles each consisting of 15 s at 95°C, 10 s of annealing at 60°C, and 15 s of extension at 72°C. The parameters were determined by melting-curve analysis. A standard curve was generated from dilutions of the wild-type strain ranging from 10−2 to 10−7. The transcription levels of the target genes were calculated using the “2ΔCT” (treated − untreated) method relative to that of the gapA gene internal normalization control.
Statistical analysis.
The significance of differences between groups was analyzed using the Prism software program (GraphPad). The Mantel-Cox log rank test was utilized to analyze chicken survival rates and the Mann-Whitney test was used to analyze bacterial colonization in tissues.
RESULTS
The smpB-ssrA mutants were constructed correctly in APEC E058.
RT-PCR analysis showed that neither the smpB nor the ssrA genes were normally transcribed in the mutant strains, whereas the upstream and downstream genes were not influenced compared to the parental strain E058 (Fig. 1). Growth curve analysis revealed no significant differences between the wild-type, mutant, and complementation strains when cultured in LB broth at 37 and 42°C (data not shown).
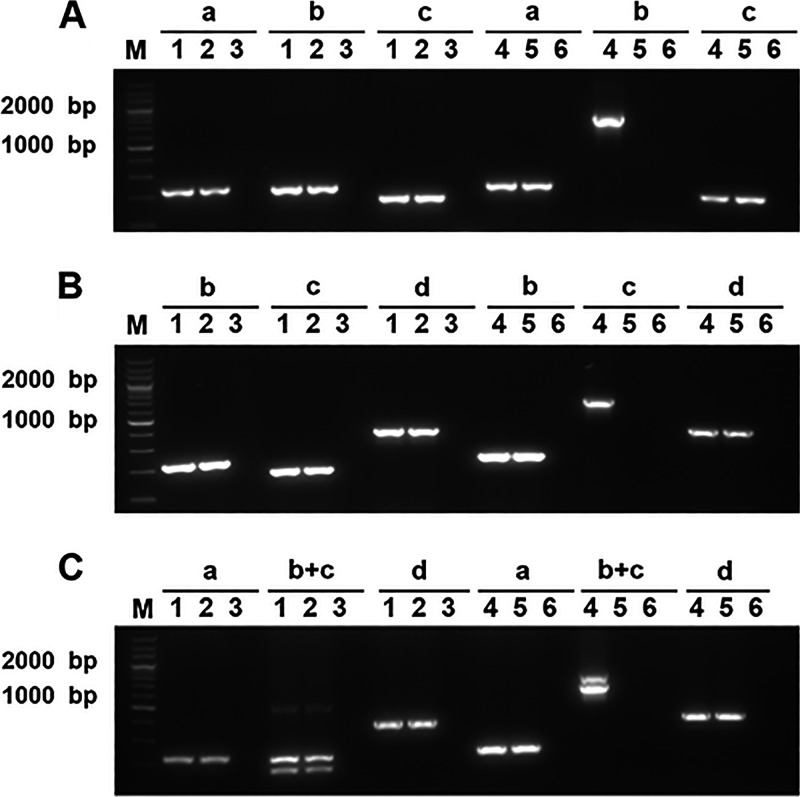
Fig 1.
RT-PCR analysis of transcription of smpB (A), ssrA (B), or both genes (C) and their upstream and downstream genes. Lanes: a, yfjG-F/yfjG-R; b, smpB-F/smpB-R; c, ssrA-F/ssrA-R; d, 3912-F/3912-R; b+c, smpB-F/smpB-R and ssrA-F/ssrA-R. Sample identities: lane 1, genomic DNA from E058 (A, B, and C); lane 2, cDNA derived from total RNA of E058 (A, B, and C); lane 3, total RNA from E058 without RT after genomic DNA was removed (A, B, and C); lane 4, genomic DNA from E058ΔsmpB (A), E058ΔssrA (B), or E058ΔsmpBΔssrA (C); lane 5, cDNA derived from total RNA of mutants E058ΔsmpB (A), E058ΔssrA (B), or E058ΔsmpBΔssrA (C); lane 6, total RNA from mutants E058ΔsmpB (A), E058ΔssrA (B), or E058ΔsmpBΔssrA (C) without RT after genomic DNA was removed. A 200-bp marker (TaKaRa) was used as the molecular size standard (lane M).
The E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants of APEC E058 are significantly attenuated.
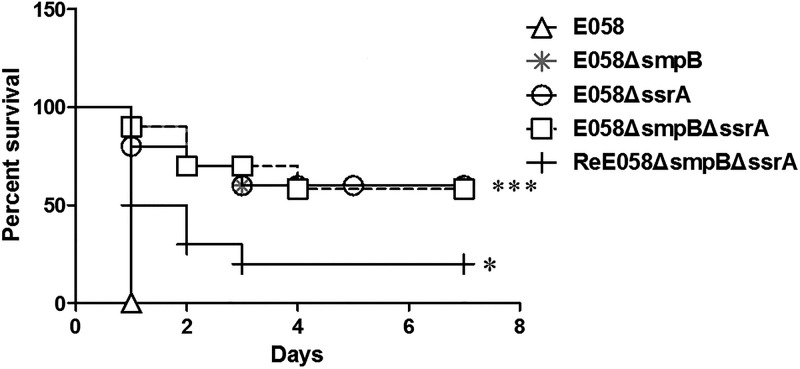
Birds infected with E058 exhibited 100% mortality within 1 day postchallenge (Fig. 2). In contrast, chickens infected with the E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants exhibited 40% mortality within 3 days postchallenge (P < 0.0001), demonstrating that the virulence of the mutants was significantly decreased compared to that of APEC E058. Chickens infected with the complementation strain exhibited symptoms of infection as early as 1 day postinfection, and the mortality reached 80% by 3 days postinfection. This indicated that the virulence phenotype of the complementation strain was restored to a significant level (P = 0.0118) (Fig. 2).
Fig 2.
Virulence of APEC E058 and its isogenic smpB and ssrA mutants in a chicken infection model. Birds (n = 10) were infected via the air sac with 107 CFU of the wild-type, mutant, and complementation strains. The survival of birds was monitored for 7 days postinfection. The data were analyzed by the Mantel-Cox log rank test (*, P < 0.02; ***, P < 0.0001).
smpB and ssrA were vital for colonization and persistence in vivo.
High levels of bacteria E058 were detected in all of the tissues after 24 h. In contrast, the levels of mutants detected in these tissues were low or undetectable. Chickens challenged with the wild-type strain E058 developed severe systemic infection while the mutants did not exhibit symptoms of disease. In addition, compared to the wild-type strain, the bacterial burden in the cardiac blood, liver, lungs, and kidneys of birds infected with the E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants was ∼105-fold lower than that of the parental E058 strain (Fig. 3A, B, D, and E), whereas that detected in the spleen was ∼104-fold lower (all P < 0.0001) (Fig. 3C). A complementation strain was generated to verify that the influence of the smpB and ssrA genes on virulence was direct and not a result of a secondary mutations. The results showed that similar numbers of colonies were recovered from the liver (Fig. 3B), spleen (Fig. 3C), and lungs (Fig. 3D) (P > 0.05) of birds inoculated with the ReE058ΔsmpBΔssrA complementation strain and the wild-type strain. However, the numbers of incomplete samples recovered from the cardiac blood and kidneys were significant (Fig. 3A and E) (P < 0.05).
Fig 3.
Tissue colonization and persistence by wild-type, mutant, and complementation strains. Fifteen 5-week-old SPF chickens were challenged with E058 (△), E058ΔsmpB (*), E058ΔssrA (○), E058ΔsmpBΔssrA (□), and ReE058ΔsmpBΔssrA (+) strains (108 CFU). Each data point represents a single sample from an individual bird, and data are presented as the log10 CFU/g of tissue. Horizontal bars indicate the mean log10 CFU/g values. The data were analyzed using a Mann-Whitney test (*, P < 0.05; ***, P < 0.0001).
smpB and ssrA were unnecessary for serum resistance of APEC E058.
E058, E058ΔsmpB, E058ΔssrA, E058ΔsmpBΔssrA, and ReE058ΔsmpBΔssrA strains all rapidly adapted to growth in the presence of chicken serum, and the number of cells almost increased by 1.6-fold compared to that of the initial inoculation (data not shown).
The E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants exhibited reduced replication ability in macrophage cells compared to E058.
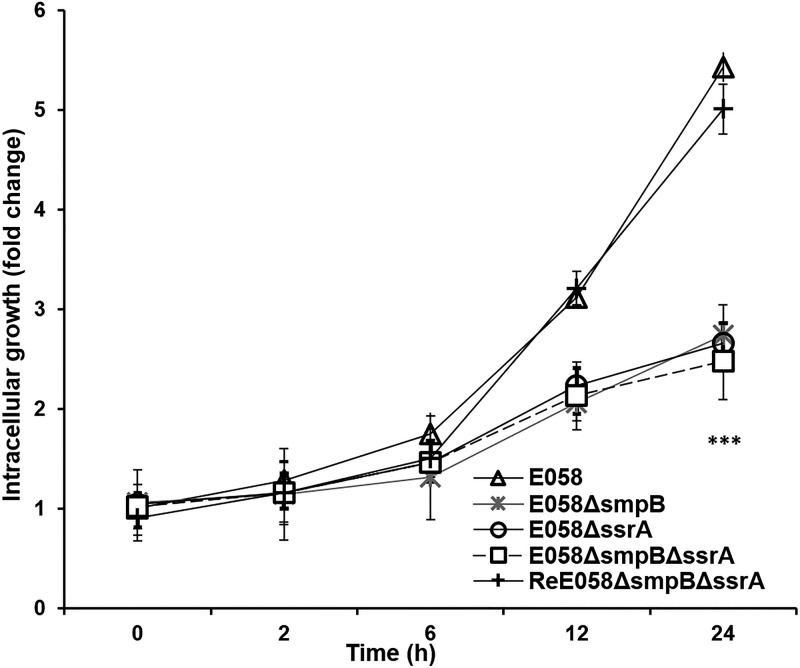
At 0 h, the control did not have bacteria in plates, which demonstrated that gentamicin can completely kill the extracellular bacteria. HD11 cells internalized almost equivalent numbers of E058 (1.58 × 104 CFU/well), E058ΔsmpB (1.69 × 104 CFU/well), E058ΔssrA (1.61 × 104 CFU/well), E058ΔsmpBΔssrA (1.65 × 104 CFU/well), and ReΔsmpBΔssrA (1.41 × 104 CFU/well) bacteria at 0 h of infection (Fig. 4).
Fig 4.
Survival and replication of E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants within the macrophage HD11 cell line. Intracellular bacterial growth is shown as the change compared to the primary internalized bacteria (0 h). The data were analyzed by unpaired t tests. Asterisks indicate statistically significant differences between the wild-type or complementation strain and the mutants (***, P < 0.0001). Each value represents the average of three independent experiments.
Subsequent enumeration of intracellular bacteria at 2, 6, 12, and 24 h postinfection revealed that wild-type bacteria adapted to growth in macrophages by 2 h postinfection, with the numbers reached to 1.28-, 1.75-, 3.12-, and 5.43-fold at 2, 6, 12, and 24 h, respectively, compared to that at 0 h. At these four time points, the numbers of E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutant cells increased slowly, reaching a 1.5-fold increase at 24 h compared to that at 0 h (Fig. 4).
The tmRNA-SmpB system affected the transcription of some virulence genes.
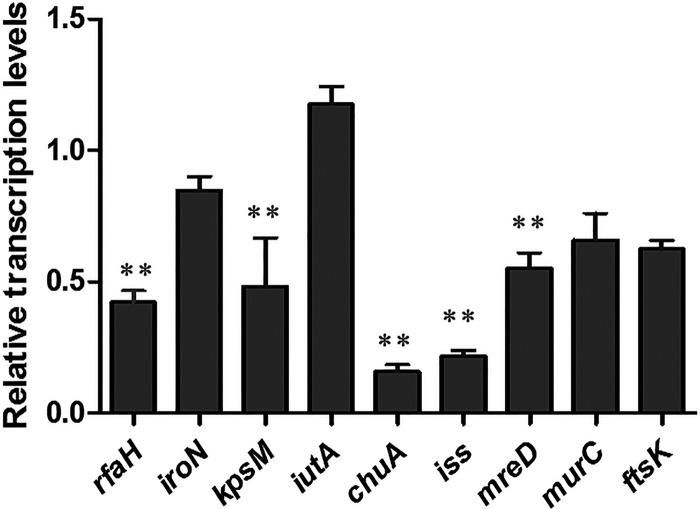
The transcription levels of rfaH, iroN, kpsM, iutA, chuA, iss, mreD, murC, and ftsK were quantified by quantitative RT-PCR (qRT-PCR). The transcription levels of rfaH, kpsM, chuA, iss, and mreD were significantly decreased in the E058ΔsmpBΔssrA mutant by 0.42, 0.48, 0.16, 0.22, and 0.55 times, respectively (P < 0.01). However, no significant differences were detected in the levels of iroN, iutA, murC, and ftsK between the two strains (Fig. 5).
Fig 5.
Quantitative RT-PCR analysis of the effects of smpB and ssrA gene mutations on the transcription of virulence genes. The relative transcription levels of rfaH, iroN, kpsM, iutA, chuA, iss, mreD, murC, and ftsK were determined by qRT-PCR. Error bars indicate the standard deviations of triplicate measurements. Asterisks indicate statistically significant differences (**, P < 0.01).
DISCUSSION
In this study, we demonstrated that the smpB and ssrA genes play an important role in the pathogenicity of avian pathogenic Escherichia coli. In a chicken challenge model, the E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants of APEC E058 were shown to be significantly attenuated. E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutant cells were defective in colonization and persistence in vivo and exhibited impaired replication ability in macrophage HD11 cells compared to the E058 parental strain. Furthermore, the smpB and ssrA genes reduced transcription of some genes.
The tmRNA-SmpB system plays a key role in intracellular protein quality control and also provides a mechanism by which aberrant mRNAs are cleared from stalled ribosomes. To date, three mechanisms have been demonstrated in Escherichia coli for the rescue of ribosomes stalled by non-stop mRNAs. The tmRNA-SmpB system is considered to be the classical mechanism. When trans-translation activity is limiting, ArfA, a small putative protein of the ydhL gene, functions as a backup system that binds non-stop translation complexes and recruits release factor 2 (RF-2) to hydrolyze the peptidyl-tRNA, thus rescuing the stalled ribosomes (21, 23, 42). The third mechanism involves the hydrolysis and release of the peptidyl-tRNA of ribosomes stalled at the 3′ ends of non-stop mRNAs by ArfB, a conserved hypothetical protein product of the yaeJ gene (25). This may explain why the ssrA gene is not essential for viability in E. coli under normal laboratory conditions, whereas it is essential in some bacterial species, which do not have alternative ribosome rescue mechanisms (43–46).
Interestingly, equivalent decreases in the virulence level of E058 were observed in colonization and persistence in vivo assays of both single- and double-deletion mutants (Fig. 3). Furthermore, although smpB is essential for the peptide-tagging activity of ssrA (12), no synergy in the effects of the deletions was observed in chickens infection assays (Fig. 2). This phenomenon remains to be explained.
E. coli has developed complex mechanisms to survive and proliferate inside the host (47–50). APEC must escape a multitude of host defense mechanisms in order to cause septicemia. Phagocytosis is one such mechanism, which is essential in guarding against, and disposing of, facultative intracellular pathogens such as Yersinia (51, 52). It is well documented that smpB-ssrA mutants of several strains are impaired in intracellular survival and/or replication within macrophages (27, 29). Our investigation revealed that, although equal numbers of wild-type and mutant bacteria were phagocytosed by HD11 macrophage cells, the E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants exhibited restricted ability to proliferate and/or survive within HD11 cells compared to the wild-type strain (Fig. 4), reaching a significant difference at 24 h. These results showed that the tmRNA-SmpB system influences the survival and/or replication in macrophages of APEC E058.
Serum complement and bactericidal effects represent another such mechanism. Indeed, serum resistance has been shown to be an important virulence trait of APEC (53). The increased serum survival gene iss has been shown to be associated with APEC complement resistance and pathogenicity (38, 54). Although smpB and ssrA mutations in most bacterial species may not be associated with growth defects under normal laboratory conditions, it is apparent that mutant cells become sensitive in their ability to adapt and survive in hostile environments. For example, destruction of the tmRNA-SmpB system renders cells more sensitive to translation-specific inhibitors, as well as oxidative and nitrosative stress (29, 55, 56). Our results showed that E058ΔsmpB, E058ΔssrA, and E058ΔsmpBΔssrA mutants can rapidly adapt to SPF chicken serum and have no significant bactericidal effects (data not shown), whereas the transcription level of iss in the E058ΔsmpBΔssrA mutant was downregulated compared to that of the wild-type strain (Fig. 5). This result suggests that other genes contribute to APEC serum resistance and that the presence of multiple alternative mechanisms mediate APEC pathogenicity.
Additional functions of the tmRNA-SmpB system in bacterial pathogenesis can be illustrated by alteration in the expression of virulence genes. smpB-ssrA mutants of Salmonella are reported to be ∼200-fold less virulent than the wild type due to the deregulated expression of several genes (28). Bacterial gene expression is frequently regulated at the level of transcription. A number of recent reports have suggested that the tmRNA-SmpB system plays a regulatory role through regulatory factors (57). These factors might include transcriptional activators and repressors (20, 29, 56, 58–60). Activators and repressors exert their control at the level of transcriptional initiation (58). Transcript elongation is controlled by intricate termination/antitermination mechanisms (61). RfaH is a bacterial virulence regulator that functions as a transcriptional antiterminator (33, 62–65). Our qRT-PCR results observed that the E058ΔsmpBΔssrA mutant had significantly reduced levels of rfaH mRNA (Fig. 5). One possible explanation for the reduced synthesis of the E058ΔsmpBΔssrA strain was the lack of requisite intracellular concentrations of essential regulatory factors, thereby reducing the ability of the regulator to influence gene expression.
We then evaluated the expression of several important genes associated with virulence in extraintestinal pathogenic E. coli that are clustered into long operons and regulated by RfaH, comprising the capsule gene kpsM, the ferric aerobactin uptake gene iutA, and the hemin uptake gene chuA. We found that the E058ΔsmpBΔssrA mutant had significantly reduced levels of kpsM and chuA mRNA transcripts. However, the level of iutA was not significantly affected compared to the wild-type strain. We found that chuA and kpsM were regulated by rfaH in APEC (33). Based on the results of the present study, we inferred that the RfaH might affect the expression of kpsM and chuA in the E058ΔsmpBΔssrA mutant. For genes that were not related to virulence, we found that the transcription levels of mreD were significantly decreased, but murC and ftsK were not affected by smpB and ssrA (Fig. 5). These results showed that the tmRNA-SmpB system influences the transcription not only of virulence genes but also other genes not related to the virulence of APEC E058.
To date, the pathway by which the tmRNA-SmpB system directly or indirectly affects the expression of some genes is unknown. It can be speculated that the tmRNA-SmpB system selectively regulates the expression of some genes (28, 29). The possible model is that the tmRNA-SmpB system exerts its effect through tagging a key factor for direct degradation (29, 56, 59), thereby controlling its intracellular concentrations. If these functions are insufficient in an E058ΔsmpBΔssrA strain, the subsequent imbalance results in downstream effects that directly or indirectly affect the expression of other genes.
The activity of tmRNA has been examined in only a few pathogens thus far; however, in E. coli, ca. 2 to 4% of all protein chains are tagged by the tmRNA system (66), indicating that the tmRNA-SmpB system plays an important and general role in bacterial pathogenesis (67). In bacteria, the tmRNA quality control system is the primary mediator of ribosome rescue and thus is critical for several aspects of bacterial fitness, including growth and development, stress responses, and pathogenesis (29, 68, 69). Further investigations are required to fully elucidate the mechanism by which the tmRNA-SmpB system influences the pathogenesis of APEC.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Xin'an Jiao of the Jiangsu Key Laboratory of Zoonosis of the Yangzhou University for kindly providing the cell line HD 11.
This study was supported by grants from the National Natural Science Foundation of China (31272559, 30972196, 30771604, and 30471281), the National Program for High Technology Research and Development in China (2003 AA 222141), the Special Fund for Agroscientific Research in the Public Interest (201303044 to S.G.), the Natural Science Foundation of China (NSFC 31101821 to H.H.), the Program for Changjiang Scholars and Innovative Research Teams in Universities (PCSIRT0978), and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Published ahead of print 6 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00628-13.
REFERENCES
- 1.Dozois CM, Curtiss R., III 1999. Pathogenic diversity of Escherichia coli and the emergence of ‘exotic' islands in the gene stream. Vet. Res. 30:157–179 [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 3.Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753–1754 [DOI] [PubMed] [Google Scholar]
- 4.Dho-Moulin M, Fairbrother JM. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299–316 [PubMed] [Google Scholar]
- 5.Lutful Kabir SM. 2010. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int. J. Environ. Res. Public Health 7:89–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orskov F, Orskov I. 1992. Escherichia coli serotyping and disease in man and animals. Can. J. Microbiol. 38:699–704 [PubMed] [Google Scholar]
- 7.Cortes P, Blanc V, Mora A, Dahbi G, Blanco JE, Blanco M, Lopez C, Andreu A, Navarro F, Alonso MP, Bou G, Blanco J, Llagostera M. 2010. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76:2799–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, Gao S, Huan H, Xu X, Zhu X, Yang W, Gao Q, Liu X. 2009. Comparison of virulence factors and expression of specific genes between uropathogenic Escherichia coli and avian pathogenic E. coli in a murine urinary tract infection model and a chicken challenge model. Microbiology 155:1634–1644 [DOI] [PubMed] [Google Scholar]
- 10.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 91:9223–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993 [DOI] [PubMed] [Google Scholar]
- 12.Karzai AW, Susskind MM, Sauer RT. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18:3793–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore SD, Sauer RT. 2005. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 58:456–466 [DOI] [PubMed] [Google Scholar]
- 14.Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, Shpanchenko O, Nierhaus KH, Spahn CM. 2012. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature 485:526–529 [DOI] [PubMed] [Google Scholar]
- 15.Rudinger-Thirion J, Giege R, Felden B. 1999. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA 5:989–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wower IK, Zwieb CW, Guven SA, Wower J. 2000. Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J. 19:6612–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore SD, Sauer RT. 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76:101–124 [DOI] [PubMed] [Google Scholar]
- 18.Gueneau de Novoa P, Williams KP. 2004. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 32:D104–D108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madrid C, Nieto JM, Juarez A. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med. Microbiol. 291:425–432 [DOI] [PubMed] [Google Scholar]
- 20.Withey JH, Friedman DI. 2002. The biological roles of trans-translation. Curr. Opin. Microbiol. 5:154–159 [DOI] [PubMed] [Google Scholar]
- 21.Chadani Y, Ito K, Kutsukake K, Abo T. 2012. ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli. Mol. Microbiol. 86:37–50 [DOI] [PubMed] [Google Scholar]
- 22.Garza-Sanchez F, Schaub RE, Janssen BD, Hayes CS. 2011. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol. Microbiol. 80:1204–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaub RE, Poole SJ, Garza-Sanchez F, Benbow S, Hayes CS. 2012. Proteobacterial ArfA peptides are synthesized from non-stop messenger RNAs. J. Biol. Chem. 287:29765–29775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chadani Y, Ono K, Kutsukake K, Abo T. 2011. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA-and ArfA-mediated pathways. Mol. Microbiol. 80:772–785 [DOI] [PubMed] [Google Scholar]
- 25.Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. 2012. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335:1370–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handa Y, Inaho N, Nameki N. 2011. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 39:1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. 1994. Salmonella typhimurium loci involved in survival within macrophages. Infect. Immun. 62:1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julio SM, Heithoff DM, Mahan MJ. 2000. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J. Bacteriol. 182:1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okan NA, Bliska JB, Karzai AW. 2006. A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2:e6. 10.1371/journal.ppat.0020006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okan NA, Mena P, Benach JL, Bliska JB, Karzai AW. 2010. The smpB-ssrA mutant of Yersinia pestis functions as a live attenuated vaccine to protect mice against pulmonary plague infection. Infect. Immun. 78:1284–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong L, Ling J, Gao Q, Zhou Y, Li T, Gao S, Liu X. 2012. Construction of iucB and iucB-iutA mutants of avian pathogenic Escherichia coli and evaluation of their pathogenicity. Vet. Microbiol. 159:420–431 [DOI] [PubMed] [Google Scholar]
- 33.Gao Q, Xu H, Wang X, Zhang D, Ye Z, Gao S, Liu X. 2013. RfaH promotes the ability of the avian pathogenic Escherichia coli O2 strain E058 to cause avian colibacillosis. J. Bacteriol. 195:2474–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bister B, Bischoff D, Nicholson GJ, Valdebenito M, Schneider K, Winkelmann G, Hantke K, Sussmuth RD. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17:471–481 [DOI] [PubMed] [Google Scholar]
- 35.Boyd EF, Hartl DL. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skyberg JA, Johnson TJ, Nolan LK. 2008. Mutational and transcriptional analyses of an avian pathogenic Escherichia coli ColV plasmid. BMC Microbiol. 8:24. 10.1186/1471-2180-8-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stojiljkovic I, Perkins-Balding D. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281–295 [DOI] [PubMed] [Google Scholar]
- 38.Nolan LK, Giddings CW, Horne SM, Doetkott C, Gibbs PS, Wooley RE, Foley SL. 2002. Complement resistance, as determined by viable count and flow cytometric methods, and its association with the presence of iss and the virulence of avian Escherichia coli. Avian Dis. 46:386–392 [DOI] [PubMed] [Google Scholar]
- 39.Vats P, Shih YL, Rothfield L. 2009. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol. Microbiol. 72:170–182 [DOI] [PubMed] [Google Scholar]
- 40.Emanuele JJ, Jr, Jin H, Jacobson BL, Chang CY, Einspahr HM, Villafranca JJ. 1996. Kinetic and crystallographic studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Protein Sci. 5:2566–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F. 2008. Asymmetry of chromosome replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet. 4:e1000288. 10.1371/journal.pgen.1000288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu Y. 2012. ArfA recruits RF2 into stalled ribosomes. J. Mol. Biol. 423:624–631 [DOI] [PubMed] [Google Scholar]
- 43.Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U. S. A. 99:966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. 69:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thibonnier M, Thiberge JM, De Reuse H. 2008. Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS One 3:e3810. 10.1371/journal.pone.0003810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramadoss NS, Zhou X, Keiler KC. 2013. tmRNA is essential in Shigella flexneri. PLoS One 8:e57537. 10.1371/journal.pone.0057537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Q, Wang X, Xu H, Xu Y, Ling J, Zhang D, Gao S, Liu X. 2012. Roles of iron acquisition systems in virulence of extraintestinal pathogenic Escherichia coli: salmochelin and aerobactin contribute more to virulence than heme in a chicken infection model. BMC Microbiol. 12:143. 10.1186/1471-2180-12-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fetherston JD, Kirillina O, Bobrov AG, Paulley JT, Perry RD. 2010. The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun. 78:2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caza M, Lepine F, Dozois CM. 2011. Secretion, but not overall synthesis, of catecholate siderophores contributes to virulence of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 80:266–282 [DOI] [PubMed] [Google Scholar]
- 50.Kwon HJ, Seong WJ, Kim JH. 2013. Molecular prophage typing of avian pathogenic Escherichia coli. Vet. Microbiol. 162:785–792 [DOI] [PubMed] [Google Scholar]
- 51.Brubaker RR. 2003. Interleukin-10 and inhibition of innate immunity to Yersiniae: roles of Yops and LcrV (V antigen). Infect. Immun. 71:3673–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heesemann J, Sing A, Trulzsch K. 2006. Yersinia's stratagem: targeting innate and adaptive immune defense. Curr. Opin. Microbiol. 9:55–61 [DOI] [PubMed] [Google Scholar]
- 53.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, III, Brown PK, Arne P, Bree A, Desautels C, Fairbrother JM. 2003. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71:536–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfaff-McDonough SJ, Horne SM, Giddings CW, Ebert JO, Doetkott C, Smith MH, Nolan LK. 2000. Complement resistance-related traits among Escherichia coli isolates from apparently healthy birds and birds with colibacillosis. Avian Dis. 44:23–33 [PubMed] [Google Scholar]
- 55.de la Cruz J, Vioque A. 2001. Increased sensitivity to protein synthesis inhibitors in cells lacking tmRNA. RNA 7:1708–1716 [PMC free article] [PubMed] [Google Scholar]
- 56.Karzai AW, Roche ED, Sauer RT. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449–455 [DOI] [PubMed] [Google Scholar]
- 57.Keiler KC, Ramadoss NS. 2011. Bifunctional transfer-messenger RNA. Biochimie 93:1993–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karmakar R. 2010. Conversion of graded to binary response in an activator-repressor system. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 81:021905. [DOI] [PubMed] [Google Scholar]
- 59.Abo T, Inada T, Ogawa K, Aiba H. 2000. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J. 19:3762–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranquet C, Geiselmann J, Toussaint A. 2001. The tRNA function of SsrA contributes to controlling repression of bacteriophage Mu prophage. Proc. Natl. Acad. Sci. U. S. A. 98:10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henkin TM. 2000. Transcription termination control in bacteria. Curr. Opin. Microbiol. 3:149–153 [DOI] [PubMed] [Google Scholar]
- 62.Artsimovitch I, Landick R. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193–203 [DOI] [PubMed] [Google Scholar]
- 63.Bailey MJ, Hughes C, Koronakis V. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845–851 [DOI] [PubMed] [Google Scholar]
- 64.Belogurov GA, Sevostyanova A, Svetlov V, Artsimovitch I. 2010. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol. Microbiol. 76:286–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagy G, Dobrindt U, Schneider G, Khan AS, Hacker J, Emody L. 2002. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect. Immun. 70:4406–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito K, Chadani Y, Nakamori K, Chiba S, Akiyama Y, Abo T. 2011. Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS One 6:e28413. 10.1371/journal.pone.0028413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. 2009. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One 4:e4809. 10.1371/journal.pone.0004809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang C, Glover JR. 2009. The SmpB-tmRNA tagging system plays important roles in Streptomyces coelicolor growth and development. PLoS One 4:e4459. 10.1371/journal.pone.0004459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ranquet C, Gottesman S. 2007. Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J. Bacteriol. 189:4872–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao S, Liu X, Zhang R, Zhang R, Jiao X, Wen Q, Wu C, Tang Y, Zhu X, Li C, Chen J, Cui H. 1999. The isolation and identification of pathogenic Escherichia coli isolates of chicken origin from some regions in China. Acta Vet. Zootechnol. Sin. 30:164–171 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.