Abstract
SUMMARY
Cyanobacteria are the globally dominant photoautotrophic lineage. Their success is dependent on a set of adaptations collectively termed the CO2-concentrating mechanism (CCM). The purpose of the CCM is to support effective CO2 fixation by enhancing the chemical conditions in the vicinity of the primary CO2-fixing enzyme, d-ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO), to promote the carboxylase reaction and suppress the oxygenase reaction. In cyanobacteria and some proteobacteria, this is achieved by encapsulation of RubisCO within carboxysomes, which are examples of a group of proteinaceous bodies called bacterial microcompartments. Carboxysomes encapsulate the CO2-fixing enzyme within the selectively permeable protein shell and simultaneously encapsulate a carbonic anhydrase enzyme for CO2 supply from a cytoplasmic bicarbonate pool. These bodies appear to have arisen twice and undergone a process of convergent evolution. While the gross structures of all known carboxysomes are ostensibly very similar, with shared gross features such as a selectively permeable shell layer, each type of carboxysome encapsulates a phyletically distinct form of RubisCO enzyme. Furthermore, the specific proteins forming structures such as the protein shell or the inner RubisCO matrix are not identical between carboxysome types. Each type has evolutionarily distinct forms of the same proteins, as well as proteins that are entirely unrelated to one another. In light of recent developments in the study of carboxysome structure and function, we present this review to summarize the knowledge of the structure and function of both types of carboxysome. We also endeavor to cast light on differing evolutionary trajectories which may have led to the differences observed in extant carboxysomes.
INTRODUCTION
Carboxysomes are specialized protein microcompartments composed of a polyhedral protein shell within which cyanobacteria concentrate CO2 around their primary carboxylating enzyme, d-ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO). All photosynthetically competent cyanobacteria, and some autotrophic bacteria, possess carboxysomes. These structures form part of what is known as the CO2-concentrating mechanism (CCM), which operates together with active CO2- and HCO3−-uptake transporters which accumulate HCO3− in the cytoplasm of the cell. Cyanobacteria (also known as blue-green algae) are predominantly aquatic organisms and are remarkably productive on a global scale, especially those from deep-sea niches. This productivity would not be possible without the CCM, which is dependent on carboxysomes. Some autotrophic bacteria are also conditionally dependent on carboxysomes and may have been the origin of some of the smaller shell proteins during evolution or may have obtained them from more distantly related bacterial microcompartments (BMC) used for unrelated biochemistry. Remarkably, two types of carboxysomes have arisen, possibly by convergent evolution, with similar designs and functions but different protein makeups. The types of carboxysomes are α-carboxysomes, found predominantly in oceanic cyanobacteria (α-cyanobacteria), and β-carboxysomes, found mainly in freshwater/estuarine cyanobacteria (β-cyanobacteria), based on their RubisCO phylogeny. Considerable progress has now been made toward understanding the structure, physiology, and evolution of carboxysomes, and these subjects are explored in this review.
Evolutionary Pressures Due to RubisCO and Altered Atmospheric Conditions
Cyanobacteria can be traced back at least 2.4 billion years ago (Gya), and possibly as far back as 3.5 Gya, but the current thinking is that they did not always have carboxysomes or a CCM. Although an ancestral cyanobacterial endosymbiont gave rise to chloroplasts in algae and plants, the evolution of the CCM appears to postdate this pivotal event. In fact, the cyanobacterial CCM, with fully functional carboxysomes, may not have evolved fully until as late as about 350 million years ago (1–3), although the adoption of carboxysomes may have predated this point to some extent (2). The inefficiencies of the RubisCO enzyme have been the driving force for these changes. As oxygen has risen, since the advent of oxygenic photosynthesis by cyanobacteria, and CO2 levels have fallen, evolutionary pressure has driven either the development of more efficient forms of RubisCO or the development of various CCMs (4). The inefficiencies of RubisCO include a low affinity for CO2, a low catalytic rate, and the ability of O2 to act as an alternative substrate. The advent of a carboxysome-based CCM in cyanobacteria ameliorated these problems with RubisCO and resulted in a high catalytic rate for carboxylation and a low oxygenation rate in vivo. As the full name of the enzyme conveys, RubisCO has the unfortunate ability to undertake the wasteful fixation of O2 into phosphoglycolate. This product is toxic and must be removed quickly from the cell or recycled via an energy-requiring process known as photorespiration (5, 6). The ability of O2 to react as a secondary substrate of RubisCO under the appropriate conditions is largely because CO2 and O2, as enzyme substrates, are remarkably similar in molecular size and lack of charge (7, 8).
When cyanobacteria first evolved, up to 3.5 Gya, the oxygenase activity of RubisCO was irrelevant, largely because early atmospheric conditions were very different, featuring high CO2 levels (up to 50-fold higher than present levels) and almost nonexistent O2 levels (9). These conditions presumably meant that RubisCO operated efficiently in its primary role as a carboxylase, fixing CO2 into phosphoglycerate (PGA), the first 3-carbon sugar product of the Calvin cycle. Paradoxically, it was the early cyanobacteria that gradually decreased CO2 and elevated O2 in the atmosphere, due to their ability to perform oxygenic photosynthesis. Initially, most O2 produced by cyanobacteria was removed by vast amounts of marine reductant, such as Fe2+, such that atmospheric changes were minimal. Approximately 1.5 Gya, O2 levels rose dramatically, to levels similar to today's atmospheric level (∼21%), and CO2 levels dropped, although they were still well above today's level of 0.04% (9, 10). This change in atmospheric conditions would have exposed RubisCO to limiting levels of CO2 and inhibitory levels of O2 and created an increase of evolutionary pressures to evolve better kinetic properties of RubisCO or different types of CCMs. These pressures were more acute in aquatic phototrophs because of the slow diffusion of CO2 in water (some 10,000 times slower than in air) and other factors, such as pH, poor mixing, and temperature, that also affect CO2 availability.
In the algal lineages that finally led to terrestrial plants known as C3 plants, the RubisCO enzymes evolved an increased affinity for CO2 and a better selectivity between CO2 and O2, but the trade-off appears to have resulted in a reduction in the catalytic turnover rate per enzyme (8, 11). This evolutionary response worked, but nevertheless, most C3 plants still deal with rates of photorespiration of some 30% of the maximum potential photosynthetic rate, and as much as 25% of leaf nitrogen is allocated to this relatively slow enzyme (5, 6). Some land plants, such as the C4 plants, eventually evolved several types of anatomically and biochemically compartmentalized CCMs that function to elevate CO2 around RubisCO, thus overcoming its inefficiencies (12).
However, in cyanobacteria and some eukaryotic microalgae, diatoms, and coccoliths, evolutionary pressure on RubisCO resulted in the evolution of other types of CCMs that are single cell based and feature active accumulation of HCO3− before delivering elevated CO2 levels to RubisCO (1, 2, 4, 10, 13, 14). While some lineages, such as the green algae, evolved RubisCO enzymes with a higher specificity for CO2 and more moderate abilities to accumulate HCO3−, cyanobacteria and some proteobacteria were able to maintain a RubisCO form that may resemble the form that was present in cyanobacteria >2.4 Gya. Cyanobacteria have retained a form 1 RubisCO with a high carboxylation rate (typical carboxylation reaction kcat [kcatc] of 12 to 13 s−1), a low affinity for CO2 as a substrate [typical Km(CO2) of 250 to 330 μM], and a poor selectivity for CO2 over O2 (typical SC/O of 43 to 53) (4, 8). The latter two attributes are hardly a problem if the organism possesses a potent CCM, and accordingly, cyanobacteria have one that is able to reach inorganic carbon (Ci) accumulation factors of up to 1,000-fold over the level in bulk medium (4, 14, 15). Among the few alpha-, beta-, and gammaproteobacteria that possess carboxysomes, it is inferred from what is known about their RubisCO kinetics (e.g., a low affinity for CO2), such as those of Halothiobacillus species (16, 17), that they also feature high HCO3− accumulation factors and a CCM. One of the few studies of carboxysome-containing bacteria, which examined the deep-sea vent bacterium Thiomicrospira crunogena, showed that a 100-fold internal overaccumulation can be achieved with moderately high affinities for CO2 and HCO3− as substrates (18).
There are two key parts of the CCM in cyanobacteria and those bacteria that possess carboxysomes. First, it is essential to transport CO2 and/or HCO3− as a substrate, enabling accumulation of the relatively membrane-impermeative HCO3− species in the cytoplasm. To prevent rapid leakage of accumulated HCO3− from the cytoplasm, the enzyme carbonic anhydrase (CA) must be absent from this compartment (19, 20) to restrict CO2 production (though this may not necessarily be true of some structurally complex cyanobacteria, such as Chlorogloeopsis fritschii and Anabaena variabilis, whose cells contain some cytoplasmic CA activity [21, 22]). Nonetheless, the accumulated HCO3− needs to be converted to CO2 and utilized by RubisCO in carboxysome microcompartments where CO2 is elevated by localized production of CO2 (1, 2, 4, 14, 23–26). The requirement of carboxysomes is known to be essential for proper CCM function, largely as a result of the analysis of a large number of characterized mutants in which carboxysome functionality is destroyed (see Table 2), leading to an inability to elevate CO2 levels in the carboxysome, even though the cells are still able to hyperaccumulate HCO3− in the cytosol (see below). In a functional carboxysome, RubisCO is substrate saturated for CO2, and the oxygenase reaction is largely eliminated under optimal conditions (i.e., sufficient CO2/HCO3− availability and light). However, it is now known that cyanobacteria possess three interconnected pathways for metabolizing phosphoglycolate (27), and thus it is likely that cyanobacteria do, from time to time, encounter photorespiratory conditions which allow some RubisCO oxygenase activity, such as with dim light (morning and evening), low environmental CO2/HCO3− levels, or acclimation to limiting CO2 levels. Photorespiration is not generally a concern when the carboxysomal CO2 level is very low or when cells are in darkness, as cyanobacterial RubisCO has a low affinity for O2, is rapidly inactivated under such conditions, and does not reactivate until sufficient CO2, Mg2+, and light are available (28, 29).
Table 2.
Carboxysome morphologies in α- and β-carboxysome mutantsa
| Carboxysome morphology | Mutant gene [reference(s)] |
|
|---|---|---|
| α-Carboxysomes | β-Carboxysomes | |
| Wild type | csoS4ABb (92) | ccmK3 (159) |
| csoS1A (177) | ccmK4 (159) | |
| csoS3 (91) | ccmK3-ccmK4 (159) | |
| csoS1Dc (122) | ccaA (73, 94, 174, 246) | |
| Elongated (vertex protein deficit) | csoS4ABb (92) | ccmL (93, 94, 140) |
| Polar body (shell recruitment deficit) | ccmK2 (140, 159) | |
| ccmO (159) | ||
| ccmN (143, 150, 151) | ||
| rbcSd (153) | ||
| Aggregated (carboxysome localization deficit) | ccmK3K4 (159) | |
| parA (181) | ||
| mreB (181) | ||
| None | ccmMe (97, 140–142, 151) | |
| ccmO (163) | ||
| rbcLSf (176) | ||
| Empty shell | rbcL (121) | |
The α-carboxysome mutants were largely generated in H. neapolitanus, and the β-carboxysome mutants in S. elongatus PCC 7942, though Synechocystis PCC 6803 and Synechococcus PCC 7002 mutants are included in this table.
Most carboxysomes in Halothiobacillus csoS4AB::Kanr are ultrastructurally normal, but the proportion that are elongated exceeds that of the wild type (92).
When the H. neapolitanus cso operon was expressed without csoS1D, the resulting α-carboxysomes were slightly aberrant, though essentially wild type (122).
Extended rbcS reading frame mutant (153).
Mutant 28 (151) is probably an insertional mutant of ccmM.
The cyanorubrum mutant replaced the native rbcLS genes with the type II RubisCO rbcM gene, abolishing carboxysomes in Synechocystis PCC 6803 (176).
The presence of a functional CCM is obligate for the survival of cyanobacteria living in most natural habitats, although less so for carboxysome-containing bacteria, but the advantages are considerable. Being able to operate RubisCO at substrate saturation results in a lower allocation of cellular nitrogen, to as low as 3 to 5% of total cellular nitrogen in the case of cyanobacteria (19, 30), whereas C3 plants (no CCM) allocate up to 25% of cellular nitrogen to RubisCO (31). Combined with the aforementioned highly reduced rates of oxygenase activity and photorespiration, cyanobacterial cells possess elements of high photosynthetic efficiency and environmental competiveness. Hence, oceanic cyanobacteria and other phytoplankton contribute almost 50% of global primary productivity (as kg C fixed per m2 per year) on an annual basis (32), with the oceanic α-cyanobacteria being one of the most productive groups of organisms, contributing as much as half of this productivity (33, 34), suggesting that >25% of all global CO2 fixation occurs within carboxysomes.
BASIC FEATURES OF THE CCM
Cyanobacterial Ci Uptake
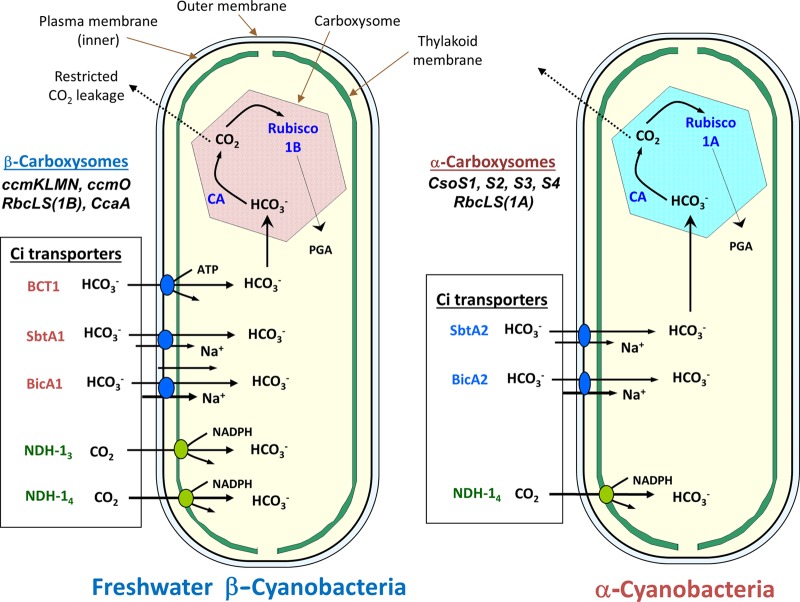
The two most biologically relevant species of dissolved inorganic carbon (Ci) for photosynthesis in aquatic environments are dissolved CO2 and HCO3−, with HCO3− being the more abundant species above a pH of approximately 6.5. However, both species serve as substrates for as many as three identified HCO3− transporters and two CO2-uptake systems (CO2-to-HCO3− converters) (Fig. 1). Physiological information linking genes to proteins for CO2- and HCO3−-uptake systems is most complete for a few model strains of the β-cyanobacteria, while for the α-cyanobacteria and bacteria that possess carboxysomes, the linkage is fragmentary. However, the availability of >120 completely sequenced cyanobacterial genomes and thousands of bacterial genomes does make it possible to track the presence of the known Ci transporter gene homologs throughout the prokaryotes (1, 2, 35, 36). Not all cyanobacteria have genes for the five known transporter types, but in general, the β-cyanobacteria have more transporter homologs than the α-cyanobacteria (1, 36).
Fig 1.
Overview of the general CCM characteristics of α- and β-cyanobacteria, showing the types of Ci transporters typically present in each cyanobacterial type. Typical β-cyanobacteria possess up to two types of CO2 pumps (green) and up to three types of HCO3− transporters (orange) and make use of β-carboxysomes (form 1B RubisCO plus ccm gene products), while typical α-cyanobacteria possess only two or three identifiable Ci transporters and make use of α-carboxysomes (form 1A RubisCO plus cso gene products). In the case of Prochlorococcus species (oceanic α-cyanobacteria), the NDH-1-based CO2 pump genes are entirely missing, and the only candidates for HCO3− transport are unproven (blue; BicA2 and SbtA2).
Cyanobacterial CO2 fixation is supported by up to three types of plasma membrane-associated bicarbonate transporters: SbtA, a high-affinity Na+/HCO3− symporter from the TC.2.A.83 family of Na+/solute symporters (37–39); BicA, a medium- to low-affinity Na+-dependent bicarbonate transporter from the SulP/SLC26 anion transporter family (40–42); and BCT1, a multimeric high-affinity bicarbonate ABC transporter (43, 44). In addition to these transporters, the homologous sbtA2 and bicA2 genes have been identified as candidates for bicarbonate transport in α-cyanobacteria (14, 35–37), though it may be the case that the SbtA2 protein has altered transport specificity with respect to the canonical SbtA transporter (45). The roles of SbtA2 and BicA2 in cyanobacterial CCMs remain unknown; however, these are the only known candidates for bicarbonate transport in the ecologically important genus Prochlorococcus, and hence, they could be the most abundant bicarbonate transporters in the biosphere (14).
In addition to bicarbonate transport, vectorial CO2 uptake and intracellular CO2 scavenging are achieved through two types of CO2-uptake complexes, namely, NDH-I3 and NDH-I4, which are modifications of the NDH-I respiratory complex (46–49). These thylakoid-bound complexes achieve CO2 uptake by vectorial CA activity, contributing to the intracellular bicarbonate pool by hydration of passively accumulated CO2 (50–52). The two types of CO2-uptake complex have different affinities for CO2 (53), with transcription of the higher-affinity NDH-I3 complex being induced in response to Ci limitation by model β-cyanobacteria (54–58). As speculated previously (20, 59–62), CO2 which has escaped the carboxysome is recycled into the bicarbonate pool by these complexes, enhancing the efficiency of the cyanobacterial CCM (49).
It is likely that cyanobacterial Ci uptake is subject to posttranslational control resulting in rapid activation of HCO3− transport (63–65). We direct readers interested in these and other aspects of cyanobacterial Ci uptake to recent and thorough reviews (14, 15).
Proteobacterial Ci Uptake
In contrast to the case for cyanobacteria, the Ci-uptake properties of carboxysome-containing proteobacteria are poorly understood (66). Despite this, it is clear that at least Thiomicrospira crunogena and Halothiobacillus neapolitanus (gammaproteobacteria) possess CCMs (18, 67), whereas Thiobacillus versutus (Paracoccus versutus; an alphaproteobacterium) does not (68). Similarly, different species appear to preferentially utilize different carbon species (CO2 and HCO3−) for Ci uptake (18, 67). The general observation that many α-carboxysome-containing proteobacteria are acidophilic (69–71), or even acidogenic, through metabolic oxidation of inorganic sulfur, suggests that the primary carbon species utilized by Ci-uptake systems for these CCMs is CO2. Indeed, CO2 is the predominant carbon species utilized by H. neapolitanus (67), whereas T. crunogena can utilize CO2 or HCO3− (18).
It has been speculated that extracellular or cytoplasmic CA enzymes may play a role in CO2 fixation by facilitating the interconversion of carbon species. However, in T. crunogena, noncarboxysomal α-CA and β-CA enzymes (note that the α-, β-, and γ-classes of CA enzymes mentioned in this review are a separate classification system from that for carboxysomes and cyanobacteria) were recently shown not to have a direct role in CO2 fixation (72). Similarly, the EcaB β-CA enzyme from Synechocystis strain PCC 6803 is probably not involved in CO2 fixation, though its periplasmic targeting suggests that it may play some role in external exchange of Ci species (73).
Critical Role for Carboxysomes
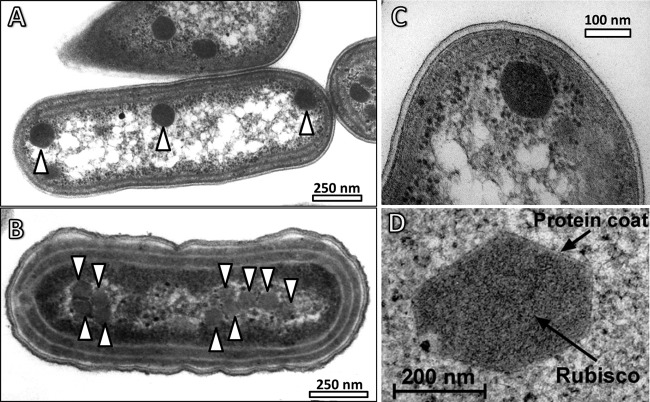
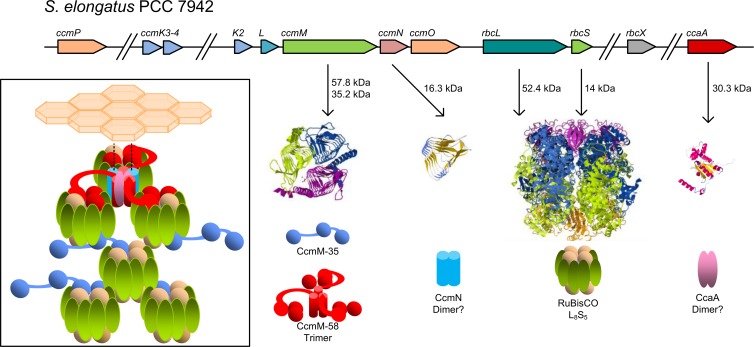
Carboxysomes were first identified as polyhedral cell inclusion bodies of unknown function in cyanobacteria and chemotrophic gammaproteobacteria (74–78) (Fig. 2 shows examples of electron micrographs of cyanobacterial carboxysomes). Later work in the cyanobacterium Anabaena cylindrica and the chemoautotroph Halothiobacillus neapolitanus showed that a cell's RubisCO content and activity were associated with these polyhedral bodies, and the term “carboxysome” was coined (16, 79–86). Subsequently, it was shown in the model organism Synechococcus elongatus PCC 7942 that virtually all of the cellular CA enzymatic activity was also associated with the carboxysome (19, 30, 87) (though this is not always the case in cyanobacteria [21, 22]), an observation that is highly consistent with this being the site of carbon fixation in S. elongatus PCC 7942 and other cyanobacteria. A putative mechanism/model for effective carboxysomal Ci fixation emerged from these data, which we summarize briefly here.
Fig 2.
Carboxysomes and their subcellular context (arrowheads in panels A and B indicate the positions of carboxysomes). (A) β-Carboxysomes present in Synechococcus elongatus PCC 7942. (B) α-Carboxysomes present in Cyanobium PCC 7001. (Courtesy of Lynne Whitehead.) (C) Close-up of a β-carboxysome from S. elongatus PCC 7942. (D) Close-up of a β-carboxysome from Anabaena variabilis M3. Note the size differences between the different types of β-carboxysomes in panels C and D.
The carboxysome encapsulates the RubisCO enzyme, with a selectively permeable shell layer that provides a diffusion barrier to CO2 efflux and O2 influx yet permits transit of ribulose-1,5-bisphosphate (RuBP), 3-PGA, Mg2+, and HCO3− between the carboxysomal and cytoplasmic pools. Within the lumen of the carboxysome, a carboxysomal CA enzyme dehydrates HCO3− to CO2, where it is fixed into 3-carbon sugars by RubisCO. Basic concepts inherent in early models of carboxysome organization and function (88–90) have largely been borne out by structure-function analyses. Reinhold and coworkers' most developed model specifically placed the CO2 diffusion resistance function at the carboxysome shell (88), and this function was subsequently confirmed in both α- and β-carboxysomes (19, 91–94). Similarly, current models of carboxysomal CA localization, where CA nuclei are present throughout the shell structure (95–99), are consistent with one model version (88) in which numerous “CA sites” are surrounded by “RubisCO zones” rather than carboxysomes which contain a monolithic CA core.
TWO TYPES OF CARBOXYSOMES WITH DISTINCT COMPONENTS AND EVOLUTION
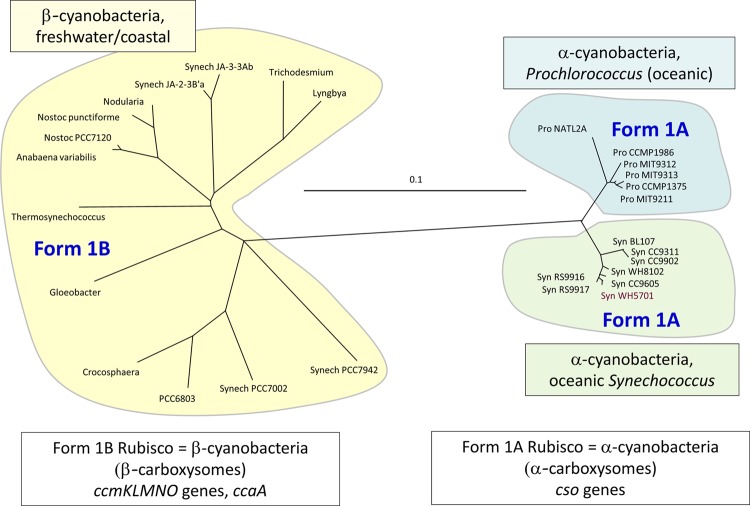
Tabita (100) showed that RubisCO form 1 sequences cluster into four distinct evolutionary groups, with oceanic cyanobacteria possessing RubisCO genes with phylogenetic affinity to those of proteobacterial species (RubisCO form 1A) and other cyanobacteria containing RubisCO genes with phylogenetic affinity to those of higher plants (RubisCO form 1B). Hence, the cyanobacteria came to be known as α-cyanobacteria if they contained form 1A RubisCO and β-cyanobacteria if they contained form 1B RubisCO (Fig. 3). This concept was extended by Badger et al. (1), who showed that the protein/gene components of the carboxysome also typify this phylogenetic divide. Thus, cyanobacteria have come to be known as α-cyanobacteria if they have form 1A RubisCO within α-type carboxysomes (encoded by the cso operon) and as β-cyanobacteria if they contain form 1B RubisCO within β-type carboxysomes (encoded primarily by the ccm operon) (Table 1; Fig. 3 to 5). The names of the carboxysome types are shortened to simply “α-carboxysomes” and “β-carboxysomes.”
Fig 3.
Phylogenies of representative sets of cyanobacteria, based on RubisCO large-subunit proteins (RbcL and CbbL proteins). Phylogenies were constructed as detailed previously (40). Note that oceanic cyanobacteria, i.e., α-cyanobacteria (Prochlorococcus and Synechococcus species), have form 1A RubisCO enzymes, fall into two minor groups, and are readily distinguished from the more diverse β-cyanobacterial group (form 1B RubisCO).
Table 1.
Relative abundances of α- and β-carboxysomal proteins in model carboxysomes from Synechococcus elongatus PCC 7942 (β-cyanobacteria), Prochlorococcus marinus MED4 (α-cyanobacteria), and Halothiobacillus neapolitanus C2 (Gammaproteobacteria)a
| Protein category | β-Carboxysomes |
α-Carboxysomes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein |
S. elongatus PCC 7942 (2% CO2) |
Protein |
Prochlorococcus MED4 |
Halothiobacillus C2 |
Structure | |||||
| n | nm | Structure | n | nm | n | nm | ||||
| Shell proteins | CcmK2-4 | 6,863 | 1,143 | Hexamer (147) | CsoS1AB | 3,232 | 539 | 540 | 90 | Hexamer (118) |
| CsoS1C | 0 | 0 | 2,970 | 495 | Hexamer (119) | |||||
| CcmO | 3,075 | 1,025b | Trimer (159) | CsoS1D | 38 | 13 | ? (112, 120) | ? | Trimer (120) | |
| CcmP | ? | ? | Trimer (161) | CsoS1E | 0 | 0 | ? (112) | ? | Trimer (112) | |
| CcmL | 60 | 12 | Pentamer (123) | CsoS4AB | 60 | 12 | 60 | 12 | Pentamer (123) | |
| CA enzymes | CcaA | 1,058 | 529 | Dimer (173) | CsoSCA | 58 | 29 | 81 | 40.5 | Dimer (168) |
| CcmM-58d | ||||||||||
| Structural proteins | CcmN | ? | ? | Monomer? (143) | CsoS2A | 0 | 0 | 143 | 329 | Monomer? |
| CcmM-58 | 2,177 | 725 | Trimer (98) | CsoS2B | 163c | 163 | 186 | |||
| CcmM-35 | 3,829 | 3,829 | Monomer | |||||||
| RubisCO enzyme | RbcL | 8,960 | 1,120 | L8S4CcmM-354 (96) | CbbL | 1,216 | 152 | 2,160 | 270 | L8S8e |
| RbcS | 6,073 | CbbS | 1,216 | 2,160 | ||||||
The protein interactions, sizes, and putative structures of carboxysomes are illustrated in Fig. 7 and 8. Data for S. elongatus PCC 7942 are from references 96 and 159, those for P. marinus MED4 are from reference 112, and those for H. neapolitanus C2 are from reference 66. Other references for data are indicated in parentheses. n, gross number of proteins per carboxysome; nm, number of protein multimers per carboxysome.
Calculated as the difference between the total number of hexagonal units required to cover beta-Cbx (2,168) (96) and the apparent number of CcmKx hexamers (1,143) (159).
CsoS2 from MED4 is presumed to be equivalent to the unglycosylated form of CsoS2B from H. neapolitanus (112).
CcmM is an active γ-CA enzyme in T. elongatus BP-1, and probably also in other species (138).
From the crystal structure deposited in the Protein Data Bank (PDB entry 1SVD).
Fig 5.
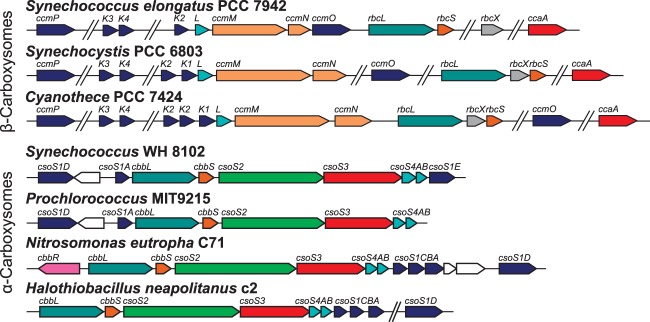
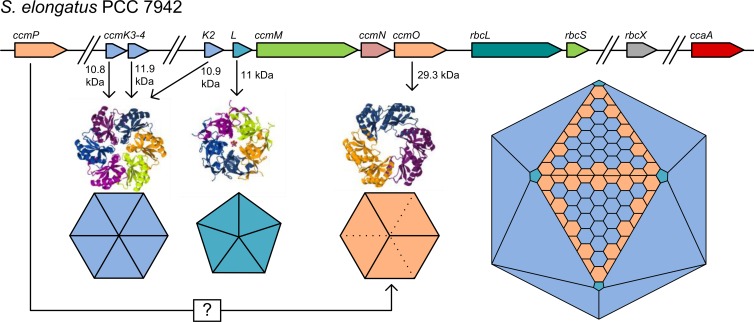
Genomic organization of representative β-carboxysomal ccm operons (top) and α-carboxysomal cso operons (bottom). Genes with structurally and/or functionally similar products are the same color. Data were adapted from the MicrobesOnline database (237).
The phyletic distributions of the two types of carboxysomes are quite well understood. β-Carboxysomes are found in all subsections of cyanobacteria sensu Rippka et al. (101) and, to date, have not been found in any other bacterial lineage. Recently, it emerged that a notable diazotrophic strain, “cyanobacterium UCYN-A,” lacks major photosynthetic pathways, which is an interesting reduction of β-cyanobacterial features in adaptation to a symbiotic lifestyle (102, 103). UCYN-A does not carry the components of a carboxysome, thus escaping the α- and β-cyanobacterial paradigm; however, it is phylogenetically affiliated with the Cyanothece genus of β-cyanobacteria (102, 104, 105).
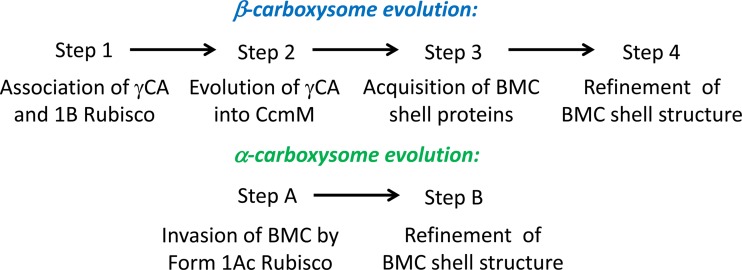
It has emerged that α-cyanobacteria are a monophyletic clade which diverged from planktonic, unicellular, freshwater β-cyanobacteria approximately 1.0 Gya (36, 106) (Fig. 4) and that they probably gained their cso operon, encoding the components of α-carboxysomes, by horizontal gene transfer from a gammaproteobacterial genus such as Nitrococcus (2, 107). Indeed, numerous species of alpha-, beta-, and gammaproteobacteria contain α-carboxysomes that support chemolithoautotrophic and mixotrophic growth, and thus their phyletic distribution is paraphyletic, probably involving horizontal gene transfer (1, 2, 11). The two types of carboxysome, while having distinct phyletic distributions, also have distinct sets of protein components, which are discussed below. Given these observations and the apparent spread of the α-carboxysomal cso operon by horizontal gene transfer into disparate cyanobacterial and proteobacterial lineages (107), we view it as possible that the two types of cyanobacterial carboxysomes arose by convergent evolution, perhaps after the divergence 1.0 Gya of α-cyanobacteria and β-cyanobacteria, since, so far at least, none of the α-cyanobacteria phylogenetically definable by core gene sequences possess β-carboxysome genes, and vice versa (36). On the other hand, it is very likely that throughout history many cyanobacterial lineages have occupied (and, indeed, still occupy) environmental niches in which intra- or extracellular O2 concentrations may limit CO2 fixation, such as microbial mats (35); thus, the requirement for a carboxysomal CCM is likely an ancient one.
Fig 4.
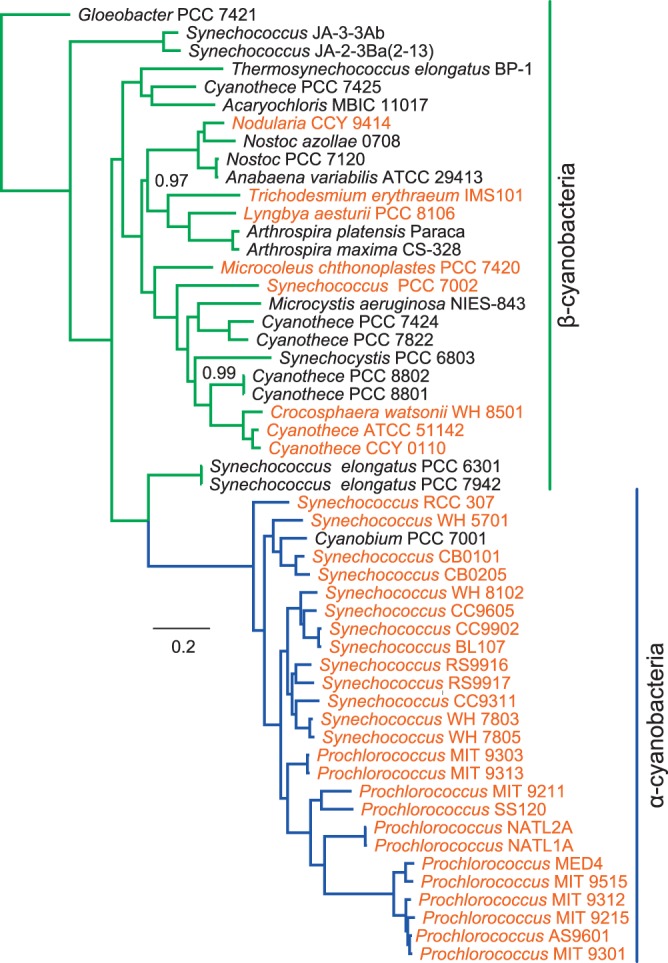
Cyanobacterial species phylogeny. α- (blue) and β-cyanobacterial (green) clades are highlighted, and marine species are shown in orange. The phylogeny shows Bayesian posterior probability values of <1.0, and AMPHORA (232), MrBayes 3.1.2 (233, 234), SeaView 4.2 (235), and GBlocks (236) were used to generate the phylogeny.
Further support for the hypothesis of convergent evolution is gained by the observation that the primary sequences of proteins comprising the shell structure of each type of carboxysome are more similar to the shell proteins of other types of bacterial microcompartments than to each other (108). Thus, despite their conserved functional role and their almost identical outward appearances, α- and β-carboxysomes have strikingly different internal structures. Nonetheless, both carboxysome types share several key features that underlie their conserved function. Below, we describe how convergent evolution resulted in these functions being achieved in remarkably different ways and through distinct structural paradigms.
Certainly all photosynthetically competent cyanobacteria, whether α-cyanobacteria or β-cyanobacteria, contain functional carboxysomes, or at least have the necessary genes. However, among other bacteria, the presence of carboxysomes is much more sporadic. α-Carboxysome-containing organisms can be found in the following groups:
α-cyanobacteria (1), including α-cyanobacterial symbionts (109);
sulfur-oxidizing bacteria of the genera Thiobacillus Kelly and Harrison (110) (now named Acidiphilium) (alphaproteobacteria), Thiobacillus and Thiomonas (both betaproteobacteria), and Halothiobacillus and Acidithiobacillus (both gammaproteobacteria) (69, 111);
Acidimicrobium (actinobacteria);
nitrifying bacteria of the genera Bradyrhizobium (alphaproteobacteria), Nitrobacter, Nitrosomonas (betaproteobacteria), and Nitrococcus (gammaproteobacteria); and
the α-cyanobacterial “chromatophore” of Paulinella chromatophora (107).
Among the autotrophic proteobacteria that possess RubisCO genes, it is possible to detect those that have carboxysomes and those that do not based on sequence differences in their RubisCO protein sequences (11). Thus, form 1A RubisCO sequences can be subdivided further into RubisCO form 1Ac, which is found associated with carboxysomes, and form 1Aq, with the putative RubisCO chaperonin gene cbbQ and without an obvious carboxysome operon.
Two Distinct RubisCO Types
As previously mentioned, α- and β-carboxysomes encapsulate RubisCO form 1A and form 1B, respectively (1). There is growing evidence that these are packaged into the carboxysome in remarkably different ways. As we describe in greater depth here, β-carboxysomes have significant internal structure, with RubisCO holoenzymes being scaffolded in three dimensions by products of the ccmM gene, whereas the interior of α-carboxysomes appears to be less ordered or possesses one or more ordered layers bound to the inner surface of the shell.
Structure of α-Carboxysomes
Study of the α-carboxysomes has revealed a complex structure composed of as few as 8 polypeptides in Prochlorococcus marinus MED4 (112) to as many as 11 polypeptides in H. neapolitanus C2 or 10 polypeptides reported by Heinhorst et al. (66), as well as CsoS1D (112). Apart from CbbL and CbbS, the large and small subunits of RubisCO form 1A, the remaining proteins fall into two categories: shell proteins of small size (CsoS1A to -E and CsoS4A and -B) and larger shell-associated proteins (CsoS2A and -B and CsoSCA).
The shell of α-carboxysomes is formed from lineage-specific subsets of the CsoS1 protein type, as well as the CsoS4A and CsoS4B proteins common to all α-carboxysomes (113, 114). All are small, 10- to 11-kDa proteins. The protein structures forming the outer shells of carboxysomes have been reviewed extensively (113–117). Briefly, crystal structures of the CsoS1A, CsoS1C, and CsoS1D proteins have been elucidated, showing that these proteins contain the characteristic bacterial microcompartment (BMC) domain and form flattened, regularly hexagonal hexamers (CsoS1A and -C) or trimers (CsoS1D) (118–120), as BMC proteins from ethanolamine (EUT) and propanediol (PDU) microcompartments do (113). An important observation is that each hexamer type has a central pore of a size and charge distribution that may allow entry of specific substrates (HCO3−, RuBP, and Mg2+) or exit of products such as PGA, while retarding entry of O2 and leakage of CO2 (113, 118, 119). These BMC oligomers form sheets in crystallographic studies, which strongly suggests that the sheet structure represents the behavior of CsoS1 proteins in vivo. Indeed, CsoS1A hexamers probably interact with one another very closely, more closely than other BMC proteins such as CcmK2 do, forming sheets which could be impermeable to CO2 or O2 (118). However, multiple CsoS1 homologues participate in the outer shell of all α-carboxysomes, potentially modifying these tight interactions.
The proposed mechanism for selective permeability of the shell relates to the presence of a conserved charged pore at the 6-fold axis of symmetry in the CsoS1x oligomers studied to date (113, 118, 119), which may be sufficient to allow preferential transit of charged molecules. Kinney et al. (113) suggested that the positively charged pores may promote passage or binding of negatively charged molecules, such as bicarbonate, while remaining indifferent to uncharged molecules, such as O2 or CO2. Furthermore, the crystal structure of the CsoS1D protein, with each protomer consisting of tandem BMC domains (120), suggests a unique, dual-layer hexameric unit. The hexameric CsoS1D structure consists of two trimeric subunits, with distinct “open” and “closed” conformations, and Klein et al. (120) proposed that interchange between these conformations could gate the entry and exit of the comparatively large RubisCO substrate RuBP. Indeed, biochemical studies support these models, with the α-carboxysome shell being a barrier to CO2 and RuBP transit (91, 92, 121, 122), but without compelling evidence for an RuBP transit role for CsoS1D (122).
The nature of icosahedral geometry requires a specialized shell protein to fit in the gaps left at the 6-fold axes of symmetry, i.e., the vertices. In α-carboxysomes, the CsoS4A and CsoS4B proteins achieve this function (123), as they were shown to close the shell, thus preventing CO2 escape (92). The CsoS4A protein formed pyramidal pentamers in crystal structure, which comfortably fit into the 12 vertices in idealized models of α-carboxysomes (123).
Empty α-carboxysome shells can form due to mutations in vivo (121, 124), and largely or partially empty shells are observed regularly in wild-type cells or α-carboxysome preparations (80, 125–129). The shell of α-carboxysomes appears to be a scaffold to which as many as two isoforms of CsoS2 attach, as well as the α-carboxysomal CA enzyme CsoSCA/CsoS3 (95, 130–133). In bacterial two-hybrid studies, the CsoS2 protein was shown to interact with many α-carboxysome proteins (131); however, whether its role is essential is currently unknown. The protein-protein interactions reported by Gonzales et al. (131) could be spurious; for instance, molecular models of the α-carboxysome shell suggest that there is little potential for interaction of the vertex protein CsoS4A or CsoS4B with luminal proteins (123). There is evidence that the csoS2 gene codes for a full-length protein (130 kDa) and a shorter protein (85 kDa) in H. neapolitanus; however, the roles of these two forms are not known (130), and only the shorter, unglycosylated form is observed in α-carboxysomes from P. marinus (112). Nonetheless, an interesting hypothesis is that CsoS2 organizes the subshell structure of α-carboxysomes in some way, although evidence for significant internal structure in α-carboxysomes is scarce (see below).
Given that there appear to be two distinct protein domains within CsoS2 (134), much as there are two distinct domains in CcmM (see below), it is appealing to apply the structural role of CcmM to CsoS2. Potentially, as RubisCO does appear to be organized into defined layers, at least in the immediate subshell layer, CsoS2 links RubisCO enzymes to the inner shell. This could allow RubisCO to take up most of the carboxysome interior, depending on the size of the carboxysome; indeed, CsoS2A and CsoS2B are present in quantities roughly equal to those of RubisCO holoenzymes (66), indicating a 1:1 stoichiometry within the α-carboxysome (Table 1). We suggest that this structure may in fact be the basis for the consistently small diameters of α-carboxysomes with respect to β-carboxysomes. In other words, if RubisCO is attached to the inner shell of α-carboxysomes by CsoS2, then this protein, with a fixed size, must thus operate over a fixed distance from the shell, especially since csoS2 does not appear to produce the same types of domain-specific isoforms as ccmM (66, 112). This structural feature would ensure that the ratio of inner surface area (CA-rich zone) to stromal volume (RubisCO-rich zone) remained high. Recent electrostatic potential maps of the form 1A RubisCO enzyme showed that this enzyme has a much lower surface charge than that of form 1B enzymes (135). Whether this is a chemistructural feature allowing electrostatic agglomeration of RubisCO enzymes within α-carboxysomes remains debatable, but we see this as a promising avenue for investigation. In fact, Holthuijzen et al. (127) showed in H. neapolitanus that the RubisCO large subunit was preferentially liberated from urea-disrupted α-carboxysomes—though this phenomenon is debated (121); thus, they posited that the small subunit was strongly bound to the shell structure. This is supported by the work of Badger and Bek (11), who showed that synapomorphies in the RubisCO small subunit are the principal distinguishing features of a carboxysomal form 1A RubisCO enzyme with respect to noncarboxysomal form 1A RubisCO. Thus, it appears that RubisCO enzyme incorporation into α-carboxysomes acts through the small subunit of RubisCO form 1A, although Menon et al. (121) have produced evidence which suggests that the large subunit may also have important assembly information.
Structure of β-Carboxysomes
To date, the majority of the work on β-carboxysomes has been undertaken in the freshwater strains S. elongatus PCC 7942 and Synechocystis PCC 6803. In the former species, two CcmM proteins (58 and 35 kDa) are expressed from the ccmM gene, and these are undoubtedly the most important structural proteins in the β-carboxysome (136, 137). The protein structures of CcmM-58 and CcmM-35 inform us of their specific roles within the β-carboxysome. The full-length CcmM-58 isoform has an N terminus with structural and primary sequence similarity to γ-carbonic anhydrase enzymes such as Cam from Methanosarcina thermophila (108, 138, 139). The C terminus of CcmM-58 contains three tandem copies of a protein domain with similarity to the RubisCO small subunit (SSU-like domains) (140). The SSU-like domains share up to 30% sequence identity with their cognate RbcS sequences, although the ∼85-amino-acid SSU-like domain does not align with the N terminus of RbcS (141). Despite this, the SSU-like domains of the proteins from S. elongatus PCC 7942, Synechococcus PCC 7002, and Synechocystis PCC 6803 have a translocated sequence motif with similarity to the absent N-terminal region (141). As many as five SSU-like repeats are evident in predicted CcmM proteins from β-cyanobacteria, though the extra SSU copy number variation has an unknown function, if any (96, 140–142).
The CcmM-35 isoform is translated from an internal ribosome entry site and consists of only the SSU-like domains (98, 141). This scheme of multiple CcmM isoforms expressed from a single gene has been observed in Synechocystis PCC 6803 (99, 141), Synechococcus PCC 7002 (141), and Acaryochloris marina MBIC11017, and all ccmM genes are expected to behave similarly (96).
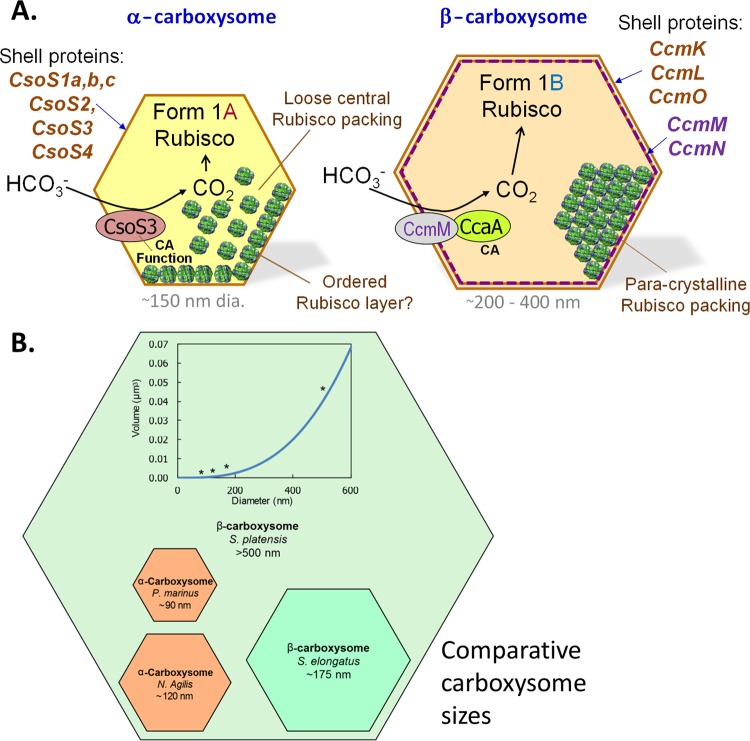
In current models of β-carboxysome organization and stoichiometry, at least 11 polypeptides form the β-carboxysomes from S. elongatus PCC 7942. These fall into two categories: proteins forming the β-carboxysome shell layers (inner shell bicarbonate dehydration/RubisCO-organizing layer, formed by CcaA, CcmM-58, CcmN, RubisCO CcmK2-4, CcmL, and CcmO; and outer shell BMC layer, formed by CcmK2-4, CcmL, and CcmO) and proteins forming the carboxysome lumen (CcmM-35, RbcL, and RbcS) (Fig. 5). CcmM-58 is restricted to the inner shell (96, 97). There, it simultaneously interlinks adjacent RubisCO molecules, recruits the carboxysomal carbonic anhydrase CcaA, and recruits the outer shell (97–99). The interaction between CcmM-58 and the outer shell is due to direct interaction with the outer shell protein CcmK2 (97, 99) and indirect interaction through the CcmN protein, which may link the outer and inner shell structures (143). In contrast, the CcmM-35 isoform is predicted to be confined to the carboxysome lumen, interlinking adjacent RubisCO enzymes in two planes (96), thereby organizing RubisCO into the paracrystalline array evident in some electron microscopic studies of β-carboxysomes (144) (Fig. 6A; see Fig. 8).
Fig 6.
(A) Comparison of the components and possible RubisCO packing within α- and β-carboxysomes. Note that the latter structures are always bigger than the former (see Fig. 4B). It is likely that the β-carboxysome has an outer layer composed of CcmK, CcmO, and CcmL, while the inner, less-dense, RubisCO-attached layer is composed of CcmM, CcaA, and CcmN; the interior appears to be paracrystalline and possibly organized by the shorter form of CcmM. For comparison, the smaller α-carboxysomes may feature a shell that is composed mostly of CsoS1 and CsoS4 forms, with less organization of internal RubisCO. (B) Comparison of the diameters and volumes of extreme carboxysomes of both types (112, 159, 238, 239). The inset shows the relationship between internal volume and maximum cross-sectional diameter, assuming that this measurement is the same as the diameter of a sphere circumscribing a perfectly icosahedral carboxysome. The volume of each carboxysome type is shown and is indicated by an asterisk on the curve.
Fig 8.
Current models of the interactions of proteins within β-carboxysomes. The CcmM-58 and CcmM-35 protein isoforms have independent roles, with the larger isoform (red) occupying the inner shell bicarbonate dehydration/RubisCO-organizing layer (inset) and recruiting the outer shell BMC layer via CcmN (blue), as well as recruiting the carboxysomal carbonic anhydrase CcaA (pink). Stoichiometric models suggest that the CcmM-35 isoform is probably localized predominantly to the interior RubisCO layers and interlinks adjacent RubisCO enzymes (green and tan) in three dimensions. Protein structure images were generated in Jmol (240), and protein threading was performed in Swiss-MODEL (241–243), using the following protein structures: CcmM-58 (PDB entry 3KWC) (138), CcmN (generated by threading the CcmN protein sequence onto PDB entry 3KWD chain A) (138), Rubisco (PDB entry 1RBL) (244), and CcaA (threaded onto PDB entry 1EKJG chain A) (245).
CcmM-mediated organization of β-carboxysomes implicitly suggests a strong interaction between the SSU-like domains of CcmM and the RubisCO enzyme. It is intuitive that these occur through occupation by the SSU-like domains of the RbcS-binding site of the RubisCO large subunit. The SSU-like domains do appear to retain many of the hydrophobic residues thought to underlie RbcL binding (141), but it is well known that perturbation of the RbcL-RbcS interaction or stoichiometry has kinetic consequences for the enzyme (121, 145). We have previously shown that the RbcL-RbcS subunit stoichiometry is greater in vivo than that reported for the L8S8 RubisCO holoenzyme, that is, the subunit stoichiometry appears to be about L8S5 in vivo (96). This was supported by our data showing that the RbcL-RbcS stoichiometry was rather more fluid than previously supposed, potentially supporting the hypothesis that the SSU-like domain occupies a varying proportion of the RbcS-binding sites. It is important that the RubisCO standard used for this work was a crystal structure-verified L8S8 stoichiometry produced by using an Escherichia coli ectopic expression system (146). The exact interaction underlying the scaffolding of RubisCO-CcmM interactions, especially the possibility that SSU-like domains in CcmM displace RubisCO SSU or could catalytically mimic the role of the SSU, is therefore worthy of further examination.
In current models, a complex outer shell structure is attached to this scaffolded array of RubisCO and CA enzymes. The outer shell is composed of oligomeric protein complexes consisting of CcmK2-4 and CcmO, which contain the BMC domain (pfam number PF00936) (113, 114, 123, 147). These proteins were identified primarily by mutant analyses of S. elongatus PCC 7942 which revealed that knockout of any of the genes in the operon upstream of rbcLS, ccmK-ccmO (ccmK-O) resulted in high-CO2-requiring mutants (93, 94, 140, 148–154). Significantly, a homologous relationship between the CcmK and CcmO genes was established, with CcmO consisting of tandem repeats of a CcmK-like unit (150, 155)—the BMC domain. With the advent of genomic sequencing, two more ccmK homologues became apparent in the cyanobacterial genome: ccmK3 and -4 (136, 156, 157). The products of these genes came to be known as CcmK3 and CcmK4, and although CcmK4 has been implicated in functioning of the CCM in Synechocystis (158), the secondary functions of these proteins in the β-carboxysome only recently became apparent (159). We showed that CcmK3 and CcmK4 are individually required for maximum β-carboxysome function in S. elongatus PCC 7942; however, the carboxysome is not entirely crippled in their absence, much in contrast to the case for other carboxysome shell proteins. Thus, the ΔccmK3-4 mutant is capable of slow growth in ambient CO2 (159).
As previously mentioned for α-carboxysomes, the icosahedral geometry of β-carboxysomes requires a unique structural element to completely close the carboxysome shell at the 5-fold axes of symmetry—the vertices. The CcmL protein, containing the PF03319 protein domain, forms pentagonal, pyramidal pentamers in crystal studies (123). These fit well into models of the outer shell of idealized β-carboxysomes, where they occupy the five-way interface between adjacent β-carboxysome facets (117, 123). Early work showing characteristic rod-like carboxysomes in ccmL mutants generally supports this hypothesis, with rod-like carboxysomes being formed due to the inability of the cell to close the vertices (93, 94, 140).
Recent work suggests that a double layer of BMC proteins makes up the outer β-carboxysome shell (160). This hypothesis is intriguing, as the reported width of a double layer of CcmK2 hexamers (4 to 7 nm) (147, 160) is generally supported by electron microscopic observation of the shell width (5 to 6 nm) (144). Moreover, the presence of two outer BMC shell layers would be supported by the potential hexameric, double-layer conformation of CcmP reported by Cai et al. (161). At this stage, however, no direct evidence for the existence of a dual shell layer, or the presence of CcmP in it, has been gathered. A criticism of this hypothesis is that there are more protein-protein interactions at play in the vicinity of CcmK2 in vivo than in protein crystals. Does the presence of CcmN and CcmM-58, which are known CcmK2 interactors (97, 99, 143), alter the self-interactivity of CcmK2 or provide a directionality to the CcmK2 pore? It is also unclear why a double shell layer is a likely structure given that, assuming a single shell layer, there is in fact a reported ∼30% shortfall of CcmK2 for β-carboxysome surface coverage (96) (Table 1). Indeed, further work is required to validate this structure. We previously speculated that CcmO could account for this shortfall (159), but the exact composition of the outer shell in our models remains speculative.
Structural insights into BMC proteins have hinted at the underlying basis for the observed diffusion resistances of the β-carboxysome shell to certain metabolites (19, 123, 147, 162). Pioneering crystal structures of CcmK1, CcmK2, and CcmK4 from Synechocystis PCC 6803 revealed their ability to form hexamers which are then capable of oligomerization, such that they form sheets or linear strips of hexamers (117, 147), the corollary being that these sheets and strips probably represent valid β-carboxysomal structures. Each hexameric unit has a positively charged pore which is thought to mediate charged-solute transit, through an unknown mechanism (147). Significantly, no crystal structure has been published for the tandem BMC domain protein CcmO; however, its role in the outer shell of the β-carboxysome is clearly established (159, 163, 164). CcmO is expected to form a trimeric unit with properties and a structure similar to those of CcmK hexamers (Fig. 7) and tandem BMC proteins from other types of microcompartment, such as CsoS1D (120), PduT (165), and EutL (166). Recently, a gene designated ccmP was identified as an orthologue of the α-carboxysomal csoS1D gene (161). CcmP has not previously been identified as an essential β-carboxysome shell protein, but it is expected to recapitulate the curious crystal structure of CsoS1D, providing a potential pore for large metabolite transit.
Fig 7.
Model for the outer shell structure of β-carboxysomes from Synechococcus elongatus PCC 7942. CcmK2, -K3, and -K4 produce flattened hexamers, and CcmK2 is by far the most abundant form. CcmO is postulated to form flattened trimers that could potentially interface with the triangular facets. The rarer CcmL pentamers would close the 5-fold vertices. The carboxysome model is not drawn to scale. Protein structure images were generated using Jmol (240), based on structures of the following proteins: CcmK (Protein Data Bank [PDB] entry 2A1B) (147), CcmO (represented by the structure of CsoS1D [PDB entry 3F56]) (120), and CcmL (PDB entry 2QW7) (147).
The ultrastructure of β-carboxysome shell mutants in S. elongatus PCC 7942 is consistent with major roles for CcmK2, CcmO, CcmL, and CcmN in the outer shell (93, 140, 143, 159). We have previously shown that CcmK3 and CcmK4 are not essential for carboxysome biogenesis; however, at least one of these proteins is required for maximum carboxysome function and correct subcellular localization (159). Given that the amount of CcmK2 present in β-cyanobacterial cells would result in a shortfall of carboxysome surface coverage (96), the protein-protein interactions between CcmO and CcmK2 (159), and the minor role taken by the CcmK3 and CcmK4 proteins, CcmO was proposed as a second major outer shell protein in β-carboxysomes (159). These observations led to speculative models for facets of the outer β-carboxysome shell (Tables 1 and 2; Fig. 6 to 8), with the bulk facet being made up of CcmK2, with various roles for the CcmO protein, but typically at the vertex, the facet-facet interface (icosahedral edge), or both (159).
An α-Carboxysomal Carbonic Anhydrase
Carboxysomal CO2 accumulation beyond the spontaneous supply rate is achieved through the action of carboxysome-specific CA enzymes. These convert HCO3− to CO2 in the vicinity of RubisCO, allowing saturation of the RubisCO active site with CO2. The CA enzymes of both types of carboxysomes are associated with the shell structure (95–97, 99, 138).
The α-carboxysomal CA enzyme, CsoSCA (formerly known as CsoS3), was identified as the product of csoS3 in H. neapolitanus (95, 132, 167). Initially identified as the type enzyme of the novel ε class of CA enzymes (132), X-ray crystallographic investigation revealed that CsoSCA was a structural analogue of the previously characterized β class of CA enzymes (133). CsoSCA, while forming a 57-kDa homodimer, contains only one active site where other β-CA enzymes contain two (133). The consequence of this unusual structure on CA activity is minimal, with the activity of CsoSCA being sufficient to saturate CO2 fixation (91, 168). CsoSCA is extremely tightly bound to the shell structure (168), but the csoS3::Kmr mutant has apparently normal carboxysomes (91), leading Cannon et al. (169) to conclude that CsoSCA is probably not an intrinsic shell component but a shell-associated component. Interestingly, CsoSCA (CsoS3) is redox inactivated under reducing conditions (132), like CcaA (see the next section), but it is not known if it is also Mg2+ dependent.
Two Types of β-Carboxysomal Carbonic Anhydrases
The canonical β-carboxysomal CA enzyme is also a member of the β-CA class. The β-carboxysomal carbonic anhydrase CcaA (ccaA; formerly known as icfA) (170, 171) is a component of the inner shell bicarbonate dehydration/RubisCO scaffolding complex of β-carboxysomes in S. elongatus PCC 7942, Synechocystis PCC 6803, and a number of other β-cyanobacterial species (30, 96–99, 170–172). As a functional dimer, CcaA is recruited to the β-carboxysome by CcmM-58 in S. elongatus PCC 7942 (98). Dimerization and enzyme activity were shown to be dependent on the extended C terminus of CcaA, which is the major point of difference between it and canonical noncarboxysomal β-CA enzymes (173). The enzyme activity of CcaA is restricted to the β-carboxysome (30, 172), probably because the presence of CA enzymes in the cytoplasm is injurious to the CCM (19). The enzyme activity of CcaA is low, however, though still sufficient to saturate β-carboxysomal CO2 fixation (30). In fact, cytoplasmic CcaA is rapidly degraded, such that Ci uptake is not short-circuited by noncarboxysomal CA activity (97, 136). A further regulatory function of CcaA is that it is inactivated under reducing conditions, suggesting that any enzyme en route to the carboxysome is effectively inactive (30), and also implying that the carboxysome interior is a thioredoxin-inaccessible compartment. CcaA is also Mg2+ dependent in a way similar to that of RubisCO (30). Since the free Mg2+ concentration may be downregulated in the dark, similar to the situation in plant chloroplasts, this suggested that CcaA is predominantly active in the light, like RubisCO (30). With respect to its role in β-carboxysome structure, inactivation mutants of ccaA possess β-carboxysomes with a wild-type appearance; however, they are physiologically impaired (73, 94, 174). This leads to the question of carboxysomal CA activity in species that lack an obvious ccaA gene (96, 99, 169, 175). As we discuss below, it appears that CcaA is the alternative β-carboxysomal carbonic anhydrase, with the γ-CA-like domain present in CcmM being most likely to be the original CA enzyme in evolutionary terms.
It has long been presumed that the N-terminal γ-CA-like domain of CcmM could provide β-carboxysomal CA activity in species that lack CcaA (99, 169, 175). However, no CA activity could be detected in the CcmM proteins from a number of species which also possess ccaA (99, 167). Recently, however, it emerged that the N terminus of CcmM from Thermosynechococcus elongatus BP-1 (which lacks ccaA) is a catalytically active γ-CA enzyme (138). The γ-CA-like domain is highly redox sensitive, suggesting once again that the carboxysome is a compartment that is inaccessible to redox equivalents—much like the way in which eukaryotic organelles operate (108). With the cytoplasm being a highly reducing environment in the light, this redox inhibition of γ-CA activity is probably an effective adaptation, like CcaA being redox inhibited and rapidly degraded in the cytoplasm, protecting the CCM from cytoplasmic CA activity (108, 138). The crystal structure of the γ-CA-like CcmM domain forms a homotrimer (138), which is consistent with observations of trimeric CcmM behavior in carboxysomes (98). Because CcmM is an integral protein to β-carboxysomes, it is present in all β-cyanobacteria. In comparison, CcaA is found in only a few β-cyanobacterial lineages (96). The phyletic distribution of CcaA broadly supports the hypothesis that ccaA arose in the freshwater genus Cyanothece, and its current distribution reflects horizontal gene transfer.
What, then, are the individual roles of CcaA and CcmM in the numerous β-cyanobacterial species that appear to possess both the ccaA gene and a CA-active CcmM enzyme (96, 108, 138)? It is perhaps likely that the CcmM protein, which appears to be the original β-carboxysomal CA enzyme, is primarily structural in these species, as it is in Synechocystis PCC 6803 and S. elongatus PCC 7942 (108). However, an identified peptide motif that correlates with γ-CA activity in the CcmM protein (138) suggests that many species contain two active carboxysomal CA enzymes. There is no obvious reason why CcaA or CcmM would be preferred over the other in β-carboxysomes. However, one possibility is that adoption of CcaA allows greater flexibility toward the fine adjustment of the CA/RubisCO activity ratio, whereas a CA-active version of CcmM would tend to be far less flexible in this ratio. Previous modeling has shown that optimal CO2 levels in the carboxysome are quite sensitive to the CA/RubisCO ratio, with too much CA activity leading to excessive leakage and too little leading to restriction of the fixation rate (94, 148).
Distinct Lumen Structures in α- and β-Carboxysomes
The current state of knowledge of the protein structures within α- and β-carboxysomes suggests a tantalizing possibility, namely, that α-carboxysomes are shell-centric structures, while β-carboxysomes are structurally lumen- or RubisCO-centric (Fig. 6A). Electron microscopic investigations support these structural hypotheses for β-carboxysomes. Kaneko et al. (144) showed that the lumen of β-carboxysomes is composed of a paracrystalline array of RubisCO molecules. This is consistent with current models of β-carboxysome structure and stoichiometry, in which RubisCO is regularly scaffolded in three dimensions by the small isoform of CcmM (96–98). Moreover, the enzymatic and structural components of β-carboxysomes appear to be part of a greater RubisCO scaffolding complex, where the lumen of the carboxysome is composed of CcmM-35 and RubisCO, with the large isoform of CcmM (CcmM-58) recruiting the carboxysomal carbonic anhydrase to the inner shell bicarbonate dehydration complex at the periphery of this structure (96–99). An important finding is that the French press/Triton-Percoll process for purifying β-carboxysomes removes about 70 to 80% of the principal shell protein CcmK2 (97, 98), yet carboxysomes from these preparations retain their icosahedral structure (30, 137). Thus, the selectively permeable outer shell appears to be associated weakly with the regularly ordered, RubisCO-centric β-carboxysome lumen; in fact, the importance of CcmM to this structure is demonstrated by the carboxysomeless phenotype of all reported ccmM mutants (97, 140–142), in which no traces of empty shell structures are apparent.
On the other hand, while RubisCO knockouts cannot be investigated because they are lethal to β-cyanobacteria, replacement of RubisCO form 1B with a noncarboxysomal form II RubisCO enzyme abolished all carboxysome structure in Synechocystis PCC 6803 (176). The primacy of the CcmM-RubisCO structure was detailed by Long et al. (97), who showed that by varying the stoichiometry of two CcmM isoforms in vivo, relevant β-cyanobacterial subcomplexes could be produced. The presence of the shell protein CcmK2 in carboxysome-rich fractions was dependent upon the presence of M58, indicating that the β-carboxysome shell is recruited to the inner RubisCO-CcmM structure (96, 97), probably via CcmN (143). Similarly, in β-cyanobacterial mutants deficient in the shell protein CcmK2, CcmO, or CcmN, carboxysome-like polar bodies in which the entire β-carboxysome shell is absent form, leaving a naked RubisCO-CcmM-CcaA complex (159). These phenotypes seem to be consistent with the RubisCO-centric hypothesis. In other words, the polar bodies arise through CcmM scaffolding of RubisCO and CcaA ad infinitum.
This regular and ordered luminal structure is not consistently apparent in α-carboxysomes. While it seems possible that CsoS2 is involved in the interior organization of the α-carboxysome (131), Iancu et al. (128) suggested that the interior of the α-carboxysome from H. neapolitanus is for the most part disordered and that much of the RubisCO is free-floating within the shell rather than being specifically anchored. Furthermore, it appears that only about half of the α-carboxysomes from Synechococcus WH 8102 and H. neapolitanus have some level of internal order; when ordered structures are apparent, these are usually within the RubisCO layers immediately below the shell (125, 129). Thus, the center of most α-carboxysomes studied by Iancu et al. (129), in addition to the entire α-carboxysome in almost half of those studied, was relatively disordered. These results match earlier studies of α-carboxysome structure in which α-carboxysomes were estimated to contain only one layer of RubisCO molecules, against the inside of the shell, and many were seen to be electron transparent beyond the immediate subshell layer (126). It is conceivable that this first RubisCO layer, representing a significant fraction of the luminal volume (especially in smaller carboxysomes), is the ordered RubisCO signal recently detected by small angle X-ray scattering (SAXS) analysis of whole Halothiobacillus carboxysomes (C. A. Kerfeld, personal communication).
These transmission electron microscopy (TEM) data are largely consistent with our knowledge of the less organized protein interactions within the α-carboxysome, in contrast to the highly ordered structure observed in β-carboxysomes (144). It appears that all of the non-RubisCO components of the α-carboxysome interact primarily with the carboxysome shell. The CsoS2 protein, which is presumed by some to organize the α-carboxysome lumen through its numerous protein-protein interactions (131), is bound to the inner surface of the shell (130), as is the α-carboxysomal CA enzyme CsoSCA (95). It is well established that empty α-carboxysome shells can form in the absence of form 1A RubisCO (121, 124) and that knockout of the csoS1A gene, encoding an α-carboxysomal shell protein, results in fewer carboxysomes per cell and reduces the proportion of cellular RubisCO localized to the carboxysome (177) rather than forming polar bodies as β-carboxysomal shell mutants do. These results, as well as the fact that crystallized sheets of the α-carboxysomal shell subunit CsoS1A are more closely bound than sheets of β-carboxysomal CcmK2 (118), are consistent with the shell-centric hypothesis of α-carboxysome structure.
Biogenesis of the Two Types of Carboxysomes
The putative differences in internal organization of the two types of carboxysome led us to propose two separate but related processes for the biogenesis of carboxysomes. The ability of β-cyanobacteria to express mutant carboxysomes with nonstandard subunit stoichiometries (96, 97), as well as shell-free polar bodies (29, 143, 150, 159), showed that the β-carboxysomal RubisCO-organizing complex, while being the principal underlying structure of the β-carboxysome, requires the simultaneous addition of the carboxysome shell for carboxysome biogenesis. That is, without the shell, a single β-carboxysome-like polar body is formed, and with the shell, about 4 or 5 carboxysomes are formed under Ci sufficiency. It is likely that the CcmN protein provides a key link between the RubisCO-organizing inner shell layer and the selectively permeable outer shell (143), but CcmM is also known to interact with the primary outer shell protein CcmK2 (97, 99) (Fig. 8). This scheme of simultaneous encapsulation of the nascent β-carboxysome is consistent with early electron microscopic studies of β-carboxysome formation (178), that is, subcarboxysomal RubisCO arrays, presumed here to be scaffolded by CcmM, form simultaneously into β-carboxysomes with a defined shell structure.
Targeting of proteins to the β-carboxysome is thus an intuitive process. The long and short forms of CcmM interact with most β-carboxysomal proteins (97–99, 143), and the only proteins that are not currently known to do so, CcmO (159) and CcmK3 (99), are expected to interact through other outer shell proteins (114, 159). Interestingly, recent work proposed that a short, C-terminal peptide sequence in CcmN is the underlying basis for its interaction with the outer shell of β-carboxysomes (143). In fact, the contribution of the 18 C-terminal residues of CcmN to outer shell recruitment appeared to exceed that of the CcmM-58 protein (143). This is similar to the schemes evident in PDU metabolosomes whereby the PduP enzyme is targeted to the interior of the PDU shell by a short N-terminal peptide sequence which interacts with the PduA and PduJ shell proteins (179, 180). It seems that this targeting scheme may be common to all types of microcompartment (143, 180).
In contrast to β-carboxysomes, it is more likely that α-carboxysomes form by infilling of the α-carboxysome shell with the enzymatic components (121, 124, 128). Menon et al. (121) are quite right to emphasize that this process is nonsequential and that the simultaneous lumen-infilling/shell construction model of Price and Badger (93) is probably more realistic for α-carboxysomes. It is possible that the CsoS2 protein is involved in this process, perhaps by direct interaction with RubisCO, CsoSCA, and the outer shell, as seen by Gonzales et al. (131). A specific RbcS synapomorphy identified by Badger and Bek (11) in form 1Aq RubisCO led to RbcS being proposed as the factor underlying specific incorporation of form 1Ac RubisCO into α-carboxysomes, a proposal supported by the work of Holthuijzen et al. (127). However, Menon et al. (121) proposed that it is the RubisCO large subunit that determines incorporation of RubisCO into the α-carboxysome in those species that have multiple types of form 1A RubisCO. The precise mechanism by which α-carboxysome-specific RubisCO enzymes are incorporated into carboxysomes remains unclear. Naturally, it is expected that a process similar to those elucidated for β-carboxysomes and PDU metabolosomes may be at work, though its molecular mechanism is still unknown.
Intracellular Localization and Partitioning of Carboxysomes
Recent work by Savage et al. (181) showed that β-carboxysomes are spatially distributed throughout the cell. Microfilaments and microtubule structures have long been observed with β-carboxysomes (182). This interaction with the bacterial cytoskeleton resolves previous questions of how carboxysomes are apportioned to daughter cells at mitosis and is reminiscent of the partitioning systems of bacterial chromosomes, plasmids, magnetosomes, chromosomes, etc. (183, 184). While a definitive link has been established between the presence of ParA, and perhaps MinD, and correct subcellular localization and mitotic partitioning of β-carboxysomes in S. elongatus PCC 7942, it was only recently proposed that the outer shell protein homologues CcmK3 and CcmK4 could also be required for this function (159). These proteins are individually required for the correct subcellular distribution of β-carboxysomes, but the ΔccmK3-4 mutant phenotype was beyond the expected deficit due to aberrant carboxysome partitioning; thus, the CcmK3 and CcmK4 proteins probably have further, as yet unidentified roles.
There is scant evidence supporting the same mechanisms for localization and mitotic partitioning of α-carboxysomes. In some genera, such as Nitrococcus and Halothiobacillus, the α-carboxysomes can seem to be distributed randomly within the cell in electron micrographs (77, 79, 80, 185), although the same can occasionally be said of the β-cyanobacterium S. elongatus PCC 7942, which has unambiguously well-ordered β-carboxysomes when imaged by fluorescence confocal microscopy (181). Nonetheless, there are some instances where α-carboxysomes are observed with filamentous structures (128), though the functional relationship between these filaments and the α-carboxysome has not been established. It should be noted that although other types of microcompartment appear to be subject to cytoskeleton-dependent subcellular localization and are associated with filament structures (186, 187), ectopic expression of the H. neapolitanus carboxysome in E. coli resulted in disordered α-carboxysomes (122). Intriguingly, though, most noncyanobacterial cso operons have a homologue of parA in the extended operon. These are perhaps most similar to the parA/parF homologues carried by mobile plasmids and prophages, and homologues of the cso-associated parA gene are generally found in prophages or integrated conjugative elements. Whether or not these contribute to partitioning or subcellular localization of α-carboxysomes remains to be seen and should provide a focus for future investigation.
GENE ORGANIZATION OF TYPICAL α- AND β-CARBOXYSOMES
The cso Operon Encodes Components of the α-Carboxysome
To date, all species with α-carboxysomes encode these components in a single operon. Depending on the species, the extended α-carboxysome (cso) operon may also contain other CCM genes, such as the CO2-uptake operon ndhF4D4-chpX in many α-cyanobacteria (36, 143) and genes for putative form 1A RubisCO chaperonins (11). In addition, the cso operons of many species contain other, seemingly extraneous genes (143); for instance, a gene with homology to the parA/parF family of bacterial actin homologues is found within some noncyanobacterial cso operons.
As a specific example, the canonical cso operon from H. neapolitanus C2 (Fig. 5) carries the genes for the large and small subunits of RubisCO form 1A (cbbL-cbbS), a protein thought to organize the interior of α-carboxysomes (csoS2), and the α-carboxysomal carbonic anhydrase enzyme (csoS3). These are followed by five short genes encoding small proteins with functions in the shell of α-carboxysomes: csoS4AB and csoS1A to -C. The products of the csoS1A to -C genes share the BMC domain, whereas the csoS4AB genes encode proteins with a distinct but functionally related domain (pfam number PF03319). Recently, a gene near the canonical cso operon in α-cyanobacteria and proteobacteria, termed csoS1D, was found to encode a protein with tandem BMC domains and that forms a quaternary structure similar to that of carboxysome shell proteins (120). In addition, the csoS1E gene, downstream of the cso operon in most α-cyanobacteria, encodes a protein that probably contains tandem BMC domains (112).
It is important that while all α-carboxysomes are thought to be transcribed from a single operon, these operons vary from species to species (Fig. 5). Roberts et al. (112) identified six types of α-cyanobacterial cso operons which had specific CCM-related and extraneous genes; one of these operon types lacked the csoS1E gene and was limited to the high-light-adapted Prochlorococcus clade. Interestingly, the six α-carboxysomal cso operon types closely match published phylogenies of α-cyanobacteria (36, 105), suggesting that the α-carboxysome was present in the ancestor of the α-cyanobacteria and has been distributed vertically by horizontal gene transfer since its initial introgression.
The ccm and Other Operons Encode the Components of the β-Carboxysome
In contrast to the conserved nature of the α-carboxysomal cso operon, the genes encoding components of the β-carboxysome are distributed throughout β-cyanobacterial genomes, although the core carboxysome component genes are most often found in the canonical ccm operon, and occasionally near the RubisCO rbcLS genes. In S. elongatus PCC 7942, the ccmK2-O, rbcLS (downstream of ccmK2-O), ccaA, and ccmK3-4 operons encode the known components of the carboxysome (Fig. 7). However, this gene set is highly variable from species to species, and β-cyanobacteria may have as many as eight homologues of ccmK (159); note that ccmO is a ccmK homologue that encodes a protein containing tandem BMC domains (150). These genes are homologues of csoS1A to -E and encode BMC proteins that are thought to form the outer shell of the β-carboxysome. In addition, ccmL is a homologue of the csoS4AB genes and contains the PF03319 domain. In addition to the known ccmK genes, Cai et al. (161) identified a β-cyanobacterial homologue of csoS1D, termed ccmP. While structural studies are forthcoming, the CcmP protein bears significant similarity to CsoS1D and may recapitulate structural peculiarities of the latter protein (161).
The ccmM and ccmN genes are always found in the core ccm operon (Fig. 5). Intriguingly, the alternative carboxysomal carbonic anhydrase gene, ccaA, is never found within the core ccm operon, and it is rare to find rbcLXS, encoding RubisCO form 1B and the assembly chaperonin RbcX, with the core ccm genes. Significantly, the β-carboxysome can contain different types of CA enzyme depending on the β-cyanobacterial lineage, with some species utilizing the carboxysomal carbonic anhydrase (CcaA) enzyme (170, 172) and some using the γ-CA-like domain in CcmM for this purpose (138). This isolated genomic position of ccaA is consistent with the view that ccaA was a late incorporation into β-cyanobacterial lineages.
Regulation of CCM/Carboxysome Genes
In general, the set of genes that make up the β-carboxysome are constitutively expressed with regard to a range of CO2 levels. However, fine adjustments in the relative expression of specific carboxysome gene transcripts have been observed, particularly at low CO2 levels. Studies using quantitative PCR analysis of transcript changes upon transfer of cells from 2 to 5% CO2 to air (Ci limitation) have found little change or transient small changes (2- to 5-fold increases) in the ccmKLMNO, ccaA, and rbcLXS genes (56, 188, 189). Changes found using microarray analysis have generally been small (190, 191). In comparison, low-CO2-responsive genes (coding for CO2- and HCO3−-uptake systems) such as bicA (except in PCC 6803), sbtAB, cmpABCD, porB (in PCC 7002), and ndhF3-ndhD3-chpY undergo large changes in transcript abundance, although sometimes these changes are transiently maximal over the first 2 h from the onset of induction (56, 188–191). An electron microscopic investigation found that carboxysome numbers per cell for cells grown under severe Ci limitation can be up to 4-fold higher than those for high-CO2-grown cells of Synechococcus UTEX 625 (equivalent to PCC 6301) (192). However, much of this extra carboxysome accumulation could occur due to a markedly increased time between cell divisions while maintaining a relatively constant synthesis of carboxysome components. Other studies have shown that there is a degree of coregulation between ccmM and ccaA expression and that there is permissible flexibility in the relative abundances of carboxysome components before reductions in carboxysome function become apparent (96).
In comparison, there is little information on the regulation of carboxysome genes in α-cyanobacteria. This has, however, been studied in some proteobacterial species, which for the most part contain distinct carboxysomal and noncarboxysomal RubisCO operons (11). Expression of the carboxysomal RubisCO operons in these organisms may not necessarily be responsive to changing Ci concentrations (66), though it has been observed that some species prefer noncarboxysomal RubisCO types during Ci-replete periods and that some repress all CO2 fixation during heterotrophic growth (193, 194). Thus, these species are in a constant process of metabolic streamlining, allowing them to forego the costly processes of α-carboxysome biogenesis and CO2 fixation.
REGULATORY ROLE FOR MICROCYSTINS IN THE β-CARBOXYSOME?
While carboxysomal RubisCO binds some proteins as a structural necessity, a number of other proteins are known to associate with RubisCO or carboxysomes. Notably, thioredoxin was observed to bind CcmM and RubisCO in Synechocystis PCC 6803 (195, 196), and the toxic nonribosomal cyclic peptide microcystin was shown to bind RubisCO in Microcystis aeruginosa PCC 7806 (197). The role of thioredoxin in the former example, or even whether this was an artifactual consequence of artificial cell fragmentation, was not discussed; however, further work in M. aeruginosa suggested that microcystin-bound RubisCO is less susceptible to proteolytic degradation. These results are generally supported by electron microscopic investigations of microcystin localization in which β-carboxysomes from M. aeruginosa PCC 7806 were hot spots of microcystin localization (198). The role of microcystins in the carboxysome is unclear; however, their production is strongly linked to the transition from high to low Ci in M. aeruginosa PCC 7806, with the microcystin maximum preceding peak Ci uptake (199). Some data suggest a relationship between microcystin production and photosynthesis (199–202). Perhaps, then, microcystins play a role in adaptation of cyanobacteria to limiting Ci in some environmental niches.
We perceive similar functional roles for microcystins and thioredoxins in cyanobacteria. These molecules could act as thioprotectants, reducing redox damage to carboxysome components during carboxysome biogenesis and maturation. Similarly, these moieties may alter carboxysomal CA enzyme activities depending on their subcellular location, i.e., as they transit the cytoplasm before incorporation into the nascent carboxysome. Thus, thioredoxins and microcystins may enhance carboxysome biogenesis and also protect the cytoplasmic HCO3− pool from CA activity during CA enzyme production, but more work is needed to determine a definitive link.
THE PYRENOID: A EUKARYOTIC ANALOGUE?
Similar to the CCMs of prokaryotes, most eukaryotic algae have structural and enzymatic features which enhance CO2 fixation: intracellular Ci accumulation by Ci pumps (sometimes assisted by extracellular carbonic anhydrase enzymes) and subsequent enzymatic conversion to CO2 in a RubisCO-rich compartment (24). While the carboxysome provides an ultrastructural focal point of the many prokaryotic CCMs, most eukaryotic algae and some basal bryophytes possess an alternative macromolecular structure that is thought to be of similar importance, namely, the pyrenoid (203–205). The pyrenoid is the region of chloroplast stroma which contains the majority of plastidic RubisCO; as such, it is the site of algal CO2 fixation (206, 207). To our current knowledge, a prerequisite for pyrenoids is the presence of an enzymatic CCM; however, pyrenoid-free eukaryotic CCMs do exist (208). Like the case for carboxysomes, multiple distinct morphotypes of pyrenoids are evident (4, 209); however, the structural components and function of these structures are currently poorly understood. It is known that some pyrenoids contain pyrenoid-localized CA enzymes (210), whereas others contain thylakoid invaginations/strands which may contain thylakoid-localized CA enzymes (211). Indeed, Markelova et al. (212) showed that the CAH3 CA enzyme was localized to both the thylakoid lumen and the pyrenoid body in Chlamydomonas reinhardtii, with this CA enzyme supporting CO2 fixation only when correctly localized to the pyrenoid-traversing thylakoids (213).
While the presence of CA activity in pyrenoids is long resolved, the structures of a potential pyrenoid shell or internal organizing proteins are unknown. Genkov et al. (214) recently established that assembly of the C. reinhardtii pyrenoid is dependent on the presence of the native RubisCO small subunit, as strains in which this had been replaced by the SSU from Helianthus annus lacked this body. Similarly, the CIA6 protein appears to play a vital, if unclear, role in pyrenoid formation, with RubisCO being distributed throughout the chloroplast stroma in a cia6 mutant (215). The latter study showed for the first time that the pyrenoid is required for proper function of the C. reinhardtii CCM, but the essential role of CIA6 remains unclear, with the intriguing possibilities being that this protein promotes pyrenoid formation through posttranslational modification of RubisCO or through linking of adjacent RubisCO holoenzymes, perhaps through their SSU as seen in α-carboxysomes (121, 215). Thus, there appear to be mechanistic similarities between carboxysomes and pyrenoids, but whether pyrenoids achieve a carboxysome-like level of structural sophistication remains an open question. It is nonetheless clear that the pyrenoid is paracrystalline at the ultrafine scale, with regular ordering of individual units apparent in early electron microscopic studies (we now interpret these units as RubisCO holoenzymes) (216–219).
The uncertainty over pyrenoid structure extends to the possibility of a pyrenoid shell structure, a concept which has been discussed extensively and seems unlikely (220). No candidate shell proteins have been identified, and early speculations suggesting a shell-like role for the starch sheaths commonly surrounding pyrenoids have proven to be unfitting (221). An important consideration in assessing the necessity of a pyrenoid shell structure is the kinetics of RubisCO enzymes from pyrenoid-containing organisms, which may account for the lack of a shell through altered substrate specificity (4). There is a possibility that a semipermeable shell structure may be unnecessary for effective pyrenoid function, especially given that the CO2/O2 specificity of most algal RubisCO enzymes is higher than that of the cyanobacterial enzymes (4, 222). Indeed, an evolutionary comparison of carboxysomes, pyrenoids, and pyrenoid-free chloroplasts with their respective RubisCO enzymes showed that the more complex structure is inversely correlated with the specificity of the cognate RubisCO enzymes (4).
The concept of pyrenoids as “skinless carboxysomes” is not an alien one. Reinhold et al. (88–90) proposed that a carboxysome (or any macromolecular RubisCO complex) need not have an exclusive shell structure if it has sufficient internal CA activity, whether it be focused at the center of the body or at multiple foci as current models of β-carboxysomes contend (96–99). Further work to investigate this hypothesis would increase our knowledge of the largely unknown structure of the pyrenoid.
RELATEDNESS OF CARBOXYSOMES TO OTHER BACTERIAL MICROCOMPARTMENTS
While it is tempting to view carboxysomes in isolation, BMC diversity is not limited to carboxysomes. Rather than a unique innovation of the CO2 fixation pathway, bacterial microcompartments are widespread in bacteria (113, 114, 155). The best studied of these are the propanediol (PDU) (223, 224) and ethanolamine (EUT) (225, 226) detoxification organelles, called metabolosomes or enterosomes. Additionally, comparative genomic approaches have revealed many putative operons encoding novel bacterial microcompartments with a wide range of functions, with some estimates suggesting that one-fifth of bacterial species possess the genetic capacity for microcompartment formation (114, 143). A common feature of these bacterial microcompartments is the sequestration of problematic metabolic pathways. For instance, while carboxysomes are thought to enhance RuBP carboxylation and to minimize oxygenation, PDU and EUT metabolosomes protect the cell from damage by sequestering reactive metabolic intermediates (227, 228).
The evolutionary relationships between different types of microcompartments are unclear, but numerous parallels are evident between them. They all contain a conserved outer shell structure, encapsulating problematic enzymatic reactions. The PDU and EUT microcompartments are reviewed in detail elsewhere (114–116).
POSSIBLE EVOLUTIONARY SEQUENCES THAT GAVE RISE TO FUNCTIONAL CARBOXYSOMES
Various mutant carboxysome phenotypes have been described which suggest an intriguing dichotomy in the evolution of the two types of carboxysome. As discussed above, β-carboxysomes appear to have high intrinsic order in three dimensions, their internal enzymatic structure could be largely self-assembled, and the outer semipermeable shell would be attached loosely to this central lumen. On the other hand, α-carboxysomes undoubtedly have a strong outer shell structure, which was shown to form even in the absence of the RubisCO enzyme (121). This could suggest the hypothesis that α-carboxysomes evolved by recruitment of the CO2 fixation machinery to existing BMC protein shells in primordial cells with the genetic ability to produce both.
In contrast, the available evidence tends to support an opposite evolutionary scheme for β-carboxysomes—the underlying bicarbonate dehydration/RubisCO-organizing complex (CcmM-58–CcaA–RbcLS–CcmM-35) has immediate appeal as an interim functional unit in the absence of the outer shell.
Simple Model for Evolution of β-Carboxysomes from a Naive Cell
The β-carboxysome may have evolved from a primordial photosynthetic CO2 fixation structure made from RubisCO and γ-CA (proto-CcmM) enzymes, while the α-carboxysome may have evolved by capture of a preexisting BMC shell structure. This highly ordered structure led to the speculative model outlining the evolution of the β-carboxysome which is presented below.
A set of possible steps in the evolution of a β-carboxysome, as we see them, is shown in Fig. 9. The first step may have involved a loose arrangement of RubisCO and CA, possibly giving a small advantage to the kinetics of RubisCO by maintaining the local supply rate of CO2 from ambient cellular Ci (Fig. 9, step 1). A second embellishment may have allowed specific colocalization of RubisCO and CA, possibly involving a third organizing protein. A contemporary example of this arrangement is in the ΔccmM ccmM-SD strain (97), and intriguing ccmM-like genes have been observed in the genera Acaryochloris and Leptolyngbya, where the γ-CA domain is fused to a single RubisCO SSU-like domain (96); once again, the kinetic advantage may have been quite small. A third development may have allowed a trimeric γ-CA to be fused to a RubisCO-organizing protein, producing an ordered RubisCO-CA aggregation (Fig. 9, step 2), or essentially a naked carboxysome looking like a ΔccmK2, ΔccmO, or ΔccmN mutant (143, 159). The most important step in the evolution of carboxysomes would have been the acquisition of the small BMC shell proteins from an existing organism with a BMC used for another purpose (Fig. 9, step 3). Although this step may have led to a significant elevation of CO2 around RubisCO, the full advantage of the system would not have become apparent until systems for active HCO3− uptake, and then CO2 uptake, had evolved (2) so that the concentration of HCO3− entering the carboxysome compartment could lead to maximal elevation of CO2 around RubisCO. Other refinements of these hypothetical primitive carboxysomes would have involved molecular evolution of the pores in CcmK-type hexamers to control HCO3− and RuBP entry (and PGA exit) while also allowing retardation of CO2 loss and O2 entry. Other refinements may have followed, such as abandoning the γ-CA function of CcmM (while retaining the RubisCO-organizing function) after the recruitment of the alternative CA enzyme CcaA (Fig. 9, step 4), potentially allowing better control over the CA/RubisCO ratio. Modeling (94, 148) has shown that too little CA or too much CA activity in the carboxysomes can have negative effects on the efficiency of CO2 fixation in the structures (too little CO2 supply in the first case and too much leakage in the second case). The possibly older strategy of employing γ-CA activity associated with CcmM trimers seems to be present in some of the filamentous cyanobacteria with large carboxysomes, such as Nostoc PCC 7120, whereas many unicellular strains (with smaller carboxysomes), such as S. elongatus PCC 7942, seem to have adopted the CcaA strategy (96) (Fig. 6B). The latter strategy might also have evolved a fine alteration of CcmN for the flexible recruitment of CcaA and the outer shell, as described previously (143).
Fig 9.
Putative evolutionary transitions from naive cells to those containing α- and β-carboxysomes. The schemes are described in the text.
Simple Model for Evolution of α-Carboxysomes from Preexisting Shell Structures
Whereas possible insights into β-carboxysome evolution have arisen from the use of β-cyanobacterial mutants with carboxysome phenotypes ranging from absent to wild-type appearance, the evolution of the α-carboxysome system is less clear, and a putative scheme is briefly described in Fig. 9. The incorporation of enzyme subunits through connection to the shell structure is a common feature of α-carboxysomes and PDU microcompartments (180), so it is conceivable that incorporation of form 1A RubisCO into the α-carboxysome may have arisen by invasion of an alternative form of microcompartment by divergent RubisCO and CA enzymes which were capable of incorporation into the forming microcompartment by using this targeting method (Fig. 9, step A) (2). This hypothesis relies on the coexistence of RubisCO and one of the alternative BMC types in the same bacterial strain, a scenario that is readily observed in database searches. Refinement of the BMC structure would then have followed (Fig. 9, step B). Interestingly, there are numerous examples of bacterial strains which contain both the pdu and eut operons simultaneously, such as Salmonella enterica serovar Typhimurium (229), and many strains, such as Rhodospirillum rubrum, which contain a RubisCO gene and a noncarboxysomal BMC operon (230). However, there is no evidence of a bacterial strain simultaneously containing an α- or β-carboxysomal operon and any other kind of BMC operon.
CONCLUSIONS
Carboxysomes form an essential part of the cyanobacterial CCM by acting as microcompartments which encapsulate the RubisCO enzyme and allow CO2 levels to be elevated around this relatively sluggish enzyme. Partly as a result of these unique polyhedral structures, cyanobacteria are remarkably productive on a global scale. Two types of carboxysomes arose by convergent evolution, namely, the α-carboxysomes, found predominantly in oceanic cyanobacteria, and the β-carboxysomes, found mainly in freshwater/estuarine cyanobacteria. Impressive progress has now been made in understanding the structure, physiology, and evolution of carboxysomes, but considerable gaps are identifiable.
The solved crystal structures of the small hexameric (CcmK1-4 and CsoS1ABC forms) and rarer pentameric (CcmL and CsoS4AB forms) shell proteins have been immensely helpful for the development of models that describe the likely structures of the outermost part of the icosahedral shells of both carboxysome types. However, provision of crystal structures for the RubisCO-organizing proteins (CcmM-58 and -35 forms), the potential “facet zipper” protein CcmO (or possibly CsoS1E in alpha types), and the large CsoS2 shell-associated protein CcmN (shell recruitment) has proved recalcitrant at this stage. Although technically difficult, a critical phase of crystal structure analyses is to also attempt to determine the fine structural basis of the functional interactions of two or more proteins, such as CcmM-58–RubisCO, CcmM-35–RubisCO, CcmK2-CcmO, CcmM-CcmN, CcmK2–CcmM-58, CsoS2-CsoSCA, etc., to allow the further refinement and validation of existing models.
Some of the questions yet to be answered are as follows:
Does the smaller RubisCO-organizing protein, CcmM-35, interact with RubisCO to the center of Synechococcus PCC 7942 β-carboxysomes as postulated?
Does the larger RubisCO-organizing protein, CcmM-58, interact with RubisCO at the inner shell layer of β-carboxysomes, as indicated by protein-protein interaction analysis?
What direct approaches can be used to confirm that models describing the icosahedral shell structure as being composed of triangular sheets composed of tiled small hexamers are accurate?
What direct approaches can be used to ascertain if the concave or convex face of CcmK2/CsoS1 hexamers face the cytoplasm?
Do specific charged and/or gated pores in the different forms of CcmK or CsoS1 hexamers really allow specificity toward specific solutes, such as RuBP, PGA, HCO3−, or Mg2+?
Can a presumed early evolutionary event, such as a naked RubisCO-CA aggregate structure, provide some advantage toward more efficient performance of RubisCO?
New Directions
There may be possible biotech applications involving the generation of new ion-selective membranes for battery applications and purification of water by osmosis. Other applications may involve the introduction of carboxysomes into the chloroplasts of crop plants in an effort to improve crop yields through the introduction of a minimal form of the cyanobacterial CCM (231).
ACKNOWLEDGMENTS
B.D.R. was supported by an ANU Ph.D. scholarship. G.D.P. and M.R.B. thank the Australian Research Council for support via the Discovery grant program.
We thank Britta Forster for helpful comments, Loraine Tucker for excellent technical assistance, and Lynne Whitehead for EM images of Cyanobium.
REFERENCES
- 1. Badger MR, Hanson D, Price GD. 2002. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 29:161–173 [DOI] [PubMed] [Google Scholar]
- 2. Badger MR, Price GD. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609–622 [DOI] [PubMed] [Google Scholar]
- 3. Raven JA. 2003. Inorganic carbon concentrating mechanisms in relation to the biology of algae. Photosynth. Res. 77:155–171 [DOI] [PubMed] [Google Scholar]
- 4. Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD. 1998. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can. J. Bot. 76:1052–1071 [Google Scholar]
- 5. Cleland WW, Andrews TJ, Gutteridge S, Hartman FC, Lorimer GH. 1998. Mechanism of RUBISCO—the carbamate as general base. Chem. Rev. 98:549–561 [DOI] [PubMed] [Google Scholar]
- 6. Spreitzer RJ, Salvucci ME. 2002. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53:449–475 [DOI] [PubMed] [Google Scholar]
- 7. Whitney SM, Houtz RL, Alonso H. 2011. Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco. Plant Physiol. 155:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tcherkez GGB, Farquhar GD, Andrews TJ. 2006. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U. S. A. 103:7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berner RA. 2006. GEOCARBSULF: a combined model for Phanerozoic atmospheric O2 and CO2. Geochim. Cosmochim. Acta 70:5653–5664 [Google Scholar]
- 10. Raven JA. 1997. The role of marine biota in the evolution of terrestrial biota: gases and genes. Atmospheric composition and evolution of terrestrial biota. Biogeochemistry 39:139–164 [Google Scholar]
- 11. Badger MR, Bek EJ. 2008. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59:1525–1541 [DOI] [PubMed] [Google Scholar]
- 12. Sage RF. 2004. The evolution of C4 photosynthesis. New Phytol. 161:341–370 [DOI] [PubMed] [Google Scholar]
- 13. Raven JA, Giordano M, Beardall J, Maberly SC. 2012. Algal evolution in relation to atmospheric CO2: carboxylases, carbon-concentrating mechanisms and carbon oxidation cycles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Price GD, Badger MR, Woodger FJ, Long BM. 2008. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59:1441–1461 [DOI] [PubMed] [Google Scholar]
- 15. Price GD. 2011. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth. Res. 109:47–57 [DOI] [PubMed] [Google Scholar]
- 16. Cannon GC, Shively JM. 1983. Characterization of a homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch. Microbiol. 134:52–59 [Google Scholar]
- 17. Holthuijzen YA, Kuenen JG, Konings WN. 1987. Activity of ribulose-1,5-bisphosphate carboxylase in intact and disrupted carboxysomes of Thiobacillus neapolitanus. FEMS Microbiol. Lett. 42:121–124 [Google Scholar]
- 18. Dobrinski KP, Longo DL, Scott KM. 2005. The carbon-concentrating mechanism of the hydrothermal vent chemolithoautotroph Thiomicrospira crunogena. J. Bacteriol. 187:5761–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price GD, Badger MR. 1989. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype. Evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol. 91:505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Volokita M, Zenvirth D, Kaplan A, Reinhold L. 1984. Nature of the inorganic carbon species actively taken up by the cyanobacterium Anabaena variabilis. Plant Physiol. 76:599–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lanaras T, Hawthornthwaite AM, Codd GA. 1985. Localization of carbonic anhydrase in the cyanobacterium Chlorogloeopsis fritschii. FEMS Microbiol. Lett. 26:285–288 [Google Scholar]
- 22. Yagawa Y, Shiraiwa Y, Miyachi S. 1984. Carbonic anhydrase from the blue-green alga (cyanobacterium) Anabaena variabilis. Plant Cell Physiol. 25:775–783 [Google Scholar]
- 23. Wang Y, Duanmu D, Spalding MH. 2011. Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynth. Res. 109:115–122 [DOI] [PubMed] [Google Scholar]
- 24. Spalding MH. 2008. Microalgal carbon-dioxide-concentrating mechanisms: Chlamydomonas inorganic carbon transporters. J. Exp. Bot. 59:1463–1473 [DOI] [PubMed] [Google Scholar]
- 25. Badger MR, Spalding MH. 2004. CO2 acquisition, concentration and fixation in cyanobacteria and algae, p 369–397 In Leegood RC, Sharkey TD, Von CaemmererS. (ed), Photosynthesis: physiology and metabolism, vol 1 Kluwer, Dordrecht, The Netherlands. [Google Scholar]
- 26. Morel FMM, Cox EH, Kraepiel AML, Lane TW, Milligan AJ, Schaperdoth I, Reinfelder JR, Tortell PD. 2002. Acquisition of inorganic carbon by the marine diatom Thalassiosira weissflogii. Funct. Plant Biol. 29:301–308 [DOI] [PubMed] [Google Scholar]
- 27. Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. 2008. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. U. S. A. 105:17199–17204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Badger MR. 1980. Kinetic-properties of ribulose 1,5-bisphosphate carboxylase-oxygenase from Anabaena variabilis. Arch. Biochem. Biophys. 201:247–254 [DOI] [PubMed] [Google Scholar]
- 29. Schwarz R, Reinhold L, Kaplan A. 1995. Low activation state of ribulose-1,5-bisphosphate carboxylase oxygenase in carboxysome-defective Synechococcus mutants. Plant Physiol. 108:183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Price GD, Coleman JR, Badger MR. 1992. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 100:784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evans JR. 1989. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78:9–19 [DOI] [PubMed] [Google Scholar]
- 32. Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere—integrating terrestrial and oceanic components. Science 281:237–240 [DOI] [PubMed] [Google Scholar]
- 33. Liu HB, Nolla HA, Campbell L. 1997. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 12:39–47 [Google Scholar]
- 34. Liu HB, Landry MR, Vaulot D, Campbell L. 1999. Prochlorococcus growth rates in the central equatorial Pacific: an application of the fmax approach. J. Geophys. Res. Oceans 104:3391–3399 [Google Scholar]
- 35. Badger MR, Price GD, Long BM, Woodger FJ. 2006. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot. 57:249–265 [DOI] [PubMed] [Google Scholar]
- 36. Rae BD, Forster B, Badger MR, Price GD. 2011. The CO2-concentrating mechanism of Synechococcus WH5701 is composed of native and horizontally-acquired components. Photosynth. Res. 109:59–72 [DOI] [PubMed] [Google Scholar]
- 37. Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, Kaplan A, Ogawa T. 2002. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria—function and phylogenetic analysis. J. Biol. Chem. 277:18658–18664 [DOI] [PubMed] [Google Scholar]
- 38. Zhang P, Battchikova N, Jansen T, Appel J, Ogawa T, Aro EM. 2004. Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16:3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Price GD, Shelden MC, Howitt SM. 2011. Membrane topology of the cyanobacterial bicarbonate transporter, SbtA, and identification of potential regulatory loops. Mol. Membr. Biol. 28:265–275 [DOI] [PubMed] [Google Scholar]
- 40. Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. 2004. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 101:18228–18233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price GD, Howitt SM. 2011. The cyanobacterial bicarbonate transporter BicA: its physiological role and the implications of structural similarities with human SLC26 transporters. Biochem. Cell Biol. 89:178–188 [DOI] [PubMed] [Google Scholar]
- 42. Shelden MC, Howitt SM, Price GD. 2010. Membrane topology of the cyanobacterial bicarbonate transporter, BicA, a member of the SulP (SLC26A) family. Mol. Membr. Biol. 27:12–23 [DOI] [PubMed] [Google Scholar]
- 43. Omata T, Takahashi Y, Yamaguchi O, Nishimura T. 2002. Structure, function and regulation of the cyanobacterial high-affinity bicarbonate transporter, BCT1. Funct. Plant Biol. 29:151–159 [DOI] [PubMed] [Google Scholar]
- 44. Omata T, Price GD, Badger MR, Okamura M, Gohta S, Ogawa T. 1999. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp strain PCC 7942. Proc. Natl. Acad. Sci. U. S. A. 96:13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Rozycki T, Schultzel MA, Saier MH., Jr 2004. Sequence analyses of cyanobacterial bicarbonate transporters and their homologues. J. Mol. Microbiol. Biotechnol. 7:102–108 [DOI] [PubMed] [Google Scholar]
- 46. Ohkawa H, Price GD, Badger MR, Ogawa T. 2000. Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp strain PCC 6803. J. Bacteriol. 182:2591–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohkawa H, Pakrasi HB, Ogawa T. 2000. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp strain PCC6803. J. Biol. Chem. 275:31630–31634 [DOI] [PubMed] [Google Scholar]
- 48. Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T. 2001. Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc. Natl. Acad. Sci. U. S. A. 98:11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maeda S, Badger MR, Price GD. 2002. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol. Microbiol. 43:425–435 [DOI] [PubMed] [Google Scholar]
- 50. Ohkawa H, Sonoda M, Hagino N, Shibata M, Pakrasi HB, Ogawa T. 2002. Functionally distinct NAD(P)H dehydrogenases and their membrane localization in Synechocystis sp PCC6803. Funct. Plant Biol. 29:195–200 [DOI] [PubMed] [Google Scholar]
- 51. Folea IM, Zhang P, Nowaczyk MM, Ogawa T, Aro E-M, Boekema EJ. 2008. Single particle analysis of thylakoid proteins from Thermosynechococcus elongatus and Synechocystis 6803: localization of the CupA subunit of NDH-1. FEBS Lett. 582:249–254 [DOI] [PubMed] [Google Scholar]
- 52. Xu M, Ogawa T, Pakrasi HB, Mi H. 2008. Identification and localization of the CupB protein involved in constitutive CO2 uptake in the cyanobacterium, Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 49:994–997 [DOI] [PubMed] [Google Scholar]
- 53. Price GD, Maeda S, Omata T, Badger MR. 2002. Modes of active inorganic carbon uptake in the cyanobacterium, Synechococcus sp PCC7942. Funct. Plant Biol. 29:131–149 [DOI] [PubMed] [Google Scholar]
- 54. McGinn PJ, Jones MJ, Macdonald AB, Campbell DA. 2005. Light is required for low-CO2-mediated induction of transcripts encoding components of the CO2-concentrating mechanism in the cyanobacterium Synechococcus elongatus: analysis by quantitative reverse transcription-polymerase chain reaction. Can. J. Bot. 83:711–720 [Google Scholar]
- 55. McGinn PJ, Price GD, Badger MR. 2004. High light enhances the expression of low-CO2-inducible transcripts involved in the CO2-concentrating mechanism in Synechocystis sp PCC6803. Plant Cell Environ. 27:615–626 [Google Scholar]
- 56. McGinn PJ, Price GD, Maleszka R, Badger MR. 2003. Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp strain PCC6803. Plant Physiol. 132:218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Woodger FJ, Badger MR, Price GD. 2005. Regulation of cyanobacterial CO2-concentrating mechanisms through transcriptional induction of high-affinity Ci-transport systems. Can. J. Bot. 83:698–710 [Google Scholar]
- 58. Woodger FJ, Badger MR, Price GD. 2005. Sensing of inorganic carbon limitation in Synechococcus PCC7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol. 139:1959–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Badger MR, Andrews TJ. 1982. Photosynthesis and inorganic carbon usage by the marine cyanobacterium, Synechococcus sp. Plant Physiol. 70:517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Espie GS, Miller AG, Canvin DT. 1989. Selective and reversible inhibition of active CO2 transport by hydrogen sulfide in a cyanobacterium. Plant Physiol. 91:387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miller AG, Espie GS, Canvin DT. 1990. Physiological aspects of CO2 and HCO3− transport by cyanobacteria: a review. Can. J. Bot. 68:1291–1302 [Google Scholar]
- 62. Fridlyand L, Kaplan A, Reinhold L. 1996. Quantitative evaluation of the role of a putative CO2-scavenging entity in the cyanobacterial CO2-concentrating mechanism. Biosystems 37:229–238 [DOI] [PubMed] [Google Scholar]
- 63. Kaplan A, Zenvirth D, Marcus Y, Omata T, Ogawa T. 1987. Energization and activation of inorganic carbon uptake by light in cyanobacteria. Plant Physiol. 84:210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sültemeyer D, Klughammer B, Badger MR, Price GD. 1998. Fast induction of high-affinity HCO3− transport in cyanobacteria. Plant Physiol. 116:183–192 [Google Scholar]
- 65. Sültemeyer D, Klughammer B, Badger MR, Price GD. 1998. Protein phosphorylation and its possible involvement in the induction of the high-affinity CO2 concentrating mechanism in cyanobacteria. Can. J. Bot. 76:954–961 [Google Scholar]
- 66. Heinhorst S, Cannon GC, Shively JM. 2006. Carboxysomes and carboxysome-like inclusions, p 141–165 In Shively J. (ed), Complex intracellular structures in prokaryotes, vol 2 Springer, Berlin, Germany [Google Scholar]
- 67. Holthuijzen YA, van Dissel-Emiliana FFM, Kuenen JG, Konings WN. 1987. Energetic aspects of CO2 uptake in Thiobacillus neapolitanus. Arch. Microbiol. 147:285–290 [Google Scholar]
- 68. Karagouni AD, Kelly DP. 1989. Carbon dioxide fixation by Thiobacillus versutus: apparent absence of a CO2-concentrating mechanism in organisms grown under carbon-limitation in the chemostat. FEMS Microbiol. Lett. 58:179–182 [Google Scholar]
- 69. Kelly D, Wood A, Stackebrandt E. 2005. Thiobacillus Beijerinck 1904b, 597AL, p 764–769 In Garrity GM, Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2, part C. The proteobacteria: the alpha-, beta-, delta-, and epsilonproteobacteria Springer-Verlag, New York, NY [Google Scholar]
- 70. Kuenen JG, Beudeker RF, Shively JM, Codd GA. 1982. Microbiology of thiobacilli and other sulphur-oxidizing autotrophs, mixotrophs and heterotrophs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 298:473–497 [DOI] [PubMed] [Google Scholar]
- 71. Harrison AP., Jr 1984. The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 38:265–292 [DOI] [PubMed] [Google Scholar]
- 72. Dobrinski KP, Enkemann SA, Yoder SJ, Haller E, Scott KM. 2012. Transcriptional response of the sulfur chemolithoautotroph Thiomicrospira crunogena to dissolved inorganic carbon limitation. J. Bacteriol. 194:2074–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. So AKC, Van Spall HGC, Coleman JR, Espie GS. 1998. Catalytic exchange of 18O from 13C18O-labelled CO2 by wild-type cells and ecaA, ecaB, and ccaA mutants of the cyanobacteria Synechococcus PCC7942 and Synechocystis PCC6803. Can. J. Bot 76:1153–1160 [Google Scholar]
- 74. Wildon DC, Mercer FV. 1963. The ultrastructure of the vegetative cell of blue-green algae. Aust. J. Biol. Sci. 16:585–596 [Google Scholar]
- 75. Hall WT, Claus G. 1961. Electron microscope studies on ultrathin sections of Oscillatoria chalybea Mertens. Protoplasma 54:355–368 [Google Scholar]
- 76. Gantt E, Conti SF. 1969. Ultrastructure of blue-green algae. J. Bacteriol. 97:1486–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shively JM, Decker GL, Greenawalt JW. 1970. Comparative ultrastructure of the thiobacilli. J. Bacteriol. 101:618–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jensen TE, Bowen CC. 1961. Organization of the centroplasm in Nostoc pruniforme. Proc. Iowa Acad. Sci. 68:89–96 [Google Scholar]
- 79. Shively JM, Ball F, Brown DH, Saunders RE. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584–586 [DOI] [PubMed] [Google Scholar]
- 80. Shively JM, Ball FL, Kline BW. 1973. Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J. Bacteriol. 116:1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Codd GA, Stewart WDP. 1976. Polyhedral bodies and ribulose 1,5-diphosphate carboxylase of blue-green alga Anabaena cylindrica. Planta 130:323–326 [DOI] [PubMed] [Google Scholar]
- 82. Beudeker RF, Cannon GC, Kuenen JG, Shively JM. 1980. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch. Microbiol. 124:185–189 [Google Scholar]
- 83. Stewart WDP, Codd GA. 1975. Polyhedral bodies (carboxysomes) of nitrogen-fixing blue-green algae. Br. Phycol. J. 10:273–278 [Google Scholar]
- 84. Cossar JD, Rowell P, Darling AJ, Murray S, Codd GA, Stewart WDP. 1985. Localization of ribulose 1,5-bisphosphate carboxylase/oxygenase in the N2-fixing cyanobacterium Anabaena cylindrica. FEMS Microbiol. Lett. 28:65–68 [Google Scholar]
- 85. Lanaras T, Codd GA. 1981. Ribulose 1,5-bisphosphate carboxylase and polyhedral bodies of Chlorogloeopsis fritschii. Planta 153:279–285 [DOI] [PubMed] [Google Scholar]
- 86. Lanaras T, Codd GA. 1981. Structural and immunoelectrophoretic comparison of soluble and particulate ribulose bisphosphate carboxylases from the cyanobacterium Chlorogloeopsis fritschii. Arch. Microbiol. 130:213–217 [Google Scholar]
- 87. Price GD, Badger MR. 1989. Ethoxyzolamide inhibition of CO2 uptake in the cyanobacterium Synechococcus PCC7942 without apparent inhibition of internal carbonic anhydrase activity. Plant Physiol. 89:37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Reinhold L, Kosloff R, Kaplan A. 1991. A model for inorganic carbon fluxes and photosynthesis in cyanobacterial carboxysomes. Can. J. Bot. 69:984–988 [Google Scholar]
- 89. Reinhold L, Zviman M, Kaplan A. 1987. Inorganic carbon fluxes and photosynthesis in cyanobacteria—a quantitative model, p 289–296 In Biggins J. (ed), Proceedings of the VIIth International Congress on Photosynthesis, Providence, Rhode Island, USA, August 10–15, 1986, vol 4 M. Nijhoff Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 90. Reinhold L, Zviman M, Kaplan A. 1989. A quantitative model for inorganic carbon fluxes and photosynthesis in cyanobacteria. Plant Physiol. Biochem. 27:945–954 [Google Scholar]
- 91. Dou Z, Heinhorst S, Williams E, Murin C, Shively J, Cannon G. 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283:10377–10384 [DOI] [PubMed] [Google Scholar]
- 92. Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4:e7521. 10.1371/journal.pone.0007521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Price GD, Badger MR. 1991. Evidence for the role of carboxysomes in the cyanobacterial CO2-concentrating mechanism. Can. J. Bot. 69:963–973 [Google Scholar]
- 94. Price GD, Badger MR. 1989. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942. Two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the carboxysome. Plant Physiol. 91:514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Baker SH, Williams DS, Aldrich HC, Gambrell AC, Shively JM. 2000. Identification and localization of the carboxysome peptide Csos3 and its corresponding gene in Thiobacillus neapolitanus. Arch. Microbiol. 173:278–283 [DOI] [PubMed] [Google Scholar]
- 96. Long BM, Rae BD, Badger MR, Price GD. 2011. Over-expression of the β-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth. Res. 109:33–45 [DOI] [PubMed] [Google Scholar]
- 97. Long BM, Tucker L, Badger MR, Price GD. 2010. Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 153:285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Long BM, Badger MR, Whitney SM, Price GD. 2007. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 282:29323–29335 [DOI] [PubMed] [Google Scholar]
- 99. Cot SSW, So AKC, Espie GS. 2008. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J. Bacteriol. 190:936–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tabita FR. 1999. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 60:1–28 [Google Scholar]
- 101. Rippka R, Deruelles J, Waterbury J, Herdman M, Stanier R. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111:1–61 [Google Scholar]
- 102. Zehr JP, Bench SR, Carter BJ, Hewson I, Niazi F, Shi T, Tripp HJ, Affourtit JP. 2008. Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science 322:1110–1112 [DOI] [PubMed] [Google Scholar]
- 103. Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D, Kuypers MMM, Zehr JP. 2012. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337:1546–1550 [DOI] [PubMed] [Google Scholar]
- 104. Larsson J, Nylander JA, Bergman B. 2011. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol. Biol. 11:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Criscuolo A, Gribaldo S. 2011. Large-scale phylogenomic analyses indicate a deep origin of primary plastids within cyanobacteria. Mol. Biol. Evol. 28:13019–13032 [DOI] [PubMed] [Google Scholar]
- 106. Blank CE, Sanchez-Baracaldo P. 2010. Timing of morphological and ecological innovations in the cyanobacteria—a key to understanding the rise in atmospheric oxygen. Geobiology 8:1–23 [DOI] [PubMed] [Google Scholar]
- 107. Marin B, Nowack ECM, Glockner G, Melkonian M. 2007. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol. Biol. 7:85. 10.1186/1471-2148-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Espie GS, Kimber MS. 2011. Carboxysomes: cyanobacterial RubisCO comes in small packages. Photosynth. Res. 109:7–20 [DOI] [PubMed] [Google Scholar]
- 109. Erwin PM, Lopez-Legentil S, Turon X. 2012. Ultrastructure, molecular phylogenetics, and chlorophyll a content of novel cyanobacterial symbionts in temperate sponges. Microb. Ecol. 64:771–783 [DOI] [PubMed] [Google Scholar]
- 110. Kelly D, Harrison A. 1989. Genus Thiobacillus, p 1842–1858 In Staley JT, Bryant MP. (ed), Bergey's manual of systematic bacteriology, vol 3 Williams & Wilkins, Baltimore, MD [Google Scholar]
- 111. Kelly DP, Wood AP. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50:511–516 [DOI] [PubMed] [Google Scholar]
- 112. Roberts EW, Cai F, Kerfeld CA, Cannon GC, Heinhorst S. 2012. Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J. Bacteriol. 194:787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kinney JN, Axen SD, Kerfeld CA. 2011. Comparative analysis of carboxysome shell proteins. Photosynth. Res. 109:21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kerfeld CA, Heinhorst S, Cannon GC. 2010. Bacterial microcompartments. Annu. Rev. Microbiol. 64:391–408 [DOI] [PubMed] [Google Scholar]
- 115. Yeates TO, Thompson MC, Bobik TA. 2011. The protein shells of bacterial microcompartment organelles. Curr. Opin. Struct. Biol. 21:223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yeates T, Crowley C, Tanaka S. 2010. Bacterial microcompartment organelles: protein shell structure and evolution. Annu. Rev. Biophys. 39:185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tanaka S, Sawaya M, Phillips M, Yeates T. 2009. Insights from multiple structures of the shell proteins from the β-carboxysome. Protein Sci. 18:108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. 2007. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 5:e144. 10.1371/journal.pbio.0050144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tsai Y, Sawaya MR, Yeates TO. 2009. Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallogr. D Biol. Crystallogr. 65:980–988 [DOI] [PubMed] [Google Scholar]
- 120. Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392:319–333 [DOI] [PubMed] [Google Scholar]
- 121. Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC. 2008. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. PLoS One 3:e3570. 10.1371/journal.pone.0003570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. 2012. Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. U. S. A. 109:478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083–1086 [DOI] [PubMed] [Google Scholar]
- 124. Baker SH, Jin SM, Aldrich HC, Howard GT, Shively JM. 1998. Insertion mutation of the form I cbbL gene encoding ribulose bisphosphate carboxylase/oxygenase (Rubisco) in Thiobacillus neapolitanus results in expression of form II Rubisco, loss of carboxysomes, and an increased CO2 requirement for growth. J. Bacteriol. 180:4133–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Schmid M, Paredes A, Khant H, Soyer F, Aldrich H, Chiu W, Shively J. 2006. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J. Mol. Biol. 364:526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Holthuijzen YA, Vanbreem JF, Konings WN, Vanbrugg EF. 1986. Electron-microscopic studies of carboxysomes of Thiobacillus neapolitanus. Arch. Microbiol. 144:258–262 [Google Scholar]
- 127. Holthuijzen YA, Vanbreem JF, Kuenen JG, Konings WN. 1986. Protein composition of the carboxysomes of Thiobacillus neapolitanus. Arch. Microbiol. 144:398–404 [Google Scholar]
- 128. Iancu CV, Morris DM, Dou ZC, Heinhorst S, Cannon GC, Jensen GJ. 2010. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. 2007. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 372:764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Baker SH, Lorbach SC, Rodriguez-Buey M, Williams DS, Aldrich HC, Shively JM. 1999. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch. Microbiol. 172:233–239 [DOI] [PubMed] [Google Scholar]
- 131. Gonzales AD, Light YK, Zhang ZD, Iqbal T, Lane TW, Martino A. 2005. Proteomic analysis of the CO2-concentrating mechanism in the open-ocean cyanobacterium Synechococcus WH8102. Can. J. Bot. 83:735–745 [Google Scholar]
- 132. So AKC, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC. 2004. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J. Bacteriol. 186:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sawaya MR, Cannon GC, Heinhorst S, Tanaka S, Williams EB, Yeates TO, Kerfeld CA. 2006. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J. Biol. Chem. 281:7546–7555 [DOI] [PubMed] [Google Scholar]
- 134. Cannon GC, Baker SH, Soyer F, Johnson DR, Bradburne CE, Mehlman JL, Davies PS, Jiang QL, Heinhorst S, Shively JM. 2003. Organization of carboxysome genes in the Thiobacilli. Curr. Microbiol. 46:115–119 [DOI] [PubMed] [Google Scholar]
- 135. Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. 2013. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 64:787–798 [DOI] [PubMed] [Google Scholar]
- 136. Price GD, Sültemeyer D, Klughammer B, Ludwig M, Badger MR. 1998. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains—a review of general physiological characteristics, genes, proteins, and recent advances. Can. J. Bot. 76:973–1002 [Google Scholar]
- 137. Long BM, Price GD, Badger MR. 2005. Proteomic assessment of an established technique for carboxysome enrichment from Synechococcus PCC7942. Can. J. Bot. 83:746–757 [Google Scholar]
- 138. Peña KL, Castel SE, de Araujo C, Espie GS, Kimber MS. 2010. Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM. Proc. Natl. Acad. Sci. U. S. A. 107:2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Alber BE, Ferry JG. 1994. A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc. Natl. Acad. Sci. U. S. A. 91:6909–6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Price GD, Howitt SM, Harrison K, Badger MR. 1993. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC 7942 involved in carboxysome assembly and function. J. Bacteriol. 175:2871–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ludwig M, Sültemeyer D, Price G. 2000. Isolation of ccmKLMN genes from the marine cyanobacterium, Synechococcus sp. PCC7002 (Cyanophyceae), and evidence that CcmM is essential for carboxysome assembly. J. Phycol. 36:1109–1119 [Google Scholar]
- 142. Ogawa T, Amichay D, Gurevitz M. 1994. Isolation and characterization of the ccmM gene required by the cyanobacterium Synechocystis PCC6803 for inorganic carbon utilization. Photosynth. Res. 39:183–190 [DOI] [PubMed] [Google Scholar]
- 143. Kinney JN, Salmeen A, Cai F, Kerfeld CA. 2012. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 287:17729–17736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kaneko Y, Danev R, Nagayama K, Nakamoto H. 2006. Intact carboxysomes in a cyanobacterial cell visualized by Hilbert differential contrast transmission electron microscopy. J. Bacteriol. 188:805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Getzoff TP, Zhu GH, Bohnert HJ, Jensen RG. 1998. Chimeric Arabidopsis thaliana ribulose-1,5-bisphosphate carboxylase/oxygenase containing a pea small subunit protein is compromised in carbamylation. Plant Physiol. 116:695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Newman J, Gutteridge S. 1993. The X-ray structure of Synechococcus ribulose-bisphosphate carboxylase oxygenase-activated quaternary complex at 2.2-angstrom resolution. J. Biol. Chem. 268:25876–25886 [PubMed] [Google Scholar]
- 147. Kerfeld C, Sawaya M, Tanaka S, Nguyen C, Phillips M, Beeby M, Yeates T. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–938 [DOI] [PubMed] [Google Scholar]
- 148. Badger MR, Price GD, Jian WY. 1991. Selection and analysis of mutants of the CO2-concentrating mechanism in cyanobacteria. Can. J. Bot. 69:974–983 [Google Scholar]
- 149. Ronen-Tarazi M, Lieman Hurwitz J, Gabay C, Orus MI, Kaplan A. 1995. The genomic region of rbcLS in Synechococcus sp. PCC 7942 contains genes involved in the ability to grow under low CO2 concentration and in chlorophyll biosynthesis. Plant Physiol. 108:1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Friedberg D, Kaplan A, Ariel R, Kessel M, Seijffers J. 1989. The 5′-flanking region of the gene encoding the large subunit of ribulose-1,5-bisphoshate carboxylase/oxygenase is crucial for growth of the cyanobacterium Synechococcus sp. strain PCC7942 at the level of CO2 in air. J. Bacteriol. 171:6069–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Friedberg D, Jager KM, Kessel M, Silman NJ, Bergman B. 1993. Rubisco but not Rubisco activase is clustered in the carboxysomes of the cyanobacterium Synechococcus sp. PCC 7942: mud-induced carboxysomeless mutants. Mol. Microbiol. 9:1193–1201 [DOI] [PubMed] [Google Scholar]
- 152. Schwarz R, Friedberg D, Kaplan A. 1988. Is there a role for the 42 kilodalton polypeptide in inorganic carbon uptake by cyanobacteria? Plant Physiol. 88:284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Lieman-Hurwitz J, Schwarz R, Martinez F, Maor Z, Reinhold L, Kaplan A. 1991. Molecular analysis of high CO2 requiring mutants: involvement of genes in the region of rbc, including rbcS, in the ability of cyanobacteria to grow under low CO2. Can. J. Bot. 69:945–950 [Google Scholar]
- 154. Schwarz R, Lieman-Hurwitz J, Marco E, Ronen-Tarazi M, Ohad N, Hassidim M, Gabay C, Reinhold L, Kaplan A. 1992. The CO2-concentrating mechanism of cyanobacteria: elucidation with the aid of high-CO2-requiring mutants, p 437–440 In Murata N. (ed), Research in photosynthesis, vol 3 Kluwer, Dordrecht, Netherlands [Google Scholar]
- 155. Shively JM, Bradburne CE, Aldrich HC, Bobik TA, Mehlman JL, Jin S, Baker SH. 1998. Sequence homologs of the carboxysomal polypeptide csoS1 of the Thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes—polyhedral bodies. Can. J. Bot. 76:906–916 [Google Scholar]
- 156. Cannon GC, Heinhorst S, Bradburne CE, Shively JM. 2002. Carboxysome genomics: a status report. Funct. Plant Biol. 29:175–182 [DOI] [PubMed] [Google Scholar]
- 157. Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ. Microbiol. 67:5351–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhang SL, Laborde SM, Frankel LK, Bricker TA. 2004. Four novel genes required for optimal photoautotrophic growth of the cyanobacterium Synechocystis sp strain PCC 6803 identified by in vitro transposon mutagenesis. J. Bacteriol. 186:875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Rae BD, Long BM, Badger MR, Price GD. 2012. Structural determinants of the outer shell of β-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL. PLoS One 7:e43871. 10.1371/journal.pone.0043871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Samborska B, Kimber MS. 2012. A dodecameric CcmK2 structure suggests β-carboxysomal shell facets have a double-layered organization. Structure 20:1353–1362 [DOI] [PubMed] [Google Scholar]
- 161. Cai F, Kerfeld CA, Sandh G. 2012. Bioinformatic identification and structural characterization of a new carboxysome shell protein, p 345–356 In Burnap R, Vermaas W. (ed), Functional genomics and evolution of photosynthetic systems, vol 33 Springer, Dordrecht, Netherlands [Google Scholar]
- 162. Satoh R, Himeno M, Wadano A. 1997. Carboxysomal diffusion resistance to ribulose 1,5-bisphosphate and 3-phosphoglycerate in the cyanobacterium Synechococcus PCC7942. Plant Cell Physiol. 38:769–775 [Google Scholar]
- 163. Marco E, Martinez I, Ronen Tarazi M, Orus MI, Kaplan A. 1994. Inactivation of ccmO in Synechococcus sp. strain PCC 7942 results in a mutant requiring high levels of CO2. Appl. Environ. Microbiol. 60:1018–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Martinez I, Orus MI, Marco E. 1997. Carboxysome structure and function in a mutant of Synechococcus that requires high levels of CO2 for growth. Plant Physiol. Biochem. 35:137–146 [Google Scholar]
- 165. Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. 2010. Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J. Biol. Chem. 285:37838–37846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sagermann M, Ohtaki A, Nikolakakis K. 2009. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc. Natl. Acad. Sci. U. S. A. 106:8883–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. So AKC, Espie GS. 2005. Cyanobacterial carbonic anhydrases. Can. J. Bot. 83:721–734 [Google Scholar]
- 168. Heinhorst S, Williams E, Cai F, Murin C, Shively J, Cannon G. 2006. Characterization of the carboxysomal carbonic anhydrase CsoSCA from Halothiobacillus neapolitanus. J. Bacteriol. 188:8087–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Cannon GC, Heinhorst S, Kerfeld CA. 2010. Carboxysomal carbonic anhydrases: structure and role in microbial CO2 fixation. Biochim. Biophys. Acta 1804:382–392 [DOI] [PubMed] [Google Scholar]
- 170. Yu JW, Price GD, Song L, Badger MR. 1992. Isolation of a putative carboxysomal carbonic anhydrase gene from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 100:794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Fukuzawa H, Suzuki E, Komukai Y, Miyachi S. 1992. A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc. Natl. Acad. Sci. U. S. A. 89:4437–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. So AKC, Espie GS. 1998. Cloning, characterization and expression of carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Plant Mol. Biol. 37:205–215 [DOI] [PubMed] [Google Scholar]
- 173. So AKC, Cot SSW, Espie GS. 2002. Characterization of the C-terminal extension of carboxysomal carbonic anhydrase from Synechocystis sp PCC6803. Funct. Plant Biol. 29:183–194 [DOI] [PubMed] [Google Scholar]
- 174. So AKC, John-McKay M, Espie GS. 2002. Characterization of a mutant lacking carboxysomal carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Planta 214:456–467 [DOI] [PubMed] [Google Scholar]
- 175. Badger MR. 2003. The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms. Photosynth Res. 77:83–94 [DOI] [PubMed] [Google Scholar]
- 176. Pierce J, Carlson TJ, Williams JG. 1989. A cyanobacterial mutant requiring the expression of ribulose bisphosphate carboxylase from a photosynthetic anaerobe. Proc. Natl. Acad. Sci. U. S. A. 86:5753–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. English RS, Jin S, Shively JM. 1995. Use of electroporation to generate a Thiobacillus neapolitanus carboxysome mutant. Appl. Environ. Microbiol. 61:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Orus MI, Rodriguez ML, Martinez F, Marco E. 1995. Biogenesis and ultrastructure of carboxysomes from wild type and mutants of Synechococcus sp strain PCC 7942. Plant Physiol. 107:1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Fan CG, Cheng SQ, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. 2010. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. U. S. A. 107:7509–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Fan CG, Cheng SQ, Sinha S, Bobik TA. 2012. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc. Natl. Acad. Sci. U. S. A. 109:14995–15000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Savage D, Afonso B, Chen A, Silver P. 2010. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327:1258. [DOI] [PubMed] [Google Scholar]
- 182. Jensen TE, Ayala RP. 1976. The fine structure of a microplate-microtubule array, microfilaments and polyhedral body associated microtubules in several species of Anabaena. Arch. Microbiol. 111:1–6 [DOI] [PubMed] [Google Scholar]
- 183. Szardenings F, Guymer D, Gerdes K. 2011. ParA ATPases can move and position DNA and subcellular structures. Curr. Opin. Microbiol. 14:712–718 [DOI] [PubMed] [Google Scholar]
- 184. Rudner DZ, Losick R. 2010. Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2:a000307. 10.1101/cshperspect.a000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Watson SW, Waterbury JB. 1971. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch. Microbiol. 77:203–230 [Google Scholar]
- 186. Parsons JB, Frank S, Bhella D, Liang MZ, Prentice MB, Mulvihill DP, Warren MJ. 2010. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol. Cell 38:305–315 [DOI] [PubMed] [Google Scholar]
- 187. Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. 2011. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J. Bacteriol. 193:1385–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Woodger FJ, Badger MR, Price GD. 2003. Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol. 133:2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Woodger FJ, Bryant DA, Price GD. 2007. Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp strain PCC 7002: role of NdhR/CcmR. J. Bacteriol. 189:3335–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Hackenberg C, Huege J, Engelhardt A, Wittink F, Laue M, Matthijs HC, Kopka J, Bauwe H, Hagemann M. 2012. Low-carbon acclimation in carboxysome-less and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 158:398–413 [DOI] [PubMed] [Google Scholar]
- 191. Wang HL, Postier BL, Burnap RL. 2004. Alterations in global patterns of gene expression in Synechocystis sp PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 279:5739–5751 [DOI] [PubMed] [Google Scholar]
- 192. McKay RML, Gibbs SP, Espie GS. 1993. Effect of dissolved inorganic carbon on the expression of carboxysomes, localization of Rubisco and the mode of inorganic carbon transport in cells of the cyanobacterium Synechococcus UTEX 625. Arch. Microbiol. 159:21–29 [Google Scholar]
- 193. Kusian B, Bowien B. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135–155 [DOI] [PubMed] [Google Scholar]
- 194. Dubbs JM, Tabita FR. 2004. Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28:353–376 [DOI] [PubMed] [Google Scholar]
- 195. Mata-Cabana A, Florencio FJ, Lindahl M. 2007. Membrane proteins from the cyanobacterium Synechocystis sp. PCC 6803 interacting with thioredoxin. Proteomics 7:3953–3963 [DOI] [PubMed] [Google Scholar]
- 196. Lindahl M, Florencio FJ. 2003. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. U. S. A. 100:16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Zilliges Y, Kehr J, Meissner S, Ishida K, Mikkat S, Hagemann M, Kaplan A, Börner T, Dittmann E. 2011. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One 6:e17615. 10.1371/journal.pone.0017615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Gerbersdorf SU. 2006. An advanced technique for immuno-labelling of microcystins in cryosectioned cells of Microcystis aeruginosa PCC 7806 (cyanobacteria): implementations of an experiment with varying light scenarios and culture densities. Toxicon 47:218–228 [DOI] [PubMed] [Google Scholar]
- 199. Jähnichen S, Ihle T, Petzoldt T, Benndorf J. 2007. Impact of inorganic carbon availability on microcystin production by Microcystis aeruginosa PCC 7806. Appl. Environ. Microbiol. 73:6994–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Sivonen K, Börner T. 2008. Bioactive compounds produced by cyanobacteria, p 159–197 In Herrero A, Flores E. (ed), The cyanobacteria: molecular biology, genomics and evolution, vol 1 Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 201. Long BM, Jones GJ, Orr PT. 2001. Cellular microcystin content in N-limited Microcystis aeruginosa can be predicted from growth rate. Appl. Environ. Microbiol. 67:278–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Deblois CP, Juneau P. 2010. Relationship between photosynthetic processes and microcystin in Microcystis aeruginosa grown under different photon irradiances. Harmful Algae 9:18–24 [Google Scholar]
- 203. Moroney JV, Somanchi A. 1999. How do algae concentrate CO2 to increase the efficiency of photosynthetic carbon fixation? Plant Physiol. 119:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Smith EC, Griffiths H. 1996. A pyrenoid-based carbon-concentrating mechanism is present in terrestrial bryophytes of the class Anthocerotae. Planta 200:203–212 [Google Scholar]
- 205. Griffiths D. 1970. The pyrenoid. Bot. Rev. 36:29–58 [Google Scholar]
- 206. Smith EC, Griffiths H. 1996. The occurrence of the chloroplast pyrenoid is correlated with the activity of a CO2-concentrating mechanism and carbon isotope discrimination in lichens and bryophytes. Planta 198:6–16 [Google Scholar]
- 207. Borkhsenious ON, Mason CB, Moroney JV. 1998. The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Plant Physiol. 116:1585–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Giordano M, Beardall J, Raven JA. 2005. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56:99–131 [DOI] [PubMed] [Google Scholar]
- 209. Bedoshvili Y, Popkova T, Likhoshway Y. 2009. Chloroplast structure of diatoms of different classes. Cell Tissue Biol. 3:297–310 [Google Scholar]
- 210. Kuchitsu K, Tsuzuki M, Miyachi S. 1991. Polypeptide composition and enzyme-activities of the pyrenoid and its regulation by CO2 concentration in unicellular green-algae. Can. J. Bot. 69:1062–1069 [Google Scholar]
- 211. Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G. 1998. A novel alpha-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J. 17:1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Markelova AG, Sinetova MP, Kupriyanova EV, Pronina NA. 2009. Distribution and functional role of carbonic anhydrase Cah3 associated with thylakoid membranes in the chloroplast and pyrenoid of Chlamydomonas reinhardtii. Russ. J. Plant Physiol. 56:761–768 [Google Scholar]
- 213. Sinetova MA, Kupriyanova EV, Markelova AG, Allakhverdiev SI, Pronina NA. 2012. Identification and functional role of the carbonic anhydrase Cah3 in thylakoid membranes of pyrenoid of Chlamydomonas reinhardtii. Biochim. Biophys. Acta Bioenerg. 1817:1248–1255 [DOI] [PubMed] [Google Scholar]
- 214. Genkov T, Meyer M, Griffiths H, Spreitzer RJ. 2010. Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits engineered RbcS cDNA for expression in Chlamydomonas. J. Biol. Chem. 285:19833–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Ma Y, Pollock SV, Xiao Y, Cunnusamy K, Moroney JV. 2011. Identification of a novel gene, CIA6, required for normal pyrenoid formation in Chlamydomonas reinhardtii. Plant Physiol. 156:884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Kowallik K. 1969. The crystal lattice of the pyrenoid matrix of Prorocentrum micans. J. Cell Sci. 5:251–269 [DOI] [PubMed] [Google Scholar]
- 217. Leadbeater BSC, Manton I. 1971. Fine structure and light microscopy of a new species of Chrysochromulina (C. acantha). Arch. Microbiol. 78:58–69 [DOI] [PubMed] [Google Scholar]
- 218. Bertagnolli BL, Nadakavukaren MJ. 1970. An ultrastructural study of pyrenoids from Chlorella pyrenoidosa. J. Cell Sci. 7:623–630 [DOI] [PubMed] [Google Scholar]
- 219. Markowitz MM, Hoffman LR. 1974. Chloroplast inclusions in zoospores of Oedocladium. J. Phycol. 10:308–315 [Google Scholar]
- 220. Ramazanov Z, Rawat M, Henk MC, Mason CB, Matthews SW, Moroney JV. 1994. The induction of the CO2-concentrating mechanism is correlated with the formation of the starch sheath around the pyrenoid of Chlamydomonas reinhardtii. Planta 195:210–216 [Google Scholar]
- 221. Villarejo A, Martinez F, Plumed MD, Ramazanov Z. 1996. The induction of the CO2 concentrating mechanism in a starch-less mutant of Chlamydomonas reinhardtii. Physiol. Plant 98:798–802 [Google Scholar]
- 222. Tortell PD. 2000. Evolutionary and ecological perspectives on carbon acquisition in phytoplankton. Limnol. Oceanogr. 45:744–750 [Google Scholar]
- 223. Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B-12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Chen P, Andersson DI, Roth JR. 1994. The control region of the pdu/cob regulon in Salmonella typhimurium. J. Bacteriol. 176:5474–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Stojiljkovic I, Baumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Bobik TA. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 70:517–525 [DOI] [PubMed] [Google Scholar]
- 228. Penrod JT, Roth JR. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188:2865–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 230. Munk AC, Copeland A, Lucas S, Lapidus A, Del Rio TG, Barry K, Detter JC, Hammon N, Israni S, Pitluck S, Brettin T, Bruce D, Han C, Tapia R, Gilna P, Schmutz J, Larimer F, Land M, Kyrpides NC, Mavromatis K, Richardson P, Rohde M, Goker M, Klenk HP, Zhang Y, Roberts GP, Reslewic S, Schwartz DC. 2011. Complete genome sequence of Rhodospirillum rubrum type strain (S1). Stand. Genomic Sci. 4:293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Price GD, Pengelly JJ, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR. 2013. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 64:753–768 [DOI] [PubMed] [Google Scholar]
- 232. Wu M, Eisen J. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9:R151. 10.1186/gb-2008-9-10-r151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 234. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 235. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 236. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 237. Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. 2005. The MicrobesOnline Web site for comparative genomics. Genome Res. 15:1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Shively JM, Bock E, Westphal K, Cannon GC. 1977. Icosahedral inclusions (carboxysomes) of Nitrobacter agilis. J. Bacteriol. 132:673–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. van Eykelenburg C. 1980. Ecophysiological studies on Spirulina platensis. Effect of temperature, light intensity and nitrate concentration on growth and ultrastructure. Antonie Van Leeuwenhoek 46:113–127 [DOI] [PubMed] [Google Scholar]
- 240. Hanson R. 2010. Jmol—a paradigm shift in crystallographic visualization. J. Appl. Crystallogr. 43:1250–1260 [Google Scholar]
- 241. Peitsch MC. 1995. Protein modeling by e-mail. Nat. Biotechnol. 13:658–660 [Google Scholar]
- 242. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 243. Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37:D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Newman J, Branden C-I, Jones TA. 1993. Structure determination and refinement of ribulose 1,5-bisphosphate carboxylase/oxygenase from Synechococcus PCC6301. Acta Crystallogr. D Biol. Crystallogr. 49:548–560 [DOI] [PubMed] [Google Scholar]
- 245. Kimber MS, Pai EF. 2000. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J. 19:1407–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Suzuki E, Fukuzawa H, Miyachi S. 1991. Identification of a genomic region that complements a temperature-sensitive, high CO2-requiring mutant of the cyanobacterium, Synechococcus sp. PCC7942. Mol. Gen. Genet. 226:401–408 [DOI] [PubMed] [Google Scholar]