Abstract
SUMMARY
Homologous recombination is a universal process, conserved from bacteriophage to human, which is important for the repair of double-strand DNA breaks. Recombination in mitochondrial DNA (mtDNA) was documented more than 4 decades ago, but the underlying molecular mechanism has remained elusive. Recent studies have revealed the presence of a Rad52-type recombination system of bacteriophage origin in mitochondria, which operates by a single-strand annealing mechanism independent of the canonical RecA/Rad51-type recombinases. Increasing evidence supports the notion that, like in bacteriophages, mtDNA inheritance is a coordinated interplay between recombination, repair, and replication. These findings could have profound implications for understanding the mechanism of mtDNA inheritance and the generation of mtDNA deletions in aging cells.
INTRODUCTION
Mitochondria produce the bulk of cellular energy in eukaryotic cells. The maintenance of mitochondrial function is crucial for cell survival, especially in highly energy-demanding tissues. Because mitochondrial DNA (mtDNA) encodes components of the respiratory complexes I, III, IV, and V in the oxidative phosphorylation pathway, the integrity of mtDNA is critical for efficient energy production (1). The faithful inheritance of mtDNA demands an incredible amount of cellular resources. A large cohort of proteins is required for mtDNA organization, replication, repair, transmission, and other transactions. These proteins are all encoded by the nuclear genome and are imported into mitochondria after their synthesis by the cytosolic ribosome.
mtDNA is organized as protein-DNA complexes, known as mitochondrial nucleoids (mt-nucleoids) (2–5). Each mitochondrion harbors 2 or 3 nucleoids which are associated with the mitochondrial inner membrane (6–9). mtDNA has a much higher mutation rate than the nuclear genome (10, 11). The constant attack by reactive oxygen species produced in close proximity of the inner membrane and the inherent error-prone feature of the mtDNA replisome are held to be largely responsible for the high mutation rate in mtDNA (12). Progressive accumulation of mtDNA mutations has been proposed to contribute to aging (13, 14). This idea is supported by studies showing that transgenic animals with increased mtDNA mutations exhibit premature-aging phenotypes (15, 16).
Much less is known about how damaged DNA is repaired in mitochondria than about how it is repaired in the nucleus. So far, base excision and mismatch repair activities have been documented in the organelle (17–22). These are specific mechanisms for fixing the base lesions and DNA mismatches, respectively. However, the most dramatic damage to the mitochondrial genome is probably the deletion of mtDNA, which occurs frequently in aged and oxyradical-rich tissues such as brain, heart, and skeletal muscle (23–26). Although the overall abundance of the mtDNA deletions is low in many aged tissues, there is experimental evidence indicating that large-scale deletions in some cell types can attain a level sufficiently high to directly affect oxidative phosphorylation. Single-cell studies have shown that deletions can occur in ∼32 to 80% of mtDNA molecules in substantia nigra neurons of brains from patients with Parkinson's disease (26–28). Extensive deletions are also seen in these neuronal cells from normally aged people. Such high levels of mtDNA deletions directly cause a cytochrome c oxidase (or complex IV) defect and respiratory deficiency in these cells.
How do the mtDNA deletions arise in aged cells? It has previously been proposed that they may be generated by replicative slippage over repeated sequences that are scattered over the mitochondrial genome (29). A newly emerging model instead posits that mtDNA deletions may be produced during the repair of double-strand breaks (DSBs) by recombination-based processes (30). The latter hypothesis is supported by abundant evidence. However, the actual mechanism of DNA recombination in mitochondria is poorly understood.
Homologous recombination (HR) is one of the most important DSB repair pathways in organisms ranging from bacteriophage to human (31). This mechanism is also critical for the repair of interstrand cross-links and the reinitiation of DNA replication from collapsed replication forks (32). Is this almost universal DNA repair mechanism also present in mitochondria for the repair and maintenance of the mitochondrial genome? In this article, I first provide a brief summary of experimental data accumulated in the last 4 decades that document homologous recombination in mitochondria of fungal and lower-animal cells. Much discussion is focused on evidence that supports or rejects the existence of mtDNA recombination in mitochondria of higher animals. I then review recent progress in the molecular dissection of the mitochondrial HR machineries and the possible implications of recombinational errors for the generation of mtDNA deletions in aged cells.
mtDNA RECOMBINATION IN YEAST AND ANIMALS
A prerequisite for homologous recombination to occur is the availability of a wild-type DNA template in the vicinity of the broken DNA molecule, which is required for informational retrieval. In the nucleus, this requirement is met when cells enter into the S and G2 phases with the sister chromatids fully replicated. The organization of mtDNA provides a natural genetic setting in favor of homologous recombination. Electron and superresolution fluorescence microscopies have revealed that each mitochondrion harbors numerous mt-nucleoids and each nucleoid contains multiple copies of mtDNA (4, 33–35). mt-nucleoids could therefore serve as an organizational scaffold for intranucleoidal recombination, provided that molecular machinery for recombination is in place.
mtDNA Recombination in Yeast
Genetic evidence.
The transmission of mtDNA is generally biparental in the model organism Saccharomyces cerevisiae. When the diploid zygotic cells are formed from two haploid strains, the parental mtDNAs are rapidly mixed and randomly transmitted into the progeny. This property has provided a powerful tool to document some early evidence for mtDNA recombination (36, 37). By crossing haploid strains carrying deleted (ρ−) forms of the mitochondrial genome with different buoyant densities or restriction maps, the progeny were found to have mtDNA with characteristics of both parents, which is indicative of recombination. Direct genetic evidence for mtDNA recombination was demonstrated when antibiotic-resistant genetic markers on mtDNA became available (38, 39). Analyses of zygotic colonies derived from crosses between chloramphenicol and erythromycin resistance markers located in different regions of mtDNA demonstrated the stable transmission of both parental and recombinant genotypes. In general, the frequency of recombinants between two unlinked markers in yeast mtDNA can reach as high as 20 to 25% (37). This indicates that extensive pairing and recombination events occur between the parental mtDNAs during zygote formation.
mtDNA recombination is not limited as an intermolecular event (37, 40). Subgenomic recombinations also take place at short and direct sequence repeats, a pattern consistent with the involvement of a recombination-excision mechanism. These events underlie the frequent generation of respiration-deficient petite mutants in S. cerevisiae, which stably transmits the extensively deleted ρ− genomes (41–43). Similar repeat-mediated recombination events were seen in Neurospora crassa, which are manifested by the generation of plasmid-like supercoiled subgenomic circles in mitochondria (44). These subgenomic circles coexist with the wild-type genome, as large mtDNA deletions are not compatible with cell viability in the petite-negative Neurospora species. For a long time, a lack of efficient recombination machinery was thought to partly account for the inability of the petite-negative yeasts to generate ρ− genomes. Studies in Kluyveromyces lactis have shown that these cells are capable of generating ρ− genomes like S. cerevisiae, provided that specific nuclear mutations that suppress cell lethality caused by the loss of mtDNA-encoded functions are present (45–48). Thus, recombination is rather an inherent property of the mitochondrial genetic system in yeast and other fungal species. The ability of yeast mtDNA to recombine was strongly supported by the observation that exogenously added DNA can be integrated into the mitochondrial genome after delivery by microprojectile bombardment (49), a procedure that is now commonly used for mitochondrial transformation (50).
Molecular evidence.
One of the landmarks in the study of mtDNA recombination was the revelation of Holliday junctions in mtDNA and the presence of junction-processing enzymes in mitochondria. The hypersuppressive ρ− genomes, which contain repeated ori/rep sequences, have a highly penetrative transmission in crosses against the wild-type (ρ+) cells. The Fangman group identified the nuclear mgt1/cce1 mutation that affects the biased transmission of hypersuppressive ρ− mtDNA (51, 52). The MTG1/CCE1 gene encodes a Holliday junction resolvase specifically localized in mitochondria (53, 54). Disruption of MTG1/CCE1 causes extensive physical linkage between mtDNA molecules and the formation of giant mt-nucleoids. This in turn reduces the number of heritable units and therefore the transmission and hypersuppressiveness of the ρ− genomes. Remarkably, in the mtg1/cce1 mutant, DNA fragments indicative of the X-shaped cross-links can be directly detected on an agarose gel after releasing from the highly repeated ρ− genomes by digestion with restriction enzymes. The presence of branched mtDNA structures representing recombination intermediates can also be visualized in the wild-type mitochondrial genome using two-dimensional agarose gel electrophoresis (51, 55). The detection of these recombination intermediates provided unequivocal molecular evidence for the presence of active recombination in yeast mtDNA. Similar recombination intermediates have been recently reported in Drosophila melanogaster mitochondria (56).
mtDNA Recombination in Mammals
The no-recombination rule.
In animals whose males naturally inherit mtDNAs from both parents, such as those in the bivalve families, the occurrence of frequent recombinations has been unambiguously documented (57, 58). However, it has long been debated whether or not homologous recombination takes place in higher animals (59, 60). In these animals, mtDNA is maternally inherited, which presents a tremendous burden for detecting recombination. Based on extensive phylogeographical studies on natural populations, there seems to be little direct support for recombination between polymorphic mtDNA molecules (61–65). This has led to the general belief that animal mtDNA does not recombine. This assumption is ostensibly logical, as the maternal inheritance of mtDNA may spare the need for recombination, whose main role is expected to be to mix genetic information from different lineages and to gain a selection advantage during evolution. However, the question has persisted as to whether rare recombination events in these species occur but are masked by factors that impede their detection. A critical challenge is that maternally inherited mtDNA is generally homoplasmic and recombination between homologous molecules gives rise to recombinants which logistically cannot be distinguished from the parental molecules. Maternal inheritance also gives little chance for polymorphic mtDNAs to mix and possibly recombine in the natural populations. Furthermore, somatic cells, which are routinely used for detecting recombination, contain up to several thousand copies of mtDNA. The frequency of recombination between potential heterologous molecules that result from newly arising mutations or from rare leakage of heterologous paternal mtDNAs (see below) may be too low to be detected by standard technologies (for a review, see reference 60).
Evidence supporting recombination in mammalian mtDNA.
Despite the fact that the no-recombination rule for animal mtDNA holds up fairly well, increasing exceptions violating this rule have emerged in the last decade. There are generally two ways to describe mtDNA recombination events. Past recombinations are detected by comparing the pattern of sequence variations among individuals in a population, whereas real-time recombinations report the novel sequence patterns in the progeny that can be directly compared with those in their parents. Numerous polymorphism analyses for past recombination have provided evidence supporting widespread mitochondrial recombination across the animal kingdom (66–68), although some skepticism has been expressed about the power of this type of analysis (63, 65, 69). Data strongly supporting past recombination events have come from the analyses of mtDNA in lizard and salmon populations. These recombination events result from the interbreeding of divergent populations, which may have increased paternal mtDNA leakage, thereby generating a heteroplasmic state for intermolecular recombination (70, 71). Paternal mtDNA leakage-induced mtDNA recombination has also been reported in nematodes (72). In contrast, in natural populations of Drosophila melanogaster, paternal mtDNA leakage can attain a level of 6%, yet recombinant haplotypes remains undetectable, suggesting that combination either is too rare to be detected or is counterselected (73).
Paternal mtDNA leakage also facilitated the capture of real-time recombination events in humans. Kraytsberg et al. defined unequivocal human mtDNA recombination products in an individual with an unusual transmission leakage of the paternal mtDNA. The paternal mtDNA leakage likely resulted from the incomplete elimination of paternal mitochondria in early embryogenesis (74, 75). In this remarkable circumstance, mtDNA recombination appears to be rather common in the skeletal muscle (76). It was proposed that the recombination products may arise from a stalled replication fork (75). This generates a recombinogenic 3′ end that invades a neighboring mtDNA and initiates recombination by means of a putative recombinase activity. Based on these data, there seems to be no natural barrier that impedes mtDNA recombination even in humans, provided that sufficient heterologous mtDNA molecules are allowed to mix in vivo.
The analysis of deleted human mtDNAs that cause mitochondrial diseases has revealed the frequent presence of direct repeats that flank the edges of the deletion points, which suggests the involvement of recombination events (77). In the 1960s and 1970s, several groups used electron microscopy to study the structural organization of mtDNA in leucocytes. They found that a large fraction of mitochondrial DNA from cases of acute leukemia and leukemic-phase poorly differentiated lymphocytic lymphoma is present in dimeric or multiple interlocked circular forms (78–80). The leukemic patients were not subjected to chemotherapy. This provided the first evidence for the multimerization property of the human mitochondrial genome. Subsequent studies by others revealed dimeric and trimeric mtDNAs in cultured human cell lines (81, 82). The generation and subsequent monomerization of these molecular species were proposed to result from homologous recombination.
Cell line and transgenic animal models provided further support for the existence of recombination in human and mouse mtDNAs. Recombinant genotypes containing mitochondrial genes from different cells can be detected when the parental cells are forced to fuse (83). In a different experimental strategy, Bacman et al. expressed a mitochondrially targeted restriction endonuclease to introduce multiple DSBs in cultured cells. Following the induction of mtDNA breakage, intramolecular recombinations were frequently detected (84). Intermolecular recombination was also identified, but at a much lower frequency. Fan et al. recently traced multiple mutations in the mtDNA of mouse L cell lines. The data suggested that a recombination-based reassortment of mutant alleles can generate recombinant mtDNA haplotypes with a proliferative advantage after a long-term maintenance of heteroplasmy (85). In a mouse model in which the restriction endonuclease is specifically expressed in neuronal cells (236), the breakage of mtDNA is followed by the formation of mtDNA deletions. Analysis of these deletion products revealed that the recombinations take place at sites with or without the presence of direct repeated sequences. This suggests that in addition to homologous recombination, other mechanisms such as homeologous recombination or even nonhomologous end joining may also be involved.
In the last few years, recombination intermediates in human mtDNA were directly visualized. By using two-dimensional agarose gel electrophoresis, an early report showed that molecular species suggestive of Holliday junctions exist in the human heart mtDNA (86). This observation has now been further corroborated by transmission electron microscopy (87). In contrast to the conventional paradigm, heart mtDNA was found to have a complex organization with abundant dimeric and oligomeric molecules, branched structures, and prominent four- and three-way junctions. These molecular species are detected only in human heart and human and mouse brains and not in other tissues examined (87). This finding suggests an active homologous recombination in the oxyradical-rich heart and brain mitochondria. The various mtDNA conformers remarkably resemble those observed in yeast (88–90) and in leukemic leukocytes as previously reported by Firkin and Clark-Walker (78).
The dramatic observation of recombination intermediates in adult human heart and brain mtDNA reaffirms the notion that recombination does occur in higher animals but is highly tissue specific. In most cell types the recombination events may be negligible or be too scarce to be molecularly detected. In this regard, it is important to note that recombination is required primarily for DNA repair. If double-strand DNA breaks occur as relatively rare events, recombination would have to be kept low. Limiting unnecessary recombination is important for preventing recombination errors that are detrimental for genome stability (see below).
GENES AFFECTING mtDNA RECOMBINATION
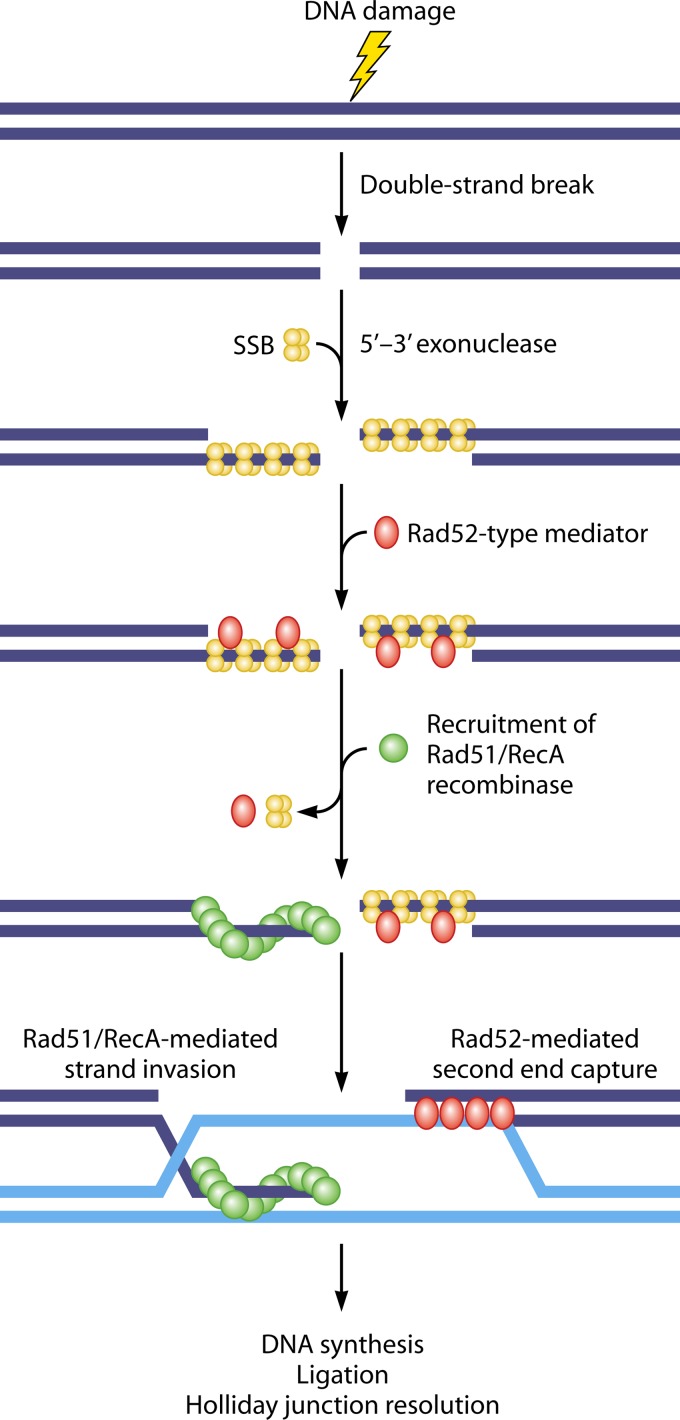
A critical challenge in the study of mtDNA recombination is to identify the molecular components promoting the recombinational reaction. A canonical recombination pathway in the eukaryotic nucleus and in bacteria (Fig. 1) involves many proteins. The most critical ones include exonucleases that process the double-stranded DNA (dsDNA) ends, single-stranded DNA binding proteins (SSBs) required for the stabilization of the resulting 3′ single-stranded DNA (ssDNA) tails, a Rad52-type recombination mediator for recombinase recruitment, and the Rad51/RecA-type recombinases that directly promote dsDNA remodeling and homologous pairing. Following strand invasion, DNA synthesis, and ligation, the Holliday junctions are formed, which are finally resolved by specific endonucleases (31, 91, 92). Among these proteins, only SSB has been clearly identified in mitochondria. The mitochondrial SSB has a prokaryotic origin and exists in a homotetramer form in both yeast and human (93–96). Yeast mitochondria also harbor the well-defined Holliday junction resolvase Mgt1/Cce1 (see above). Several other proteins that have been identified to affect mtDNA recombination are listed in Table 1.
Fig 1.
Simplified schematic of the early steps in a canonical homologous recombination pathway in the eukaryotic nucleus and in bacteria. Upon DNA damage, the free ends of a DSB are first processed by an exonuclease. The exposed 3′ ssDNA tails are coated by single-strand DNA binding proteins (SSBs) to prevent the formation of secondary structures. The recombination mediator, Rad52 (or RecO in bacteria), then displaces SSB and recruits the Rad51/RecA-type recombinase to form presynaptic helical nucleoprotein filaments. These filaments then initiate homology search and catalyze ATP-dependent strand invasion within duplex DNA templates. The invading strand provides a free 3′ end for priming DNA replication that allows the restoration of genetic information missing from the dsDNA breaks. The Rad52 protein has a second function in this pathway, which is to capture the second end through its single-strand annealing activity. After DNA synthesis and ligation, double Holliday junctions are formed. The Holliday junctions can be resolved by different molecular strategies with or without DNA strand crossover. Without the capturing of the second end by Rad52, the invading strand may be dissociated from the D-loop and reannealed to the ssDNA on the second end, a process known as synthesis-dependent strand annealing (not shown).
Table 1.
Proteins known to affect DNA recombination in yeast, mammalian, and plant mitochondria
| Organism and protein | Molecular function | Mutant phenotype on recombination |
|---|---|---|
| Yeast | ||
| Abf2 | High-mobility-group protein primarily for mtDNA packaging | Increases Holliday junctions, reduces repeat-mediated recombination |
| Exo5 | 5′-3′ exonuclease | Unknown |
| Hmi1 | 3′-5′ DNA helicase | mtDNA fragmentation |
| Mgm101 | Single-strand annealing protein | Decreases repeat-mediated recombination |
| Mhr1 | Homologous pairing in vitro | Decreases gene conversion |
| MRX complex | 5′-3′ exonuclease | Decreases repeat-mediated recombination |
| Msh1 | Mismatch repair-like protein | Increases repeat-mediated recombination |
| Nuc1 | Endo/exonuclease | Decreases mtDNA recombination |
| Pif1 | 5′-3′ DNA helicase | Reduces recombination in ρ+ × ρ− crosses |
| Rim1 | Single-stranded DNA binding protein | Suppresses pif1 mutation |
| Mammal | ||
| mtTFA | mtDNA transcription and packaging, binding to four-way junctions | Increases Holliday junctions |
| Rad51 | Recombinase | Unknown |
| SSB | Single-stranded DNA binding protein | Unknown |
| Twinkle | DNA helicase | Overexpression increases Holliday junctions |
| Plant | ||
| Msh1 | Mismatch repair-like protein | Increases mtDNA rearrangements |
| RecA3 | RecA ortholog | Increases mtDNA rearrangements |
| Rad52-1B | Single-strand annealing protein | Unknown |
DNA Helicases
Foury and Kolodynski devised a genetic screen in yeast for nuclear mutations that affect mtDNA recombination (97). In this system, a tandemly repeated ρ− genome carrying the oligomycin resistance marker can be efficiently integrated into a ρ+ DNA by recombination. This gives rise to drug-resistant colonies on plates containing a nonfermentable carbon source. It was found that mutations in a nuclear locus named PIF1 (petite integration frequency 1) specifically affect the integration of the ρ− mtDNA into the ρ+ genome. The mtDNA in the pif1 mutants is hypersensitive to UV light, suggesting a defect in mtDNA repair. The PIF1 gene encodes a 5′ → 3′ DNA helicase that unwinds DNA duplexes (98, 99). DNA helicases are known to participate in various stages of homologous recombination, which include double-strand DNA end resection, nucleoprotein filament stabilization, regulation of crossover formation, and the branch migration in Holliday junctions (92, 100, 101). Foury and coworkers discovered two classes of ρ− mtDNAs which are dependent or independent of Pif1 for recombining with ρ+ mtDNAs (102). Pif1-dependent ρ− mtDNAs normally have high recombination frequencies in ρ− × ρ+ crosses, and recombination is drastically reduced in pif1 mutants. These ρ− mtDNAs contain a dyad symmetry capable of forming cruciform structures. These secondary structures, apparently promoted or stabilized by Pif1, may stimulate recombination. As Pif1 preferentially unwinds forked DNA-DNA substrates, it is possible that the processing of the cruciform structures by Pif1 generates single-stranded regions that favor strand invasion/annealing and recombination. In contrast, Pif1-independent ρ− mtDNAs are characterized by the presence of inverted repeats. The repeated units may form large palindromic structures that promote recombination independent of Pif1.
Yeast pif1 mutants are respiratory deficient at the nonpermissive temperature. This phenotype is suppressed by overexpressing the mitochondrial single-strand binding protein Rim1 (93). Mitochondrial SSB is known to stimulate DNA helicase activity (96). Indeed, Rim1 stimulates Pif1 activity by 4- to 5-fold, and these two proteins physically interact with each other (103). In the absence of a functional Pif1 helicase, the suppression of the respiration-deficient phenotype by Rim1 also suggests that the latter may stimulate the activity of a second DNA helicase that functionally overlaps with Pif1. In fact, yeast mitochondria express a second DNA helicase activity encoded by the HMI1 gene (104). A defect in the Hmi1 function leads to mtDNA fragmentation, suggesting that it has a role in mtDNA repair (105). HMI1 is required for the maintenance of the ρ+ but not the highly repeated ρ− genomes (105–107), like other recombination genes, including MHR1 and MGM101 (see below). Hmi1 functionally overlaps with the Pif1 helicase with regard to the maintenance of some ρ− genomes (106). One report showed that the helicase activity of Hmi1 is not essential for its mtDNA maintenance function (108). The nature of this cryptic function remains unsettled. Biochemical analysis has shown that Hmi1 unwinds dsDNA with 3′-ssDNA overhangs. It efficiently unwinds forked DNA such as the flaps resembling the chain displacement structures generated by the recombination process (104). This further supports the idea that Hmi1 plays a role in mtDNA recombination.
Multiple DNA helicases have been detected in human mitochondria, including Twinkle, Dna2, and Pif1 (109). The Twinkle helicase is a key component of the mtDNA replisome whose mutation causes multiple deletions in mtDNA (110). Twinkle is specifically targeted into the mt-nucleoids. Interestingly, overexpression of the mouse Twinkle helicase increases Holliday junctions in heart mtDNA (87), suggesting that it may also play a role in recombination. Dna2 is a nuclease/helicase present in mitochondria to process flap intermediates during DNA replication and base excision repair (19, 111). Human cells also express a variant of the Pif1 helicase in mitochondria known as Pif1β, in addition to a nuclear version derived from a splicing variant (112). This protein is poorly characterized, and it is unclear whether it plays a role in recombination like its counterpart in yeast mitochondria.
Abf2 and TFAM
By using the microprojectile bombardment approach, Sia and coworkers developed an elegant genetic assay for measuring the frequency of repeat-mediated mtDNA recombination in yeast (113). In this scheme, expression of the mitochondrial COX2 gene, which encodes the subunit 2 of cytochrome c oxidase, is interrupted by the insertion of ARG8m (114). ARG8m is flanked by directly repeated sequences. Recombination between the repeats results in the deletion of ARG8m and the restoration of COX2 expression, which can be readily scored by the formation of respiration-competent colonies. Using this system, they found that mutations in the nuclear ABF2 gene increased repeat-mediated recombination in mtDNA by 5-fold (115). Abf2 is a high-mobility-group like protein involved in mtDNA packaging and the protection of mtDNA against oxidative damage (116–119). The loss of Abf2 may enhance accessibility of mtDNA for homology search and therefore homologous recombination. Abf2 inactivation may also increase double-strand breaks and therefore recombination frequency, due to increased oxidative damage to mtDNA (120). An early study showed that Abf2 stabilizes recombination intermediates and that overexpression of Abf2 increases the Holliday junctions (55). A decrease of mtDNA recombination intermediates was confirmed in Candida albicans disrupted in the GCF1 gene, encoding an Abf2 homolog (121). The binding by Abf2 may suppress the processing of recombination intermediates. Likewise, the mouse homolog of Abf2, TFAM, also stabilizes the Holliday junctions in heart mtDNA (87). TFAM is known to bind four-way DNA junctions (122).
Exonucleases
Using the recombination reporter system developed by the Sia group, it was also found that loss of the MRX complex decreases repeat-mediated recombination by 4.5-fold (123). The MRX complex is composed of Mre11, Rad50, and Xrs2. It defines an exonuclease activity required for resecting DSB ends and to generate the recombinogenic 3′ single-stranded DNA in the nucleus (100). This complex also seems to affect DNA repair in mitochondria. Loss of the MRX complex does not affect mtDNA stability under physiological conditions. It remains unknown whether this protein complex is required for mtDNA repair under DNA-damaging conditions. Alternatively, it is possible that a second exonuclease activity that functionally overlaps with MRX is present in mitochondria. Yeast mitochondria harbor a robust endo/exonuclease known as Nuc1 (124), a homolog of the mammalian EndoG. Nuc1 possesses a 5′-exonuclease activity on dsDNA substrates (125). Disruption of NUC1 reduces recombination between linked mtDNA markers and decreases the frequency of gene conversion in the mitochondrial genome (126). However, loss of the Nuc1 function, like that of the MRX complex, has little effect on mitochondrial function, suggesting that its role in mtDNA maintenance is rather limited.
Much attention has recently been focused on the study of the Din7 protein. Din7 shares sequence homology with several exonucleases, including the Schizosaccharomyces pombe ExoI protein. Despite the facts that Din7 is specifically targeted into mitochondria and that its expression is induced by DNA-damaging agents (127, 128), disruption of the DIN7 gene has little effect on mtDNA stability. It has been shown that overexpression of Din7 destabilizes poly(GT) in mtDNA and enhances gross rearrangements in mtDNA, presumably due to enhanced recombination (129). The Sia group has shown that loss of the Din7 function does not significantly affect repeat-mediated mtDNA deletions, but it synergizes with the G776D allele of MSH1 (see below) to increase this particular recombination-based process (130). This observation seems to support a role of Din7 in suppressing rather than promoting this type of recombination. No recombinant Din7 has been successfully prepared so far. Ling and coworkers have recently demonstrated that mitochondrial extracts overexpressing Din7 have increased 5′-exodeoxyribonuclease activity in an in vitro assay using dsDNA as a substrate (131). It was further shown that loss of Din7 function leads to the overaccumulation of dsDNA breaks in mtDNA and to a defect in increasing mtDNA copy number in response to oxidative stress. Increased expression of Din7 appears to enhance homologous recombination in the Endo.SceI-induced polarized recombination assay. These data support a potential role of Din7 in generating 3′-ssDNA tails for initiating recombinational repair of dsDNA breaks. Again, given that the key recombination proteins such as Mhr1 and Mgm101 (see below) are essential for mtDNA maintenance, it remains unexplained why disruption of DIN7 has little effect on mtDNA stability if its primary role is to promote recombination. Currently, it may be premature to conclude that Din7 is the major exonuclease that promotes the resection of dsDNA ends prior to recombination.
A 5′ DNA exonuclease activity, defined by the EXO5 gene, has been recently identified in yeast mitochondria (132, 133). EXO5 is essential for the maintenance of ρ+ but not the ori-containing hypersuppressive ρ− genomes. This property is reminiscent of other mutations affecting mtDNA recombination proteins such as mhr1 and mgm101 (see below). Exo5 is distantly related to the bacterial RecB, a subunit of the RecBCD enzyme involved in dsDNA end processing and HR. However, Exo5 seems to specifically degrade ssDNA and does not process double-stranded DNA ends. One possible scenario for a potential role in recombination is that a 3′-5′ helicase (e.g., Hmi1) is first recruited to generate a single-stranded 5′-ended substrate, which is subsequently degraded by Exo5. Such a helicase/exonuclease partnership in dsDNA end processing in mitochondria is yet to be proved. The human MGME1 protein was recently identified to have an ssDNA-specific 5′-3′ exonuclease activity like the yeast Exo5 (134). MGME1 is proposed to play a role in processing DNA flap substrates during mtDNA replication and repair. Mutations in MGME1 cause multisystemic mitochondrial disease manifested by mtDNA depletion and multiple deletions.
Msh1
Msh1 is homologous to the bacterial mismatch repair protein MutS. It specifically localizes to mitochondria and is essential for mtDNA maintenance in yeast (135, 136). There is so far no clear evidence for the involvement of Msh1 in mismatch repair in mitochondria, although overexpression of MSH1 appears to have an antimutator phenotype (137). Moderate overexpression of MSH1 seems to stimulate homologous recombination (138). The mechanism for this effect remains unknown. Specific mutations in Msh1 predicted to affect mismatch repair activity appear to retain mitochondrial respiration, suggesting that this protein may have an additional function (139). The exact nature of this novel function is not well understood. In bacteria, MutS plays a role in recombination editing, an activity for heteroduplex rejection and the inhibition of recombination between mismatched substrates (140). This may also be true for its mitochondrial counterpart. Disruption of the MSH1 gene in yeast increases repeat-mediated recombination by 50- to 170-fold (130). The MutS homolog in plant mitochondria has been clearly demonstrated to contribute to recombination surveillance, and its disruption results in enhanced mitochondrial genome recombination at numerous repeated sequences (141). In lower animals such as corals, MutS is encoded by mtDNA (142). This highlights the importance of maintaining such an activity in mitochondria during evolution. In human mitochondria, the presence of Msh5, which is normally targeted to the nucleus for mismatch repair in the form of the Msh4-Msh5 heterodimeric complex, has been recently reported (143). Interestingly, another recent study has demonstrated that human mitochondria have a robust mismatch repair activity which is independent of the classic mismatch repair enzymes (22). Instead, the mitochondrial mismatch binding and repair activities may involve the Y-box binding protein 1 (YB-1). This is in line with the assumption that the MutS homologs in mitochondria may carry out functions other than the conventional mismatch repair. A comprehensive functional characterization of these proteins is required in the future, given the critical role of these proteins in the maintenance of mtDNA stability.
Mhr1
Mhr1 and gene conversion in mtDNA.
Extensive studies of the core molecular machinery for mtDNA recombination have come from the Ling and Shibata group. These investigators screened for nuclear mutations in yeast that sensitize mtDNA to UV irradiation (144). A recessive mutation termed mhr1 (for mitochondrial homologous recombination) was identified. The mhr1-1 allele reduces the highly polarized homing of the ω intron from 98.2% in the wild-type cells to 37.8% in crosses involving two mhr1-1 parents. The homing of the ω intron occurs by a site-specific gene conversion mechanism, following a DSB introduced by the intron-encoded Endo.SceI endonuclease. The mhr1-1 mutation has only a slight effect on intergenic recombination between two antibiotic resistance markers, which is dependent on crossing over. It appears that MHR1 has a specific role in homologous gene conversion and that homologous crossing over instigated by other double-strand breaks may be dependent on an additional recombination pathway. This explanation is consistent with the observation that mtDNA loss in the mgt1/cce1 mutant is accelerated in the mhr1-1 background (145). Mgt1/Cce1 is a Holliday junction resolvase, and its inactivation results in the overaccumulation of Holliday junctions, mtDNA clustering, and, ultimately, reduced transmission efficiency (see above). If Mhr1 is involved in the classic homologous crossover, it would be expected that a partial defect in Mhr1 should reduce cross-links between mtDNA molecules, which in turn improves transmission and increases the stability of mtDNA in the mgt1/cce1 background. This turned out to not be the case.
Biochemical properties.
MHR1 encodes a DNA binding protein of 27 kDa in the mitochondrial matrix that lacks sequence similarity to any known recombinases (144, 145). The Mhr1 protein binds double- and single-stranded DNAs with comparable affinities (146). Ling and Shibata found that Mhr1 promotes homologous pairing between ssDNA and a homologous dsDNA duplex in an in vitro D-loop formation assay (145). This activity is independent of ATP. This led to the assumption that Mhr1 is an ATP-independent DNA recombinase that mediates the strand invasion mode of homologous recombination like the conventional RecA/Rad51-type recombinases (Fig. 1). In addition to its ATP-independent characteristic, several features of Mhr1 distinguish it from the classic strand invasion-type recombination proteins. Unlike the currently known DNA recombinases, which all form oligomers and distinct helical nucleoprotein filaments on ssDNA substrates, Mhr1 is monomeric in solution (146). There is no indication that it forms helical filaments similar to those for RecA/Rad51. In vitro assays have shown that like the RecA/Rad51 recombinases, Mhr1 promotes the homologous pairing between single-stranded DNA and a negatively supercoiled dsDNA template (145). In addition, Mhr1 seems to preferably use relaxed rather than supercoiled dsDNA as a substrate for homologous pairing (147). The dsDNA was proposed to wrap Mhr1 in a right-handed manner, which results in DNA untwisting to compensate for the negative supercoils generated by the wrapping step. Interestingly, Mhr1 shares limited sequence similarity with Rad54 (148), a dsDNA-dependent helicase that regulates homologous recombination in the nucleus (92, 101). Whether or not Mhr1 is related to Rad54 and affects recombination by fulfilling some of the Rad54-related functions is yet to be investigated. An Mhr1-related function that is unequivocally defined so far is the rolling-circle replication of mtDNA (see below), which is likely initiated after the invasion of mtDNA circles by a single-stranded 3′ tail. This is in accordance with a role of Mhr1 in recombination.
RecA/Rad51 Homologs
Extensive efforts have been directed toward the identification of evolutionarily conserved proteins in mitochondria that catalyze strand invasion, a central reaction in the classic recombination mode. In an early study, the presence of an ATP-dependent RecA-type activity in human mitochondria was postulated (149). This was based on the observation that rat liver mitochondrial extracts stimulate the recombination between double- and single-stranded DNA molecules in vitro. The DNA substrates contain two defective alleles in the kanamycin resistance (kan) gene. Recombination restores the function of kan and gives rise to kanamycin-resistant colonies after transformation into Escherichia coli. The recombination efficiency was therefore measured by scoring the frequency of kanamycin-resistant colonies. One possible caveat for such an assay is that the mitochondrial extracts may be contaminated by the nuclear and cytosolic Rad51-type recombinases. More recently, the Knight group used a stringent fractionation protocol to demonstrate that a significant fraction of Rad51 and several related proteins such as Rad51C and Xrcc3 is associated with mitochondria in carcinoma cell lines (150). The levels of these proteins in the mitochondrial fractions are increased in response to oxidative stress. Furthermore, it was shown that cells depleted of these Rad51-type proteins are unable to maintain mtDNA copy number after DNA damage. These results would argue that mitochondria may have a homologous recombination activity similar to that of the nucleus. However, the presence of recombinational repair by the conventional strand invasion mode would also entail that other components in the pathway that are compatible with Rad51, including Rad52 and RPA, should also be present in mitochondria. No experimental data are currently available to support this. Thus, whether these proteins are actually involved in homologous recombination in human mitochondria needs to be further substantiated. Interestingly, a subsequent study by the same group has shown that the recruitment of Rad51 to mitochondria is increased in cells recovering from mtDNA depletion (151). This finding supports the idea that Rad51 is required for sustaining mtDNA synthesis under conditions of replicative stress. It may participate in the reinitiation of mtDNA replication from stalled or collapsed replication forks. Besides immortalized cell lines, the presence of Rad51 in mitochondria of normal tissues is yet to be proved.
Several RecA-like proteins, including RecA2 and RecA3, have been unequivocally identified in plant mitochondria. These proteins are speculated to participate in mtDNA repair especially under DNA-damaging conditions (152–154). Heterologous expression of RECA2 and RECA3 partially complements E. coli mutants defective in DNA repair. Interestingly, loss of the RecA3 function in Arabidopsis thaliana mitochondria increases rather than suppresses mtDNA rearrangements and recombination between intermediate-size repeats (154, 155). Suppression of repeat-mediated genome rearrangements by a RecA ortholog has also been shown in the mitochondria of Physcomitrella patens (156). The RecA orthologs in plant mitochondria seem to have acquired novel properties that, together with Msh1, reject the invading strand from the D loop and prevent crossing over at short repeats in mtDNA. This specific function directs the recombination event into a gene conversion or synthesis-dependent strand annealing mode (155). Thus, determination of whether or not the organellar RecA orthologs promote conventional strand invasion-type recombination in plant mitochondria also awaits further experimental tests.
Mgm101
The MGM101 (for mitochondrial genome maintenance) gene was identified in 1993 (157), but the biochemical function of the Mgm101 protein has remained largely unknown. Recent studies have revealed that Mgm101 is a Rad52-related protein of bacteriophage origin. This finding suggested the presence of a single-strand annealing (SSA)-based recombination system for the repair of mtDNA.
Mgm101 as a Rad52-related protein essential for mtDNA maintenance.
Chen et al. initially discovered the MGM101 gene in a forward genetic screen for temperature-sensitive mutations affecting mtDNA maintenance in S. cerevisiae (157). Cells expressing the mgm101-1ts allele lose mtDNA at the nonpermissive temperature. The MGM101 gene was cloned by complementation of the mgm101-1ts allele for respiratory growth at 37°C and was found to encode a protein of 269 amino acids. Mgm101 is positively charged and has a predicted isoelectric point of 10.08, suggesting that it may bind to DNA. Disruption of MGM101 in the petite-negative Kluyveromyces lactis yeast is lethal because of the loss of mtDNA (158). The Nunnari group has shown that Mgm101 is a component of mt-nucleoids (159). More importantly, mtDNA in the mgm101-2 mutant is hypersensitive to γ ray irradiation and oxidative damage, suggesting that it plays a role in mtDNA repair. Although MGM101 is essential for the maintenance of ρ+ and most ori-devoid ρ− genomes, the inheritance of hypersuppressive petite genomes that contain highly repeated ori sequences is not affected by the disruption of MGM101 (107). The ori sequences contain GC-rich clusters with inverted repeats capable of forming cruciform structures. These sequences are known to be hyperrecombinogenic (41). These phenotypes underscored a possible role of Mgm101 in recombination which can be bypassed by the presence of the highly recombinogenic mtDNA sequences. A relatively mild mutant allele of Mgm101, mgm101N150A, has been identified. Although this allele has only a moderate effect on mtDNA stability, it reduces repeat-mediated deletions in mtDNA by 8.1-fold (146). This finding further suggests that Mgm101 may be involved in mtDNA recombination.
The mature form of Mgm101 contains 247 amino acids after proteolytic processing in the mitochondrial matrix. The biochemical function of Mgm101 has remained unknown for more than a decade since the first characterization of the MGM101 gene. Zuo et al. found that the central domain of Mgm101, which is highly conserved among Mgm101 homologs from fungal species and lower animals (Fig. 2), shares low (17%) but recognizable sequence similarity with the N-terminal single-strand annealing (SSA) domain of the yeast Rad52 (Rad52-N-ter) (160). The Mgm101 core is predicted to have the β3-β4-β5-α3 fold which is characteristic of the SSA domain in Rad52 (161, 162) (Fig. 3A). Recent studies by the Chen group have shown that several amino acids in the Mgm101 core (e.g., N150, F153, and F235), which are conserved in Rad52, are essential for mtDNA maintenance in vivo (146). These observations lend important support for possible functional similarities between Mgm101 and Rad52.
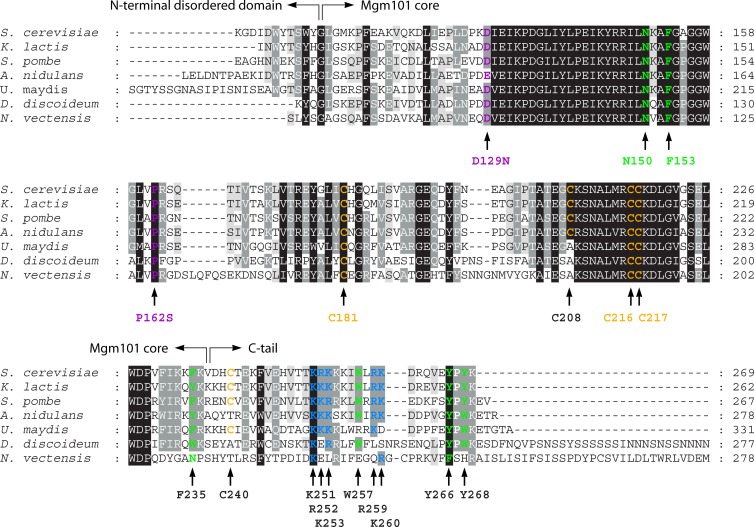
Fig 2.
Sequence conservation among closely related Mgm101 orthologs from the fungal (Saccharomyces cerevisiae, Kluyveromyces lactis, Schizosaccharomyces pombe, Aspergillus nidulans, Neurospora crassa, and Ustilago maydis), animal (Nematostella vectensis), and Dictyostelids (Dictylostelium discoideum) lineages within the unikont clade of the eukaryotic tree. Highlighted are the residues analyzed by mutagenesis in the S. cerevisiae Mgm101. The P162S and D129N alleles are temperature sensitive for mtDNA maintenance in vivo (157, 159). N150, F153, and F235 are conserved in Rad52, and their replacement by alanine affects protein folding and destabilize mtDNA (146). Similar mutations in Rad52 affect DNA repair in the nucleus. The C216/C217 cysteine pair is speculated to sense the redox state or other types of signals in mitochondria, which regulates Mgm101 activity (167). The K251/R252/K253 triad, Y266, and Y268 are required for ssDNA binding, and R259 is essential for the maintenance of the ring structure (179).
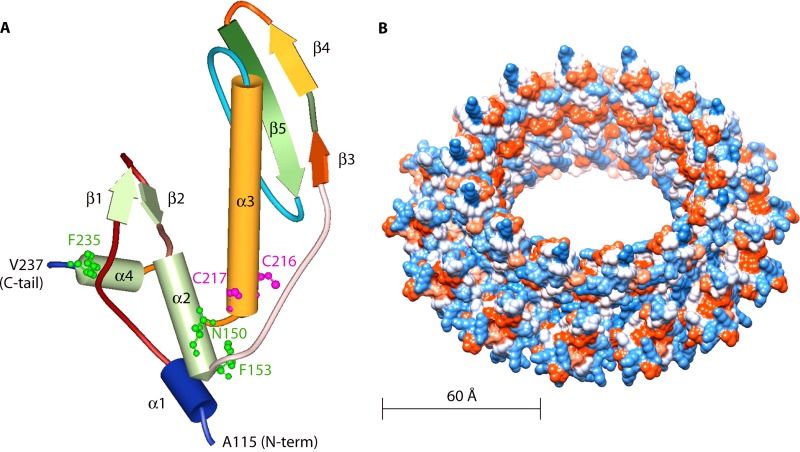
Fig 3.
Molecular modeling of the Mgm101 core domain (from A115 to V237) based on the Rad52 structure (1H2I). (A) The positions of N150, F153, and F235, which are highly conserved in Rad52-related proteins, are highlighted. Also indicated are C216 and C217, which form a putative redox sensor for regulating Mgm101 function. (B) Surface representation model of a 14-mer ring formed by Mgm101115-237. (Adapted from reference 165 [© The American Society for Biochemistry and Molecular Biology].)
Biochemical, functional, and structural properties.
Another serious challenge in the study of Mgm101 is the insolubility of the protein when expressed in Escherichia coli, which has greatly hindered its biochemical characterization. This issue has recently been overcome by expression of Mgm101 in a maltose binding protein (MBP)-fused form, which allows the production of large quantities of the protein for biochemical studies (146, 163). This achievement has greatly contributed to the rapid progress in the understanding of Mgm101 function. Indeed, it was found that Mgm101 shares biochemical, structural, and functional properties with Rad52-N-ter (146). Mgm101 preferentially binds to ssDNA over dsDNA, with Kd (dissociation constant) values of 192 nM and 1.068 μM, respectively. Functionally, Mgm101 catalyzes the annealing of ssDNAs, even when the ssDNA substrates are precomplexed with the mitochondrial single-stranded DNA binding protein Rim1. This is consistent with the model that free ssDNA is first bound by Rim1 to prevent the formation of secondary structure. Mgm101 then displaces Rim1 and forms nucleoprotein filaments competent for strand annealing.
Mgm101 also shares structural similarities with Rad52-related proteins. It forms oligomeric rings of ∼14-fold symmetry with a large central channel (Fig. 3B). The Mgm101 rings have a diameter of ∼200 Å, as revealed by negative-stain transmission electron microscopy (146). More importantly, all the Rad52-related proteins so far known form homo-oligomeric rings of 10- to 14-fold symmetry (161, 162, 164–166). This finding further supports a common evolutionary origin between Mgm101 and Rad52-related proteins, despite rather limited similarities in their primary sequences. A solution structure of Mgm101 has recently been solved by small-angle X-ray scattering analysis, which confirms the ring-shaped higher-order structural organization (167).
Structural dynamics and functional regulation.
Although freshly prepared Mgm101 forms rings uniformly, the higher-order structure of Mgm101 is rather dynamic (146). The protein forms highly compressed helical filaments with a pitch of only ∼50 in vitro after storage at 4°C. The mechanism of filamentation is currently unknown. One possibility is that it is stimulated by slow oxidation during storage. It also is not clear whether filamentation has some functional implications in vivo or is merely a nonphysiological property in vitro. The wild-type filaments have an ssDNA binding activity comparable to that of the rings. It is important to note that when bound to ssDNA, no Mgm101 rings remain visible. Instead, Mgm101 forms highly condensed nucleoprotein complexes on the ssDNA substrate. This finding supports the idea that the Mgm101 rings or filaments are subject to conformational remodeling when exposed to ssDNA.
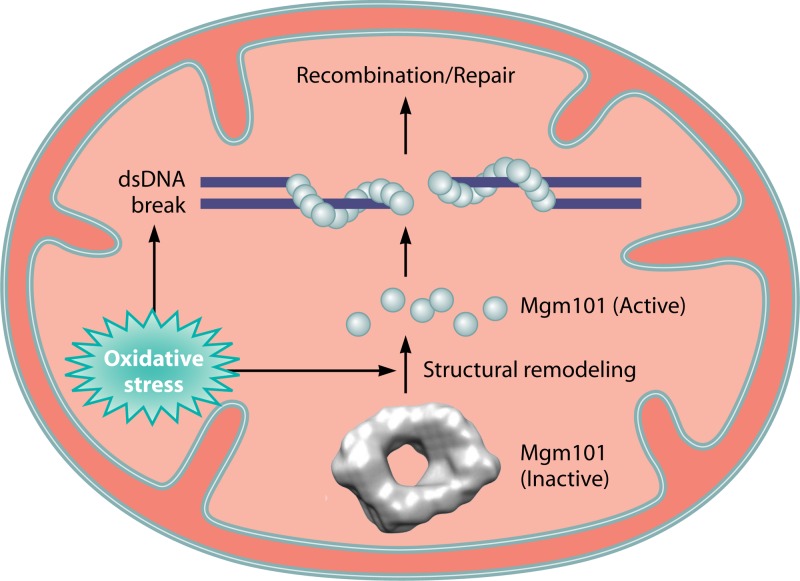
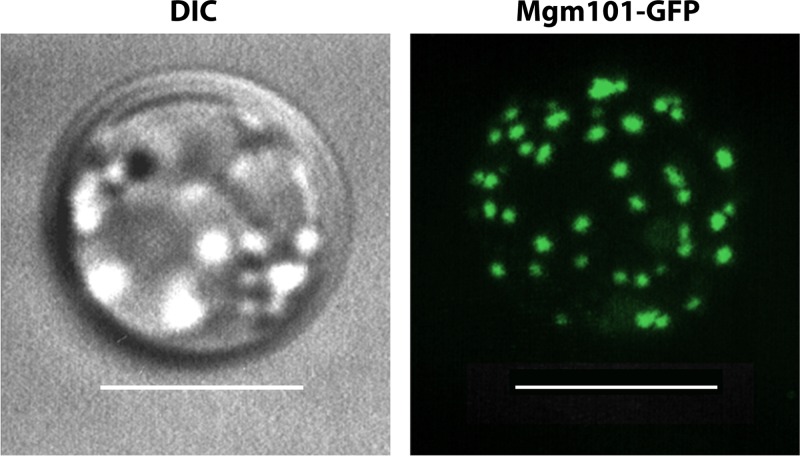
The higher-order structure of Mgm101 may be subject to regulation as a function of the redox state in mitochondria. Mutations in a vicinal pair of cysteines (C216 and C217) (Fig. 2 and 3A), which either disassemble the ring structure or induce the formation of extended polymers/filaments, drastically destabilize mtDNA in vivo (167). These cysteine residues are proposed to be part of a molecular device that senses the redox or other types of physiological stresses (Fig. 4). Modification of the cysteines may reconfigure the Mgm101 rings and activate its mtDNA repair activity. It was shown that the C216A C217A double mutant protein exists in a “broken-washer” instead of a closed-ring conformation (167). This specific conformation stimulates the formation of helical filaments in vitro, in parallel with the loss of mtDNA maintenance function in vivo. On the other hand, replacement of C216 and C217 by aspartic acid leads to protein aggregation. Aggregate formation may result from excessive disassembly of the ring structure, which produces the otherwise highly unstable protomers of Mgm101. These findings further strengthen evidence for a potential role of these two cysteines in regulating the higher-order structural organization and function of Mgm101 through a redox-based mechanism. The current model maintains that the rings/filaments may be the storage form that concentrates Mgm101 in the mt-nucleoids (Fig. 4). In response to stress, the ring structure is remodeled to activate ssDNA binding activity. The dsDNA breaks can therefore be rapidly repaired without de novo synthesis of Mgm101 in the cytosol and subsequent import into mitochondria. In respiring cells overexpressing an Mgm101-green fluorescent protein (GFP) fusion, up to ∼40 to 50 bright foci can be observed (Fig. 5). This is close to the average number of mt-nucleoids per cell. These distinct structures remarkably resemble the Rad52 foci formed in the nuclear DNA repair center under DNA-damaging conditions (168). It remains unknown how exactly the potential regulatory cysteines are modified under stress conditions to activate the mtDNA repair function of Mgm101.
Fig 4.
A model for activation of Mgm101 for mtDNA repair. The Mgm101 rings are proposed to be the storage form of the protein in mt-nucleoids. In response to oxidative stress or other signals, the C216/C217 cysteine pair is modified. This may induce structural remodeling in the rings, which stimulates ssDNA binding and the repair of dsDNA breaks.
Fig 5.
Confocal microscopy showing that overexpressed Mgm101-GFP forms bright structures resembling the Rad52 foci in the nuclear DNA repair center. Mgm101-GFP was expressed from the constitutive ADH1 promoter. Scale bar, 5 μm. DIC, differential interference contrast.
Evolutionary origin and functional implications.
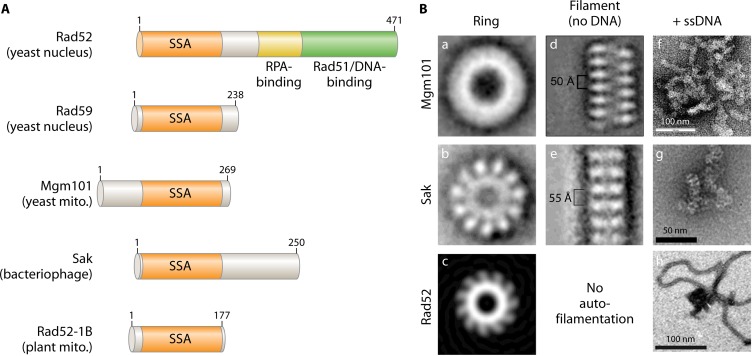
The Rad52-related proteins are widespread in the temperate bacteriophages (165, 169, 170), as represented by Redβ and Erf from the bacteriophages λ and P22, RecT from the prophage rac, and Sak from the lactococcal phage ul36. These proteins are probably better described as single-strand annealing proteins (SSAPs) based on their unique biochemical activity (169). Unlike the eukaryotic Rad52, which contains a large C-terminal domain with multiple activities, including the recruitment of the Rad51 core recombinase, Mgm101 and its bacteriophage counterparts lack such a domain (Fig. 6A). Accordingly, the SSAPs in bacteriophages catalyze recombination by the single-strand annealing mechanism independent of the bacterial RecA recombinase (171). The higher-order structure of Mgm101 is closer to that of the bacteriophage enzymes than to that of the eukaryotic Rad52, in that they all self-assemble into macromolecular filaments and form condensed nucleoprotein complexes in the presence of ssDNA (Fig. 6B). Thus, Mgm101 may have evolved from an ancestral protein of bacteriophage origin.
Fig 6.
Structural organization of Mgm101 in comparison with the nuclear Rad52, the Sak protein from the lactococcal phage ul36, and the mitochondrial Rad52-1B variant of A. thaliana. (A) Rad52 is a central recombination protein for mediating Rad51-catalyzed strand invasion of dsDNA in the nucleus. It has retained the single-strand annealing (SSA) activity on its N terminus. Rad52 has acquired a large C-terminal domain that interacts with the nuclear single-strand binding protein RPA. It recruits Rad51 onto ssDNA and enhances Rad51 nucleation through a second DNA binding activity (229–233). The yeast Rad52 paralog, Rad59, lacks the C-terminal Rad51-interacting domain (234). (B) The higher-order structural organization of Mgm101 in comparison with Sak and the SSA domain of Rad52. (a to c) Transmission electron microscopy shows that Mgm101 forms rings of ∼14-fold symmetry (a) and that Sak (b) and Rad52 (c) form rings with 11 subunits. These rings have diameters of 200 Å for Mgm101, 150 Å for Sak, and 130 Å for the SSA domain of Rad52. (d and e) Mgm101 and Sak, but not Rad52, also form highly compressed filaments with pitches of only 50 and 55 Å, respectively. (f to h) In the presence of ssDNA, Mgm101 and Sak form condensed nucleoprotein complexes, in contrast to Rad52, which forms uniform thin filaments on ssDNA. (Panels a, d, and f are reprinted from reference 146 with permission of the publisher [© The American Society for Biochemistry and Molecular Biology]. Panels b, c, e, and g are reprinted from reference 166 with permission of Elsevier. Panel h is adapted from reference 235 with permission of the publisher [© The American Society for Biochemistry and Molecular Biology].)
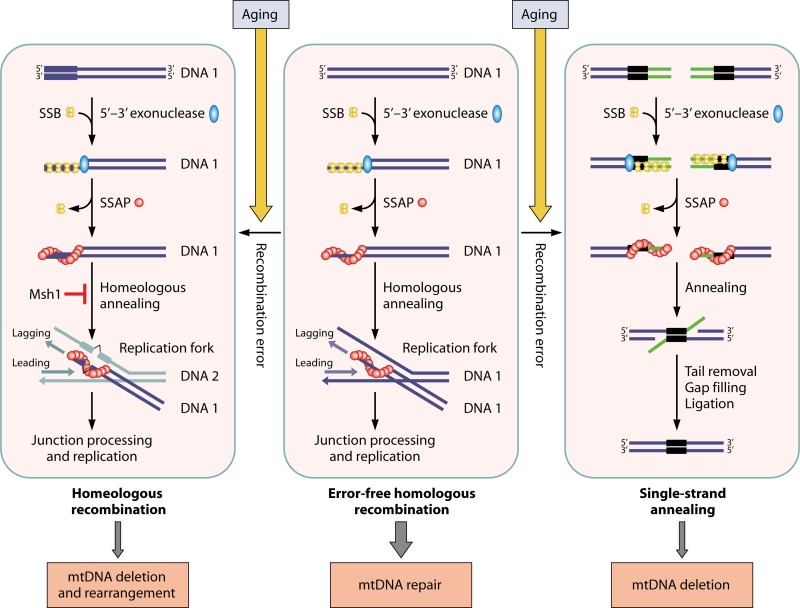
In light of the structural and biochemical similarities between Mgm101 and SSAPs in the bacteriophages, it is proposed that the mitochondrial recombination apparatus may operate in the same way as its bacteriophage counterpart. The Redβ protein in phage λ initiates strand annealing by preferentially targeting on the lagging strand of a replication fork (172, 173). By analogy, Mgm101 may well function in the same manner (Fig. 7, middle panel). Instead of invading dsDNA duplexes like the ATP-dependent Rad51/RecA type-recombinases, the Mgm101-ssDNA nucleoprotein complexes may be directly annealed to those homologous single-stranded donor sequences, thereby mediating error-free recombinational repair of mtDNA. In addition to the lagging strand, other singled-stranded sources may also serve as templates. For instance, mitochondrial transcription can displace the nontranscribed strand, which gives rise to abundant and long single-stranded species (174). Likewise, the processing of RNA/DNA hybrids and the formation of cruciform structures may also expose ssDNA stretches that are targeted for homologous annealing and the initiation of recombination. Like in the bacteriophages, the recombination in mtDNA may be nonreciprocal, which produces single rather than double Holliday junctions. These recombination intermediates are finally processed by the Mgt1/Cce1 endonuclease.
Fig 7.
Models for the operation mode of a recombination system of bacteriophage origin in mitochondria and its implications for the generation of mtDNA deletions in aged cells. A single-stranded DNA-annealing protein such as Mgm101 plays a central role in the repair of double-strand DNA breaks (middle panel). After the resection of a dsDNA end by a 5′-3′ exonuclease, SSB is bound to the exposed ssDNA to prevent the formation of secondary structures. SSB is then displaced by an SSAP like Mgm101. The SSAP/ssDNA nucleoprotein filaments initiate recombination by annealing to homologous single-stranded DNA on the lagging strand of a replication fork, like the Redβ protein in the phage λ (172, 173). The Mgt1/Cce1 endonuclease may process the recombination junction before the loading of the mtDNA replisome. However, recombinational errors may occasionally occur, either by a classic single-strand annealing between intramolecularly repeated sequences, which generates unrepairable mtDNA deletions (right panel), or by homeologous annealing to a nonhomologous lagging strand, which causes mtDNA deletions and rearrangements (left panel). The mismatch repair protein Msh1 may play a role in recombination editing by rejecting homeologous annealing. In young cells, mtDNA recombination is kept at a very low level to clear the rarely arising DSBs. In aged cells, mtDNA recombination increases as a result of oxidative stress and elevated mtDNA damage. This may inevitably increase erroneous recombinational events, which leads to the time-dependent accumulation of unfixable deleted and rearranged mtDNAs.
Molecular and functional organization.
The mechanistic details of Mgm101-catalyzed recombination are yet to be established. This is a rather challenging task because recombination may be tightly coupled to mtDNA replication (see below). Mutations affecting the recombination activity of Mgm101 would directly affect replication and result in a ρo state, which prevents the capture of recombination events in the mitochondrial genome. As mentioned above, the only piece of direct genetic evidence so far available for the involvement of Mgm101 in recombination is that the N150A allele of Mgm101, which has a very mild effect on mtDNA stability, has reduced repeat-mediated recombination compared with that of a wild-type control (146).
A pertinent question is how Mgm101 interacts with other proteins in the recombination pathway, including Rim1, Mhr1, and DNA helicases (e.g., Pif1 and Hmi1). Mgm101 has a unique long N-terminal region (Fig. 6A), which contains a 68-amino-acid domain predicted to be intrinsically unstructured or disordered. This domain is essential for mtDNA maintenance in a species-specific manner. Hayward et al. generated chimeric proteins from yeast and the coral Acropora millepora by swapping the N-terminal disordered domain and the central SSA core (175). It was found that the core domain of A. millepora Mgm101 can functionally replace the yeast sequences, but the N-terminal disordered domain of the coral protein cannot substitute for its yeast counterpart for mtDNA maintenance. It is speculated that the N-terminal disordered domain of Mgm101 is the interaction target of a specific mt-nucleoid protein in yeast which is not recognized by the corresponding domain in A. millepora Mgm101. The N-terminal disordered domain seems to direct the protein into the mt-nucleoids, possibly through association with another mt-nucleoid protein. It would be predicted that additional protein-protein interactions may be important for modulating the strand-annealing reaction catalyzed by the SSA core domain.
Another important question is how Mgm101 initiates interaction with ssDNA and promotes strand annealing. In the case of the eukaryotic Rad52, it is proposed that ssDNA binds to a central groove along the surface of the ring formed by the SSA core (162). Homology search is achieved through successive interactions between two separate ssDNA-wrapped rings until the formation of a stable duplex occurs (176, 177). Indeed, mutations in the presumptive central binding groove affect ssDNA binding activity (161, 178). Surprisingly, the Chen group has recently found that the major ssDNA binding activity in Mgm101 resides in a 32-amino-acid carboxyl-terminal tail instead of the central SSA core (179). The C-terminal tail is not conserved as well as the core SSA domain among the closely related Mgm101 homologs (Fig. 2), but several conserved amino acids have been identified to play roles in ssDNA binding. The data supported the model that the positively charged 251-KRK-253 triad in the C-terminal tail may initiate the contact with the phosphate backbone of ssDNA by electrostatic interactions. The highly conserved 266-YPY-268 motif at the extreme end of the protein may stabilize the interactions by base stacking. These interactions could disrupt the ring structure maintained by salt bridges involving Arg259. This may facilitate the deployment of additional subunits in the ring for interactions with ssDNA. As the C-terminal tail is conserved in Mgm101 homologs that function in mitochondria but not in the nuclear Rad52 or bacteriophage SSAPs, it is possible that it may have arisen from convergent evolution as a successful adaptation to catalyze recombination and DNA repair in the specific genetic setting of mitochondria.
In addition to ssDNA binding, the C-terminal tail is also required for the stabilization of the Mgm101 rings in vitro and for protein stability in vivo (179). Mutant proteins unable to maintain the ring structure in vitro are degraded in vivo. The degradation of unassembled Mgm101 monomers may be necessary for the formation of ring-based mtDNA repair centers in the mt-nucleoids.
Mgm101 homologs.
Several Mgm101 homologs have been functionally characterized. Like in yeast, mitochondria in Arabidopsis thaliana also express a short version of a Rad52-related protein similar to Mgm101, known as Rad52-1B (Fig. 6A) (180). Gualberto and coworkers independently identified Rad52-1B as ODB1 (organellar DNA binding protein 1) based on its DNA binding activity. Rad52-1B/ODB1 shares similar properties with Mgm101, including its preferential binding to ssDNA and the ability to promote single-strand annealing (181). More interestingly, these investigators also found that under genotoxic stress, the recombination in mtDNA repeats is significantly reduced in odb1 mutant plants, reminiscent of the phenotype observed with the N150A allele of MGM101 (146). In contrast, microhomology-mediated recombination is increased in the absence of ODB1. This suggests a critical role of the protein in effective repair of double-strand breaks. A defect in such a function directs the repair to the microhomology-mediated recombination mechanism. Plants lacking both ODB1 and RecA3 seem to be nonviable. This would support the idea that ODB1 and RecA3 may operate in separate but functionally redundant mtDNA repair pathways. Based on the molecular organization of Rad52-1B/ODB1, which lacks an apparent RecA binding domain as in Rad52, it would be predicted that it may function in a RecA-independent manner like Mgm101 and its bacteriophage counterparts.
The Sasaki group identified the Mgm101 homolog Glom2 in Physarum polycephalum (182). Glom2 has a core domain highly homologous to that of Mgm101, but it also has a 218-amino-acid long C-terminal domain containing three polyproline tracts. The exact function of the long C-terminal domain is unknown. It would be interesting to know whether or not it interacts with a RecA-type protein, if such a conventional recombinase exists in Physarum mitochondria. Downregulation of Glom2 does not have an obvious effect on mtDNA maintenance. When combined with the loss of the Abf2 homolog Glom, the downregulation of Glom2 further reduces the size of mt-nucleoids and mtDNA copy number. A potential role of Glom2 in mtDNA recombination and repair remains to be investigated.
OTHER FACTORS AFFECTING mtDNA RECOMBINATION
In addition to a potential difference in the basic machineries for HR, several additional factors have been proposed to account for the wide variation of recombination frequencies in different cell types and species. For instance, nucleoid organization, mitochondrial dynamics, mtDNA content, and mitochondrial density may all affect recombination. In cultured human cells, the nucleoidal organization is highly autonomous (183, 184). Schon and coworkers fused two cell lines which were homoplasmic for deletions in two nonoverlapping regions affecting protein synthesis. It was found that the two mtDNAs can transcomplement defects in protein synthesis but remain in independent nucleoids without stable intermixing. Human mitochondria in most cell lines are rather fragmented, and each nucleoid contains three or fewer copies of the 16.5-kb mtDNA (33–35). The high autonomy of nucleoids and low mtDNA content may set physical barriers for inter- and intranucleoidal recombinations, respectively. In contrast, the mtDNA copy number in the myocardium is ∼1.9- to 3.8- and ∼10-fold higher than those in skeletal muscle and epithelial cells, respectively (185–187). Data on the exact copy number of mtDNA in each nucleoid and mitochondrion in these tissues are currently unavailable. It would be predicted that the high mtDNA content, together with a highly oxidative environment, may contribute to the high levels of recombination in human heart mtDNA (see above). Likewise, the relatively high frequency of recombination in yeast mtDNA may also be partially explained by differences in genome size, nucleoid organization, and mitochondrial dynamics. The yeast mitochondrial genome is 4.5 times larger than human mtDNA. Under normal growth conditions on glucose medium, which was used for most recombination assays, yeast cells develop an elongated mitochondrial reticulum that contains multiple giant nucleoids, with each harboring up to 20 copies of mtDNA (188). These conditions may facilitate both intranucleoidal recombination and internucleoidal exchanges.
LINKING mtDNA RECOMBINATION, REPAIR, AND REPLICATION
DSBs are the most detrimental damage to any genome. It is now increasingly appreciated that HR may have evolved as a molecular strategy for the repair of DSBs. For a long time, the importance of recombination between homologous mtDNA molecules in the mostly homoplasmic cells has been undervalued. As DSBs arise as rare events in most cell types under physiological conditions, mtDNA recombination should be expected to be low. It is now argued that a low level of recombination may be sufficient for mutational clearance in mtDNA. Evolutionarily, maintaining homologous recombination at a low level is necessary to protect mtDNA against invasion of selfish elements (189, 190). A direct link between DNA recombination and repair has been supported by the observations that the recombination proteins Mgm101 and Mhr1 are both required for the maintenance of the mitochondrial genome when yeast cells are exposed to DNA-damaging conditions (146, 159, 191). Consistent with this idea, mtDNA-carried genes from mammals accumulate more mutations than those in yeast (192). A low recombination activity of mammalian mitochondria may account for the lower rate of mutation correction than in yeast, in which recombination is high.
The implications of homologous recombination could be beyond mtDNA repair. Early studies showed that immediately following the inactivation of MGM101, the mtDNA copy number is halved by every cell division in yeast (107). This severe phenotype led to the assumption that Mgm101 might be involved in mtDNA replication, possibly by generating recombination intermediates with free 3′ ends that are used for priming replication. This mode of replication initiation may be followed by a rolling-circle mechanism initially proposed by the Clark-Walker group (193). In fact, recombination-based replication has been proposed as an alternative mechanism for mtDNA synthesis in several organisms (55, 194, 195). The recombination-based mtDNA replication model has recently gained strong support from studies suggesting that the replicative forks in Candida albicans mtDNA are generated from recombination intermediates (196). The presence of recombination-based replication in human heart and brain (87) and in plant mitochondria (197–199) has also been proposed. The physiological impact of this specific mechanism vis-à-vis the conventional RNA-primed replication mode is yet to be substantiated.
Ling and Shibata have provided some key evidence for the involvement of the Mhr1 protein in the initiation of the rolling-circle replication of yeast mtDNA (145). Rolling-circle replication generates mtDNA concatemers. Mutation in MHR1 reduces concatemer formation, which subsequently affects the partitioning of newly synthesized mtDNA into the buds and delays the establishment of mtDNA homoplasmy in the daughter cells (200, 201). Mhr1 is also required for the replicative advantage of the ori5 hypersuppressive ρ− genome, which has apparently frequent double-strand breaks (202), consistent with its role in recombinational repair of mtDNA. It was initially proposed that Mhr1 may have a Rad52-type activity like Redβ in the phage λ. Redβ is known to initiate rolling-circle replication by annealing the Redβ-ssDNA filaments preferentially on the lagging strand of a replication fork, followed by template switch and DNA synthesis (172, 173, 203). The recent finding of Mgm101 as a Rad52-type protein raises the possibility that Mgm101 may fulfill such a role, in a manner dependent on or independent of Mhr1 (146, 167). The Nunnari laboratory has shown that Mgm101 is preferentially associated with actively replicating mtDNA nucleoids in vivo, and that Mgm101 coimmunoprecipitates with the mitochondrial DNA polymerase Mip1 (204). Thus, Mgm101 may directly recruit the mtDNA replisome following the strand-annealing reaction. Future studies are required to support a role of Mgm101 in initiating the strand-annealing reaction prior to the rolling-circle replication. It is important to note that mhr1 and mgm101 mutants share similar phenotypes: both genes are essential for the stability of ρ+ but not the highly recombinogenic ρ− genomes (107, 144). The loss of Mhr1 has a less severe effect on the maintenance of the N1 ρ− genome than the disruption of MGM101 (107). These two proteins likely function in the same pathway in initiating recombination-based mtDNA replication. Mhr1 does not stimulate the single-strand annealing activity of Mgm101, and Mgm101 has little effect on the D-loop formation activity of Mhr1 in vitro (our unpublished observation). How these two proteins cooperate in initiating recombination-based mtDNA repair and replication is a pressing issue in the field.
HOMOLOGOUS VERSUS HOMEOLOGOUS RECOMBINATION: IMPLICATIONS FOR mtDNA DELETION/REARRANGEMENT
No biological system is perfect. A controlled recombination activity benefits mtDNA repair, but hyperrecombination could be detrimental for mitochondrial genome stability. Extensive mtDNA deletions in dopaminergic neurons from aged individuals and those suffering from Parkinson's disease have been reported (26, 27). Krishnan et al. proposed that these deletions arise by a mechanism resembling the classic SSA (30) (Fig. 7, right panel). After symmetrical resection of dsDNA ends by a 5′-3′ exonuclease, repeated sequences exposed on the other strands misanneal. The removal of the unpaired tails by exonucleases followed by gap filling and ligation generates intramolecular deletions in mtDNA. The Rad52-related SSAPs are well known for their activity in promoting single-strand annealing (205–207), which generates deletions of genomic sequences between directly repeated regions.
In light of its functional similarity to SSAPs, Mgm101 may be involved in single-strand annealing and mtDNA deletion. However, this operation mode would require an extensive resection of dsDNA ends in order to expose the homologous sequences. This could become a limiting factor when the homologous sequences are separated by large intervening regions. Alternatively, it is possible that homeologous recombination, which occurs between mismatched sequences, may play a major role in generating mtDNA deletions/rearrangements (Fig. 7, left panel). It has been well recognized that the SSAPs catalyze recombination between related but diverged DNA sequences. For example, a sequence divergence of 4% between the target genes has little effect on the recombination catalyzed by the Redβ protein in phage λ (208). This is in sharp contrast to recombination in the E. coli chromosome catalyzed by RecA, in which a 4% divergence reduces recombination by 270-fold (209). Redβ can tolerate a sequence divergence in the recombination target genes of as high as 22%, although the recombination efficiency is reduced by ∼100-fold. Interestingly, detailed analysis of human mtDNA suggested that deletions most likely happen between imperfectly matched DNA sequences that are capable of forming long and stable duplexes (210). Homeologous recombination is the simplest way to promote these events.
To what extent can Mgm101 tolerate sequence divergence in the recombination targets and how homeologous recombination is controlled remain to be determined. The annealing of mismatched single-stranded DNA by the bacteriophage Redβ is suppressed by the host mismatch repair system (211). It may be expected that its counterpart in mitochondria, Msh1, could also play a role in the suppression of homeologous recombination. This may explain the dramatic hyperrecombinogenic phenotype associated with mutations in MSH1 and the rapid disintegration of mtDNA in an msh1 null mutant (130). In plant mitochondria, loss of Msh1 function leads to DNA exchange between sequences sharing only 85% identity (212). The barrier for preventing homeologous recombination in mtDNA seems to be naturally low in some animal species. In sea mussels, recombination between maternal and paternal mtDNA regions in the obligatorily heteroplasmic males that are diverged by 20% can be readily detected (213).
mtDNA RECOMBINATION IN AGING
Recombination errors, introduced by either homeologous recombination or SSA, could have important implications for mtDNA integrity in aging cells. Aging seems to increase mtDNA recombination, concomitant with increased mtDNA deletions/rearrangements. This has been shown by experiments carried out in Podospora anserina (214). Mitochondria in this filamentous ascomycete are inherited in a strictly uniparental manner (215, 216). However, heteroplasmons may be formed in the contact zone of heterokaryon incompatible strains, in which leakage of paternal mitochondria into the maternal mycelium occurs after transient hyphal fusion. This property provided a unique opportunity for examining the frequency of mtDNA recombination in the heteroplasmons derived from cells at different time points in their life spans. In reciprocal confrontational crosses between strains harboring different mtDNA markers, van Diepeningen et al. demonstrated that the levels of recombinant mtDNA remain low in crosses involving young maternal mycelium. Juvenile strains have little to no recombination in mtDNA. The recombination rate only increases marginally over the first 3/4 of the life span. Remarkably, in crosses generated from the maternal mycelium in the last 1/4 of its life span, the percentage of offspring with recombined mtDNA increases to nearly 50% (214). These observations strongly suggest that mtDNA becomes hyperrecombinogenic in aged cells.
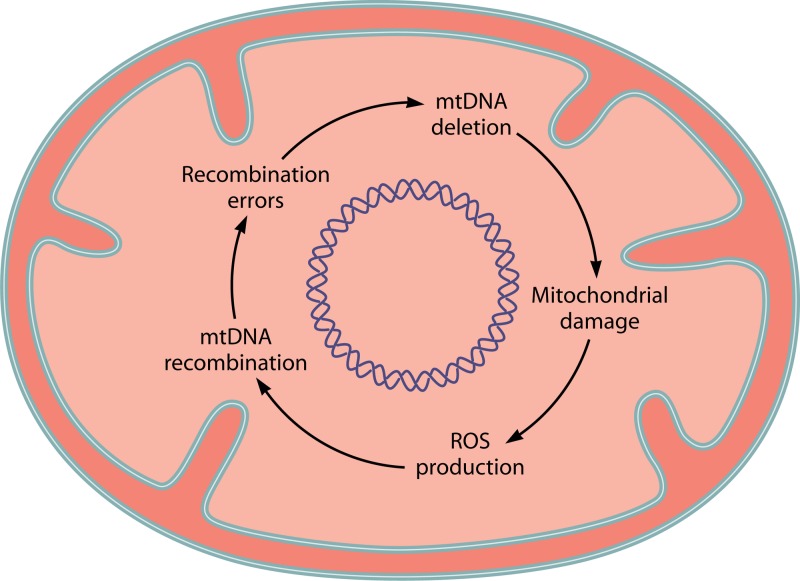
The senescence of Podospora is associated with the accumulation of various classes of mtDNA derivatives which can be both circularly and tandemly arranged, with or without coding function. Some of these mtDNA species are thought to contribute to cellular senescence (217). These “senescent” mtDNA molecules result from intramolecular recombination between short direct repeats (218). This raises the possibility that the hyperrecombinogenic state in aged cells may become a significant source of mtDNA instability. In young cells, because both DNA damage and recombination activity are low, mtDNA deletions also remain low. In contrast, the high mtDNA recombination activity in aged cells may result in the increased incidence of recombinational errors, which overwhelms the cell's capacity to clear damaged mitochondria by processes such as mitophagy. The mechanism of mtDNA recombination in Podospora and exactly how this activity is elevated in aged cells are unknown. It is possible that the recombinase is directly activated by reactive oxygen species. A closely related Mgm101 homolog is present in Podospora anserine (our unpublished observation), and its role in mtDNA recombination is yet to be established. Genetic runaway may occur. Mitochondrial dysfunction in aged cells increases the production of reactive oxygen species (ROS), which activates the Mgm101-based recombination mechanism. Hyperactive recombination increases the overall recombination errors and mtDNA deletions, which further damages mitochondria and increases ROS production (Fig. 8).
Fig 8.
A vicious-cycle model for homeologous recombination-mediated mtDNA deletions in aged cells. See the text for details.
Age-dependent increases in mtDNA recombination in other organisms have yet to be explored. Replicatively aged yeast cells have destabilized mtDNA (219, 220); however, it is unknown whether or not mtDNA disintegration is caused by hyperrecombination. Interestingly, four-way junctions in human heart mtDNA are not detected in newborns. They are acquired during early childhood concomitant with increased mtDNA copy number (221). This has been interpreted as a cellular adaptive process that responds to increased energy demand, ROS production, and double-strand DNA breaks (222). In humans, a common deletion in mtDNA is the 4,977-bp deletion which occurs between two 13-bp direct repeats. This deletion likely results from a recombination-excision mechanism and is increased in an age-dependent manner, with a correlation coefficient of 0.82 in specific tissues such as the anterior wall of the left ventricle in the heart (23). Extensive large-scale mtDNA deletions have been reported in neuronal cells during aging (26). It remains unknown whether this results from hyperrecombination.
CONCLUSION AND PERSPECTIVES
Accumulating evidence indicates that mtDNA recombination occurs not only in the traditionally studied yeast and lower animals that have biparental inheritance but also in fungal and animal species with uniparental inheritance and even in specific human tissues, including the heart and brain. Although controversy persists, increasing evidence suggests that mtDNA recombination is more widespread than was previously thought. In animals with homoplasmic mtDNA, recombinant products are indistinguishable from their progenitor molecules. Unequivocally capturing the actual mtDNA recombination events in most cell types is still a challenge in the field. Next-generation sequencing technology is already showing its power in detecting rare alleles and subgenomes of mtDNA (212, 223, 224). These technologies would be expected to detect possible recombinations between the newly arising rare mutations. This may also help to answer the question regarding the universality of mtDNA recombination across different biota (225).
Despite the fact that mtDNA recombination has been reported for over 4 decades, the study of the underlying molecular machinery has just made its debut. The field of mtDNA recombination is gaining strong momentum following the recent discovery of a bacteriophage-type SSAP in mitochondria as defined by Mgm101. Understanding the functional relationship between Mgm101 and other components in the mtDNA recombination pathway, particularly Mhr1, will be critical for advancing this field. In the coming years, a critical question to be addressed is whether human mitochondria express a similar mtDNA recombination system, instead of using a canonical recombination pathway involving a RecA/Rad51-type recombinase. Human mitochondria also harbor SSA activity conferred by proteins ostensibly unrelated to SSAPs (e.g., the Twinkle helicase [226]). Close orthologs of Mgm101 have been identified in lower animals (e.g., Nematostella vectensis) using current bioinformatic tools. However, the presence of a distantly related SSAP in higher animals cannot be completely ruled out. The SSAPs are often conserved at the structural level but not in their primary sequences. For instance, Mgm101 shares structural and functional similarities with the SSAPs in bacteriophages such as Redβ, Erf, and Sak, but sequence similarity between these proteins is hardly recognizable. Nonetheless, mitochondria and bacteriophages seem to share more molecular machineries than previously thought. Both the mitochondrial DNA and RNA polymerases are phylogenetically originated from the T-odd lineage of bacteriophages instead of alphaproteobacteria, the ancestor of mitochondria (227). The experimental data now also show that mitochondria and bacteriophages share similar molecular mechanisms for mtDNA recombination and repair. It is possible that these enzymes were acquired together from an infecting phage at the onset of endosymbiosis. Future studies should be focused on understanding how the processes of recombination, repair, and replication are interlinked and coordinated in mitochondria, which is a salient feature of DNA metabolism in bacteriophages (228).
ACKNOWLEDGMENTS
I thank George Desmond Clark-Walker (Australian National University) and the Chen lab members for comments and critical reading of the manuscript, the anonymous reviewers for their valuable suggestions, Stephan Wilkens for help in constructing the molecular models of Mgm101, and Xiaoming Zuo for generating the Mgm101-GFP construct.
I am supported by grant AG023731 from the National Institute on Aging, National Institutes of Health.
Biography

Xin Jie Chen is an Associate Professor in the Department of Biochemistry and Molecular Biology, State University of New York Upstate Medical University. He received his B.S. degree in animal science from Zhejiang Agricultural University (now Zhejiang University), his D.E.A. from Université Nancy I, and his Ph.D from Université Paris-Sud (XI). His Ph.D work was focused on yeast molecular genetics in the laboratory of Dr. Hiroshi Fukuhara at Institut Curie (Orsay, France). He conducted postdoctoral work with Dr. George Desmond Clark-Walker at the Australian National University, studying mitochondrial DNA inheritance. He also served as a faculty member at the Australian National University, Nanyang Technological University, and University of Texas Southwestern Medical Center at Dallas. For the last 2 decades, Dr. Chen's research interests have been focused on mitochondrial genetics and aging.
REFERENCES
- 1. Wallace DC. 2007. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 76:781–821 [DOI] [PubMed] [Google Scholar]
- 2. Chen XJ, Butow RA. 2005. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6:815–825 [DOI] [PubMed] [Google Scholar]
- 3. Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN. 2007. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion 7:311–321 [DOI] [PubMed] [Google Scholar]
- 4. Bogenhagen DF. 2011. Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta 1819:914–920 [DOI] [PubMed] [Google Scholar]
- 5. Spelbrink JN. 2010. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62:19–32 [DOI] [PubMed] [Google Scholar]
- 6. Nass MM. 1969. Mitochondrial DNA. I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J. Mol. Biol. 42:521–528 [DOI] [PubMed] [Google Scholar]
- 7. Albring M, Griffith J, Attardi G. 1977. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replicaton. Proc. Natl. Acad. Sci. U. S. A. 74:1348–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prachar J. 2010. Mouse and human mitochondrial nucleoid—detailed structure in relation to function. Gen. Physiol. Biophys. 29:160–174 [PubMed] [Google Scholar]
- 9. Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. 2012. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl. Acad. Sci. U. S. A. 109:6136–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown WM, George M, Jr., Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. U. S. A. 76:1967–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Howell N. 1996. Mutational analysis of the human mitochondrial genome branches into the realm of bacterial genetics. Am. J. Hum. Genet. 59:749–755 [PMC free article] [PubMed] [Google Scholar]
- 12. Larsson NG. 2010. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 79:683–706 [DOI] [PubMed] [Google Scholar]
- 13. Harman D. 1972. The biological clock: the mitochondria? J. Am. Geriatr. Soc. 20:145–147 [DOI] [PubMed] [Google Scholar]
- 14. Linnane AW, Marzuki S, Ozawa T, Tanaka M. 1989. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet i:642–645 [DOI] [PubMed] [Google Scholar]
- 15. Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309:481–484 [DOI] [PubMed] [Google Scholar]
- 16. Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417–423 [DOI] [PubMed] [Google Scholar]
- 17. Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. 2007. DNA repair, mitochondria, and neurodegeneration. Neuroscience 145:1318–1329 [DOI] [PubMed] [Google Scholar]
- 18. Wilson DM, III, Bohr VA. 2007. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst.) 6:544–559 [DOI] [PubMed] [Google Scholar]
- 19. Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. 2008. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 32:325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kazak L, Reyes A, Holt IJ. 2012. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat. Rev. Mol. Cell Biol. 13:659–671 [DOI] [PubMed] [Google Scholar]
- 21. Sykora P, Wilson DM, III, Bohr VA. 2012. Repair of persistent strand breaks in the mitochondrial genome. Mech. Ageing Dev. 133:169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. 2009. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst.) 8:704–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T, Oehmichen M. 2008. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp. Gerontol. 43:645–652 [DOI] [PubMed] [Google Scholar]
- 24. Cortopassi GA, Arnheim N. 1990. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 18:6927–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. 2006. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 79:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. 2006. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 38:518–520 [DOI] [PubMed] [Google Scholar]
- 27. Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. 2006. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 38:515–517 [DOI] [PubMed] [Google Scholar]
- 28. Bender A, Schwarzkopf RM, McMillan A, Krishnan KJ, Rieder G, Neumann M, Elstner M, Turnbull DM, Klopstock T. 2008. Dopaminergic midbrain neurons are the prime target for mitochondrial DNA deletions. J. Neurol. 255:1231–1235 [DOI] [PubMed] [Google Scholar]
- 29. Shoffner JM, Lott MT, Voljavec AS, Soueidan SA, Costigan DA, Wallace DC. 1989. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc. Natl. Acad. Sci. U. S. A. 86:7952–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. 2008. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 40:275–279 [DOI] [PubMed] [Google Scholar]
- 31. San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77:229–257 [DOI] [PubMed] [Google Scholar]
- 32. Gangavarapu V, Prakash S, Prakash L. 2007. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:7758–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iborra FJ, Kimura H, Cook PR. 2004. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA. 2011. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 31:4994–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. 2011. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U. S. A. 108:13534–13539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Birky CW., Jr 1978. Transmission genetics of mitochondria and chloroplasts. Annu. Rev. Genet. 12:471–512 [DOI] [PubMed] [Google Scholar]
- 37. Dujon B, Slonimski PP, Weill L. 1974. Mitochondrial genetics IX: a model for recombination and segregation of mitochondrial genomes in Saccharomyces cerevisiae. Genetics 78:415–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas DY, Wilkie D. 1968. Recombination of itochondrial drug resistance factors in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 30:368–372 [DOI] [PubMed] [Google Scholar]
- 39. Rank GH, Bech-Hansen NT. 1972. Somatic segretation, recombination, asymmetrical distribution and complementation tests of cytoplasmically-inherited antibiotic-resistance mitochondrial markers in S. cerevisiae. Genetics 72:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Michaelis G, Petrochilo E, Slonimski PP. 1973. Mitochondrial genetics. 3. Recombined molecules of mitochondrial DNA obtained from crosses between cytoplasmic petite mutants of Saccharomyces cerevisiae: physical and genetic characterization. Mol. Gen. Genet. 123:51–65 [DOI] [PubMed] [Google Scholar]
- 41. Gaillard C, Strauss F, Bernardi G. 1980. Excision sequences in the mitochondrial genome of yeast. Nature 283:218–220 [DOI] [PubMed] [Google Scholar]
- 42. de Zamaroczy M, Faugeron-Fonty G, Bernardi G. 1983. Excision sequences in the mitochondrial genome of yeast. Gene 21:193–202 [DOI] [PubMed] [Google Scholar]
- 43. Clark-Walker GD. 1989. In vivo rearrangement of mitochondrial DNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 86:8847–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gross SR, Levine PH, Metzger S, Glaser G. 1989. Recombination and replication of plasmid-like deriviatives of a short section of the mitochondrial genome of Neurospora crassa. Genetics 121:693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen XJ, Clark-Walker GD. 1993. Mutations in MGI genes convert Kluyveromyces lactis into a petite-positive yeast. Genetics 133:517–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen XJ, Clark-Walker GD. 1995. Specific mutations in α- and γ-subunits of F1-ATPase affect mitochondrial genome integrity in the petite-negative yeast Kluyveromyces lactis. EMBO J. 14:3277–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen XJ, Clark-Walker GD. 1996. The mitochondrial genome integrity gene, MGI1, of Kluyveromyces lactis encodes the beta-subunit of F1-ATPase. Genetics 144:1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen XJ, Clark-Walker GD. 2000. The petite mutation in yeasts: 50 years on. Int. Rev. Cytol. 194:197–238 [DOI] [PubMed] [Google Scholar]
- 49. Johnston SA, Anziano PQ, Shark K, Sanford JC, Butow RA. 1988. Mitochondrial transformation in yeast by bombardment with microprojectiles. Science 240:1538–1541 [DOI] [PubMed] [Google Scholar]
- 50. Bonnefoy N, Fox TD. 2007. Directed alteration of Saccharomyces cerevisiae mitochondrial DNA by biolistic transformation and homologous recombination. Methods Mol. Biol. 372:153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lockshon D, Zweifel SG, Freeman-Cook LL, Lorimer HE, Brewer BJ, Fangman WL. 1995. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell 81:947–955 [DOI] [PubMed] [Google Scholar]
- 52. Zweifel SG, Fangman WL. 1991. A nuclear mutation reversing a biased transmission of yeast mitochondrial DNA. Genetics 128:241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kleff S, Kemper B, Sternglanz R. 1992. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 11:699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ezekiel UR, Zassenhaus HP. 1993. Localization of a cruciform cutting endonuclease to yeast mitochondria. Mol. Gen. Genet. 240:414–418 [DOI] [PubMed] [Google Scholar]
- 55. MacAlpine DM, Perlman PS, Butow RA. 1998. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl. Acad. Sci. U. S. A. 95:6739–6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Joers P, Jacobs HT. 2013. Analysis of replication intermediates indicates that Drosophila melanogaster mitochondrial DNA replicates by a strand-coupled theta mechanism. PLoS One 8:e53249. 10.1371/journal.pone.0053249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ladoukakis ED, Zouros E. 2001. Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 18:1168–1175 [DOI] [PubMed] [Google Scholar]
- 58. Burzynski A, Zbawicka M, Skibinski DO, Wenne R. 2003. Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Mol. Biol. Evol. 20:388–392 [DOI] [PubMed] [Google Scholar]
- 59. Hagelberg E. 2003. Recombination or mutation rate heterogeneity? Implications for Mitochondrial Eve. Trends Genet. 19:84–90 [DOI] [PubMed] [Google Scholar]
- 60. White DJ, Wolff JN, Pierson M, Gemmell NJ. 2008. Revealing the hidden complexities of mtDNA inheritance. Mol. Ecol. 17:4925–4942 [DOI] [PubMed] [Google Scholar]
- 61. Berlin S, Smith NG, Ellegren H. 2004. Do avian mitochondria recombine? J. Mol. Evol. 58:163–167 [DOI] [PubMed] [Google Scholar]
- 62. Ballard JW, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13:729–744 [DOI] [PubMed] [Google Scholar]
- 63. Elson JL, Andrews RM, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. 2001. Analysis of European mtDNAs for recombination. Am. J. Hum. Genet. 68:145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Innan H, Nordborg M. 2002. Recombination or mutational hot spots in human mtDNA? Mol. Biol. Evol. 19:1122–1127 [DOI] [PubMed] [Google Scholar]
- 65. Piganeau G, Eyre-Walker A. 2004. A reanalysis of the indirect evidence for recombination in human mitochondrial DNA. Heredity (Edinb.) 92:282–288 [DOI] [PubMed] [Google Scholar]
- 66. Piganeau G, Gardner M, Eyre-Walker A. 2004. A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 21:2319–2325 [DOI] [PubMed] [Google Scholar]
- 67. Ladoukakis ED, Eyre-Walker A. 2004. Evolutionary genetics: direct evidence of recombination in human mitochondrial DNA. Heredity 93:321. [DOI] [PubMed] [Google Scholar]
- 68. Tsaousis AD, Martin DP, Ladoukakis ED, Posada D, Zouros E. 2005. Widespread recombination in published animal mtDNA sequences. Mol. Biol. Evol. 22:925–933 [DOI] [PubMed] [Google Scholar]
- 69. Eyre-Walker A, Awadalla P. 2001. Does human mtDNA recombine? J. Mol. Evol. 53:430–435 [DOI] [PubMed] [Google Scholar]
- 70. Ciborowski KL, Consuegra S, Garcia de Leaniz C, Beaumont MA, Wang J, Jordan WC. 2007. Rare and fleeting: an example of interspecific recombination in animal mitochondrial DNA. Biol. Lett. 3:554–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ujvari B, Dowton M, Madsen T. 2007. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol. Lett. 3:189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hoolahan AH, Blok VC, Gibson T, Dowton M. 2012. Evidence of animal mtDNA recombination between divergent populations of the potato cyst nematode Globodera pallida. Genetica 140:19–29 [DOI] [PubMed] [Google Scholar]
- 73. Nunes MD, Dolezal M, Schlotterer C. 2013. Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster. Mol. Ecol. 22:2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schwartz M, Vissing J. 2002. Paternal inheritance of mitochondrial DNA. N. Engl. J. Med. 347:576–580 [DOI] [PubMed] [Google Scholar]
- 75. Kraytsberg Y, Schwartz M, Brown TA, Ebralidse K, Kunz WS, Clayton DA, Vissing J, Khrapko K. 2004. Recombination of human mitochondrial DNA. Science 304:981. [DOI] [PubMed] [Google Scholar]
- 76. Zsurka G, Kraytsberg Y, Kudina T, Kornblum C, Elger CE, Khrapko K, Kunz WS. 2005. Recombination of mitochondrial DNA in skeletal muscle of individuals with multiple mitochondrial DNA heteroplasmy. Nat. Genet. 37:873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mita S, Rizzuto R, Moraes CT, Shanske S, Arnaudo E, Fabrizi GM, Koga Y, DiMauro S, Schon EA. 1990. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 18:561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Firkin FC, Clark-Walker GD. 1979. Abnormal mitochondrial DNA in acute leukaemia and lymphoma. Br. J. Haematol. 43:201–206 [DOI] [PubMed] [Google Scholar]
- 79. Clayton DA, Vinograd J. 1967. Circular dimer and catenate forms of mitochondrial DNA in human leukaemic leucocytes. Nature 216:652–657 [DOI] [PubMed] [Google Scholar]
- 80. Clayton DA, Vinograd J. 1969. Complex mitochondrial DNA in leukemic and normal human myeloid cells. Proc. Natl. Acad. Sci. U. S. A. 62:1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tang Y, Manfredi G, Hirano M, Schon EA. 2000. Maintenance of human rearranged mitochondrial DNAs in long-term cultured transmitochondrial cell lines. Mol. Biol. Cell 11:2349–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Holt IJ, Dunbar DR, Jacobs HT. 1997. Behaviour of a population of partially duplicated mitochondrial DNA molecules in cell culture: segregation, maintenance and recombination dependent upon nuclear background. Hum. Mol. Genet. 6:1251–1260 [DOI] [PubMed] [Google Scholar]
- 83. D'Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, Moraes CT, Manfredi G. 2004. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 13:3171–3179 [DOI] [PubMed] [Google Scholar]
- 84. Bacman SR, Williams SL, Moraes CT. 2009. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 37:4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fan W, Lin CS, Potluri P, Procaccio V, Wallace DC. 2012. mtDNA lineage analysis of mouse L-cell lines reveals the accumulation of multiple mtDNA mutants and intermolecular recombination. Genes Dev. 26:384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT. 2001. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2:1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pohjoismaki JL, Goffart S, Tyynismaa H, Willcox S, Ide T, Kang D, Suomalainen A, Karhunen PJ, Griffith JD, Holt IJ, Jacobs HT. 2009. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 284:21446–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sena EP, Revet B, Moustacchi E. 1986. In vivo homologous recombination intermediates of yeast mitochondrial DNA analyzed by electron microscopy. Mol. Gen. Genet. 202:421–428 [DOI] [PubMed] [Google Scholar]
- 89. Williamson D. 2002. The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 3:475–481 [DOI] [PubMed] [Google Scholar]
- 90. Bendich AJ. 1996. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. 255:564–588 [DOI] [PubMed] [Google Scholar]
- 91. Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Symington LS. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Van Dyck E, Foury F, Stillman B, Brill SJ. 1992. A single-stranded DNA binding protein required for mtiochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 11:3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. 1994. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur. J. Biochem. 221:435–443 [DOI] [PubMed] [Google Scholar]
- 95. Yang C, Curth U, Urbanke C, Kang C. 1997. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat. Struct. Biol. 4:153–157 [DOI] [PubMed] [Google Scholar]
- 96. Oliveira MT, Kaguni LS. 2010. Functional roles of the N- and C-terminal regions of the human mitochondrial single-stranded DNA-binding protein. PLoS One 5:e15379. 10.1371/journal.pone.0015379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Foury F, Kolodynski J. 1983. Pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 80:5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lahaye A, Leterme S, Foury F. 1993. PIF1 DNA helicase from Saccharomyces cerevisiae—biochemical characterization of the enzyme. J. Biol. Chem. 268:26155–26161 [PubMed] [Google Scholar]
- 99. Lahaye A, Stahl H, Thines-Sempoux D, Foury F. 1991. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 10:997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mimitou EP, Symington LS. 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst.) 8:983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. 2010. Rad54, the motor of homologous recombination. DNA Repair (Amst.) 9:286–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Foury F, Dyck EV. 1985. A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. EMBO J. 4:3525–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ramanagoudr-Bhojappa R, Blair LP, Tackett AJ, Raney KD. 2013. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 41:1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kuusk S, Sedman T, Joers P, Sedman J. 2005. Hmi1p from Saccharomyces cerevisiae mitochondria is a structure-specific DNA helicase. J. Biol. Chem. 280:24322–24329 [DOI] [PubMed] [Google Scholar]
- 105. Sedman T, Joers P, Kuusk S, Sedman J. 2005. Helicase Hmi1 stimulates the synthesis of concatemeric mitochondrial DNA molecules in yeast Saccharomyces cerevisiae. Curr. Genet. 47:213–222 [DOI] [PubMed] [Google Scholar]
- 106. Sedman T, Kuusk S, Kivi S, Sedman J. 2000. A DNA helicase required for maintenance of the functional mitochondrial genome in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:1816–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zuo XM, Clark-Walker GD, Chen XJ. 2002. The mitochondrial nucleoid protein, Mgm101p, of Saccharomyces cerevisiae is involved in the maintenance of rho+ and ori/rep-devoid petite genomes but is not required for hypersuppressive rho− mtDNA. Genetics 160:1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Monroe DS, Jr, Leitzel AK, Klein HL, Matson SW. 2005. Biochemical and genetic characterization of Hmi1p, a yeast DNA helicase involved in the maintenance of mitochondrial DNA. Yeast 22:1269–1286 [DOI] [PubMed] [Google Scholar]
- 109. de Souza-Pinto NC, Aamann MD, Kulikowicz T, Stevnsner TV, Bohr VA. 2010. Mitochondrial helicases and mitochondrial genome maintenance. Mech. Ageing Dev. 131:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. 2001. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 28:223–231 [DOI] [PubMed] [Google Scholar]
- 111. Copeland WC, Longley MJ. 2008. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol. Cell 32:457–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Futami K, Shimamoto A, Furuichi Y. 2007. Mitochondrial and nuclear localization of human Pif1 helicase. Biol. Pharm. Bull. 30:1685–1692 [DOI] [PubMed] [Google Scholar]
- 113. Phadnis N, Sia RA, Sia EA. 2005. Analysis of repeat-mediated deletions in the mitochondrial genome of Saccharomyces cerevisiae. Genetics 171:1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Steele DF, Butler CA, Fox TD. 1996. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. U. S. A. 93:5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Sia RA, Carrol S, Kalifa L, Hochmuth C, Sia EA. 2009. Loss of the mitochondrial nucleoid protein, Abf2p, destabilizes repetitive DNA in the yeast mitochondrial genome. Genetics 181:331–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Diffley JF, Stillman B. 1991. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl. Acad. Sci. U. S. A. 88:7864–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Diffley JFX, Stillman B. 1992. DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem. 267:3368–3374 [PubMed] [Google Scholar]
- 118. Friddle RW, Klare JE, Martin SS, Corzett M, Balhorn R, Baldwin EP, Baskin RJ, Noy A. 2004. Mechanism of DNA compaction by yeast mitochondrial protein Abf2p. Biophys. J. 86:1632–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Brewer LR, Friddle R, Noy A, Baldwin E, Martin SS, Corzett M, Balhorn R, Baskin RJ. 2003. Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p. Biophys. J. 85:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. O'Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. 2002. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol. Cell. Biol. 22:4086–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Visacka K, Gerhold JM, Petrovicova J, Kinsky S, Joers P, Nosek J, Sedman J, Tomaska L. 2009. Novel subfamily of mitochondrial HMG box-containing proteins: functional analysis of Gcf1p from Candida albicans. Microbiology 155:1226–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ohno T, Umeda S, Hamasaki N, Kang D. 2000. Binding of human mitochondrial transcription factor A, an HMG box protein, to a four-way DNA junction. Biochem. Biophys. Res. Commun. 271:492–498 [DOI] [PubMed] [Google Scholar]
- 123. Kalifa L, Quintana DF, Schiraldi LK, Phadnis N, Coles GL, Sia RA, Sia EA. 2012. Mitochondrial genome maintenance: roles for nuclear nonhomologous end joining proteins in Saccharomyces cerevisiae. Genetics 190:951–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zassenhaus HP, Hofmann TJ, Uthayashanker R, Vincent RD, Zona M. 1988. Construction of a yeast mutant lacking the mitochondrial nuclease. Nucleic Acids Res. 16:3283–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Dake E, Hofmann TJ, McIntire S, Hudson A, Zassenhaus HP. 1988. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 263:7691–7702 [PubMed] [Google Scholar]
- 126. Zassenhaus HP, Denniger G. 1994. Analysis of the role of the Nuc1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr. Genetics 25:142–149 [DOI] [PubMed] [Google Scholar]
- 127. Mieczkowski PA, Fikus MU, Ciesla Z. 1997. Characterization of a novel DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, which is a structural homolog of the RAD2 and RAD27 DNA repair genes. Mol. Gen. Genet. 253:655–665 [DOI] [PubMed] [Google Scholar]
- 128. Fikus MU, Mieczkowski PA, Koprowski P, Rytka J, Sledziewska-Gojska E, Ciesla Z. 2000. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics 154:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Koprowski P, Fikus MU, Dzierzbicki P, Mieczkowski P, Lazowska J, Ciesla Z. 2003. Enhanced expression of the DNA damage-inducible gene DIN7 results in increased mutagenesis of mitochondrial DNA in Saccharomyces cerevisiae. Mol. Genet. Genomics 269:632–639 [DOI] [PubMed] [Google Scholar]
- 130. Mookerjee SA, Sia EA. 2006. Overlapping contributions of Msh1p and putative recombination proteins Cce1p, Din7p, and Mhr1p in large-scale recombination and genome sorting events in the mitochondrial genome of Saccharomyces cerevisiae. Mutat. Res. 595:91–106 [DOI] [PubMed] [Google Scholar]
- 131. Ling F, Hori A, Yoshitani A, Niu R, Yoshida M, Shibata T. 17 April 2013. Din7 and Mhr1 expression levels regulate double-strand-break-induced replication and recombination of mtDNA at ori5 in yeast. Nucleic Acids Res. [Epub ahead of print.] 10.1093/nar/gkt273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Burgers PM, Bauer GA, Tam L. 1988. Exonuclease V from Saccharomyces cerevisiae. A 5′-3′ deoxyribonuclease that produces dinucleotides in a sequential fashion. J. Biol. Chem. 263:8099–8105 [PubMed] [Google Scholar]
- 133. Burgers PM, Stith CM, Yoder BL, Sparks JL. 2010. Yeast exonuclease 5 is essential for mitochondrial genome maintenance. Mol. Cell. Biol. 30:1457–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kornblum C, Nicholls TJ, Haack TB, Scholer S, Peeva V, Danhauser K, Hallmann K, Zsurka G, Rorbach J, Iuso A, Wieland T, Sciacco M, Ronchi D, Comi GP, Moggio M, Quinzii CM, Dimauro S, Calvo SE, Mootha VK, Klopstock T, Strom TM, Meitinger T, Minczuk M, Kunz WS, Prokisch H. 2013. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 45:214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chi NW, Kolodner RD. 1994. Purification and characterization of MSH1, a yeast mitochondrial protein that binds to DNA mismatches. J. Biol. Chem. 269:29984–29992 [PubMed] [Google Scholar]
- 136. Reenan RA, Kolodner RD. 1992. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics 132:963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Dzierzbicki P, Koprowski P, Fikus MU, Malc E, Ciesla Z. 2004. Repair of oxidative damage in mitochondrial DNA of Saccharomyces cerevisiae: involvement of the MSH1-dependent pathway. DNA Repair (Amst.) 3:403–411 [DOI] [PubMed] [Google Scholar]
- 138. Kaniak A, Dzierzbicki P, Rogowska AT, Malc E, Fikus M, Ciesla Z. 2009. Msh1p counteracts oxidative lesion-induced instability of mtDNA and stimulates mitochondrial recombination in Saccharomyces cerevisiae. DNA Repair (Amst.) 8:318–329 [DOI] [PubMed] [Google Scholar]
- 139. Mookerjee SA, Lyon HD, Sia EA. 2005. Analysis of the functional domains of the mismatch repair homologue Msh1p and its role in mitochondrial genome maintenance. Curr. Genet. 47:84–99 [DOI] [PubMed] [Google Scholar]
- 140. Rayssiguier C, Thaler DS, Radman M. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396–401 [DOI] [PubMed] [Google Scholar]
- 141. Abdelnoor RV, Yule R, Elo A, Christensen AC, Meyer-Gauen G, Mackenzie SA. 2003. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. U. S. A. 100:5968–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Pont-Kingdon GA, Okada NA, Macfarlane JL, Beagley CT, Wolstenholme DR, Cavalier-Smith T, Clark-Walker GD. 1995. A coral mitochondrial mutS gene. Nature 375:109–111 [DOI] [PubMed] [Google Scholar]
- 143. Bannwarth S, Figueroa A, Fragaki K, Destroismaisons L, Lacas-Gervais S, Lespinasse F, Vandenbos F, Pradelli LA, Ricci JE, Rotig A, Michiels JF, Vande Velde C, Paquis-Flucklinger V. 2012. The human MSH5 (MutS Homolog 5) protein localizes to mitochondria and protects the mitochondrial genome from oxidative damage. Mitochondrion 12:654–665 [DOI] [PubMed] [Google Scholar]
- 144. Ling F, Makishima F, Morishima N, Shibata T. 1995. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 14:4090–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ling F, Shibata T. 2002. Recombination-dependent mtDNA partitioning: in vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J. 21:4730–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Mbantenkhu M, Wang X, Nardozzi JD, Wilkens S, Hoffman E, Patel A, Cosgrove MS, Chen XJ. 2011. Mgm101 is a Rad52-related protein required for mitochondrial DNA recombination. J. Biol. Chem. 286:42360–42370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Ling F, Yoshida M, Shibata T. 2009. Heteroduplex joint formation free of net topological change by Mhr1, a mitochondrial recombinase. J. Biol. Chem. 284:9341–9353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Masuda T, Ling F, Shibata T, Mikawa T. 2010. Analysis of DNA-binding sites on Mhr1, a yeast mitochondrial ATP-independent homologous pairing protein. FEBS J. 277:440–452 [DOI] [PubMed] [Google Scholar]
- 149. Thyagarajan B, Padua RA, Campbell C. 1996. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 271:27536–27543 [DOI] [PubMed] [Google Scholar]
- 150. Sage JM, Gildemeister OS, Knight KL. 2010. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J. Biol. Chem. 285:18984–18990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Sage JM, Knight KL. 2013. Human Rad51 promotes mitochondrial DNA synthesis under conditions of increased replication stress. Mitochondrion 13:350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Khazi FR, Edmondson AC, Nielsen BL. 2003. An Arabidopsis homologue of bacterial RecA that complements an E. coli recA deletion is targeted to plant mitochondria. Mol. Genet. Genomics 269:454–463 [DOI] [PubMed] [Google Scholar]
- 153. Odahara M, Inouye T, Fujita T, Hasebe M, Sekine Y. 2007. Involvement of mitochondrial-targeted RecA in the repair of mitochondrial DNA in the moss, Physcomitrella patens. Genes Genet. Syst. 82:43–51 [DOI] [PubMed] [Google Scholar]
- 154. Miller-Messmer M, Kuhn K, Bichara M, Le Ret M, Imbault P, Gualberto JM. 2012. RecA-dependent DNA repair results in increased heteroplasmy of the Arabidopsis mitochondrial genome. Plant Physiol. 159:211–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. 2007. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19:1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Odahara M, Kuroiwa H, Kuroiwa T, Sekine Y. 2009. Suppression of repeat-mediated gross mitochondrial genome rearrangements by RecA in the moss Physcomitrella patens. Plant Cell 21:1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Chen XJ, Guan MX, Clark-Walker GD. 1993. MGM101, a nuclear gene involved in maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3473–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Clark-Walker GD, Chen XJ. 1996. A vital function for mitochondrial DNA in the petite-negative yeast Kluyveromyces lactis. Mol. Gen. Genet. 252:746–750 [DOI] [PubMed] [Google Scholar]
- 159. Meeusen S, Tieu Q, Wong E, Weiss E, Schieltz D, Yates JR, Nunnari J. 1999. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 145:291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Zuo X, Xue D, Li N, Clark-Walker GD. 2007. A functional core of the mitochondrial genome maintenance protein Mgm101p in Saccharomyces cerevisiae determined with a temperature-conditional allele. FEMS Yeast Res. 7:131–140 [DOI] [PubMed] [Google Scholar]
- 161. Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. 2002. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell 10:359–371 [DOI] [PubMed] [Google Scholar]
- 162. Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. 2002. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. U. S. A. 99:13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wang X, Mbantenkhu M, Wierzbicki S, Chen XJ. Preparation of the Mgm101 recombination protein by MBP-based tagging strategy J. Vis. Exp., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Poteete AR, Sauer RT, Hendrix RW. 1983. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J. Mol. Biol. 171:401–418 [DOI] [PubMed] [Google Scholar]
- 165. Passy SI, Yu X, Li Z, Radding CM, Egelman EH. 1999. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc. Natl. Acad. Sci. U. S. A. 96:4279–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Ploquin M, Bransi A, Paquet ER, Stasiak AZ, Stasiak A, Yu X, Cieslinska AM, Egelman EH, Moineau S, Masson JY. 2008. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr. Biol. 18:1142–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Nardozzi JD, Wang X, Mbantenkhu M, Wilkens S, Chen XJ. 2012. A properly configured ring structure is critical for the function of the mitochondrial DNA recombination protein, Mgm101. J. Biol. Chem. 287:37259–37268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Lisby M, Rothstein R, Mortensen UH. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. U. S. A. 98:8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Iyer LM, Koonin EV, Aravind L. 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. 2010. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 38:3952–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Muyrers JP, Zhang Y, Buchholz F, Stewart AF. 2000. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 14:1971–1982 [PMC free article] [PubMed] [Google Scholar]
- 172. Lim SI, Min BE, Jung GY. 2008. Lagging strand-biased initiation of red recombination by linear double-stranded DNAs. J. Mol. Biol. 384:1098–1105 [DOI] [PubMed] [Google Scholar]
- 173. Poteete AR. 2008. Involvement of DNA replication in phage lambda Red-mediated homologous recombination. Mol. Microbiol. 68:66–74 [DOI] [PubMed] [Google Scholar]
- 174. MacAlpine DM, Kolesar J, Okamoto K, Butow RA, Perlman PS. 2001. Replication and preferential inheritance of hypersuppressive petite mitochondrial DNA. EMBO J. 20:1807–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hayward DC, Dosztanyi Z, Clark-Walker GD. 2013. The N-terminal intrinsically disordered domain of Mgm101p is localized to the mitochondrial nucleoid. PLoS One 8:e56465. 10.1371/journal.pone.0056465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Rothenberg E, Grimme JM, Spies M, Ha T. 2008. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. U. S. A. 105:20274–20279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M. 2010. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 38:2917–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lloyd JA, McGrew DA, Knight KL. 2005. Identification of residues important for DNA binding in the full-length human Rad52 protein. J. Mol. Biol. 345:239–249 [DOI] [PubMed] [Google Scholar]
- 179. Mbantenkhu M, Wierzbicki S, Wang X, Guo S, Wilkens S, Chen XJ. 2013. A short carboxyl-terminal tail is required for ssDNA-binding, higher order structural organization and stability of the mitochondrial single strand annealing protein Mgm101. Mol. Biol. Cell 24:1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Samach A, Melamed-Bessudo C, Avivi-Ragolski N, Pietrokovski S, Levy AA. 2011. Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 23:4266–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Janicka S, Kuhn K, Le Ret M, Bonnard G, Imbault P, Augustyniak H, Gualberto JM. 2012. A RAD52-like single-stranded DNA binding protein affects mitochondrial DNA repair by recombination. Plant J. 72:423–435 [DOI] [PubMed] [Google Scholar]
- 182. Itoh K, Izumi A, Mori T, Dohmae N, Yui R, Maeda-Sano K, Shirai Y, Kanaoka MM, Kuroiwa T, Higashiyama T, Sugita M, Murakami-Murofushi K, Kawano S, Sasaki N. 2011. DNA packaging proteins Glom and Glom2 coordinately organize the mitochondrial nucleoid of Physarum polycephalum. Mitochondrion 11:575–586 [DOI] [PubMed] [Google Scholar]
- 183. Gilkerson RW, Schon EA. 2008. Nucleoid autonomy: an underlying mechanism of mitochondrial genetics with therapeutic potential. Commun. Integr. Biol. 1:34–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Gilkerson RW, Schon EA, Hernandez E, Davidson MM. 2008. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J. Cell Biol. 181:1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Barthelemy C, Ogier de Baulny H, Diaz J, Cheval MA, Frachon P, Romero N, Goutieres F, Fardeau M, Lombes A. 2001. Late-onset mitochondrial DNA depletion: DNA copy number, multiple deletions, and compensation. Ann. Neurol. 49:607–617 [PubMed] [Google Scholar]
- 186. Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. 2003. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 31:e61. 10.1093/nar/gng060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K. 2002. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc. Natl. Acad. Sci. U. S. A. 99:5521–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Shiiba D, Fumoto SI, Miyakawa I, Sando N. 1997. Isolation of giant mitochondrial nucleoids from the yeast Saccharomyces cerevisiae. Protoplasma 198:177–185 [Google Scholar]
- 189. Neiman M, Taylor DR. 2009. The causes of mutation accumulation in mitochondrial genomes. Proc. Biol. Sci. 276:1201–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168:39–50 [DOI] [PubMed] [Google Scholar]
- 191. Ling F, Morioka H, Ohtsuka E, Shibata T. 2000. A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontaneously introduced in yeast mtDNA. Nucleic Acids Res. 28:4956–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Clark-Walker GD. 1991. Contrasting mutation rates in mitochondrial and nuclear genes of yeasts versus mammals. Curr. Genet. 20:195–198 [DOI] [PubMed] [Google Scholar]
- 193. Maleszka MR, Skelly PJ, Clark-Walker GD. 1991. Rolling circle replicaton of DNA in yeast mitochondria. EMBO J. 10:3923–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Fangman WL, Henly JW, Brewer BJ. 1990. RPO41-independent maintenance of ρ− mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Preiser PR, Wilson RJM, Moore PW, McCready SM, Hajibagheri M, Blight AN, Strath KJ, Williamson DH. 1996. Recombination associated with replication of malarial mitochondrial DNA. EMBO J. 15:684–693 [PMC free article] [PubMed] [Google Scholar]
- 196. Gerhold JM, Aun A, Sedman T, Joers P, Sedman J. 2010. Strand invasion structures in the inverted reseat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell 39:851–861 [DOI] [PubMed] [Google Scholar]
- 197. Backert S. 2002. R-loop-dependent rolling-circle replication and a new model for DNA concatemer resolution by mitochondrial plasmid mp1. EMBO J. 21:3128–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Backert S, Borner T. 2000. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37:304–314 [DOI] [PubMed] [Google Scholar]
- 199. Backert S, Dorfel P, Lurz R, Borner T. 1996. Rolling-circle replication of mitochondrial DNA in the higher plant Chenopodium album (L.). Mol. Cell. Biol. 16:6285–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Ling F, Shibata T. 2004. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol. Biol. Cell 15:310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Shibata T, Ling F. 2007. DNA recombination protein-dependent mechanism of homoplasmy and its proposed functions. Mitochondrion 7:17–23 [DOI] [PubMed] [Google Scholar]
- 202. Ling F, Hori A, Shibata T. 2007. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast rho− mitochondrial DNA that contains the replication origin ori5. Mol. Cell. Biol. 27:1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Silberstein Z, Maor S, Berger I, Cohen A. 1990. Lambda Red-mediated synthesis of plasmid linear multimers in Escherichia coli K12. Mol. Gen. Genet. 223:496–507 [DOI] [PubMed] [Google Scholar]
- 204. Meeusen S, Nunnari J. 2003. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Davis AP, Symington LS. 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159:515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Feng Q, During L, de Mayolo AA, Lettier G, Lisby M, Erdeniz N, Mortensen UH, Rothstein R. 2007. Rad52 and Rad59 exhibit both overlapping and distinct functions. DNA Repair (Amst.) 6:27–37 [DOI] [PubMed] [Google Scholar]
- 207. Wu Y, Sugiyama T, Kowalczykowski SC. 2006. DNA annealing mediated by Rad52 and Rad59 proteins. J. Biol. Chem. 281:15441–15449 [DOI] [PubMed] [Google Scholar]
- 208. Martinsohn JT, Radman M, Petit MA. 2008. The lambda red proteins promote efficient recombination between diverged sequences: implications for bacteriophage genome mosaicism. PLoS Genet. 4:e1000065. 10.1371/journal.pgen.1000065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Elez M, Radman M, Matic I. 2007. The frequency and structure of recombinant products is determined by the cellular level of MutL. Proc. Natl. Acad. Sci. U. S. A. 104:8935–8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Guo X, Popadin KY, Markuzon N, Orlov YL, Kraytsberg Y, Krishnan KJ, Zsurka G, Turnbull DM, Kunz WS, Khrapko K. 2010. Repeats, longevity and the sources of mtDNA deletions: evidence from ‘deletional spectra'. Trends Genet. 26:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Costantino N, Court DL. 2003. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. U. S. A. 100:15748–15753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Davila JI, Arrieta-Montiel MP, Wamboldt Y, Cao J, Hagmann J, Shedge V, Xu YZ, Weigel D, Mackenzie SA. 2011. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Ladoukakis ED, Theologidis I, Rodakis GC, Zouros E. 2011. Homologous recombination between highly diverged mitochondrial sequences: examples from maternally and paternally transmitted genomes. Mol. Biol. Evol. 28:1847–1859 [DOI] [PubMed] [Google Scholar]
- 214. van Diepeningen AD, Goedbloed DJ, Slakhorst SM, Koopmanschap AB, Maas MF, Hoekstra RF, Debets AJ. 2010. Mitochondrial recombination increases with age in Podospora anserina. Mech. Ageing Dev. 131:315–322 [DOI] [PubMed] [Google Scholar]
- 215. Belcour L, Begel O. 1977. Mitochondrial genes in Podospora anserina: recombination and linkage. Mol. Gen. Genet. 153:11–21 [DOI] [PubMed] [Google Scholar]
- 216. Belcour L. 1975. Cytoplasmic mutations isolated from protoplasts of Podospora anserina. Genet. Res. 25:155–161 [DOI] [PubMed] [Google Scholar]
- 217. Osiewacz HD. 2002. Genes, mitochondria and aging in filamentous fungi. Ageing Res. Rev. 1:425–442 [DOI] [PubMed] [Google Scholar]
- 218. Jamet-Vierny C, Boulay J, Briand JF. 1997. Intramolecular cross-overs generate deleted mitochondrial DNA molecules in Podospora anserina. Curr. Genet. 31:162–170 [DOI] [PubMed] [Google Scholar]
- 219. Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. 2009. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137:1247–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wang X, Zuo X, Kucejova B, Chen XJ. 2008. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nat. Cell Biol. 10:1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Pohjoismaki JL, Goffart S, Taylor RW, Turnbull DM, Suomalainen A, Jacobs HT, Karhunen PJ. 2010. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS One 5:e10426. 10.1371/journal.pone.0010426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Pohjoismaki JL, Goffart S. 2011. Of circles, forks and humanity: topological organisation and replication of mammalian mitochondrial DNA. Bioessays 33:290–299 [DOI] [PubMed] [Google Scholar]
- 223. He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. 2010. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 464:610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Williams SL, Huang J, Edwards YJ, Ulloa RH, Dillon LM, Prolla TA, Vance JM, Moraes CT, Zuchner S. 2010. The mtDNA mutation spectrum of the progeroid Polγ mutator mouse includes abundant control region multimers. Cell Metab. 12:675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Rokas A, Abbot P. 2009. Harnessing genomics for evolutionary insights. Trends Ecol. Evol. 24:192–200 [DOI] [PubMed] [Google Scholar]
- 226. Sen D, Nandakumar D, Tang GQ, Patel SS. 2012. Human mitochondrial DNA helicase TWINKLE is both an unwinding and annealing helicase. J. Biol. Chem. 287:14545–14556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Shutt TE, Gray MW. 2006. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 22:90–95 [DOI] [PubMed] [Google Scholar]
- 228. Mosig G. 1998. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 32:379–413 [DOI] [PubMed] [Google Scholar]
- 229. Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U. S. A. 93:10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Sung P. 1997. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272:28194–28197 [DOI] [PubMed] [Google Scholar]
- 231. New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391:407–410 [DOI] [PubMed] [Google Scholar]
- 232. Plate I, Hallwyl SC, Shi I, Krejci L, Muller C, Albertsen L, Sung P, Mortensen UH. 2008. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283:29077–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P, Krejci L. 2008. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J. Biol. Chem. 283:12166–12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Bai Y, Symington LS. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025–2037 [DOI] [PubMed] [Google Scholar]
- 235. Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. 2001. Homologous pairing promoted by the human Rad52 protein. J. Biol. Chem. 276:35201–35208 [DOI] [PubMed] [Google Scholar]
- 236. Fukui H, Moraes CT. 2009. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 18:1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]