Abstract
Streptococcus suis serotype 2 (S. suis 2) is an important zoonotic pathogen that causes considerable economic losses to the pig industry and significantly threatens public health worldwide. The highly pathogenic S. suis 2, which contains the 89K pathogenicity island (PAI), has caused large-scale outbreaks of infections in humans, resulting in high mortality rates. In this study, we established two loop-mediated isothermal amplification (LAMP)-based assays that can rapidly detect S. suis 2 and the 89K PAI and can be performed simultaneously under the same conditions. Further, based on the findings of these two LAMP assays and using the same set of serially diluted DNA samples, we compared the sensitivities of different LAMP product detection methods, including SYBR green detection, gel electrophoresis, turbidimetry, calcein assays, and hydroxynaphthol blue detection. The results suggest that target genes can be amplified and detected within 48 min under 63°C isothermal conditions. The sensitivity of tests for S. suis 2 detection varies between detection methods and reaction systems, indicating that for each LAMP reaction system, multiple detection methods should be performed to select the optimal one. The sensitivities of the optimized methods (7.16 copies/reaction) in the present study were identical to those of the real-time PCR assay, and the test results for reference strains and clinical samples showed that these LAMP systems have high specificities. Thus, since the LAMP systems established in this study are simple, fast, and sensitive, they may have good clinical potential for detecting the highly pathogenic S. suis 2.
INTRODUCTION
Streptococcus suis is an important zoonotic pathogen found worldwide (1). It is made up of 35 serotypes (types 1/2 and 1 to 34) according to its capsular polysaccharide (CPS) antigens (2). Of these 35 serotypes, S. suis serotype 2 (S. suis 2) is the most virulent and prevalent strain, with the highest clinical isolation rate in most countries, and it can cause a variety of life-threatening infections, including meningitis, septicemia, endocarditis, and even sudden death in both pigs and humans (3); thus, this pathogen poses a great danger to both the pig industry and public health.
In two large-scale outbreaks of human S. suis 2 infection in China (14 deaths of 25 cases in Jiangsu province in 1998 and 38 deaths of 215 cases in Sichuan province in 2005) (4), a high proportion of infected patients showed the rarely seen streptococcal toxic shock syndrome (STSS), which is characterized by rapid disease progression and high mortality rates (5). Chen et al. (6) performed a comparative genomics analysis on S. suis 2 isolated from these two outbreaks and suggested that a specific pathogenicity island (PAI) called 89K PAI might account for the high virulence of S. suis 2. Subsequent studies confirmed that knockout of the salK or salR gene on 89K PAI resulted in the loss of bacterial virulence, while salK or salR complementary strains displayed recovered virulence, indicating that 89K PAI is associated with the high virulence of S. suis 2 (7). Thus, 89K PAI is an important indicator for evaluating the virulence of S. suis 2 (8, 9).
The rapid and accurate detection of S. suis 2 and 89K PAI is very important in the early diagnosis and treatment of this infection, as it helps control epidemic situations and improve patient outcomes. The commonly used bacterial culture methods require about a week for bacterial isolation and identification, but a patient's condition may have already deteriorated by then due to the resistance of S. suis 2 to multiple antibiotics that are used in empirical treatments for pathogenic infections (10). The cps2J gene-based PCR and real-time PCR assays are highly sensitive and specific assays for S. suis 2, but they require expensive apparatuses, sophisticated techniques, and tedious operations (11, 12). Further, their use is not suitable for on-site detection in the field or in primitive clinical laboratories, particularly in developing countries. Therefore, the development and evaluation of a new simple, rapid, and cost-effective assay for detecting highly pathogenic S. suis 2 are urgently needed.
Loop-mediated isothermal amplification (LAMP) is an option for rapid DNA amplification (13). LAMP employs Bst DNA polymerase with strand displacement activity and a set of 4 to 6 specially designed primers that recognize a total of 6 to 8 distinct sequences on the target DNA under 60 to 65°C isothermal conditions; the cycling reaction results in the accumulation of 109 to 1010 copies of the target in less than an hour. Since the LAMP assay is quick and easy to perform and requires only a thermostatic incubator, it has been widely used for the detection of various pathogens (14–16).
Huy et al. (17) designed primers according to the common 16S rRNA sequences of four bacterial species (Staphylococcus aureus, Streptococcus pneumoniae, S. suis, and Streptococcus agalactiae) and established a single-tube LAMP technique to amplify bacterial nucleic acid. However, this method requires a 90-min period for enzyme digestion of the amplified product and two rounds of gel electrophoresis to identify bacterial species, and it cannot distinguish between bacterial serotypes. To our knowledge, there has been no report to date on the use of LAMP to detect S. suis 2.
SYBR green I and agar gel electrophoresis are normally used in LAMP product detection (18), and a turbidimeter is also used to monitor the turbidity of magnesium pyrophosphate, the by-product of LAMP amplification, in real time (19). Tomita et al. (20) and Goto et al. (21) recently reported that the visual detection of LAMP products can be realized by the use of preadded calcein or hydroxynaphthol blue (HNB) and that the use of these two methods can prevent the nucleic acid contamination that is introduced during lid opening. However, some studies claimed that the inhibition of calcein in the LAMP reaction significantly affects product detection sensitivity (22, 23), while several reports indicated that the sensitivity of turbidimetry is lower than that of the calcein assay (24). In contrast, other studies have found no difference in the sensitivities of these detection methods (25, 26). Thus, the sensitivities of the different LAMP product detection methods have yet to be confirmed, and to our knowledge, no reports of a systematic comparison of the sensitivities of the five most commonly used LAMP product detection methods have been published thus far.
Based on the cps2J gene of S. suis 2 and the salK or salR gene in 89K PAI, we established and evaluated two LAMP assays (Cps2J-LAMP and SalK-LAMP) that can be performed simultaneously under the same conditions and can rapidly identify S. suis 2 to distinguish whether it is a highly virulent strain containing 89K PAI. Further, based on the two S. suis detection systems, we used the same set of serially diluted DNA samples as a template and compared the sensitivities of different visual detection methods (SYBR green I detection, gel electrophoresis, turbidimetry, calcein assays, and HNB detection) for the detection of LAMP products.
MATERIALS AND METHODS
Bacterial strains.
We used a total of 55 bacterial strains to evaluate and optimize the sensitivity and specificity of the LAMP reaction (Table 1). S. suis serotypes 1/2, 1, and 3 to 34 were kindly provided by Marcelo Gottschalk. S. suis 2 strains P1/7, S735, S10, 7996, and T15 were obtained from Astrid de Creeff. The highly pathogenic S. suis 2 strains 05ZYH33, SC84, and 98HAH12 were isolated from patients with STSS. The S. suis 2 strains 05JYS68 and 07NJH06 were stored in our laboratory. All the S. suis strains were grown in Todd-Hewitt broth (THB) (Difco Laboratories, Detroit, MI) or on THB plates with 5% sheep blood. Other reference bacterial strains were purchased from the American Type Culture Collection (ATCC) (Manassas, VA).
Table 1.
Loop-mediated isothermal amplification (LAMP) and real-time PCR detection of the 55 bacterial strains used in this study
| Strain and/or serotype | Strain source or ATCC no. | Results by gene and assay |
|||
|---|---|---|---|---|---|
| LAMP |
Real-time PCR |
||||
| cps2J | salK | cps2J | salK | ||
| Streptococcus suis 2 strains | |||||
| 05ZYH33 | China (Sichuan, 2005), from a dead patient with STSSa | + | + | + | + |
| SC84 | China (Sichuan, 2005), from a patient with STSS | + | + | + | + |
| 98HAH12 | China (Jiangsu, 1998), from a patient with STSS | + | + | + | + |
| 07NJH06 | China (Jiangsu, 2007), from a sporadic human case without STSS | + | − | + | − |
| 05JYS68 | China (Jiangsu, 2005), from a healthy pig | + | − | + | − |
| P1/7 | Britain | + | − | + | − |
| S10 | Netherlands | + | − | + | − |
| S735 | Canada | + | − | + | − |
| T15 | Netherlands | + | − | + | − |
| 7996 | Netherlands | + | − | + | − |
| Other Streptococcus suis serotypes | |||||
| 2651 (S. suis 1/2) | Netherlands | + | − | + | − |
| 22083 (S. suis 9) | Denmark | − | + | − | + |
| S. suis 1 | Netherlands | − | − | − | − |
| S. suis 3–8, 10–13, and 16b | Denmark | − | − | − | − |
| S. suis 14, 15 | Netherlands | − | − | − | − |
| S. suis 17–19 and 21–34 | Canada | − | − | − | − |
| S. suis 20 | United States | − | − | − | − |
| Other Streptococcus spp. | |||||
| S. pneumoniae | ATCC 49619 | − | − | − | − |
| S. pyogenes | ATCC 19615 | − | − | − | − |
| S. agalactiae | ATCC 12386 | − | − | − | − |
| S. bovis | ATCC 49147 | − | − | − | − |
| Other strains | |||||
| Neisseria meningitidis | ATCC 13102 | − | − | − | − |
| Staphylococcus aureus | ATCC 27217 | − | − | − | − |
| Haemophilus influenzae | ATCC 49247 | − | − | − | − |
| Escherichia coli | ATCC 8739 | − | − | − | − |
| Klebsiella pneumoniae | ATCC 700603 | − | − | − | − |
| Pseudomonas aeruginosa | ATCC 27853 | − | − | − | − |
| Legionella pneumophila | ATCC 33152 | − | − | − | − |
STSS, streptococcal toxic shock syndrome.
S. suis 3–8, 10–13, and 16 indicate the 11 bacterial strains from S. suis serotypes 3, 4, 5, 6, 7, 8, 10, 11, 12, 13, and 16.
DNA extraction.
A Wizard genomic DNA purification kit (Promega, Madison, WI) was used for bacterial DNA extraction. All procedures were performed according to the manufacturer's instructions.
Primer design.
According to the conserved sequence of the S. suis 2 cps2J gene and the conserved sequence of salK or salR on the 89K PAI of the highly virulent S. suis 05ZYH33 strain, we designed three sets of primers for each LAMP system using the PrimerExplorer V4 software (http://primerexplorer.jp/elamp4.0.0/index.html). Each primer set included two outer primers (F3 and B3) and two inner primers (FIP and BIP). To reduce the reaction times, we also designed LB and LF loop primers. The real-time PCR primers and the probe sequence for cps2J are as described previously (12). The real-time PCR primers and the probe sequences for salK or salR were designed by Primer Express 3.0 (Life Technologies, Carlsbad, CA). All other primers and probes were synthesized by Invitrogen (Shanghai, China). The optimized primers that were used for the LAMP reaction and real-time PCR procedures are listed in Table 2.
Table 2.
Primers used in LAMP and real-time PCR in this study
| Assay and primera | Position | Length (bp) | Primer sequence (5′ to 3′)b | Target and GenBank accession no. |
|---|---|---|---|---|
| Cps2J-LAMP | cps2J gene of S. suis 2, DQ410854 | |||
| B3 | 409–430 | 22 | GCACCTCTTTTATCTCTTCCAA | |
| F3 | 203–222 | 20 | GTGTTTCAAACGAAGGAAT | |
| BIP | B1c: 321–344 | 43 | AGAGAATGATAGTGATTTGTCGGG | |
| B2: 376–394 | TTTGCAGCTCAGATTCTTG | |||
| FIP | F1c: 225–245 | 46 | GTTGCCGTCAACAATATCATCAGAA | |
| F2: 267–291 | CGGTATCAAAAATAGCACAGC | |||
| LB | 345–368 | 24 | AGGGTTACTTGCTACTTTTGATGG | |
| Cps2J-qPCR | ||||
| Cps2J-F | 347–373 | 27 | GGTTACTTGCTACTTTTGATGGAAATT | |
| Cps2J-R | 409–431 | 23 | CGCACCTGTTTTATCTCTTCCAA | |
| Cps2J-Probe | 375–407 | 33 | FAM-TCAAGAATCTGAGCTGCAAAAGTGTCAAATTGA-BHQ1 | |
| SalK-LAMP | salK or salR gene of 89K PAI, CP000407 and SSU05_0944 | |||
| B3 | 1103–1120 | 18 | TAGAGTCCGCTTGCTCAA | |
| F3 | 912–932 | 21 | AGAGCTCGTTAATAACGCTTA | |
| BIP | B1c: 1032–1054 | 41 | ACCATTTAAACATGGACACGGAT | |
| B2: 1080–1097 | ATAGTCCCCCCTACTGAC | |||
| FIP | F1c: 938–957 | 45 | CCATAACTGATAAAGAGAGTTCCCG | |
| F2: 982–1006 | ATTCCAATGCTCAAACGGTT | |||
| LF | 958–981 | 24 | CCCATCTCTAGCCAATTTTACAGT | |
| SalK-qPCR | ||||
| SALK-P | 940–962 | 23 | TCCAATGCTCAAACGGTTACTGT | |
| SALK-R | 1060–1088 | 29 | CCTACTGACTTTACTTGTTCTTCCAAGAC | |
| SALK-Probe | 967–982 | 16 | FAM-TTGGCTAGAGATGGGC-BHQ1 |
FIP is a long primer with two recognition sequences, F1c and F2; BIP is also a long primer with two recognition sequences, B1c and B2.
FAM, 6-carboxyfluorescein; BHQ1, black hole quencher 1.
LAMP reaction.
To optimize the LAMP reaction conditions, we used DNA extracted from the S. suis 2 strain 05ZYH33 as a template, and an LA-320C turbidimeter (Eiken Chemical Co. Ltd., Tokyo, Japan) was used to measure the turbidity of LAMP reactions using different sets of primers, incubation temperatures (60 to 65°C), reaction times (10 to 90 min), magnesium ion concentrations (3 to 10 mM), and primer concentrations (0.1 to 1.8 μM). The conditions that took the least amount of time to reach a turbidity at an optical density (OD) of 0.1 and had a high peak turbidity curve value were considered optimal. The optimized Cps2J-LAMP and SalK-LAMP reactions were performed in a total volume of 25 μl comprising 20 mmol/liter Tris-HCl (pH 8.8), 10 mmol/liter KCl, 10 mmol/liter (NH4)2SO4, 0.1% Triton X-100, 8 mmol/liter MgSO4, 0.8 mmol/liter betaine (Sigma, St. Louis, MO), 1.4 mmol/liter each deoxynucleoside triphosphate (dNTP) (Promega), 8 U Bst DNA polymerase (New England BioLabs, Beverly, MA), a corresponding primer set, including 1.6 μM FIB and BIP, 0.2 μM primers F3 and B3, 0.8 μM primers LF or LB, and 1 μl of the template. In the calcein and HNB detection methods, 1 μl of the calcein mixture (0.625 mM calcein and 12.5 mM MnCl2) or 3 mM HNB was preadded to the reaction mixture systems to create a total volume of 25 μl. Nucleic acid amplifications were carried out at 63°C for 46 min and were terminated at 85°C for 2 min. Distilled water was used as a control.
Analyses of LAMP products.
Five methods were used to detect the amplification of the products of the Cps2J-LAMP and SalK-LAMP systems.
(i) Turbidity detection.
The LA-320C turbidimeter was used to monitor the turbidity of LAMP products in real time (measured every 6 s at a 650-nm wavelength), and the samples were considered positive if the turbidity value exceeded 0.1.
(ii) Gel electrophoresis detection.
After the LAMP reactions, 1 μl of each product was used for 2% agar gel electrophoresis (100 V constant for 40 min), and a Gel Doc XR+ imaging system (Bio-Rad, Hercules, CA) was used to observe the band patterns. The samples were considered positive if they showed a characteristic ladder-like band pattern.
(iii) SYBR green I detection.
After the LAMP reactions, 1 μl of SYBR green I diluted 1:10 (BioWhittaker Molecular Applications, Rockland, ME) was added to observe the solution color. The samples were considered positive if the solution turned green and negative if the solution turned orange.
(iv) Calcein detection.
Calcein and MnCl2 were preadded to the LAMP reaction systems at a concentration of 25 mM and 0.5 mM, respectively, and the color changes of the reaction mixtures were detected after amplification. The samples were considered positive if the solution turned green and negative if the solution turned orange.
(v) HNB detection.
HNB was preadded to the LAMP systems at a concentration of 120 mM to detect postamplification color changes. The samples that turned sky blue were considered positive, while those that turned violet were considered negative.
Real-time PCR.
The real-time PCR system and conditions for the detection of the S. suis 2 cps2J gene (Cps2J-quantitative PCR [qPCR]) were as described previously (12). The real-time PCR for salK or salR in 89K PAI (SalK-qPCR) was performed in a total volume of 20 μl and contained 10 μl of 2× Premix Ex Taq (TaKaRa, Dalian, China), 0.2 μl of 50× ROX reference dye II (TaKaRa, China), 0.2 μl of primers (SALK-F, SALK-R, and probe SALK-Probe), and 1 μl of the template. An ABI 7500 Fast real-time PCR system (Life Technologies) was used for the real-time PCR, and the reaction steps consisted of predenaturation at 95°C for 30 s, denaturation at 95°C for 30 s, annealing at 56°C for 10 s, and elongation at 72°C for 30 s for 45 cycles. On validation of the positive and negative controls, the samples were considered positive if the 6-carboxyfluorescein (FAM) threshold cycle (CT) values were <40.
Sensitivity comparisons of LAMP product detection methods.
We used a NanoDrop 2000 (Thermo Scientific, Rockford, IL) to quantify the DNA extracted from the 05ZYH33 strain (16.19 ng/μl) and then calculated the copy numbers (7.16 × 106 copies/μl) from the 2,096.309-kb full-length genome. DNA was serially diluted 1:10 up to 10−7-fold dilutions (16.19 ng/μl to 1.619 fg/μl), and 1 μl of each serial dilution was used as a template in the Cps2J-LAMP and SalK-LAMP systems. The LAMP reactions were divided into three groups and detected by the turbidimeter (for the normal LAMP assay, no dyes were added before the reaction), calcein method, and HNB method. All reactions were repeated three times. Next, 1 μl of the LAMP product was used for agar gel electrophoresis to observe the band patterns. Finally, 1 μl of SYBR green I was added to the remaining products from the turbidimeter group for observation of color changes.
A total of 1 μl of each DNA serial dilution was also used as a template for the Cps2J-qPCR and SalK-qPCR assays. Both reactions were repeated twice.
Evaluation of the LAMP reaction using clinical samples.
A total of 66 clinical samples were used in this study to evaluate the feasibility of LAMP-based S. suis 2 and 89K PAI detection. Nine liquid nitrogen-preserved blood serum samples were obtained from S. suis 2-infected patients (from the 2005 Sichuan outbreak) who had typical STSS (samples were appropriately coded for anonymity, and local ethical board approval was obtained). Twelve cerebrospinal fluid samples and 22 blood serum samples were aseptically collected from pigs with suspected S. suis infection, while 23 nasal swab samples were collected from healthy pigs in Jiangsu Province between October and December 2012. The samples were cultivated in THB for 16 h, followed by DNA isolation. The Cps2J-LAMP and SalK-LAMP systems were incubated in a SC25 metal bath (Torrey Pines Scientific, Carlsbad, CA), and the products were visually detected using calcein and HNB, respectively. All samples were also tested by Cps2J-qPCR and SalK-qPCR. MedCalc 11.4.2 (MedCalc software, Mariakerke, Belgium) was used for data processing, and Cohen's kappa coefficient (κ) was used to measure interrater agreement between the LAMP and the qPCR assays.
RESULTS
Primer and temperature selection for the LAMP reaction.
To optimize the LAMP reaction, we conducted the procedures under different conditions. Of the three sets of primers for the cps2J amplifications, those whose sequences are listed in Table 2 for Cps2J-LAMP had the highest amplification rates and the shortest peak appearance times; similarly, the three sets of primers for the salK or salR amplifications whose sequences are listed in Table 2 for SalK-LAMP had the highest amplification rates and the shortest peak appearance times (data not shown). Thus, these primers were selected for future amplifications. The optimized temperature for Cps2J-LAMP and SalK-LAMP, at a template concentration of 16.19 ng/μl, was 63°C, and positive results were detected at 18 min and 16.5 min, respectively (data not shown).
Specificity of the LAMP reaction.
To evaluate the specificity of the LAMP assays, optimized Cps2J-LAMP and SalK-LAMP systems were used to amplify DNA extracted from the 55 strains listed in Table 1. All S. suis 2 strains and the S. suis 1/2 strain tested positive in the Cps2J-LAMP system, while the other strains tested negative (as expected), indicating that Cps2J-LAMP is highly specific to S. suis 2. Highly virulent 89K PAI-containing S. suis 2 (05ZYH33, 98HAH12, and SC84 strains) tested positive in the SalK-LAMP system, while the other S. suis strains and reference strains, excluding S. suis 9, tested negative, indicating good specificity of SalK-LAMP to 89K PAI. The results of the Cps2J-LAMP and SalK-LAMP tests were consistent with those of the Cps2J-qPCR and SalK-qPCR assays (Table 1).
Sensitivity of the different detection methods. (i) Gel electrophoresis and SYBR green detection.
In the Cps2J-LAMP and SalK-LAMP reactions without preadded dyes, gel electrophoresis (data not shown) and the SYBR green test (Fig. 1A and B) showed the highest detection sensitivities (7.16 copies/reaction), which were identical to those of the real-time PCR (Table 3). This finding suggests that direct detection of the amplified DNA products should be the most reliable method if the tedious electrophoresis steps and contamination introduced during lid opening are not taken into consideration.
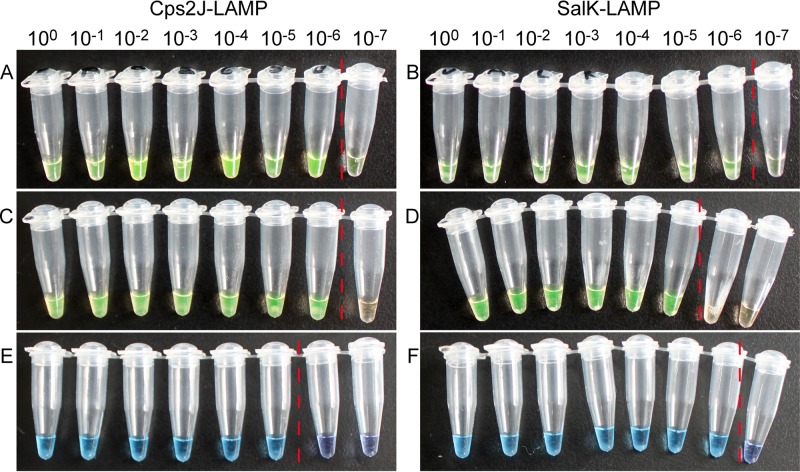
Fig 1.
Detection of the same set of serial dilutions of 16.19 ng/μl S. suis strain 05ZYH33 DNA using three different visible detection methods in Cps2J-LAMP and SalK-LAMP. (A and B) SYBR green I; (C and D) calcein with MnCl2; (E and F) hydroxynaphthol blue. The red vertical dotted lines indicate the cutoffs of positive versus negative.
Table 3.
Detection limits of different methods evaluated using 10-fold serial dilutions of DNA from strain 05ZYH33a
| Evaluation method | Detection limits for gene (CT) |
|
|---|---|---|
| cps2J | salK | |
| Normal LAMP assay | ||
| Turbidity | 1 × 10−5 | 1 × 10−6 |
| Gel electrophoresis | 1 × 10−6 | 1 × 10−6 |
| SYBR green I | 1 × 10−6 | 1 × 10−6 |
| Calcein with MnCl2 | ||
| Color changes | 1 × 10−6 | 1 × 10−5 |
| Gel electrophoresis | 1 × 10−6 | 1 × 10−5 |
| Hydroxynaphthol blue | ||
| Color changes | 1 × 10−5 | 1 × 10−6 |
| Gel electrophoresis | 1 × 10−5 | 1 × 10−6 |
| qPCR (TaqMan) | 1 × 10−6 (37.6) | 1 × 10−6 (38.1) |
Initial concentration, 16.19 ng/μl; 7.16 × 106 copies/μl.
(ii) Turbidity detection.
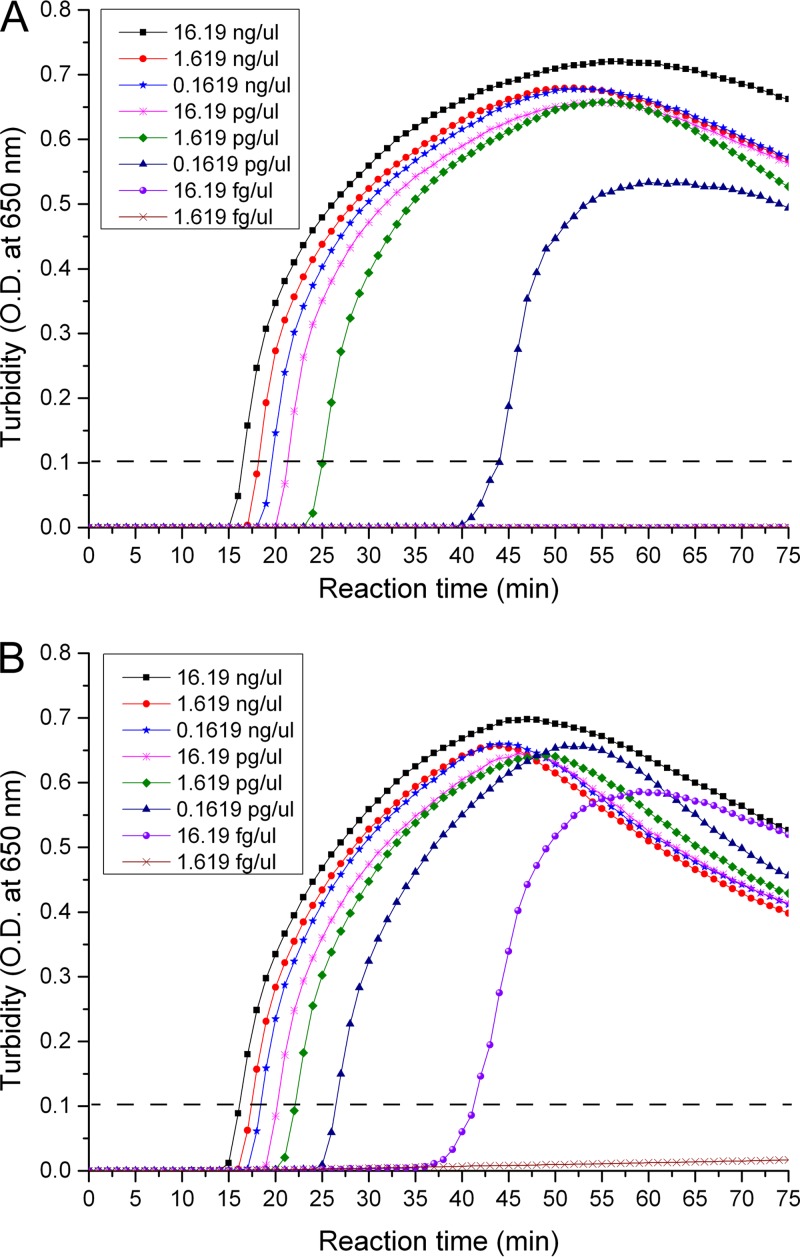
In the Cps2J-LAMP reaction, turbidity detection showed a sensitivity of 71.6 copies/reaction (Fig. 2A), which is lower than that of gel electrophoresis. This result was consistent with that of another published study (27). However, in the SalK-LAMP reaction, the turbidity detection showed a sensitivity of 7.16 copies/reaction (Fig. 2B), which was similar to that of gel electrophoresis (Table 3).
Fig 2.
Sensitivities of the two loop-mediated isothermal amplification (LAMP)-based systems. Serial dilutions of 16.19 ng/μl S. suis strain 05ZYH33 DNA were amplified by Cps2J-LAMP (A) and SalK-LAMP (B).
(iii) Calcein detection.
The sensitivities of calcein detection seemed to be paradoxical in the two different LAMP systems. In the CpsJ-LAMP test, the sensitivity was 7.16 copies/reaction (Fig. 1C), which is higher than that of the HNB and turbidity methods. However, in the SalK-LAMP reactions, the sensitivity decreased to 71.6 copies/reaction (Fig. 1D), which is the lowest among the detection methods used. The amplified products with preadded calcein were separated using gel electrophoresis, and the results were consistent with the color changes (Table 3). This finding indicates that preadded dyes had inhibitive effects on the SalK-LAMP reaction, resulting in a 10-fold-lower sensitivity, although they had no negative effects on the Cps2J-LAMP system.
(iv) HNB detection.
In the two LAMP systems, the sensitivities of HNB detection were identical to those of turbidity detection. In the SalK-LAMP reaction, the HNB sensitivity was 7.16 copies/reaction, which is 10-fold higher than that of the calcein detection (Fig. 1F). However, in the Cps2J-LAMP reaction, HNB showed lower sensitivity (Fig. 1E). This finding suggests that this recently widely used and highly appraised detection method might also have an inhibitory effect in partial LAMP systems.
Comparison of detection limits.
Cps2J-qPCR and SalK-qPCR were used to detect serially diluted DNA from the S. suis 2 05ZYH33 strain. The detection limit of both systems was 7.16 copies/reaction (data not shown), which indicates that the detection limits of calcein, gel electrophoresis, and SYBR green for Cps2J-LAMP products are the same as those of the Cps2J-qPCR. Similarly, the detection limits of turbidity, gel electrophoresis, SYBR green, and HNB detection for the SalK-LAMP products were identical to those of the SalK-qPCR (Table 3).
Clinical sample detection.
Both LAMP and real-time PCR were used to test the 66 clinical samples. Of the 23 nasal swab samples from healthy pigs, 7 tested positive in Cps2J-qPCR and 6 tested positive in Cps2J-LAMP, but all samples tested negative in SalK-LAMP and SalK-qPCR. The results of sample testing by LAMP were identical to those of real-time PCR, and the cps2J and salK or salR genes were both detectable in the 9 serum samples from patients with STSS. Thus, Cps2J-LAMP showed a sensitivity of 96.3% and a specificity of 100%, while SalK-LAMP showed a sensitivity and specificity of 100%. Compared to real-time PCR, Cps2J-LAMP (κ = 0.968; 95% confidence interval [CI], 0.91 to 1.00) and SalK-LAMP (κ = 1; 95% CI, 1.00 to 1.00) showed high degrees of consistency, though the LAMP reaction required less detection time (Table 4).
Table 4.
Results of LAMP amplification using clinical specimens
| Result | No. (%) of samples using: |
Kappaa | No. (%) of samples using: |
Kappa | ||
|---|---|---|---|---|---|---|
| Cps2J-LAMP | Cps2J-qPCRb | SalK-LAMP | SalK-qPCR | |||
| Positive | 26 (39.4) | 27 (40.9)c | 0.968 | 9 (13.6) | 9 (13.6) | 1 |
| Negative | 40 (60.6) | 39 (59.1) | 57 (86.4) | 57 (86.4) | ||
| Total | 66 | 66 | 66 | 66 | ||
| Reaction time (min) | 18–48 | 95 | 17–48 | 65 | ||
Statistical analysis was performed using MedCalc (11.4.2.0) software.
Amplification performed using an ABI 7500 Fast system.
CT < 40 cycles.
DISCUSSION
Pork, a very important meat product, is one of the largest sources of protein for humans. However, S. suis infection has become widespread in large-scale pig farms, causing huge economic losses and threatening public health due to its rapid spread and high mortality rates (28). So far, S. suis 2 has caused >95% of reported human S. suis infections (29) and it is a major pathogen that causes life-threatening bacterial meningitis in developing countries (30, 31). In recent years, several virulence-associated biomarkers, such as CPS, muramidase-released protein, and extracellular factor, have been used for the detection of S. suis 2 infection. However, whether these factors are associated with bacterial pathogenicity remains controversial (32, 33).
The identification of 89K PAI is an important factor for evaluating S. suis 2 virulence. Other studies have indicated that 89K PAI may undergo excision, cyclization, and horizontal transfer within the genome (34) to increase the risk of international transmission and spread. The rapid detection of S. suis 2 and 89K PAI are imperative for the clinical diagnosis and epidemiological surveillance of S. suis 2 infections. Since LAMP is a rapid detection technique that does not require expensive thermocyclers and its results can be seen with the naked eye (35), it is a preferred method for clinical and on-site diagnosis of S. suis 2 infection.
In this study, we established two LAMP assays for the detection of S. suis 2 and 89K PAI. We also designed five primers to detect the cps2J gene, which encodes a glycosyltransferase that participates in the synthesis of S. suis 2 CPS. Since the S. suis 2 cps2J gene shares high homology with S. suis 1/2 (no human infections reported) but low homology with the other 33 serotypes, we used the cps2J gene as a target in the detection of S. suis 2 by LAMP. In this study, the Cps2J-LAMP assay successfully identified all S. suis 2 strains from bacterial and clinical samples, showing good clinical potential.
We also designed five primers and used LAMP to detect the salK or salR gene, which encodes a two-component signal transduction system. The highly virulent 89K PAI-containing 05ZYH33, 98HAH12, and SC84 strains and blood serum samples from patients with STSS all tested positive in SalK-LAMP, while S. suis 2 strains without 89K PAI and other S. suis strains (excluding S. suis 9, of which no human infections were reported) tested negative. In fact, if dual detection is conducted on single colonies using Cps2J-LAMP and SalK-LAMP, the negative Cps2J-LAMP result and positive SalK-LAMP result may indicate the presence of S. suis 9. Thus, the minor defect in SalK-LAMP specificity in fact revealed a novel method to identify S. suis 9.
In the present study, we performed Cps2J-LAMP and SalK-LAMP testing on the same set of serially diluted 05ZYH33 DNA templates and evaluated the sensitivities of the different product detection methods. The results suggested that for each LAMP reaction system that is designed even for single-pathogen detection, multiple product detection methods should be evaluated to ensure the selection of an optimal method. Magnesium pyrophosphate-based turbidity detection or color-based (metal ion indicator) calcein and HNB detection identified only concentration changes in the chemical compositions of the reaction systems rather than directly reflecting DNA amplification levels. Thus, the results of the detection may be affected by various factors. A “tailor-made” method and a proper comparison are essential to LAMP product detection. For example, with calcein and HNB detection, the Cps2J-LAMP and SalK-LAMP systems showed the highest sensitivities (7.16 copies/reaction) and results that were identical to those of the real-time PCR, and the different colors even made it easier to identify bacterial types and virulence. In one aspect, the comparisons of different detection methods in our study optimized the LAMP assays for detecting the highly pathogenic S. suis 2, and in another aspect, they helped explain the inconsistencies in detection sensitivities that have been reported in other studies (22–26).
In conclusion, this study established a rapid LAMP-based method for the identification of S. suis 2 strains containing 89K PAI that demonstrated high sensitivity and specificity. We also used this method to compare the sensitivities of various product detection methods and applied it to clinical sample detection. The results indicate that LAMP-based detection is a rapid, simple, reliable, and sensitive method with the potential for use in field conditions for epidemic prevention and entry-exit inspection.
ACKNOWLEDGMENTS
This work was supported by the National Science and Technology Project for Infectious Diseases Control (grant no. 2013ZX10004-103, 2013ZX10004-104, 2013ZX10004-203, and 2013ZX10004-218), the National Natural Science Foundation of China (grant no. 31170124, 81171527, and 81172794), the Key Project of the Military Twelfth-Five Year Research Program of PLA (no. AWS11C001 and AWS11C009), the Key Technology R&D Program of Jiangsu Province, China (no. BE2012609 and BE2013603), the Natural Science Foundation of Jiangsu Province, China (no. BK2011097 and BK2011098), and the Key Problems Project in Science and Technology of Nanjing Command, China (no. 10Z039 and 11Z040).
Footnotes
Published ahead of print 24 July 2013
REFERENCES
- 1.Gottschalk M, Xu JG, Calzas C, Segura M. 2010. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5:371–391 [DOI] [PubMed] [Google Scholar]
- 2.Ftika L, Maltezou HC. 2013. Viral haemorrhagic fevers in healthcare settings. J. Hosp. Infect. 83:185–192 [DOI] [PubMed] [Google Scholar]
- 3.Ju CX, Gu HW, Lu CP. 2012. Characterization and functional analysis of atl, a novel gene encoding autolysin in Streptococcus suis. J. Bacteriol. 194:1464–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Wang Y, Jin S, Wu Z, Chin DP, Koplan JP, Wilson ME. 2008. Emergence and control of infectious diseases in China. Lancet 372:1598–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang J, Wang C, Feng Y, Yang W, Song H, Chen Z, Yu H, Pan X, Zhou X, Wang H, Wu B, Wang H, Zhao H, Lin Y, Yue J, Wu Z, He X, Gao F, Khan AH, Wang J, Zhao GP, Wang Y, Wang X, Chen Z, Gao GF. 2006. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. 10.1371/journal.pmed.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, Song Y, Zhu X, Sun H, Feng T, Guo Z, Ju A, Ge J, Dong Y, Sun W, Jiang Y, Wang J, Yan J, Yang H, Wang X, Gao GF, Yang R, Wang J, Yu J. 2007. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2:e315. 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Wang C, Feng Y, Pan X, Cheng G, Wang J, Ge J, Zheng F, Cao M, Dong Y, Liu D, Wang J, Lin Y, Du H, Gao GF, Wang X, Hu F, Tang J. 2008. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3:e2080. 10.1371/journal.pone.0002080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, Zhang H, Ma Y, Gao GF. 2010. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol. 18:124–131 [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Liu P, Li C, Tan Y, Cai X, Zhou D, Jiang Y. 2012. Isolation and characterization of 89K pathogenicity island-positive ST-7 strains of Streptococcus suis serotype 2 from healthy pigs, northeast China. ScientificWorldJournal 2012:302386. 10.1100/2012/302386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holden M, Hauser H, Sanders M, Ngo T, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail M, Price C, Rabbinowitsch E, Sharp S, Croucher N, Chieu T, Mai N, Diep T, Chinh N, Kehoe M, Leigh J, Ward P, Dowson C, Whatmore A, Chanter N, Iversen P, Gottschalk M, Slater J, Smith H, Spratt B, Xu J, Ye C, Bentley S, Barrell B, Schultsz C, Maskell D, Parkhill J. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4:e6072. 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marois C, Bougeard S, Gottschalk M, Kobisch A. 2004. Multiplex PCR assay for detection of Streptococcus suis species and serotypes 2 and 1/2 in tonsils of live and dead pigs. J. Clin. Microbiol. 42:3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nga TV, Nghia HD, Tu le TP, Diep TS, Mai NT, Chau TT, Sinh DX, Phu NH, Nga TT, Chau NV, Campbell J, Hoa NT, Chinh NT, Hien TT, Farrar J, Schultsz C. 2011. Real-time PCR for detection of Streptococcus suis serotype 2 in cerebrospinal fluid of human patients with meningitis. Diagn. Microbiol. Infect. Dis. 70:461–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. 10.1016/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayawardena S, Cheung CY, Barr IG, Chan KH, Chen H, Guan Y, Peiris JSM, Poon LLM. 2007. Loop-mediated isothermal amplification for influenza A (H5N1) virus. Emerg. Infect. Dis. 13:899–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong QM, Lu SH, Tong QB, Lou D, Chen R, Zheng B, Kumagai T, Wen LY, Ohta N, Zhou XN. 2012. Loop-mediated isothermal amplification (LAMP): early detection of Toxoplasma gondii infection in mice. Parasites Vectors 5. 10.1186/1756-3305-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Jiang L, Ge B. 2012. Loop-mediated isothermal amplification assays for detecting Shiga toxin-producing Escherichia coli in ground beef and human stools. J. Clin. Microbiol. 50:91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huy NT, Hang le TT, Boamah D, Lan NT, Van Thanh P, Watanabe K, Huong VT, Kikuchi M, Ariyoshi K, Morita K, Hirayama K. 2012. Development of a single-tube loop-mediated isothermal amplification assay for detection of four pathogens of bacterial meningitis. FEMS Microbiol. Lett. 337:25–30 [DOI] [PubMed] [Google Scholar]
- 18.Iwamoto T, Sonobe T, Hayashi K. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori Y, Kitao M, Tomita N, Notomi T. 2004. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59:145–157 [DOI] [PubMed] [Google Scholar]
- 20.Tomita N, Mori Y, Kanda H, Notomi T. 2008. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 3:877–882 [DOI] [PubMed] [Google Scholar]
- 21.Goto M, Honda E, Ogura A, Nomoto A, Hanaki KI. 2009. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46:167–172 [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Li M, Cui Y, Zhao J, Cui Z, Li Q, Qu K. 2012. Electrochemical behavior of calcein and the interaction between calcein and DNA. Electroanalysis 24:1878–1886 [Google Scholar]
- 23.Wastling SL, Picozzi K, Kakembo AS, Welburn SC. 2010. LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl. Trop. Dis. 4:e865. 10.1371/journal.pntd.0000865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosaka N, Ndembi N, Ishizaki A, Kageyama S, Numazaki K, Ichimura H. 2009. Rapid detection of human immunodeficiency virus type 1 group M by a reverse transcription-loop-mediated isothermal amplification assay. J. Virol. Methods 157:195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das A, Babiuk S, McIntosh MT. 2012. Development of a loop-mediated isothermal amplification assay for rapid detection of capripoxviruses. J. Clin. Microbiol. 50:1613–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatesan G, Bhanuprakash V, Balamurugan V, Singh RK, Pandey AB. 2012. Development of loop-mediated isothermal amplification assay for specific and rapid detection of camelpox virus in clinical samples. J. Virol. Methods 183:34–39 [DOI] [PubMed] [Google Scholar]
- 27.Enomoto Y, Yoshikawa T, Ihira M, Akimoto S, Miyake F, Usui C, Suga S, Suzuki K, Kawana T, Nishiyama Y, Asano Y. 2005. Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. J. Clin. Microbiol. 43:951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle B, Vaillancourt K, Bonifait L, Charette SJ, Gottschalk M, Grenier D. 2012. Genome sequence of the swine pathogen Streptococcus suis serotype 2 strain S735. J. Bacteriol. 194:6343–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wertheim HF, Nghia HD, Taylor W, Schultsz C. 2009. Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48:617–625 [DOI] [PubMed] [Google Scholar]
- 30.Mai NT, Hoa NT, Nga TV, Linh le D, Chau TT, Sinh DX, Phu NH, Chuong LV, Diep TS, Campbell J, Nghia HD, Minh TN, Chau NV, de Jong MD, Chinh NT, Hien TT, Farrar J, Schultsz C. 2008. Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46:659–667 [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi D, Kerdsin A, Pienpringam A, Loetthong P, Samerchea S, Luangsuk P, Khamisara K, Wongwan N, Areeratana P, Chiranairadul P, Lertchayanti S, Petcharat S, Yowang A, Chaiwongsaen P, Nakayama T, Akeda Y, Hamada S, Sawanpanyalert P, Dejsirilert S, Oishi K. 2012. Population-based study of Streptococcus suis infection in humans in Phayao province in northern Thailand. PLoS One 7:e31265. 10.1371/journal.pone.0031265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pian Y, Gan S, Wang S, Guo J, Wang P, Zheng Y, Cai X, Jiang Y, Yuan Y. 2012. Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect. Immun. 80:2402–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houde M, Gottschalk M, Gagnon F, Van Calsteren MR, Segura M. 2012. Streptococcus suis capsular polysaccharide inhibits phagocytosis through destabilization of lipid microdomains and prevents lactosylceramide-dependent recognition. Infect. Immun. 80:506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Shen X, Yan J, Han H, Zheng B, Liu D, Cheng H, Zhao Y, Rao X, Wang C, Tang J, Hu F, Gao GF. 2011. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79:1670–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Njiru ZK. 2012. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl. Trop. Dis. 6:e1572. 10.1371/journal.pntd.0001572 [DOI] [PMC free article] [PubMed] [Google Scholar]