Abstract
Japanese encephalitis virus (JEV) was isolated from the cerebrum of a calf which showed severe neurological symptoms in late September 2009, and the JEV isolate was revealed to be of genotype 1. This is the first report describing the isolation of genotype 1 JEV from cattle.
CASE REPORT
In late September 2009, a 141-day-old female Japanese black calf showed decreased appetite and depression in a farm located in the central area of Miyazaki Prefecture, Japan. Four days after the onset of these symptoms, the calf showed circling and disordered consciousness. Despite palliative treatments, no improvement was seen in the calf, and the calf then became unable to stand and was euthanized. A necropsy was performed, and organs, including the brain, spinal cord, heart, lung, liver, kidney, and spleen, were collected for virological and pathological investigations. No clinical symptoms were observed in the other 3 calves and 8 cows on the farm.
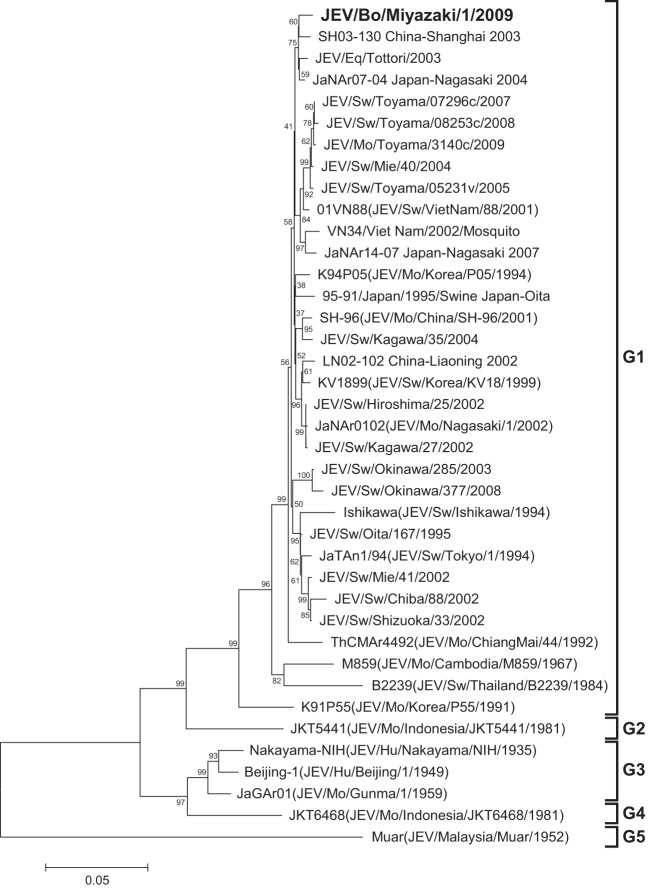
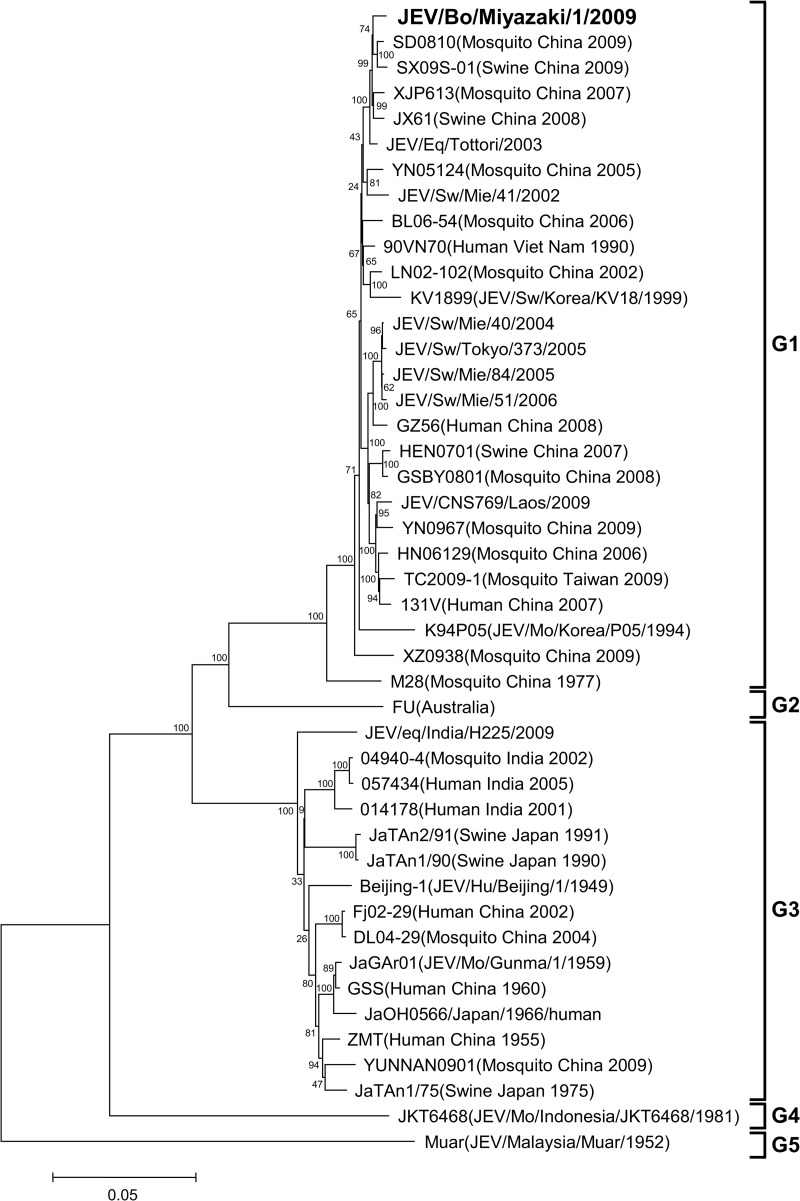
From the cerebrum and medulla oblongata of the affected calf, a 10% homogenate was prepared. The homogenate and cerebrospinal fluid were subjected to extraction of RNA with the use of a High Pure viral RNA kit (Roche Applied Science, Mannheim, Germany). Then, reverse transcription-PCR (RT-PCR) and nested PCR were performed for detection of Japanese encephalitis virus (JEV) RNA with the use of a Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA, USA) and the Qiagen Taq PCR master kit (Qiagen), respectively (1, 2). The cerebrum tested positive for JEV, but the medulla oblongata and cerebrospinal fluid tested negative. The homogenate of the cerebrum was then subjected to virus isolation by intracranial inoculation into suckling mice (1). As a result, JEV was isolated from the brains of the mice. From the JEV isolate, which we named JEV/bovine (Bo)/Miyazaki/1/2009, cDNAs containing the E region and 3′ untranslated region (3′UTR) were amplified by RT-PCR with the use of a Qiagen OneStep RT-PCR kit and the primer sets that were originally designed for amplifying the E region and the 3′UTR of JEV isolates in Japan in 2002 to 2004 (3). In addition, RT-PCR was performed with several other primer sets for determination of the complete genome sequence. The 5′-terminal sequence was determined by using the rapid amplification of 5′ cDNA ends (5′RACE) system (Invitrogen, Carlsbad, CA, USA). The 3′-terminal sequences were determined with viral RNA, to which we added a poly(A) tail at the 3′ end with a poly(A) tailing kit (Ambion, Austin, TX, USA) by using the 3′RACE system (Invitrogen) (4). All the primers used for the RT-PCR, nested PCR, 5′RACE, 3′RACE, and sequencing are shown in Table 1. The nucleotide sequences of JEV/Bo/Miyazaki/1/2009 and other isolates of JEV were aligned by the Clustal W program (5); then phylogenetic trees of the E region and the complete genome were constructed with MEGA5 using the neighbor-joining method, and the reliability of the branching orders was evaluated by the bootstrap test (n = 1,000) (6). As a result, the JEV isolate was found to contain 10,965 nucleotides and clustered with other isolates belonging to JEV genotype 1 (G1) (Fig. 1 and 2). Based on the phylogenetic analysis of the E region, the JEV isolate, JEV/Bo/Miyazaki/1/2009, was most closely related to SH03-130, an isolate from Culex tritaeniorhynchus in Shanghai, China (7), and also closely related to JaNAr07-04 and JEV/equine (Eq)/Tottori/2003, isolates from a mosquito in Nagasaki, Japan, and an affected horse in Tottori, Japan, respectively (8, 9) (Fig. 1). The phylogenetic analysis of the complete genome sequence reveled that JEV/Bo/Miyazaki/1/2009 was closely related to several other isolates in China identified in 2007 to 2009, such as SD0810, SX09S-01, XJP613, and JX61 (Fig. 2). The nucleotide sequences of the 3′UTRs of JEV/Bo/Miyazaki/1/2009 and other JEV isolates belonging to genotypes 1 to 5 available in GenBank were then compared. The nucleotide alignment revealed that JEV/Bo/Miyazaki/1/2009 has the same deletion in its 3′UTR (nucleotides 5 to 6, 14 to 26, 35, 46, and 58 to 59) as several other isolates of JEV G1 in Japan, China, and South Korea identified in 1994 to 2008, such as Ishikawa, JEV/Eq/Tottori/2003, JEV/swine (Sw)/Okinawa/377/2008, LN02-102, and K94P05 (data not shown). The samples collected from the affected calf were also screened for orthobunyaviruses (10), bovine herpesvirus 1, and Borna disease virus (11, 12), but all the samples tested negative (data not shown). Protocols for the animal experimentation involved in this report were approved by the Institutional Animal Care and Use Committee of the National Institute of Animal Health.
Table 1.
Oligonucleotide primers used for the cDNA amplification and sequencing of JEV/Bo/Miyazaki/1/2009
| Primer | Sequence (5′–3′) | Nucleotide positions | Purpose | Reference |
|---|---|---|---|---|
| JE8K-S | ATGGAACCCCCCTTC | 2098–2112 | 1st RT-PCR | NIID and Japan Association of Prefectural and Municipal Public Health Institutes (2) |
| JEER | AGCAGGCACATTGGTCGCTA | 2478–2459 | ||
| JE8K-inner-S | ATCGTGGTTGGGAGGGGAGA | 2125–2146 | Nested PCR | |
| JEER inner-C | AGCACACCTCCTGTGGCTAA | 2450–2431 | ||
| JE955f | TGYTGGTCGCTCCGGCTTA | 956–974 | RT-PCR (E region) | Nerome et al. (3) |
| JE2536r | AAGATGCCACTTCCACAYCTC | 2537–2517 | ||
| JE10141f | TGGATTGAAGAAAATGAATGGATG | 10141–10164 | RT-PCR (3′UTR) | Nerome et al. (3) |
| JE10965r | AGATCCTGTGTTCTTCCTCTC | 10965–10945 | ||
| JEV9-2193F | ATCTGTGTGAACTTCTTGGC | 9–28 | RT-PCR and sequencing | This study |
| JEV9-2193R | TTTACCCAGCGTGCTTCCAGC | 2193–2173 | ||
| JEV1850-3845F | TGGACAAACTGGCTCTGAAGGG | 1850–1871 | ||
| JEV1850-3845R | TTCTCTTGGTTCGTCCATCTCG | 3845–3824 | ||
| JEV3655-5606F | CTACTTGTGCTGATGCTTGG | 3655–3674 | ||
| JEV3655-5606R | ATTGGGGCATTTGAGTC | 5606–5590 | ||
| JEV5409-7958F | CCATAGACTAATGTCACCAAAC | 5409–5430 | ||
| JEV5409-7958R | AGAGTTGCTGCGTAGTAG | 7958–7941 | ||
| JEV7543-9421F | GACAATGGAGCCAGTGC | 7543–7559 | ||
| JEV7543-9421R | CCTTGACCACTTTGTGCCTG | 9421–9402 | ||
| JEV9234-10965F | CATTCTCCGTGACATAGCAGG | 9234–9254 | ||
| JEV9234-10965R | AGATCCTGTGTTCTTCCTCACC | 10965–10944 | ||
| JEV1108R | GGACATCTAGTGTTGGTTTG | 1108–1089 | 5′RACE | This study |
| JEV1030R | CTCCACTGGCTCCTTCTATG | 1030–1011 | ||
| JEV157R | ATCCGCGTTTCAGCATATTGATGG | 157–134 | ||
| JEV9568F | GTCATCGGACCACAACACTTG | 9568–9588 | 3′RACE | This study |
| JEV10757F | CCGTGGAAACAACATTATGC | 10757–10776 | ||
| JEV2312F | TTGGCGGTGCATTCAGAAC | 2312–2330 | Sequencing | This study |
| JEV3305R | TCAAGGACAATGCCGTTCTC | 3305–3286 | ||
| JEV4293F | CGAATCTATGTCAATACCCTTCATG | 4293–4317 | ||
| JEV5124R | CTCTTGACGGTCGCCTTGC | 5124–5106 | ||
| JEV5976F | CCAACGGAGAGGTAGAGTAGGC | 5976–5997 | ||
| JEV7367R | TCCACGACGGCATTCTTCATTAT | 7367–7345 | ||
| JEV7819F | AACATAGTGGGAGGACATC | 7819–7837 | ||
| JEV8993R | GCATTGACCATCTCCCAGAAC | 8993–8973 | ||
| JEV9578F | CACAACACTTGGAACAG | 9578–9594 | ||
| JEV10306R | CCCTCACTTGGTTTATTGCCG | 10306–10286 | ||
| JEV10746R | AACCTCTAGTCCTTACACC | 10746–10728 |
Fig 1.
Phylogenetic profile showing the relationships among JEV isolates based on a comparison of their E regions (1,500 nucleotides). The bootstrap percentages calculated from 1,000 replications are indicated around the internal nodes. The scale represents 0.05% sequence divergence. Mo, mosquito; Hu, human.
Fig 2.
Phylogenetic profile showing the relationships among JEV isolates based on a comparison of the complete genome (approximately 11,000 nucleotides). The bootstrap percentages calculated from 1,000 replications are indicated around the internal nodes. The scale represents 0.05% sequence divergence.
No gross lesions were observed in the brain, spinal cord, or other organs of the calf; however, the histological examination revealed nonsuppurative encephalomyelitis (Table 2). In the cerebrum, diffuse neuronal degeneration and necrosis were observed mainly in the gray matter, and neuronophagia was also occasionally detected. Perivascular infiltration of lymphocytes (Fig. 3A) and glial nodules (Fig. 3B) was detected widely in the gray and white matter of the cerebrum, and diffuse lymphocytic infiltration was detected in the cerebral medulla. Also, nonsuppurative meningitis was observed in the cerebrum, cerebellum, and midbrain. Histological findings in other parts of the central nervous system included neuronal necrosis, microglial infiltration, perivascular infiltration, and glial nodules, and all of these were detected in the cerebellum, midbrain, pons, medulla oblongata, and spinal cord, except that glial nodules were not detected in the cerebellum. No histological findings were observed in the heart, lung, liver, kidney, or spleen, except for slightly thickened alveolar septa in the lungs.
Table 2.
Histological findings and JEV antigen detected by the IHC assay in the cerebrum, cerebellum, midbrain, pons, medulla oblongata, and spinal cord of the affected calfa
| Sample type | Neuronal degeneration | Perivascular infiltration of lymphocytes | Glial nodules | Nonsuppurative meningitis | JEV antigen |
|---|---|---|---|---|---|
| Cerebrum | |||||
| Frontal lobe | +++a | +++ | + | + | +++ |
| Parietal lobe | +++ | +++ | ++ | ++ | +++ |
| Temporal lobe | ++ | +++ | + | ++ | +++ |
| Occipital lobe | ++ | +++ | + | ++ | ++ |
| Hippocampus | +++ | +++ | + | − | +++ |
| Diencephalon | + | ++ | + | − | − |
| Cerebellum | + | + | − | + | + |
| Midbrain | + | ++ | + | + | − |
| Pons | + | ++ | ++ | − | + |
| Medulla oblongata | + | ++ | ++ | − | − |
| Spinal cord | |||||
| Cervical | + | + | + | − | + |
| Thoracic | + | + | + | − | + |
| Lumbar | + | + | + | − | + |
Subjective determinations were made for the presence or absence of histological lesions and JEV antigen and are indicated as + (minimal) to +++ (abundant) or − (not observed).
Fig 3.
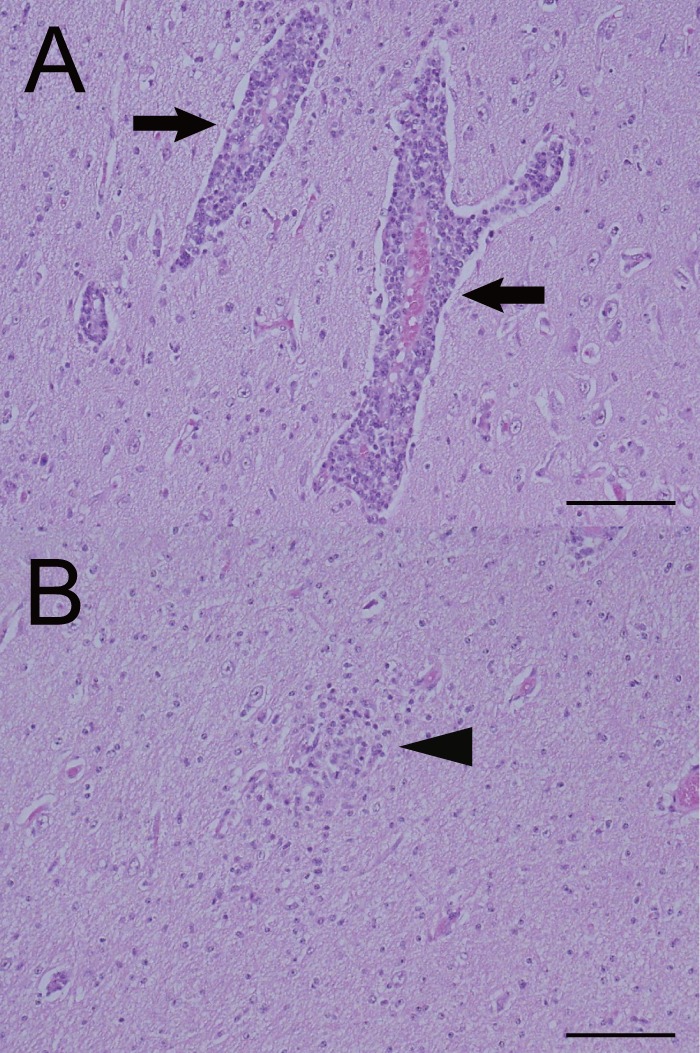
Perivascular infiltration of lymphocytes (A) and a glial nodule (B) in the cerebrum of the affected calf. Arrows indicate the perivascular infiltration of lymphocytes, and an arrowhead points out the glial nodule. Hematoxylin and eosin stain. Bar = 200 μm.
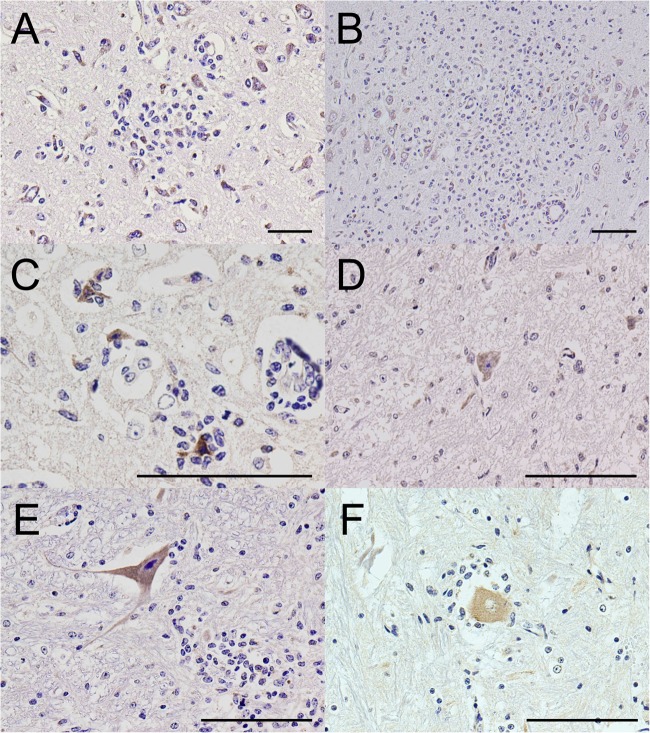
An immunohistochemical (IHC) assay was performed using a Histofine SAB-PO kit (Nichirei, Tokyo). Anti-JEV polyclonal mouse serum (kindly provided by the Chuo Livestock Hygiene Service Center of Chiba Prefecture, Japan), which was produced in a mouse immunized with whole inactivated virus (Nakayama-Yakken strain of JEV) (13) diluted 1:2,000, was used as the primary antibody for detection of JEV antigen. As a result, strong immunoreactivity for JEV antigens was revealed mainly within the cytoplasm of neurons and nerve axons in the cerebrum (Fig. 4A to C and Table 2). The immunoreactivity for JEV was also observed occasionally in the cerebellum, pons, and spinal cord, including the cervical, thoracic, and lumbar spinal cord (Fig. 4D to F and Table 2).
Fig 4.
IHC assay results. Detection of JEV antigen in the cerebrum (A to C), cerebellum (D), pons (E), and spinal cord (F) of the affected calf. JEV-positive granules (dark brown) are observed mainly in the cytoplasm of neurons and occasionally in nerve axons. (C) Positively labeled neurons are undergoing neuronophagia. Bar = 100 μm.
The histopathological diagnosis of the calf's case was neuronal necrosis and neuronophagia with nonsuppurative encephalomyelitis and meningitis. These lesions are consistent with a neuronotropic viral infection, such as Akabane virus (AKAV) and JEV (14–16). We detected JEV RNA in the homogenate of the cerebrum of the affected calf, and we detected JEV antigen in the cerebrum, cerebellum, pons, and spinal cord. We isolated JEV from the cerebrum, and therefore, we diagnosed this case as Japanese encephalitis (JE) of a calf. This diagnosis was also supported by serological evidence of a JEV epidemic in the central area of Miyazaki Prefecture that showed the prevalence of anti-JEV hemagglutination inhibition (HI) antibodies in porcine sera (5 to 8 months old) collected form slaughterhouses in Miyazaki Prefecture in August 2009 (information obtained at http://idsc.nih.go.jp/yosoku/JE/2009JEsw/JE09_6.html [in Japanese]).
This is the first report describing the isolation of G1 JEV from cattle. Among the five genotypes of JEV based on the sequence of the E region, which encodes envelope protein (17), the dominant genotype shifted from 3 to 1 in Japan in the mid-1990s (3, 8, 18, 19). It is thus thought to be JEV genotype 3 (G3) that caused natural infection in cattle or was used for an experimental infection of calves in the 1940 to 1950s (20–22), and it was also the G3 P20778 strain which was used for an experimental infection of cattle in India (23). A natural case of JE in an 18-month-old cow was reported in Chiba Prefecture, Japan, in 1996 (16), and JEV was isolated from the affected cow in this case; however, there has been no report that described the genotype of the isolate.
Although our data clearly indicated that in the calf's case the causative agent was JEV, it remains unclear why the calf developed the disease, since cattle usually have undetectable or no viremias after JEV infection (22, 23). Among humans, children are at high risk for a fatal outcome of JE (24), and the age of the calf may have contributed to the outcome in this case, but the number of bovine JE cases is quite low and seems not to be enough to discuss the age dependency of JE in cattle (16, 20, 21).
The immune status of the calf might not have been good, because a pathological change—slightly thickened alveolar septa—was observed in the lungs, but there was no available information about indicators of the calf's immune status. The main factors for the development of the disease might have been in the JEV isolate, JEV/Bo/Miyazaki/1/2009; however, the JEV isolate was not very unique but similar to other isolates of JEV found in recent years with regard to the deletion observed in the 3′UTR, which was suggested to influence viral replication (4, 24–26), and the phylogenetic characteristics of its E region and its complete genome.
Further studies are needed for the elucidation of viral replication in vitro and in vivo. Also, we desire to clarify the pathogenicity of the JEV/Bo/Miyazaki/1/2009 isolate in cattle. Furthermore, testing for JEV is recommended in bovine cases that exhibit nonsuppurative encephalomyelitis or encephalitis but are negative for other viruses that cause neurological disorders, such as AKAV and Chuzan virus (14, 15, 27), in areas of JEV endemicity for a better understanding of the relationship between JEV infection and its pathogenicity in cattle.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this work were deposited in the DNA Data Bank of Japan (DDBJ) with the accession numbers AB795032 (E region), AB795033 (3′UTR), and AB830335 (complete genome).
ACKNOWLEDGMENTS
We thank Tomokazu Kitano and Miho Miura (Miyazaki Prefectural Institute for Public Health and Environment) for providing detailed information about the prevalence of anti-JEV hemagglutination inhibition (HI) antibodies in porcine sera collected at slaughterhouses in Miyazaki Prefecture.
Footnotes
Published ahead of print 24 July 2013
REFERENCES
- 1.OIE 2008. Japanese encephalitis, p 231–239 In OIE Biological Standards Commission (ed), Manual of diagnostic tests and vaccines for terrestrial animals, 6th ed, vol 1 Office International des Epizooties, Paris, France [Google Scholar]
- 2.National Institute of Infectious Diseases (NIID) and Japan Association of Prefectural and Municipal Public Health Institutes 2003. Manuals for the detection of pathogens. NIID, Tokyo, Japan: http://www0.nih.go.jp/niid/reference/pathogen-manual-60.pdf Accessed 12 July 2013 (In Japanese.) [Google Scholar]
- 3.Nerome R, Tajima S, Takasaki T, Yoshida T, Kotaki A, Lim CK, Ito M, Sugiyama A, Yamauchi A, Yano T, Kameyama T, Morishita I, Kuwayama M, Ogawa T, Sahara K, Ikegaya A, Kanda M, Hosoya Y, Itokazu K, Onishi H, Chiya S, Yoshida Y, Tabei Y, Katsuki K, Tabata K, Harada S, Kurane I. 2007. Molecular epidemiological analyses of Japanese encephalitis virus isolates from swine in Japan from 2002 to 2004. J. Gen. Virol. 88:2762–2768 [DOI] [PubMed] [Google Scholar]
- 4.Tajima S, Nukui Y, Takasaki T, Kurane I. 2007. Characterization of the variable region in the 3′ non-translated region of dengue type 1 virus. J. Gen. Virol. 88:2214–2222 [DOI] [PubMed] [Google Scholar]
- 5.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang HY, Takasaki T, Fu SH, Sun XH, Zhang HL, Wang ZX, Hao ZY, Zhang JK, Tang Q, Kotaki A, Tajima S, Liang XF, Yang WZ, Kurane I, Liang GD. 2007. Molecular epidemiological analysis of Japanese encephalitis virus in China. J. Gen. Virol. 88:885–894 [DOI] [PubMed] [Google Scholar]
- 8.Nabeshima T, Loan HT, Inoue S, Sumiyoshi M, Haruta Y, Nga PT, Huoung VT, del Carmen Parquet M, Hasebe F, Morita K. 2009. Evidence of frequent introductions of Japanese encephalitis virus from south-east Asia and continental east Asia to Japan. J. Gen. Virol. 90:827–832 [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka T, Tsujimura K, Kondo T, Yasuda W, Okada A, Noda K, Okumura T, Matsumura T. 2006. Isolation and genetic analysis of Japanese encephalitis virus from a diseased horse in Japan. J. Vet. Med. Sci. 68:293–295 [DOI] [PubMed] [Google Scholar]
- 10.Ohashi S, Yoshida K, Yanase T, Kato T, Tsuda T. 2004. Simultaneous detection of bovine arboviruses using single-tube multiplex reverse transcription-polymerase chain reaction. J. Virol. Methods 120:79–85 [DOI] [PubMed] [Google Scholar]
- 11.Schynts F, Baranowski E, Lemaire M, Thiry E. 1999. A specific PCR to differentiate between gE negative vaccine and wildtype bovine herpesvirus type 1 strains. Vet. Microbiol. 66:187–195 [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y, Nakaya T, Hagiwara K, Momiyama N, Kagawa Y, Taniyama H, Ishihara C, Sata T, Kurata T, Ikuta K. 1999. High susceptibility of Mongolian gerbil (Meriones unguiculatus) to Borna disease virus. Vaccine 17:480–489 [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi Y, Hasegawa H, Oyama T, Tamai T, Kusaba T. 1984. Antigenic analysis of Japanese encephalitis virus by using monoclonal antibodies. Infect. Immun. 44:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kono R, Hirata M, Kaji M, Goto Y, Ikeda S, Yanase T, Kato T, Tanaka S, Tsutsui T, Imada T, Yamakawa M. 2008. Bovine epizootic encephalomyelitis caused by Akabane virus in southern Japan. BMC Vet. Res. 4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazato S, Miura Y, Hase M, Kubo M, Goto Y, Kono Y. 1989. Encephalitis of cattle caused by Iriki isolate, a new strain belonging to Akabane virus. Nihon Juigaku Zasshi 51:128–136 [DOI] [PubMed] [Google Scholar]
- 16.Katayama K, Ikeda A, Saika K, Ishizaka M, Onodera M, Ito T. 2000. A case of Japanese encephalitis in a black Japanese Heifer. J. Jpn. Vet. Med. Assoc. 53:293–296 (In Japanese, with English summary.) [Google Scholar]
- 17.Mackenzie JS, Gubler DJ, Petersen LR. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98–S109 [DOI] [PubMed] [Google Scholar]
- 18.Pan XL, Liu H, Wang HY, Fu SH, Liu HZ, Zhang HL, Li MH, Gao XY, Wang JL, Sun XH, Lu XJ, Zhai YG, Meng WS, He Y, Wang HQ, Han N, Wei B, Wu YG, Feng Y, Yang DJ, Wang LH, Tang Q, Xia G, Kurane I, Rayner S, Liang GD. 2011. Emergence of genotype I of Japanese encephalitis virus as the dominant genotype in Asia. J. Virol. 85:9847–9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takegami T, Ishak H, Miyamoto C, Shirai Y, Kamimura K. 2000. Isolation and molecular comparison of Japanese encephalitis virus in Ishikawa, Japan. Jpn. J. Infect. Dis. 53:178–179 [PubMed] [Google Scholar]
- 20.Yamamoto S, Tsubahara H, Yoshida T, Harada K. 1949. First observation on case of Japanese B encephalitis of cattle. Bull. Natl. Inst. Anim. Health 22:197–203 (In Japanese, with English summary.) [Google Scholar]
- 21.Shimizu T, Mochizuki H, Sugawa Y, Okazaki K, Matumoto M. 1951. Studies on Japanese encephalitis of cattle—1. Bovine encephalitis caused by natural infection with Japanese encephalitis virus. Natl. Inst. Anim. Health Q. 23:111–118 [Google Scholar]
- 22.Shimizu T, Mochizuki H, Sugawa Y, Okazaki K, Matumoto M. 1951. Studies on Japanese encephalitis of cattle—2. Experimental infection of calves with virus of Japanese encephalitis. Natl. Inst. Anim. Health Q. 23:119–128 [Google Scholar]
- 23.Ilkal MA, Dhanda V, Rao BU, George S, Mishra AC, Prasanna Y, Gopalkrishna S, Pavri KM. 1988. Absence of viraemia in cattle after experimental infection with Japanese encephalitis virus. Trans. R. Soc. Trop. Med. Hyg. 82:628–631 [DOI] [PubMed] [Google Scholar]
- 24.Gubler DJ, Kuno G, Markoff L. 2007. Flaviviruses, p 1153–1252 In Kinipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. 2005. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200–212 [DOI] [PubMed] [Google Scholar]
- 26.Yun SI, Choi YJ, Song BH, Lee YM. 2009. 3′ cis-acting elements that contribute to the competence and efficiency of Japanese encephalitis virus genome replication: functional importance of sequence duplications, deletions, and substitutions. J. Virol. 83:7909–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamakawa M, Kubo M, Furuuchi S. 1999. Molecular analysis of the genome of Chuzan virus, a member of the Palyam serogroup viruses, and its phylogenetic relationships to other orbiviruses. J. Gen. Virol. 80:937–941 [DOI] [PubMed] [Google Scholar]