Abstract
Amikacin is a major drug used for the treatment of Mycobacterium avium complex (MAC) disease, but standard laboratory guidelines for susceptibility testing are not available. This study presents in vitro amikacin MICs for 462 consecutive clinical isolates of the MAC using a broth microdilution assay. Approximately 50% of isolates had amikacin MICs of 8 μg/ml, and 86% had MICs of ≤16 μg/ml. Of the eight isolates (1.7%) with MICs of 64 μg/ml, five had an MIC of 32 μg/ml on repeat testing. Ten isolates (2.1%) had an initial amikacin MIC of >64 μg/ml, of which seven (1.5%) had MICs of >64 μg/ml on repeat testing. These seven isolates had a 16S rRNA gene A1408G mutation and included M. avium, Mycobacterium intracellulare, and Mycobacterium chimaera. Clinical data were available for five of these seven isolates, all of which had received prolonged (>6 months) prior therapy, with four that were known to be treated with amikacin. The 16S mutation was not detected in isolates with MICs of ≤64 μg/ml. We recommend primary testing of amikacin against isolates of the MAC and propose MIC guidelines for breakpoints that are identical to the CLSI guidelines for Mycobacterium abscessus: ≤16 μg/ml for susceptible, 32 μg/ml for intermediate, and ≥64 μg/ml for resistant. If considered and approved by the CLSI, this will be only the second drug recommended for primary susceptibility testing against the MAC and should facilitate its use for both intravenous and inhaled drug therapies.
INTRODUCTION
Amikacin is considered to be a major drug used for the treatment of Mycobacterium avium complex (MAC) lung disease, especially for patients who have cavitary disease or whose isolate is macrolide resistant (1–3). Its use is difficult in part because standard laboratory guidelines for susceptibility testing have not been established. The American Thoracic Society (ATS) and the 2011 Clinical and Laboratory Standards Institute (CLSI) guidelines currently recommend reporting primary susceptibilities only for clarithromycin and secondarily for linezolid and moxifloxacin against isolates of the MAC (1, 4).
In vitro and in a murine model, the bactericidal and chemotherapeutic efficacies of amikacin and other aminoglycosides against MAC have been known for >20 years; however, drug toxicities and adverse events, including hearing loss, have limited the chemotherapeutic use of intravenous (i.v.) or intramuscular (i.m.) amikacin, especially in elderly patients (1, 5, 6). The introduction of aerosolized amikacin has offered a potentially effective, practical, and less toxic alternative to parenteral administration of the antibiotic (7). The aerosolized form of amikacin has been touted as safer and easier to tolerate than the parenterally administrated form. Additionally, it may be possible with the parenteral form to use a higher and thus presumably more effective dose than is usually tolerable with the i.v. or i.m. modalities (7). This formulation is being increasingly used in patients with MAC and with Mycobacterium abscessus infections.
We initiated a retrospective study of the in vitro MICs of amikacin in 462 consecutive isolates of the MAC submitted for susceptibility testing to the Mycobacteria/Nocardia Laboratory at UT Health Northeast (formerly The University of Texas Health Science Center at Tyler).
The aim of our study was 2-fold. We retrospectively examined the in vitro MICs of amikacin in 462 consecutive isolates of the MAC that were submitted to our laboratory for susceptibility testing to determine a resistance breakpoint. Additionally, prior amikacin use was examined for patients with initial and repeat amikacin MICs of >64 μg/ml and those with an rrn gene mutation to help establish guidelines for an additional primary drug for treating MAC disease.
MATERIALS AND METHODS
Organisms.
We studied 462 consecutive clinical isolates of the MAC that had been submitted for susceptibility testing to the Mycobacteria/Nocardia Research Laboratory at UT Health Northeast between 2011 and 2012. This laboratory receives isolates for susceptibility testing from all over the United States. Isolates were submitted as being of the MAC, most of which had been identified as MAC by a commercial probe assay, which according to the package insert has a sensitivity of 99.9% and specificity of 100%; however, the manufacturer states that rare isolates of MAC may not test positive by the probe assay. Isolates with initial and repeat amikacin MICs of >64 μg/ml were identified as species of the MAC by molecular identification using multiplex PCR and internal transcribed spacer (ITS) sequencing as previously described (8, 9).
For isolates with MICs of ≥64 μg/ml, stored culture samples were screened for additional isolates recovered before or after the ≥64 μg/ml isolate.
Susceptibility testing.
Antimicrobial susceptibility testing was performed using the CLSI-recommended method of broth microdilution in cation-adjusted Mueller-Hinton broth plus oleic acid-albumin-dextrose-catalase (OADC) using commercially available MIC panels (Thermo Fisher, formerly Trek Diagnostics, Cleveland, OH). Transparent colonies of the MAC (if present) were selected from Middlebrook 7H10 agar plates. The colonies were inoculated into cation-adjusted Mueller-Hinton broth plus 5% OADC. The inoculum was standardized to match a 0.5 McFarland standard by using a nephelometer. Serial dilutions were performed to deliver final concentrations of approximately 1 × 105 to 5 × 105 CFU/ml in a 0.1-ml volume. Susceptibilities were read using a mirrored light box (Trek Diagnostics) after incubation at 35°C in room air for 7 days. The endpoint (MIC) was complete inhibition of growth. Concentrations of amikacin tested ranged from ≤1 to 64 μg/ml. Isolates with amikacin MICs of 64 μg/ml or >64 μg/ml underwent repeat testing. For isolates with MICs of 64 or >64 μg/ml, the growth in the next lowest MIC well was reviewed and compared to the control growth.
Patients.
Clinical information about amikacin use and the testing of any additional isolates was sought for patients with MAC isolates with amikacin MICs of 64 μg/ml or >64 μg/ml. This study was approved by the UT Health Northeast and Duke University institutional review boards.
Variable-number tandem repeat typing.
Variable-number tandem repeat (VNTR) typing of the MAC isolates with MICs of >64 μg/ml and prior isolates with MICs of ≤32 μg/ml from the same patient were performed using previously described methods (8, 10, 11). Isolates of Mycobacterium chimaera were tested using Mycobacterium intracellulare primers (8).
Quality control.
Quality control was performed using M. avium strain ATCC 700898 and incubation for 7 days as for the clinical isolates. The acceptable range of MICs was derived by using values ± one dilution from the modal value in a series of 34 test values. Additional quality control was provided using Pseudomonas aeruginosa strain ATCC 27853. After the established 18- to 24-h incubation period for bacteria, the acceptable MIC range for P. aeruginosa was 1 to 4 μg/ml, as previously defined by the CLSI (12).
PCR restriction endonuclease analysis of the 16S rRNA gene.
A sequence of approximately 520 bp of the 16S rRNA gene that included bp position 1408 was selected for study and was amplified by PCR. Briefly, 25 μl of reaction mixture, which consisted of 12.5 μl of FailSafe PreMix I, 2 μmol of each primer, 0.625 U of FailSafe PCR enzyme (Epicentre, Madison, WI), and 1 μl of template DNA was placed in a thermocycler for 35 cycles. Primer 261, corresponding to 16S rRNA Escherichia coli positions 1520 to 1539 (5′-AAGGAGGTGATCCAGCCGCA-3′), and primer 297, corresponding to 16S rRNA E. coli positions 1056 to 1075 (5′-TCCCTTGTGGCCTGTGTGCA-3′), were used (13). The amplicon size was approximately 520 and the readable sequence was 480 bp.
The restriction endonuclease Tsp45I (New England BioLabs, Ipswich, MA) failed to cut the 520-bp amplicon in the presence of the A1408G mutation but cut the wild-type sequence into two fragments of approximately 130 and 390 bp. PCR restriction endonuclease analysis (PRA) was performed on isolates with amikacin MICs of ≥64 μg/ml. The use of this technique allows for the detection of a mixed population of isolates with and without the A1408G mutation.
16S rRNA gene sequencing.
Isolates were subjected to sequencing of the 16S rRNA gene region, which was previously shown to confer resistance to amikacin in other mycobacteria (positions 1408, 1409, and 1411, Escherichia coli numbering system) (13).
RESULTS
Organisms.
A total of 462 consecutive isolates from multiple states within the United States were tested. Isolates with initial and repeat amikacin MICs of >64 were reconfirmed as being of the MAC using a 16S multiplex PCR or commercially available DNA probes (AccuProbe; Hologic Gen-Probe, San Diego, CA). All seven of the repeat isolates were identified as species of the M. avium complex by ITS sequencing (8). These results demonstrated the presence of one isolate of M. avium, six isolates of M. intracellulare, and one isolate of M. chimaera.
An additional 16 isolates from five of the seven patients with MAC isolates with initial and repeat MICs of >64 μg/ml from different time periods before and after the MIC of >64 μg/ml were identified and available for amikacin MIC and 16S rRNA gene analysis comparisons (Table 1).
Table 1.
Correlation of amikacin MICs and prior amikacin exposure available on five of the seven patients with a Mycobacterium avium complex isolate with an initial and repeat amikacin MIC of >64 μg/ml and a 16S rRNA gene A1408G mutation
| Case | Culturea | Species Identification | VNTR type | Amikacin MIC (μg/ml) (date) | Exposure to amikacin | rrn at bp 1408b | Clinical history |
|---|---|---|---|---|---|---|---|
| 1 | Pre-Rx | Inhaled × 9 mo (2012) | MAC disease × 11 yr; improved AFB culture positivity and chest CT on inhaled amikacin, then relapsed microbiologically (see Fig. 1) | ||||
| MA-2778 no. 4 | 32 (30 December 2005) | WT | |||||
| MA-5225 | M. intracellulare | No. 16 | 8 (20 May 2011) | WT | |||
| On-Rx | |||||||
| MA-5812 | 16 (8 February 2012) | WT | |||||
| MA-5826 | >64 (July 2012) | Mut. | |||||
| MA-5278 | M. intracellulare | No. 16 | >64 (20 October 2012) | Mut. | |||
| 2 | Pre-Rx | 2 yr | November 2008 to 20 January 2010, on i.v.c amikacin with other drugs; 20 February 2010 to 11 May 2011, on inhaled amikacin; drugs/doses at another institution; 11 May 2011, returned and had worsened clinically; cultures and CT improved with switch to streptomycin | ||||
| MA-3675 | Not available | ||||||
| On-Rx | |||||||
| MA-4437 | 8 (11 January 2010) | WT | |||||
| MA-5263 | M. intracellulare | No. 40 | 8 (20 January 2010) | WT | |||
| MA-5263 | 8 (20 January 2010) | WT | |||||
| MA-5846 | >64 (11 May 2011) | Mut. | |||||
| MA-5222 | M. intracellulare | No. 40 | >64 (5 December 2011) | Mut. | |||
| 3 | Pre-Rx | Not available | On/off over 5 yr (2005–2009) | CF,d severe cavitary disease; also frequent use of inhaled tobramycin; worsening, on lung transplant list | |||
| On-Rx | |||||||
| MA-3675 | >64 (25 July 2007) | Mut. | |||||
| Post-Rx | |||||||
| MA-4481 | >64 (12 March 2010) | Mut. | |||||
| MA-4777 | >64 (8 December 2010) | Mut. | |||||
| MA-5200 | >64 (14 November 2011) | Mut. | |||||
| MA-5505 | >64 (1 June 2012) | Mut. | |||||
| MA-5845 | M. intracellulare | >64 (30 January 2013) | |||||
| 4 | Pre-Rx | 2 mo i.v., 13 mo inhaled over 4 yr; start amikacin August 2005; stop 23 November 2009 | WT | Improved, then relapsed | |||
| MA-2799 | No. 15, new | 16 (7 April 2003) | |||||
| Post-Rx | |||||||
| MA-2799 no. 2 | >64 (23 October 2006) | Mut. | |||||
| MA-5319 | No. 15 | >64 (23 January 2012) | Mut. | ||||
| 5 | Pre-Rx | Unknown | History of treated M. avium-M. intracellulare “treatment failure” | ||||
| MA-5215 | M. chimaera | No. 41 | 8 (November 2011) | WT | |||
| Post-Rx | |||||||
| MA-5582 | M. chimaera | No. 41 | >64 (June 2012) | Mut. |
Pre-Rx, pretreatment; Post-Rx, posttreatment.
WT, wild-type rrn at bp 1408; Mut., A→G mutation rrn at bp 1408.
i.v., intravenous administration.
CF, cystic fibrosis.
Susceptibility testing.
Almost one-half (48.9%) of the 462 consecutive clinical isolates had amikacin MICs of ≤8 μg/ml, and almost 90% (85.7%) had MICs of ≤16 μg/ml. The MIC50 was 16 μg/ml and MIC90 was 32 μg/ml. Only 1.7% of the isolates had amikacin MICs of 64 μg/ml, and only 2.1% of isolates had MICs of >64 μg/ml (Table 2). All eight isolates with amikacin MICs of 64 μg/ml had minimal growth (±growth) in the 32-μg/ml well. On repeat testing, five of eight isolates had MICs of 32 μg/ml. Of the eight patients with MAC MICs of 64 μg/ml, clinical histories were inadequate to determine if there was a history of prior amikacin treatment.
Table 2.
Initial broth microdilution amikacin MICs of 462 consecutive clinical isolates of M. avium complexa
| Initial amikacin MIC (μg/ml) | No. of isolates | Cumulative % of isolates |
|---|---|---|
| <1 | 7 | 1.5 |
| 2 | 18 | 5.4 |
| 4 | 57 | 17.7 |
| 8 | 144 | 48.9 |
| 16 | 171 | 85.9 |
| 32 | 46 | 95.9 |
| 64 | 9 | 97.8 |
| >64 | 10 | 100 |
These data were determined with the CLSI-approved broth microdilution method (4).
MIC mode, 16 μg/ml; MIC50, 16 μg/ml; MIC90, 32 μg/ml.
Of the 10 isolates with initial MICs of >64 μg/ml, seven were reproducible on repeat testing (see Table 2). The amounts of growth in the 32-μg/ml and 64-μg/ml wells were equivalent to the control growth. The remaining three isolates had repeat MICs of 64 μg/ml (±growth at 32 μg/ml).
Patients.
Of the seven patients with MAC isolates with MICs of >64 μg/ml, on both initial and repeat testing, clinical histories were available for five. All five patients (Table 1, cases 1 to 5) had histories of prolonged (>6 months) prior drug therapy. Four patients were known to have received amikacin (three had inhaled and i.v. forms, one had an inhaled form only), and all were considered treatment failures. Not surprisingly, 5/7 isolates were also macrolide resistant, with clarithromycin MICs of >64 μg/ml. The case summary of one patient is listed below.
Case summary.
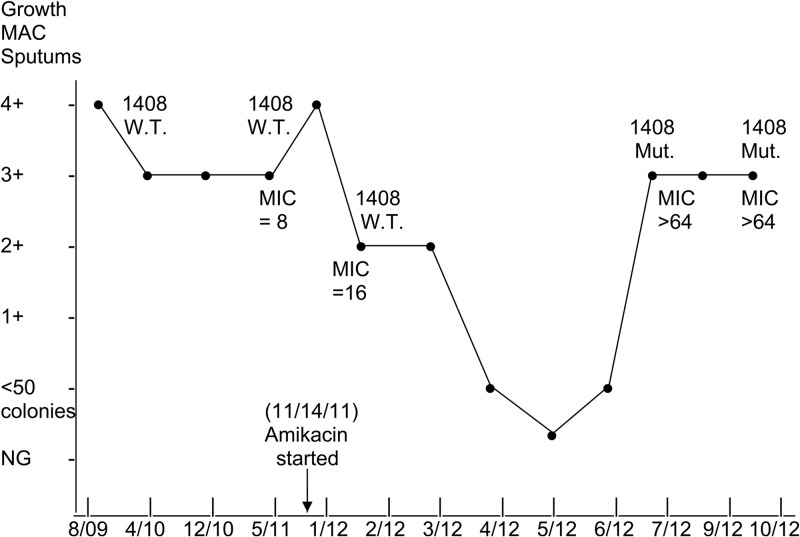
Patient one was a 59-year-old female with nodular MAC lung disease. She was treated for 5 years with clarithromycin, ethambutol, and rifabutin with subsequent cessation of treatment for approximately 5 years when the isolate infecting the patient became macrolide resistant. Subsequently, the patient worsened clinically and showed marked worsening of her nodular disease on pulmonary computed tomography (CT) scan. Multiple sputum samples (6) over the past 3 years had 3+ to 4+ colony counts on 7H10 agar for MAC. The patient was started on three-times-weekly 300 mg rifabutin and 35 mg ethambutol/kg of body weight (see Fig. 1). The patient had never been on amikacin but had been on the latter two drugs at the same doses for the first 5 years. A daily regimen of 500 mg aerosolized amikacin was then added. The amikacin MIC was 8 μg/ml, and the isolate had a wild-type 16S rRNA gene at position 1408. At 6 months following initiation of the aerosolized amikacin regimen, the patient's sputum MAC colony counts on 7H10 agar decreased to <50 colonies, and subsequently, the patient had her first negative acid-fast bacillus (AFB) culture in many years. However, at approximately 7 months after the initiation of amikacin therapy, the patient's MAC colony counts on sputum cultures rose to 3+ to 4+ on 7H10 Middlebrook agar, and repeat amikacin MICs were >64 μg/ml. Follow-up amikacin MICs on additional isolates were also >64 μg/ml on both initial and repeat testing. Sequencing of the 16S rRNA gene showed the presence of an A→G mutation at position 1408 in the two relapse isolates. Clinically, however, the patient remained much improved and her chest CT scan showed dramatic clearing of her nodular lung disease. The patient remains stable as of the time of this report. A graphic representation of the clinical treatment with amikacin related to the amikacin MICs and rrn mutations is shown in Fig. 1.
Fig 1.
Graphic representation of the microbiological response of patient 1 (case 1), with Mycobacterium intracellulare lung disease, to the addition of inhaled amikacin. The semiquantitation of MAC sputum growth on 7H10 agar has been described (1). The cultures were in the 3+ to 4+ range in the 3 years from 2009 to 2011. Note the drop in the colony counts of MAC over a 5-month period, with a return to a 3+ culture associated with a rise in the amikacin MIC to >64 μg/ml and the development of the ribosomal mutation. WT, wild-type 16S rRNA gene at bp position 1408. Mut., mutation at position 1408.
Quality control.
The amikacin MICs for the M. avium control strain ranged from 4 to 16 μg/ml. The strain M. avium ATCC 700898 was tested 34 times. The modal MIC was 8 μg/ml (28 values), with five values at 4 μg/ml and 1 value at 16 μg/ml. The MICs for P. aeruginosa ATCC 27853 were within the established CLSI acceptable range of 1 to 4 μg/ml (12).
VNTR typing.
Pretreatment isolates with amikacin MICs of 8 μg/ml and posttherapy isolates of >64 μg/ml on initial and repeat testing in four patients (Table 1, cases 1, 2, 4, and 5), and one isolate obtained 4 months after an MIC of >64 μg/ml, which was 32 μg/ml, underwent VNTR typing. All paired isolates belonged to the same species, and all had the same VNTR type, excluding the possibility of infection with a new recent isolate with an MIC of >64 μg/ml or specimen contamination. Case 4 had two VNTR genotypes on initial testing (Table 1).
PRA of the 16S rRNA gene.
A total of 40 patient isolates with amikacin MICs of ≤32 μg/ml underwent PRA. All exhibited a wild-type bp at position 1408. Of the eight isolates with initial amikacin MICs of 64 μg/ml (all with ±growth at 32 μg/ml), all exhibited a wild-type bp at position 1408. Of the 10 isolates with initial amikacin MICs of >64 μg/ml, seven isolates from six patients had the same MIC on repeat testing. All seven of these isolates exhibited a mutation at 16S rRNA position 1408. The three isolates with a repeat MIC of ≤64 μg/ml were also wild type.
16S rRNA gene sequencing.
A 16S rRNA gene mutation of bp A1408G was observed in all seven of the isolates with a mutation pattern on PRA testing. Upon sequencing the 530-bp region, no other mutations were observed. The sequencing of seven isolates with MICs of 32 μg/ml and 64 μg/ml, and those with >64 μg/ml on only one of two MIC determinations also showed a wild-type 530-bp region.
DISCUSSION
This study demonstrates that 96.2% of the clinical MAC isolates submitted for susceptibility testing have MICs of ≤32 μg/ml using the CLSI susceptibility testing method (4).
The amikacin MIC values are consistent within the achievable serum and inhaled levels and with the clinical response to MAC that is seen when amikacin is used, especially in patients with cavitary disease or macrolide resistance (2). Prolonged exposure to amikacin was present in isolates with initial and repeat MICs of >64 μg/ml, which correlated with the development of a 16S rRNA gene A→G mutation at position 1408. This was clearly demonstrated in case 1 (Fig. 1).
The amikacin MIC results reported in each well for each isolate with MICs of ≥64 μg/ml were compared to the control growth in each panel. There are several possible explanations for the minimal amounts of growth seen at 32 μg/ml with isolates with amikacin MICs of 64 μg/ml (interpreted as ±growth). These include the presence of an aminoglycoside-modifying enzyme that is weakly inducible, such as the erm gene that is seen in rapidly growing mycobacteria (14). Other possibilities are related to the presence of inoculum effects in drugs, such as ethambutol or sulfonamides, which need carefully controlled inocula to prevent trailing endpoints (3), or that the prior concentration with growth (i.e., 32 μg/ml) is very close to the real MIC, which still might be within achievable drug levels (especially for inhaled amikacin) (13, 15, 16).
The ±growth interpretation at 32 μg/ml might also be attributed to reader interpretation. Multiple individuals read the amikacin MICs over the course of the study. A comparison of readings performed by readers with little experience (≤2 years) in reading MAC susceptibilities compared to readers with considerable experience (>5 years) showed that the wells recorded as “±” by the less-experienced reader would have been recorded as negative by the more experienced reader (B. Brown-Elliott, unpublished data). This was supported by the finding that most of the isolates with initial amikacin MICs of 64 μg/ml had MICs of 32 μg/ml on repeat testing (5/8). Thus, just as CLSI guidelines currently recommend repeating amikacin MIC determinations for isolates of M. abscessus with MICs of ≥64 μg/ml, it is important to confirm amikacin MICs of ≥64 μg/ml in isolates of the MAC by either repeating the MIC or by sending the isolates to a qualified reference laboratory with specific experience in MAC susceptibility testing (4).
The assertion that an elevated amikacin MIC should be deemed amikacin resistant (with implications for subsequent treatment) may seem to contradict earlier works that found no association between aminoglycoside (streptomycin in this study) MICs and clinical outcomes in MAC pulmonary disease (17–19). However, the differences between the present findings and the earlier works are readily explicable. The patients in the prior studies had not been previously treated for MAC, and a relatively small number of isolates had streptomycin MICs of ≥64 μg/ml. If the resistance cutoff for streptomycin is similar to that for amikacin (which might be expected given their similar pharmacokinetics and mechanisms of action), a very small number of patients in these studies would be in the resistant range, providing limited power to discern differences. A variation in the MICs within a susceptible range would not necessarily be expected to be associated with differences in clinical outcomes. Furthermore, all patients in these studies were treated with multiple drugs, so susceptibility or resistance to a single drug might have had a limited impact on outcomes. Furthermore, heterogeneity in the severity and extent of clinical disease reduces the power to detect significant associations between antimicrobial susceptibility and treatment outcomes. Larger prospective studies will be needed to address these questions.
Additional mutations related to amikacin resistance other than rrn A1408G have been demonstrated in other mycobacterial species (16). It is possible that MAC isolates with amikacin MICs of ≥64 μg/ml may harbor a mutation other than the position 1408 mutation, and this question remains under investigation.
We recommend that isolates of the MAC be routinely tested against amikacin by broth microdilution assay. We further propose that the amikacin breakpoints for M. abscessus susceptibility be adopted for isolates of the MAC: MICs of ≤16 μg/ml would be considered susceptible, 32 μg/ml, intermediate, and ≥64 μg/ml, resistant. These values may allow for better usage of amikacin with either i.v. or inhaled administration, and they highlight the finding that MICs of >64 μg/ml on initial and repeat testing may develop with prolonged drug therapy as a consequence of a ribosomal mutation that is not seen in isolates with lower amikacin MICs. There are no data at present to suggest that the breakpoints for inhaled amikacin for both M. abscessus and MAC may be different than those approved or proposed for intravenous administration. There currently are no CLSI breakpoints for two other inhaled antibiotics (tobramycin and aztreonam) that are FDA approved for the treatment of P. aeruginosa in patients with cystic fibrosis. The single case report (case 1) and other isolates from patients with available treatment histories suggest that higher concentrations of inhaled amikacin are not effective in the presence of the ribosomal mutation described here. The efficacy of inhaled therapy for isolates with MICs of 64 μg/ml will need evaluation, as their response for therapy is not known.
ACKNOWLEDGMENTS
We thank Joanne Woodring for her excellent secretarial assistance. We also appreciate the laboratory assistance of Ravikiran Vasireddy, Sruthi Vasireddy, and Steven McNulty for performing molecular identification and Nicholas Parodi, Victoria Gee, Shannon O'Neil, Anita Strong, and Paula Johnson for their help with the broth microdilution testing.
Financial support for this study was provided by in-house funding and a grant from the Amon G. Carter Foundation.
Footnotes
Published ahead of print 14 August 2013
REFERENCES
- 1.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367–416 [DOI] [PubMed] [Google Scholar]
- 2.Griffith DE, Brown-Elliott BA, Langsjoen B, Zhang Y, Pan X, Girard W, Nelson K, Caccitolo J, Alvarez J, Shepherd S, Wilson R, Graviss EA, Wallace RJ., Jr 2006. Clinical and molecular analysis of macrolide resistance in Mycobacterium avium complex lung disease. Am. J. Respir. Crit. Care Med. 174:928–934 [DOI] [PubMed] [Google Scholar]
- 3.van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. 2012. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist. Updat. 15:149–161 [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute 2011. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; approved standard, 2nd ed. CLSI document M24-A2 Clinical and Laboratory Standards Institute, Wayne, PA: [PubMed] [Google Scholar]
- 5.Heifets LB, Lindholm-Levy P. 1989. Comparison of bactericidal activities of streptomycin, amikacin, kanamycin, and capreomycin against Mycobacterium avium and M. tuberculosis. Antimicrob. Agents Chemother. 33:1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangadharam PRJ, Perumal VK, Podapati NR, Kesavalu L, Iseman MD. 1988. In vivo activity of amikacin alone or in combination with clofazimine or rifabutin or both against acute experimental Mycobacterium avium complex infections in beige mice. Antimicrob. Agents Chemother. 32:1400–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis KK, Kao PN, Jacobs SS, Ruoss SJ. 2007. Aerosolized amikacin for treatment of pulmonary Mycobacterium avium infections: an observational case series. BMC Pulm. Med. 7:2. 10.1186/1471-2466-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iakhiaeva E, McNulty S, Brown-Elliott BA, Falkinham JO, III, Williams MD, Vasireddy R, Wilson RW, Turenne C, Wallace RJ., Jr 2013. Mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) genotyping of Mycobacterium intracellulare for strain comparison with establishment of a PCR database. J. Clin. Microbiol. 51:409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilton S, Cousins D. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1:269–273 [DOI] [PubMed] [Google Scholar]
- 10.Wallace RJ, Jr, Iakhiaeva E, Williams MD, Brown-Elliott BA, Vasireddy S, Vasireddy R, Lande L, Peterson DD, Sawicki J, Kwait R, Tichenor WS, Turenne C, Falkinham JO., III 2013. Absence of Mycobacterium intracellulare and the presence of Mycobacterium chimaera in household water and biofilm samples of patients in the United States with Mycobacterium avium complex respiratory disease. J. Clin. Microbiol. 51:1747–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagaki T, Nishimori K, Yagi T, Ichikawa K, Moriyama M, Nakagawa T, Shibayama T, Uchiya K, Nikai T, Ogawa K. 2009. Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J. Clin. Microbiol. 47:2156–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S21 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13.Prammananan T, Sander P, Brown BA, Frischkorn K, Onyi GO, Zhang Y, Böttger EC, Wallace RJ., Jr 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177:1573–1581 [DOI] [PubMed] [Google Scholar]
- 14.Nash KA, Brown-Elliott BA, Wallace RJ., Jr 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53:1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown-Elliott BA, Nash KA, Wallace RJ., Jr 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin. Microbiol. Rev. 25:545–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nessar N, Reyrat JM, Murray A, Gicquel B. 2011. Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J. Antimicrob. Chemother. 66:1719–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobashi Y, Matsushima T, Oka M. 2007. A double-blind randomized study of aminoglycosides infusion with combined therapy for pulmonary Mycobacterium avium complex disease. Respir. Med. 101:130–138 [DOI] [PubMed] [Google Scholar]
- 18.Kobashi Y, Yoshida K, Miyashita N, Oka M. 2006. Relationship between clinical efficacy of treatment of pulmonary Mycobacterium avium complex disease and drug-sensitivity testing of Mycobacterium avium complex isolates. J. Infect. Chemother. 12:195–202 [DOI] [PubMed] [Google Scholar]
- 19.Kobashi Y, Abe M, Mouri K, Obase Y, Kato Y, Oka M. 2012. Relationship between clinical efficacy for pulmonary MAC and drug-sensitivity test for isolated MAC in recent 6-year period. J. Infect. Chemother. 18:436–443 [DOI] [PubMed] [Google Scholar]