Abstract
To characterize Streptococcus pneumoniae “serotype 6E,” complete cps loci were sequenced. The capsular genes of “serotype 6E” isolates differed much from those of serotypes 6A and 6B. We identified 10 additional “serotype 6E” isolates, which are not confined to a restricted geographic locality. Most of these “serotype 6E” isolates belonged to sequence type 90 and its single-locus variants. The homogeneity of their genetic background and cps loci suggests a recent origin of the “serotype 6E” isolates.
TEXT
In a previous study, we identified a distinct group, group 6X, among serogroup 6 Streptococcus pneumoniae isolates (1). These group 6X strains were hypothesized to be similar to class 2 isolates in previous reports (2, 3) and capsular subtype 6B-III in a study by Elberse et al. (4). Elberse et al. (4) proposed that subtype 6B-III may represent a different serotype that cross-reacts with 6B-specific antiserum and predicted that it might come to be designated “serotype 6E” on the basis of serological and biochemical characterization. In the present study, we sequenced and compared the genes of the cps locus and the region around it in S. pneumoniae serotype 6A and 6B isolates, as well as isolates of “serotype 6E,” to identify the evolutionary scenario resulting in “serotype 6E.”
A total of 15 S. pneumoniae isolates were subjected to sequencing of genes of the cps locus, which is positioned between the dexB and aliA genes, and the regions upstream and downstream of it (Tables 1 and 2). Five isolates, HK02-14, J01-5, M12-6, K13-21, and V03-9, were identified as belonging to “serotype 6E” (1), six were identified as belonging to serotype 6A, and four were identified as belonging to serotype 6B (Table 1). The wzy gene, which encodes a polymerase that catalyzes the polymerization of oligosaccharide repeating units in capsular synthesis, was also sequenced in an additional 45 isolates (Table 1). The serotype was determined by using the capsular Quellung reaction with commercial antisera (Statens Serum Institut, Copenhagen, Denmark). A previously described serotype-specific PCR method was also used to identify serotypes 6C and 6D (4). Multilocus sequencing typing (MLST) of “serotype 6E” isolates was performed as described previously (5).
Table 1.
Strains investigated in this study
| Serotype and isolate | Locality | Yr | Specimen | ST in MLST (allelic profileb) | Sequencing |
|---|---|---|---|---|---|
| “6E” | |||||
| HK02-14 | Hong Kong | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | Whole cps locus |
| J01-5 | Japan | 2008 | Sputum | 1165 (5-6-1-8-6-3-4) | Whole cps locus |
| M12-6 | Malaysia | 2008 | Sputum | 95 (5-6-33-2-6-3-4) | Whole cps locus |
| K13-21 | South Korea | 2008 | Sputum | 1624 (5-6-1-2-6-3-1) | Whole cps locus |
| V03-9 | Vietnam | 2008 | Sputum | 1404 (7-25-4-16-6-20-28) | Whole cps locus |
| K08-92 | South Korea | 2009 | Sputum | 3418 (7-11-19-1-6-1-50) | wzy gene |
| K13-2 | South Korea | 2008 | Sputum | 4759 (5-4-1-2-6-3-4) | wzy gene |
| K13-47 | South Korea | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| K13-86 | South Korea | 2009 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| HK01-15 | Hong Kong | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| HK01-57 | Hong Kong | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| HK01-90 | Hong Kong | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| HK01-95 | Hong Kong | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| HK01-99 | Hong Kong | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| HK01-101 | Hong Kong | 2008 | Sputum | 90 (5-6-1-2-6-3-4) | wzy gene |
| 6A | |||||
| K13-75 | South Korea | 2009 | Sputum | Whole cps locus | |
| K10-25 | South Korea | 2000 | NAa | Whole cps locus | |
| Kor 146 | South Korea | 1998 | NA | Whole cps locus | |
| K13-59 | South Korea | 2008 | Sputum | Whole cps locus | |
| K08-52 | South Korea | 2008 | Sputum | Whole cps locus | |
| J01-16 | Japan | 2008 | Sputum | Whole cps locus | |
| K07-13 | South Korea | 2008 | Sputum | wzy gene | |
| K13-17 | South Korea | 2008 | Sputum | wzy gene | |
| K13-50 | South Korea | 2008 | Sputum | wzy gene | |
| K13-67 | South Korea | 2008 | Sputum | wzy gene | |
| K13-70 | South Korea | 2009 | Sputum | wzy gene | |
| K13-71 | South Korea | 2009 | Sputum | wzy gene | |
| K13-72 | South Korea | 2009 | Sputum | wzy gene | |
| K13-73 | South Korea | 2009 | Sputum | wzy gene | |
| K13-74 | South Korea | 2008 | Sputum | wzy gene | |
| K13-78 | South Korea | 2009 | Sputum | wzy gene | |
| K13-87 | South Korea | 2009 | Sputum | wzy gene | |
| K13-98 | South Korea | 2009 | Sputum | wzy gene | |
| K13-102 | South Korea | 2009 | Sputum | wzy gene | |
| K13-120 | South Korea | 2009 | Sputum | wzy gene | |
| K13-130 | South Korea | 2009 | Sputum | wzy gene | |
| K16-94 | South Korea | 2008 | Blood | wzy gene | |
| HK01-44 | Hong Kong | 2008 | Sputum | wzy gene | |
| HK02-77 | Hong Kong | 2008 | Sputum | wzy gene | |
| J01-2 | Japan | 2008 | Sputum | wzy gene | |
| J01-18 | Japan | 2008 | Sputum | wzy gene | |
| M13-25 | Malaysia | 2008 | Pleural fluid | wzy gene | |
| M13-38 | Malaysia | 2009 | CSFc | wzy gene | |
| 6B | |||||
| M12-8 | Malaysia | 2008 | Sputum | Whole cps locus | |
| J01-4 | Japan | 2008 | Sputum | Whole cps locus | |
| Tw03-308 | Taiwan | 2009 | Sputum | Whole cps locus | |
| K13-51 | South Korea | 2009 | Sputum | Whole cps locus | |
| HK01-2 | Hong Kong | 2008 | Sputum | wzy gene | |
| HK01-79 | Hong Kong | 2008 | Sputum | wzy gene | |
| HK01-31 | Hong Kong | 2008 | Sputum | wzy gene | |
| 6C | |||||
| K04-4 | South Korea | 2008 | Sputum | wzy gene | |
| M06-2 | Malaysia | 2008 | Sputum | wzy gene | |
| V07-123 | Vietnam | 2007 | Middle ear fluid | wzy gene | |
| 6D | |||||
| 07-056 | South Korea | 2007 | Sputum | wzy gene | |
| 07-077 | South Korea | 2007 | Sputum | wzy gene | |
| 07-107 | South Korea | 2007 | Transtracheal aspirate | wzy gene | |
| K13-22 | South Korea | 2008 | Sputum | wzy gene | |
| K13-108 | South Korea | 2009 | Sputum | wzy gene | |
| K13-109 | South Korea | 2009 | Sputum | wzy gene | |
| K13-110 | South Korea | 2009 | Sputum | wzy gene |
NA, not available.
aroE-gdh-gki-recP-spi-xpt-ddl.
CSF, cerebrospinal fluid.
Table 2.
Primers used in this study
| Gene | Primer | Sequence (5′–3′) | Reference |
|---|---|---|---|
| dexB | dexB-F | AGGACACGAGCAGCGTGGAAAT | This study |
| dexB-R | CTGCACTAAGGACACGCTTCTT | This study | |
| dexB-F2 | TCCATGGGATGCTTTCTGTGTGG | This study | |
| orf1-orf4 | ORF-F1 | CGTTTCCGACGAATTCGAGCTGT | This study |
| ORF-R1 | GCTTCCAAATTGGCTGGCGCA | This study | |
| ORF-F2 | GCCACCTCAGTCCGTAAGCGT | This study | |
| ORF-R2 | GCCGACGAACCTGAATCAGCCC | This study | |
| ORF-F3 | GCCTTCGTTCATATACAGTCCC | This study | |
| ORF-F4 | ACGACTCTCTCCCGCCAGTCT | This study | |
| ORF-F5 | CAGGAAGTTTTCAGCTGTCGG | This study | |
| wzg | 5122F | TTCGTCCATTCACACCTTAG | 8 |
| 3120R | CGAAGTGAAGTTCAATCGCAC | This study | |
| wzh | 5122F-1 | ATGTAGATGACGGTCCCA | This study |
| 5122F-2 | ATACGGAGAAGCGAAGGCT | This study | |
| wzd | 3121R | GATTGCGATTCACTACG | 8 |
| wze | 5113F | GGGAAAAATAAAAAATAGGTCGGG | 8 |
| wchA | 5106F | TACCATGCAGGGTGGAATGT | 8 |
| 3122R2 | CGTCCAAGCTAGTCTTCCGTAT | 1 | |
| wciN | 5101F | ATTTGGTGTACTTCCTCC | 8 |
| wciO | 5103F | AAACATGACATCAATTACA | 8 |
| 5103F2 | GGAACTTACTAGATGGAGTAG | 1 | |
| 3101R | CCATCCTTCGAGTATTGC | 3 | |
| wciP | 5123F | TGCCTATATCTGGGGGTGTA | 8 |
| wzy | 5140F | CCTAAAGTGGAGGGAATTTCG | 3 |
| 5140F-1 | GGTTACCATTGGATATGGAATAC | This study | |
| 3103R | AACCCCTAACAATATCAAAC | 8 | |
| 3143R | CCTCCCATATAACGAGTGATG | 3 | |
| wzx | 5141F | TTCGAATGGGAATTCAATGG | 3 |
| 3144R-2 | GATAGGCTGGCAACCAGAGGTC | This study | |
| rlmA | 3123R-2 | TGGATCCTTCACTTGGTAGCC | This study |
| 5124F-2 | GCTCCGTTTGATTGGAGAAGCA | This study | |
| rlmC | 3124R-1 | CCAGTAATCATTGACCAGATAGC | This study |
| 5125F | AGTGATTGATGCGAGTAAGG | 8 | |
| rlmB | 5126F | GAGTCAAGGCAACGATTTCCA | This study |
| 3124R-2 | GGTATGAATCCAGTCACGAACG | This study | |
| rlmD | 5127F | ACCATTGTGCAGCCTACACCG | This study |
| 5128F | GCGAACCGCTAGAAGCTTATCG | This study | |
| 3125R | TCTCACATGTGCCCAACATCC | This study | |
| aliA | 5128-F2 | GGAAACAGCTAAACGTCATCA | This study |
| 3126R-2 | GTCTTGAGCTTTGACTGCCGC | This study |
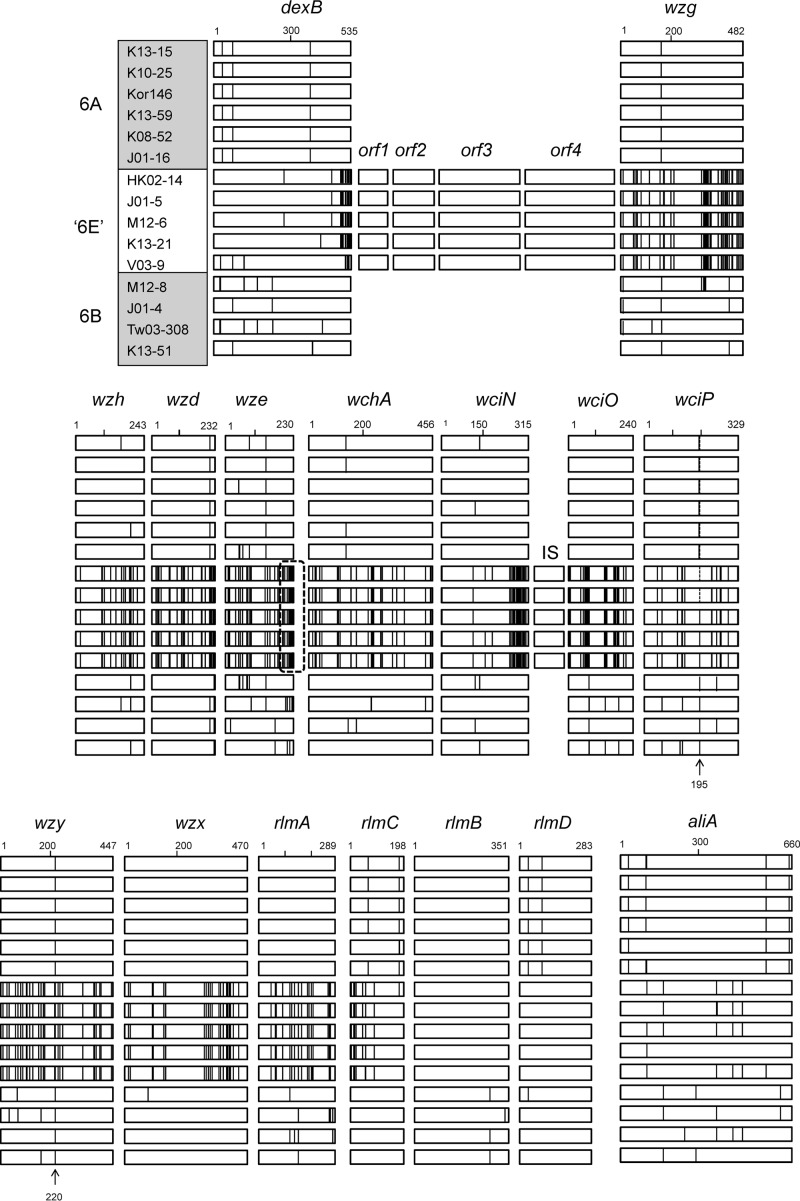
Figure 1 shows the polymorphism of genes of the cps locus and the regions upstream and downstream of it in 15 S. pneumoniae isolates. First of all, four open reading frames (orf1 to orf4) are present between the dexB and wzg genes of “serotype 6E” isolates. A BLAST search indicates that orf1, orf3, and orf4 encode transposases and that orf2 may be a conserved hypothetical protein gene. In addition, three amino acids, positions 220 to 222 of the wze gene, are deleted in “serotype 6E” isolates. As indicated previously (1, 3), an approximately 300-bp insertion was identified between the wciN and wciO genes in all “serotype 6E” isolates.
Fig 1.
Schematic overview of the cps loci and the genes upstream and downstream of it in serotype 6A, 6B, and “6E” isolates. Polymorphic amino acid sites are marked with vertical lines to indicate divergences from the consensus sequence. In wze genes, a dashed box indicates the deletion region of three amino acids in the “serotype 6E” isolates. Arrows indicate the sites of differentiation between serotypes 6A and 6B (wciP195) and among serotypes 6A, 6B, and “6E” (wzy220), respectively.
With respect to amino acid sequences, “serotype 6E” isolates diverge from serotype 6A and 6B isolates, particularly in the region from the 3′ region of dexB to the 5′ region of rlmC. However, amino acid sequences of genes of the cps locus among five “serotype 6E” isolates are very homogeneous, except at a few sites. Of these, amino acid 195 encoded by wciP has been regarded as a site differentiating serotype 6A from serotype 6B (3, 6). Amino acid 195 encoded by wciP of two of five “serotype 6E” pneumococcal isolates (HK02-14 and J01-5) is serine, corresponding to that of serotype 6A. However, the other isolates (M12-6, K13-21, and V03-9) have asparagine at this position, corresponding to that of serotype 6B.
In contrast to the homogeneity of genes in the cps locus, genes upstream and downstream of it showed amino acid sequence divergence among five “serotype 6E” isolates. Although serotype 6A pneumococcal isolates showed very homogeneous amino acid sequences in all genes, including those of the cps locus, serotype 6B isolates showed sequence divergences.
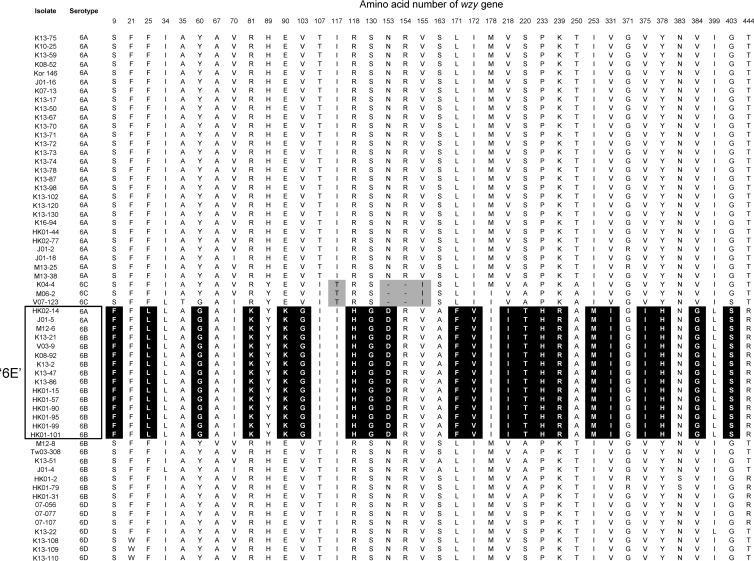
Comparison of the amino acid sequences encoded by the cps loci of 15 S. pneumoniae isolates showed that amino acid 220 of the wzy gene differentiates among serotype 6A, 6B, and “6E” isolates, with serine in serotype 6A, alanine in serotype 6B, and threonine in “serotype 6E.” Figure 2 shows the amino acid polymorphisms of the wzy gene among 60 S. pneumoniae isolates. Comparison of wzy gene sequences showed that an additional 10 pneumococcal isolates serotyped as 6B by the Quelling method showed the same amino acid sequences as the “serotype 6E” isolates. Thus, we tentatively regarded them as “serotype 6E.” They were confirmed to have an additional four genes (orf1 to orf4) by PCR.
Fig 2.
Amino acid polymorphisms of the wzy gene among 60 serogroup 6 isolates. The amino acids unique to “serotype 6E” isolates are indicated by a black background, and those unique to serotype 6C isolates are indicated by a gray background.
By MLST analysis, nine isolates from Hong Kong and South Korea were typed as belonging to sequence type 90 (ST90) and four isolates from Japan, Malaysia, and South Korea were typed as ST1165, ST95, ST1624, and ST4759, respectively, which are single-locus variants of ST90, among 15 “serotype 6E” isolates (Table 1). Thus, all of the “serotype 6E” isolates but two belonged to the same clonal complex, CC90. V03-9 from Vietnam and K08-92 from South Korea were distinct STs, ST1404 and ST3418.
In S. pneumoniae, evidence of vaccine escape through serotype switching has been provided by whole-genome sequencing of pneumococcal isolates (7). The sequence homogeneity of cps loci among the “serotype 6E” isolates, in contrast to the relative diversity in regions upstream and downstream of the cps locus, may suggest that horizontal transfer of the cps locus into the ancestor of “serotype 6E” isolates was a relatively recent event. However, it cannot be determined whether the emergence of “serotype 6E” was a response to pneumococcal vaccine because it is not known when the recombination event occurred. In addition, the origin of the cps locus of “serotype 6E” has not been identified.
We also discovered that the new serotype, “serotype 6E,” may not be rare. “Serotype 6E” isolates may not be restricted to a specific geographic locality. In our study, “serotype 6E” isolates from five Asian locales were identified. Pneumococcal strains with class 2 sequences in a study by Mavroidi et al. (3) from six countries were identified, and capsular subtype 6B-III was identified in isolates from The Netherlands (4). Thus, “serotype 6E” isolates may be prevalent worldwide.
In this study, we identified remarkable amino acid sequence divergence of the cps locus of “serotype 6E” isolates from those of serotype 6A and 6B isolates. We also identified an insertion of four genes, including transposase genes, upstream of the cps locus. Many serotype 6B isolates from South Korea and Hong Kong qualify as being of “serotype 6E,” which may suggest that the new serotype is widespread. Further study of the prevalence of serotype 6E and the effectiveness of vaccine against it is required.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers KC522422 to KC522601 and KC686719 to KC686825.
ACKNOWLEDGMENTS
The S. pneumoniae isolates used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID, Seoul, South Korea).
This study was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry of Health, Welfare, and Family Affairs, Seoul, Republic of Korea (A111251).
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Song JH, Baek JY, Ko KS. 2011. Comparison of capsular genes of Streptococcus pneumoniae serotype 6A, 6B, 6C, and 6D isolates. J. Clin. Microbiol. 49:1758–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bratcher PE, Park IH, Oliver MB, Hortal M, Camilli R, Hollingshead SK, Camou T, Nahm MH. 2011. Evolution of the capsular gene locus of Streptococcus pneumoniae serogroup 6. Microbiology 157:189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mavroidi A, Godoy D, Aanensen DM, Robinson DA, Hollingshead SK, Spratt BG. 2004. Evolutionary genetics of the capsular locus of serotype 6 pneumococci. J. Bacteriol. 186:8181–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elberse K, Witteveen S, van der Heide H, van de Pol I, Schot C, van der Ende Berbers G, Schouls L. 2011. Sequence diversity within the capsular genes of Streptococcus pneumoniae serogroup 6 and 19. PLoS One 6:e25018. 10.1371/journal.pone.0025018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3069 [DOI] [PubMed] [Google Scholar]
- 6.Ko KS, Baek JY, Song JH. 2012. Multidrug-resistant Streptococcus pneumoniae serotype 6D clones in South Korea. J. Clin. Microbiol. 50:818–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambersten LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park IH, Pritchard DG, Cartee R, Brandao A, Brandileone MC, Nahm MH. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]