Abstract
A total of 1,021 extended-spectrum-β-lactamase-producing Escherichia coli (ESBLEC) isolates obtained in 2006 during a Spanish national survey conducted in 44 hospitals were analyzed for the presence of the O25b:H4-B2-ST131 (sequence type 131) clonal group. Overall, 195 (19%) O25b-ST131 isolates were detected, with prevalence rates ranging from 0% to 52% per hospital. Molecular characterization of 130 representative O25b-ST131 isolates showed that 96 (74%) were positive for CTX-M-15, 15 (12%) for CTX-M-14, 9 (7%) for SHV-12, 6 (5%) for CTX-M-9, 5 (4%) for CTX-M-32, and 1 (0.7%) each for CTX-M-3 and the new ESBL enzyme CTX-M-103. The 130 O25b-ST131 isolates exhibited relatively high virulence scores (mean, 14.4 virulence genes). Although the virulence profiles of the O25b-ST131 isolates were fairly homogeneous, they could be classified into four main virotypes based on the presence or absence of four distinctive virulence genes: virotypes A (22%) (afa FM955459 positive, iroN negative, ibeA negative, sat positive or negative), B (31%) (afa FM955459 negative, iroN positive, ibeA negative, sat positive or negative), C (32%) (afa FM955459 negative, iroN negative, ibeA negative, sat positive), and D (13%) (afa FM955459 negative, iroN positive or negative, ibeA positive, sat positive or negative). The four virotypes were also identified in other countries, with virotype C being overrepresented internationally. Correspondingly, an analysis of XbaI macrorestriction profiles revealed four major clusters, which were largely virotype specific. Certain epidemiological and clinical features corresponded with the virotype. Statistically significant virotype-specific associations included, for virotype B, older age and a lower frequency of infection (versus colonization), for virotype C, a higher frequency of infection, and for virotype D, younger age and community-acquired infections. In isolates of the O25b:H4-B2-ST131 clonal group, these findings uniquely define four main virotypes, which are internationally distributed, correspond with pulsed-field gel electrophoresis (PFGE) profiles, and exhibit distinctive clinical-epidemiological associations.
INTRODUCTION
In recent years, extended-spectrum β-lactamase (ESBL) production in Enterobacteriaceae, particularly Escherichia coli, has significantly increased in many countries, including Spain (1). In 2000, the first nationwide study of ESBL-producing E. coli (ESBLEC) in Spain was developed (GEIH-BLEE-2000) (2). The overall prevalence of ESBL production was 0.5%, with CTX-M-9, SHV-12, and CTX-M-14 predominating. No CTX-M-15-producing E. coli isolates were detected. In contrast, in 2006, a similarly designed nationwide study (GEIH-BLEE-2006) showed that in just 6 years, the prevalence of ESBLEC in Spain had increased 8-fold, to 4% (3, 4). In the 2006 study, it was found that CTX-M-15 had joined CTX-M-14, SHV-12, and CTX-M-9 as a prevalent ESBL type. Thus, the predominant ESBL type had changed quickly, primarily due to the introduction and dissemination of a single clonal group characterized by CTX-M-15, serotype O25b:H4, phylogenetic group B2, and sequence type 131 (ST131), i.e., the international O25b:H4-B2-ST131 clonal group (5–9).
Unlike most other antimicrobial-resistant E. coli isolates, most of which derive from non-B2 phylogenetic groups (i.e., A, B1, and D) but are similar to other group B2 clonal groups, O25b:H4-B2-ST131 E. coli isolates typically exhibit multiple virulence factors, including adhesins, toxins, siderophores, and group 2 capsules. Thus, this clonal group combines both resistance and virulence genes, which in classical extraintestinal pathogenic E. coli (ExPEC) isolates have been mutually exclusive (6, 10–14).
In the report of the GEIH-BLEE-2006 study isolates (4), only the 37 CTX-M-15-positive isolates were screened for O25b-ST131 status, and all but five corresponded to this clonal group; the virulence genotypes of the O25b-ST131 isolates were not assessed. Here, we screened all 1,021 ESBLEC isolates from the GEIH-BLEE-2006 study for O25b-ST131 status and investigated a subset of the detected O25b-ST131 isolates for virulence genotypes and pulsed-field gel electrophoresis (PFGE) profiles. The main objective was to characterize the virulence profile diversity of the ESBL-producing E. coli human O25b-ST131 isolates from Spain, thereby identifying prominent virotypes. Secondarily, we screened for these virotypes among ST131 isolates from other countries and sought to find associations between individual virotypes and epidemiological and clinical features.
MATERIALS AND METHODS
Bacterial isolates.
Forty-four hospitals representing all Spanish regions participated in the GEIH-BLEE-2006 project. During the study period (1 February to 30 March 2006), 1,021 ESBL isolates were obtained from clinical samples (4). Species identification was performed with the API 20E system (bioMérieux, Marcy l'Etoile, France). ESBL production was confirmed by broth microdilution according to the CLSI guidelines (15).
For comparison, 52 international O25b:H4-B2-ST131 isolates (50 of human and 2 of avian origin), representing eight countries and three continents and taken from the reference collections of Nicolas-Chanoine et al. (6) and Johnson et al. (16), were analyzed to detect globally spread clonal variants in Spain. These isolates included the most prevalent ST131-associated XbaI pulsotypes (800, 812, 905, and 968) and others (699, 788, 797, 806, 903, 904, 909, 910, 911, 913, 915, 916, 917, 919, 979, 981, 1160, 1201, and 1202), as previously described by Johnson et al. (16).
Identification of O25b:H4-B2-ST131 isolates: serotyping, phylogenetic grouping, and multilocus sequence typing.
The 1,021 Spanish study isolates were screened using a triplex PCR that was previously developed for the presumptive identification of O25b-ST131 isolates (5). Presumptive O25b-ST131 isolates were confirmed as O25:H4 by serotyping, using type-specific O and H antisera. The major E. coli phylogenetic group was determined by triplex PCR (17). Multilocus sequencing typing (MLST) relied on a sequence analysis of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) according to the protocol and primers that are specified on the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli) (18).
Antibiotic susceptibility testing and molecular characterization of resistance mechanisms.
MICs were determined by the MicroScan WalkAway automated system (Siemens, Spain), used according to the manufacturer's instructions, and were interpreted as specified by the CLSI (15). ESBL production was screened for by using cephalosporin resistance and the double-disk synergy test. ESBL genotype was determined by PCR using published TEM-, SHV-, CTX-M-1-, and CTX-M-9-group-specific primers (19). The resistance genes blaOXA-1 and aac(6′)-Ib-cr were detected by PCR screening and bidirectional sequencing of amplicons (10). The genetic environment of blaCTX-M-15 was investigated by a specific PCR for upstream insertion elements (ISEcp1 and IS26) in isolates of clone O25b-ST131 (20).
Virulence genotyping.
The detection of 40 ExPEC-associated virulence genes was done by multiplex PCR (21, 22). The genes we sought to detect included fimH, fimAvMT78, F10 papA, papA, papC, papEF (positive isolates were tested for the papG I, papG II, and papG III alleles), sfa/focDE (positive isolates were tested for sfaS and focG), afa/draBC, afa operon FM955459, iha, bmaE, gafD, sat, cdtB, cnf1, hlyA, iucD, iutA, iroN, fyuA, chuA, kpsM II (with its neuC-K1, K2, and K5 variants), kpsMT III, cvaC, iss, traT, ibeA, malX, usp, tsh, and ompT (Table 1). Isolates were classified as ExPEC if they carried ≥2 of papEF (P fimbriae), sfa/focDE (S/F1C fimbriae), afa/draBC (Afa/Dr adhesins), iutA (aerobactin receptor), and kpsM II (group 2 capsule synthesis) (23). The virulence score was the number of virulence genes that were detected in an isolate. PCR screening of the fimB insertion sequence (ISL3-like transposase) was performed as described elsewhere (24). Investigation of the fim operon sequence in the E. coli EC958 ST131 strain revealed a 1,895-bp insertion in the fimB gene, which encodes the FimB recombinase that switches on the expression of type 1 fimbriae (24).
Table 1.
Comparison of virulence gene prevalences of 130 representatives of O25b:H4-B2-ST131 ESBLEC isolates in relation to their virotypes
| Virulence gene | Isolates in virotype:a |
O25b-ST131 isolates | Pb | |||
|---|---|---|---|---|---|---|
| A | B | C | D | |||
| Total no. | 29 | 40 | 41 | 17 | 130 | |
| Adhesins (no. [%]) | ||||||
| fimH | 29 (100) | 40 (100) | 40 (98) | 17 (100) | 129 (99) | |
| ISL3-like in fimB | 29 (100) | 40 (100) | 41 (100) | 0 | 110 (85) | <0.001 |
| fimAvMT78 | 0 | 0 | 0 | 1 (6) | 1 (1) | |
| F10 papA | 27 (93) | 31 (78) | 41 (100) | 0 | 100 (77) | <0.001 |
| papA | 0 | 0 | 0 | 12 (71) | 12 (9) | <0.001 |
| papC | 0 | 0 | 2 (5) | 9 (53) | 11 (8) | <0.001 |
| papEF | 0 | 0 | 0 | 12 (71) | 12 (9) | <0.001 |
| papG I | 0 | 0 | 0 | 0 | 0 | |
| papG II | 0 | 0 | 0 | 0 | 0 | |
| papG III | 0 | 0 | 0 | 12 (71) | 12 (9) | <0.001 |
| sfa/focDE | 0 | 0 | 0 | 0 | 0 | |
| sfaS | 0 | 0 | 0 | 0 | 0 | |
| focG | 0 | 0 | 0 | 0 | 0 | |
| afa/draBC | 29 (100) | 0 | 1 (2) | 0 | 30 (23) | <0.001 |
| afa FM955459 | 29 (100) | 0 | 0 | 0 | 29 (22) | <0.001 |
| iha | 27 (93) | 29 (73) | 41 (100) | 0 | 98 (75) | <0.001 |
| bmaE | 0 | 0 | 0 | 0 | 0 | |
| gafD | 0 | 0 | 0 | 0 | 0 | |
| Toxins (no. [%]) | ||||||
| sat | 28 (97) | 30 (75) | 41 (100) | 0 | 99 (76) | <0.001 |
| cdtB | 0 | 0 | 0 | 5 (29) | 5 (4) | <0.001 |
| cnf1 | 0 | 0 | 0 | 10 (59) | 10 (8) | <0.001 |
| hlyA | 0 | 0 | 0 | 10 (59) | 10 (8) | <0.001 |
| Siderophores (no. [%]) | ||||||
| iucD | 27 (93) | 39 (98) | 41 (100) | 13 (76) | 121 (93) | |
| iutA | 28 (97) | 40 (100) | 41 (100) | 13 (76) | 122 (94) | |
| iroN | 0 | 40 (100) | 0 | 13 (76) | 53 (41) | <0.001 |
| fyuA | 29 (100) | 40 (100) | 41 (100) | 17 (100) | 130 (100) | |
| chuA | 29 (100) | 40 (100) | 41 (100) | 17 (100) | 130 (100) | |
| Capsula (no. [%]) | ||||||
| kpsM II | 29 (100) | 31 (78) | 28 (68) | 17 (100) | 106 (82) | |
| kpsM II-K2 | 29 (100) | 0 | 0 | 0 | 29 (22) | <0.001 |
| kpsM II-K5 | 0 | 31 (78) | 28 (68) | 15 (88) | 75 (58) | <0.001 |
| neuC-K1 | 0 | 0 | 0 | 2 (12) | 2 (2) | |
| kpsM III | 0 | 0 | 0 | 0 | 0 | |
| Miscellaneous (no. [%]) | ||||||
| cvaC | 0 | 1 (3) | 0 | 9 (53) | 10 (1) | <0.001 |
| iss | 0 | 39 (98) | 0 | 16 (94) | 55 (42) | <0.001 |
| traT | 21 (72) | 21 (53) | 36 (88) | 16 (94) | 96 (74) | |
| ibeA | 0 | 0 | 0 | 17 (100) | 17 (13) | <0.001 |
| malX (PAI) | 29 (100) | 40 (100) | 41 (100) | 17 (100) | 130 (100) | |
| usp | 29 (100) | 40 (100) | 41 (100) | 17 (100) | 130 (100) | |
| tsh | 0 | 0 | 0 | 0 | 0 | |
| ompT | 29 (100) | 40 (100) | 41 (100) | 17 (100) | 130 (100) | |
| ExPEC status (no. [%]) | 29 (100) | 31 (78) | 28 (68) | 17 (100) | 106 (82) | |
| Range of virulence genes (no.) | 10–15 | 11–17 | 12–14 | 9–20 | 9–20 | NA |
| Mean no. of virulence genes | 14.4 | 14.5 | 13.6 | 16.8 | 14.4 | NA |
Significant differences are indicated in bold.
P values (by Fisher's exact test) are shown where there was a P value of <0.05. NA, not applicable.
fimH subtyping.
The fimHTR allele was identified based on sequence variation in the E. coli type-1 fimbrial adhesion gene (positions 64 to 552). Amplification and sequencing were performed as previously described (25).
Pulsed-field gel electrophoresis.
XbaI PFGE analysis was performed as previously described (21). Profiles were analyzed with the BioNumerics fingerprinting software (Applied Maths, St-Martens-Latem, Belgium). Cluster analysis of the Dice similarity indices based on the unweighted-pair group method using average linkages (UPGMA) was done to generate a dendrogram describing the relationship among the PFGE profiles.
Epidemiological and clinical features.
Epidemiological and clinical features were collected by using a structured questionnaire based on the following data: age, sex, health care environment, underlying conditions, invasive procedures performed during the preceding week, antimicrobial use during the preceding month, whether the isolate represented colonization or infection (and, if infection, the type of infection), and outcome. Isolates were classified as nosocomially acquired (NA) if obtained 48 h after hospital admission, as health care-associated (HCA) if the patient had been admitted to an acute or long-term care center or had received hemodialysis, specialized home care, or day hospital care during the preceding 3 months, and as community-acquired (CA) if none of these applied. The ethics committee of each participating center approved the study.
Statistical analysis.
Comparisons of proportions and scores were tested using Fisher's exact test, chi-square test, and the Mann-Whitney U test, as appropriate. The criterion for statistical significance was set at a P value of <0.05.
Nucleotide sequence accession number.
The sequence for the CTX-M-103 gene was deposited in EMBL under accession number HG423149.
RESULTS
Prevalence and distribution of the O25b:H4-B2-ST131 clonal group.
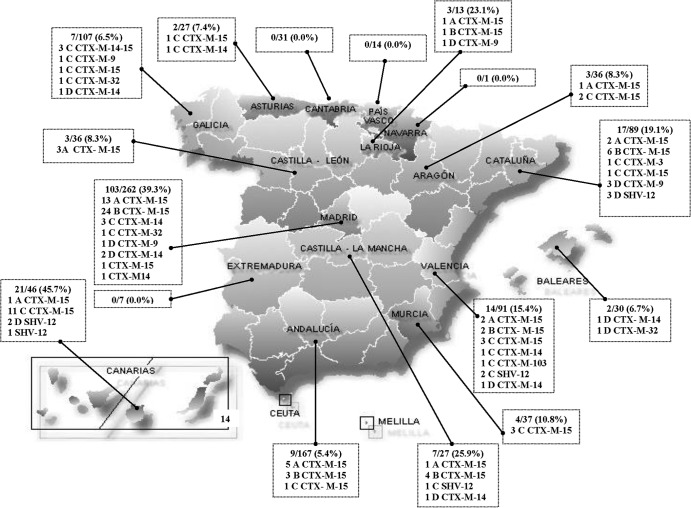
According to PCR-based detection, the E. coli O25b-ST131 clonal group accounted for 195 (19%) of the 1,021 ESBLEC isolates from the Spanish GEIH-BLEE-2006 project. The O25b-ST131 isolates were widely distributed across Spain and were recovered from 30 of the 44 participating hospitals, including in 13 of the 17 autonomous communities. By hospital, the prevalence of ST131 among the local ESBL study isolates ranged from 0% to 52% (see Table S1 in the supplemental material), with the highest values observed in Madrid and the Canary Islands (Fig. 1).
Fig 1.
Distribution of O25b-ST131 isolates in Spain. The O25b-ST131 isolates were widely distributed across Spain, being recovered from 30 of the 44 participant hospitals, including 13 of the 17 autonomous communities of Spain.
ESBL enzymes produced by O25b:H4-B2-ST131 isolates.
From the 195 total O25b-ST131 isolates, 130 (a maximum of 10 per hospital) were selected randomly for further characterization. Of these, 96 (74%) were positive for CTX-M-15, 15 (12%) for CTX-M-14, 9 (7%) for SHV-12, 6 (5%) for CTX-M-9, and 5 (4%) for CTX-M-32, with the remaining two isolates being positive, respectively, for CTX-M-3 and a new ESBL enzyme, CTX-M-103, which was first detected in this study. CTX-M-103 (EMBL accession no. HG423149) has a single amino acid change compared to CTX-M-15 and CTX-M-3, and it may be grouped in cluster CTX-M-1 (26). Three O25b-ST131 isolates from three different hospitals in the Galicia autonomous community exhibited both CTX-M-14 and CTX-M-15 (Fig. 1; see also Table S1 in the supplemental material).
Virulence genes and virotypes of O25b:H4-B2-ST131 isolates.
Of the 40 studied ExPEC-associated virulence genes, 13 were detected by PCR in a majority of the 130 selected O25b-ST131 isolates (Table 1). These 13 genes included fimH (in 99%), F10 papA (77%), iha (75%), sat (76%), iucD (93%), iutA (94%), fyuA (100%), chuA (100%), kpsM II (82%), traT (74%), malX (100%), usp (100%), and ompT (100%). In contrast, nine virulence genes were not detected in any O25b-ST131 isolate; these included papG I, papG II, sfa/focDE, sfaS, focG, bmaE, gafD, kpsM III, and tsh. Overall, the 130 O25b-ST131 isolates exhibited relatively high aggregate virulence scores (mean, 14.4; range, 9 to 20), and 106 (82%) satisfied the molecular criteria for ExPEC.
Although the virulence profiles were fairly homogeneous overall, four discrete virotypes (labeled arbitrarily as A, B, C, and D) were resolved based on the presence or absence of four distinctive virulence genes, including afa FM955459 (specific for an ST131 clone encoding an Afa/Dr adhesin), iroN (catecholate siderophore receptor), ibeA (invasion of brain endothelium), and sat (secreted autotransporter toxin). The patterns were as follows: virotype A (afa FM955459 positive, iroN negative, ibeA negative, sat positive or negative), virotype B (afa FM955459 negative, iroN positive, ibeA negative, sat positive or negative), virotype C (afa FM955459 negative, iroN negative, ibeA negative, sat positive), and virotype D (afa FM955459 negative, iroN positive or negative, ibeA positive, sat positive or negative). The 130 isolates were distributed among the four virotypes as follows: A, 29 (22%), B, 40 (31%), C, 41 (32%), and D, 17 (13%), with three isolates remaining unclassified as to virotype since they lacked all four virotype-defining genes.
Interestingly, the 17 virotype D isolates exhibited significantly higher virulence scores than did those of other virotypes (mean, 16.8 for D versus 14.4 for A, 14.5 for B, and 13.6 for C) (Table 1).
All four virotypes identified for the Spanish O25b:H4-B2-ST131 isolates were also represented in 51 of the 52 studied international O25b:H4-B2-ST131 isolates. Virotype C was by far the most prevalent (65%), followed by virotypes A (17%), B (10%), and D (6%).
Insertion of ISL3-like transposase gene in fimB in relation to virotype.
The 130 selected Spanish O25b-ST131 isolates were also analyzed for an insertion in fimB, similar to the ISL3 transposase gene observed in E. coli strain Nissle 1917 (GenBank accession no. AF188737). The fimB insertion was detected in all 113 isolates of virotypes A, B, and C but in none of the 17 virotype D isolates (P < 0.001 for all comparisons versus virotype D) (Table 1).
fimH alleles and O25b:H4-B2-ST131 virotypes.
fimH sequence analysis was done for 40 of the Spanish O25b-ST131 isolates, including 10 each for the four virotypes, which were selected to include a representation of all virulence gene profiles and PFGE groups encountered within a particular virotype. Two different fimH alleles were identified among these 40 isolates. All representatives of virotypes A, B, and C contained fimH30, whereas all representatives of virotype D contained fimH22 (P < 0.001 for all comparisons versus virotype D).
Virotype versus ESBL variants, CTX-M-15-associated genes, and antimicrobial resistance.
Among the 130 selected O25b:H4-B2-ST131 isolates, all 69 isolates from virotypes A and B and 63% (26/41) of the virotype C isolates produced CTX-M-15; however, no virotype D isolates produced CTX-M-15 (P < 0.001 for all comparisons versus virotype D). In contrast, virotype D isolates produced CTX-M-9 (5 isolates), CTX-M-14 (6 isolates), SHV-12 (5 isolates), and CTX-M-32 (1 isolate) (Table 2).
Table 2.
ESBL enzymes, genes associated with CTX-M-15 plasmids, and antimicrobial resistance of 130 representatives of O25b:H4-B2-ST131 ESBLEC isolates in relation to their virotypes
| Characteristic | No. (%) of isolates by virotypea: |
No. (%) of O25b-ST131 isolates | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Total no. | 29 | 40 | 41 | 17 | 130 |
| ESBL enzymes | |||||
| CTX-M-3 | 0 | 0 | 1 (2) | 0 | 1 (0.8) |
| CTX-M-9 | 0 | 0 | 1 (2) | 5 (29) | 6 (5) |
| CTX-M-14 | 0 | 0 | 8 (20) | 6 (35) | 15 (12) |
| CTX-M-15 | 29 (100) | 40 (100) | 26 (63) | 0 | 96 (74) |
| CTX-M-32 | 0 | 0 | 4 (10) | 1 (6) | 5 (4) |
| CTX-M-103 | 0 | 0 | 1 (2) | 0 | 1(0.8) |
| SHV-12 | 0 | 0 | 3 (7) | 5 (29) | 9 (7) |
| Genes associated with CTX-M-15 | |||||
| blaOXA-1 | 20 (69) | 35 (88) | 21 (51) | 0 | 85 (65) |
| aac(6′)-Ib-cr | 29 (100) | 35 (88) | 21 (51) | 0 | 85 (65) |
| IS26 element | 29 (100) | 0 | 0 | 0 | 30 (23) |
| Antimicrobial resistance | |||||
| Ciprofloxacin | 29 (100) | 40 (100) | 40 (98) | 5 (29) | 117 (90) |
| Gentamicin | 0 | 27 (68) | 16 (39) | 1 (6) | 46(35) |
| Tobramycin | 29 (100) | 33 (83) | 28 (68) | 3 (18) | 95 (73) |
| Trimethoprim-sulfamethoxazole | 29 (100) | 31 (78) | 20 (50) | 10 (59) | 92 (71) |
| Amoxicillin-clavulanic acid | 10 (34) | 6 (15) | 9 (22) | 0 | 26 (20) |
| Fosfomycin | 5 (17) | 3 (8) | 2 (5) | 0 | 11 (9) |
Significant differences are indicated in bold. P values are shown in Results.
As expected, the blaOXA-1 and aac(6′)-Ib-cr genes were associated with CTX-M-15-producing isolates. These genes were detected in 69% and 100% of virotype A isolates, respectively, 88% (both genes) of virotype B isolates, and 51% (both genes) of virotype C isolates but in 0% of virotype D isolates (P < 0.001 for all comparisons versus virotype D) (Table 2).
Only the 29 virotype A isolates showed an IS26 element within the terminal inverted repeat of the ISEcp1-like element upstream of blaCTX-M-15, separating the blaCTX-M-15 allele from its usual promoter, as found in the epidemic ST131 “strain A” that is prevalent in the United Kingdom (5, 20) (P < 0.001 for comparisons with virotypes B, C, and D) (Table 2).
Different patterns of associated antimicrobial resistances were detected in relation to virotype. Specifically, ciprofloxacin and tobramycin resistance were significantly associated with virotypes A, B, and C (P < 0.001 for all comparisons versus virotype D). In contrast, virotypes B and C were significantly associated with gentamicin resistance (P < 0.05 for all comparisons versus virotypes A and D), and virotype A was significantly associated with trimethoprim-sulfamethoxazole resistance (P < 0.05 for all comparisons versus virotypes B, C, and D) (Table 2).
PFGE profiles of O25b:H4-B2-ST131 isolates in relation to virotype.
In the PFGE-based dendrogram, 116 of the 130 Spanish O25b-ST131 isolates were distributed among 4 large virotype-specific clusters, defined at similarity levels of approximately 72% (11 virotype D isolates), 76% (28 virotype A isolates), 77% (33 virotype C isolates), and 74% (39 virotype B isolates) (see Fig. S1 in the supplemental material).
A similar clustering of PFGE profiles by virotype was detected when the 130 Spanish study isolates were compared with the 52 international isolates (see Fig. S2 in the supplemental material). Four main clusters, each largely virotype specific, comprised 162 of the 182 total isolates. Each of these clusters, defined at similarity levels of approximately 80% (47 isolates, 40 of virotype B), 78% (60 isolates, 58 of virotype C), 82% (37 isolates, 36 of virotype A), and 67% (18 isolates, 16 of virotype D), included isolates from multiple countries.
In the present study, among 13 international isolates representing the top four pulsotypes described by Johnson et al. (16), we found that virotype A corresponded with pulsotype 812, virotype B with pulsotype 905, and virotype C with all four pulsotypes (968, 800, 905, and 812), whereas none of the virotype D isolates belonged to these top four pulsotypes.
Epidemiological and clinical associations of ESBLEC O25b-ST131.
Associated clinical data were compared for O25b-ST131 and non-O25b-ST131 Spanish ESBLEC isolates (Table 3). In univariate analyses, compared with the non-O25b-ST131 isolates, the O25b-ST131 isolates were significantly associated with older age, nursing home residents, a health care-related origin, asymptomatic bacteriuria, and bacteremia. However, after adjustment for acquisition type and age by logistic regression analysis, the association of O25b-ST131 isolates with bacteremia was not statistically significant (odds ratio [OR], 1.58; 95% confidence interval [CI], 0.91 to 2.75) (P = 0.10).
Table 3.
Epidemiological and clinical data of patients with colonization or infection due to ESBLEC O25b:H4-B2-ST131 compared to those with non-ST131 infection
| Patient variable | Value for isolatea: |
Pe | |
|---|---|---|---|
| ST131 | Non-ST131 | ||
| Total no. | 190 | 818 | |
| Male gender (no. [%]) | 70 (37) | 328 (40) | |
| Median age (IQR) (yr) | 75 (68–84) | 69 (50–79) | 0.01b |
| No. (%) of pediatric patients (age ≤ 14 yr) | 4 (2) | 53 (6) | 0.01c |
| Acquisition type (no. [%]) | |||
| Nosocomial | 66 (35) | 238 (29) | |
| Health care related | 47 (25) | 115 (14) | <0.001 |
| Community | 77 (41) | 465 (57) | <0.001 |
| Nursing home resident | 37 (19) | 39 (5) | <0.001 |
| Infection site (no. [%])d | 136 (72) | 651 (80) | 0.02 |
| Urinary tract | 102 (75) | 485 (75) | |
| Soft tissue | 16 (12) | 71 (11) | |
| Digestive tract | 6 (4) | 37 (6) | |
| Other types | 20 (15) | 116 (18) | |
| Bacteremia (primary or secondary) | 22 (16) | 56 (9) | 0.01 |
| Crude 30-day mortality (no. [%])d | 5 (4) | 12 (2) | |
Data from 5 patients with ST131 and from 8 patients with non-ST131 isolates were unavailable. P values were calculated by chi-square test except where otherwise noted.
Calculated by Mann-Whitney U test.
Calculated by Fisher's exact test.
Only patients with infection were considered.
P values are shown where P < 0.05.
Associations of the four virotypes with demographic data, acquisition type, and type of infection also were explored (Table 4). Virotype B was significantly associated with older patients and a lower likelihood of symptomatic infection, specifically urinary tract infection, but a higher likelihood of respiratory tract infection. In contrast, virotype C was significantly associated with a higher likelihood of symptomatic infection, whereas virotype D was significantly associated with younger patients and community acquisition. Additionally, virotypes A and B were more highly associated with nursing home residents (17/69 [25%]) than were virotypes C and D (4/61 [7%]) (P = 0.005).
Table 4.
Association of virotype with epidemiological and clinical data among 130 O25b:H4-B2-ST131 ESBLEC isolates
| Patient variable | Isolates by virotypea: |
|||
|---|---|---|---|---|
| A | B | C | D | |
| Total no. | 29 | 40 | 41 | 17 |
| Male gender (no. [%]) | 10 (35) | 16 (40) | 22 (54) | 7 (41) |
| Age ≤ 14 yr (no. [%]) | 1 (3) | 0 | 1 (2) | 2 (12) |
| Median age (IQR) (yr) | 77 (63–82) | 75 (64–83)a | 64 (56–79) | 60 (34–78)b |
| Acquisition type (no. [%]) | ||||
| Nosocomial | 9 (31) | 18 (45) | 18 (44) | 4 (24) |
| Health care related | 9 (31) | 9 (23) | 7 (17) | 1 (6) |
| Community | 11 (38) | 13 (32) | 16 (39) | 12 (71)c |
| Nursing home resident | 7 (24) | 10 (25) | 3 (7) | 1 (6) |
| Infection type | 22 (85) | 21 (57)d | 36 (92)e | 13 (77) |
| Urinary tract | 20 (69) | 13 (33)f | 25 (61) | 13 (77) |
| Respiratory tract | 0 | 4 (10)g | 1 (2) | 0 |
| Digestive tract | 0 | 0 | 2 (5) | 0 |
| Primary bacteremia | 0 | 2 (5) | 3 (7) | 0 |
| Other types | 2 (7) | 2 (5) | 5 (12) | 0 |
| Bacteremia (primary or secondary) | 5 (17) | 8 (20) | 8 (15) | 0 |
| Crude 30-day mortality (no. [%]) | 0 | 6 (15) | 4 (10) | 1 (6) |
| Infection-related deaths (no. [%]) | 0 | 3 (8) | 1 (2) | 0 |
P = 0.05 (calculated by Mann-Whitney U test). All comparisons other than those indicated by a footnote have a P value of >0.05.
P = 0.03 (calculated by Mann-Whitney U test).
P = 0.03 (calculated by Fisher's exact test).
P = 0.009.
P = 0.01.
P = 0.0001.
P = 0.03 (calculated by Fisher's exact test).
Triplex PCR for virotyping.
A triplex PCR based on the detection of sat, iroN, and ibeA was designed for virotyping of the O25b:H4-B2-ST131 isolates. When applied to the present 130 Spanish and 52 international O25b-ST131 isolates, this assay exhibited 100% specificity and sensitivity (not shown). This assay can be combined with a previously described triplex PCR (based on detection of the afa operon FM955459, the O25b rfb allele, and the 3′ end of blaCTX-M-15) to establish the virotypes of the O25b-ST131 isolates (Table 5; see also Fig. S3 in the supplemental material).
Table 5.
Primers included in the two triplex PCR assays used for specific identification of clonal group O25b:H4-B2-ST131 and virotyping
| Gene | Primer | Oligonucleotide sequence (5′ to 3′) | Fragment size (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| afa FM955459a | AFA-O25F | GAGTCACGGCAGTCGCGGCGG | 207 | 55 | 5 |
| AFA-O25R | TTCACCGGCGACCAGCCATCTCC | ||||
| rfbO25ba | rfb.1bis | ATACCGACGACGCCGATCTG | 300 | 55 | 45 |
| rfbO25b.r | TGCTATTCATTATGCGCAGC | ||||
| blaCTX-M-15a | CTX-M-F1 | ATAAAACCGGCAGCGGTG | 483 | 55 | 19 |
| CTX-M-F2 | GAATTTTGACGATCGGGG | ||||
| iroNb | IRON-F | AAGTCAAAGCAGGGGTTGCCCG | 667 | 60 | 46 |
| IRON-R | GACGCCGACATTAAGACGCAG | ||||
| satb | SAT-F | ACTGGCGGACTCATGCTGT | 387 | 60 | 47 |
| SAT-R | AACCCTGTAAGAAGACTGAGC | ||||
| ibeAb | IBEA 10F | AGGCAGGTGTGCGCCGCGTAC | 170 | 60 | 22 |
| IBEA 10R | TGGTGCTCCGGCAAACCATGC |
Triplex PCR used for specific identification of O25b-ST131 isolates (5).
Triplex PCR used for virotyping of O25b-ST131 isolates (developed in this study).
DISCUSSION
The prevalence and epidemiology of ESBLEC are changing rapidly. In recent years, ESBL production in E. coli has increased significantly, due primarily to the spread of CTX-M types. In fact, the prevalence of ESBLEC strains in Spain increased 8-fold between 2000 and 2006, from 0.5% to 4% (4). The emergence and dissemination of ESBLEC have two possible explanations, that it occurs through dissemination of mobile genetic elements between non-clonally related strains or through clonal spread. The two mechanisms might occur simultaneously, thereby contributing to the rapid dissemination of ESBLEC strains. Until a few years ago, most ESBLEC strains were clonally unrelated; however, recently, the O25b:H4-B2-ST131 intercontinental E. coli clonal group that produces CTX-M-15 with high virulence potential has been reported worldwide, representing a major public health problem (6, 10).
In the present study, 195 (19%) of the 1,021 total ESBLEC study isolates were of the O25b-ST131 group. The O25b-ST131 isolates were widely distributed across Spain, as they were at 30 of the 44 participating centers and accounted for up to 52% of ESBLEC strains per hospital. This contrasts with the 9% prevalence of ST131 among the 92 analyzed ESBLEC isolates from a similar study from 2004 done in 11 Spanish hospitals (7 of which were included in the present study) (8). More recent studies found even higher prevalences of O25b-ST131 among ESBLECs in various Spanish cities, including Lugo in 2008 (23%) (5) and in 2012 (61%) (27), Barcelona (32% in 2008) (28), Seville (13% in 2010) (29), and Madrid (21% in 2008) (30). Other countries have also had high reported prevalences of ST131 among the ESBLEC strains, including Denmark (38%) (31), Japan (41%) (32), the United States (47%) (33), and Canada (78%) (34).
The Spanish O25b-ST131 isolates in this study carried not only CTX-M-15 but also CTX-M-14, SHV-12, CTX-M-9, CTX-M-32, CTX-M-3, and the new ESBL enzyme CTX-M-103 first described here. Although in most countries, ST131 is associated mainly with CTX-M-15 (6, 33–35), exceptions do occur, such as in Japan, where O25b-ST131 is frequently associated with CTX-M-14 (36), or in Ireland, where it is associated with CTX-M-3 (37). In the present study, we showed that association between E. coli O25b-ST131 and CTX-M types other than CTX-M-15 was mainly identified in the isolates with virotype D.
ExPEC isolates have specialized virulence factors enabling them to colonize host surfaces, injure host tissues, and avoid host defense systems. Thirteen genes (fimH, F10 papA, iha, sat, iucD, iutA, fyuA, chuA, kpsM II, traT, malX, usp, and ompT) were detected in most of the 130 Spanish O25b-ST131 ESBLEC isolates in this study. In two recent studies, Coelho et al. (28) in Spain and Johnson et al. (38) in the United States found that these virulence genes were significantly more prevalent among ST131 than non-ST131 isolates. Therefore, they might be important in the worldwide dissemination of E. coli O25b-ST131. Moreover, the present 130 Spanish O25b-ST131 isolates exhibited high virulence scores, and most qualified molecularly as being ExPEC (23).
After analyzing all the virulence gene profiles together with the PFGE pulsotypes, we observed that the virotypes we established corresponded well with the PFGE clusters. Although ST131 isolates share a large set of putative virulence factors, we selected the four virulence genes (afa FM955459, iroN, ibeA, and sat) that clearly define the four virotypes within the clonal group O25b-ST131. Notably, the four virotypes, most prominently virotype C, were also present in other countries (France, Portugal, Switzerland, United States, Canada, Korea, and Lebanon). The XbaI PFGE dendrogram revealed four major clusters that corresponded closely with the virotypes, suggesting a clonal basis for the virotypes. Isolates of virotype D exhibited significantly higher virulence scores, which might explain the association of this virotype with younger patients and community-acquired infections.
Like Banerjee et al. (39), we found that patients with O25b-ST131 isolates were older and more frequently had health care-associated acquisition (particularly through nursing home residency) than those with non-O25b-ST131 ESBLEC isolates. Although the O25b-ST131 isolates were associated with bacteremia in univariate analyses, this association was not significant when age and acquisition type were considered in multivariate analysis. Chung et al. (40) found that patients with clone ST131 were more likely to have secondary bacteremia than those with non-ST131 isolates. As has been found in other studies (39–41), we did not find that O25b-ST131 isolates were associated with increased mortality. Further studies are needed to more fully assess the clinical implications of the comparatively high virulence scores and the group B2 background of the O25b-ST131 ESBLEC isolates. Of note, virotype D was linked with community-acquired infections and virotypes A and B with nursing home residents.
In a recent PFGE analysis of 579 ST131 isolates from diverse years, hosts, and locales, Johnson et al. (16) found that the four most prevalent pulsotypes (among 170 total) accounted for 46% of the population and tended to occur in more recent years, which is consistent with recent emergence and expansion and implies greater fitness of these pulsotypes. In the present study, among 13 international isolates representing the top four pulsotypes of Johnson et al. (16), we found virotypes A, B, and C.
Seven distinct fimH-based putative clonal lineages (H15, H22, H27, H30, H35, H41, and H49) were found among 352 historical and recent ST131 isolates obtained primarily from the United States (42). The H22 subclone was dominant (73%) among the historical isolates (those from 1967 to 1999), whereas the H30 subclone was the most prevalent (75%) among the current clinical isolates (those from 2000 to 2011) and accounted for nearly all fluoroquinolone-resistant isolates. Interestingly, in the present study, the H30 subclone accounted for all analyzed Spanish O25b-ST131 isolates of virotypes A, B, and C, whereas the H22 subclone accounted for all analyzed virotype D isolates. Consistent with the association of the H30 ST131 subclone with fluoroquinolone resistance, we found the prevalence of ciprofloxacin resistance to be significantly higher among the (H30-derived) virotype A, B, and C isolates (100%, 100%, and 98%, respectively) than among the (non-H30) virotype D isolates (29%) (P < 0.001).
Diverse explanations have been proposed for the rapid and successful dissemination of E. coli O25b-ST131 among humans, including possible transmission through animal contact and food consumption (43). Mora et al. (44) reported an association of E. coli O25b-ST131 with clinical disease in poultry and identified this clonal group in retail chicken meat in Spain. In that study, the 19 avian O25b-ST131 isolates (of which 7 produced CTX-M-9) were of virotype D, defined according to the criteria specified here. However, in the present study, only 2 of the 17 Spanish human isolates of virotype D (isolates 16.30 and 16.31; fimH, iucD, iroN, kpsMII-K1, iss, traT, ibeA, malX, usp, and SHV-12) and the 2 avian virotype D isolates from the United States (isolates CD285 and CD287; fimH, iucD, iroN, kpsMII-K1, iss, traT, ibeA, malX, usp, and tsh) showed virulence gene profiles that were highly similar to those of the Spanish avian isolates (44).
In conclusion, we found a high prevalence of E. coli O25b:H4-B2-ST131 among (mostly CTX-M-15-producing) ESBLEC isolates from hospitals across Spain in 2006. We newly defined four main virulence gene profile variants within ST131, which we termed virotypes A to D, and showed these to be prevalent also among isolates from other countries. The four ST131 virotypes, which appeared to represent clonal variants within ST131, exhibited distinctive distributions and clinical and epidemiological associations. Future studies of the conventional and molecular epidemiology of ST131 should address relevant within-ST divisions, including fimH-based subclones and virotypes. Such studies should improve our understanding of the basis for the worldwide dissemination and emergence of the E. coli ST131 clonal group.
Supplementary Material
ACKNOWLEDGMENTS
We thank Veronika Tchesnokova and Mariya Billig (University of Washington School of Medicine, Seattle, WA) for their help during the fimH subtyping. We thank Monserrat Lamela for skillful technical assistance.
A.M. acknowledges the Ramón y Cajal program from the Spanish Ministerio de Economía y Competitividad, Gobierno de España. R.M. acknowledges the grant of the Agencia Española de Cooperación Internacional (AECI) (Ministerio de Asuntos Exteriores y de Cooperación). This work was partially supported by the Red Española de Investigación en Patología Infecciosa (REIPI) (no. RD06/0008/1018-1016, RD12/0015) and grant no. PI09/01273, 070190, 10/02021, 10/01955, 10/00795, and PI11/01117 (Instituto de Salud Carlos III, Fondo de Investigación Sanitaria, Ministerio de Economía y Competitividad, Gobierno de España), CN2012/303 09TAL007261PR and 10MRU261023PR (Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia and the European Regional Development Fund [ERDF]), 0048/2008 and CTS-5259 (Junta de Andalucía), BFU2011-26608 (Spanish Ministry of Education), 282004/FP7-HEALTH.2011.2.3.1-2 (European VII Framework Program), and FEDER-INNTERCONECTA-COLIVAC (CDTI, Ministerio de Economía y Competitividad, Gobierno de España; Consellería de Economía e Industria, Xunta de Galicia; ERDF). This material also is based partly upon work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant no. 1 I01 CX000192 01.
The Spanish GEIH-BLEE 2006 study group includes C. Martínez Peinado (Villajoyosa), J. F. Ordás (Cangas de Nancea), E. Garduño (Badajoz), M. A. Domínguez (Barcelona), F. Navarro (Barcelona), G. Prats (Barcelona), F. Marco (Barcelona), E. Ojeda (Burgos), P. Marín (Cádiz), R. Carranza (Alcazar de San Juan), F. Rodríguez (Cordova), C. García Tejero (Figueras), F. Artiles (Gran Canaria), B. Palop (Granada), I. Cuesta (Jaén), M. Cartelle (A Coruña), M. D. Rodríguez (Ferrol), I. Fernández (León), E. Ugalde (Logroño), R. Cantón (Madrid), E. Cercenado (Madrid), F. Chaves (Madrid), J. J. Picazo (Madrid), A. Delgado (Alcorcón), C. Guerrero (Murcia), B. Fernández (Orense), A. Fleites (Oviedo), A. Oliver (Palma de Mallorca), J. J. García (Pamplona), M. García (Pontevedra), J. Elías (Salamanca), J. Calvo (Santander), M. Treviño (Santiago de Compostela), M. Ruiz (Seville), M. A. Díaz and J. R. Hernández-Bello (Seville), M. Lara (Tenerife), L. Torres (Teruel), E. García (Toledo), D. Navarro (Valencia), M. Gobernado (Valencia), A. Tenorio (Valladolid), I. Otero (Vigo), L. Michaus (Vitoria), and J. Castillo (Zaragoza).
Footnotes
Published ahead of print 7 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01555-13.
REFERENCES
- 1.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165–174 [DOI] [PubMed] [Google Scholar]
- 2.Hernández JR, Martínez-Martínez L, Cantón R, Coque TM, Pascual A, Spanish Group for Nosocomial Infections (GEIH) 2005. Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum beta-lactamases in Spain. Antimicrob. Agents. Chemother. 49:2122–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Díaz MA, Hernández JR, Martínez-Martínez L, Rodríguez-Baño J, Pascual A, Grupo de Estudio de Infección Hospitalaria (GEIH) 2009. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Spanish hospitals: 2nd multicenter study (GEIH-BLEE project, 2006). Enferm. Infecc. Microbiol. Clin. 27:503–510. (Article in Spanish.) [DOI] [PubMed] [Google Scholar]
- 4.Díaz MA, Hernández-Bello JR, Rodríguez-Baño J, Martínez-Martínez L, Calvo J, Blanco J, Pascual A, Spanish Group for Nosocomial Infections (GEIH) 2010. Diversity of Escherichia coli strains producing extended-spectrum β-lactamases in Spain: second nationwide study. J. Clin. Microbiol. 48:2840–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco M, Alonso MP, Nicolas-Chanoine MH, Dahbi G, Mora A, Blanco JE, López C, Cortés P, Llagostera M, Leflon-Guibout V, Puentes B, Mamani R, Herrera A, Coira MA, García-Garrote F, Pita JM, Blanco J. 2009. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63:1135–1141 [DOI] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281 [DOI] [PubMed] [Google Scholar]
- 7.Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Blanco JE, Alonso MP, García-Garrote F, Chaves F, Orellana MÁ, Martínez-Martínez L, Calvo J, Prats G, Larrosa MN, González-López JJ, López-Cerero L, Rodríguez-Baño J, Pascual A. 2011. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J. Antimicrob. Chemother. 66:2011–2021 [DOI] [PubMed] [Google Scholar]
- 8.Oteo J, Diestra K, Juan C, Bautista V, Novais A, Pérez-Vázquez M, Moyá B, Miró E, Coque TM, Oliver A, Cantón R, Navarro F, Campos J, Spanish Network in Infectious Pathology Project (REIPI) 2009. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34:173–176 [DOI] [PubMed] [Google Scholar]
- 9.Oteo J, Navarro C, Cercenado E, Delgado-Iribarren A, Wilhelmi I, Orden B, García C, Miguelañez S, Pérez-Vázquez M, García-Cobos S, Aracil B, Bautista V, Campos J. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J. Clin. Microbiol. 44:2359–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321 [DOI] [PubMed] [Google Scholar]
- 12.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 13.Horcajada JP, Soto S, Gajewski A, Smithson A, Jiménez de Anta MT, Mensa J, Vila J, Johnson JR. 2005. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J. Clin. Microbiol. 43:2962–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Da Silva GJ, Mendonça N. 2012. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 3:18–28 [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI M100-S19 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 16.Johnson JR, Nicolas-Chanoine MH, DebRoy C, Castanheira M, Robicsek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA, MASTER Investigators 2012. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg. Infect. Dis. 18:598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leflon-Guibout V, Jurand C, Bonacorsi S, Espinasse F, Guelfi MC, Duportail F, Heym B, Bingen E, Nicolas-Chanoine MH. 2004. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 48:3736–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, Pearson A, Harry S, Leach JB, Loughrey A, Lowes JA, Warren RE, Livermore DM. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 54:735–743 [DOI] [PubMed] [Google Scholar]
- 21.Mora A, López C, Dabhi G, Blanco M, Blanco JE, Alonso MP, Herrera A, Mamani R, Bonacorsi S, Moulin-Schouleur M, Blanco J. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 9:132. 10.1186/1471-2180-9-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 23.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Totsika M, Beatson SA, Sarkar S, Phan MD, Petty NK, Bachmann N, Szubert M, Sidjabat HE, Paterson DL, Upton M, Schembri MA. 2011. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One 6:e26578. 10.1371/journal.pone.0026578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 78:1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L, Gniadkowski M, Nordmann P. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031–1034 [DOI] [PubMed] [Google Scholar]
- 27.Dahbi G, Mora A, López C, Alonso MP, Mamani R, Marzoa J, Coira A, García-Garrote F, Pita JM, Velasco D, Herrera A, Viso S, Blanco JE, Blanco M, Blanco J. 2013. Emergence of new variants of ST131 clonal goup in extraintestinal pathogenic Escherichia coli producing extended-spectrum β-lactamases. Int. J. Antimicrob. Agents. 10.1016/j.ijantimicag.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 28.Coelho A, Mora A, Mamani R, López C, González-López JJ, Larrosa MN, Quintero-Zarate JN, Dahbi G, Herrera A, Blanco JE, Blanco M, Alonso MP, Prats G, Blanco J. 2011. Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J. Antimicrob. Chemother. 66:517–526 [DOI] [PubMed] [Google Scholar]
- 29.López-Cerero L, Bellido MD, Serrano L, Liró J, Cisneros JM, Rodríguez-Baño J, Pascual A. 2012. Escherichia coli O25b:H4/ST131 are prevalent in Spain and are often not associated with ESBL or quinolone resistance. Enferm. Infecc. Microbiol. Clin. 31:385–388 [DOI] [PubMed] [Google Scholar]
- 30.Novais Â, Viana D, Baquero F, Martínez-Botas J, Cantón R, Coque TM. 2012. Contribution of IncFII and broad-host IncA/C and IncN plasmids to the local expansion and diversification of phylogroup B2 Escherichia coli ST131 clones carrying blaCTX-M-15 and qnrS1 genes. Antimicrob. Agents Chemother. 56:2763–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olesen B, Hansen DS, Nilsson F, Frimodt-Møller J, Leihof RF, Struve C, Scheutz F, Johnston B, Krogfelt KA, Johnson JR. 2013. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J. Clin. Microbiol. 51:1779–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumura Y, Yamamoto M, Higuchi T, Komori T, Tsuboi F, Hayashi A, Sugimoto Y, Hotta G, Matsushima A, Nagao M, Takakura S, Ichiyama S. 2012. Prevalence of plasmid-mediated AmpC β-lactamase-producing Escherichia coli and spread of the ST131 clone among extended-spectrum β-lactamase-producing E. coli in Japan. Int. J. Antimicrob. Agents. 40:158–162 [DOI] [PubMed] [Google Scholar]
- 33.Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS, Jr, Hansen G, Edelstein PH, Robicsek A, Cleary T, Adachi J, Paterson D, Quinn J, Hanson ND, Johnston BD, Clabots C, Kuskowski MA, AMERECUS Investigators 2012. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob. Agents Chemother. 56:2364–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peirano G, van der Bij AK, Gregson DB, Pitout JD. 2010. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J. Clin. Microbiol. 50:294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brisse S, Diancourt L, Laouénan C, Vigan M, Caro V, Arlet G, Drieux L, Leflon-Guibout V, Mentré F, Jarlier V, Nicolas-Chanoine MH, Coli β Study Group 2012. Phylogenetic distribution of CTX-M- and non-extended-spectrum-β-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J. Clin. Microbiol. 50:2974–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. 2009. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63:72–79 [DOI] [PubMed] [Google Scholar]
- 37.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppé E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277 [DOI] [PubMed] [Google Scholar]
- 38.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 39.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. 2013. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect. Control Hosp. Epidemiol. 34:361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung HC, Lai CH, Lin JN, Huang CK, Liang SH, Chen WF, Shih YC, Lin HH, Wang JL. 2012. Bacteremia caused by extended-spectrum-β-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob. Agents Chemother. 56:618–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodríguez-Baño J, Mingorance J, Fernández-Romero N, Serrano L, López-Cerero L, Pascual A, ESBL-REIPI Group 2013. Outcome of bacteraemia due to extended-spectrum β-lactamase-producing Escherichia coli: impact of microbiological determinants. J. Infect. 67:27–34 [DOI] [PubMed] [Google Scholar]
- 42.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 207:919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet. Microbiol. 153:99–108 [DOI] [PubMed] [Google Scholar]
- 44.Mora A, Herrera A, Mamani R, López C, Alonso MP, Blanco JE, Blanco M, Dahbi G, García-Garrote F, Pita JM, Coira A, Bernárdez MI, Blanco J. 2010. Recent emergence of clonal group O25b:K1:H4-B2-ST131 ibeA strains among Escherichia coli poultry isolates, including CTX-M-9-producing strains, and comparison with clinical human isolates. Appl. Environ. Microbiol. 76:6991–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028 [DOI] [PubMed] [Google Scholar]
- 46.Johnson JR, Russo TA, Tarr PI, Carlino U, Bilge SS, Vary JC, Jr, Stell AL. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN (E. coli), among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JR, Gajewski A, Lesse AJ, Russo TA. 2003. Extraintestinal pathogenic Escherichia coli as a cause of invasive nonurinary infections. J. Clin. Microbiol. 41:5798–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.