Abstract
Cox1, the core subunit of the cytochrome c oxidase, receives two heme a cofactors during assembly of the 13-subunit enzyme complex. However, at which step of the assembly process and how heme is inserted into Cox1 have remained an enigma. Shy1, the yeast SURF1 homolog, has been implicated in heme transfer to Cox1, whereas the heme a synthase, Cox15, catalyzes the final step of heme a synthesis. Here we performed a comprehensive analysis of cytochrome c oxidase assembly intermediates containing Shy1. Our analyses suggest that Cox15 displays a role in cytochrome c oxidase assembly, which is independent of its functions as the heme a synthase. Cox15 forms protein complexes with Shy1 and also associates with Cox1-containing complexes independently of Shy1 function. These findings indicate that Shy1 does not serve as a mobile heme carrier between the heme a synthase and maturing Cox1 but rather cooperates with Cox15 for heme transfer and insertion in early assembly intermediates of cytochrome c oxidase.
INTRODUCTION
The mitochondrial respiratory chain receives electrons from NADH and FADH2 and utilizes these to reduce molecular oxygen to water. During this process, protons are translocated across the inner mitochondrial membrane to generate a pH gradient that drives ATP synthesis by the F1Fo-ATPase. Four multisubunit protein complexes, harboring different types of redox centers for electron transport, form the respiratory chain. The terminal enzyme of this electron transport chain is the cytochrome c oxidase, which oxidizes cytochrome c and delivers electrons to O2. Therefore, the core subunit of the complex, Cox1, contains two heme molecules (a and a3) and a single copper-containing CuB site. A second copper site comprising two copper ions, the CuA site, is present in the Cox2 subunit (1–4). These redox sites are established during assembly of the cytochrome c oxidase in a process that requires assistance by multiple assembly-promoting factors. One challenge in this assembly process is the fact that the core subunits (Cox1, Cox2, and Cox3) of cytochrome c oxidase are synthesized within mitochondria, while all other subunits are translated in the cytoplasm and imported into mitochondria (5). This process is best studied in the yeast Saccharomyces cerevisiae. The assembly process of cytochrome c oxidase initiates with the synthesis of Cox1, which engages with the assembly factors Coa3 and Cox14 in the inner membrane of mitochondria (6–9). This association affects translation of the COX1 mRNA as the translational activator, Mss51, is recruited into the Cox1-containing assembly intermediate and inactivated (for a review, see reference 10). Subsequently, the assembly factors Coa1 and Shy1, as well as the first nucleus-encoded subunits, associate with the Cox1 assembly intermediate before Mss51 is released to initiate new rounds of Cox1 translation (11–14). During this process, the heme groups are thought to be incorporated into Cox1; however, the exact stage of heme incorporation and the underlying mechanisms remain unresolved (4) despite being a critical step. Work by Khalimonchuk et al. revealed that once heme is inserted into Cox1, the a3 site needs to be protected by assembly factors in order to prevent the uncontrolled generation of reactive oxygen species through unrestricted access of O2 to the redox active heme group (15).
Heme a is synthesized in mitochondria from heme o. Cox10 represents the heme o synthase that transfers a farnesyl diphosphate to protoheme. One possible reaction scheme is that the hydroxyethyl farnesyl heme o, which is generated by Cox10, represents an intermediate, which is subsequently oxidized by the heme a synthase Cox15 to form heme a. In this reaction, a methyl group of the pyrrole ring is oxidized to a formyl group (16, 17). Both enzymatic functions are conserved from yeast to humans, and defects in both enzymes lead to severe human disorders with cytochrome c oxidase deficiency (18–20). Besides Cox10 and Cox15, SURF1 (termed Shy1 in yeast) is also a conserved protein and required for cytochrome c oxidase biogenesis. Yeast and human mitochondria lacking functional Shy1/SURF1 display a severe reduction in cytochrome c oxidase complexes (for a review, see references 2, 3, 21, and 22). This defect leads to Leigh syndrome in humans, a fatal neuromuscular disorder (23, 24). Shy1/SURF1 is required for Cox1 biogenesis and hence is part of early assembly intermediates in yeast and human mitochondria, termed COA and MITRAC complexes, respectively (7, 11, 12, 25–27). A study of Paracoccus denitrificans SURF1 indicated that SURF1 is capable of binding heme a, thus linking its function to the heme incorporation step for Cox1 during cytochrome c oxidase biogenesis (28).
Here we performed mass spectrometric analyses of isolated Shy1 complexes after blue-native PAGE (BN-PAGE) to define their protein composition in an unbiased manner. Unexpectedly, we found that the heme a synthase Cox15 associates with Shy1 in distinct complexes. Moreover, both proteins are also present in early cytochrome c oxidase assembly intermediates (COA complexes). C-terminal epitope tagging of Cox15 selectively affects its association to COA complexes. Cox15 also forms complexes with maturing Cox1, the heme receiving subunit of cytochrome c oxidase, in the absence of Shy1. Based on our findings, we suggest that heme transfer, from the heme a synthase to Cox1, occurs in early assembly intermediates in a Shy1-assisted manner.
MATERIALS AND METHODS
Yeast strains and molecular cloning.
All S. cerevisiae strains used in this study are listed in Table 1. With the exception of the cox1− (derivative of 777-3A) and cox2− (derivative of AB1-4D) strains (29), all strains are derived from YPH499 (30). Chromosomal deletions of COA1, COX10, and COX15, as well as tagged versions of COX15, were generated by introduction of the TRP1 or His3MX6 cassettes (31). COX15 was cloned with its endogenous promoter and terminator from yeast (YPH499) genomic DNA into pRS416 (Stratagene). The H368M point mutation was introduced by site-directed mutagenesis (QuikChange; Stratagene), and the resulting plasmid was verified by sequencing (GATC Biotech, Germany). Yeast strains were transformed with PCR-amplified integration cassettes or with plasmids, using the lithium acetate method, and confirmed by PCR or Western blot analysis.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| YPH499 | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 | 30 |
| Shy1ProtA strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; shy1::shy1-TEV-ProtA-7HIS-HIS3MX6 | 12 |
| shy1Δ strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; shy1::HIS3MX6 | 12 |
| cox1− strain | MATα ade1 op1; cox1-G421 | 29 |
| cox2− strain | MATα ade1 op1 met3; cox2-V25 | 50 |
| Coa3HA strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; coa3::coa3-3HA-HIS3MX6 | 6 |
| Cox15ProtA (BBY02) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox15::cox15-TEV-ProA-7HIS-HIS3MX6 | This study |
| Cox15FLAG (BBY32) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox15::cox15-FLAG-HIS3MX6 | This study |
| cox15Δ (BBY14) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox15::klTRP1 | This study |
| cox15Δ Shy1ProtA (BBY22) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox15::klTRP1 shy1::shy1-TEV-ProtA-7HIS-HIS3MX6 | This study |
| cox10Δ (BBY15) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox10::klTRP1 | This study |
| cox10Δ Shy1ProtA (BBY27) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox10::klTRP1 shy1::shy1-TEV-ProtA-7HIS-HIS3MX6 | This study |
| cox15Δ + pRS416 (BBY16) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox15::klTRP1 [pRS416] | This study |
| cox15Δ + Cox15WT (BBY17) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox15::klTRP1 [pRS416-COX15] | This study |
| cox15Δ + Cox15H368M (BBY18) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; cox15::klTRP1 [pRS416-COX15H368M] | This study |
| Shy1ProtA cox15Δ + Cox15H368M (BBY25) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; shy1::shy1-TEV-ProtA-7HIS-HIS3MX6 cox15::klTRP1 [pRS416-COX15H368M] | This study |
| Shy1ProtA/rho0 (BBY47) strain | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; shy1::shy1-TEV-ProtA-7HIS-HIS3MX6/rho0 | This study |
Yeast handling and mitochondrial preparation.
Yeast cells were grown in liquid medium containing 1% yeast extract, 2% peptone, and either 2% glucose or galactose or 3% glycerol (YPD, YPGal, or YPG, respectively). Strains containing plasmid-borne wild-type or mutant forms of Cox15 were grown on synthetic medium (0.67% yeast nitrogen base and 0.07% CSM lacking uracil [MP Biomedicals]) and 2% galactose. If not indicated otherwise, yeast cells were grown at 30°C with shaking (150 to 220 rpm). For growth tests, precultures were adjusted to an optical density at 600 nm (OD600) of 0.3, and serial 1:10 dilutions were spotted onto YPD, YPG, or SD or SG lacking His (MP Biomedicals) solid medium plates and incubated for 3 to 5 days at the indicated temperatures. Mitochondria were isolated essentially as previously described (32). For steady-state analyses of mitochondrial proteins, different amounts were subjected to SDS-PAGE, followed by Western blotting.
Generation of rho0 yeast strains.
Cells were kept growing in YPD medium containing 25 μg/ml ethidium bromide (EtBr) and diluted (1:60) in fresh YPD with EtBr every day. After 3 days, cells were plated onto YPD solid medium plates without EtBr and incubated for 2 days, and single colonies were picked. The absence of mitochondrial DNA was verified by the absence of growth on glycerol plates (nonfermentable carbon source) and the absence of various mitochondrion-encoded proteins.
IgG chromatography.
For native complex isolation, human IgGs (Sigma-Aldrich) were coupled to CNBr-activated Sepharose (GE Healthcare) according to the manufacturer's specifications. Mitochondria from wild-type (WT) and protein A-tagged strains were solubilized on ice in 20 mM Tris-HCl (pH 7.4), 0.1 M NaCl, 10% glycerol, 5 mM EDTA, 2 mM phenylmethylsulfonyl fluoride (PMSF), and 1% digitonin for 30 min. After removal of unsolubilized material by centrifugation (15 min, 20,000 × g, 4°C), a sample was taken and the mitochondrial extract was applied to IgG-Sepharose for 2 h at 4°C with mild agitation. After extensive washing with wash buffer (20 mM Tris-HCl [pH 7.4], 0.1 M NaCl, 10% glycerol, 5 mM EDTA, 2 mM PMSF, and 0.3% digitonin), AcTEV (tobacco etch virus [TEV]) protease (Invitrogen) was added and incubated overnight at 4°C to cleave protein A-tagged protein from the Sepharose. TEV protease, carrying a polyhistidine, was removed by addition of nickel-nitrilotriacetic acid (Ni-NTA) (Qiagen) preequilibrated with wash buffer. The released native complexes were mixed with appropriate loading buffer and further analyzed by SDS- or BN-PAGE. Samples that were analyzed only by SDS-PAGE were not treated with TEV protease but instead were eluted directly with SDS loading buffer.
Coimmunoprecipitation.
Coa3/Coa1 or, as a control, Ylh47-specific antisera were bound to protein A-Sepharose (GE Healthcare) in 0.1 M potassium phosphate buffer (pH 7.4) and subsequently cross-linked with 5 mg/ml dimethyl pimelimidate (DMP) solution in 0.1 M sodium borate (pH 9.0) for 30 min. Mitochondria were solubilized, and the lysate was cleared as described above. An input sample (total) was taken. The mitochondrial lysate was split and either bound to anti Coa3/Coa1-coupled Sepharose, respectively, or to a control Sepharose coupled with an unrelated antiserum for 90 min at 4°C under mild agitation. After washing with wash buffer as described above, proteins were eluted with 0.1 M glycine (pH 2.8) and immediately neutralized with 1 M Tris (pH 11.5). Eluates and the total sample were analyzed by SDS-PAGE and Western blotting.
Mass spectrometry.
For interactome analysis, eluted proteins were mixed with 10× BN loading dye and separated on a 4 to 13% gradient BN gel. Directly following the gel run, the corresponding gel lane was cut into 23 equal gel slices. Gel slices were washed three times in 5 mM ammonium bicarbonate–50% acetonitrile buffer (pH 8.0), and proteins therein were in-gel digested with trypsin (Promega) as described previously (33). Chymotrypsin was used as the protease instead of trypsin in replicate experiments. Tryptic/chymotryptic peptides from each gel slice were analyzed as described previously (34) by nanoflow high-performance liquid chromatography (HPLC) (Agilent 1100; Agilent Technologies) coupled to nanoelectrospray LTQ-Orbitrap XL mass spectrometer (Thermo Fischer Scientific). The raw mass spectrometry (MS) files from the mass spectrometer were analyzed by the software programs MaxQuant (35) and Andromeda (36) using the UniProt S. cerevisiae protein database (version 29.11.11). Razor and unique peptide numbers from MaxQuant were normalized for each protein and visualized using R as described previously (37). The “match between runs” option in MaxQuant was selected in replicate analysis to detect peptides of Cox15.
Miscellaneous.
Standard techniques were used for SDS-PAGE and Western blotting on polyvinylidene fluoride (PVDF) membranes. For detection and visualization of antibody-protein complexes, peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) and enhanced chemiluminescence reagent (GE Healthcare) were used. For BN-PAGE, mitochondria were solubilized in 1% digitonin, 20 mM Tris-HCl (pH 7.4), 5 mM EDTA, 100 mM NaCl, 10% glycerol, and 2 mM PMSF for 30 min at 4°C. The lysate was cleared at 20,000 × g at 4°C for 15 min. After addition of 10× loading dye (5% Coomassie G-250, 500 mM 6-aminohexanoic acid, and 0.1 M Bis-Tris, pH 7.0), the supernatant was separated on a 4 to 13% polyacrylamide gradient gel (38). Antibodies used as control were directed against late COX subunits (Cox4, Cox13), subunits of cytochrome bc1 complex (Rip1 and Qcr8), a subunit of ATP synthase (Atp5), subunits of the TOM complex (translocase of the outer membrane) (Tom40 and Tom70), a subunit of the translocase of the inner mitochondrial membrane, TIM23 complex (Tim23), mitochondrial porin of the outer membrane (Por1), aconitase, a soluble protein of the mitochondrial matrix (Aco1), and a component of the mitochondrial inner membrane organizing system, MINOS (Mio10). Note that in BN-PAGE, purified COA complexes display slightly faster migration than if analyzed in whole mitochondrial extracts, probably due to the smaller amount of protein present in the lane. Multiple sequence alignment of Cox15 was performed using the ClustalW2 software program.
RESULTS
Mapping of Shy1 protein complexes.
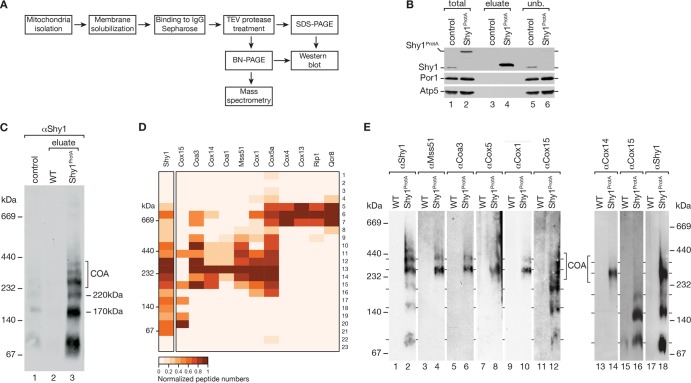
Shy1 plays a central role in early assembly steps of Cox1 during cytochrome c oxidase maturation. Previous analyses found Shy1 in several distinct mitochondrial protein complexes, most of which have been resolved by Blue native PAGE (BN-PAGE) analyses using different detergents for solubilization (6, 7, 11, 12, 14, 27, 39). However, a comprehensive analysis of the protein composition of these complexes was still lacking. To close this gap, we used a functional fusion construct of Shy1 (Shy1ProtA), which carries a protein A tag at the C terminus that can be cleaved from the Shy1 portion by TEV (tobacco etch virus) protease treatment to release native Shy1-containing complexes (12). The Shy1ProtA protein was expressed from the chromosomal SHY1 locus under the control of the authentic SHY1 promoter. We purified Shy1ProtA by IgG affinity chromatography from isolated mitochondria after solubilization in digitonin-containing buffer. As a control, we used mitochondria from a strain expressing wild-type Shy1. Shy1 and its associated proteins were released from the resin by TEV protease treatment (Fig. 1A). This approach allowed efficient purification of Shy1 from mitochondrial extracts, which we assessed by SDS-PAGE and Western blotting of the different fractions (Fig. 1B). Based on this small-scale isolation, we purified Shy1-containing complexes in preparative scale and separated protein complexes by BN-PAGE. An aliquot of the eluate was used for Western blot analysis, revealing a variety of distinct Shy1-containing complexes in a size range from 80 to 440 kDa (Fig. 1C). To determine the protein composition of these complexes, a gel lane containing the majority of the eluate was cut into 23 slices, which were subjected to in-gel trypsin digestion prior to mass spectrometric analysis (see Table S1 in the supplemental material). A heat map representation of selected proteins showed that the slower migrating Shy1 complexes, of 270 to 350 kDa, contained the early assembly factors of Cox1, such as Coa1, Coa3, Cox14, the translational activator Mss51, and the structural subunits Cox1 and Cox5a, termed COA complexes (Fig. 1D). In addition, respiratory chain supercomplexes, which have previously been reported to associate with Shy1 (12, 40), were recovered, as indicated by the high-molecular-weight cluster containing Cox4, Cox13 of the cytochrome c oxidase, and Rip1 and Qcr8 of the bc1 complex (Fig. 1D, right side). Interestingly, we also identified the heme a synthase, Cox15, in distinct Shy1 protein complexes. This finding agreed with our previous observation that Cox15 could be identified by mass spectrometry in Shy1 complexes purified from digitonin-solubilized membranes (12). However, the BN-PAGE/MS mapping approach revealed an unexpected result: Cox15 was recovered in COA complexes that represent the early assembly intermediates of Cox1, yet it also formed two distinct complexes with Shy1, of 170 and 220 kDa, that did not contain any COA subunits (Fig. 1D).
Fig 1.
Isolation and characterization of complexes containing Shy1ProtA. (A) Schematic illustration of the native purification of TEV-protein A-7His-tagged Shy1 (Shy1ProtA)-containing complexes by IgG chromatography. (B) SDS-PAGE and Western blot analysis of eluates from Shy1ProtA IgG chromatography. Amount of protein loaded in the total and unbound (unb.) samples correspond to 5.5% of the eluate. (C) Native eluted proteins (as in panel B) were analyzed by BN-PAGE for Shy1ProtA-containing complexes. As a control, solubilized wild-type mitochondria were loaded (6.5% of eluate). (D) Preparative isolation of Shy1ProtA-containing complexes was analyzed by BN-PAGE, followed by mass spectrometry as described in Materials and Methods. Heat map illustrating normalized peptide numbers of proteins coimmunoprecipitated with Shy1ProtA in individual gel slices. Positions of marker bands were estimated from their running distances in a Coomassie-stained gel lane. (E) Western blot analysis of protein complexes that were isolated with Shy1ProtA and separated by BN-PAGE. Two independent experiments are shown.
To support these findings by an independent approach, we isolated Shy1-containing complexes as described above and analyzed the complex pattern with available antibodies. In agreement with previous reports and the mass spectrometric data, Shy1, Mss51, Coa3, and the structural subunits Cox5 and Cox1 were detected in the COA complexes but not in the 170-kDa or 220-kDa complex, suggesting that these complexes represent novel species composed of Cox15 and Shy1 but lacking known assembly factors or structural subunits (Fig. 1E). To detect the authentic Cox15 protein, we generated a polyclonal antiserum that specifically recognizes Cox15 in purified mitochondria. In agreement with the mass spectrometric data, we detected Cox15 primarily in the 170- and 220-kDa complexes that were purified with Shy1ProtA. However, a fraction was present in a higher-molecular-weight range, seemingly comigrating with COA complexes containing assembly factors and Cox1 (Fig. 1E). These findings contradict previous reports that epitope-tagged Cox15 forms a single high-molecular-weight complex in mitochondria and does not appear to complex with Cox1 (41, 42).
The heme a synthase Cox15 is associated with COA complexes.
To further support the association of Cox15 with cytochrome c oxidase assembly intermediates, we integrated a protein A- or FLAG tag-encoding cassette into the chromosomal COX15 locus to generate tagged versions of Cox15 for purification of Cox15-containing complexes. To assess the functionality of the fusion proteins, we analyzed growth of the strains on nonfermentable and fermentable carbon sources. Both strains displayed wild-type-like growth behavior, indicating that Cox15ProtA and Cox15FLAG are functional (Fig. 2A). Thus, we purified mitochondria from wild-type and Cox15ProtA-expressing cells. After solubilization of mitochondria in digitonin buffer, we purified Cox15ProtA-containing complexes by IgG chromatography and analyzed samples by Western blotting. Cox15ProtA was efficiently isolated from mitochondrial extracts. Besides Cox15, we also recovered Shy1 in the eluate of the purification. However, we were unable to identify subunits of early Cox1 assembly intermediates, e.g., Cox14 or Coa1 (Fig. 2B). Moreover, a BN-PAGE analysis of purified Cox15 complexes did not reveal high-molecular-mass Cox15 protein complexes but instead an abundant 130-kDa complex and a less-abundant 170-kDa complex (Fig. 2C). The absence of the COA complexes from Cox15ProtA isolations obviously contradicted the mass spectrometric data on Shy1 complexes yet was in agreement with previous analyses using tagged Cox15 for complex analyses (41, 42).
Fig 2.
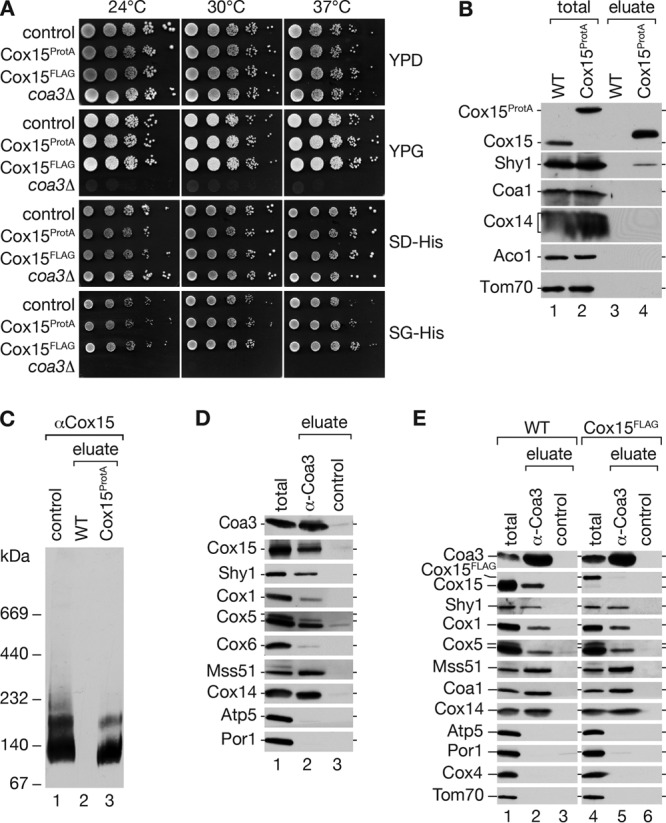
A C-terminal tag on Cox15 affects association with COA complexes. (A) Growth test on fermentable and nonfermentable full and synthetic media. Cells were spotted in serial 10-fold dilutions and incubated at indicated temperatures. As a control, Coa3HA genomically integrated with the His3MX6 cassette was used. (B) Mitochondria from wild-type (WT) and protein A-tagged Cox15 (Cox15ProtA) were solubilized in 1% digitonin buffer and subjected to IgG chromatography. After TEV protease cleavage, the eluate was split, and proteins were separated by SDS-PAGE and analyzed by Western blotting (total, 6.25%; eluate, 100%) or by BN-PAGE, followed by Western blotting using antiserum against Cox15 (C). (D) Coimmunoprecipitation from digitonin-solubilized mitochondria. After solubilization, samples were incubated with Coa3-specific or control antisera. Bound material was eluted and analyzed by SDS-PAGE and Western blotting (total, 8%; eluate, 100%). (E) Coimmunoprecipitation as described for panel D using wild-type (WT) or Cox15FLAG mitochondria.
To clarify this discrepancy, we performed immunoprecipitations of COA complexes using an antibody against Coa3 (6). Solubilized mitochondria were subjected to immunoprecipitation with Coa3 or control antibodies, and precipitates were analyzed by Western blotting (this approach does not allow a native analysis of isolated complexes by BN-PAGE). The Coa3 antibody efficiently immunoprecipitated COA complexes, as indicated by the presence of Coa3, Cox14, Cox1, Mss51, and Shy1 in the eluate (Fig. 2D). However, in agreement with the mass spectrometric data of Shy1 complex purifications, we also specifically recovered Cox15 in the COA complexes.
Thus, we speculated that a C-terminal tag on Cox15 compromises its association with COA complexes, leading to a loss of cytochrome c oxidase assembly intermediates during the isolation procedure. To address this hypothesis directly, we performed Coa3 immunoprecipitations in wild-type and Cox15FLAG mitochondrial extracts. To avoid interaction of Cox15ProtA with the Coa3 antibodies via protein A, we used Cox15FLAG mitochondria for these experiments. While we were able to immunoprecipitate COA complex constituents from wild-type and Cox15FLAG mitochondria, Cox15 was coprecipitated only in the wild-type samples and was selectively lacking when precipitation was performed from Cox15FLAG mitochondria (Fig. 2E). Thus, we conclude that a C-terminal tag on Cox15 affects the stability of the association between Cox15 and COA complexes but not between Cox15 and Shy1 (as seen in Fig. 2C).
Cox15 interaction with Cox1 assembly intermediates occurs independently of its enzymatic function.
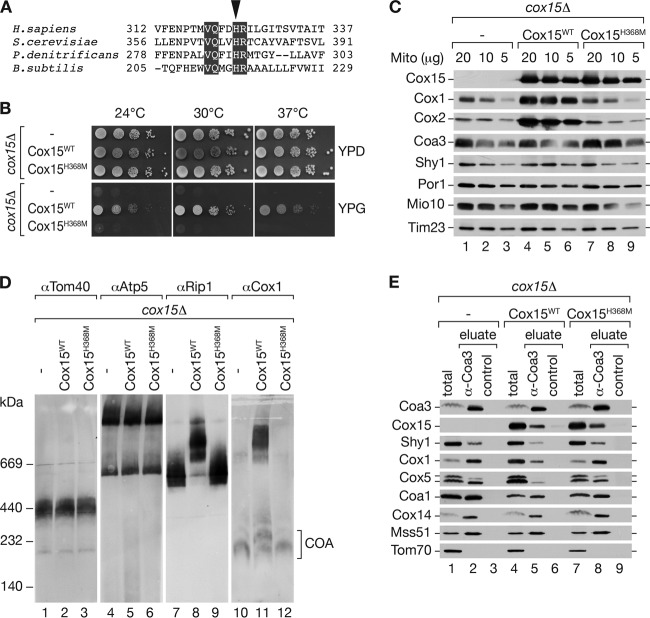
Cox15 catalyzes the conversion of heme o to heme a by hydroxylation of its methyl group at position C-8. Further oxidation of the alcohol generates the characteristic aldehyde (17, 43). We asked if the enzymatic activity of Cox15 was required for its association with COA complexes. Thus, we made use of a previously characterized point mutation in a conserved histidine of the heme a synthase of Bacillus subtilis (CtaA), which displays severely reduced activity (Fig. 3A) (44). Using site-directed mutagenesis, we generated a plasmid-borne mutant, COX15, encoding the protein with a H368M exchange. cox15Δ mutant yeast cells were transformed with a plasmid encoding Cox15H368M or wild-type Cox15. Transformants were analyzed for growth on fermentable and nonfermentable carbon sources. As expected, Cox15H368M was unable to rescue the cox15Δ mutant growth phenotype, while wild-type Cox15 cells grew well under all tested conditions (Fig. 3B). To assess that the observed growth defect was not due to instability of the mutant Cox15H368M protein, we purified mitochondria from cox15Δ cells and cox15Δ cells expressing Cox15 or Cox15H368M and performed Western blot analyses. These analyses confirmed that Cox15H368M was present at wild-type levels in mitochondria (Fig. 3C). As expected, cox15Δ and Cox15H368M mitochondria displayed a severe reduction of Cox1 and Cox2 in a steady-state protein analysis of selected proteins. However, Coa3 and Shy1 were present at wild-type levels (Fig. 3C). Next, we analyzed mitochondrial protein complexes in these mitochondria. The amount and stability of the TOM complex and the F1Fo ATPase were not affected in the mutant mitochondria compared to results for the wild-type control. However, a lack of mature cytochrome c oxidase in cox15Δ and Cox15H368M mitochondria led to formation of bc1-complex dimers and a concomitant lack of larger supercomplexes (Fig. 3D). Despite the reduced Cox1 levels, the cox15Δ and Cox15H368M mitochondria displayed residual amounts of COA complexes that we detected with an anti Cox1 antibody (Fig. 3D). Based on this finding, we addressed the association of Cox15 and Cox15H368M with assembly intermediates by Coa3 immunoprecipitation. Despite the small amount of Cox1, the remaining protein was present in Coa3-containing complexes, indicating the accumulation of Cox1-containing assembly intermediates in the mutants. Western blot analysis further revealed that Cox15 and Cox15H368M were coprecipitated with similar efficiency from Cox15- and Cox15H368M - containing mitochondria. Accordingly, the catalytically compromised Cox15 was able to associate with assembly intermediates independently of the formation of its final product, heme a (Fig. 3E).
Fig 3.
Cytochrome c oxidase (COX) assembly in yeast strains is affected in heme biogenesis. (A) Alignment of a region in Cox15 homologues from different species (ClustalW2). Black boxes indicate identical residues in all species shown. The arrowhead indicates a conserved histidine at position 216 in B. subtilis, corresponding to H368 in S. cerevisiae. (B) Growth test on fermentable or nonfermentable medium. cox15Δ cells expressing either wild-type Cox15 (Cox15WT) or a catalytically inactive Cox15 (Cox15H368M) were spotted in serial 10-fold dilutions and incubated at the indicated temperatures (—, plasmid without insert). (C and D) For analysis of steady-state protein levels, mitochondria (Mito) isolated from the indicated strains were separated by SDS-PAGE or BN-PAGE and analyzed by Western blotting. (E) Coimmunoprecipitation of Coa3 from digitonin-solubilized mitochondria isolated from the indicated strains. Solubilized mitochondria were incubated with Coa3-specific or control antisera, and eluates were analyzed by SDS-PAGE and Western blotting. The amounts of protein loaded in the total samples correspond to 8% of the eluate.
Cox15 interaction with Cox1 assembly intermediates occurs independently of the heme o synthase Cox10.
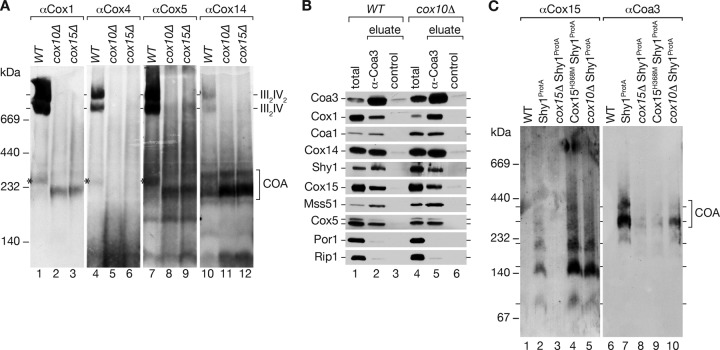
In a complementary approach, we utilized cox10Δ cells to address if Cox15 interacts with Cox1-containing complexes in the absence of heme o. Cox10 is the heme o synthase in mitochondria that generates the substrate for Cox15 (16, 45). To assess if COA complexes were still present in cox10Δ mitochondria, we purified mitochondria from wild-type, cox10Δ, and cox15Δ cells. After solubilization in digitonin buffer, protein complexes were separated by BN-PAGE. As expected, cox10Δ and cox15Δ mitochondria lacked mature cytochrome c oxidase complexes, as indicated by the absence of respiratory chain supercomplexes (Fig. 4A). However, in comparison to results for wild-type mitochondria, COA complexes accumulated in both mutants, as indicated by the presence of Cox1, Cox14, and Cox5 in comigrating complexes (Fig. 4A, lanes 8 and 9 versus lanes 11 and 12). Interestingly, the COA complexes migrated similarly in BN-PAGE analysis of both cox10Δ and cox15Δ mutant mitochondria. Thus, a lack of Cox15 apparently does not affect COA complex size, indicating that Cox15 is not a stoichiometric subunit of these complexes (Fig. 4A, lane 2 versus lane 3). This finding agrees with the fact that only small amounts of Cox15 are present in isolated COA complexes as judged by Western blotting (Fig. 1E).
Fig 4.
Analysis of cytochrome c oxidase assembly in the absence of heme o or heme a synthase. (A) Proteins from mitochondria isolated from wild-type, cox10Δ, and cox15Δ cells were separated by BN-PAGE and analyzed by Western blotting using antiserum against different structural subunits and an assembly factor. The asterisk indicates monomeric COX. (B) Solubilized wild-type (WT) and cox10Δ mitochondria were subjected to immunoprecipitation using Coa3 or control antisera (total, 8%; eluate, 100%). (C) Comparison of complexes purified with Shy1ProtA from solubilized mitochondria from the cox15Δ mutant, the cox15Δ mutant expressing Cox15H368M from a plasmid (Cox15H368M Shy1ProtA), or the cox10Δ mutant on BN-PAGE.
To directly determine if Cox15 associates with COA complexes in cox10Δ mitochondria, we subjected detergent extracts to Coa3 immunoprecipitation. In cox10Δ mitochondria, Coa3 efficiently coimmunoprecipitated the assembly factors (Coa3, Cox14, Coa1, and Shy1), Mss51, the structural subunits Cox1 and Cox5, and Cox15 (Fig. 4B). When purified Shy1ProtA complexes from mutant mitochondria were analyzed by BN-PAGE, we found that COA complexes were still present in the cox10Δ mutant although slightly decreased in abundance compared to the wild-type situation, and that the 170- and 220-kDa Cox15 complexes accumulated, probably due to the reduction in Cox1 amounts (Fig. 4C). These findings show that the association of Cox15 with COA complexes does not require its substrate heme o.
Shy1 is dispensable for Cox15 association with COA complexes.
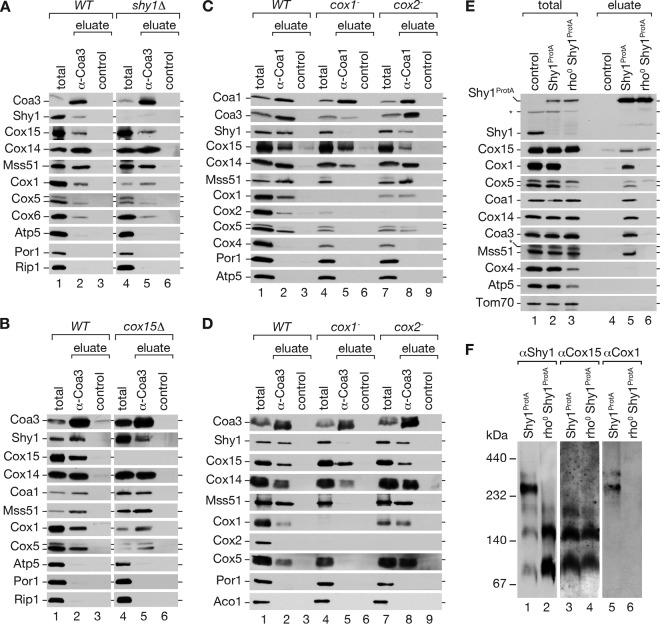
Shy1 and its homologs (SURF1) have been implicated in heme a insertion into Cox1 (28). The association of the terminal enzyme in the biosynthesis of heme a, Cox15, with Shy1 and with COA complexes, led us to investigate if the recruitment of Cox15 into COA complexes depends on Shy1. Therefore, we performed Coa3 immunoprecipitations from wild-type and shy1Δ mitochondria. Despite the absence of Shy1, Cox15 was coimmunoprecipitated with Coa3 and the assembly factor Cox14, the translational regulator Mss51, and the early assembling structural subunits Cox1, Cox5, and Cox6 (Fig. 5A). Note that in the absence of Shy1, only a small fraction of mature cytochrome c oxidase is present in mitochondria, thus reducing the total amount of Cox1 (Fig. 5A, lane 1 versus lane 4). Accordingly, Shy1 was not required to associate Cox15 with assembly intermediates of Cox1. In analogy, we asked if Cox15 is necessary for association of Shy1 with COA complexes. Hence, we performed Coa3 immunoprecipitations from wild-type and cox15Δ mitochondria (Fig. 5B). Despite the overall reduction of Cox1 levels in the cox15Δ mutant (Fig. 3C), immunoprecipitation of Coa3 led to coprecipitation of significant amounts of COA complexes, as judged by the presence of Cox1, Coa1, Cox14, and Mss51 in the eluate (Fig. 5B, lane 2 versus land 5). Moreover, Shy1 was coimmunoprecipitated in cox15Δ mutant mitochondria (Fig. 5B, lane 5). In these analyses, we repeatedly observed a significant increase in the amounts of Shy1 and Coa3 in cox15Δ mutant mitochondria, possibly indicating a compensatory upregulation of Shy1 expression (Fig. 5B, lane 1 versus land 4). However, despite the increased amount of Shy1, we did not observe an increase in association of COA and Shy1. Thus, we conclude that Shy1 and Cox15 are able to associate with cytochrome c oxidase assembly intermediates independently of each other.
Fig 5.
Association of Shy1 and Cox15 with COA complexes. (A) Coimmunoprecipitation of Coa3 from digitonin-solubilized mitochondria isolated from wild-type (WT) and shy1Δ cells were analyzed by SDS-PAGE and Western blotting (total, 8%; eluate, 100%). (B) Coimmunoprecipitation experiments, as for panel A, were performed from wild-type and cox15Δ mitochondrial extracts. (C and D) Mitochondria from wild-type, cox1−, and cox2− cells were subjected to Coa3 (C) or Coa1 (D) immunoprecipitation. (E) Purification of Shy1ProtA-containing complexes by IgG chromatography from wild-type mitochondria compared to results for mitochondria lacking mitochondrial DNA (rho0) (total, 3%; eluate, 100%). The asterisk indicates a cross-reactive signal. (F) BN gel of Shy1ProtA purified complexes from wild-type and rho0 mitochondria.
Cox15 associates with Coa3 and Coa1 in the absence of Cox1.
The heme a that is generated by Cox15 is integrated only into one cellular protein, Cox1. Based on the structural information available on the bovine cytochrome c oxidase, this insertion is considered to occur prior to association of Cox2 with Cox1 (1, 43). We therefore asked if Cox15 interacts with Cox1 assembly factors in the absence of Cox1 or Cox2. For these analyses, we isolated mitochondria from wild-type, cox1−, and cox2#x2212; cells (29, 50). To analyze protein interactions of Cox15 with cytochrome c oxidase assembly factors, we performed immunoprecipitations using anti-Coa1 and anti-Coa3 antibodies, since Coa1 is considered to associate later with Cox1 than with Coa3 and Cox14 (6, 7, 11, 12). Coa3 immunoprecipitates from digitonin-solubilized mitochondria were analyzed by Western blotting for the presence or absence of Cox15 and other selected mitochondrial proteins. As expected from previous analyses, in the absence of Cox1, Coa3 immunoprecipitated Cox14 but displayed reduced coimmunoprecipitation of Mss51 and Shy1 compared to results for wild-type or cox2#x2212; mitochondria (Fig. 5D) (6). Interestingly, Cox15 was efficiently coimmunoprecipitated with Coa3 despite the absence of Cox1 (Fig. 5D, lane 5). The analogous immunoprecipitation experiments using Coa1 (Fig. 5C) antibodies led to a similar coimmunoprecipitation pattern, as observed in the case of Coa3 immunoprecipitation experiments. Hence, Cox15 was able to associate with at least Coa1 and Coa3 in the absence of Cox1 with efficiency similar to that with the wild-type control. In contrast, the amount of Shy1 associated to Coa1 or Coa3 under these conditions was drastically reduced.
The differential behavior of Cox15 and Shy1 with regard to their association with Coa1 and Coa3 in the absence of Cox1 led us to investigate if the Shy1/Cox15 complex was formed in the absence of Cox1. Therefore, we generated a yeast strain expressing Shy1ProtA but lacking mitochondrial DNA and consequently Cox1. (The absence of Cox2 did not have a significant influence on these associations [Fig. 5C and D].) Isolated mitochondria from wild-type cells and cells expressing Shy1ProtA in the presence or absence of mitochondrial DNA were solubilized in digitonin buffer and subjected to IgG chromatography. As expected, the association of Shy1 with Coa1, Cox14, Coa3, and Mss51 was lost in rho0 mitochondria (Fig. 5E and F). In contrast, in the absence of mitochondrial translation products, Cox15 still forms a complex with Shy1 (Fig. 5E, lane 5 versus lane 6). In summary, the association of Cox15 with Shy1 can occur independently of Cox1, indicating that this complex is not a pure dissociation product from COA complexes. Moreover, Cox15 is able to bind to Coa1 and Coa3 independently of Cox1 or Shy1.
DISCUSSION
Shy1/SURF1 is a conserved protein that has been implicated in Cox1 maturation due to its association with early assembly intermediates of cytochrome c oxidase (11, 12, 27, 46). Recent work on SURF1 from Paracoccus denitrificans provided evidence for a heme a-binding activity and defined conserved histidine residues that contribute to the observed heme a association. This observation led to the hypothesis that Shy1/SURF1 could act as a mobile heme a shuttle between the heme a synthase and Cox1 (28). In contrast to this report, mutagenesis of the corresponding conserved histidines of yeast Shy1 does not affect protein function, indicating that if Shy1 binds heme a in yeast mitochondria, this activity is not required for cytochrome c oxidase assembly or functionality (26). Here we find that Shy1 is in complex with the heme a synthase, Cox15, indicating a close functional link for Shy1 between heme a synthesis and cofactor incorporation into Cox1 (Fig. 6). In addition, Cox15 was found in assembly intermediates of Cox1, containing early assembly factors such as Coa3, Cox14, and Coa1. This finding is especially interesting in light of the phenotype of the shy1Δ mutant, since these mitochondria still display residual cytochrome c oxidase activity, which requires proper heme a insertion (12, 13, 26, 27). Similarly, Leigh syndrome patients bearing SURF1 mutations that result in the absence of the protein or SURF1 knockout mice display small amounts of cytochrome c oxidase activity (23, 24, 47, 48). Thus, despite a lack of Shy1/SURF1, heme a incorporation into Cox1 can still occur, although with drastically reduced efficiency. The finding that Cox15 associates with Cox1 assembly intermediates might provide an explanation for heme a incorporation in the absence of Shy1. In agreement with this, we observed Shy1-independent association of Cox15 with assembly intermediates. Thus, even in the absence of Shy1, Cox15 is in complex with the heme a-receiving Cox1 protein.
Fig 6.
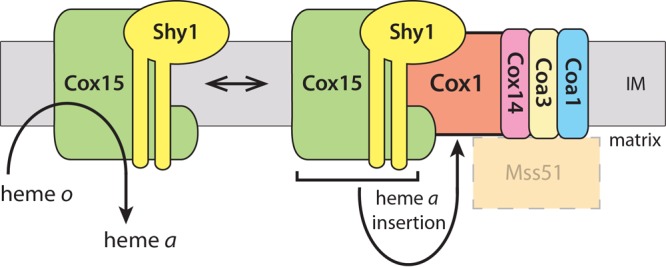
Heme a synthase Cox15 forms complexes with Shy1 and associates with cytochrome c oxidase assembly intermediates. Cox15 is required for hydroxylation of heme o to form heme a, an essential prosthetic group of COX. Cox15 is also found to be a component of Cox1 assembly intermediates together with the assembly factors Coa3, Cox14, Coa1, and Shy1, the potential stage of heme insertion into Cox1. Mss51 associates with COA complexes; however, the presence of Cox15 and Mss51 in a common complex was not addressed. IM, inner mitochondrial membrane.
In contrast to Shy1, the absence of functional Cox15 in yeast eliminates active cytochrome c oxidase (49) (see also Fig. 3D). The observed association of Cox15 with COA intermediates might indicate an additional chaperone-like function for the protein. The cytochrome c oxidase deficiency observed in the absence of Cox15 can be attributed to the lack of heme a production and incorporation. Nevertheless, an enzymatically compromised Cox15 is still able to associate with Cox1 assembly intermediates, indicating that complex formation is not linked to enzyme activity. Moreover, wild-type Cox15 associates with intermediates in the absence of the heme a precursor heme o. These findings could indicate a Cox15 function, which is independent of its role in heme a formation. Surprisingly, the association of both Shy1 and Cox15 to Cox1 assembly intermediates is independent of heme a or its precursor heme o.
The observation, that Shy1 and Cox15 are present in COA complexes suggests that the final step of heme a synthesis could be spatially closely associated with its insertion into the assembling enzyme and might take place (almost simultaneously) in the Cox1 assembly intermediates. This scenario also explains why mitochondria deficient in subsequent assembly steps, such as in cox2− mitochondria, display wild-type-like association of Shy1 and Cox15 to assembling Cox1, while COA components associated with Cox1 accumulate (Fig. 5C) (6, 12). This finding indicates that the heme a insertion into Cox1 occurs normally in these mutants but that the resulting intermediates require subsequent shielding by COA components to protect the cell from the generation of reactive oxygen species (15).
We find that Shy1 and Cox15 are present in common protein complexes that can be separated by BN-PAGE. The association of Cox15 with the COA complexes seems to be substoichiometric, but the distinct Shy1 and Cox15 complexes, of 170 and 220 kDa, suggest that both proteins form heterooligomers and cooperate tightly in their functions. However, the interaction of COA with Shy1 is lost in the absence of Cox1, whereas Cox15 can still associate with Coa3. In this context, it is tempting to hypothesize that the association of Shy1 with a productive Cox1 assembly intermediate might prevent premature progression of the Cox1 intermediate until Cox15 has inserted newly synthetized heme a. In the absence of Shy1, premature progression would generate a heme-deficient and hence malfunctioning cytochrome c oxidase that is degraded by the mitochondrial quality control system, leaving only a fraction of active enzyme behind. This explains why an increase in Cox1 synthesis, such as during Mss51 overexpression, can suppress the absence of Shy1 (13), since it raises the probability of generation of productive heme-inserted intermediates by Cox15, which functions as a heme a synthase and insertase during cytochrome c oxidase assembly.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to D. Winge for helpful discussions and S. Callegari for critical reading of the manuscript. We thank M. Raabe for expert technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft, SFB860, the Göttingen Graduate School for Neurosciences and Molecular Biosciences (DFG grant GSC 226/1 and grant GSC 226/2) (to B.B.) and the Max Planck Society (to H.U. and P.R.).
We declare that we have no competing financial interests.
Footnotes
Published ahead of print 26 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00747-13.
REFERENCES
- 1.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272:1136–1144 [DOI] [PubMed] [Google Scholar]
- 2.Mick DU, Fox TD, Rehling P. 2011. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12:14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carr HS, Winge DR. 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36:309–316 [DOI] [PubMed] [Google Scholar]
- 4.Soto IC, Fontanesi F, Liu J, Barrientos A. 2012. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta 1817:883–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrmann JM, Woellhaf MW, Bonnefoy N. 2013. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim. Biophys. Acta 1833:286–294 [DOI] [PubMed] [Google Scholar]
- 6.Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. 2010. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 191:141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontanesi F, Clemente P, Barrientos A. 2011. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 286:555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrientos A, Zambrano A, Tzagoloff A. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23:3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shingú-Vázquez M, Camacho-Villasana Y, Sandoval-Romero L, Butler CA, Fox TD, Pérez-Martínez X. 2010. The carboxyl-terminal end of Cox1 is required for feedback assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J. Biol. Chem. 285:34382–34389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox TD. 2012. Mitochondrial protein synthesis, import, and assembly. Genetics 192:1203–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. 2007. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26:4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. 2007. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26:4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrientos A, Korr D, Tzagoloff A. 2002. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 21:43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McStay GP, Su CH, Tzagoloff A. 2013. Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell 24:440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalimonchuk O, Bird A, Winge DR. 2007. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282:17442–17449 [DOI] [PubMed] [Google Scholar]
- 16.Moraes CT, Diaz F, Barrientos A. 2004. Defects in the biosynthesis of mitochondrial heme c and heme a in yeast and mammals. Biochim. Biophys. Acta 1659:153–159 [DOI] [PubMed] [Google Scholar]
- 17.Hederstedt L. 2012. Heme A biosynthesis. Biochim. Biophys. Acta 1817:920–927 [DOI] [PubMed] [Google Scholar]
- 18.Antonicka H, Leary SC, Guercin G-H, Agar JN, Horvath R, Kennaway NG, Harding CO, Jaksch M, Shoubridge EA. 2003. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12:2693–2702 [DOI] [PubMed] [Google Scholar]
- 19.Antonicka H, Mattman A, Carlson CG, Glerum DM, Hoffbuhr KC, Leary SC, Kennaway NG, Shoubridge EA. 2003. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 72:101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rotig A. 2000. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 9:1245–1249 [DOI] [PubMed] [Google Scholar]
- 21.Shoubridge EA. 2001. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106:46–52 [DOI] [PubMed] [Google Scholar]
- 22.Barrientos A, Barros MH, Valnot I, Rötig A, Rustin P, Tzagoloff A. 2002. Cytochrome oxidase in health and disease. Gene 286:53–63 [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert A, Newbold R, Wang J, Chevrette M, Brown G, Brown R, Shoubridge E. 1998. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20:337–343 [DOI] [PubMed] [Google Scholar]
- 24.Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviani M. 1998. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63:1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B, Rehling P. 2012. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 151:1528–1541 [DOI] [PubMed] [Google Scholar]
- 26.Bestwick M, Jeong M-Y, Khalimonchuk O, Kim H, Winge DR. 2010. Analysis of Leigh syndrome mutations in the yeast SURF1 homolog reveals a new member of the cytochrome oxidase assembly factor family. Mol. Cell. Biol. 30:4480–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nijtmans LG, Artal Sanz M, Bucko M, Farhoud MH, Feenstra M, Hakkaart GA, Zeviani M, Grivell LA. 2001. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 498:46–51 [DOI] [PubMed] [Google Scholar]
- 28.Bundschuh F, Hannappel A, Anderka O, Ludwig B. 2009. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. J. Biol. Chem. 284:25735–25741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Netter P, Carignani G, Jacq C, Groudinsky O, Clavilier L, Slonimski PP. 1982. The cytochrome oxidase subunit I split gene in Saccharomyces cerevisiae: genetic and physical studies of the mtDNA segment encompassing the “cytochrome b-homologous” intron. Mol. Gen. Genet. 188:51–59 [DOI] [PubMed] [Google Scholar]
- 30.Sikorski R, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972 [DOI] [PubMed] [Google Scholar]
- 32.Meisinger C, Pfanner N, Truscott K. 2006. Isolation of yeast mitochondria. Methods Mol. Biol. 313:33–39 [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860 [DOI] [PubMed] [Google Scholar]
- 34.Thakar K, Karaca S, Port SA, Urlaub H, Kehlenbach RH. 2013. Identification of CRM1-dependent nuclear export cargos using quantitative mass spectrometry. Mol. Cell. Proteomics 12:664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- 36.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805 [DOI] [PubMed] [Google Scholar]
- 37.Nikolov M, Stützer A, Mosch K, Krasauskas A, Soeroes S, Stark H, Urlaub H, Fischle W. 2011. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol. Cell. Proteomics 10:M110.005371. 10.1074/mcp.M110.005371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dekker P, Martin F, Maarse A, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinhold R, Bareth B, Balleininger M, Wissel M, Rehling P, Mick DU. 2011. Mimicking a SURF1 allele reveals uncoupling of cytochrome c oxidase assembly from translational regulation in yeast. Hum. Mol. Genet. 20:2379–2393 [DOI] [PubMed] [Google Scholar]
- 40.Vukotic M, Oeljeklaus S, Wiese S, Vögtle F-N, Meisinger C, Meyer HE, Zieseniss A, Katschinski DM, Jans DC, Jakobs S, Warscheid B, Rehling P, Deckers M. 2012. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 15:336–347 [DOI] [PubMed] [Google Scholar]
- 41.Bestwick M, Khalimonchuk O, Pierrel F, Winge DR. 2010. The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30:172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khalimonchuk O, Kim H, Watts T, Pérez-Martínez X, Winge DR. 2012. Oligomerization of heme o synthase in cytochrome oxidase biogenesis is mediated by cytochrome oxidase assembly factor Coa2. J. Biol. Chem. 287:26715–26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soto IC, Fontanesi F, Myers RS, Hamel P, Barrientos A. 2012. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metab. 16:801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hederstedt L, Lewin A, Throne-Holst M. 2005. Heme A synthase enzyme functions dissected by mutagenesis of Bacillus subtilis CtaA. J. Bacteriol. 187:8361–8369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nobrega MP, Nobrega FG, Tzagoloff A. 1990. COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. J. Biol. Chem. 265:14220–14226 [PubMed] [Google Scholar]
- 46.Barrientos A, Gouget K, Horn D, Soto IC, Fontanesi F. 2009. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta 1793:97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coenen M, van den Heuvel L, Nijtmans L, Morava E, Marquardt I, Girschick H, Trijbels F, Grivell L, Smeitink J. 1999. SURFEIT-1 gene analysis and two-dimensional blue native gel electrophoresis in cytochrome c oxidase deficiency. Biochem. Biophys. Res. Commun. 265:339–344 [DOI] [PubMed] [Google Scholar]
- 48.Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. 2007. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum. Mol. Genet. 16:431–444 [DOI] [PubMed] [Google Scholar]
- 49.Glerum DM, Muroff I, Jin C, Tzagoloff A. 1997. COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J. Biol. Chem. 272:19088–19094 [DOI] [PubMed] [Google Scholar]
- 50.Kruszewska A, Szczesniak B, Claisse M. 1980. Recombinational analysis of OXI1 mutants and preliminary analysis of their translational products in S. cerevisiae. Curr. Genet. 2:45–51 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.