Abstract
A special group of mitochondrial outer membrane proteins spans the membrane once, exposing soluble domains to both sides of the membrane. These proteins are synthesized in the cytosol and then inserted into the membrane by an unknown mechanism. To identify proteins that are involved in the biogenesis of the single-span model protein Mim1, we performed a high-throughput screen in yeast. Two interesting candidates were the cytosolic cochaperone Djp1 and the mitochondrial import receptor Tom70. Our results indeed demonstrate a direct interaction of newly synthesized Mim1 molecules with Tom70. We further observed lower steady-state levels of Mim1 in mitochondria from djp1Δ and tom70 tom71Δ cells and massive mislocalization of overexpressed GFP-Mim1 to the endoplasmic reticulum in the absence of Djp1. Importantly, these phenotypes were observed specifically for the deletion of DJP1 and were not detected in mutant cells lacking any of the other cytosolic cochaperones of the Hsp40 family. Furthermore, the djp1Δ tom70Δ tom71Δ triple deletion resulted in a severe synthetic sick/lethal growth phenotype. Taking our results together, we identified Tom70 and Djp1 as crucial players in the biogenesis of Mim1. Moreover, the involvement of Djp1 provides a unique case of specificity between a cochaperone and its substrate protein.
INTRODUCTION
Mitochondria harbor between 800 (in budding yeast) and 1,500 (in mammals) different proteins. In cells where it has been tested, such as the fungi Saccharomyces cerevisiae and Neurospora crassa, about 5% (∼40 different proteins) of the mitochondrial proteome resides in the mitochondrial outer membrane (MOM) (1, 2). These MOM proteins include a diverse set of enzymes, components of protein import machineries, pore-forming elements, and mitochondrial fusion- and fission-mediating proteins. Thus, the outer membrane plays a crucial role in the biogenesis, inheritance, and dynamics of the organelle. All of these outer membrane proteins are encoded by nuclear genes and synthesized on cytosolic ribosomes. Hence, they have to bear appropriate signals that ensure both their correct import into the organelle and their ability to acquire the desired topologies in the lipid bilayer. None of the known MOM proteins contain a canonical cleavable N-terminal presequence, and their targeting and sorting signals seem to be internal and noncleavable. Despite their well-recognized importance, the diverse molecular mechanisms by which MOM proteins are specifically targeted to the organelle and inserted into the target membrane remain incompletely defined.
A special class of proteins of the outer membrane comprises those that contain a single transmembrane segment and assume an orientation where the N-terminal domain is in the cytosol and the C-terminal part is facing the mitochondrial intermembrane space (IMS). Known members of this family in fungi include Mim1, Mim2, Tom22, and Atg32 (3–7). The mechanism by which these proteins are integrated into the outer membrane is largely unknown. Tom22, the only protein from this group whose import mechanism has been studied, was reported to require for its own biogenesis Tom import receptors as well as the TOB/SAM complex and the MOM protein Mdm10 (8, 9). However, as Tom22 is a core component of the TOM complex, its biogenesis mechanism probably reflects a specific case and does not provide a general paradigm for other proteins from this group. Previous studies have shown that the cytosolic domains of Tom22 and Mim1 are not required for their targeting to and insertion into the MOM (8, 10, 11). Following these findings, it has been assumed that the transmembrane segment and its flanking regions serve as a targeting signal to the organelle. However, which factors are involved in the biogenesis process of these proteins, what part of the substrate protein they bind, and where those factors are located are questions waiting to be answered.
Due to their synthesis in the cytosol, MOM proteins with a single membrane-spanning segment, like other hydrophobic mitochondrial precursor proteins, present the cell with the major task of keeping such mitochondrial precursor proteins upon their synthesis in an import-competent cytosolic soluble form. Accordingly, many studies reported on the involvement of cytosolic chaperones and other cofactors in mitochondrial import. Such cytosolic factors have been shown to include Hsp70s, their Hsp40 cochaperones, nascent polypeptide-associated complex (NAC), ribosome-associated complex (RAC), and mitochondrial import stimulation factor (MSF) (reviewed in references 12 and 13). For example, it was shown that presequence peptides can bind in vitro to yeast cytosolic Hsp70 (Ssa1) (14).
Although cytosolic chaperones are clearly involved in the import of precursor proteins into mitochondria, the specificity of this process is still poorly understood. Convincing evidence for a direct cooperation between Hsp70, Hsp90, and the import receptor Tom70 has been presented only for the family of mitochondrial metabolite carriers (15). It is unknown whether the chaperones only protect their substrate proteins from aggregation or if they also participate in the targeting to the MOM. Additionally, the determinants guiding the binding are not identified yet. Similarly unclear is the role of the cochaperones from the Hsp40 family. Although the yeast Hsp40 protein Ydj1 was shown to play an undefined role in protein import into mitochondria (16), a specific role for a cytosolic J protein in modulating the import of a subset of mitochondrial precursor proteins was not reported.
In the present study, we used a chimeric protein, Ura3-Mim1-degron, as a probe for correct membrane insertion of the model single-span protein Mim1. We systematically scanned a collection containing mutants in every yeast gene and searched for candidates in which the degron did not reach its anticipated location in the IMS; therefore, it was exposed to the cytosol. In these mutants, the Ura3-Mim1-degron fusion protein was degraded, creating a requirement for uracil for normal growth. The results of this screen and further biochemical analyses demonstrate a specific requirement for the cytosolic cochaperone Djp1 and no other cytosolic Hsp40 in the biogenesis of such single-span proteins of the MOM. This is the first indication for an involvement of Djp1 in the import of mitochondrial proteins, although the protein was reported to play an indefinite role in the biogenesis of peroxisomes (17). We further show that Djp1 works with Hsp70 to enable targeting through the Tom70 receptor. Collectively, our results highlight the essential role of Hsp40 in substrate matching for their Hsp70 chaperone partners and provide a unique case of specificity between a cochaperone and its substrate protein.
MATERIALS AND METHODS
Construction of Mim1 variants and yeast strains.
Unless stated otherwise, yeast strains in this study are based on the BY4741 laboratory strain. The tom70Δ tom71Δ strain was kindly provided by J. Shaw and K. Okamoto. Strains used in this study are listed in Table 1.
Table 1.
List of yeast strains used in this study
| Name | Mating type | Genetic background | Source or reference |
|---|---|---|---|
| BY4741 | MATa | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 18 |
| YMS721 | MATα | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 can1Δ::STE2pr-spHIS5 lyp1Δ::STE3pr-LEU2 | 19 |
| YSNK01 | MATα | YMS721 his3Δ1 leu2Δ0 met15Δ0 ura3Δ::natR TEF2pr-URA3-MIM1-SL17-ADH1term can1Δ::STE2pr-spHIS5 lyp1Δ::STE3pr-LEU2 | This study |
| YMS1258 | MATa | BY4741 NatR::ADHp-GFP-Mim1 | This study |
| YMS1305 | MATa | BY4741 NatR::ADHp-GFP-Mim1 djp1Δ::KANR | This study |
| YMS1273 | MATa | BY4741 KAN::Gal1p-Djp1 NatR::ADHp-GFP-Mim1 | This study |
| YMS1306 | MATa | BY4741 NatR::ADHp-GFP-Mim1 apj1Δ::KANR | This study |
| YMS1307 | MATa | BY4741 NatR::ADHp-GFP-Mim1 zuo1Δ::KANR | This study |
| YMS1304 | MATa | BY4741 NatR::ADHp-GFP-Mim1 xdj1Δ::KANR | This study |
| YMS1303 | MATa | BY4741 NatR::ADHp-GFP-Mim1 caj1Δ::KANR | This study |
| YMS1302 | MATa | BY4741 NatR::ADHp-GFP-Mim1 jid1Δ::KANR | This study |
| YMS1301 | MATa | BY4741 NatR::ADHp-GFP-Mim1 jem1Δ::KANR | This study |
| YMS1300 | MATa | BY4741 NatR::ADHp-GFP-Mim1 jjj3Δ::KANR | This study |
| YMS1299 | MATa | BY4741 NatR::ADHp-GFP-Mim1 jjj2Δ::KANR | This study |
| YMS1298 | MATa | BY4741 NatR::ADHp-GFP-Mim1 jjj1Δ::KANR | This study |
| YMS1387 | MATa | BY4741 NatR::ADHp-GFP-Mim1 hlj1Δ::KANR | This study |
| YMS1388 | MATa | BY4741 NatR::ADHp-GFP-Mim1 swa2Δ::KANR | This study |
| SUB62 | MATa | lys2-801 leu2-3 2-112 ura3-52 his3-Δ200 trp1-1amber | 20 |
| Lee1 | MATa | SUB62 lys2-801 leu2-3 2-112 ura3-52 his3-Δ200 trp1-1amber ubc6::HIS3 ubc7::TRP1 | 20 |
| JSY7452 | MATα | ade2-1 leu2-3 his15,15 trp1-1 ura3-1 can1-100 | 21 |
| tom70Δ tom71Δ (JSY8283) | MATα | JSY7254 tom70::TRP1 tom71::HIS3 | 21 |
| YPH499 | MATa | ade2-101 his3-Δ200 trp1-Δ63 ura3-52 lys2-801 | 22 |
| mas37Δ | MATa | YPH499 mas37::HIS3 | 23 |
| mim1Δ | MATa | YPH499 mim1::HIS3 | 10 |
| GAL10-His8-TOB55 | MATa | YPH499 tob55::HIS3-pGAL-His8-TOB55 | 24 |
For cloning of degron constructs, URA3 was amplified from pRS426, MIM1 from pGEM4-Mim1s.c., and the SL17 degron from pGEMT-SL17. Inserts were sequentially assembled into the yeast expression vector pYX142. The resulting URA3-MIM1-SL17 sequence was amplified from this vector and cloned into pFA6a-TEF2pr-eGFP-ADH1term-NATMX4 so that it replaced the enhanced green fluorescent protein (EGFP) fragment. For the construction of the YSNK01 strain, the DNA fragment from pFA6a-TEF2pr-URA3-MIM1-SL17-ADH1term-NATMX4 was amplified by PCR. The primers were designed to flank the cassette to be integrated with 40 bp of homology each to regions in the 5′ and 3′ sequences of the URA3 locus. The PCR product was transformed into a synthetic genetic array (SGA)-compatible strain (YMS721), and positive colonies were selected on yeast extract-peptone-dextrose plus ClonNAT (Nourseothricin) plates and verified by PCR.
The functionality of the various Mim1 variants was monitored by their capacity to complement the phenotype of mim1Δ cells in the previously described deletion strain (10). All J protein deletion strains were verified by PCR for the lack of the corresponding Hsp40.
Yeast library construction and screening.
To introduce the TEF2pr-URA3-MIM1-SL17-ADH1term-NATMX4 sequence into the yeast deletion collection, we used the SGA technique. The SGA technique allows efficient introduction of a trait (mutation or marker) into systematic yeast libraries. SGA was performed as previously described (25–27), employing the BY4741 strain that was used as the background strain for the yeast deletion and hypomorphic allele libraries (19, 28). Briefly, using a RoToR benchtop colony arrayer (Singer Instruments, United Kingdom) to manipulate libraries in high-density formats (384 or 1536 colonies per plate), haploid strains from opposing mating types, each harboring a different genomic alteration, were mated on rich medium plates. Diploid cells were selected on plates containing all selection markers found on both parent haploid strains. Sporulation was then induced by transferring cells to nitrogen starvation plates. Haploid cells containing all desired mutations were selected for by transferring cells to plates containing all selection markers alongside the toxic amino acid derivatives canavanine and thialysine to select against remaining diploids. The new yeast libraries, in which each colony harbored the URA3-MIM1-SL17 locus on the genetic background of a single mutation, were spotted on synthetic medium in the presence or absence of uracil. Colony size was then quantified using the Balony free software for the analysis of images of plates containing arrays of yeast (the software package is maintained by Barry Young at the University of British Columbia, Vancouver, Canada).
Fluorescence microscopy.
Microscopy was performed using an Olympus IX71 microscope controlled by the Delta Vision SoftWoRx 3.5.1 software with oil lenses at magnifications of ×60 or ×100. Images were captured by a Photometrics Coolsnap HQ camera with excitation at 490/20 nm and emission at 528/38 nm (GFP) or excitation at 555/28 nm and emission at 617/73 nm (mCherry/red fluorescent protein [RFP]). Images were transferred to Adobe Photoshop CS3 for slight contrast and brightness adjustments.
Drop-dilution assay.
Cells were grown to logarithmic phase in appropriate liquid media, harvested by centrifugation, and resuspended in sterile water to an optical density at 600 nm (OD600) of 1. Cell suspensions were serially diluted 5-fold in water, and 5-μl aliquots of the various suspensions were spotted on the appropriate plates, which were then incubated at different temperatures.
Peptide scan assay.
Peptides corresponding to Mim1 with a length of 15 amino acids each were synthesized on a cellulose membrane as described previously (29, 30). The amino acid sequence was shifted by 3 amino acids from one spot to the next. The membrane was incubated in methanol for 1 min at room temperature and subsequently washed twice for 5 min in washing buffer (100 mM KCl, 30 mM Tris-HCl, pH 7.6). The membrane next was incubated in 150 nM solution of purified recombinant Djp1his dissolved in binding buffer (30 mM Tris-HCl, 0.5% bovine serum albumin [BSA], 0.05% [vol/vol] Tween 20, 5% [wt/vol] sucrose, 100 mM KCl, pH 7.6). The incubation occurred at 4°C in the first hour, followed by 30 min at room temperature. The membrane was washed for 3 min with washing buffer at room temperature with gentle shaking, after which it was blotted on a nitrocellulose membrane by applying 1 mA per cm2. Bound Djp1 was determined through incubation with anti-Djp1 antibody (kindly provided by A. van der Zand), followed by visualization via the enhanced chemiluminescence (ECL) method. The intensity of the signals was quantified with AIDA software.
Biochemical procedures.
Mitochondria were isolated from yeast cells by differential centrifugation as previously described (31). Subcellular fractionation was performed according to published procedures (32). Protein samples were analyzed by SDS-PAGE and blotting to nitrocellulose membranes, followed by Ponceau staining, autoradiography, or incubation with antibodies and visualization by the ECL method. The intensity of the observed bands was quantified with AIDA software. Unless stated otherwise, each presented experiment represents at least three independent repetitions.
Blue native PAGE.
Mitochondria were lysed in 50 μl digitonin buffer (1% digitonin, 20 mM Tris-HCl, 0.1 mM EDTA, 50 mM NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], pH 7.4). After incubation for 15 min at 4°C and a clarifying spin (30,000 × g, 15 min, 2°C), 5 μl sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM Bis-Tris, 500 mM 6-aminocaproic acid, pH 7.0) was added, and the mixture was analyzed by electrophoresis in a 6 to 13% polyacrylamide gradient blue native gel (33). Gels were blotted to polyvinylidene fluoride membranes, and proteins were further analyzed by immunodecoration.
Coimmunoprecipitation.
Aliquots of protein G-Sepharose beads were washed with water and equilibrated in immunoprecipitation (IP) buffer (10 mM Tris, 300 mM NaCl, 1% Triton X-100, pH 7.5). One aliquot was left intact, whereas the other was incubated in IP buffer at 4°C for 1 h with antibody against Hsc70. In the following step, unspecific binding sites on the beads were blocked by incubation with a solution of 1% (wt/vol) BSA in IP buffer. Rabbit reticulocyte lysate (TNT; Promega) containing newly synthesized radiolabeled Mim1 and ADP (2 mM) then were added to both aliquots and incubated with the beads for 4 h at 4°C. Bound material was eluted by a 10-min incubation at 37°C in Laemmli buffer without β-mercaptoethanol but supplemented with 0.05% (vol/vol) H2O2. The beads were spun down and proteins in the supernatant were analyzed by SDS-PAGE, followed by blotting to the nitrocellulose membrane. The membrane was first analyzed by autoradiography and then by immunodecoration with an antibody against Hsc70.
Pulldown of GST-tagged proteins.
Aliquots of glutathione-Sepharose beads were equilibrated in basic buffer (20 mM HEPES, 100 mM NaCl, 1.5 mM MgCl2, pH 7.2), after which they were left untreated or were incubated with 200 μg of glutathione S-transferase (GST) or GST-tagged proteins dissolved in basic buffer. Unspecific binding sites on the beads next were blocked by incubation with rabbit reticulocyte lysate. Rabbit reticulocyte lysate with radiolabeled Mim1 then was incubated with the beads for 1 h at 4°C. Bound material was eluted by a 10-min incubation at 95°C in Laemmli buffer. The beads were spun down, and the proteins contained in the supernatant were separated by SDS-PAGE and further analyzed by blotting to nitrocellulose membrane, Ponceau staining, and autoradiography.
RESULTS
Ura3-Mim1-degron can be used to monitor the insertion of Mim1 in vivo.
To identify proteins that are involved in the biogenesis of single-span outer membrane proteins, we performed a high-throughput screen in yeast. As a probe we used a hybrid protein that is based on the 113-residue protein Mim1. This hybrid protein contains the Ura3 enzyme fused to the cytosol-facing N terminus of Mim1 and the degron sequence SL17 (34) attached to the C-terminal domain of the protein that faces the IMS. URA3 encodes the cytosolic enzyme orotidine-5′-phosphate (OMP) decarboxylase (267 amino acid residues), which catalyzes one of the steps in uridine-5′-triphosphate (UTP) synthesis. SL17 is a degron sequence of 50 amino acids whose ubiquitination is facilitated by the E2-conjugating enzymes Ubc6 and Ubc7. As a consequence, SL17 marks fusion proteins for degradation via the proteasome (34). SL17 was used before to specifically deplete the cytosolic pool of the dual-localized protein aconitase (20).
Ubc6 and Ubc7 cannot access the inner parts of mitochondria, as they are localized only to the cytosol and the cytosolic surface of the ER. Thus, SL17 fusion proteins are protected from degradation if the degron is localized to the IMS or mitochondrial matrix. Our rationale was that proper insertion of Mim1 would result in the correct orientation of the degron in the IMS, where it will be shielded from degradation (Fig. 1A). Such a correct insertion will enable the cytosol-facing Ura3 enzyme to complement the uracil auxotrophy of the yeast strains in the mutants' collection. However, if Mim1 insertion is abolished, then the degron region will be exposed to the cytosol, where it will cause degradation of the entire chimeric protein. Consequently, the cells will become auxotrophic for uracil (Fig. 1A).
Fig 1.

In vivo assay to monitor the biogenesis of Mim1. (A) Schematic representation of the various constructs used to determine the validity of our screening approach. The potential of these proteins to support growth on synthetic medium without uracil is indicated. (B) Wild-type or ubc6Δ ubc7Δ cells transformed with the indicated constructs were analyzed at 30°C by drop-dilution assay on synthetic medium lacking leucine and uracil. (C) Wild-type or mim1Δ cells transformed with an empty plasmid (Ø) or mim1Δ cells expressing the indicated constructs were analyzed at 30°C by drop-dilution assay on rich medium containing either glucose (YPD) or glycerol (YPG).
To validate the principles of the screen, a construct encoding the full-length fusion protein Ura3-Mim1-SL17, as well as constructs encoding Ura3, Ura3-Mim1, or Ura3-SL17, were introduced into the yeast expression vector pYX142, which carries the LEU selection cassette. These constructs were transformed into the wild-type (wt) yeast strain (SUB62) that was previously used for such assays (20), and the capacity of the transformed cells to grow in the absence of leucine and uracil was tested. As we hypothesized, Ura3 alone, Ura3-Mim1, and Ura3-Mim1-SL17 supported growth under these conditions, demonstrating that Ura3 can function even when localized to the mitochondrial surface. In contrast, Ura3-SL17, which is expected to be constantly degraded, indeed did not support growth. The deletion of Ubc6 and Ubc7 restored the growth capacity of the Ura3-SL17-harboring strain, demonstrating that, as anticipated, degradation was indeed cytosolic (Fig. 1B). The functionality of the fusion proteins was tested by their ability to rescue the phenotype of mim1Δ cells. Since all examined variants could rescue the growth phenotype of mim1Δ cells, it seems that these Mim1 variants are correctly localized in the MOM and are functional (Fig. 1C).
A systematic screen uncovers Djp1 and Tom70 as mediators of Mim1 biogenesis.
To systematically screen the fusion protein on the background of mutations in all yeast genes, we integrated the Ura3-Mim1-SL17 construct into the URA3 locus of a strain that is compatible with synthetic genetic array (SGA) methodology (25–27). This query strain (YSNK01) was then crossed against two independent derivatives of the yeast deletion library harboring deletions in every nonessential yeast gene (28). Two derivative libraries were used in order to enable sieving out of cross-contaminated colonies that give false-positive hits. In addition, the query strain also was crossed against a decreased abundance by an mRNA perturbation (DAmP) library, which consists of ∼1,000 strains, each expressing a hypomorphic allele of an essential gene (19, 35). Following SGA, we retrieved haploids that harbored the URA3-Mim1-degron chimeric protein on the background of every mutation in a ura3Δ background. We then spotted the mutant strains on a synthetic medium in the presence or absence of uracil (schematically depicted in Fig. 2A). The initial plates (Fig. 2B, value 1) were replicated on new plates that were incubated for a further 2 days (Fig. 2B, value 2). The sizes of colonies on all plates were quantified, and mutants where the absence of uracil in the medium caused clear growth retardation were considered potential hits (Fig. 2B).
Fig 2.
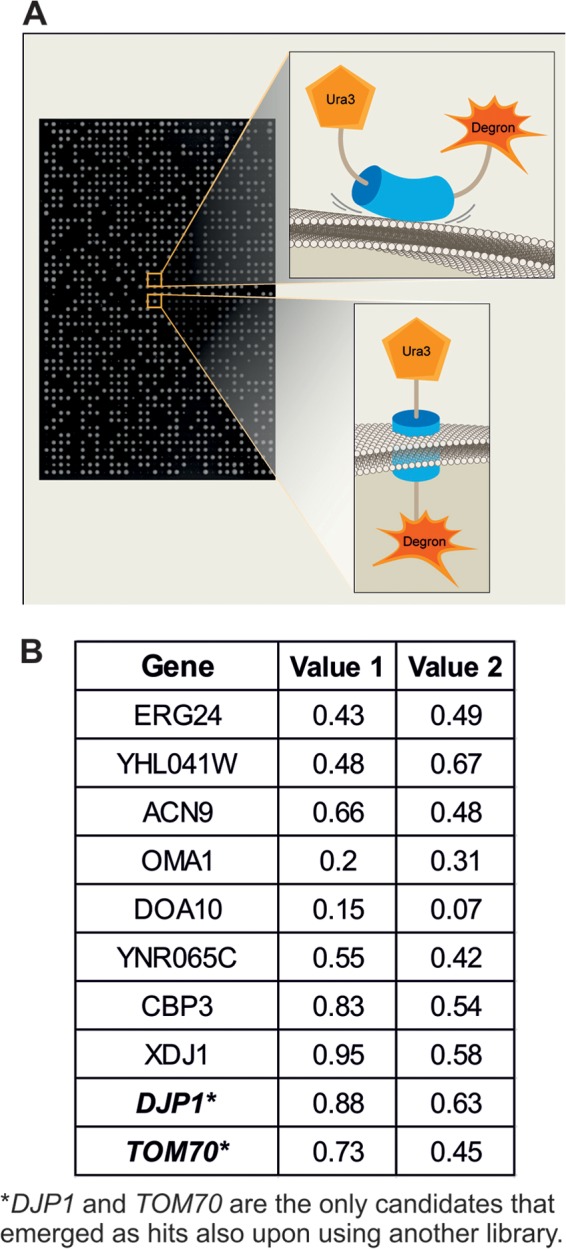
Systematic screen uncovers Djp1 and Tom70 as mediators of Mim1 biogenesis. (A) Schematic representation of either noninserted (top) or correctly inserted (bottom) Ura3-Mim1-degron fusion protein. An example of the corresponding growth of colonies harboring these proteins on medium lacking uracil is presented. (B) Candidates obtained in the genetic high-throughput screen. The plates were analyzed for the first time after 2 days on the corresponding medium (value 1). The colonies then were replicated to new plates that were incubated for a further 2 days before their evaluation (value 2). The plates were scanned, and sizes of colonies were quantified. The values in the table represent the ratio of the sizes of colonies of the corresponding deletion strain on medium without uracil compared to their size on medium with uracil.
As expected, mutations in various genes (URA1, URA2, URA4, and URA5) whose protein products are required for the biosynthesis of uracil were identified as such hits. Several other mutant backgrounds displayed a reduced ability to grow in the absence of uracil. However, only the cytosolic J domain-containing (Hsp40) cochaperone Djp1 and the mitochondrial import receptor Tom70 were identified as high-confidence candidates using both versions of the deletion library (Fig. 2B). The identification of the tom70Δ strain in our screen is in line with our recent observation that tom70Δ tom71Δ cells harbor reduced steady-state levels of Mim1 (36).
Loss of Djp1 affects biogenesis of Mim1 and Mim2.
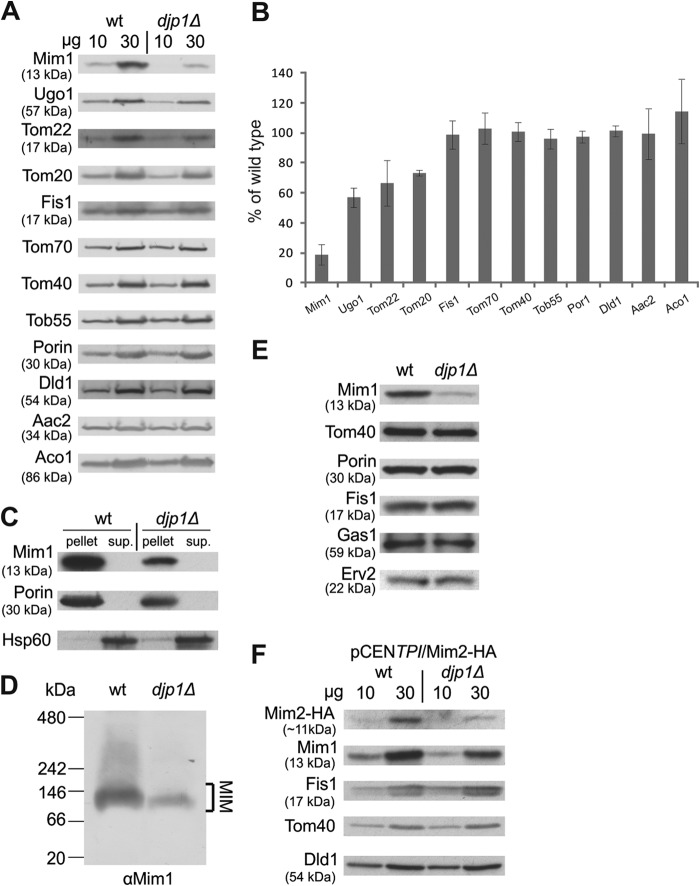
Since djp1Δ cells reproducibly emerged in our screen, we decided to investigate its role in Mim1 biogenesis. First, we isolated mitochondria from wild-type and djp1Δ cells and monitored the steady-state levels of various proteins known to follow different import pathways. The levels of Mim1 were dramatically reduced to ∼20% of those found in control cells (Fig. 3A and B). Of note, the levels of the outer membrane proteins Ugo1 and Tom20 (known substrates of Mim1) and Tom22 were moderately reduced (Fig. 3A and B). Importantly, the levels of other MOM proteins, like the tail-anchored protein Fis1, the signal-anchored protein Tom70, and the β-barrel proteins like Tom40, Tob55, and Porin, were not affected. Similarly, marker proteins for other mitochondrial compartments, like Dld1 (IMS), Aac2 (inner membrane), and aconitase (matrix), were observed in normal amounts. These observations suggest a specific effect of Djp1 on Mim1 biogenesis rather than one on mitochondrial import in general.
Fig 3.
Djp1 mediates the biogenesis of Mim1 and Mim2. (A) Mitochondria isolated from either wt or djp1Δ strains were analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated mitochondrial proteins. (B) The intensity of the bands in three independent experiments was quantified, and the amounts of proteins in mutant mitochondria are expressed as mean percentages (±SD) of their levels in wt organelles. (C) Mitochondria isolated from either wt or djp1Δ strains were subjected to carbonate extraction. The pellet and the supernatant (sup.) fractions were analyzed by SDS-PAGE and immunodecoration with antibodies against Mim1 as well as the outer membrane protein Porin and the soluble matrix protein Hsp60. (D) Mitochondria isolated from either wt or djp1Δ strains were analyzed by blue native PAGE and immunodecoration with antibody against Mim1. The MIM complex is indicated. (E) Cell lysates were separated by centrifugation into total cellular compartments in the pellet and the cytosolic fraction in the supernatant. Pellets obtained from either wt or djp1Δ cells were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies. Tom40, Porin, and Fis1 are MOM proteins. Gas1 and Erv2 are marker proteins for the plasma membrane and ER, respectively. (F) Mitochondria isolated from either wt or djp1Δ strains overexpressing Mim2-HA were analyzed by SDS-PAGE and immunodecoration with antibodies against the HA tag, Mim1, and the indicated mitochondrial proteins.
To test whether Djp1 is required for the membrane insertion per se, we performed alkaline extraction on mitochondrial membranes. Although the absence of Djp1 resulted in reduced levels of Mim1, the detected molecules all were in the membrane pellet fraction (Fig. 3C), suggesting that the cochaperone is not involved in the insertion into the membrane but most probably in targeting and/or chaperoning Mim1.
We next aimed to rule out that Djp1 is affecting Mim1 due to an unknown role of the former as part of the MIM complex that contains both Mim1 and Mim2 (7). To that end, we analyzed the MIM complex by blue native gels in the presence or absence of Djp1 and found that although the complex has reduced levels, its migration behavior is not altered (Fig. 3D). Thus, Djp1 does not affect the composition of the MIM complex.
The lower steady-state mitochondrial levels of Mim1 can result from either mistargeting or inability to retain the molecule in an import-competent state. Mistargeting would be displayed as a mislocalization to other cellular membranes, whereas loss of chaperoning should result in enhanced degradation of noninserted molecules. To address the first option, we separated by centrifugation total cellular compartments from the cytosolic fraction and analyzed this membrane-containing pellet by immunodecoration. Even when measuring Mim1 levels in such a pellet that represents all cellular compartments, djp1Δ cells were still significantly reduced in their Mim1 levels, demonstrating that under normal expression, the low levels in mitochondria do not result from another population of Mim1 molecules in other competing cellular membranes (Fig. 3E).
We then asked whether the role of Djp1 was restricted to Mim1 or is a more general factor for insertion of MOM proteins with a central single membrane-spanning segment. To that end, we analyzed the levels of the recently identified Mim2, which has a membrane topology (Ncyt-Cims [N terminus in the cytosol, C terminus in the mitochondrial intermembrane space]) very similar to that of Mim1 (7). Indeed, we found that the amounts of Mim2-HA expressed from an overexpression plasmid were reduced in cells lacking Djp1 relative to wt cells (Fig. 3F). These findings suggest that Mim2 is also a substrate of Djp1. The lower content of Mim2-HA in djp1Δ cells is probably not a secondary effect of the reduced levels of Mim1 in these cells, because Mim2-HA was observed in normal amounts in cells completely lacking Mim1 (7).
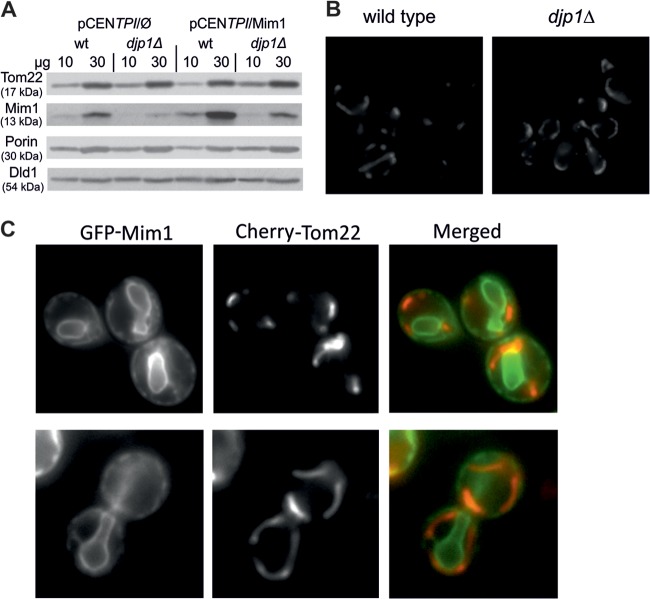
Since Atg32 and Tom22 also have membrane topology similar to that of Mim1, we monitored their levels and localization in cells lacking Djp1. Interestingly, the steady-state levels of Tom22 in cells deleted for DJP1 were similar to those in wt cells (Fig. 4A). The reduced amounts of Tom22 observed in Fig. 3A and B probably resulted from growth under conditions where the amounts of Mim1 were so dramatically reduced that it caused secondary effects, like the subsequently altered content of the Mim1 substrate Tom20. The latter protein is a known stabilizer and an interaction partner of Tom22; therefore, its absence is known to cause reduction in Tom22 content (3, 37). As we did not have antibodies against native Atg32, we decided to monitor the fluorescently tagged forms of both Atg32 and Tom22 to further investigate the effect of Djp1 on their biogenesis. The absence of the cochaperone did not alter the mitochondrial location of GFP-Atg32 and Cherry-Tom22, suggesting that both proteins are not bona fide substrates for Djp1 (Fig. 4B and C).
Fig 4.
(A) Wild-type (wt) and djp1Δ cells were transformed with either an empty plasmid (pCENTPI/Ø) or plasmid overexpressing Mim1 (pCENTPI/Mim1). Mitochondria isolated from these strains were analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated mitochondrial proteins. (B) Fluorescence images of wt or djp1Δ yeast cells expressing GFP-Atg32. (C) Fluorescence images of djp1Δ yeast cells expressing GFP-Mim1 and Cherry-Tom22.
The TOB complex is involved in the biogenesis of Mim1.
Previous studies have demonstrated that Mdm10 and the TOB complex are involved in the biogenesis of single-span MOM proteins like Tom22 and the small TOM subunits (9, 38). Therefore, although components of this pathway were not identified as hits in our screen, we next asked whether these factors are also required for the biogenesis of Mim1. To that end, we investigated the steady-state levels of Mim1 in strains mutated for MDM10, MAS37, and TOB55. The absence of Mdm10 did not affect the levels of Mim1 (Fig. 5A). In contrast, when cells lacking Mas37 were grown at 37°C to induce the mas37Δ phenotype, a moderate reduction in the amounts of steady-state Mim1 was observed (Fig. 5B). Tob55 is the central component of the TOB complex and an essential protein for the viability of yeast cells. Thus, to study its contribution, we utilized a strain where the expression of Tob55 is under the control of the inducible GAL10 promoter (24, 39). As expected, gradual depletion of Tob55 resulted in reduced levels of the β-barrel proteins Tom40 and Porin as well as in the amounts of Tom22 (Fig. 5C). Of note, similar to the aforementioned proteins and in parallel to the reduction of assembled TOM complex (Fig. 5C, lower), Mim1 levels were dramatically reduced after 21 h of depletion (Fig. 5C, upper). Hence, it appears that a functional TOB complex is required for proper biogenesis of Mim1, although it remains to be determined whether this effect is direct or indirect.
Fig 5.
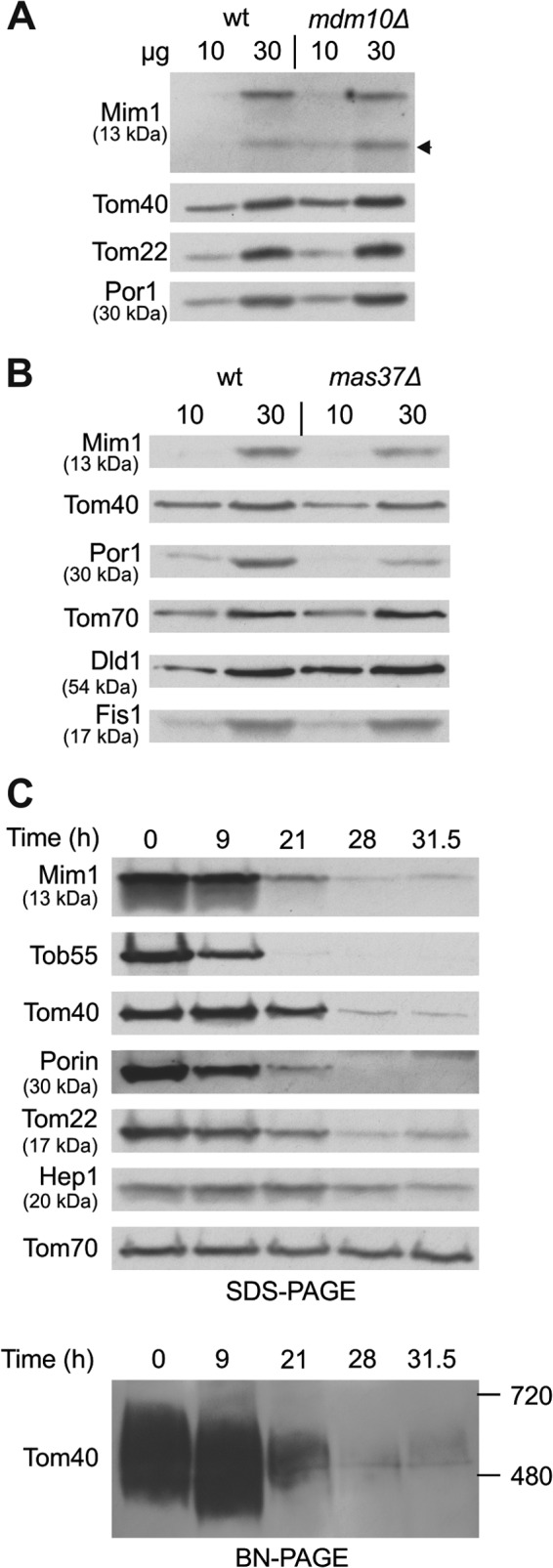
Mim1 requires TOB complex for its biogenesis. (A) Mitochondria were isolated from mdm10Δ and its corresponding wt strain. Mitochondrial proteins (10 and 30 μg) were analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated proteins. A proteolytic fragment of Mim1 is indicated with an arrow. (B) mas37Δ and its corresponding wt strain were grown to log phase at 24°C, after which the cultures were shifted to 37°C for 6 h. Mitochondria were isolated and analyzed as described for panel A. (C) Cells from a strain expressing Tob55 under the control of the GAL10 promoter were harvested at the indicated time points after a shift from galactose- to glucose-containing medium. Mitochondria were isolated and proteins were analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated proteins (upper). Alternatively, mitochondria were analyzed by blue native-PAGE (BN-PAGE) and immunodecoration with antibody against Tom40 (lower). The assembled TOM complex is shown.
Genetic analysis supports a role for Djp1 in mitochondrial biogenesis.
The absence of Mim1 results in a severe growth retardation phenotype (4, 5). In contrast, neither the deletion of Tom70 and its paralogue Tom71 nor the absence of Djp1 results in a major growth phenotype (Fig. 6A). However, when we created a triple deletion strain, we observed a clear synthetic genetic interaction that resulted in a severe growth retardation phenotype (Fig. 6A). The specificity of this genetic interaction is demonstrated by the fact that no synthetic phenotype is observed upon the deletion of TOM70 and TOM71 with another cytosolic cochaperone, XDJ1. This genetic interaction and the more severe phenotype on a nonfermentable carbon source (Fig. 6A, YPG), where fully functional mitochondria are required, underscore the importance of Djp1 in the biogenesis of the organelle. Of note, these genetic interactions cannot be directly correlated with the reduced amounts of Mim1 in the mutated cells. Whereas Mim1 content in mitochondria isolated from the triple deletion strain was almost similar to that in djp1Δ cells, other mitochondrial proteins, like ADP/ATP carrier or aconitase, were even more affected than they were in the single or double mutants (Fig. 6B). Thus, it seems that the severe growth phenotype of the triple deletion strain resulted from the additive effects of reduced Mim1 content together with compromised levels of other mitochondrial proteins, like the carrier family and/or matrix components.
Fig 6.
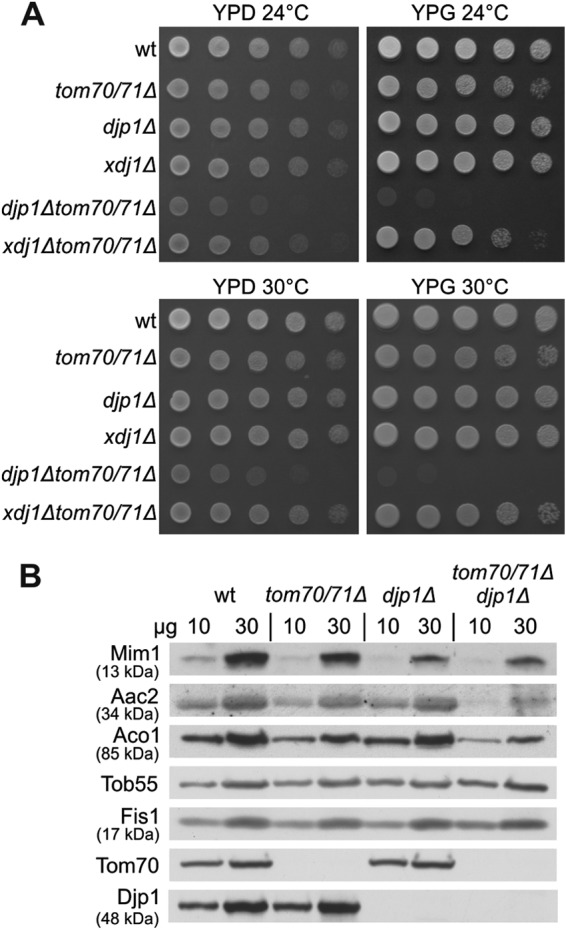
Synthetic sick genetic interaction between djp1Δ and tom70Δ tom71Δ strains. (A) Indicated strains were analyzed at either 24 or 30°C by drop-dilution assay on rich medium containing either glucose (YPD) or glycerol (YPG). (B) Mitochondria were isolated from the indicated strains, and mitochondrial proteins (10 and 30 μg) were analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated mitochondrial proteins. Aac2, ADP/ATP translocator (inner membrane); Aco1, aconitase (matrix). Tob55 and Fis1 are from the outer membrane.
Djp1 is a highly specific factor involved in targeting of single-span MOM proteins.
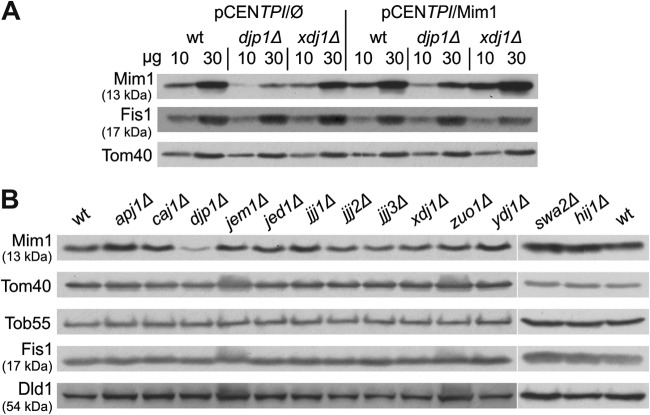
Djp1 is one of more than a dozen different DnaJ-like cochaperones in the yeast cytosol (40). In our original screen, we also found an additional DnaJ cochaperone, Xdj1, as a potential hit. To investigate whether the effect of Djp1 on Mim1 biogenesis was unique or if other Hsp40s can also aid in Mim1 insertion, we overexpressed Mim1 in cells lacking either Djp1 or Xdj1. Whereas the absence of Djp1 resulted in diminished amounts of mitochondrial Mim1, no such effect was observed in mitochondria from xdj1Δ cells (Fig. 7A). To further test for the specificity of Djp1 in Mim1 biogenesis, we employed verified mutant strains for all cytosolic DnaJ-like proteins. We next probed each strain for the steady-state levels of Mim1 in mitochondria. Strikingly, Djp1 was the only J protein whose absence significantly affected Mim1 biogenesis, demonstrating a unique dependence on this cochaperone (Fig. 7B).
Fig 7.
Involvement of Djp1 is unique among the cytosolic cochaperones. (A) Wild type (wt) or djp1Δ or xdj1Δ mutant cells were transformed with either empty plasmid (pCENTPI/Ø) or with plasmid overexpressing Mim1 (pCENTPI/Mim1). Mitochondria isolated from these strains were analyzed by SDS-PAGE and immunodecoration with antibodies against Mim1 and the indicated mitochondrial proteins. (B) Mitochondria isolated from either wt or deletion strains of 13 cytosolic Hsp40s were analyzed by SDS-PAGE and immunodecoration with antibodies against Mim1 and the other indicated mitochondrial proteins.
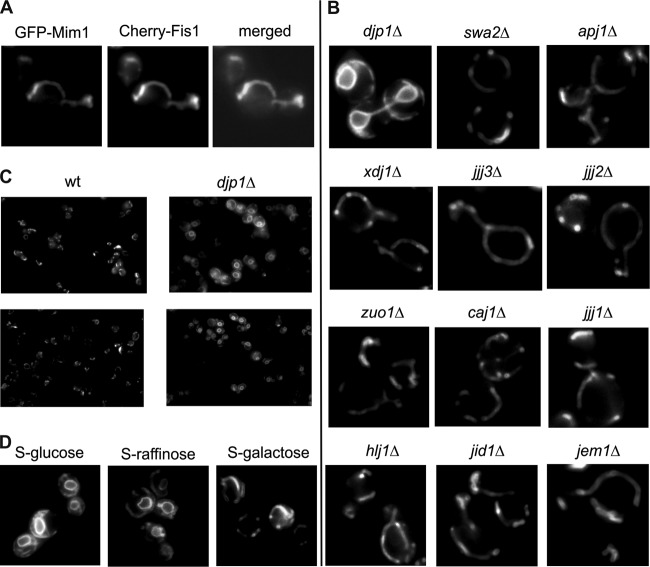
To substantiate this specificity, we constructed an N-terminal fusion of green fluorescent protein (GFP) with Mim1 (GFP-Mim1) and monitored the subcellular localization of the overexpressed protein by fluorescence microscopy. This construct stained mitochondrial structures in wild-type cells (Fig. 8A). In contrast, we observed a massive mislocalization of the overexpressed GFP-Mim1 to the endoplasmic reticulum (ER) in the absence of Djp1 (Fig. 8B and C). Importantly, such ER staining was not observed in cells lacking any of the other cytosolic cochaperones (Fig. 8B), revealing again that Djp1 uniquely contributes to the specific mitochondrial targeting of Mim1.
Fig 8.
Overexpressed GFP-Mim1 is mislocalized to the ER in djp1Δ cells. (A) Fluorescence images of wt yeast cells expressing GFP-Mim1 and Cherry-Fis1 (as a mitochondrial marker protein). (B) GFP-Mim1 was transformed into yeast cells deleted in each of the cytosolic Hsp40 cochaperones, and fluorescence images were taken. (C) Fluorescence images of wild-type or djp1Δ yeast cells expressing GFP-Mim1. (D) Yeast cells expressing Djp1 under the control of the GAL1 promoter were transformed with the GFP-Mim1 expressing vector. Cells were grown on synthetic medium containing glucose (to repress expression of Djp1), raffinose (to enable low levels of Djp1 expression), or galactose (to overexpress Djp1), and fluorescence images were taken.
To demonstrate the direct correlation between the level of Djp1 and the correct mitochondrial targeting of Mim1, we constructed a strain where the expression of Djp1 is under the control of the inducible GAL1 promoter and expressed GFP-Mim1 in these cells. Growth on glucose that represses Djp1 expression resulted in a clear mislocalization of the Mim1 variant to the ER. Growth on raffinose, which only partially represses the expression, resulted in a minor targeting phenotype, whereas growth on galactose, which facilitates high expression levels of Djp1, allowed full mitochondrial localization of Mim1 (Fig. 8D). Hence, these results clearly indicate that the low expression level of Djp1 is the sole cause for the biogenesis phenotype of Mim1 and underscores the specificity of the Djp1 requirement.
Import receptors, Djp1, and Hsp70 physically interact with Mim1.
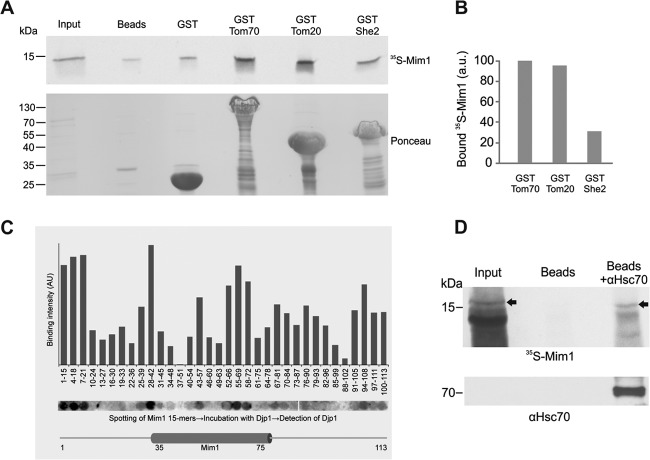
The second candidate protein, Tom70, being a mitochondrial import receptor, is a natural candidate to recognize precursor molecules of Mim1 upon their arrival at the surface of the organelle. To substantiate such an ability, we incubated radiolabeled Mim1 molecules newly synthesized in reticulocyte lysate with recombinant GST-tagged versions of the cytosolic domains of Tom70 and Tom20 or with GST fused to the unrelated protein She2 as a control. All GST-tagged proteins were expressed in Escherichia coli cells. Although there is a residual binding of Mim1 to the beads with GST alone or with the control protein She2, we observed more than 3-fold stronger binding in the case of GST-Tom70 or GST-Tom20 (Fig. 9A and B). These findings support the ability of Tom70 and Tom20 to bind Mim1 directly and are in line with previous reports suggesting an overlapping binding capacity of both receptor proteins (41, 42).
Fig 9.
Mim1 can physically bind Tom70, Tom20, Djp1, and Hsp70. (A) The cytosolic domain of Tom70 and Tom20 can recognize newly synthesized Mim1 molecules. Radiolabeled Mim1 was mixed with glutathione beads or with beads harboring GST or GST fused to either the cytosolic domain of mitochondrial proteins Tom70 (GST-Tom70) and Tom20 (GST-Tom20) or to the cytoplasmic protein She2 (GST-She2). The beads were washed, after which bound material was eluted with sample buffer. Aliquots of the input (5%) and bound material (100%) were analyzed by SDS-PAGE followed by Ponceau staining (lower panel) and autoradiography (upper panel). The samples with GST-tagged Tom70, Tom20, and She2 contain degradation products of the fusion proteins (Ponceau staining [lower panel]). (B) The intensity of the radioactive bands corresponding to 35S-Mim1 pulled down with the three different GST-tagged proteins was quantified and is presented as a percentage of the amount of protein bound by Tom70. (C) A peptide library on a cellulose membrane covering the entire sequence of Mim1 was incubated with purified Djp1his. Bound protein was detected with antibodies against Djp1, and binding was quantified by scanning densitometry of the spots. Different segments of Mim1 are displayed below the corresponding peptides. The putative transmembrane domain (amino acid residues 35 to 75) is indicated. (D) Mim1 physically interacts with Hsc70. Protein G-Sepharose beads were left untreated or were preincubated with anti-Hsc70 antibody before addition of rabbit reticulocyte lysate containing radiolabeled Mim1. The beads were washed and bound material was eluted. The major portion of the eluted material (85%) was analyzed via SDS-PAGE followed by autoradiography (upper panel), whereas the remaining 15% was analyzed by SDS-PAGE and immunodecorated with antibody against Hsc70 (lower panel). Arrows in the upper panel indicate the bands corresponding to radiolabeled Mim1. A very strong band at 14 kDa represents hemoglobin that is extremely abundant in the reticulocyte lysate. Input refers to 1% of the volume of lysate used in the pulldown samples.
We next asked whether there are distinct Djp1-binding sites within the Mim1 structure. To address this point, we synthesized a peptide library corresponding to the sequence of Mim1 consisting of 15mers with an overlap of 12 residues and spotted these peptides on a membrane. The peptides were scanned for their ability to bind recombinant Djp1 expressed in E. coli cells. The results indicate that the sequence of Mim1 contains several binding sites for Djp1, a binding behavior that is typical of chaperones and cochaperones. (Fig. 9C).
Members of the Hsp70 family are the common functional partners of Hsp40 proteins. Thus, we were interested in examining whether newly synthesized Mim1 molecules are indeed associated with cytosolic Hsp70. To that end, radiolabeled Mim1 synthesized in rabbit reticulocyte lysate was incubated with beads coupled to antibody against mammalian Hsc70. As expected, these antibodies could pull down Hsc70 molecules present in the reticulocyte lysate and, importantly, radiolabeled Mim1 (Fig. 9D). These findings demonstrate the ability of cytosolic Hsp70 to engage newly synthesized Mim1 molecules.
DISCUSSION
In this study, we performed a high-throughput screen designed to identify proteins involved in the biogenesis of the MOM protein Mim1. We found the cytosolic protein Djp1 and mitochondrial Tom70 to be the most reliable candidates in our screen. Indeed, reduced steady-state levels of Mim1 were observed in the absence of either Djp1 (this study) or Tom70 (36). We noticed that the biogenesis of Mim2, which has membrane topology similar to that of Mim1, is also affected by the absence of Djp1. Djp1 belongs to the Hsp40 family of molecular cochaperones, the members of which are known to regulate the activity of Hsp70s and have been suggested to confer the binding specificity to the complex (40, 43, 44). The other candidate protein, Tom70, is an import receptor for mitochondrial hydrophobic proteins with internal targeting signals. Among its previously known substrates are multispan proteins of the inner and outer membranes (36, 45, 46). These hydrophobic proteins are probably protected from premature aggregation by interactions with cytosolic chaperones. Indeed, Tom70 was found to be a docking site for cytosolic Hsp70 and Hsp90 chaperones that stabilize newly synthesized inner membrane carrier proteins (15). Although the cytosolic domains of both Tom20 and Tom70 could bind in vitro newly synthesized Mim1 molecules, in contrast to tom70Δ tom71Δ cells, cells lacking Tom20 harbor normal levels of Mim1 (36). Thus, we propose that the importance of Tom70 in the biogenesis of Mim1 results not only from an ability, which is shared by Tom20, to recognize Mim1 substrate molecules but also from its unique function as an anchor site for cytosolic chaperones.
Although cytosolic Hsp70 appears to be the common denominator of the two candidate proteins, and even though mammalian cytosolic Hsc70 interacts in vitro with newly synthesized Mim1 molecules, such a chaperone was not identified in our screen. We believe that this stems from the redundancy in the function of such chaperones. The various cytosolic Hsp70s of the Ssa family (Ssa1, Ssa2, Ssa3, and Ssa4), which share very high amino acid identity, probably have overlapping activities in this process; thus, a single deletion does not cause a clear biogenesis defect. Accordingly, it was previously reported that mitochondrial import defects were not observed in single deletion strains but only in a strain where all four Ssa genes are deleted and one of them is reintroduced on a conditionally expressed allele (47). Moreover, it has been well documented that Hsp70 binding to substrate is quite promiscuous, and it is the J domain-containing cochaperones that create the specificity. Thus, as probably the vast majority of the cytoplasmic J proteins in yeast interact with Ssa1/2 (40), and since the expression of Ssa3/4 is only induced under stress conditions (48), we speculate that it is Ssa1/2 that are involved in the early stages of Mim1 import.
The importance of Djp1 for proper biogenesis of Mim1 is reflected by the heavily reduced levels of Mim1 in djp1Δ cells. This reduction suggests that in the absence of Djp1, newly synthesized Mim1 molecules are mislocalized to other compartments and/or become import incompetent. Both alternatives can lead to degradation of the precursor proteins. Indeed, upon overexpression of GFP-Mim1 in these mutated cells an ER staining is observed, suggesting mislocalization of the protein. Thus, it seems that Djp1 not only is involved in the general stabilization of newly synthesized Mim1 molecules but also contributes to the specific targeting of the substrate protein to mitochondria. The capacity of Djp1 to mediate the biogenesis of Mim1 is extremely specific. Among all cytosolic J proteins that we investigated, Djp1 was the only one whose deletion caused alterations in the targeting and steady-state levels of Mim1. Djp1 is a type II J protein, with a classical J domain in its N-terminal domain and a short glycine-rich segment following it (40). Hsp40s of this type were suggested to bind to nonnative substrates in order to present the latter to their partner, Hsp70. Since these features are shared by other cytosolic Hsp40s that do not affect Mim1 targeting, we suggest that, as proposed for other Hsp40s (49), the unique binding to Mim1 is mediated by Djp1's C-terminal region. In an effort to identify the binding site on Mim1 for Djp1, we performed a peptide scan but did not find a clear defined single site but rather several binding domains. This finding is common to chaperones and their cochaperones that do not bind to well-defined binding signals but rather to nonnative unfolded hydrophobic patches.
An interesting question is why Djp1 is required for the biogenesis of Mim1 but not for that of the other two MOM proteins with similar topology, namely, Atg32 and Tom22. In an attempt to shed light on this issue, we analyzed by the ProtParam tool (http://web.expasy.org/protparam) the sequences of Mim1, Atg32, and Tom22 with respect to both hydrophobicity (grand average of hydropathicity [GRAVY]) and thermostability (aliphatic index). Both parameters indicate that Mim1 is more hydrophobic and less thermostable than the other two proteins. Thus, we can speculate that Mim1 is more prone to aggregation in the aqueous environment of the cytosol; hence, it requires the association with chaperones as a protection from misfolding and aggregation. Naturally, Atg32 and Tom22 may also be associated with Djp1. However, in their case, such an interaction is not absolutely necessary for proper delivery.
Djp1 was previously found to be required for the biogenesis of peroxisomes. In its absence, peroxisomal matrix proteins were mislocalized to the cytoplasm, and peroxisomal structures failed to grow to full size (17). However, the proximal cause for the altered peroxisome biogenesis in cells lacking Djp1 is not known. Recently, several reports indicated possible relations between the biogenesis of both peroxisomes and mitochondria either via vesicles transport between these organelles or by sharing similar fission machineries (50, 51). Thus, Djp1 provides a new example of a protein that is involved in the biogenesis of both compartments. Based on our current results, it seems likely that Djp1 is required for import of a certain important peroxin protein, maybe with features similar to those of Mim1, and reduced levels of this peroxin could result in the various peroxisome phenotypes.
In summary, based on our results, we suggest that newly synthesized Mim1 molecules are recognized in the cytosol by the J protein Djp1, which then also engages the corresponding Hsp70. The mitochondrial precursor protein and its chaperones next associate with the import receptor Tom70. The substrate is released from the chaperones and gets inserted into the outer membrane in a process that requires the TOB complex. Decoding the mechanism of the downstream membrane integration awaits future studies.
ACKNOWLEDGMENTS
We thank E. Kracker and R. Wombacher for technical support, K. Krumpe for help with pulldown assays, A. van der Zand for Djp1 antibody and constructs, J. Buchner for antibodies, O. Pines for the degron construct, and A. Azem, K. S. Dimmer, and T. Ulrich for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (RA 1048/2-2 and RA 1048/5-1 to D.R.), the Feinberg Foundation Visiting Faculty Program (D.R.); the Marie Curie Reintegration grant (239224), and an ERC starting grant (260395) to the Schuldiner laboratory.
Footnotes
Published ahead of print 19 August 2013
REFERENCES
- 1.Schmitt S, Prokisch H, Schlunck T, Camp DG, Jr, Ahting U, Waizenegger T, Scharfe C, Meitinger T, Imhof A, Neupert W, Oefner PJ, Rapaport D. 2006. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics 6:72–80 [DOI] [PubMed] [Google Scholar]
- 2.Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schonfisch B, Guiard B, Pfanner N, Meisinger C. 2006. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17:1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lithgow T, Junne T, Suda K, Gratzer S, Schatz G. 1994. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc. Natl. Acad. Sci. U. S. A. 91:11973–11977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J. Cell Biol. 166:621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waizenegger T, Schmitt S, Zivkovic J, Neupert W, Rapaport D. 2005. Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep. 6:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto K, Kondo-Okamoto N, Ohsumi Y. 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17:87–97 [DOI] [PubMed] [Google Scholar]
- 7.Dimmer KS, Papic D, Schumann B, Sperl D, Krumpe K, Walther DM, Rapaport D. 2012. A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J. Cell Sci. 125:3464–3473 [DOI] [PubMed] [Google Scholar]
- 8.Court DA, Nargang FE, Steiner H, Hodges RS, Neupert W, Lill R. 1996. Role of the intermembrane space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell. Biol. 16:4035–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornton N, Stroud DA, Milenkovic D, Guiard B, Pfanner N, Becker T. 2010. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of alpha-helical outer membrane proteins. J. Mol. Biol. 396:540–549 [DOI] [PubMed] [Google Scholar]
- 10.Popov-Celeketic J, Waizenegger T, Rapaport D. 2008. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J. Mol. Biol. 376:671–680 [DOI] [PubMed] [Google Scholar]
- 11.Lueder F, Lithgow T. 2009. The three domains of the mitochondrial outer membrane protein Mim1 have discrete functions in assembly of the TOM complex. FEBS Lett. 583:1475–1480 [DOI] [PubMed] [Google Scholar]
- 12.Beddoe T, Lithgow T. 2002. Delivery of nascent polypeptides to the mitochondrial surface. Biochim. Biophys. Acta 1592:35–39 [DOI] [PubMed] [Google Scholar]
- 13.Hoogenraad NJ, Ward LA, Ryan MT. 2002. Import and assembly of proteins into mitochondria of mammalian cells. Biochim. Biophys. Acta 1592:97–105 [DOI] [PubMed] [Google Scholar]
- 14.Endo T, Mitsui S, Nakai M, Roise D. 1996. Binding of mitochondrial presequences to yeast cytosolic heat shock protein 70 depends on the amphiphilicity of the presequence. J. Biol. Chem. 271:4161–4167 [DOI] [PubMed] [Google Scholar]
- 15.Young JC, Hoogenraad NJ, Hartl F-U. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41–50 [DOI] [PubMed] [Google Scholar]
- 16.Caplan AJ, Cyr DM, Douglas MG. 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71:1143–1155 [DOI] [PubMed] [Google Scholar]
- 17.Hettema EH, Ruigrok CC, Koerkamp MG, van den Berg M, Tabak HF, Distel B, Braakman I. 1998. The cytosolic DnaJ-like protein djp1p is involved specifically in peroxisomal protein import. J. Cell Biol. 142:421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 19.Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5:711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shlevin L, Regev-Rudzki N, Karniely S, Pines O. 2007. Location-specific depletion of a dual-localized protein. Traffic 8:169–176 [DOI] [PubMed] [Google Scholar]
- 21.Kondo-Okamoto N, Shaw JM, Okamoto K. 2008. Tetratricopeptide repeat proteins Tom70 and Tom71 mediate yeast mitochondrial morphogenesis. EMBO Rep. 9:63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habib SJ, Waizenegger T, Lech M, Neupert W, Rapaport D. 2005. Assembly of the TOB complex of mitochondria. J. Biol. Chem. 280:6434–6440 [DOI] [PubMed] [Google Scholar]
- 24.Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426:862–866 [DOI] [PubMed] [Google Scholar]
- 25.Tong AH, Boone C. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313:171–192 [DOI] [PubMed] [Google Scholar]
- 26.Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321–324 [DOI] [PubMed] [Google Scholar]
- 27.Cohen Y, Schuldiner M. 2011. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 781:127–159 [DOI] [PubMed] [Google Scholar]
- 28.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 29.Frank R. 1992. Spot synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217–9232 [Google Scholar]
- 30.Kramer A, Schneider-Mergener J. 1998. Synthesis and screening of peptide libraries on continuous cellulose membrane supports. Methods Mol. Biol. 87:25–39 [DOI] [PubMed] [Google Scholar]
- 31.Daum G, Gasser S, Schatz G. 1982. Import of proteins into mitochondria: energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J. Biol. Chem. 257:13075–13080 [PubMed] [Google Scholar]
- 32.Walther DM, Papic D, Bos MP, Tommassen J, Rapaport D. 2009. Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 106:2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schägger H, Cramer WA, von Jagow G. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220–230 [DOI] [PubMed] [Google Scholar]
- 34.Gilon T, Chomsky O, Kulka RG. 1998. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17:2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507–519 [DOI] [PubMed] [Google Scholar]
- 36.Papic D, Krumpe K, Dukanovic J, Dimmer KS, Rapaport D. 2011. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J. Cell Biol. 194:397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T. 2011. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. U. S. A. 108:15179–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stojanovski D, Guiard B, Kozjak-Pavlovic V, Pfanner N, Meisinger C. 2007. Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J. Cell Biol. 179:881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulrich T, Gross LE, Sommer MS, Schleiff E, Rapaport D. 2012. Chloroplast beta-barrel proteins are assembled into the mitochondrial outer membrane in a process that depends on the TOM and TOB complexes. J. Biol. Chem. 287:27467–27479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5:567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keil P, Weinzierl A, Kiebler M, Dietmeier K, Söllner T, Pfanner N. 1993. Biogenesis of the mitochondrial receptor complex. Two receptors are required for binding of MOM38 to the outer membrane surface. J. Biol. Chem. 268:19177–19180 [PubMed] [Google Scholar]
- 42.Ramage L, Junne T, Hahne K, Lithgow T, Schatz G. 1993. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 12:4115–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahi C, Craig EA. 2007. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. U. S. A. 104:7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Summers DW, Douglas PM, Ramos CH, Cyr DM. 2009. Polypeptide transfer from Hsp40 to Hsp70 molecular chaperones. Trends Biochem. Sci. 34:230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiedemann N, Pfanner N, Ryan MT. 2001. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20:951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker T, Wenz LS, Kruger V, Lehmann W, Muller JM, Goroncy L, Zufall N, Lithgow T, Guiard B, Chacinska A, Wagner R, Meisinger C, Pfanner N. 2011. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 194:387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. 1988. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332:800–805 [DOI] [PubMed] [Google Scholar]
- 48.Werner-Washburne M, Stone DE, Craig EA. 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan CY, Lee S, Ren HY, Cyr DM. 2004. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol. Biol. Cell 15:761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrade-Navarro MA, Sanchez-Pulido L, McBride HM. 2009. Mitochondrial vesicles: an ancient process providing new links to peroxisomes. Curr. Opin. Cell Biol. 21:560–567 [DOI] [PubMed] [Google Scholar]
- 51.Delille HK, Alves R, Schrader M. 2009. Biogenesis of peroxisomes and mitochondria: linked by division. Histochem. Cell Biol. 131:441–446 [DOI] [PubMed] [Google Scholar]