Abstract
Dysregulated Raf/MEK/extracellular signal-regulated kinase (ERK) signaling, a common hallmark of tumorigenesis, can trigger innate tumor-suppressive mechanisms, which must be inactivated for carcinogenesis to occur. This innate tumor-suppressive signaling may provide a potential therapeutic target. Here we report that mortalin (HSPA9/GRP75/PBP74) is a novel negative regulator of Raf/MEK/ERK and may provide a target for the reactivation of tumor-suppressive signaling of the pathway in cancer. We found that mortalin is present in the MEK1/MEK2 proteome and is upregulated in human melanoma biopsy specimens. In different MEK/ERK-activated cancer cell lines, mortalin depletion induced cell death and growth arrest, which was accompanied by increased p21CIP1 transcription and MEK/ERK activity. Remarkably, MEK/ERK activity was necessary for mortalin depletion to induce p21CIP1 expression in B-RafV600E-transformed cancer cells regardless of their p53 status. In contrast, in cell types exhibiting normal MEK/ERK status, mortalin overexpression suppressed B-RafV600E- or ΔRaf-1:ER-induced MEK/ERK activation, p21CIP1 expression, and cell cycle arrest. Other HSP70 family chaperones could not effectively replace mortalin for p21CIP1 regulation, suggesting a unique role for mortalin. These findings reveal a novel mechanism underlying p21CIP1 regulation in MEK/ERK-activated cancer and identify mortalin as a molecular switch that mediates the tumor-suppressive versus oncogenic result of dysregulated Raf/MEK/ERK signaling. Our study also demonstrates that p21CIP1 has dual effects under mortalin-depleted conditions, i.e., mediating cell cycle arrest while limiting cell death.
INTRODUCTION
The Raf/MEK/extracellular signal-regulated kinase (ERK) pathway is a highly specific three-layered kinase cascade that consists of the Ser/Thr kinase Raf, the dual-specificity kinases MEK1 and its homologue MEK2 (collectively referred to as MEK1/2), and the ubiquitously expressed Ser/Thr kinases ERK1 and ERK2 (1). Upon activation, Raf phosphorylates MEK1/2, which in turn sequentially phosphorylate Tyr and Thr on the activation loop of their only known substrates, ERK1/2. ERK1/2 then activate/inactivate many proteins that mediate diverse cellular processes, thereby serving as the focal point of the pathway signaling. The Raf/MEK/ERK pathway plays pivotal roles in controlling cell survival, cell cycle progression, and differentiation (2). Therefore, dysregulated Raf/MEK/ERK signaling is a key etiologic factor in many cancers, including melanoma, thyroid cancer, and colon cancer, in which the B-RafV600E mutation is common (3).
Paradoxically, sustained activation of the Raf/MEK/ERK pathway elicits senescence-like growth arrest responses, referred to as “oncogene-induced senescence,” in primary cultured normal cells (4–6) and premalignant lesions (7–9). These phenomena are now interpreted as innate tumor-suppressive responses, which are triggered as a fail-safe antitumorigenic mechanism by aberrant cell proliferation signals (10). Knowing this, it is important to understand how these tumor-suppressive mechanisms become inactivated in the course of tumorigenesis. In different cell types, Raf/MEK/ERK-mediated growth inhibition is mediated mainly by inhibition of the Rb/E2F cell cycle machinery via cyclin-dependent kinase inhibitors p16INK4A and p21CIP1, and/or by activation of the tumor suppressor p53, which induces DNA damage responses and p21CIP1 expression (11, 12). These ostensibly straightforward mechanisms are mediated by various regulators and effectors, whose alterations can affect tumor-suppressive responses (11). Identification of a key regulator that can be exploited to reactivate the tumor-suppressive responses to Raf/MEK/ERK signaling in cancer could provide a novel therapeutic strategy.
In this study, using proteomic analysis of the MEK1/2 complex, we report the identification of mortalin (HSPA9/GRP75/PBP74) as a regulator of Raf/MEK/ERK-mediated tumor-suppressive signaling. Mortalin is a member of the heat shock protein 70 (HSP70) family (13), which is often overexpressed in different tumor types, including colon, liver, brain, and breast cancers (14–16), and is known to antagonize cellular senescence (17, 18). Although originally identified as a mitochondrial chaperone, mortalin is also detected in different subcellular compartments, especially in cancer, where it controls key regulators of cell growth and survival, such as p53 (19–21). We demonstrate that mortalin is upregulated in human melanoma biopsy specimens and that its expression is inversely correlated overall with p21CIP1 expression in different cancer lines exhibiting high MEK/ERK activity. We then investigate whether mortalin depletion in MEK/ERK-activated cancer cells can reactivate MEK/ERK-mediated p21CIP1 expression and growth arrest, and whether p53 is required for this regulation. Conversely, we also investigate whether mortalin overexpression can suppress Raf-induced MEK/ERK activation and growth arrest signaling in cells in which Raf/MEK/ERK activity is not deregulated. Furthermore, we investigate the role of p21CIP1 in cell cycle arrest and cell death under mortalin-depleted conditions. Our results suggest that mortalin is a novel negative regulator of Raf/MEK/ERK and a target exploitable for the reactivation of tumor-suppressive signaling in cancer.
MATERIALS AND METHODS
Cell culture, generation of stable lines, and reagents.
The human melanoma lines SK-MEL-28 (ATCC), SK-MEL-1 (ATCC), SK-MEL-2 (ATCC), MeWo (ATCC), and RPMI-7951 (ATCC) were maintained in minimal essential medium (MEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin, and 100 μg of streptomycin per ml. The human melanoma line A375 (ATCC) was grown in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% FBS. The human thyroid cancer lines 8505C and BCPAP (obtained from Barry Nelkin at Johns Hopkins University), and the human colon cancer line HT29 (ATCC) were grown in DMEM with 10% FBS. The K-RasG13D-transformed colon cancer line HCT116 and its p21CIP1-deficient (p21−/−) progeny (obtained from Bert Vogelstein at Johns Hopkins University) were grown in McCoy's 5A medium (Invitrogen) supplemented with 10% FBS. Table S1 in the supplemental material lists the statuses of the mitogen-activated protein kinase (MAPK) pathway and TP53 in these cell lines. The normal primary human fibroblast lines HS27 (ATCC), BJ (ATCC), and IMR90 (ATCC), as well as E1A-immortalized IMR90 (IMR90-E1A) cells (obtained from Yuri Lazebnik at Cold Spring Harbor Laboratory [CSHL]), were grown in DMEM supplemented with 10% FBS, 1% sodium pyruvate, and 1% nonessential amino acids. The human prostate cancer line LNCaP (ATCC) was maintained in phenol red-deficient RPMI 1640 medium (Invitrogen) with 10% FBS. The LNCaP line stably expressing ΔRaf-1:ER (LNCaP-Raf:ER) has been described previously (22). ΔRaf-1:ER is a CR3 catalytic domain of Raf-1 fused to the hormone binding domain of the estrogen receptor (23), and it was activated with 1 μM 4-hydroxytamoxifen (Sigma, St. Louis, MO). Cisplatin and AZD6244 were obtained from Sigma and Selleck Chemicals (Houston, TX), respectively.
Cell proliferation, death, SA β-gal, and cell cycle assays.
For cell growth curves, cells were seeded in 24-well plates (Corning, Corning, NY) at a density of 5,000 per well. Cell proliferation was measured by the colorimetric 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay, as described previously (22). Cell death was determined by counting trypan blue-stained cells using a hemocytometer. Annexin V (Invitrogen) staining was carried out according to the manufacturer's instructions. Senescence-associated acidic β-galactosidase (SA β-gal) staining was performed using the SA β-gal staining kit (Sigma) according to the manufacturer's instructions. For cell cycle analysis, cells were washed with ice cold 0.2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) and were resuspended in a 250 mM sucrose–40 mM citrate buffer (pH 7.6) containing 0.5% dimethyl sulfoxide (DMSO). Nuclei were prepared, stained with propidium iodide (24), and analyzed with an LSR II flow cytometer (Becton Dickinson, Franklin Lakes, NJ) with a gate that selects single nuclei within a normal size range. The cell cycle parameters from 10,000 gated nuclei were determined, and subsequent analysis was conducted using FCS Express software (De Novo Software, Los Angeles, CA).
Recombinant lentiviral constructs.
The primers used for the construction of the gene expression systems used in this study are listed in Table S2 in the supplemental material. Untagged or N-terminally hemagglutinin (HA)-tagged mortalin in pHAGE (pHAGE-HA-Mort) was generated using a full-length human mortalin cDNA obtained from Koichiro Muta at Kyushu University. The non-small hairpin RNA (shRNA)-targetable plasmid pHAGE-HA-Mort-shfree was generated using the QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). pHAGE-HA-HSC70, pHAGE-HA-BiP, and pHAGE-HA-HSP72 were generated using cDNAs prepared by reverse transcriptase PCR from LNCaP cells and were verified by sequencing. The mouse p21CIP1 virus (pHAGE-p21) was generated using the full-length cDNA obtained from Mariano Barbacid at the Centro Nacional de Investigaciones Oncológicas (CNIO). MEK1 with the K97M mutation (MEK1-K97M) in pHAGE (MEK1DN) was generated using the MEK1-K97M cDNA obtained from Natalie Ahn at the University of Colorado. ERK1 with the K71R, T202A, and Y204F mutations (ERK1-K71R/T202A/Y204F; referred to below as ERK1DN) in pHAGE was generated by mutagenizing ERK1-K71R, obtained from Melanie Cobb at the University of Texas Southwestern Medical Center. Bcl-2 in pHAGE was generated using the full-length cDNA obtained from Addgene (plasmid 18003).
RNA interference.
Mortalin was depleted using two independent lentiviral shRNA expression systems constructed in pLL3.7 (ATCC). pLL3.7-shMort #1 and pLL3.7-shMort #2 target two different regions of human mortalin mRNA, GGCGAUAUGAUGAUCCUGAA and GCACAUUGUGAAGGAGUUCAA, respectively. p53 was depleted using two independent lentiviral shRNA systems constructed in pLKO.1 (TRCN3754 and TRCN3756; Sigma-Aldrich). p21CIP1 was depleted using two independent lentiviral shRNA systems constructed in pLKO.1 (TRCN40123 and TRCN40125; Sigma-Aldrich). pLL3.7-shERK1 and pLL3.7-shERK2 have been described previously (22). Specific knockdown of target proteins was confirmed by Western blotting.
Viral infection.
For lentivirus production, 293T cells were cotransfected with pHAGE, pLKO.1, or pLL3.7 and a packaging vector, as described previously (25, 26). Viral supernatants were collected after 48 to 72 h and were mixed with Polybrene (Sigma) at 4 to 8 μg/ml before use. Viral titers were determined by scoring cells expressing green fluorescent protein (GFP).
TAP.
For the generation of lentiviral TAP-tagged constitutively active MEK1 (TAP-MEK1ca), a cDNA encoding MEK1-S218E/S222D was ligated into the pNTAP vector (Stratagene) for N-terminal tagging of MEK with calmodulin binding peptide and streptavidin binding peptide. The engineered MEK1-S218E/S222D was moved into the pHAGE lentiviral expression vector. The tandem affinity purification (TAP) procedure was performed according to the protocol for the InterPlay mammalian TAP system (Stratagene).
p21CIP1 promoter-luciferase reporters.
The p21CIP1 promoter-luciferase reporters H2320 and S2260 have been described previously (27). S1008, a p21CIP1 promoter-luciferase reporter harboring 1,019 bp (bp −1008 to +16) of p21CIP1 promoter DNA, was generated by partial digestion of S2260 with SmaI. Cells were transfected using Lipofectamine LTX (Invitrogen) or Effectene (Qiagen) in triplicate in 24-well plates. Cell lysates were analyzed by using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions.
qPCR.
Quantitative real-time PCR (qPCR) was performed by reverse transcription of 0.25 μg total RNA and subsequent PCR using the Mx3005P instrument and the Brilliant SYBR green QPCR core reagent kit (both from Stratagene, La Jolla, CA) according to the manufacturer's protocol. Thermocycling conditions consisted of 10 min at 95°C as the first denaturation step, followed by 40 cycles at 95°C for 30 s, 55°C for 60 s, and 72°C for 30 s. For normalization, the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or β-actin (ACTB) was measured. The primers used for qPCR are listed in Table S3 in the supplemental material. The gene copy number was calculated using the ΔΔCT method (28) as described previously (29).
Immunoblot analysis.
Cells harvested at various times were lysed and were analyzed using the bicinchoninic acid (BCA) reagent to determine the protein concentration (Pierce, Rockford, IL), as we described previously (22). Briefly, 50 to 100 μg of protein was resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane filter (Bio-Rad, Hercules, CA), and stained with fast green reagent (Thermo Fisher Scientific, Waltham, MA). Membrane filters were then blocked in 0.1 M Tris (pH 7.5)–0.9% NaCl–0.05% Tween 20 with 5% nonfat dry milk and were incubated with appropriate antibodies. Antibodies were diluted as follows: anti-B-Raf, 1:1,000; anti-MEK1/2, 1:2,500; anti-phospho-MEK1/2 (MEK1 phosphorylated at Ser217 and Ser221 [Ser217/221] and MEK2 phosphorylated at Ser222 and Ser226 [Ser222/226]), 1:2,500; anti-ERK1/2, 1:2,500; anti-phospho-ERK1/2 (Thr202/Tyr204 for ERK1 and Thr183/Tyr185 for ERK2), 1:2,500; anti-phospho-p90RSK (Thr359/Ser363), 1:2,500; anti-phospho-Rb (Ser780), 1:2,500; anti-GAPDH, 1:5,000; anti-Bcl-2, 1:2,500 (Cell Signaling Technology, Danvers, MA); anti-poly(ADP-ribose) polymerase, 1:1,000 (Thermo Fisher Scientific); anti-p21CIP1, 1:2,500; antimortalin, 1:1,000; anti-p53, 1:1,000; anti-MEK1, 1:1,000; anti-MEK2, 1:1,000; anti-HA, 1:1,000 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p16INK4A, 1:2,500 (BD Bioscience, San Jose, CA). The SuperSignal West Pico and Femto chemiluminescence kits (Pierce) were used for visualization of the signal. For densitometry, immunoblots were scanned and analyzed using Image Lab (Bio-Rad, Hercules, CA).
IP.
For immunoprecipitation (IP), cells were lysed in NP-40 lysis buffer. Immunocomplexes were then pulled down using Protein A/G Plus-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) or protein A- and protein G-coupled Dynabeads (Invitrogen). Normal IgG derived from mouse or rabbit (Santa Cruz Biotechnology) was used as the control. For the in vitro binding assay, 0.5 μM N-terminally glutathione S-transferase (GST)-tagged mortalin (Sigma) and N-terminally His-tagged MEK2 (ProSpec, East Brunswick, NJ) were incubated in 50 mM Tris (pH 7.4) containing 100 mM KCl, 100 mM NaCl, 5 mM MgCl2, 5% glycerol, and 10 mM β-mercaptoethanol for 30 min at room temperature. Glutathione-Sepharose 4B was then added to the mixture for 1 h at 4°C with gentle agitation. After an extensive wash with the binding buffer, the proteins bound to the beads were resolved by SDS-PAGE for immunoblotting.
Immunohistochemistry.
Formalin-fixed, paraffin-embedded 5-μm sections of a melanoma tissue microarray (ME1004a), containing specimens from 100 cases (56 melanoma, 20 metastatic melanoma, and 24 nevus specimens), were purchased from US Biomax (Rockville, MD). The specimens were first deparaffinized in xylene and rehydrated in ethanol. Heat-activated antigen retrieval was performed in sodium citrate buffer (pH 6.0). Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 10 min at room temperature. After blocking with 2% normal goat serum, the specimen was incubated with the monoclonal antimortalin antibody D-9 (sc-133137; Santa Cruz Biotechnology) overnight in a humidified chamber at 4°C. The specimen was then stained with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) for 60 min at room temperature. Diaminobenzidine (Vector Laboratories, Burlingame, CA) was used as the chromogen, and color development was stopped by dipping the slides in distilled water. The nuclei were then counterstained with hematoxylin (Sigma). As the negative control, normal IgG type 2 (Santa Cruz Biotechnology) was used.
Statistical analysis.
Unless otherwise specified, a two-tailed unpaired Student t test was used to assess the statistical significance of two data sets. The significance of immunohistochemistry data for human melanoma tissues was determined by the Mann-Whitney test. P values of <0.05 were considered statistically significant.
RESULTS
Identification of mortalin by TAP.
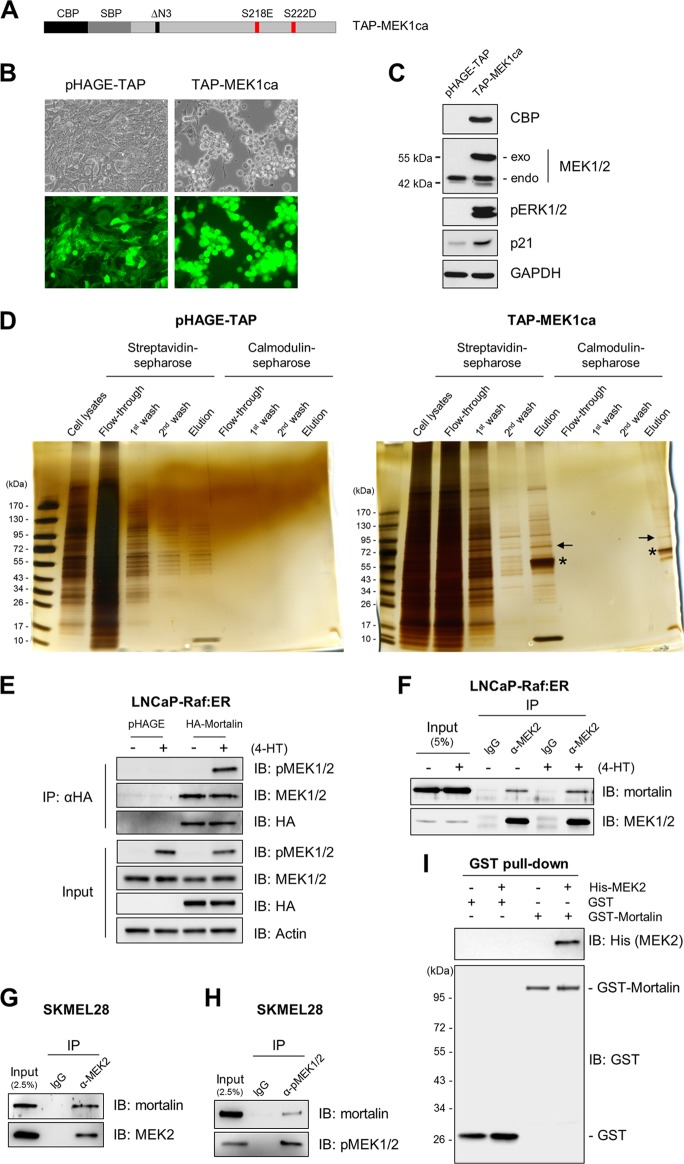
To identify regulators/mediators of Raf/MEK/ERK growth arrest signaling, we conducted TAP screening using N-terminally TAP-tagged constitutively active MEK1 (TAP-MEK1ca) (diagrammed in Fig. 1A) as a bait in LNCaP cells. These cells are well suited for MEK/ERK signaling studies because of their relatively low basal MEK/ERK activity and high sensitivity to MEK/ERK activation compared with those of other cell types (22). TAP-MEK1ca, lentivirally expressed to a level similar to the endogenous level, induced ERK1/2 phosphorylation, p21CIP1 expression, and morphological changes (Fig. 1B and C), findings consistent with previously reported growth-inhibitory effects of HA-tagged MEK1ca in this cell line (22).
Fig 1.
Tandem affinity purification of mortalin. (A) Schematic of TAP-MEK1ca engineered with N-terminal calmodulin binding protein (CBP) and streptavidin binding protein (SBP) tags. (B) Two-day infection of LNCaP cells with lentiviral TAP-MEK1ca and the control pHAGE-TAP. GFP expression indicates the efficiency of infection. (C) Western blot analysis of lysates from cells shown in panel B. exo, exogenous MEK; endo, endogenous MEK; pERK1/2, phosphorylated ERK1/2. (D) Silver-staining analysis of fractions from different purification steps run on SDS-PAGE gels. Arrows and stars indicate mortalin and TAP-MEK, respectively. (E) LNCaP-Raf:ER cells infected with lentiviral pHAGE-HA-Mort were treated with 1 μM 4-hydroxytamoxifen (4-HT) for 1 h. HA-tagged mortalin was pulled down using anti-HA–agarose beads. Pulldown fractions were then analyzed by Western blotting to determine co-IP of endogenous MEK1/2. IB, immunoblot. (F and G) Total-cell lysates from LNCaP-Raf:ER cells treated with 4-HT for 1 h (F) or SK-MEL-28 cells (G) were subjected to IP using an anti-MEK2 antibody. The endogenous MEK2 pulldown fractions were analyzed by Western blotting to determine the co-IP of endogenous mortalin. (H) Total-cell lysates of SK-MEL-28 cells were subjected to IP using an anti-phospho-MEK1/2 (α-pMEK1/2) antibody. The endogenous pMEK1/2 pulldown fractions were analyzed by Western blotting to determine the co-IP of endogenous mortalin. (I) In vitro binding assay of recombinant mortalin and MEK2. The GST pulldown fractions were analyzed by Western blotting.
TAP-MEK protein complexes from LNCaP cell lysates were purified by two-step affinity purification using streptavidin-Sepharose and calmodulin-Sepharose resins. Silver staining of the fractions from different purification steps run on SDS-PAGE gels revealed significant decreases in nonspecific binding but highly enriched specific binding (Fig. 1D). Tryptic digestion and nanospray-liquid chromatography (LC)-mass spectrometry (MS) analysis of the protein bands that emerged from the second round of affinity purification revealed a 75-kDa protein that was identified as mortalin. The interaction of MEK with mortalin was confirmed by a reverse IP using HA-tagged mortalin as the bait (Fig. 1E). Subsequent co-IP studies of endogenous proteins indicated that MEK1/2 can interact with mortalin regardless of their activation status, as determined in LNCaP cells expressing tamoxifen-inducible ΔRaf-1:ER (Fig. 1F) and the B-RafV600E mutated melanoma line SK-MEL-28 (Fig. 1G and H). Moreover, an in vitro binding assay of recombinant proteins indicated that mortalin may interact directly with MEK (Fig. 1I).
Mortalin levels are upregulated in melanoma and are inversely correlated with p21CIP1 levels in cancer cells exhibiting high Raf/MEK/ERK activity.
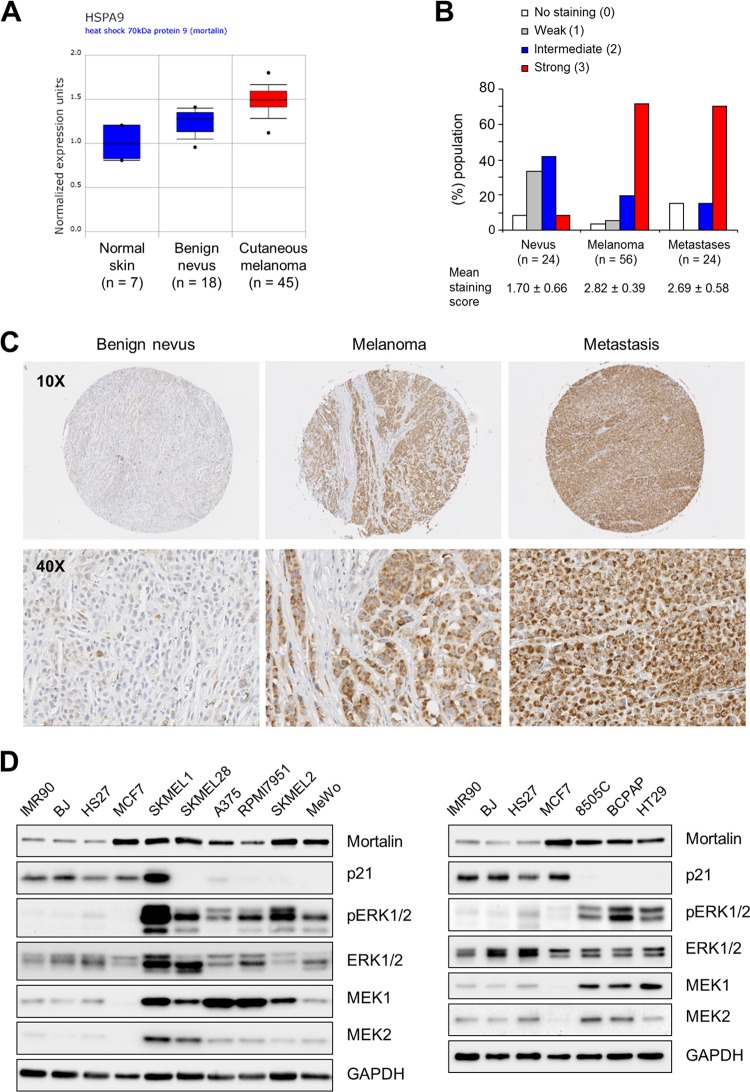
By searching the Onocomine (http://www.oncomine.org/) cancer microarray database, we found that mortalin mRNA levels may increase during the progression of melanoma (Fig. 2A). Indeed, our immunohistochemical analysis of 100 specimens of benign and malignant human melanoma tissues revealed a strong correlation between mortalin protein levels and melanoma tumor malignancy (Fig. 2B and C). In agreement with this, relatively high mortalin levels were detected in a panel of melanoma cell lines compared with those in normal primary human fibroblasts (Fig. 2D, left) (Table S1 in the supplemental material lists the mutational statuses of these cells). MCF7 breast cancer cells were used as a positive control. These cells have been shown previously to express higher levels of mortalin than other cell lines derived from human breast, prostate, colon, lung, and brain tumors (16). Moreover, papillary thyroid carcinoma and colon carcinoma cell lines, which also exhibit high MEK/ERK activity, showed relatively high mortalin levels (Fig. 2D, right). Intriguingly, p21CIP1 levels were relatively low, and were inversely correlated with mortalin levels, in the cancer cell lines that exhibited high MEK/ERK activity, except for SK-MEL-1 (Fig. 2D). In contrast, levels of MEK1/2, especially those of MEK1, were relatively high in these cells. These data led us to investigate whether mortalin has a role as an oncogenic switch in the context of aberrant Raf/MEK/ERK signaling.
Fig 2.
Mortalin is upregulated in melanoma and tumor cell lines exhibiting high MEK/ERK activity. (A) Microarray analysis of mortalin mRNA in patient biopsy specimens (data from www.oncomine.org). The boxes, whiskers, and asterisks indicate the interquartile range (25 to 75%), the 10 to 90% range, and the minimums and maximums, respectively. For benign melanocytic skin nevi versus normal skin, the fold change was 2.181 (P, 1.44E−4; t, 5.560); for cutaneous melanoma versus normal skin, the fold change was 3.207 (P, 1.29E−5; t, 8.785). (B and C) Immunohistochemical analysis of mortalin protein in patient tissue biopsy specimens. (B) Graph of mortalin protein levels in melanocytic skin nevi, cutaneous melanoma, and metastatic melanoma. For the difference between benign melanocytic skin nevi and cutaneous melanoma, the P value was <0.0001 by the Mann-Whitney test; for the difference between melanocytic skin nevi and metastatic melanoma, the P value was 0.0021 by the Mann-Whitney test. (C) Representative images of a benign nevus, melanoma, and metastasis. Figure S1 in the supplemental material validates the specificity of this staining. Figure S2 in the supplemental material shows corresponding hematoxylin-and-eosin staining. (D) Western blot analysis of total lysates of various cancer cell lines (see Table S1 in the supplemental material for mutational status). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control.
Mortalin depletion in B-Raf- or K-Ras-mutated cancer cells induces p21CIP1, G2/M-phase cell cycle arrest, and cell death.
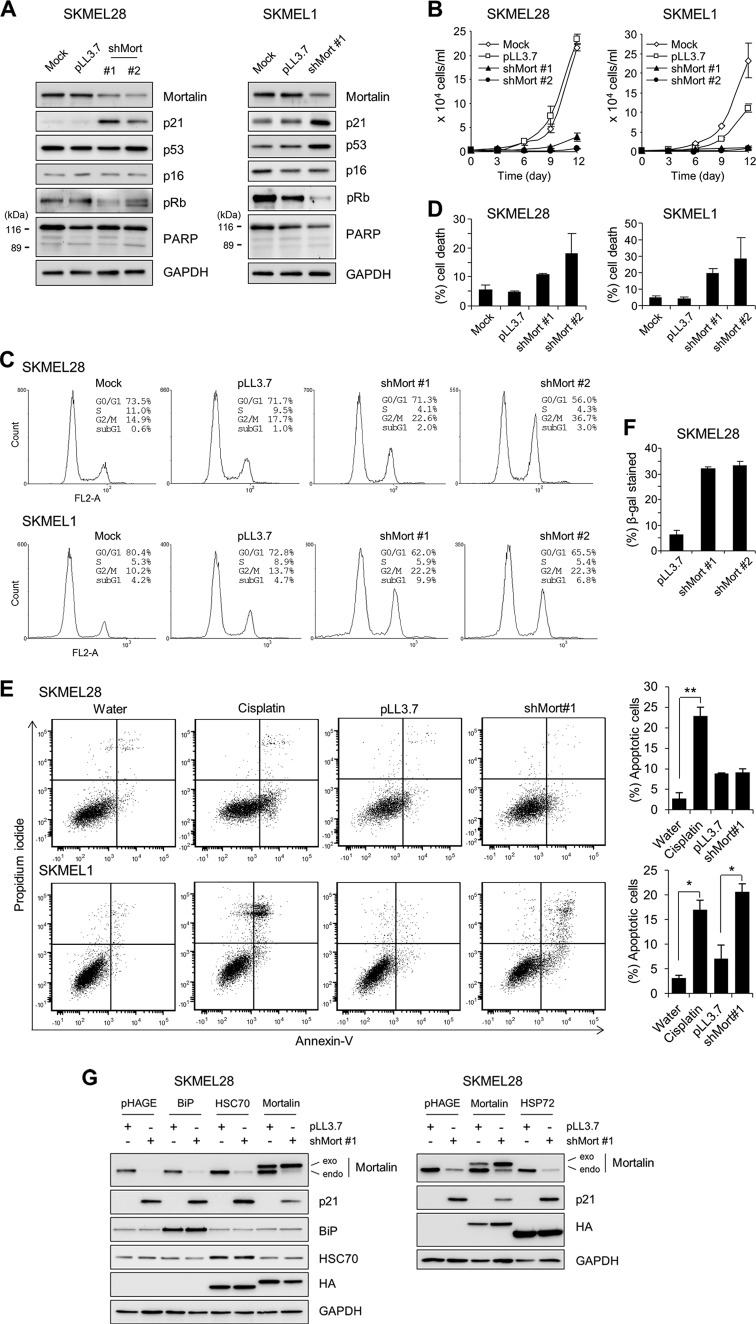
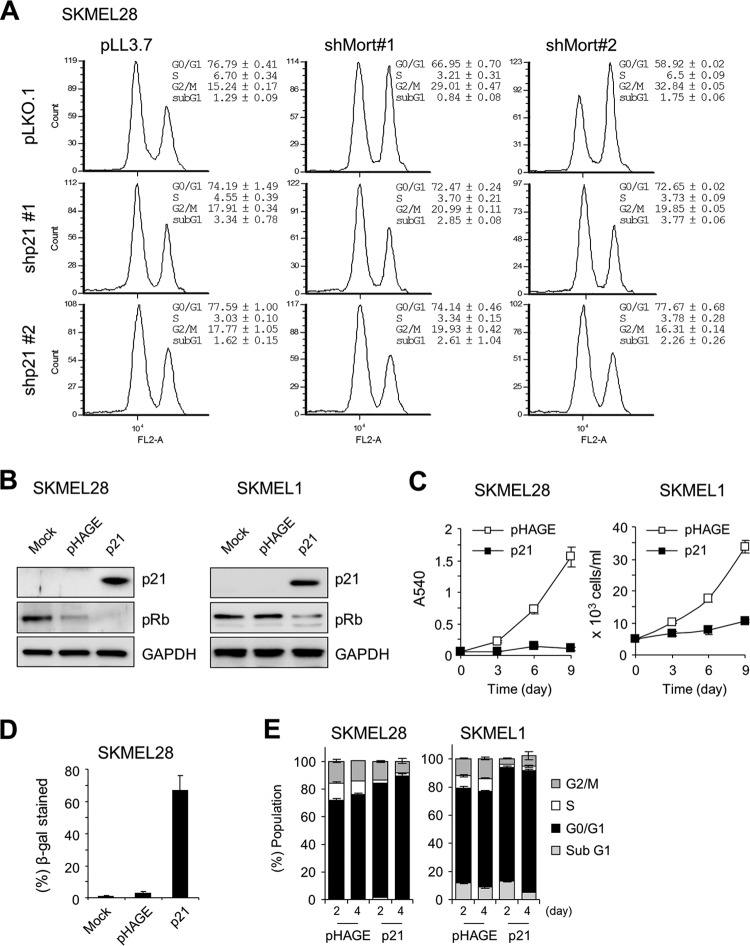
To characterize the role of mortalin in MEK/ERK-activated cancer cells, we depleted mortalin in the SK-MEL-28 and SK-MEL-1 melanoma lines by using two lentiviral shRNA constructs that specifically target different mRNA regions of mortalin (shMort). In these two cell lines, substantial mortalin knockdown was accompanied by significantly increased p21CIP1 levels and decreased Rb phosphorylation (Fig. 3A; see also Fig. S3 in the supplemental material). However, p16INK4A levels were not affected, indicating that mortalin may regulate the cell cycle via a p21CIP1/cyclin-dependent kinase/Rb axis.
Fig 3.
Mortalin knockdown induces growth inhibition in B-RafV600E-transformed melanoma lines. (A to E) SK-MEL-28 and SK-MEL-1 cells were infected with pLL3.7 viruses expressing two different shRNAs that target mortalin mRNA (shMort#1 and shMort#2). Uninfected (mock) or empty pLL3.7 virus-infected cells were used for comparison. (A) Western blot analysis of total lysates of cells infected for 5 days. pRb, phosphorylated Rb; PARP, poly(ADP-ribose) polymerase. Figure S3 in the supplemental material shows the effects of shMort#2 on SK-MEL-1 cells. (B) Proliferation rates were monitored by cell counting. (C and D) Percentages of cells in the various phases of the cell cycle (C) and percentages of cell death (D) were determined at day 5 postinfection, using propidium iodide and trypan blue, respectively. (E) Annexin V staining at day 5 postinfection. The graphs (right) show the percentages of annexin V-positive and propidium iodide-positive cell populations. Treatment with 50 μM cisplatin for 24 h to induce apoptosis was used as a positive control. Cisplatin was dissolved in water. *, P < 0.05; **, P < 0.01. (F) Cells infected for 10 days were scored for SA β-gal staining (for images, see Fig. S4A in the supplemental material). (G) SK-MEL-28 cells were coinfected for 5 days with shMort#1 and pHAGE lentivirus expressing Mort-shfree (not targetable by shMort#1), BiP, HSC70, or HSP72. Empty pHAGE and pLL3.7 are the controls for HSP and shMort viruses, respectively. Total-cell lysates were examined for the expression of the indicated proteins by Western blotting. HA indicates the relative expression levels of HSPs, except for BiP. Data (means ± standard errors of the means) are from a representative experiment performed in triplicate.
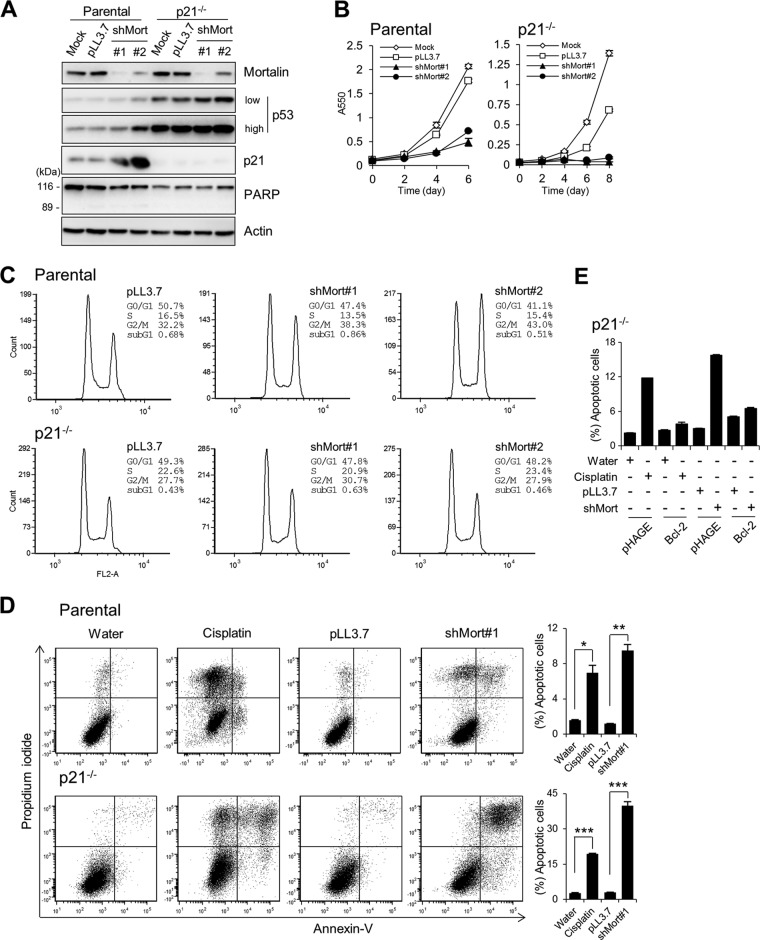
We next determined the cellular effects of mortalin depletion in these B-RafV600E-transformed cancer cells. Consistent with the changes observed in p21CIP1 and Rb, mortalin knockdown hindered the proliferation of SK-MEL-28 and SK-MEL-1 cells (Fig. 3B) and induced cell cycle arrest, most significantly in the G2/M phase (Fig. 3C). Although no significant cleavage of poly(ADP-ribose) polymerase was induced by mortalin knockdown (Fig. 3A), trypan blue (Fig. 3D) and annexin V (Fig. 3E) staining revealed that mortalin knockdown can also induce caspase-independent apoptotic cell death, particularly in SK-MEL-1 cells. Moreover, prolonged mortalin knockdown (10 days) increased the expression of SA β-gal in SK-MEL-28 cells (Fig. 3F), suggesting an onset of senescence-like responses. Mortalin knockdown also induced similar effects in BCPAP cells, a papillary thyroid cancer line carrying B-RafV600E (see Fig. S5A to C in the supplemental material). In addition, in agreement with these results from B-RafV600E-mutated cancer lines, mortalin knockdown also induced p21CIP1 expression (Fig. 4A), growth inhibition (Fig. 4B), G2/M arrest (Fig. 4C), and caspase-independent apoptotic cell death (Fig. 4D) in the K-RasG13D-transformed colon cancer line HCT116.
Fig 4.
Mortalin knockdown induces growth in the HCT116 colon cancer line and its p21−/− progeny. (A to D) HCT116 parental and p21−/− cells were infected with lentiviral shMort#1 or shMort#2. pLL3.7 is the control virus. (A) Western blot analysis of total lysates of cells infected for 5 days. PARP, poly(ADP-ribose) polymerase. (B) Proliferation rates were monitored by an MTT assay. (C) Cell cycle analysis using propidium iodide at day 5 postinfection. (D) Annexin V staining at day 5 postinfection. The graphs (right) indicate annexin V-positive and propidium iodide-positive cell populations. Cisplatin (50 μM) was used as a control to induce apoptosis. Data (means ± standard errors of the means) are from a representative experiment performed in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) HCT116 p21−/− cells infected with lentiviral pHAGE-puro-FLAG-Bcl-2 were either treated with 50 μM cisplatin for 24 h or infected with lentiviral shMort for 4 days before annexin V staining. Bcl-2 expression and annexin V staining histograms are shown in Fig. S7 in the supplemental material.
These shMort effects were mortalin specific; when expressed at a level similar to the endogenous level, a mortalin gene engineered to avoid shMort targeting (Mort-shfree) significantly abolished shMort-induced effects in SK-MEL-28 cells (see Fig. S6 in the supplemental material). Of note, in this “rescue” experiment, overexpression of other HSP70 family members—HSP72, HSC70, and the endoplasmic reticulum stress mediator BiP/GRP78 (13)—could not replace the expression of Mort-shfree in preventing p21CIP1 induction in mortalin-depleted SK-MEL-28 cells (Fig. 3G). These findings suggest that mortalin is important for the proliferation and survival of Ras/Raf-transformed tumor cells.
p21CIP1 is required for G2/M-phase cell cycle arrest while limiting cell death under mortalin-depleted conditions.
Although p21CIP1 is known mainly for its role in G0/G1-phase cell cycle arrest, it is also known to have roles in mediating G2/M-phase arrest (30) and in suppressing cell death (31). We thus investigated the role of p21CIP1 in shMort-induced G2/M arrest and cell death. In the p21CIP1-deficient (p21−/−) HCT116 progeny, mortalin knockdown did not robustly induce G2/M arrest (Fig. 4C). Nevertheless, mortalin knockdown still suppressed the growth of p21CIP1-deficient cells (Fig. 4B), and this growth suppression was correlated with significantly increased cell death, as determined by annexin V staining (Fig. 4D). In agreement with this, two independent shRNA-mediated p21CIP1 knockdowns blocked shMort-induced G2/M arrest in SK-MEL-28 cells, while they mildly increased sub-G1 populations in the cultures, an indication of cell death (Fig. 5A).
Fig 5.
Requirement of p21CIP1 for mortalin knockdown-induced growth inhibition in SK-MEL-28 and SK-MEL-1 cells. (A) Cell cycle analysis of SK-MEK-28 cells coinfected for 5 days with lentiviral shMort#1 and pLKO.1 expressing one of two different p21CIP1-targeting shRNAs (shp21#1 and shp21#2). (B to E) Cells infected either with lentiviral pHAGE expressing p21CIP1 or with empty pHAGE were examined for the indicated proteins by Western blotting of total-cell lysates at day 3 (B), for cell proliferation for 9 days by an MTT assay or cell counting (C), or for SA β-gal staining at day 5 (D), or were subjected to cell cycle analysis at day 5 (E). Images of SA β-gal staining are shown in Fig. S4B in the supplemental material.
It has been known that p21CIP1 protects cells from different stress conditions by regulating pro- and antiapoptotic Bcl-2 family proteins (32–34). Further, it has been shown that steady-state levels of Bcl-2 and Bcl-XL are downregulated in HCT116 p21−/− cells (32, 33). Indeed, lentiviral overexpression of Bcl-2 completely rescued HCT116 p21−/− cells from shMort-induced cell death, as determined by annexin V staining (Fig. 4E; see also Fig. S7 in the supplemental material). Moreover, Bcl-2 overexpression rendered SK-MEL-28 cells strongly resistant to shMort-induced cell death (see Fig. S8 in the supplemental material), suggesting that mortalin and p21CIP1 may regulate cell survival via mechanisms overlapping at the level of Bcl-2 family protein regulation. These data suggest that p21CIP1 has dual effects under mortalin-depleted conditions, i.e., mediating cell cycle arrest while limiting cell death.
We also evaluated the effects of p21CIP1 overexpression in SK-MEL-28 and SK-MEL-1 cells. Consistent with mortalin knockdown, ectopic expression of p21CIP1 significantly decreased Rb phosphorylation in these cells, as well as decreasing their proliferation (Fig. 5B and C). Moreover, prolonged p21CIP1 expression (10 days) induced SA β-gal activity in SK-MEL-28 cells (Fig. 5D). However, unlike mortalin knockdown, p21CIP1 expression did not induce any significant cell death or G2/M arrest, although it induced G0/G1 arrest (Fig. 5E). Therefore, p21CIP1 alone is not sufficient to mediate all the effects of mortalin depletion in these cells.
Mortalin depletion induces p21CIP1 expression via p53-dependent as well as p53-independent mechanisms in B-RafV600E-transformed cancer cells.
Knowing the pivotal involvement of p21CIP1 in mortalin effects, we investigated the mechanism by which mortalin depletion upregulates p21CIP1. p53 is a well-known regulator of p21CIP1 transcription (35). It has been shown previously that mortalin can sequester p53 in the cytosol to limit its activity (19, 21). Because SK-MEL-28 cells harbor a homozygous L145R mutation in the DNA binding domain of p53, whereas SK-MEL-1 cells have wild-type p53 (Cancer Genome Project at Sanger Institute [http://www.sanger.ac.uk/]), and because mortalin knockdown upregulated p53 levels only in SK-MEL-1 cells (Fig. 3A; see also Fig. S3 in the supplemental material), we determined whether p53 is required for mortalin to regulate p21CIP1 in these cells.
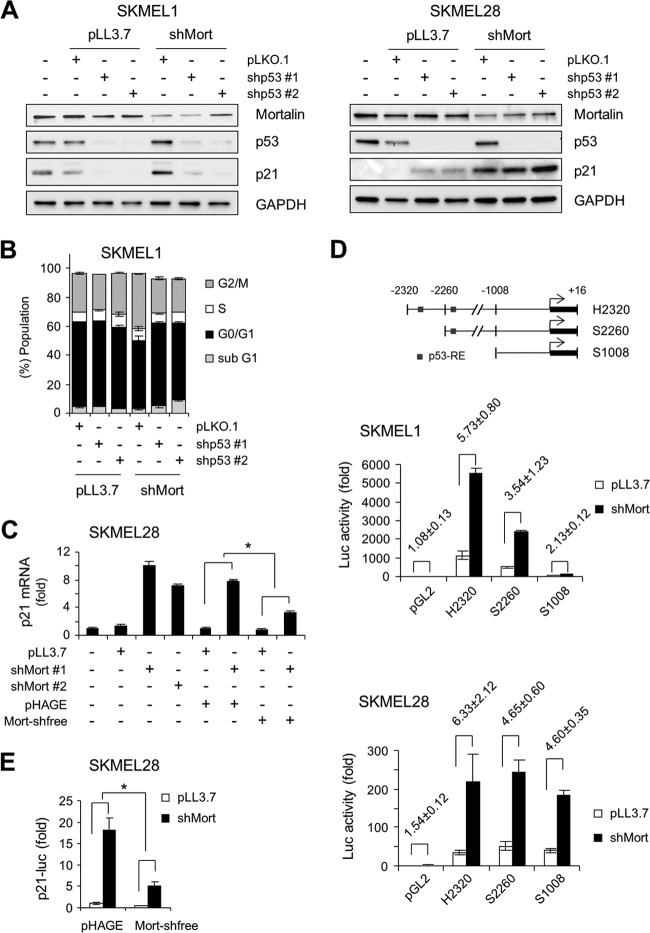
In SK-MEL-1 cells, two independent shRNA-mediated p53 knockdowns consistently blocked basal and shMort-induced p21CIP1 levels (Fig. 6A, left) and G2/M arrest (Fig. 6B; see also Fig. S9 in the supplemental material), indicating the requirement of wild-type p53 for shMort to induce p21CIP1 and growth arrest. In contrast, in SK-MEL-28 cells, p53 depletion did not block shMort-induced p21CIP1 upregulation; rather, it slightly increased the basal levels of p21CIP1 (Fig. 6A, right), indicating that mortalin depletion can also induce p21CIP1 independently of p53.
Fig 6.
Mortalin knockdown induces p21CIP1 transcription in a manner independent of p53 in SK-MEL-28 cells but dependent on p53 in SK-MEL-1 cells. (A) Western blot analysis of total lysates of cells coinfected for 5 days with lentiviral shMort#1 and pLKO.1 expressing one of two different p53-targeting shRNAs (shp53#1 and shp53#2). pLKO.1 and pLL3.7 are controls for shp53 and shMort, respectively. (B) Cell cycle analysis of SK-MEL-1 cells treated as described for panel A. Cell cycle histograms are shown in Fig. S9 in the supplemental material. (C) qPCR analysis of p21CIP1 mRNA expression in SK-MEL-28 cells coinfected with lentiviral shMort and pHAGE-Mort-shfree for 5 days. Data are expressed as fold changes from the basal levels in uninfected cells. *, P < 0.05. (D) SK-MEL-1 and SK-MEL-28 cells were cotransfected for 3 days with pLL3.7-shMort#1 and different p21CIP1 promoter-luciferase reporters (diagrammed at the top) before the determination of luciferase activity. pGL2 is the control for the reporters. Bars in graphs represent fold changes in luciferase activity from cells cotransfected with pGL2 and pLL3.7. Values for fold differences in luciferase activity between cells cotransfected with a reporter and pLL3.7 and those transfected with a reporter and pLL3.7-shMort#1 are given above the bars. (E) Luciferase assay of SK-MEL-28 cells cotransfected for 3 days with the p21CIP1 promoter-luciferase reporter S1008, pLL3.7-shMort#1, and pHAGE-Mort-shfree. Data are expressed as fold changes in luciferase activity from that for cells cotransfected with S1008, pLL3.7, and pHAGE. Data (means ± standard errors of the means) are from a representative experiment performed in triplicate.
To understand the mechanism(s) by which mortalin mediates p53-independent p21CIP1 regulation, we carried out qPCR analysis. Mortalin depletion upregulated p21CIP1 mRNA levels in SK-MEL-28 cells 7- to 10-fold within 4 days, and this upregulation was significantly attenuated by the expression of Mort-shfree (Fig. 6C). To investigate whether this resulted from p53-independent p21CIP1 transcription, we generated a luciferase reporter of the p21CIP1 promoter lacking the two known p53-responsive elements (diagrammed in Fig. 6D, top). This reporter responded to mortalin depletion in SK-MEL-28 cells at levels similar to those of reporters containing p53-responsive elements, although its response was much weaker than those of the others in SK-MEL-1 cells (Fig. 6D, center and bottom). This induction was significantly inhibited by the expression of Mort-shfree (Fig. 6E), supporting the specificity of the shMort effect. Moreover, shMort induced p21CIP1 promoter-luciferase reporter activity in BCPAP cells, which also express an inactivated form of p53, p53D259Y (see Fig. S5D in the supplemental material). These data indicate that mortalin can control p21CIP1 levels in B-RafV600E-transformed cancer cells by regulating p21CIP1 transcription in a p53-dependent as well as a p53-independent manner.
Mortalin depletion upregulates MEK/ERK activity in B-RafV600E-transformed cancer cells.
Previous studies demonstrated that Raf/MEK/ERK activation can induce p21CIP1 transcription not only via p53-dependent mechanisms but also via p53-independent mechanisms (27, 36). This ability of Raf/MEK/ERK, and our findings of mortalin interaction with MEK and its increased expression in cancer cells exhibiting high MEK/ERK activity, led us to investigate whether mortalin depletion affects MEK/ERK activity to induce p21CIP1 in B-RafV600E-mutated cancer cells.
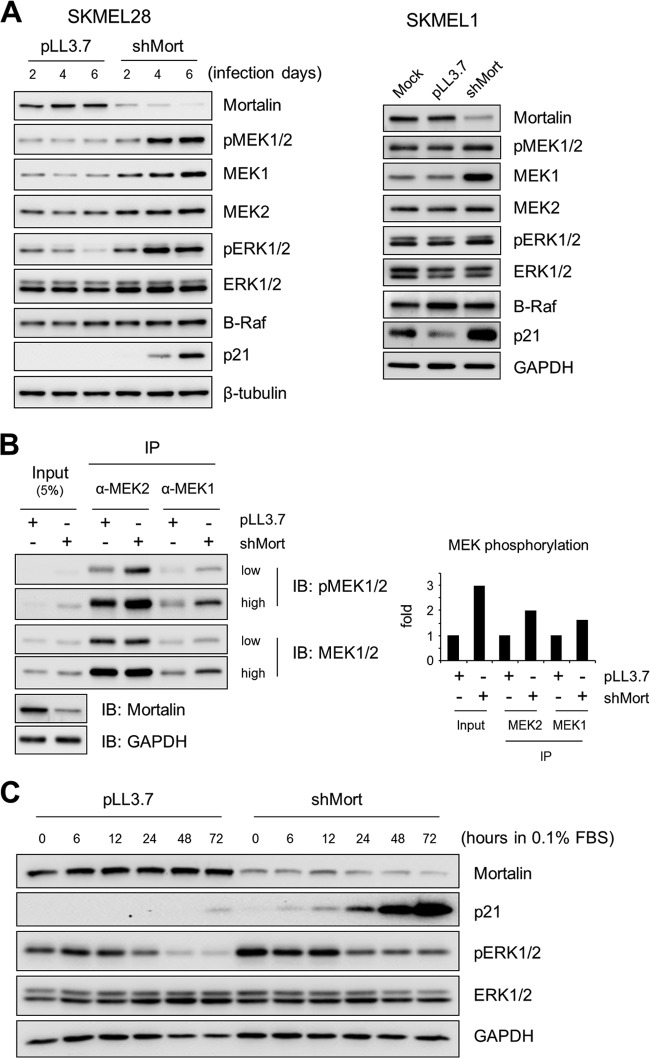
Intriguingly, mortalin knockdown substantially upregulated MEK1 protein levels without significantly affecting B-Raf or ERK1/2 levels in SK-MEL-28 and SK-MEL-1 cells (Fig. 7A; see also Fig. S3 in the supplemental material). Mortalin knockdown also upregulated MEK2 levels, but to a much lesser extent, in SK-MEL-28 cells. Consistent with this effect, mortalin knockdown increased the Western blot signals detected by an antibody specific to MEK1/2 activated by phosphorylation (at Ser217/221 for MEK1 and Ser222/226 for MEK2). Moreover, when MEK1 and MEK2 in SK-MEL-28 cells were individually immunoprecipitated and analyzed by Western blotting using the antibody, the phosphorylation levels of both proteins were significantly increased upon mortalin knockdown (Fig. 7B). Therefore, mortalin can affect MEK1/2 protein levels as well as their activation status in B-RafV600E-transformed cancer cells. In agreement with these effects, mortalin knockdown also upregulated ERK1/2 phosphorylation in association with p21CIP1 expression in SK-MEL-28 cells, as determined under the normal culture condition (Fig. 7A) and under a low-serum culture condition, where the effects of mitogenic signals other than B-RafV600E were minimal (Fig. 7C). These data suggest that mortalin can regulate MEK/ERK activity in B-RafV600E-transformed cancer.
Fig 7.
Mortalin knockdown upregulates MEK/ERK activity in SK-MEL-28 and SK-MEL-1 cells. (A) Western blot analysis of total lysates of cells infected with shMort#1 or pLL3.7 virus for the periods indicated (SK-MEL-28 cells) or for 5 days (SK-MEL-1 cells). Figure S3 in the supplemental material shows the effects of shMort#2 on SK-MEL-1 cells. (B) Total lysates of SK-MEL-28 cells infected with shMort#1 or pLL3.7 virus for 3 days were immunoprecipitated with an anti-MEK1 or anti-MEK2 antibody. (Left) Immunoprecipitates were analyzed by Western blotting to detect MEK1/2 and phosphorylated MEK1/2 (MEK1 at Ser217/221 and MEK2 at Ser222/226). (Right) To determine the degree of phosphorylation, the densitometry data of Western blot images of phospho-MEK1 and phospho-MEK2 were normalized by the data for MEK1 and MEK2, respectively. (C) SK-MEL-28 cells infected with shMort#1 or pLL3.7 virus for 2 days were switched into fresh medium containing 0.1% FBS. The expression of the indicated proteins under conditions of serum starvation was monitored by Western blotting of total-cell lysates.
MEK/ERK activity is necessary for mortalin depletion to induce p21CIP1 expression and G2/M arrest but not cell death in B-RafV600E-transformed cancer cells.
On the basis of these findings, we hypothesized that mortalin depletion can induce p21CIP1 expression in B-RafV600E-transformed cancer cells by reactivating MEK/ERK-mediated growth-inhibitory signaling. This possibility was tested in several sets of experiments aimed at determining whether MEK/ERK activity is required for mortalin depletion to induce p21CIP1 expression.
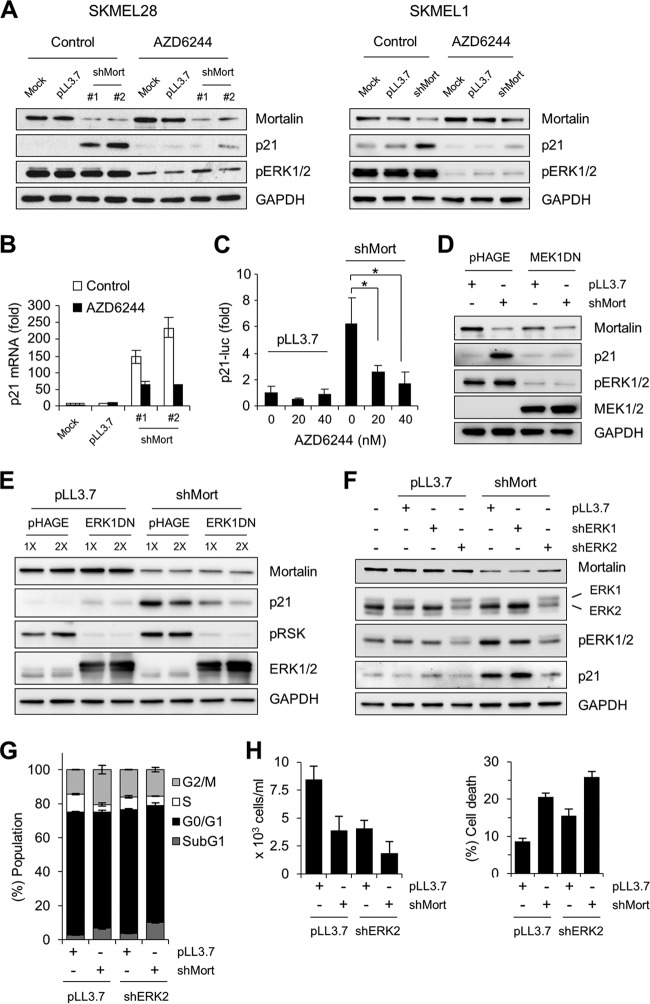
First, we determined whether the MEK1/2-specific inhibitor AZD6244 could block shMort-induced p21CIP1 expression in SK-MEL-28 and SK-MEL-1 cells. Short-term treatment of these cells with a low dose of AZD6244 significantly reduced ERK1/2 phosphorylation (Fig. 8A). Under this condition, shMort-induced p21CIP1 expression was significantly attenuated in both cell lines, indicating that mortalin requires MEK1/2 activity to regulate p21CIP1, regardless of p53 status. In agreement with this result, AZD6244 treatment also suppressed shMort-induced p21CIP1 mRNA expression (Fig. 8B) and p21CIP1 promoter-luciferase reporter activity (Fig. 8C) in SK-MEL-28 cells. Second, when overexpressed at a level sufficient to substantially deplete ERK1/2 phosphorylation, a dominant negative MEK1 construct effectively blocked shMort-induced p21CIP1 expression in SK-MEL-28 cells (Fig. 8D). Likewise, overexpression of a dominant negative ERK1 construct at a level sufficient to inhibit the phosphorylation of p90RSK1, a bona fide ERK1/2 substrate (37), also blocked shMort-induced p21CIP1 expression in these cells (Fig. 8E). Finally, knockdown of ERK2, which is expressed predominantly in SK-MEL-28 cells, was sufficient to block shMort-induced p21CIP1 expression, whereas ERK1 knockdown was not effective (Fig. 8F). Taken together, these data indicate that upregulation of MEK/ERK activity is a necessary mechanism for mortalin depletion to induce p21CIP1 expression in B-RafV600E-transformed cancer cells.
Fig 8.
MEK/ERK activity is necessary for mortalin knockdown to induce p21CIP1 expression in SK-MEL-28 and SK-MEL-1 cells. (A) Western blot analysis of total lysates of cells infected with lentiviral shMort for 3 days and then treated with 40 nM AZD6244 for 2 days. An equal volume of DMSO was used as the control for AZD6244. (B) qPCR analysis of p21CIP1 mRNA expression in SK-MEL-28 cells treated as described for panel A. (C) SK-MEL-28 cells expressing the p21CIP1 promoter-luciferase reporter H2320 were infected with lentiviral shMort#1 or pLL3.7 for 2 days. Cells were then treated with increasing doses of AZD6244 for 2 days before luciferase assays. Data (means ± standard errors of the means) from a representative experiment performed in triplicate are expressed as fold changes in luciferase activity from untreated pLL3.7-infected cells. *, P < 0.05. (D) Western blot analysis of total lysates of SK-MEL-28 cells coinfected with lentiviral shMort#1 and pHAGE expressing dominant negative MEK1-K97M (MEK1DN) for 5 days. (E) Western blot analysis of total lysates of SK-MEL-28 cells coinfected with shMort#1 and one of two different doses of pHAGE virus expressing dominant negative ERK1-K71R/T202A/Y204F (ERK1DN) for 5 days. pRSK, phosphorylated p90RSK1. (F) Western blot analysis of total lysates of SK-MEL-28 cells coinfected with lentiviral shMort#1 and pLL3.7 expressing shRNA that targets ERK1 (shERK1) or ERK2 (shERK2) for 5 days. (G and H) Cell cycle analysis was performed (G), and rates of cell death were determined, at day 5 postinfection, using propidium iodide and trypan blue, respectively.
We also examined the effects of Raf/MEK/ERK inhibition on shMort-induced cell death in SK-MEL-28 cells. We found that ERK2 depletion, at a level sufficient to substantially block p21CIP1 induction by shMort, could not rescue cells from shMort-induced cell death and sub-G0/G1-phase accumulation, although it could suppress shMort-induced G2/M arrest (Fig. 8G and H). Therefore, mortalin depletion appears to induce cell death independently of Raf/MEK/ERK. This observation is also consistent with the observation that p21CIP1 induction limits cell death under mortalin-depleted conditions.
Mortalin overexpression downregulates MEK/ERK activity and suppresses Raf-induced growth arrest signaling.
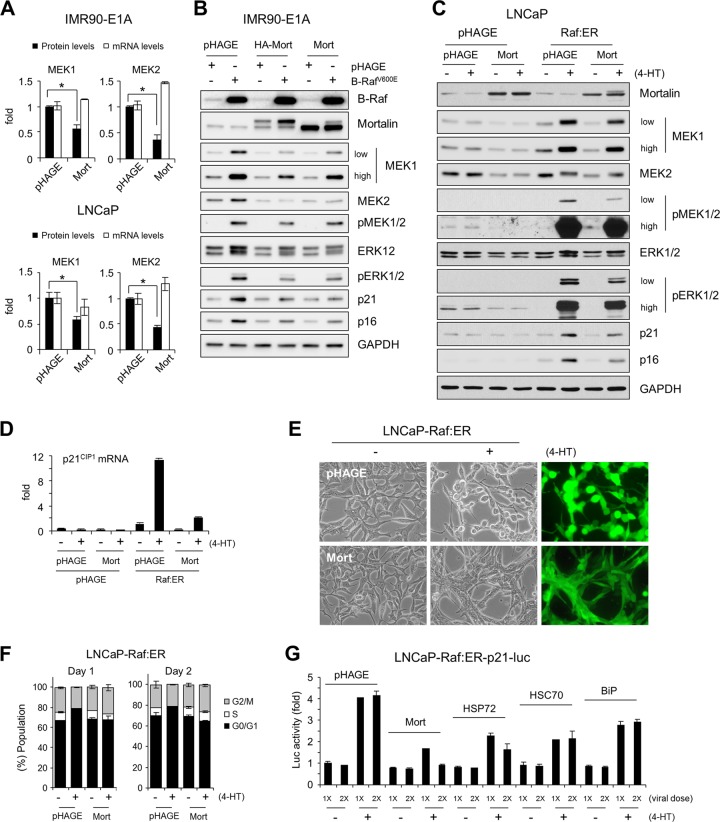
The findings presented above strongly suggested that mortalin could suppress B-RafV600E-mediated growth-inhibitory signaling, mainly by modulating MEK/ERK activity. To further evaluate this possibility, we examined the effects of mortalin overexpression in different cell types in which Raf/MEK/ERK signaling is not deregulated and p21CIP1 induction is a primary response to the pathway activation. This was addressed using the E1A-immortalized normal fibroblast line IMR90-E1A and LNCaP cells.
Mortalin overexpression was sufficient to induce substantial decreases in the basal levels of MEK1/2 proteins without decreasing MEK1/2 mRNA levels in IMR90-E1A and LNCaP cells (Fig. 9A). In these cells, mortalin overexpression also downregulated ERK1/2 levels, although not as substantially as MEK1/2 levels (Fig. 9B and C). These changes were correlated with decreased basal phosphorylation of MEK1/2 and ERK1/2, as noted in LNCaP cells (Fig. 9C). When the MEK/ERK pathway was activated in IMR90-E1A and LNCaP cells using B-RafV600E or ΔRaf-1:ER, not only phosphorylation levels but also protein levels of MEK1/2, especially MEK1, were significantly increased (Fig. 9B and C). This is consistent with the increased MEK1 levels in cancer lines exhibiting high MEK/ERK activity (Fig. 2D). Mortalin overexpression consistently attenuated these increases and inhibited Raf-induced p21CIP1 and p16INK4A expression in these cells (Fig. 9B to D). Mortalin overexpression also significantly suppressed Raf-induced morphological changes and cell cycle arrest in LNCaP cells (Fig. 9E and F). In addition, compared with that of other HSP70 family chaperones, mortalin overexpression was most effective at suppressing Raf-induced p21CIP1 promoter-luciferase reporter activity in LNCaP cells (Fig. 9G). These findings are consistent with those obtained from B-RafV600E-transformed tumor cells and strongly support the ability of mortalin to operate as a molecular switch to inactivate Raf/MEK/ERK-mediated growth-inhibitory signaling during tumorigenesis.
Fig 9.
Mortalin overexpression downregulates MEK1/2 levels, MEK/ERK activity, and Raf-induced growth arrest signaling. (A) IMR90-E1A and LNCaP cells were infected with lentiviral pHAGE-mortalin for 3 days before Western blot analysis of total-cell lysates and qPCR analysis of total RNA for MEK1 and MEK2 expression. Western blot results were analyzed by densitometry. Data (means ± standard errors of the means) are from three independent experiments. *, P < 0.05. (B) Western blot analysis of total lysates of IMR90-E1A cells coinfected with lentiviral pHAGE-mortalin (HA tagged or untagged; 4 days) and pHAGE-B-RafV600E (2 days). (C and D) LNCaP-pHAGE and LNCaP-Raf:ER cells, infected with pHAGE-Mort (untagged) for 2 days, were treated with 4-HT for 2 days. Total-cell lysates were analyzed by Western blotting (C), and RNA was analyzed by qPCR (D). (E) LNCaP-Raf:ER cells, infected with pHAGE-Mort for 2 days, were treated with 4-HT for 1 day. GFP expression indicates the efficiency of infection. (F) Cell cycle analysis of cells shown in panel E after 4-HT treatment for 1 day or 2 days. Data (means ± standard errors of the means) are from a representative experiment performed in triplicate. (G) LNCaP-Raf:ER cells stably expressing the p21CIP1 promoter-luciferase reporter S2260 were infected with increasing doses of pHAGE virus expressing mortalin, HSP72, HSC70, or BiP for 2 days. Cells were then treated with 4-HT for 2 days before the determination of luciferase activity. Data (means ± standard errors of the means) from a representative experiment performed in triplicate are expressed as fold changes from luciferase activity in untreated pHAGE-infected cells.
DISCUSSION
Our evaluation of mortalin in different cell models representing the two opposing contexts of Raf/MEK/ERK signaling (i.e., proliferation versus growth inhibition) establishes mortalin as a negative regulator of pathway-mediated growth-inhibitory signaling. This ability is critical for mortalin to regulate p21CIP1 expression, whether in a p53-dependent or a p53-independent manner, in B-RafV600E-transformed cancer cells. These functions of mortalin, together with its upregulated expression in human melanoma tissues and multiple cancer cell lines exhibiting aberrant Raf/MEK/ERK activity, suggest that mortalin may act as a molecular switch in determining the physiological outcomes of dysregulated Raf/MEK/ERK signaling. Our study also demonstrates that mortalin depletion can, independently of Raf/MEK/ERK, induce cell death, which is limited by p21CIP1 induction.
Our data indicate that mortalin can modulate MEK/ERK activity under Raf-activated conditions. Accordingly, one may speculate that some of the tumor-suppressive signaling of Raf/MEK/ERK becomes inactive as upregulated mortalin limits the extent of pathway signaling. Therefore, modulation of pathway activity below the threshold that triggers tumor-suppressive responses, yet above the threshold that stimulates cell proliferation, may also be a strategy for cancer to bypass tumor-suppressive responses. In agreement with this notion, it has been shown previously that upregulated AKT3 in melanoma downregulates MEK/ERK activity via Ser364/428 phosphorylation of B-RafV600E to facilitate cell proliferation (38). It appears that a main mechanism by which mortalin modulates MEK/ERK activity is the regulation of MEK protein levels. Intriguingly, a recent study demonstrated that varying the relative concentration of MEK in a synthetic model system of Raf/MEK/ERK signaling is most effective at conferring flexibility on the system response and priming the cascade for ultrasensitivity (39), suggesting the significance of MEK stoichiometry in the regulation of Raf/MEK/ERK signaling. Therefore, fine-tuning of MEK levels may be important in cells harboring B-RafV600E or other upstream alterations, and mortalin may be a key regulator in this process. Given the interaction of mortalin with MEK1/2, it is tantalizing to speculate that as a molecular chaperone, mortalin may regulate both protein maturation and protein degradation, a function that has recently been characterized for the related molecular chaperone HSP70 (40).
The present study provides a novel insight into p21CIP1 regulation in Raf/MEK/ERK-activated cancer. In response to mortalin depletion, B-RafV600E-transformed cancer cells expressed p21CIP1 regardless of p53 status. In contrast to the pattern for p16INK4A (41), genetic or epigenetic alteration of p21CIP1 is rare in cancer, and thus p21CIP1 silencing in cancer, including melanomas (42), has been attributed mainly to p53 inactivation. Our data suggest that mortalin is also an important determinant of p21CIP1 silencing in B-RafV600E-transformed cancers because, unless mortalin is upregulated, MEK/ERK can still mediate p21CIP1 transcription independently of p53. It is therefore conceivable that not only p53 inactivation but also mortalin upregulation is crucial for the evasion of p21CIP1-mediated tumor suppression in the course of B-RafV600E carcinogenesis. The operation of these two mechanisms may render genetic or epigenetic alteration of p21CIP1 unnecessary.
The requirement for p53 presents a contrast with the regulation of p21CIP1 by mortalin. It has been shown previously that mortalin can affect the localization and stability of p53 (19–21). This regulation may be intact in SK-MEL-1 cells, given that mortalin knockdown increased p53 levels in these cells. Accordingly, the requirement of MEK/ERK activity for this event in SK-MEL-1 cells may indicate a role for MEK/ERK in p53 regulation. Indeed, it has been reported previously that the Raf/MEK/ERK pathway can increase the stability of p53 via p38-regulated/activated protein kinase (43). On the other hand, in the p53-independent mechanism, MEK/ERK may activate transcription factors other than p53 to induce p21CIP1 transcription. In support of this hypothesis, the Raf/MEK/ERK pathway can mediate p21CIP1 transcription via not only p53-dependent but also p53-independent mechanisms (27, 36). Indeed, p53-independent p21CIP1 transcription can be mediated by different transcription factors, including ETS transcription factors, GATA, and SP (36, 44, 45). Cancer cells may stochastically develop an alteration that affects p53 or these transcription factors, which may cause the difference between SK-MEL-28 and SK-MEL-1 cells with regard to p21CIP1 regulation.
The ability of mortalin to regulate Raf/MEK/ERK-mediated growth-inhibitory signaling may have potential clinical significance. Mortalin targeting in B-RafV600E-transformed cancer would be unique in exploiting aberrant Raf/MEK/ERK activity to induce tumor suppression, in contrast to the current strategies that aim to inhibit the pathway activity in tumors. Although initially effective, pharmacological inhibition of B-RafV600E or MEK1/2 eventually leads to the emergence of resistance in cancer, creating a demand for additional therapeutic strategies (46). Because these recurring tumors still rely on MEK/ERK activity, it may be possible to divert their Raf/MEK/ERK signaling toward a growth-inhibitory context via mortalin targeting. Additional potential is also predicted. Inactivation of p53 is a hallmark of cancer, and its reactivation has been proposed as a novel therapeutic strategy (47, 48). For example, pharmacological inhibition of the p53 E3-ligase HDM2 could reactivate p53, increase p21CIP1 transcription, and induce growth arrest in some melanoma cell lines (42). This strategy is limited to tumors carrying wild-type p53 (35). Because mortalin depletion could activate p53-independent p21CIP1 transcription via MEK/ERK, mortalin targeting may be potent in B-RafV600E-transformed tumors deficient in wild-type p53.
Our data may also provide a rationale for hypothesizing that simultaneous targeting of mortalin and p21CIP1 would effectively induce cell death in certain tumor types, because mortalin depletion-induced cell death is limited by p21CIP1 induction. For that reason, it is important to understand the nature of cell death mediated by the depletion of mortalin and p21CIP1. Because overexpression of the antiapoptotic factor Bcl-2 could rescue cells from cell death induced by mortalin or p21CIP1 depletion, it is conceivable that mortalin and p21CIP1 may regulate cell survival via mechanisms overlapping at the level of Bcl-2 family protein regulation. Nevertheless, the question remains how Bcl-2 rescues mortalin-depleted SK-MEL-28 cells, because mortalin depletion in this cell line did not significantly increase the level of annexin V staining, suggesting that the cell death response of this cell line may not be apoptotic. Indeed, it is known that Bcl-2 family members also have “nonapoptotic” functions (49).
Mortalin expression was not affected by Raf/MEK/ERK signaling in any of the cell lines tested in our study. Therefore, mortalin upregulation in cancer may occur independently of B-RafV600E. Because mortalin is inducible by nutritional stresses, such as glucose deprivation (50), it is possible that mortalin upregulation is caused by a metabolic reprogramming that occurs at an early stage of tumorigenesis (51). Importantly, other HSP70 family members tested in this study could not effectively replace mortalin for p21CIP1 regulation, although some of them, i.e., HSC70 and HSP72, were previously noted for their antagonistic effects on cellular senescence (52–56). Therefore, mortalin may have a nonredundant role in the context of Raf/MEK/ERK-mediated p21CIP1 regulation. Although mortalin largely shares the characteristics of HSP70 molecular chaperones (40, 57), it is possible that an alteration in its specific clients develops a unique requirement for mortalin. Indeed, increasing evidence suggests that molecular chaperones can facilitate tumorigenesis by altering the stability or activity of important kinases and tumor suppressors (58). Elucidation of the mechanisms that underlie mortalin function and its upregulation in cancer is expected to advance our understanding of Raf/MEK/ERK-mediated carcinogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bassam Wakim (Protein and Nucleic Acid Core, Medical College of Wisconsin) for mass spectrometric analysis, Daniel Eastwood (Division of Biostatistics, Medical College of Wisconsin) for statistical analysis, and Alexandra F. Lerch-Gaggl (Pediatric BioBank & Analytical Tissue Core, Medical College of Wisconsin) for imaging immunohistochemistry data. We also thank Albert Girotti, Andrew Chan, and Barry Nelkin for critical review of the manuscript.
This work was supported by the National Cancer Institute (R01CA138441), the American Cancer Society (RSGM-10-189-01-TBE), and a FAMRI Young Investigator Award (062438) to J.-I.P.
We declare no conflict of interest.
Footnotes
Published ahead of print 19 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00021-13.
REFERENCES
- 1.Shaul YD, Seger R. 2007. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta 1773:1213–1226 [DOI] [PubMed] [Google Scholar]
- 2.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin RA. 2007. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 1773:1263–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts PJ, Der CJ. 2007. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26:3291–3310 [DOI] [PubMed] [Google Scholar]
- 4.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602 [DOI] [PubMed] [Google Scholar]
- 5.Zhu J, Woods D, McMahon M, Bishop JM. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 12:2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436:720–724 [DOI] [PubMed] [Google Scholar]
- 8.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. 2005. Tumour biology: senescence in premalignant tumours. Nature 436:642. [DOI] [PubMed] [Google Scholar]
- 9.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660–665 [DOI] [PubMed] [Google Scholar]
- 10.Mooi WJ, Peeper DS. 2006. Oncogene-induced cell senescence—halting on the road to cancer. N. Engl. J. Med. 355:1037–1046 [DOI] [PubMed] [Google Scholar]
- 11.Courtois-Cox S, Jones SL, Cichowski K. 2008. Many roads lead to oncogene-induced senescence. Oncogene 27:2801–2809 [DOI] [PubMed] [Google Scholar]
- 12.McDuff FK, Turner SD. 2011. Jailbreak: oncogene-induced senescence and its evasion. Cell. Signal. 23:6–13 [DOI] [PubMed] [Google Scholar]
- 13.Daugaard M, Rohde M, Jaattela M. 2007. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581:3702–3710 [DOI] [PubMed] [Google Scholar]
- 14.Deocaris CC, Widodo N, Ishii T, Kaul SC, Wadhwa R. 2007. Functional significance of minor structural and expression changes in stress chaperone mortalin. Ann. N. Y. Acad. Sci. 1119:165–175 [DOI] [PubMed] [Google Scholar]
- 15.Kaul SC, Deocaris CC, Wadhwa R. 2007. Three faces of mortalin: a housekeeper, guardian and killer. Exp. Gerontol. 42:263–274 [DOI] [PubMed] [Google Scholar]
- 16.Wadhwa R, Takano S, Kaur K, Deocaris CC, Pereira-Smith OM, Reddel RR, Kaul SC. 2006. Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int. J. Cancer 118:2973–2980 [DOI] [PubMed] [Google Scholar]
- 17.Wadhwa R, Takano S, Taira K, Kaul SC. 2004. Reduction in mortalin level by its antisense expression causes senescence-like growth arrest in human immortalized cells. J. Gene Med. 6:439–444 [DOI] [PubMed] [Google Scholar]
- 18.Wadhwa R, Takano S, Kaur K, Aida S, Yaguchi T, Kaul Z, Hirano T, Taira K, Kaul SC. 2005. Identification and characterization of molecular interactions between mortalin/mtHsp70 and HSP60. Biochem. J. 391:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadhwa R, Takano S, Robert M, Yoshida A, Nomura H, Reddel RR, Mitsui Y, Kaul SC. 1998. Inactivation of tumor suppressor p53 by mot-2, a hsp70 family member. J. Biol. Chem. 273:29586–29591 [DOI] [PubMed] [Google Scholar]
- 20.Wadhwa R, Yaguchi T, Hasan MK, Taira K, Kaul SC. 2003. Mortalin-MPD (mevalonate pyrophosphate decarboxylase) interactions and their role in control of cellular proliferation. Biochem. Biophys. Res. Commun. 302:735–742 [DOI] [PubMed] [Google Scholar]
- 21.Lu WJ, Lee NP, Kaul SC, Lan F, Poon RT, Wadhwa R, Luk JM. 2011. Mortalin-p53 interaction in cancer cells is stress dependent and constitutes a selective target for cancer therapy. Cell Death Differ. 18:1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong SK, Yoon S, Moelling C, Arthan D, Park JI. 2009. Noncatalytic function of ERK1/2 can promote Raf/MEK/ERK-mediated growth arrest signaling. J. Biol. Chem. 284:33006–33018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samuels ML, Weber MJ, Bishop JM, McMahon M. 1993. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human Raf-1 protein kinase. Mol. Cell. Biol. 13:6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vindelov LL, Christensen IJ, Nissen NI. 1983. A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry 3:323–327 [DOI] [PubMed] [Google Scholar]
- 25.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. 2006. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc. Natl. Acad. Sci. U. S. A. 103:16406–16411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401–406 [DOI] [PubMed] [Google Scholar]
- 27.Beier F, Taylor AC, LuValle P. 1999. The Raf-1/MEK/ERK pathway regulates the expression of the p21Cip1/Waf1 gene in chondrocytes. J. Biol. Chem. 274:30273–30279 [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 29.Hong SK, Kim JH, Lin MF, Park JI. 2011. The Raf/MEK/extracellular signal-regulated kinase 1/2 pathway can mediate growth inhibitory and differentiation signaling via androgen receptor downregulation in prostate cancer cells. Exp. Cell Res. 317:2671–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497–1501 [DOI] [PubMed] [Google Scholar]
- 31.Abbas T, Dutta A. 2009. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9:400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Javelaud D, Besançon F. 2002. Inactivation of p21WAF1 sensitizes cells to apoptosis via an increase of both p14ARF and p53 levels and an alteration of the Bax/Bcl-2 ratio. J. Biol. Chem. 277:37949–37954 [DOI] [PubMed] [Google Scholar]
- 33.Staversky RJ, Vitiello PF, Gehen SC, Helt CE, Rahman A, Keng PC, O'Reilly MA. 2006. p21Cip1/Waf1/Sdi1 protects against hyperoxia by maintaining expression of Bcl-XL. Free Radic. Biol. Med. 41:601–609 [DOI] [PubMed] [Google Scholar]
- 34.Vitiello PF, Staversky RJ, Gehen SC, Johnston CJ, Finkelstein JN, Wright TW, O'Reilly MA. 2006. p21Cip1 protection against hyperoxia requires Bcl-XL and is uncoupled from its ability to suppress growth. Am. J. Pathol. 168:1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan KD, Gallant-Behm CL, Henry RE, Fraikin JL, Espinosa JM. 2012. The p53 circuit board. Biochim. Biophys. Acta 1825:229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin SY, Kim CG, Lim Y, Lee YH. 2011. The ETS family transcription factor ELK-1 regulates induction of the cell cycle-regulatory gene p21Waf1/Cip1 and the BAX gene in sodium arsenite-exposed human keratinocyte HaCaT cells. J. Biol. Chem. 286:26860–26872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazar DF, Wiese RJ, Brady MJ, Mastick CC, Waters SB, Yamauchi K, Pessin JE, Cuatrecasas P, Saltiel AR. 1995. Mitogen-activated protein kinase kinase inhibition does not block the stimulation of glucose utilization by insulin. J. Biol. Chem. 270:20801–20807 [DOI] [PubMed] [Google Scholar]
- 38.Cheung M, Sharma A, Madhunapantula SV, Robertson GP. 2008. Akt3 and mutant V600E B-Raf cooperate to promote early melanoma development. Cancer Res. 68:3429–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. 2011. Tunable signal processing in synthetic MAP kinase cascades. Cell 144:119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, Clark WH, Jr, Tucker MA, Dracopoli NC. 1994. Germline p16 mutations in familial melanoma. Nat. Genet. 8:15–21 [DOI] [PubMed] [Google Scholar]
- 42.Ji Z, Njauw CN, Taylor M, Neel V, Flaherty KT, Tsao H. 2012. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J. Invest. Dermatol. 132:356–364 [DOI] [PubMed] [Google Scholar]
- 43.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, Dong MQ, Frangou CG, Yates JR, III, Wright PE, Han J. 2007. PRAK is essential for ras-induced senescence and tumor suppression. Cell 128:295–308 [DOI] [PubMed] [Google Scholar]
- 44.Papetti M, Wontakal SN, Stopka T, Skoultchi AI. 2010. GATA-1 directly regulates p21 gene expression during erythroid differentiation. Cell Cycle 9:1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chew YC, Adhikary G, Wilson GM, Xu W, Eckert RL. 2012. Sulforaphane induction of p21Cip1 cyclin-dependent kinase inhibitor expression requires p53 and Sp1 transcription factors and is p53-dependent. J. Biol. Chem. 287:16168–16178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solit DB, Rosen N. 2011. Resistance to BRAF inhibition in melanomas. N. Engl. J. Med. 364:772–774 [DOI] [PubMed] [Google Scholar]
- 47.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445:661–665 [DOI] [PubMed] [Google Scholar]
- 48.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonneau B, Prudent J, Popgeorgiev N, Gillet G. 2013. Non-apoptotic roles of Bcl-2 family: the calcium connection. Biochim. Biophys. Acta 1833:1755–1765 [DOI] [PubMed] [Google Scholar]
- 50.Merrick BA, Walker VR, He C, Patterson RM, Selkirk JK. 1997. Induction of novel Grp75 isoforms by 2-deoxyglucose in human and murine fibroblasts. Cancer Lett. 119:185–190 [DOI] [PubMed] [Google Scholar]
- 51.Ward PS, Thompson CB. 2012. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21:297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabai VL, Sherman MY, Yaglom JA. 2010. HSP72 depletion suppresses γH2AX activation by genotoxic stresses via p53/p21 signaling. Oncogene 29:1952–1962 [DOI] [PubMed] [Google Scholar]
- 53.Hupp TR, Meek DW, Midgley CA, Lane DP. 1992. Regulation of the specific DNA binding function of p53. Cell 71:875–886 [DOI] [PubMed] [Google Scholar]
- 54.O'Callaghan-Sunol C, Gabai VL, Sherman MY. 2007. Hsp27 modulates p53 signaling and suppresses cellular senescence. Cancer Res. 67:11779–11788 [DOI] [PubMed] [Google Scholar]
- 55.Walerych D, Kudla G, Gutkowska M, Wawrzynow B, Muller L, King FW, Helwak A, Boros J, Zylicz A, Zylicz M. 2004. Hsp90 chaperones wild-type p53 tumor suppressor protein. J. Biol. Chem. 279:48836–48845 [DOI] [PubMed] [Google Scholar]
- 56.Walerych D, Olszewski MB, Gutkowska M, Helwak A, Zylicz M, Zylicz A. 2009. Hsp70 molecular chaperones are required to support p53 tumor suppressor activity under stress conditions. Oncogene 28:4284–4294 [DOI] [PubMed] [Google Scholar]
- 57.Bukau B, Horwich AL. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366 [DOI] [PubMed] [Google Scholar]
- 58.Khalil AA, Kabapy NF, Deraz SF, Smith C. 2011. Heat shock proteins in oncology: diagnostic biomarkers or therapeutic targets? Biochim. Biophys. Acta 1816:89–104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.