Abstract
Ubiquitination plays important and diverse roles in modulating protein functions. As a C2-WW-HECT-type ubiquitin ligase, Smad ubiquitination regulatory factor 1 (Smurf1) commonly serves to regulate ubiquitin-dependent protein degradation in a number of signaling pathways. Here, we report a novel function of Smurf1 in regulating Wnt/β-catenin signaling through targeting axin for nonproteolytic ubiquitination. Our data unambiguously demonstrate that Smurf1 ubiquitinates axin through Lys 29 (K29)-linked polyubiquitin chains. Unexpectedly, Smurf1-mediated axin ubiquitination does not lead to its degradation but instead disrupts its interaction with the Wnt coreceptors LRP5/6, which subsequently attenuates Wnt-stimulated LRP6 phosphorylation and represses Wnt/β-catenin signaling. The inhibitory function of Smurf1 on Wnt/β-catenin signaling is further evidenced by analysis with Smurf1 knockout murine embryonic fibroblasts. We next identified K789 and K821 in axin as the ubiquitination sites by Smurf1. Consistently, Smurf1 could neither disrupt the interaction of an axinK789/821R double mutant with LRP5/6 nor attenuate the phosphorylation of LRP6 in axinK789/821R-expressing cells. Collectively, our studies uncover Smurf1 as a new regulator for the Wnt/β-catenin signaling pathway via modulating the activity of axin.
INTRODUCTION
During the past decade, ubiquitination has been discovered to regulate a variety of cellular processes, such as cell signaling transduction, cell cycle regulation, and gene transcription (1–3). The cascade of ubiquitination is composed of three steps: ATP-dependent activation of ubiquitin (Ub) by ubiquitin-activating enzyme (E1), delivery of Ub to ubiquitin-conjugating enzyme (E2), and ligation of Ub to the substrates via ubiquitin ligase (E3). In this process, the E3 ubiquitin ligases are mainly responsible for substrate selection and the specificity of Ub linkage (3). Briefly, most monoubiquitination and multiple monoubiquitination events are related to receptor internalization, histone regulation, and so on (4), while polyubiquitination, accomplished by sequential conjugation of Ub to a distinct lysine residue (or to the amino-terminal Met1 residue [5]) of the preceding one, regulates a wide range of protein functions and activities. There are seven lysines (K6, K11, K27, K29, K33, K48, and K63) in the Ub molecule. Generally, proteins marked by K48-linked poly-Ub chains are destined for proteasome-dependent degradation, whereas other types of Ub chains could regulate a variety of cellular processes by both proteolytic and nonproteolytic functions (5).
The E3 ubiquitin ligases have been classified into three major categories: the HECT-type E3s, the RING-type E3s, and the U box E3s. The HECT E3s are further divided into three subgroups, in which the C2-WW-HECT E3s are the best studied. The E3s of this subfamily are usually composed of an N-terminal C2 domain, two to four WW domains for substrate interactions, and a C-terminal HECT domain for maintaining the E3 catalytic activity (6). The representatives of this type of E3 include the neuronal precursor cell-expressed developmentally downregulated 4 (Nedd4) family proteins, as well as Smad ubiquitination regulatory factor 1 (Smurf1) and Smurf2. Both Smurf1 and Smurf2 antagonize the transforming growth factor β (TGF-β) pathway by targeting its crucial components for degradation (7, 8) and participate in the Wnt-PCP (planar cell polarity) signaling pathway by degrading the PCP protein Prickle1 (9). Despite sharing similarities in sequence and some functions, these two E3 ligases target distinct substrates and employ different regulatory mechanisms in some other situations. For example, Smurf1 but not Smurf2 is a key regulator for cell polarity (10) and osteoblast activity (11) by promoting the degradation of RhoA and phospho-MEKK2, respectively. Smurf2 also could trigger ubiquitin-dependent degradation of Smurf1, while Smurf1 is unable to induce Smurf2's degradation (12). These discrepancies strongly indicate that these two E3s have nonredundant functions.
The Wnt/β-catenin signaling pathway plays crucial roles in embryogenesis, and its deregulation has been correlated with tumorigenesis and many other human diseases (13, 14). Accumulating evidence demonstrated that ubiquitin-mediated regulation participates in multiple steps of the Wnt/β-catenin signaling pathway. An example is axin. Previous work has established that the RING-type E3 ligase RNF146 is able to ubiquitinate axin and promote its degradation (15), and the HECT-type E3 ligase Smurf2 could also target axin for degradation (16). Increasing findings regarding nonproteolytic ubiquitination through atypical ubiquitin linkages also have noted this pathway. For example, K63-linked polyubiquitination of Dvl is considered to promote Wnt signaling transduction (17), while HectD1-mediated antigen-presenting cell (APC) ubiquitination through K63 ubiquitin linkage inhibits Wnt signaling by facilitating axin-APC interaction (18). The E3 ligase EDD, targeting β-catenin via K11- or K29-linked polyubiquitination, was reported to stabilize β-catenin instead of triggering its degradation (19). Overall, modification of proteins by ubiquitin plays diverse roles in regulating Wnt/β-catenin signaling.
In this work, we discovered a nonproteolytic function of Smurf1. As a novel E3 ligase of axin, Smurf1 ubiquitinates axin at the K789 and K821 sites mainly through K29 ubiquitin linkage, which disrupts the association of axin with LRP5/6 and inhibits Wnt signaling transduction.
MATERIALS AND METHODS
Plasmids, siRNA, and antibodies.
Bacterially expressed Smurf1 and axin were created by introducing full-length human Smurf1 and axin into pET-28C and pGEX-4T2, respectively. Mouse Smurf1 and Smurf1-CA PCR products were produced from the cDNA library of mouse R1 embryonic stem cells and cloned into pFUGW-IRES-EGFP for lentiviral infection. Point mutations of axin and Ub (including Ub-K6, Ub-K11, Ub-K27, Ub-K29, and Ub-K33) were generated using a Stratagene QuikChange site-directed mutagenesis kit. Other plasmids have been applied previously (20–23). Specific short interfering RNA (siRNA) oligonucleotides against human Smurf1, Smurf2, and axin were synthesized by GenePharma. Their sequences were the following: Smurf1 siRNA-1, CCGACACUGUGAAAAACACTT (10, 24); Smurf1 siRNA-2, GCGUUUGGAUCUAUGCAAATT; Smurf2 siRNA, GAUGAGAACACUCCAAUUATT (25); and axin siRNA, CGAGAGCCAUCUACCGAAATT (26). The following commercial antibodies were used for Western blotting and immunoprecipitation: hemagglutinin (HA) and Myc (Covance); glutathione S-transferase (GST) and Flag (F1804) (Sigma-Aldrich); Smurf1, Smad7, β-actin, and glycogen synthase kinse 3α/β (GSK3α/β; Santa Cruz Biotechnology); axin1 (C76H11), LRP6 (C5C7), LRP6 (C47H12), phospho-LRP6 (Ser1490), Smad1, and Flag (no. 2368) (Cell Signaling Technology); Smurf2 (Abcam); and β-catenin (BD Biosciences). LRP6 (C5C7) antibody was used for assays in HEK293T cells and axin-stable cells, and LRP6 (C47H12) antibody was for assays in murine embryonic fibroblasts (MEFs). Flag-tagged rabbit antibody (no. 2368) was used to detect LRP5C3-Flag through Western blotting, while under other conditions, a Flag-tagged mouse antibody (F1804) was applied.
Cell culture, transfection, and luciferase assay.
HEK293 cells, HEK293T cells, WT MEFs, and Smurf1−/− MEFs were maintained in Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS). Axin-stable cells (HEK293T cells stably transfected with Flag-axin) were cultured in HEK293T medium plus 50 μg/ml hygromycin. Plasmid or siRNA transfection was performed using Lipofectamine plus reagent or Lipofectamine RNA interference (RNAi) max reagent (Invitrogen), respectively, and cotransfection of plasmids and siRNA was achieved using Lipofectamine 2000 reagent (Invitrogen). Luciferase assay was carried out as previously described (22).
Co-IP assay and in vivo ubiquitination assay.
Coimmunoprecipitation (co-IP) and Western blotting were performed as described before (27). The results of Western blotting were viewed by the Odyssey 9120 infrared imaging system (LI-COR) or FujiFilm Las 4000 (FujiFilm).
For in vivo ubiquitination assays, cells harvested from 35-mm dishes were lysed in 100 μl radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% [vol/vol] Triton X-100, 5 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, with phosphatase and protease inhibitors). The lysates were centrifuged at 16,000 × g at 4°C for 10 min. After that, the supernatants were switched to new tubes and 10 μl of 10% SDS was added. The samples were boiled at 95°C for 10 min to break protein-protein interactions and then diluted 10-fold with RIPA buffer. The diluted samples then were incubated with antibodies and protein A/G plus agarose (Santa Cruz Biotechnology, Inc.) at 4°C for 1.5 h. The precipitates were washed three times using RIPA buffer and denatured in SDS loading buffer at 95°C for 10 min.
In vitro binding.
Recombinant proteins were expressed and purified from Escherichia coli. In vitro binding was applied as previously described (21).
In vitro ubiquitination assay.
An in vitro ubiquitination assay was carried out in 30 μl of reaction mixture containing 125 ng of human E1, 500 ng of UbcH5C, 10 μg of HA-tagged Ub protein (Boston Biochem), 500 ng of GST-axin or GST, and 300 ng of 6His-Smurf1. Three microliters of the reaction buffer and 3 μl of ATP-Mg2+ (ubiquitin conjugation reaction buffer kit; Boston Biochem) were added as well. The reaction mixtures were incubated at 30°C for 2 h and terminated by the addition of the stop buffer (Boston Biochem). For the assay shown in Fig. 3B, axin-Flag was immunoprecipitated from transfected HEK293T cells, and the precipitates (with protein A/G plus agarose) were used as substrates for the following in vitro ubiquitination assay. 6His-tagged Ub or its mutant proteins were added as indicated. After a 2-h reaction with shaking, the samples were washed 3 times with RIPA buffer and analyzed by Western blotting.
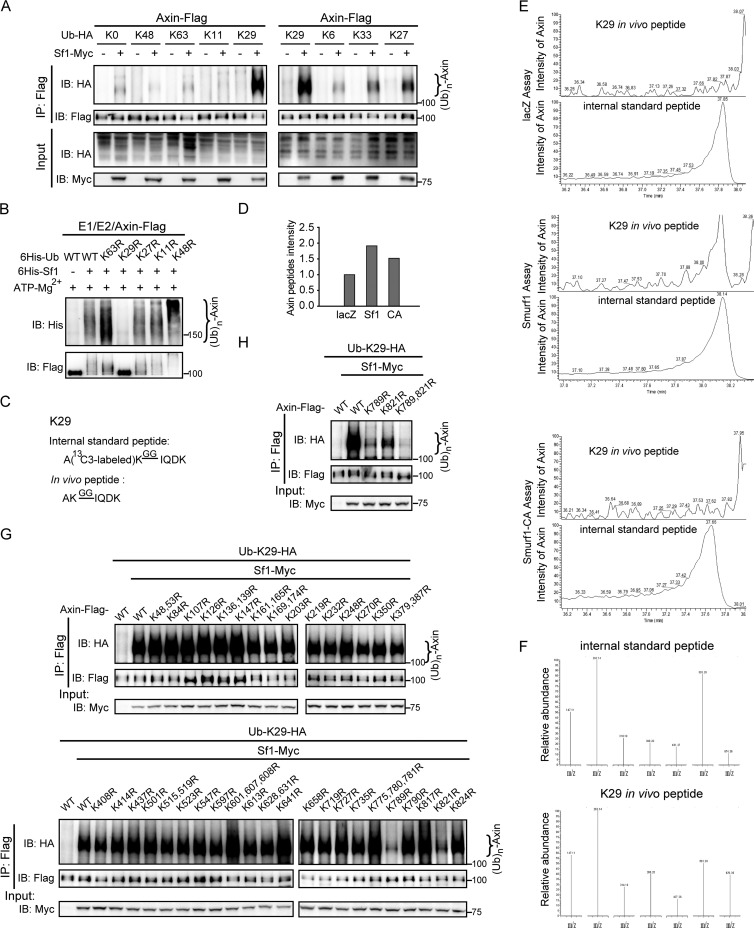
Fig 3.
Smurf1 promotes K29-linked polyubiquitination of axin. (A) Axin was ubiquitinated by Smurf1 in the presence of Ub-K29 in vivo. HEK293T cells were transfected with axin, Smurf1, and various Ub mutant plasmids, and after 24 h, an in vivo ubiquitination assay was performed. The samples were checked by Western blotting. (B) Axin ubiquitination by Smurf1 was largely reduced in the presence of Ub-K29R mutant protein compared to that with Ub-WT in vitro. The in vitro ubiquitination assay was applied using immunoprecipitated axin-Flag as the substrate, and the detailed process is described in Materials and Methods. (C) Amino acid sequence of the unique tryptic peptides digested from K29-linked polyubiquitinated substrates (light peptides), as well as its corresponding internal standard peptides (heavy peptides). Since isopeptide bond-modified lysine is relieved from trypsin digestion, the digestion between ubiquitin-ubiquitin linkage yields a missing lysine cleavage and generates a specific branched GG signature peptide. (D) Shotgun proteomic analysis of the intensity of axin protein in each SRM sample. Axin intensity of lacZ, 1.7E+07; axin intensity of Smurf1, 3.25E+07; axin intensity of Smurf1-CA, 2.58E+07. (E) The MS spectra of heavy/light K29-linked poly-Ub peptides in lacZ, Smurf1, and Smurf1-CA SRM assays are presented. (F) Associated MS/MS spectrum pattern of 7 SRM transitions derived from the heavy/light peptides in the Smurf1 SRM assay. (G and H) Smurf1 ubiquitinated axin at the sites of K789 and K821. In vivo ubiquitination assays were applied in transfected HEK293T cells and analyzed by Western blotting.
Cytoplasmic β-catenin assay.
Cytoplasmic β-catenin was extracted by following a previously described procedure (20).
Target gene assay.
Extraction of total RNAs and the subsequent assays of reverse transcription-PCR and quantitative real-time PCR (qPCR) were performed as previously described (27). The sequences of specific primers were the following: human GAPDH, 5′-GCACCACCAACTGCTTA-3′ and 5′-AGTAGAGGCAGGGATGA-3′; human AXIN2, 5′-AGTGTGAGGTCCACGGAAAC-3′ and 5′-CTTCACACTGCGATGCATTT-3′; human NKD1, 5′-GTCAACCACTCCCCAACATC-3′ and 5′-AATGGTGGTAGCA GCCAGAC-3′; mouse GAPDH, 5′-AACTTTGGCATTGTGGAAGG-3′ and 5′-CACAT TGGGGGTAGGAACAC-3′; mouse LEF1, 5′-CGATGAGATGATCCCCTTCA-3′ and 5′-TTGTCGTGGTAGGGCTCC-3′; and mouse NKD1, 5′-AGAACATTGAGAGGAGAAAC CAC-3′ and 5′-AGCCTGGGTCCACACCTT-3′.
Lentiviral infection in Smurf1−/− MEFs.
Lentiviral vectors were generated by cotransfecting pFUGW transfer vector, pCMVΔR9, and pVSV-G into HEK293T cells. Forty-eight hours later, cell medium containing the packaged viruses was collected and added to Smurf1−/− MEFs in the presence of 8 μg/ml Polybrene. After 8 h, the supernatant was switched to fresh growth medium, and 48 h later the infected MEFs were used for the following assays.
Shotgun proteomic assay.
To estimate the amount of axin protein in each sample used in the SRM (selected reaction monitoring) assay, an aliquot (1/5 for each sample) was used for the shotgun proteomic assay. The tryptic peptides were separated by nanoflow liquid chromatography and analyzed by tandem mass spectrometry (MS/MS; Thermo Electron Finnigan) as described previously (28). Briefly, an LTQ-Orbitrap equipped with an NSI nanospray source (1.7 kV) was operated in data-dependent mode in which the normalized collision energy was 35%. The full scan was performed in an Orbitrap analyzer (resolution = 100,000 at m/z 400), followed by MS/MS acquisition of the 10 most intense ions in the LTQ. Mass calibration used an internal lock mass (m/z 445.12057), the dynamic exclusion repeat count was 2, the repeat duration was 30 s, and the exclusion duration window was 90 s. Raw Orbitrap full-scan MS and ion trap MS/MS spectra were processed by MaxQuant 1.0.14.14 (29).
All identified MS/MS spectra were searched against an in-house-created target/decoy database (human IPI database, version 3.61) using Mascot version 2.1 (Matrix Science). Spectra were initially searched with a mass tolerance (7 ppm in MS and 0.5 Da in MS/MS) and strict trypsin specificity allowing up to 2 missed cleavages. Cysteine carbamidomethylation was searched as a fixed modification, whereas N-acetyl protein and oxidized methionine were searched as variable modifications. The resulting Mascot html output files were loaded into MaxQuant and fixed to the estimated false discovery rate (FDR) of all peptide and protein identifications at a maximum of 1% (29). Extracted ion currents (XIC) of all isotopic clusters associated with all axin peptides were reported as the summarized intensity to estimate the amount of axin protein.
SRM assay.
The stable isotope-labeled internal standard peptide A[13C-3]KGGIQDK (actual purity, 96.06%) was synthesized by GL Biochem (Shanghai), Ltd. Reverse-phase high-performance liquid chromatography (RP-HPLC) fractionation was performed on a Picofrit column (75 μm by 100 mm; length of C18 section of column, 5 μm; tip length, 15 μm; New Objective) using gradient B (0.1% formic acid in acetonitrile) from 0 to 40% in 2 min at a flow rate of approximately 300 nl/min after the split. A triple-quadrupole mass spectrometer (TSQ Vantage; ThermoFisher Scientific) was used to acquire SRM data. For ionization, a spray voltage of 1,800 V and a capillary temperature of 200°C were set. The selectivity of Q1 and Q3 was set at 0.2 and 0.7 Da (FWHM), respectively, and the collision gas pressure of Q2 was set at 1.2 mTorr argon. The collision energy (CE) of each theoretic SRM transition for isotope-labeled internal standard peptide was optimized, and seven better SRM transitions were selected. In all 3 SRM assays, equal amounts of isotope-labeled internal standard peptides were spiked in each in vivo sample to analyze the A[13C-3]KGGIQDK internal standard peptide and in vivo AKGGIQDK peptide. For the K29-linked polyubiquitinated heavy/light peptides, seven SRM transitions were monitored and a scan time of 20 ms was used. The best transition was used to quantify the corresponding peptide. It was considered not available if the signal-to-noise ratio was less than 3 (30).
RESULTS
Smurf1 is a novel E3 ubiquitin ligase for axin.
As described in the introduction, ubiquitination plays important roles in Wnt/β-catenin signaling. To discover potential novel E3 ligases for the Wnt/β-catenin pathway key component axin, we performed an in vivo ubiquitination assay using anti-Flag antibody in HEK293T cells transfected with Flag-tagged axin, HA-tagged Ub, and a group of E3 ligases, including HECT-type E3s (E6-AP, Smurf1, Smurf2, ITCH, WWP1, WWP2, NEDD4, and NEDD4L) and RING-type E3s (RBCK1, Siah1a, and RNF31). Among these E3 ligases, Smurf1 remarkably induced axin ubiquitination, as did its homologous protein Smurf2, a known E3 ligase of axin which destabilizes axin (16) (Fig. 1A). We then focused our attention on Smurf1-mediated axin ubiquitination.
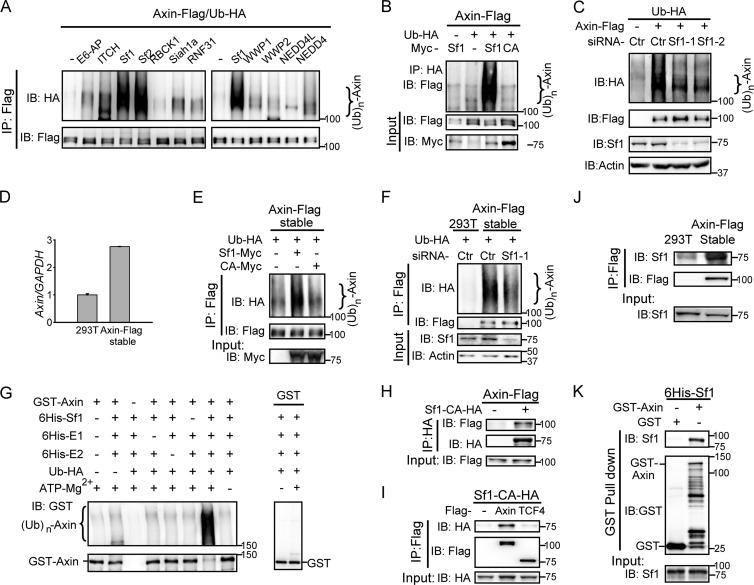
Fig 1.
Smurf1 is a novel E3 ubiquitin ligase for axin. (A) Smurf1 promoted exogenous axin ubiquitination. HEK293T cells were transfected with axin-Flag, Ub-HA, and a series of E3 ligases. Twenty-four hours later, cells were harvested in RIPA buffer with high SDS concentrations, and then the cell lysates were boiled for 10 min to break protein-protein interactions so as to exclude the possibility that the observed ubiquitination stemmed from axin's associated proteins. After that, the samples were diluted 10-fold with RIPA buffer, followed by immunoprecipitation (IP) using anti-Flag antibody. Processes are described in Materials and Methods. Sf1, Smurf1; Sf2, Smurf2. IB, immunoblot. (B) Smurf1, but not Smurf1-CA, induced axin ubiquitination. Cell lysates of transfected HEK293T cells were subjected to in vivo ubiquitination assay by immunoprecipitating Ub-HA, and the ubiquitinated axin-Flag was measured by anti-Flag antibody. CA, Smurf1-CA. (C) Knockdown of Smurf1 decreased the ubiquitination of exogenous axin. An in vivo ubiquitination assay was performed in HEK293T cells transfected with the indicated plasmids and siRNAs. The precipitates were analyzed by Western blotting. Actin, β-actin. (D) The mRNA level of axin in axin-Flag stable cells was about three times that of endogenous axin in HEK293T cells. Total RNA in HEK293T cells and axin-stable cells was extracted, and the expression of Axin mRNA was detected by qPCR and normalized by human GAPDH mRNA. (E and F) Smurf1 promoted axin ubiquitination in axin-stable cells. In vivo ubiquitination assays were applied in axin-stable cells transfected with the indicated plasmids or siRNAs. (G) In vitro ubiquitination of axin by Smurf1. The in vitro ubiquitination assay was carried out and analyzed by Western blotting. (H and I) Cotransfected axin and Smurf1 interacted with each other. Co-IP assays were carried out in HEK293T cells transfected with the indicated plasmids, and the precipitates were checked by Western blotting. (J) Endogenous Smurf1 was coimmunoprecipitated with axin in axin-stable cells. A co-IP assay was performed in axin-stable cells, and the precipitates were analyzed by Western blotting. (K) Smurf1 interacted with axin in vitro. The GST pulldown assay was performed and examined by Western blotting.
To further confirm the ubiquitination of axin by Smurf1, we pulled down Ub-HA by anti-HA antibody and tested axin ubiquitination using Western blotting with anti-Flag antibody. In addition, an E3 ligase-inactive mutant of Smurf1, Smurf1-CA (Cys699 is replaced by Ala [24]), was introduced to examine whether the E3 catalytic activity of Smurf1 is required. As Fig. 1B showed, a significant ubiquitination of axin was observed when coexpressed with Smurf1 but not with Smurf1-CA. Meanwhile, depletion of endogenous Smurf1 by Smurf1-specific siRNAs (Smurf1 siRNA-1 and siRNA-2) led to decreased ubiquitination of axin (Fig. 1C). We also examined the effect of Smurf1 on axin ubiquitination in axin-stable cells stably expressing axin-Flag at a level about three times that of endogenous axin in HEK293T cells (Fig. 1D). In vivo ubiquitination assay showed that overexpression of Smurf1 in axin-stable cells significantly increased axin ubiquitination (Fig. 1E), whereas knockdown (KD) of Smurf1 led to an obvious decrease of axin ubiquitination (Fig. 1F). To further confirm that axin is the substrate of Smurf1, we performed an in vitro ubiquitination assay using recombinant purified GST-tagged axin and 6His-tagged Smurf1. As shown in Fig. 1G, Smurf1 dramatically induced the ubiquitination of axin. Together, these results indicate that Smurf1 is an E3 ligase for axin.
To test whether Smurf1 interacts with axin physically, we performed IP assays in HEK293T cells using ectopically expressed axin and Smurf1-CA. As shown in Fig. 1H and I, axin interacted with Smurf1, and their interaction was independent of Smurf1 E3 ligase activity. As a control, Smurf1 did not associate with TCF4, a component in the Wnt/β-catenin signaling pathway (Fig. 1I). Furthermore, the interaction was also observed for endogenous Smurf1 in axin-stable cells (Fig. 1J). Finally, GST pulldown assay using recombinant purified proteins of axin and Smurf1 suggests that the interaction between axin and Smurf1 is direct (Fig. 1K).
Smurf1-mediated axin ubiquitination does not affect axin stability.
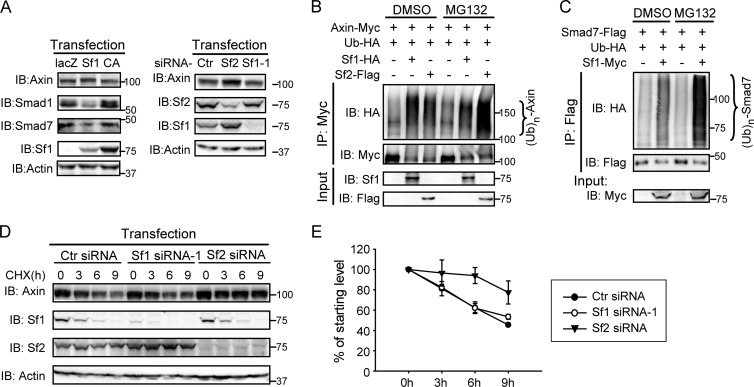
Smurf2 has been reported to target axin for its degradation. Therefore, we set out to investigate whether Smurf1-promoted axin ubiquitination plays a role similar to that of Smurf2. Surprisingly, no apparent decrease of axin levels was detected when Smurf1 was overexpressed (Fig. 2A). In contrast, the protein levels of Smad1 and Smad7 were reduced significantly by the addition of Smurf1, consistent with previous reports (8, 31) (Fig. 2A, left). As mentioned before, despite sharing high sequence similarity (∼75%), Smurf1 and Smurf2 display distinct properties in some situations. Thus, we focused our attention on exploring a possible distinct role of Smurf1-mediated axin ubiquitination. First, as shown in Fig. 2A, knockdown of Smurf1 had no effect on axin levels, whereas knockdown of Smurf2 dramatically increased the steady-state levels of axin. We next tested whether Smurf1-mediated axin ubiquitination accumulates when proteasome-dependent degradation is blocked by the proteasome inhibitor MG132. As shown in Fig. 2B and C, MG132 enhanced the accumulation of axin ubiquitination triggered by Smurf2 and also the Smad7 ubiquitination by Smurf1, but it had no effect on Smurf1-mediated axin ubiquitination, indicating that the latter is not subject to proteasome-dependent degradation. We then measured whether the turnover rate of axin is modulated by Smurf1 using cycloheximide (CHX) treatment, a common method of blocking protein synthesis. As shown in Fig. 2D and E, the half-life of axin was prolonged in Smurf2-KD but not in Smurf1-KD cells. With these results combined, we conclude that Smurf1, unlike Smurf2, does not target axin for degradation.
Fig 2.
Smurf1-mediated axin ubiquitination does not affect axin stability. (A) No apparent change of axin protein levels was detected in Smurf1-overexpressing or Smurf1-depleted cells. HEK293T cells were transfected with the indicated plasmids (left) or siRNAs (right), and endogenous proteins were examined by Western blotting. (B and C) Axin ubiquitination by Smurf1 did not accumulate under MG132 treatment, whereas that by Smurf2 or the Smad7 ubiquitination by Smurf1 was increased. In vivo ubiquitination assays were performed in HEK293T cells transfected with the indicated plasmids. Cells were treated with DMSO or MG132 (15 μM) for 4 h before harvest. (D and E) Smurf1 did not affect the turnover of axin protein. HEK293T cells transfected with the indicated siRNAs were treated with 15 μg/ml cycloheximide (CHX) for various times, and endogenous axin was analyzed by Western blotting. The graph in panel E represents the results from three independent experiments. Ctr, control.
Smurf1 promotes K29-linked polyubiquitination of axin.
To better understand the characteristics of Smurf1-mediated axin ubiquitination, we set out to investigate the types of ubiquitin linkage using a panel of Ub mutant plasmids in which no or only one lysine was retained. HEK293T cells were transfected with axin-Flag and one of these Ub mutants with or without Smurf1, followed by in vivo ubiquitination assays. Intriguingly, like Ub-K0, Ub-K6, Ub-K11, and Ub-K48, Ub-K63 did not promote Smurf1-mediated axin ubiquitination, while in the presence of Ub-K29, axin was strongly ubiquitinated by Smurf1 (Fig. 3A, left). Ub-K27 and Ub-K33 were also observed to induce ubiquitination of axin, but at a much weaker level (Fig. 3A, right). We next carried out an in vitro assay using Ub and its mutants, including Ub-K63R, Ub-K29R, Ub-K27R, Ub-K11R, and Ub-K48R. As shown in Fig. 3B, Ub-K29R dramatically reduced Smurf1-mediated axin ubiquitination, whereas other Ub mutants, including Ub-K27R, maintained their abilities to be linked to axin by Smurf1, as did wild-type Ub (Ub-WT).
The key role of Ub-K29 in Smurf1-induced axin ubiquitination was further confirmed by a targeted proteomic technology, SRM (selected reaction monitoring) assay, in which isotope-labeled internal standard peptide was used to quantify the tryptic peptides digested from the ubiquitinated substrates. The sequences of K29-internal standard peptide (heavy peptide) and its coeluting in vivo peptide (light peptide) derived from the digested proteins are illustrated in Fig. 3C. In our approaches, in vivo ubiquitination assay using anti-Flag antibody was first carried out in axin-stable cells transfected with lacZ, Smurf1, or Smurf1-CA, and the precipitates were then digested with trypsin for the following SRM assays. The intensity of axin in the 3 SRM samples was relatively equal (Fig. 3D). As shown in Fig. 3E, although the MS spectra of internal standard peptides (heavy peptides) were observed in all 3 SRM assays, those of in vivo peptides (light peptides) was only detected for Smurf1. Moreover, the associated MS/MS spectrum pattern of 7 SRM transitions derived from the heavy/light peptides in lysates of cells overexpressing Smurf1 further substantiates our conclusion that Smurf1 ubiquitinates axin primarily through K29 ubiquitin linkage.
Smurf1 ubiquitinates axin at the sites of K789 and K821.
There are 48 lysines in axin, and to identify the ubiquitination sites conferred by Smurf1, a series of axin K-to-R mutants were generated by replacing each of the 48 lysines with arginine. In vivo ubiquitination assays were performed in HEK293T cells transfected with Ub-K29, Smurf1, and wild-type axin (axinWT) or one of the axin mutants. Among all of these mutants, two of them, axinK789R and axinK821R, exhibited resistance to Smurf1-mediated ubiquitination (Fig. 3G). Additionally, among axinWT, axinK789R, axinK821R, and axinK789/821R, the double mutant displayed the lowest Ub-conjugated levels (Fig. 3H), indicating that K789 and K821 are the key Smurf1 ubiquitination sites in axin.
Smurf1 inhibits Wnt/β-catenin signaling.
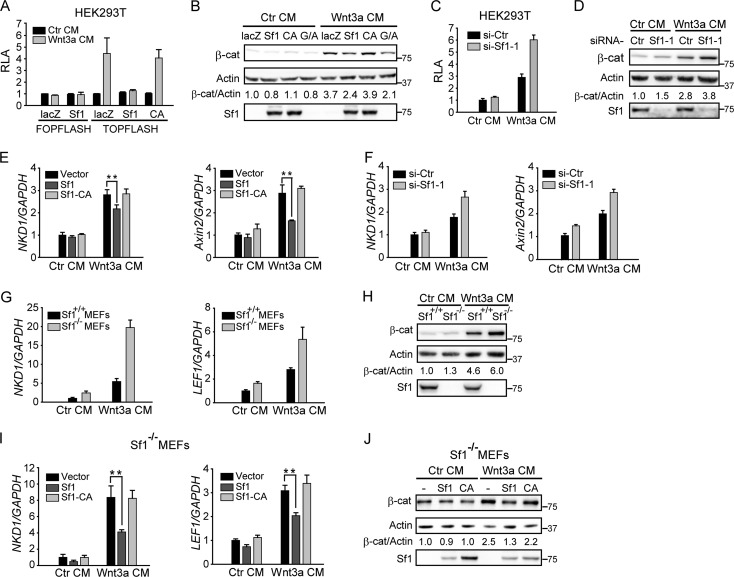
Considering that axin is a critical player in regulating Wnt/β-catenin signaling, we investigated the potential role of Smurf1 in the Wnt pathway. First, we used a TOPFlash reporter system and found that Smurf1, but not Smurf1-CA, dramatically decreased Wnt3a-stimulated TOPFlash reporter activity without detectable activity on FOPFlash (Fig. 4A). Second, overexpression of Smurf1 reduced cytoplasmic β-catenin levels, as did axin and GSK3β (Fig. 4B). We next carried out loss-of-function assays using Smurf1-specific siRNA as shown in Fig. 4C and D. The depletion of Smurf1 increased Wnt reporter activity and cytoplasmic β-catenin accumulation. Finally, we assessed the effects of Smurf1 on expression levels of two endogenous Wnt target genes, Axin2 and NKD1. Consistent results were obtained indicating that overexpression of Smurf1 decreased the Wnt-promoted increase in Axin2 and NKD1 levels (Fig. 4E), while knockdown of Smurf1 increased them (Fig. 4F). To exclude possible off-target effects of Smurf1 siRNA, Smurf1-knockout MEF cells (Smurf1−/− MEFs) were examined. Compared to wild-type MEFs, an increment of both Wnt target gene expression (mouse LEF1 and NKD1) and cytoplasmic β-catenin levels was detected in Smurf1−/− MEFs (Fig. 4G and H), and introduction of Smurf1 into Smurf1−/− MEFs by lentiviral infection reversed the effects (Fig. 4I and J). Taking these results together, we conclude that Smurf1 negatively regulates Wnt/β-catenin signaling.
Fig 4.
Smurf1 inhibits Wnt/β-catenin signaling. (A) Smurf1, but not Smurf1-CA, inhibited Wnt-induced TOPFlash activity. HEK293T cells were transfected with GFP, the Wnt reporter gene (TOPFlash or FOPFlash), and Smurf1 or Smurf1-CA. Cells were stimulated by control conditional medium (Ctr CM) or Wnt3a CM for 6 h, and then relative luciferase activity (RLA) was measured. (B) Smurf1 inhibited Wnt3a-induced accumulation of cytoplasmic β-catenin. The cytoplasmic fraction was extracted from the transfected HEK293T cells and then subjected to Western blotting. Cells were stimulated by Ctr CM or Wnt3a CM for 4 h before being lysed. G/A, cotransfection of GSK3β and axin; β-cat, β-catenin. (C and D) Knockdown of Smurf1 promoted Wnt3a-induced TOPFlash activity and cytoplasmic β-catenin levels. HEK293T cells were transfected with the indicated siRNAs. In panel C, GFP and the TOPFlash reporter plasmid were cotransfected. Forty-eight hours later, cells were treated with Wnt3a CM, and then luciferase activity and cytoplasmic β-catenin levels were tested as described for panels A and B. (E and F) Smurf1 inhibited the expression of Wnt target genes Axin2 and NKD1. qPCR analysis of human NKD1 mRNA and Axin2 mRNA was applied to HEK293 cells transfected with the indicated plasmids (E) or siRNAs (F). Cells were stimulated by Wnt3a CM for 8 h before harvest. All results were normalized by human GAPDH mRNA. (G and H) Higher expression of Wnt target gene and higher levels of cytoplasmic β-catenin were detected in Smurf1−/− MEFs. In panel G, WT MEFs and Smurf1−/− MEFs were treated with Ctr CM or Wnt3a CM for 8 h, and qPCR analyses of mouse NKD1 and LEF1 mRNA were performed. The data were normalized by mouse GAPDH mRNA. In panel H, the cytosolic fraction of WT MEFs and Smurf1−/− MEFs was tested by Western blotting. Cells were harvested after 4 h of incubation of Ctr CM or Wnt3a CM. (I and J) Introduction of Smurf1 to Smurf1−/− MEFs decreased NKD1 and LEF1 expression as well as cytoplasmic β-catenin accumulation. Smurf1 or Smurf1-CA was transfected to Smurf1−/− MEFs by lentiviral infection, and 48 h later, assays for qPCR measurement and cytoplasmic β-catenin analysis were performed as described for panels G and H. Data shown here are presented as means ± standard deviations (SD) from three independent assays. Statistical significance was determined using Student's t test. **, P < 0.01.
Smurf1-mediated axin ubiquitination inhibits axin-LRP5/6 interaction and LRP6 phosphorylation.
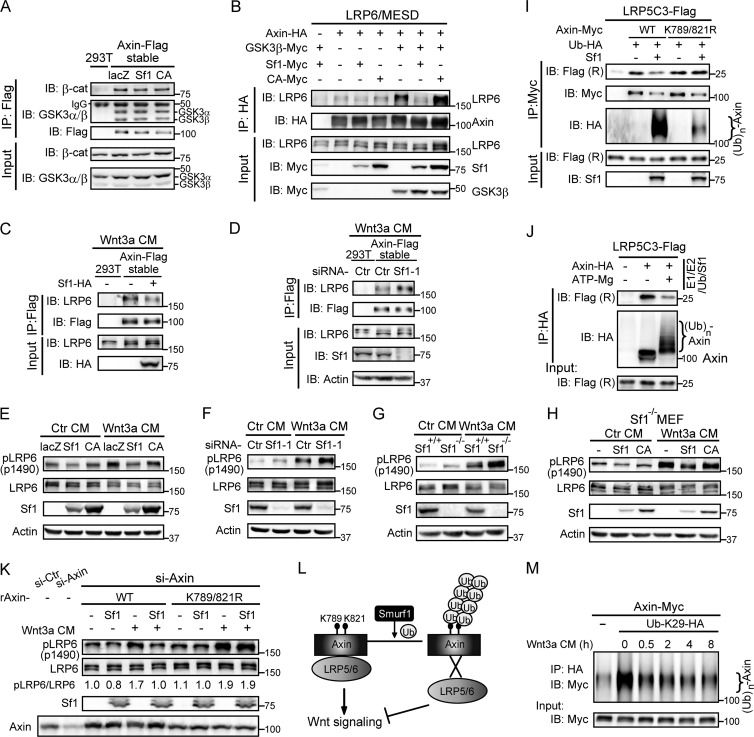
We next examined how Smurf1-mediated axin ubiquitination downregulated Wnt/β-catenin signaling. Previous studies indicate that axin plays a dual role in Wnt signaling. As a scaffold protein for the β-catenin destruction complex, axin inhibits Wnt signaling through promoting β-catenin degradation (13). On the other hand, through interacting with LRP6, axin facilitates the recruitment of GSK3 to the plasma membrane, which promotes LRP5/6 phosphorylation and Wnt signaling transduction (23, 32). To elucidate the role of Smurf1-mediated axin ubiquitination, we first examined the effects of Smurf1 on axin's function as a scaffold for the β-catenin destruction complex. As displayed in Fig. 5A, a co-IP assay performed on axin-stable cells showed that the association of axin with β-catenin or GSK3β, two major components of the destruction complex, was not affected by Smurf1.
Fig 5.
Smurf1-mediated axin ubiquitination inhibits axin-LRP5/6 interaction and LRP6 phosphorylation. (A) Smurf1 did not affect the interaction between axin and β-catenin or GSK3. A co-IP assay was performed in axin-stable cells transfected with the indicated plasmids. The precipitates were measured by Western blotting. (B) Smurf1 inhibited the interaction between exogenous axin and LRP6. A co-IP assay was performed in HEK293T cells, and the samples were analyzed by Western blotting. (C and D) Smurf1 inhibited Wnt3a-induced axin-LRP6 interaction in axin-stable cells. Co-IP assays were applied to axin-stable cells transfected with the indicated plasmids or siRNAs. Cells were treated with Wnt3a CM for 3 h before harvest. (E and F) Smurf1 inhibited Wnt3a-induced LRP6 phosphorylation. HEK293T cells were transfected with the indicated plasmids (E) or siRNAs (F), and endogenous protein levels were analyzed by Western blotting. Cells were cultured in Ctr CM or Wnt3a CM for 1 h before harvest. (G) Higher level of phospho-LRP6 was detected in Smurf1−/− MEFs. WT MEFs and Smurf1−/− MEFs were treated with Ctr CM or Wnt3a CM for 1 h, and phospho-LRP6 was measured by Western blotting. (H) Overexpression of Smurf1 decreased the level of phospho-LRP6 in Smurf1−/− MEFs. Lentiviral infection was applied in Smurf1−/− MEFs. Forty-eight hours later, cells were lysed and the samples were analyzed by Western blotting. (I and J) Smurf1-mediated axin ubiquitination decreased the binding affinity between axin and LRP5C3. In panel I, axin was coexpressed with Smurf1 in HEK293T cells, while LRP5C3 was overexpressed separately. A two-step co-IP assay was performed as follows. Exogenous axin was immunoprecipitated first, and the precipitates were committed to a second IP process with the cell lysates containing LRP5C3. The final samples were checked by Western blotting. Flag (R), Flag (no. 2368) antibody. In panel J, exogenous axin precipitated from HEK293T cells was subjected to in vitro ubiquitination initiated by the addition of ATP/Mg2+ and then monitored by a co-IP experiment with LRP5C3. The precipitates were analyzed by Western blotting. (K) Cells in which endogenous axin was replaced by raxinK789/821R were resistant to Smurf1-induced attenuation of LRP6 phosphorylation. Axin siRNA was transfected along with the RNAi-resistant form of axinWT or axinK789/821R (raxinWT or raxinK789/821R), and Smurf1 was cotransfected as indicated. Forty-eight hours later, cells were treated with Ctr CM or Wnt3a CM for 1 h. Phospho-LRP6 and LRP6 were checked by Western blotting. (L) A proposed model for the role of Smurf1-mediated axin ubiquitination in Wnt/β-catenin signaling. (M) Wnt3a stimulation decreased K29 polyubiquitination of axin in a time-dependent manner. HEK293T cells were transfected with axin-Myc and Ub-K29-HA, and 24 h later, cells were treated with Wnt3a CM for the indicated times. In vivo ubiquitination assay was then carried out by anti-HA antibody, and the samples were analyzed by Western blotting.
We next examined whether Smurf1 plays a role in regulating axin-LRP5/6 interaction. HEK293T cells were transfected with axin, LRP6, MESD (molecular chaperone of LRP6 [33]), and Smurf1 or Smurf1-CA, followed by co-IP assays. GSK3β was introduced for its positive role in regulating axin-LRP5 interaction (23). As expected, GSK3β alone elicited strong interaction between axin and LRP6. However, addition of Smurf1 led to a significant attenuation of axin-LRP6 association (Fig. 5B). Consistent with this, in axin-stable cells, Smurf1 overexpression also disrupted Wnt3a-stimulated axin-LRP6 interaction (Fig. 5C); conversely, depletion of Smurf1 facilitated their interaction (Fig. 5D). We next examined Smurf1's effect on the phosphorylation levels of LRP6 by utilizing a phospho-LRP6 antibody specifically detecting GSK3-mediated p1490 phosphorylation. As shown in Fig. 5E, overexpression of Smurf1 did reduce the basal as well as Wnt3a-stimulated phosphorylation levels of LRP6. In contrast, knockdown of Smurf1 increased LRP6 phosphorylation, which showed a synergistic effect with Wnt3a stimulation (Fig. 5F). In line with this, measurable elevation of phospho-LRP6 was also observed in Smurf1−/− MEFs (Fig. 5G), which could be decreased by introduction of Smurf1 into Smurf1−/− MEFs through lentiviral infection (Fig. 5H).
We next investigated whether the effects of Smurf1 on axin-LRP5/6 interaction, as well as LRP6 phosphorylation levels, could be attributed to Smurf1-mediated ubiquitination of axin. To address this question, we carried out two parallel assays. First, axin was overexpressed with Smurf1, while LRP5C3 (the intracellular domain of LRP5 [23]) was expressed separately. The co-IP assay was then performed to test the effect of Smurf1-mediated axin ubiquitination on binding of axin with LRP5C3. Second, exogenous axin precipitated from HEK293T cells was subjected to in vitro ubiquitination initiated by the addition of ATP/Mg2+ and followed by a co-IP experiment with LRP5C3. As illustrated in Fig. 5I and J, attenuated axin-LRP5C3 association was observed only when axin was ubiquitinated by Smurf1. Meanwhile, the interaction of the axinK789/821R double mutant with LRP5C3 was barely affected by Smurf1, suggesting that Smurf1-mediated axin ubiquitination is required for disrupting axin-LRP5/6 association. Furthermore, when endogenous axin was knocked down by axin-specific siRNA and replaced by RNAi-resistant axinWT or axinK789/821R (raxinWT and raxinK789/821R, respectively), introduction of Smurf1 decreased LRP6 phosphorylation in raxinWT-expressing cells but not in raxinK789/821R ones (Fig. 5K). Collectively, these results demonstrate that Smurf1 inhibits Wnt signaling transduction by targeting axin for ubiquitination.
DISCUSSION
Research on Smurf1 began at the end of the last century. Smurf1 is a C2-WW-HECT E3 ubiquitin ligase and plays critical roles in the bone morphogenetic protein (BMP), Wnt-PCP, and MEKK-JNK pathways by promoting ubiquitin-dependent degradation of their key regulators, such as Smad1, Smad7, Prickle1, and phospho-MEKK2 (8, 9, 11, 31). However, in the present study, we provided compelling evidence to demonstrate that axin, as a novel substrate of Smurf1, was not subjected to degradation (Fig. 2), suggesting that Smurf1 executes a broader range of functions than was expected. One possible explanation for the nondegradation fate of axin is that Smurf1 functions differently via targeting different lysines on ubiquitin. So far, three-dimensional structures of five ubiquitin chains (including K6-, K11-, K48-, K63-, and Met1-linked chains) have been resolved which exhibit diverse conformational states (5). Such conformational diversity might direct targeted proteins to distinct partners or enzymes, by which these substrates are either degraded or execute other functions. In our case, Smurf1 ubiquitinates axin primarily through K29 ubiquitin linkage. Recent work has revealed some roles of K29 ubiquitin linkage involved in various cellular processes, including Wnt/β-catenin signaling (19): EDD ubiquitinates and stabilizes β-catenin through K11- or K29-ubiquitin linkage. However, this type of linkage has not been revealed for Smurf1 before. Previously, it was reported that Smurf1 promoted STAT1 degradation through K48 ubiquitin linkage (34). Thus, it is tempting to speculate that the novel K29 ubiquitin linkage in Smurf1-mediated axin ubiquitination prefers to regulate axin-LRP5/6 interaction rather than induce its degradation. More information about the types of ubiquitin linkage in other Smurf1-mediated ubiquitinations is required to confirm this point.
Another interesting question is why Smurf1 selectively attaches K29 ubiquitin chains to axin instead of K48. It has been reported that activities of Smurf1 are regulated by various proteins, which are represented by the examples of Cdh1 (35) and protein kinase A (PKA) (36). Cdh1 promotes Smurf1 activity by disturbing its autoinhibitory homodimer, while PKA-dependent phosphorylation of Smurf1 alters its substrates preference. Therefore, one possible answer is that some partners complexed with Smurf1 or particular modifications on Smurf1 facilitate K29 polyubiquitination of axin. It is also possible that a unique conformation adopted by the Smurf1-axin complex prefers the assembly of K29 poly-Ub chains. More studies concerning the effects of Smurf1 interaction partners as well as the three-dimensional structure of Smurf1-axin will help to elucidate this problem.
More importantly, our present work reveals the essential role of Smurf1 in regulating Wnt/β-catenin signaling. Smurf1 inhibits Wnt/β-catenin signaling by attenuating axin-LRP5/6 association and LRP6 phosphorylation (Fig. 4 and 5). In contrast, its related protein, Smurf2, promotes Wnt/β-catenin signaling by targeting axin for degradation (16), indicating that these two Smurf proteins have different effects on axin even though both ubiquitinate axin. There is actually a precedent for different roles of Smurf1 and Smurf2, which were reported by Fukunaga et al. for the regulation of breast cancer cell migration. They discovered that Smurf2 was an E3 ligase of Smurf1, and that knockdown of Smurf2 increased Smurf1 expression and promoted the migration of breast cancer cells, while knockdown of Smurf1 reduced cell migration (12). Considering the close relationship between Smurf1 and Smurf2, we believe that a precise mechanism exists in cells to modify their distinct functions in the Wnt/β-catenin signaling pathway; however, this needs to be further elucidated. Additionally, we provide solid evidence that the function of Smurf1 in Wnt/β-catenin signaling depends on Smurf1-mediated axin ubiquitination at K789 and K821 (Fig. 5I to K). Based on our findings, we propose a model in which Smurf1 ubiquitinates axin at the sites of K789 and K821 mainly through K29 ubiquitin linkage, and such modification does not trigger the degradation of axin. Instead, it disrupts axin-LRP5/6 interaction and inhibits Wnt signaling transduction (Fig. 5L). Previously, we identified Caprin2 as a positive regulator in Wnt/β-catenin signaling by promoting axin-LRP5 interaction (37), and more recently, a small molecule, HLY78, was found to activate Wnt/β-catenin signaling through modulating the interaction between axin and LRP6 (38). Here, we demonstrate that protein ubiquitination provides an alternative way to regulate axin-LRP5/6 interaction.
Interestingly, we discovered that Wnt3a stimulation led to a significant decrease of K29 polyubiquitination of axin in a time-dependent manner (Fig. 5M), suggesting that Smurf1-mediated axin ubiquitination is a constitutive event in resting cells that serves as an inhibitory mechanism to prevent the interaction between axin and LRP6 and to maintain low Wnt activity. In this hypothesis, Wnt3a treatment releases axin from Smurf1-mediated K29-linked polyubiquitination and facilitates Wnt signaling transduction. However, the mechanism for how Wnt3a decreases K29 ubiquitination of axin is still obscure. Some deubiquitination events may be involved in the process. Further exploration is required to fully understand the mechanism.
Smurf1−/− mice have been generated and were reported to show an increased bone mass phenotype, which is probably due to the enhanced osteoblast activity (11). Yamashita et al. had attributed the bone phenotype to altered MEKK-JNK signaling via a Smurf1-mediated ubiquitin-dependent mechanism (11). However, it is known that hyperactivated Wnt/β-catenin signaling can also lead to increased bone mass. For example, Wnt3a and BMP9 cooperate to induce alkaline phosphatase in mesenchymal stem cells (39), and DKK1 and the Wnt receptor antagonist sclerostin negatively regulate bone mass by inhibiting Wnt/β-catenin signaling, the dysfunction of which caused bone mass increase (40). Thus, the bone phenotype in Smurf1−/− mice might also be attributed to the increased activity of Wnt signaling, which would be consistent with our findings that Smurf1 acts as an inhibitor of Wnt/β-catenin signaling. Furthermore, although Smurf1 and Smurf2 double knockout mice are embryonic lethal, these embryos display two typical phenotypes: type I mutants have gastrulation defects with an abnormal posterior structure, while type II mutants display PCP defects (9). Wnt/β-catenin and TGF-β signaling are known to be important for establishing the anterior-posterior embryonic axis and specifying the anterior or posterior fate of epiblast cells. Therefore, it is possible that the observed type I phenotype is the result of alterations in both Wnt/β-catenin and TGF-β pathways.
ACKNOWLEDGMENTS
We thank Ying E. Zhang for generously providing Smurf1−/− MEFs, Rong-Gui Hu for series of E3 ligase plasmids, Joan Massagué (Memorial Sloan-Kettering Cancer Center, USA) for Smurf1-HA and Smurf1-CA-HA plasmids, Di Chen (University of Rochester) for the Smurf1-Myc construct, Xi He (Children's Hospital Boston) for the plasmids of LRP6 and MESD, Ye-Guang Chen (Tsinghua University, China) for Smad7-Flag, and Long Yu for the plasmids of Ub-WT, Ub-K0, Ub-K48, and Ub-K63. We are grateful to Dang-Sheng Li and Xiao-Min Song for critical reading of the manuscript.
This work was supported by a Ministry of Science and Technology of China grant (2010CB912100) and National Natural Science Foundation of China grants (31230044, 30930052, and 91213304).
Footnotes
Published ahead of print 19 August 2013
REFERENCES
- 1.Chen ZJ, Sun LJ. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275–286 [DOI] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229 [DOI] [PubMed] [Google Scholar]
- 3.Tauriello DV, Maurice MM. 2010. The various roles of ubiquitin in Wnt pathway regulation. Cell Cycle 9:3700–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicke L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195–201 [DOI] [PubMed] [Google Scholar]
- 5.Kulathu Y, Komander D. 2012. Atypical ubiquitylation–the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13:508–523 [DOI] [PubMed] [Google Scholar]
- 6.Bernassola F, Karin M, Ciechanover A, Melino G. 2008. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14:10–21 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. 2001. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 98:974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. 1999. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400:687–693 [DOI] [PubMed] [Google Scholar]
- 9.Narimatsu M, Bose R, Pye M, Zhang L, Miller B, Ching P, Sakuma R, Luga V, Roncari L, Attisano L, Wrana JL. 2009. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell 137:295–307 [DOI] [PubMed] [Google Scholar]
- 10.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. 2003. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302:1775–1779 [DOI] [PubMed] [Google Scholar]
- 11.Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. 2005. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121:101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukunaga E, Inoue Y, Komiya S, Horiguchi K, Goto K, Saitoh M, Miyazawa K, Koinuma D, Hanyu A, Imamura T. 2008. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J. Biol. Chem. 283:35660–35667 [DOI] [PubMed] [Google Scholar]
- 13.Clevers H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127:469–480 [DOI] [PubMed] [Google Scholar]
- 14.Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20:781–810 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, Myer VE, Finan PM, Porter JA, Huang SM, Cong F. 2011. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 13:623–629 [DOI] [PubMed] [Google Scholar]
- 16.Kim S, Jho EH. 2010. The protein stability of Axin, a negative regulator of Wnt signaling, is regulated by Smad ubiquitination regulatory factor 2 (Smurf2). J. Biol. Chem. 285:36420–36426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauriello DV, Haegebarth A, Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H, Maurice MM. 2010. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol. Cell 37:607–619 [DOI] [PubMed] [Google Scholar]
- 18.Tran H, Bustos D, Yeh R, Rubinfeld B, Lam C, Shriver S, Zilberleyb I, Lee MW, Phu L, Sarkar AA, Zohn IE, Wertz IE, Kirkpatrick DS, Polakis P. 2013. HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC-axin interaction. J. Biol. Chem. 288:3753–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay-Koren A, Caspi M, Zilberberg A, Rosin-Arbesfeld R. 2011. The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol. Biol. Cell 22:399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T, Li M, Ding Y, Zhang LS, Xi Y, Pan WJ, Tao DL, Wang JY, Li L. 2009. Identification of zinc-finger BED domain-containing 3 (Zbed3) as a novel Axin-interacting protein that activates Wnt/beta-catenin signaling. J. Biol. Chem. 284:6683–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YL, Liu B, Zhou ZN, Hu RY, Fei C, Xie ZH, Ding X. 2009. Smad6 inhibits the transcriptional activity of Tbx6 by mediating its degradation. J. Biol. Chem. 284:23481–23490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Yuan H, Weaver CD, Mao J, Farr GH, III, Sussman DJ, Jonkers J, Kimelman D, Wu D. 1999. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. EMBO J. 18:4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao J, Wang J, Liu B, Pan W, Farr GH, III, Flynn C, Yuan H, Takada S, Kimelman D, Li L, Wu D. 2001. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7:801–809 [DOI] [PubMed] [Google Scholar]
- 24.Li S, Lu K, Wang J, An L, Yang G, Chen H, Cui Y, Yin X, Xie P, Xing G, He F, Zhang L. 2010. Ubiquitin ligase Smurf1 targets TRAF family proteins for ubiquitination and degradation. Mol. Cell. Biochem. 338:11–17 [DOI] [PubMed] [Google Scholar]
- 25.Osmundson EC, Ray D, Moore FE, Gao Q, Thomsen GH, Kiyokawa H. 2008. The HECT E3 ligase Smurf2 is required for Mad2-dependent spindle assembly checkpoint. J. Cell Biol. 183:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan W, Choi SC, Wang H, Qin Y, Volpicelli-Daley L, Swan L, Lucast L, Khoo C, Zhang X, Li L, Abrams CS, Sokol SY, Wu D. 2008. Wnt3a-mediated formation of phosphatidylinositol 4,5-bisphosphate regulates LRP6 phosphorylation. Science 321:1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei W, Li M, Wang J, Nie F, Li L. 2012. The E3 ubiquitin ligase ITCH negatively regulates canonical Wnt signaling by targeting dishevelled protein. Mol. Cell. Biol. 32:3903–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai J, Jin WH, Sheng QH, Shieh CH, Wu JR, Zeng R. 2007. Protein phosphorylation and expression profiling by Yin-yang multidimensional liquid chromatography (Yin-Yang MDLC) mass spectrometry. J. Proteome Res. 6:250–262 [DOI] [PubMed] [Google Scholar]
- 29.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- 30.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. 2007. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6:2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikauchi Y, Imamura T, Miyazono K. 2002. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 277:39919–39925 [DOI] [PubMed] [Google Scholar]
- 32.Zeng X, Huang H, Tamai K, Zhang X, Harada Y, Yokota C, Almeida K, Wang J, Doble B, Woodgett J, Wynshaw-Boris A, Hsieh JC, He X. 2008. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development 135:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines ME, Rosenquist T, Holdener BC. 2003. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell 112:355–367 [DOI] [PubMed] [Google Scholar]
- 34.Yuan C, Qi J, Zhao X, Gao C. 2012. Smurf1 protein negatively regulates interferon-gamma signaling through promoting STAT1 protein ubiquitination and degradation. J. Biol. Chem. 287:17006–17015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan L, Zou W, Gao D, Inuzuka H, Fukushima H, Berg AH, Drapp R, Shaik S, Hu D, Lester C, Eguren M, Malumbres M, Glimcher LH, Wei W. 2011. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Mol. Cell 44:721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng PL, Lu H, Shelly M, Gao H, Poo MM. 2011. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron 69:231–243 [DOI] [PubMed] [Google Scholar]
- 37.Ding Y, Xi Y, Chen T, Wang JY, Tao DL, Wu ZL, Li YP, Li C, Zeng R, Li L. 2008. Caprin-2 enhances canonical Wnt signaling through regulating LRP5/6 phosphorylation. J. Cell Biol. 182:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, Yin J, Chen D, Nie F, Song X, Fei C, Miao H, Jing C, Ma W, Wang L, Xie S, Li C, Zeng R, Pan W, Hao X, Li L. 2013. Small-molecule modulation of Wnt signaling via modulating the Axin-LRP5/6 interaction. Nat. Chem. Biol. [Epub ahead of print.] 10.1038/nchembio.1309 [DOI] [PubMed] [Google Scholar]
- 39.Tang N, Song WX, Luo J, Luo X, Chen J, Sharff KA, Bi Y, He BC, Huang JY, Zhu GH, Su YX, Jiang W, Tang M, He Y, Wang Y, Chen L, Zuo GW, Shen J, Pan X, Reid RR, Luu HH, Haydon RC, He TC. 2009. BMP-9-induced osteogenic differentiation of mesenchymal progenitors requires functional canonical Wnt/beta-catenin signalling. J. Cell. Mol. Med. 13:2448–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamiya N, Kobayashi T, Mochida Y, Yu PB, Yamauchi M, Kronenberg HM, Mishina Y. 2010. Wnt inhibitors Dkk1 and Sost are downstream targets of BMP signaling through the type IA receptor (BMPRIA) in osteoblasts. J. Bone Miner. Res. 25:200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]