Abstract
Streptococcus pneumoniae is a major human pathogen responsible for massive global morbidity and mortality. The pneumococcus attaches a variety of proteins to its cell surface, many of which contribute to virulence; one such family are the polyhistidine triad (Pht) proteins PhtA, PhtB, PhtD, and PhtE. In this study, we have examined the mechanism of Pht surface attachment using PhtD as a model. Analysis of deletion and point mutants identified a three-amino-acid region of PhtD (Q27-H28-R29) that is critical for the process. The analogous region in PhtE was also necessary for its attachment to the cell surface. Furthermore, we show that a large proportion of the total amount of each Pht protein is released into bacterial culture supernatants. Other surface proteins were also released, albeit to lesser extents, and this was not due to pneumococcal autolysis. The extent of release of surface proteins was strain dependent and was not affected by the capsule. Lastly, we compared the fitness of wild-type and ΔphtABDE pneumococci in vivo in a mouse coinfection model. Release of Pht proteins by the wild type did not complement the mutant strain, consistent with surface-attached rather than soluble forms of the Pht proteins playing the major role in virulence. The significant degree of release of Pht proteins from intact bacteria may have implications for the use of these proteins in novel vaccines.
INTRODUCTION
Streptococcus pneumoniae is a human pathogen able to cause a wide spectrum of diseases, such as pneumonia, sepsis, meningitis, and otitis media, and these result in significant global morbidity and mortality (1). A large number of pneumococcal surface proteins have now been identified, chiefly through searches for molecules suitable for inclusion in next-generation vaccines, and these proteins make major contributions to the ability of this organism to colonize and cause disease (2, 3).
Pneumococcal surface proteins can be classified based on their mechanism of attachment to the bacterial surface, and four major groups have been identified to date (4). The first two, both of which are common to many Gram-positive bacteria, are the lipoproteins, which attach to the cell membrane via a posttranslationally added N-acyl-diacylglyceryl group, and the sortase-dependent proteins, which are covalently anchored to the cell wall via an LPxTG motif (5). S. pneumoniae also produces choline binding proteins, which are attached via noncovalent interactions of conserved choline binding domains with phosphorylcholine moieties in teichoic acids of the cell wall or lipoteichoic acids embedded in the cell membrane. Lastly, there is a group of nonclassical surface proteins that lack conventional secretion or anchoring motifs but are nonetheless exposed on the cell surface (4, 6–8).
While most leading protein vaccine candidates fit into one of these groups, a notable exception is the polyhistidine triad (Pht) proteins. This family, which has four members (PhtA, PhtB, PhtD, and PhtE) in S. pneumoniae, are surface-exposed antigens characterized by the presence of five or six histidine triad (HxxHxH) motifs and are highly conserved among pneumococcal strains (9–11). A number of functions have been proposed for the proteins, including defense against complement deposition (12, 13), scavenging of zinc ions (10, 14), and promoting adherence to host cell surfaces (15). All four Pht proteins have been shown to elicit a degree of protective immunity in models of pneumococcal disease against diverse strains when given in vaccine formulations (9, 16, 17). However, despite significant examination of their vaccine potential, it is not clear how the proteins are attached to the bacterial surface, since they do not appear to fit into any of the groups described above. The proteins contain a type II signal peptidase cleavage site consensus motif (LxxC) but do not appear to be attached to the membrane and are not lipoproteins (9, 14, 18). They do not contain choline binding domains or any motifs thought to be recognized by sortases. The presence of a signal peptide also excludes them from the nonclassical surface protein group.
A further issue is that while a number of flow cytometry studies have confirmed that the proteins are surface exposed (9, 19, 20) and that they are embedded in the cell wall (9, 14, 18), Pht proteins have also been detected in culture supernatant, suggesting the possibility of active release from the bacterial surface (19, 21). In the present study, we have investigated the region(s) of the Pht proteins that is required for attachment to the bacterial surface, as well as the dynamics of their secretion/release into the culture supernatant. Using PhtD as a model for the family, we identified a three-amino-acid region that is critical for the attachment of the protein to the bacterial surface and discovered that a considerable proportion of the total amount of each Pht protein is released in a strain-dependent manner.
MATERIALS AND METHODS
Strains and growth media.
The strains used in this study are listed in Table 1. Bacteria were routinely grown at 37°C in a CO2-enriched atmosphere either in Todd-Hewitt broth supplemented with 1% yeast extract (THY), on Columbia agar supplemented with 5% (vol/vol) horse blood, or, for challenge of mice, in serum broth (10% heat-inactivated horse serum in nutrient broth).
Table 1.
Strains used in this study
| S. pneumoniae strain | Description | Source or reference |
|---|---|---|
| D39 | Serotype 2 | NCTC 7466 |
| D39ΔphtABDE | D39 mutant lacking all four pht genes | 12 |
| N full | D39 ΔphtABDE mutant complemented with full-length phtD | This study |
| N50 | D39 ΔphtABDE mutant complemented with phtD lacking S40 to P95 (inclusive) | This study |
| PhtD SP+ | D39 ΔphtABDE mutant complemented with phtD lacking V32 to V39 (inclusive) | This study |
| PhtD SP− | D39 ΔphtABDE mutant complemented with phtD lacking R26 to V39 (inclusive) | This study |
| PhtD 3 Ala | D39 ΔphtABDE mutant complemented with phtD with amino acid substitutions R26A, H27A, and Q28A | This study |
| PhtE N full | D39 ΔphtABDE mutant complemented with full-length phtE | This study |
| PhtE 6− | D39 ΔphtABDE mutant complemented with phtE lacking Q27 to E32 (inclusive) | This study |
| PhtE 3 Ala | D39 ΔphtABDE mutant complemented with phtE with amino acid substitutions Q27A, H28A, and R29A | This study |
| WCH43 | Serotype 4 | 26 |
| WU2 | Serotype 3 | 40 |
| D39 ΔlytA | D39 mutant lacking lytA | This study |
| D39 Δcps | Mutant lacking cps biosynthesis genes | This study |
| ST232/1 | Serotype 3 | 41 |
| ST180/2 | Serotype 3 | 41 |
| ST233/3 | Serotype 3 | 41 |
| ST232/11 | Serotype 3 | 41 |
| ST233/13 | Serotype 3 | 41 |
Construction of mutants.
To construct mutants with altered forms of PhtD or PhtE, primers were designed to amplify a region starting approximately 2 kb upstream of phtD or -E and extending the desired amount into the gene; separately, a region starting within phtD or -E and extending approximately 2 kb downstream was also amplified. The primers within phtD or -E were designed to allow the two PCR products to be joined by overlap extension PCR, as described previously (22). The positions and/or sequences of these primers were designed to remove or alter specific amino acids within PhtD or -E as required (Table 1). For PhtD, the overlap PCR products were used to transform the ΔphtABDE strain, which was described in a previous study (12). In this strain, phtD has been replaced by a tetracycline resistance gene; hence, transformants were selected for loss of resistance to this antibiotic. Transformation and selection were performed as described previously (23), and transformed colonies were checked by PCR and DNA sequencing. For PhtE, the Janus cassette was used as described previously (24).
The Δcps mutant strain was constructed by replacing genes from cps2A to spd_0333 (inclusive) with a chloramphenicol resistance gene by overlap extension PCR as described previously (22). Deletion of this gene locus has been used previously to generate unencapsulated strains of pneumococci (25).
Primer sequences are shown in Table 2.
Table 2.
Primers used in this study
| Primer | Sequence (5′→3′) | Description |
|---|---|---|
| PhtD Flank F | GCCTATTCTTGTCTTGGTCTTGGT | phtD forward primer |
| PhtD R | AACTCGATTAGACTCTTTCTTAACCTG | Amplification of phtD upstream flank |
| PhtD N50 F | CAGGTTAAGAAAGAGTCTAATCGAGTTTATGATGCCATCATCAGTGAAGA | Generation of N50 mutant |
| PhtD Flank R | GACTTTCTACTTCTTCCTTGCTATCTGT | phtD reverse primer |
| PhtD SP+ R | CTGACCAGCTTGGTGACGACCA | Generation of SP+ mutant |
| PhtD SP− R | ACCAAGTTCATAGGAACAAACACTTAG | Generation of SP− mutant |
| PhtD SP+ F | TGGTCGTCACCAAGCTGGTCAGTCTTATATAGATGGTGATCAGGCTG | Generation of SP+ mutant |
| PhtD SP− F | CTAAGTGTTTGTTCCTATGAACTTGGTTCTTATATAGATGGTGATCAGGCTG | Generation of SP− mutant |
| PhtD 3 Ala F | CCTATGAACTTGGTGCTGCCGCAGCTGGTCAGGTT | Generation of 3 Ala mutant |
| PhtD 3 Ala R | AACCTGACCAGCTGCGGCAGCACCAAGTTCATAGG | Generation of 3 Ala mutant |
| PhtD Janus X | CATTATCCATTAAAAATCAAACGGTCTTTCCTCACTTTAATTCTTCTGC | Replacement of phtDE with Janus cassette |
| PhtE Janus Y | GGAAAGGGGCCCAGGTCTCTGTCTGAATCAAAAATGAAGTTCTCTC | Replacement of phtDE with Janus cassette |
| PhtE 6− F | GTCTATGTGCCTATGCACTAAACAATAAGGACAATAATCGTGTCTCTT | Generation of PhtE 6− mutant |
| PhtE 6− R | GTTTAGTGCATAGGCACATAGAC | Generation of PhtE 6− mutant |
| PhtE 3 Ala F | TGTGCCTATGCACTAAACGCGGCTGCTTCGCAGGAAAATAAGGACAA | Generation of PhtE 3 Ala mutant |
| PhtE 3 Ala R | TTGTCCTTATTTTCCTGCGAAGCAGCCGCGTTTAGTGCATAGGCACA | Generation of PhtE 3 Ala mutant |
| CPS Flank F | GTTCCATGGGATGCTTTCTGTG | Generation of Δcps mutant |
| CPS Flank R | GAAAGTGCAATGCTTAACCCTG | Generation of Δcps mutant |
| CPS CmlX | CAATTTTCGGTCACGCTGTTGGTGAACGTGATTTTTTAAAACGT | Generation of Δcps mutant |
| CPS CmlY | GACGTTGAGCCTCGGAACCCGAAATCTCCAGATTAGGAACTATCCG | Generation of Δcps mutant |
| CAT F | CAACAGCGTGACCGAAAATTG | Amplification of chloramphenicol acetyltransferase |
| CAT R | GGGTTCCGAGGCTCAACGTC | Amplification of chloramphenicol acetyltransferase |
Preparation of bacterial lysates and precipitation of proteins from culture supernatants.
For comparison of proportions of proteins in bacterial lysates and culture supernatants, bacteria were grown in 5 ml THY to an optical density at 600 nm (OD600) of 0.5 and centrifuged at 3,000 × g for 10 min. After one wash with phosphate-buffered saline (PBS), the bacterial pellet was resuspended in PBS with 0.1% sodium deoxycholate and incubated for 1 h at 37°C. One milliliter of culture supernatant was centrifuged again at 18,000 × g for 10 min to ensure that no contaminating bacteria remained; 900 μl of this was collected, and trichloroacetic acid was added to a final concentration of 10% (wt/vol). After 1 h at 4°C, precipitated proteins were pelleted by centrifugation at 18,000 × g for 10 min and resuspended in PBS. Precipitated proteins were resuspended in one-fifth of the volume that was used for the cell pellets such that equal volumes could be loaded into SDS-polyacrylamide gels to represent equivalent proportions of bacterial cells and supernatant from the entire 5-ml culture. One-tenth volume of SDS-PAGE loading buffer was added to bacterial lysates and supernatant proteins; 1 μl of 2 M sodium hydroxide was added to the latter to neutralize any remaining acid. All samples were then heated to 95°C for 5 min.
Western blotting.
Samples were loaded into 4 to 12% polyacrylamide gels, and electrophoresis was performed at 180 V using the NuPAGE SDS-PAGE system (Life Technologies). Proteins were transferred to a nitrocellulose membrane using the iBlot system (Life Technologies) according to the manufacturer's instructions, followed by blocking with 5% (wt/vol) skim milk powder dissolved in Tris-buffered saline and 0.5% Tween 20 (TTBS) for 30 min at room temperature (RT). Primary antibodies were incubated in TTBS overnight at 4°C, followed by a 1:50,000 dilution of anti-mouse IRDye 800 in TTBS (Li-Cor Biosciences, NE, USA) for 1 h at RT. Blots were dried at 37°C and scanned using an Odyssey infrared imaging system (Li-Cor Biosciences). Band intensities were measured after subtraction of background using the manufacturer's application software. It should be noted that calculations of percentages of protein in the supernatant are based on the assumption of 100% efficiency of recovery of bacteria and supernatant proteins during all centrifugation/protein precipitation steps.
Flow cytometry.
Bacteria were grown as for Western blotting. Bacteria from 1 ml of culture were incubated with murine anti-PhtD serum for 1 h at 37°C, followed by Alexa Fluor 488 rabbit anti-mouse IgG(H+L) (Life Technologies) for 30 min at 4°C. Three washes with 1 ml PBS were performed between steps, and all antibodies were diluted 1:100 (vol/vol) in PBS. Fluorescence measurements from 10,000 events were collected using a BD FACSCanto flow cytometer (BD Biosciences, NSW, Australia). Data were analyzed using the software package FlowJo (TreeStar, OR, USA).
In vivo competition assay.
Mutant and wild-type bacteria were grown separately in serum broth and then mixed at an input ratio of 1:1, and 50 μl of bacterial suspension was used to challenge mice intranasally under anesthesia. Nasal wash, nasal tissue, lung, and blood samples were collected and processed as described previously (26, 27). Each sample was serially diluted and plated on blood agar and on blood agar with erythromycin to determine the ratio of mutant to wild-type bacteria. Competitive indices were calculated as the ratio of mutant to wild-type bacteria recovered from each niche adjusted by the input ratio.
RESULTS
Amino acids R26 to Q31 are required for the association of PhtD with the cell surface.
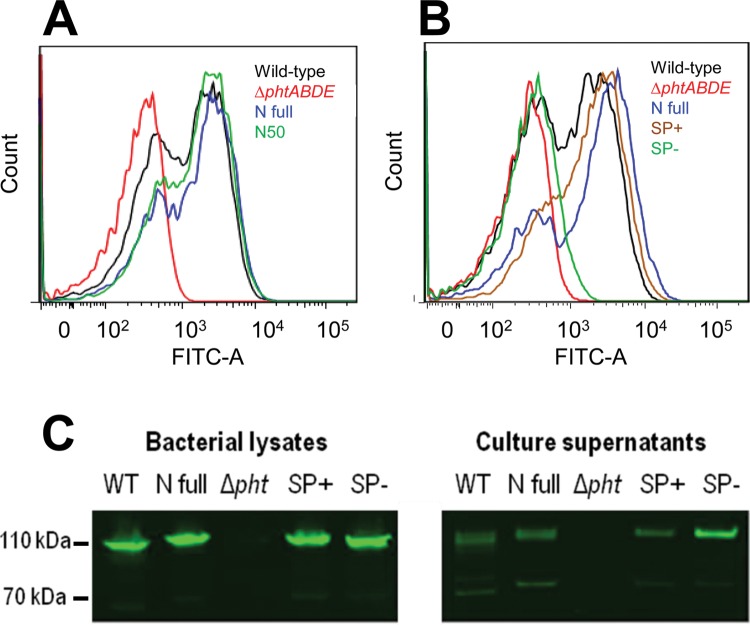
There is some evidence that when attached to the cell surface, the C termini of the Pht proteins are more exposed to the extracellular milieu than the N termini, as judged by flow cytometry studies of the ability of antibodies raised against different regions of PhtB and PhtE to bind to pneumococci (18). This led us to hypothesize that regions near the N terminus, but outside the signal peptide, may be involved in mediating attachment. To address this, we used overlap extension PCR to clone forms of phtD lacking nucleotides encoding amino acids of interest and reinserted these constructs into the phtD locus of an S. pneumoniae D39 derivative in which all four pht genes had previously been deleted (12), as described in Materials and Methods. In order to choose which nucleotides to remove, the online programs SignalP and LipoP were used to predict which amino acids of PhtD constituted its signal peptide (28, 29); however, neither program could give a confident or accurate prediction. We therefore initially chose to delete amino acids S40 to P95 (inclusive). The surface exposure of PhtD in a D39 mutant encoding this form of the protein (referred to as N50), as well as in wild-type D39 and a D39 ΔphtABDE mutant complemented with full-length PhtD (referred to as N full), was assessed by flow cytometry (Fig. 1A). The N50 strain did not show a significant difference in surface exposure of PhtD compared to the wild-type or N full strain, indicating that amino acids S40 to P95 are not required for association of the protein with the cell surface.
Fig 1.
Surface attachment of PhtD. (A and B) Flow cytometric measurement of the presence of PhtD at the bacterial surface. A representative histogram of fluorescence profiles for each strain is shown. (C) Western blots assessing the amounts of PhtD in samples of the bacterial lysate and culture supernatant. WT, wild type. Approximate molecular sizes are indicated on the left; these were estimated based on the mobilities of standard markers (not shown).
Subsequently, two more mutant strains that expressed PhtD lacking amino acids closer to the N terminus than S40 were created. The first (SP+) lacked V32 to V39, while the second (SP−) lacked R26 to V39. Flow cytometry was performed as before, as well as Western blotting of bacterial lysates and culture supernatants to look for the presence of PhtD in these fractions. The results (shown in Fig. 1B and C, respectively) indicate that PhtD could be detected at the surface of the SP+ strain at levels similar to those for the wild-type and N full strains but that the SP− strain did not have PhtD present at its surface. Interestingly, the Western blots showed that all strains except the D39 ΔphtABDE mutant had PhtD present in the culture supernatant, indicating a degree of secretion and/or release of the protein from the cells. The difference in surface availability of PhtD in the SP+ and SP− strains indicated that the amino acid stretch RHQAGQ that was absent in PhtD in SP− but present in SP+ could be required to mediate attachment of the protein to the bacterial surface. The Western blots also revealed a small amount of protein with an estimated molecular mass of approximately 70 kDa that was reactive with the anti-PhtD antiserum. This was absent in the ΔphtABDE mutant and presumably reflects proteolysis of PhtD.
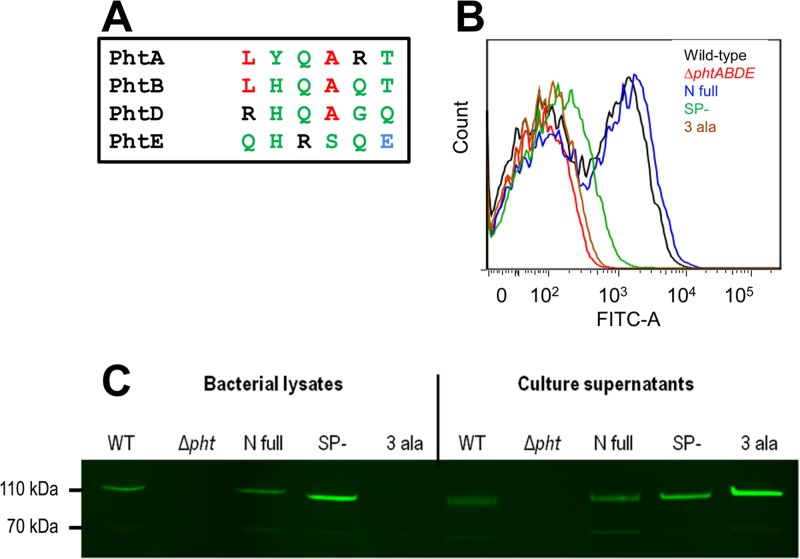
Replacement of R26, H27, and Q28 with alanine leads to loss of surface attachment of PhtD.
To confirm that the RHQAGQ amino acid stretch is indeed critical for attachment of PhtD to the bacterial surface, strain 3 Ala was generated, in which PhtD was altered to contain the amino acid substitutions R26A, H27A, and Q28A. These sites were chosen for mutagenesis based on an alignment of this region across the four pneumococcal Pht proteins (Fig. 2A), which showed that the histidine and first glutamine were conserved in three of the proteins, while the arginine was also chosen because of its charged side chain. The localization of PhtD in 3 Ala was tested as for the previous mutants. As can be seen in Fig. 2B, the PhtD in strain 3 Ala could not be detected on the bacterial surface by flow cytometry and was found almost exclusively in the culture supernatant, consistent with a defect in attachment to the cell surface. This indicates that R26, H27, and Q28 are critical for such attachment. Interestingly, the phenotypes of the SP− and 3 Ala mutants were not identical, with SP− (which lacks residues R26 to V39) showing a small amount of surface-localized PhtD by flow cytometry and a considerable amount of the protein in the bacterial lysate, where 3 Ala had none.
Fig 2.
Surface attachment of PhtD after targeted mutagenesis. (A) Alignment of the putative attachment motif in PhtD with homologous regions in PhtA, PhtB, and PhtE. The Pht proteins of S. pneumoniae D39 were aligned using ClustalW2 (available online at http://www.ebi.ac.uk/Tools/msa/clustalw2/). Only the six amino acids corresponding to RHQAGQ from PhtD are shown. Colors indicate physiochemical properties of classes of amino acids (red, small/hydrophobic; blue, acidic; black, basic; green, hydroxyl/sulfhydryl/amine). (B) Flow cytometric measurement of the presence of PhtD at the bacterial surface. A representative histogram of fluorescence profiles for each strain is shown. (C) Western blot assessing the amounts of PhtD in samples of the bacterial lysate and culture supernatant.
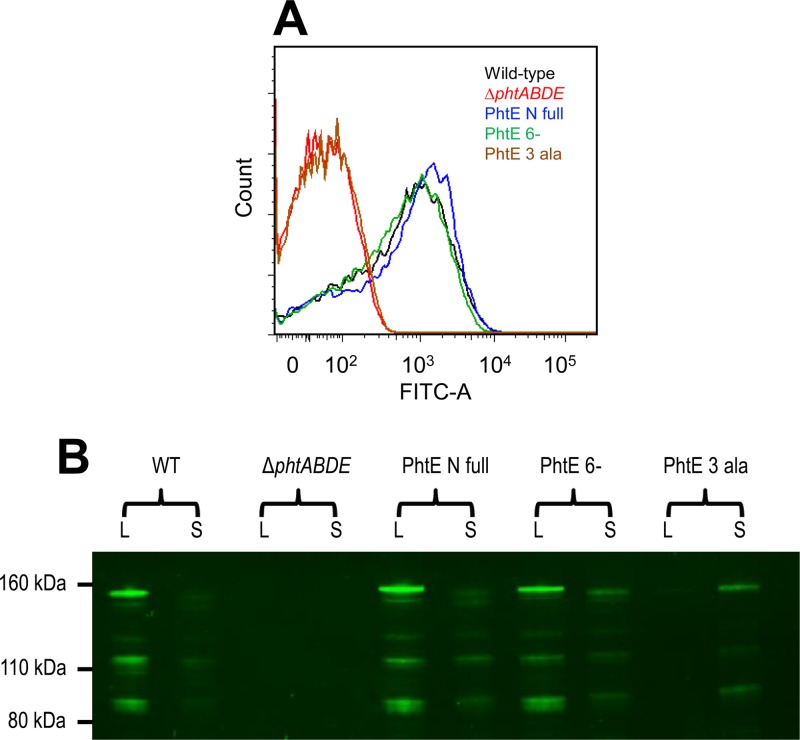
To investigate the role of these residues in another Pht protein, mutations were introduced into the analogous region of phtE. The first mutant (PhtE 6−) lacked Q27 to E32, while the second (PhtE 3 Ala) had the amino acid substitutions Q27A, H28A, and R29A. Flow cytometry (Fig. 3A) revealed that the PhtE 3 Ala mutant did not have detectable PhtE on its surface, consistent with the findings for PhtD 3 Ala. Interpretation of Western blot analysis of bacterial lysate and culture supernatant samples (Fig. 3B) was complicated by extensive proteolysis of PhtE. It was also notable that there was a band with an apparent molecular size of approximately 150 kDa, which is larger than the predicted molecular mass of PhtE (115 kDa), implying that the protein may migrate anomalously in SDS-PAGE. Nevertheless, it was apparent that the majority of the anti-PhtE-reactive material for the PhtE 3 Ala mutant was found in the culture supernatant, with very little in the cell lysate fraction, confirming that Q27, H28, and R29 are important for surface attachment of the protein in a fashion similar to that for the analogous residues (R26, H27, and Q28) in PhtD. Somewhat surprisingly, the surface exposure of PhtE in the PhtE 6− mutant was similar to that for wild-type PhtE and PhtE N full, despite it lacking residues Q27, H28, and R29. This is not dissimilar to the findings for PhtD SP− referred to above, for which surface exposure was intermediate between those of the respective positive and negative controls. These 6- and 14-amino-acid deletions in the respective Pht proteins may have conformational effects which impact attachment.
Fig 3.
Surface attachment of PhtE after targeted mutagenesis. (A) Flow cytometric measurement of the presence of PhtE at the bacterial surface. A representative histogram of fluorescence profiles for each strain is shown. (B) Western blot assessing the amounts of PhtE in samples of the bacterial lysate and culture supernatant.
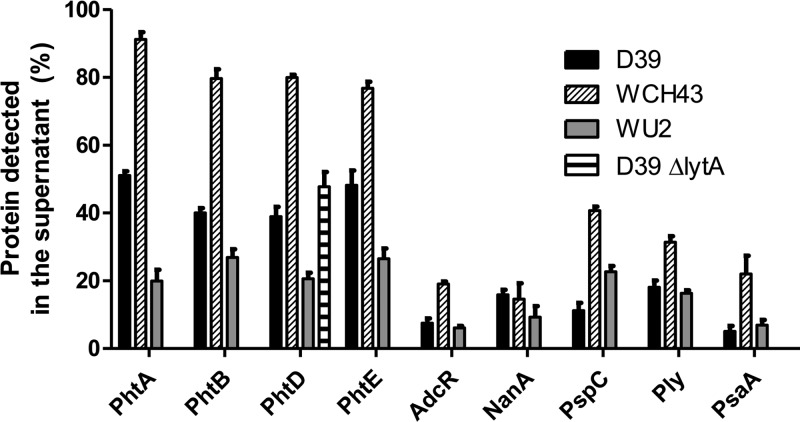
Pht proteins are found in bacterial culture supernatants of multiple strains.
The experiments described above revealed the presence of a considerable amount of PhtD in the culture supernatant fractions, even for wild-type D39. To quantify the relative amounts of soluble and cell-associated Pht proteins, wild-type bacteria of three different strains (D39, WCH43, and WU2) were grown to mid-log phase, and samples of bacterial lysate and culture supernatant representing equal proportions of the total culture were analyzed by Western blotting. Band intensities were quantified, and the proportion of protein in the supernatant was calculated. For the purpose of comparison with other surface proteins that use different mechanisms of attachment, this was also performed for neuraminidase A (NanA), pneumococcal surface protein C (PspC), pneumolysin (Ply), and pneumococcal surface antigen A (PsaA); the intracellular transcriptional regulator AdcR (12) was measured as a control to show autolysis had not occurred. The results (Fig. 4) indicate that all of the surface proteins examined can be detected in the culture supernatants but that the proportions of Pht proteins in this fraction are higher than those of any of the other surface proteins measured. The proportions of AdcR in the supernatants were low for all strains (less than 10% for D39 and WU2 and less than 20% for WCH43). To confirm that cellular autolysis was not responsible for the release of PhtD, the proportion of this protein only was measured in the supernatant of a D39 ΔlytA mutant, and no significant difference was found compared to the wild type (Fig. 4).
Fig 4.
Proportions of pneumococcal surface protein in culture supernatants of wild-type D39, WCH43, and WU2, calculated after quantitation from Western blots. Results are presented as mean and standard error from samples grown in biological triplicate. Measurement of PhtD only was made for the D39 ΔlytA mutant strain.
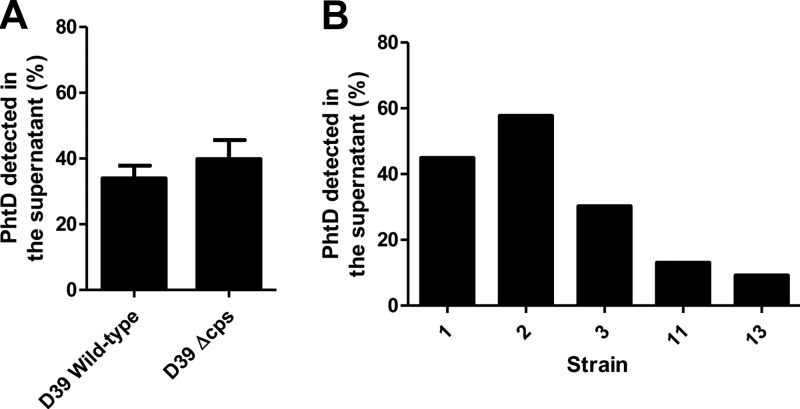
The capsule does not play a role in the secretion or release of PhtD.
The greatest secretion and/or release of Pht proteins was shown by the serotype 4 strain WCH43 and the least by the serotype 3 strain WU2. It was hypothesized that the thickness and/or chemical composition of the capsule layer could play a role in retarding the secretion or release of Pht proteins. This was tested by measuring the proportion of PhtD in culture supernatants of a mutant strain of D39 in which the cps (capsule biosynthesis) locus had been replaced with an antibiotic resistance cassette (25). However, as shown in Fig. 5A, there was no significant difference in PhtD release between the unencapsulated strain and the wild type. Furthermore, analysis of five serotype 3 clinical isolates, which might have been predicted to show consistently low proportions of PhtD in the supernatant, revealed a considerable degree of variation in the amount of PhtD secreted/released (ranging from 10% to 60%) (Fig. 5B), demonstrating that a serotype 3 capsule does not preclude the release of a large proportion of PhtD.
Fig 5.
The capsule does not affect secretion or release of PhtD. (A) Percentage of PhtD in supernatants of D39 wild-type and Δcps cultures. Results are representative of three independent experiments. (B) Percentage of PhtD in the supernatants of cultures of five serotype 3 clinical isolate strains.
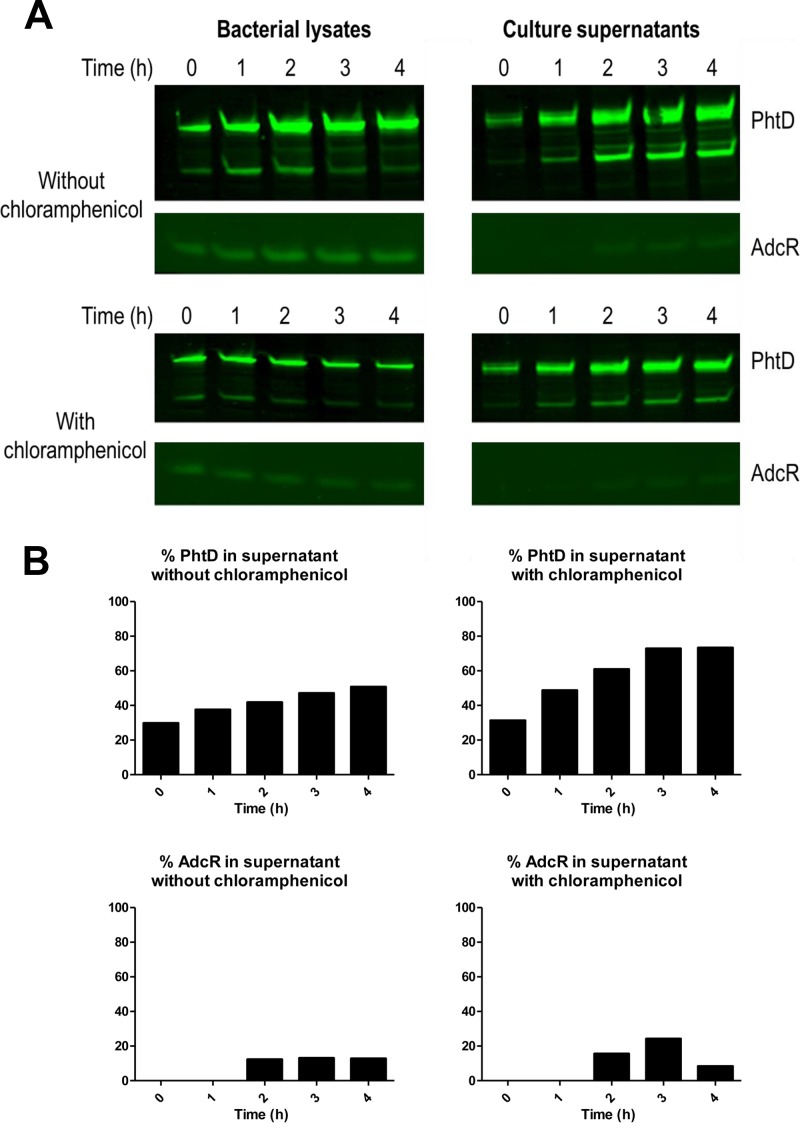
PhtD is released from the pneumococcal surface over time.
From the above-described experiments, it was not clear whether the Pht proteins found in culture supernatants originated from molecules that were previously attached to the cell surface or whether the proteins were secreted directly without being transiently attached. To address this, wild-type pneumococci were grown to an OD600 of 0.3. The culture was split in two, chloramphenicol was added to one replicate culture to inhibit protein synthesis, and samples of bacterial lysate and supernatant were collected every hour thereafter for 4 h. Western blotting was performed to detect PhtD as well as the intracellular transcriptional regulator AdcR to control for pneumococcal autolysis. Total amounts of PhtD and AdcR (i.e., the sum of amounts present in bacterial lysates and culture supernatants) were quantified from the Western blots; both proteins increased in abundance over time when chloramphenicol was absent but remained at a stable level when chloramphenicol was present, indicating that treatment with the antibiotic had been effective at inhibiting new protein synthesis (data not shown). As shown in Fig. 6A and B, there was a trend of an increasing proportion of PhtD in the culture supernatant for bacteria incubated either with or without chloramphenicol. While AdcR was detectable in culture supernatants after 2 h, the majority of the protein remained associated with the bacterial lysates, indicating that autolysis could not account for the increasing proportion of PhtD in the supernatant. Critically, the increase in the proportion of PhtD in the supernatant for bacteria incubated with chloramphenicol indicated that PhtD was being released from the cell surface, rather than originating from newly synthesized protein molecules being secreted directly into the culture medium. However, this does not preclude the possibility that a portion of newly synthesized PhtD could be directly released in the absence of chloramphenicol.
Fig 6.
Release of PhtD over time. (A) Western blots for PhtD and AdcR in bacterial lysates and culture supernatants of bacteria grown in the presence or absence of chloramphenicol. (B) Percentages of PhtD and AdcR in culture supernatants based on quantitations of band intensities. The results are representative of two independent experiments.
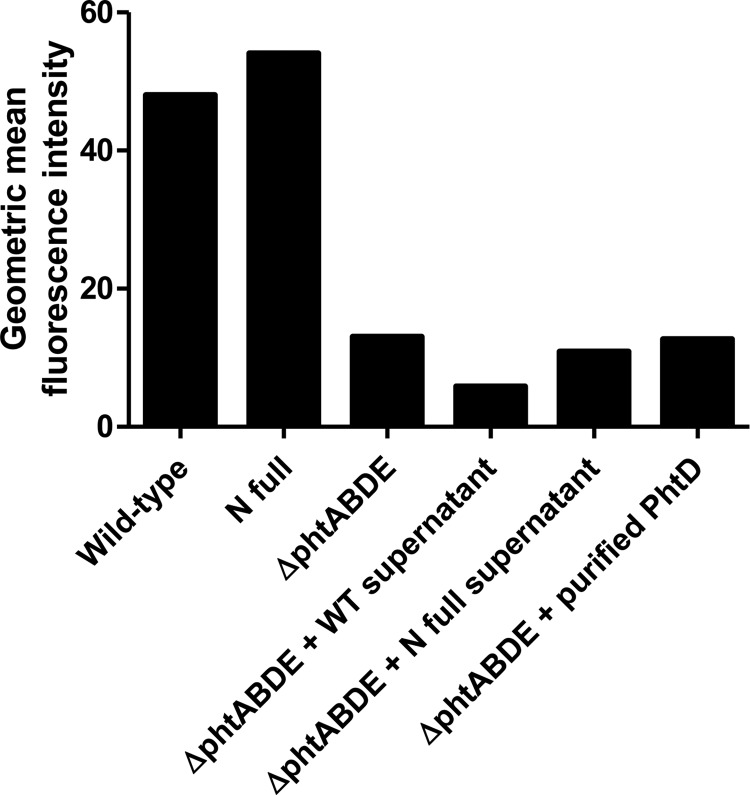
PhtD does not reattach to the bacterial surface.
To test whether PhtD can reattach to the bacterial surface after being released, cultures of the wild-type, N full, and ΔphtABDE strains were grown. D39 ΔphtABDE bacteria were then incubated with filtered supernatant from the wild-type or N full cultures or with recombinantly expressed purified PhtD for 1 h at 37°C. After three washes, flow cytometry was performed to detect any PhtD attached to the surface of the strains (Fig. 7). This indicated that either PhtD from the culture supernatants or the recombinantly expressed purified protein did not reassociate with the bacteria, since the mean fluorescence intensities of these samples were similar to those for the untreated ΔphtABDE strain.
Fig 7.
Flow cytometric measurement of PhtD at the bacterial surface of the indicated strain. ΔphtABDE bacteria were incubated with supernatant from wild-type or N full strains or purified PhtD as shown. Geometric mean fluorescence intensities from one experiment are shown; results are representative of two independent experiments.
Surface-localized forms of Pht proteins play the major role(s) of the proteins in vivo.
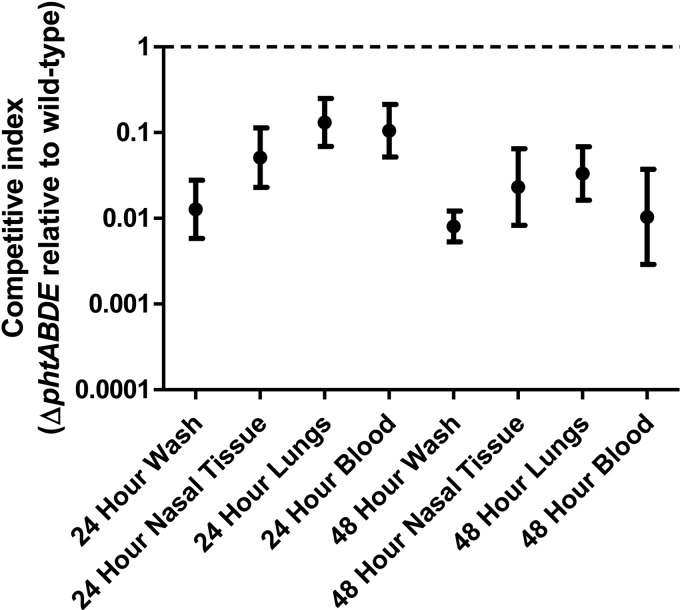
The importance of released forms of the Pht proteins for pneumococcal virulence was assessed using an in vivo competition experiment. Mice were infected intranasally with a mixed culture of wild-type and ΔphtABDE bacteria. After 24 and 48 h, nasal wash, nasal tissue, lung, and blood samples were harvested, and pneumococci were enumerated from each sample. Competitive indices (ratios of ΔphtABDE to wild-type bacteria) were calculated for each sample. If the released forms of the Pht proteins performed their major role(s), the Pht protein molecules released by wild-type bacteria would be expected to complement the mutant strain, leading to competitive indices of approximately one. However, as can be seen in Fig. 8, the competitive indices were significantly lower than one (P < 0.0001 for all groups [one-sample t tests, two tailed]), indicating that the ΔphtABDE strain was severely attenuated compared to the wild type in all niches tested. This implies that surface-localized forms of the Pht proteins are critical for full virulence and thus are likely to perform the major role(s) of the proteins in vivo.
Fig 8.
In vivo competition between wild-type and ΔphtABDE bacteria. Mice were infected intranasally with a mixed culture of the two strains and samples collected after 24 and 48 h. Geometric mean competitive indices and 95% confidence intervals are shown. A competitive index of 1 (dashed line) would indicate no difference in fitness between the two strains. Values for all groups were significantly different from the theoretical mean of 1 (P < 0.0001 [one-sample t tests, two tailed]).
DISCUSSION
The Pht proteins are an important family of pneumococcal surface virulence factors notable for their promising efficacy in preventing pneumococcal colonization and disease in preclinical vaccine trials (9, 16, 17, 30). However, they lack any recognizable motifs thought to mediate surface attachment, and they presumably use a novel mechanism to attach to the cell surface.
In this study, the amino acid region R26-H27-Q28 of PhtD was found to be critical for the association of PhtD with the bacterial surface. This was shown by flow cytometry, which demonstrated a lack of surface exposure of PhtD in the SP− and 3 Ala mutants. Importantly, PhtD was still released into the culture supernatants of these strains, indicating that the loss of surface exposure of PhtD was not due to any defect in the signal peptide, which would have interfered with secretion. Furthermore, Western blotting after extended electrophoresis did not reveal any consistent differences in the migration of cell-associated and released forms of PhtD, providing a further indication that differences in efficiency of signal peptide cleavage were not impacting release of the protein by the wild-type or mutant strains. Interestingly, as shown in Fig. 2A, neither R26, H27, nor Q28 of PhtD is conserved among all of the other Pht proteins of S. pneumoniae. This may indicate either that each protein's mechanism of attachment is different or, alternatively, that only amino acids with similar chemical properties are required; for instance, all four proteins contain positively charged and/or polar amino acids in this region. Alanine replacement mutagenesis of Q27, H28, and R29 of PhtE also resulted in loss of surface attachment of this protein, but surprisingly, deletion of Q27 to E32 did not. It is possible that the residues downstream of E32 in PhtE were able to compensate for the loss of residues in the PhtE 6− mutant. Indeed, comparison of the deleted residues (QHRSQE) with those immediately downstream (NKDNNR) shows that all of them contain charged or polar side chains. While the deletion of amino acids in the PhtD SP− and PhtE 6− mutants could have introduced conformational changes in the proteins leading to confounding effects on their surface attachment, the less disruptive changes made in the 3 Ala mutants in both PhtD and PhtE provide strong evidence that the mutated residues are critical for attachment.
A further observation from the Western blots was the presence of a protein of around 70 kDa that was reactive with the anti-PhtD antiserum. This was found for all strains (except the ΔphtABDE mutant) and was more prominent in the culture supernatant fractions. This suggests that PhtD is susceptible to proteolysis at some stage after its production. PhtE also appeared to be highly susceptible to degradation. The processing of Pht proteins, and whether this has any impact on their function(s) during infection, will require further investigation.
During the course of these experiments, it became apparent that considerable quantities of PhtD were present in culture supernatants, including those of wild-type strains. It was also notable that in the flow cytometry experiments, a population of wild-type bacteria (around 30% of the total) was negative for PhtD exposure, consistent with release of the protein into the supernatant. To examine this phenomenon in more detail, the proportions of several surface proteins found in culture supernatants were examined in three pneumococcal strains. This revealed that large quantities of all four Pht proteins are released from the bacterial surface, particularly in the serotype 4 strain WCH43. This was not due to autolysis, since only small quantities of the intracellular protein AdcR were found in culture supernatants. Moreover, mutagenesis of lytA in D39 had no impact on PhtD release. It was interesting to note that the sortase-anchored protein NanA (31), the choline binding protein PspC, and the toxin Ply, which has been found to be present in the bacterial cell wall (32, 33), could all be detected in culture supernatants as well. It is feasible that proteins anchored in the bacterial cell wall may all be released into the supernatant to various extents as a natural consequence of the actions of cell wall hydrolases. These proteins (reviewed in reference 34) specifically cleave covalent bonds in the cell wall and are thought to contribute to several critical processes, such as remodeling of the peptidoglycan during cell division to allow separation of daughter cells. Thus, surface-anchored proteins could be released, perhaps still tethered to small cell wall fragments. According to this hypothesis, surface proteins that are anchored to the cell membrane should not be affected and would not be expected to be released. Consistent with this, the lipoprotein PsaA was essentially undetectable in culture supernatants of both strains D39 and WU2. While this hypothesis will require further experimental investigation, it is nonetheless apparent that the proportions of Pht proteins released into the culture supernatant are higher than those of any of the other surface proteins measured, particularly in strains D39 and WCH43, indicating that their interaction with the bacterial surface is more easily disrupted than sortase-dependent covalent linkages or binding to phosphorylcholine.
It is also notable that within each strain, the proportions of the four Pht proteins found in the supernatant were similar. This indicates that common features of the strain background (such as the thickness and composition of the cell wall and capsule layers and/or the presence of other proteins) may have a strong effect on the amount of protein that is released. Interestingly, previous measurements of capsule size have indicated that serotype 4 strains (such as WCH43, which released the largest amount of Pht proteins) have relatively small capsules, whereas serotype 3 strains (such as WU2, which released the least) are thought to produce large quantities of capsular polysaccharide (35, 36). However, using an unencapsulated mutant strain in the D39 background, we found that there was no significant difference between the proportion of PhtD in the supernatant and that in supernatant from the wild type. Furthermore, there was significant variation in the amount of PhtD in the culture supernatants of five clinical isolates of serotype 3, indicating that capsule does not have a significant impact on the amount of PhtD that is released. We also examined the deduced amino acid sequence of the N-terminal region of PhtD in each of the strains in this study, focusing especially on positions 26, 27, and 28. However, this did not reveal any differences that consistently correlated with small or large amounts of PhtD release (data not shown). Thus, the factors that contribute to the strain-dependent differences in the proportion of PhtD that is released remain to be determined.
After showing that PhtD cannot reattach to the bacterial surface (as previously shown for the pneumococcal alpha-enolase [37]), we sought to address whether soluble forms of the Pht proteins play a role during infection, using an in vivo competition experiment. If the Pht proteins performed their major function when released rather than when surface attached, we would expect the wild-type strain to complement the mutant (which has previously been shown to have a decreased virulence phenotype in vivo [12]). However, the mutant was found to be extremely attenuated compared to the wild type. While this does not preclude the possibility that released forms of PhtD could play a minor role during infection, the very low competitive indices (less than 0.01 in some cases) strongly suggest that surface association is critical for the Pht proteins to contribute to pneumococcal fitness in vivo. This therefore raises the issue of why the Pht proteins are released, since it would seem metabolically wasteful to produce large amounts of the proteins only to release them into the surrounding environment where they are of minimal benefit. Humans are known to produce antibodies against Pht proteins as a result of natural exposure to S. pneumoniae (38, 39). Thus, Pht molecules released from the bacterium during infection might act as a sink for circulating antibodies, thereby minimizing the potential for opsonophagocytosis via surface-attached Pht. This argument could also be extended to the other important surface molecules NanA, PspC, and Ply that were examined in this study and that were also found in the culture supernatants (albeit to a lesser extent than the Pht proteins). Such a role for the released Pht proteins would not be detectable in our in vivo competition experiment, as the mice would not have had preexisting anti-Pht antibodies. While this hypothesis will require further experimental investigation, the presence of Pht proteins in the extracellular environment is nonetheless significant and may have implications for their use as vaccine antigens.
ACKNOWLEDGMENTS
We thank Andrew Lawrence for providing the serotype 3 clinical isolates and Chris McDevitt for useful discussions.
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia (Program Grant 565526 to J.C.P.) and the Australian Research Council (Discovery Project Grant DP120101432 to J.C.P.). C.D.P. holds a Northcote Graduate Scholarship; J.C.P. is an NHMRC Senior Principal Research Fellow.
Footnotes
Published ahead of print 22 July 2013
REFERENCES
- 1.Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O'Brien KL, Campbell H, Black RE. 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381:1405–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jedrzejas MJ. 2007. Unveiling molecular mechanisms of bacterial surface proteins: Streptococcus pneumoniae as a model organism for structural studies. Cell. Mol. Life. Sci. 64:2799–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288–301 [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Dorado I, Galan-Bartual S, Hermoso JA. 2012. Pneumococcal surface proteins: when the whole is greater than the sum of its parts. Mol. Oral Microbiol. 27:221–245 [DOI] [PubMed] [Google Scholar]
- 5.Paterson GK, Mitchell TJ. 2004. The biology of Gram-positive sortase enzymes. Trends Microbiol. 12:89–95 [DOI] [PubMed] [Google Scholar]
- 6.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79:3476–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergmann S. 2006. Versatility of pneumococcal surface proteins. Microbiology 152:295–303 [DOI] [PubMed] [Google Scholar]
- 9.Adamou JE, Heinrichs JH, Erwin AL, Walsh W, Gayle T, Dormitzer M, Dagan R, Brewah YA, Barren P, Lathigra R, Langermann S, Koenig S, Johnson S. 2001. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 69:949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rioux S, Neyt C, Di Paolo E, Turpin L, Charland N, Labbe S, Mortier MC, Mitchell TJ, Feron C, Martin D, Poolman JT. 2011. Transcriptional regulation, occurrence and putative role of the Pht family of Streptococcus pneumoniae. Microbiology 157:336–348 [DOI] [PubMed] [Google Scholar]
- 11.Plumptre CD, Ogunniyi AD, Paton JC. 2012. Polyhistidine triad proteins of pathogenic streptococci. Trends Microbiol. 20:485–493 [DOI] [PubMed] [Google Scholar]
- 12.Ogunniyi AD, Grabowicz M, Mahdi LK, Cook J, Gordon DL, Sadlon TA, Paton JC. 2009. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 23:731–738 [DOI] [PubMed] [Google Scholar]
- 13.Melin M, Tikkanen E DIPL, Jarva H, Neyt C, Kayhty H, Meri S, Poolman J, Vakevainen M. 2010. Interaction of pneumococcal histidine triad proteins with human complement. Infect. Immun. 78:2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loisel E, Chimalapati S, Bougault C, Imberty A, Gallet B, Di Guilmi AM, Brown J, Vernet T, Durmort C. 2011. Biochemical characterization of the histidine triad protein PhtD as a cell surface zinc-binding protein of pneumococcus. Biochemistry 50:3551–3558 [DOI] [PubMed] [Google Scholar]
- 15.Khan MN, Pichichero ME. 2012. Vaccine candidates PhtD and PhtE of Streptococcus pneumoniae are adhesins that elicit functional antibodies in humans. Vaccine 30:2900–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godfroid F, Hermand P, Verlant V, Denoël P, Poolman JT. 2011. Preclinical evaluation of the Pht proteins as potential cross-protective pneumococcal vaccine antigens. Infect. Immun. 79:238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denoël P, Philipp MT, Doyle L, Martin D, Carletti G, Poolman JT. 2011. A protein-based pneumococcal vaccine protects rhesus macaques from pneumonia after experimental infection with Streptococcus pneumoniae. Vaccine 29:5495–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamel J, Charland N, Pineau I, Ouellet C, Rioux S, Martin D, Brodeur BR. 2004. Prevention of pneumococcal disease in mice immunized with conserved surface-accessible proteins. Infect. Immun. 72:2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Masi AW, Barniak V, Mountzouros K, Hostetter MK, Green BA. 2001. Recombinant PhpA protein, a unique histidine motif-containing protein from Streptococcus pneumoniae, protects mice against intranasal pneumococcal challenge. Infect. Immun. 69:3827–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wizemann TM, Heinrichs JH, Adamou JE, Erwin AL, Kunsch C, Choi GH, Barash SC, Rosen CA, Masure HR, Tuomanen E, Gayle A, Brewah YA, Walsh W, Barren P, Lathigra R, Hanson M, Langermann S, Johnson S, Koenig S. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi C-W, Lee YG, Kwon S-O, Kim H-Y, Lee JC, Chung Y-H, Yun C-Y, Kim SI. 2012. Analysis of Streptococcus pneumoniae secreted antigens by immuno-proteomic approach. Diagn. Microbiol. Infect. Dis. 72:318–327 [DOI] [PubMed] [Google Scholar]
- 22.Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270–279 [DOI] [PubMed] [Google Scholar]
- 23.McAllister LJ, Tseng HJ, Ogunniyi AD, Jennings MP, McEwan AG, Paton JC. 2004. Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol. Microbiol. 53:889–901 [DOI] [PubMed] [Google Scholar]
- 24.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearce BJ, Iannelli F, Pozzi G. 2002. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153:243–247 [DOI] [PubMed] [Google Scholar]
- 26.Ogunniyi AD, Mahdi LK, Trappetti C, Verhoeven N, Mermans D, Van der Hoek MB, Plumptre CD, Paton JC. 2012. Identification of genes that contribute to pathogenesis of invasive pneumococcal disease by in vivo transcriptomic analysis. Infect. Immun. 80:3268–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAllister LJ, Ogunniyi AD, Stroeher UH, Leach AJ, Paton JC. 2011. Contribution of serotype and genetic background to virulence of serotype 3 and serogroup 11 pneumococcal isolates. Infect. Immun. 79:4839–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen TN, Brunak S, Von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 30.Seiberling M, Bologa M, Brookes R, Ochs M, Go K, Neveu D, Kamtchoua T, Lashley P, Yuan T, Gurunathan S. 2012. Safety and immunogenicity of a pneumococcal histidine triad protein D vaccine candidate in adults. Vaccine 30:7455–7460 [DOI] [PubMed] [Google Scholar]
- 31.Kharat AS, Tomasz A. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 71:2758–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price KE, Camilli A. 2009. Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J. Bacteriol. 191:2163–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price KE, Greene NG, Camilli A. 2012. Export requirements of pneumolysin in Streptococcus pneumoniae. J. Bacteriol. 194:3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López R, García E. 2004. Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol. Rev. 28:553–580 [DOI] [PubMed] [Google Scholar]
- 35.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Müller E, Rohde M. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73:4653–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberger DM, Trzciński K, Lu Y-J, Bogaert D, Brandes A, Galagan J, Anderson PW, Malley R, Lipsitch M. 2009. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 5:e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2001. Alpha-enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273–1287 [DOI] [PubMed] [Google Scholar]
- 38.Pichichero ME, Kaur R, Casey JR, Xu Q, Almudevar A, Ochs M. 2012. Antibody response to Streptococcus pneumoniae proteins PhtD, LytB, PcpA, PhtE and Ply after nasopharyngeal colonization and acute otitis media in children. Hum. Vaccin. Immunother. 8:799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagerman A, Posfay-Barbe KM, Grillet S, Ochs MM, Brookes RH, Greenberg D, Givon-Lavi N, Dagan R, Siegrist C-A. 2013. Influence of age, social patterns and nasopharyngeal carriage on antibodies to three conserved pneumococcal surface proteins (PhtD, PcpA and PrtA) in healthy young children. Eur. J. Clin. Microbiol. Infect. Dis. 32:43–49 [DOI] [PubMed] [Google Scholar]
- 40.Briles DE, Nahm M, Schroer K, Davie J, Baker P, Kearney J, Barletta R. 1981. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J. Exp. Med. 153:694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trappetti C, Van der Maten E, Amin Z, Potter AJ, Chen AY, Van Mourik PM, Lawrence AJ, Paton AW, Paton JC. 2013. Site of isolation determines biofilm formation and virulence phenotypes of serotype 3 Streptococcus pneumoniae clinical isolates. Infect. Immun. 81:505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]