Abstract
Apical membrane antigen 1 (AMA-1) is a leading blood-stage malaria vaccine candidate. Consistent with a key role in erythrocytic invasion, AMA-1-specific antibodies have been implicated in AMA-1-induced protective immunity. AMA-1 is also expressed in sporozoites and in mature liver schizonts where it may be a target of protective cell-mediated immunity. Here, we demonstrate for the first time that immunization with AMA-1 can induce sterile infection-blocking immunity against Plasmodium sporozoite challenge in 80% of immunized mice. Significantly higher levels of gamma interferon (IFN-γ)/interleukin-2 (IL-2)/tumor necrosis factor (TNF) multifunctional T cells were noted in immunized mice than in control mice. We also report the first identification of minimal CD8+ and CD4+ T cell epitopes on Plasmodium yoelii AMA-1. These data establish AMA-1 as a target of both preerythrocytic- and erythrocytic-stage protective immune responses and validate vaccine approaches designed to induce both cellular and humoral immunity.
INTRODUCTION
The Plasmodium apical membrane antigen-1 (AMA-1) is an integral membrane protein which is essential for the development of apicomplexan parasites in their host environment (1). The AMA-1 mRNA half-life peaks in blood-stage trophozoites, and expression of AMA-1 protein is maximized in the late asexual schizont stage (www.plasmoDB.org) (2). Although it is structurally conserved across apicomplexa, some domains of AMA-1 show high levels of amino acid polymorphism (3). It is thought that the host immune response provides the predominant selective pressure for these interstrain variations, with the parasite varying key targets to evade host immunity (1).
Upon contact with the host cell, the 83-kDa AMA-1 protein is proteolytically processed into its 66-kDa mature form, which is transported to the cell surface membrane (4). In blood-stage merozoites, AMA-1 is concentrated at the apical pole and potentially participates in the reorientation and attachment of merozoites to red blood cells (RBC) (5, 6). Recently, a direct interaction between AMA-1 on the merozoite surface and the rhoptry neck protein (RON) complex inserted into the RBC membrane prior to invasion has been described (7, 8). This AMA-1–RON complex is conserved in apicomplexans, suggesting functional importance for host cell invasion (6, 9). Upon cell entry, the AMA-1 ectodomain is shed; this process appears to be essential for invasion, since antibodies that inhibit shedding also inhibit invasion (1). The remaining cytoplasmic AMA-1 tail plays an essential role in triggering and maintaining intracellular replication of the parasite, which is distinct from its role in invasion of RBC, but the exact function of AMA-1 remains unknown (10–12).
The mechanisms of protection against malaria are also not completely understood, but they include the generation of a humoral response that blocks parasite entry into host cells and inhibits intracellular parasite growth, as well as the induction of parasite-targeted cellular immune responses that directly and indirectly promote the killing of intracellular parasites and mediate protection from reinfection (13–17).
Studies have shown that immunization with correctly folded, parasite-derived or heterogeneously expressed AMA-1 protein can protect against blood-stage parasite challenge in rodent (P. berghei, P. yoelii, or P. chabaudi) and nonhuman primate (P. knowlesi) models (1, 18–25). Additionally, AMA-1-induced protection has been associated with species-specific invasion and growth-inhibiting antibodies against AMA-1, providing protection upon passive transfer in mice (21). In humans naturally exposed to malaria, there is a high prevalence of AMA-1 antibodies, with titers correlating to age (26, 27). Accordingly, efforts to develop an AMA-1-based vaccine have focused on the induction of persistent, high-titer antibody responses and have almost exclusively used recombinant proteins formulated in various adjuvants (1, 22, 24, 26, 28, 29). However, although strong antibody responses and in vitro functionality have been reported in some studies, AMA-1-based vaccines have failed to confer significant protection in humans (30–33). There is evidence that AMA-1-specific CD4+ T cells may play a role in blood-stage immunity, since the efficacy of AMA-1 immunization depends on the presence of CD4+ T cells and adoptive transfer of AMA-1 specific CD4+ T cell lines could protect nude mice against parasitized red blood cell (pRBC) challenge (34–37). Furthermore, blood-stage vaccine trials of AMA-1 as a protein/adjuvant formulation have reportedly elicited T cell responses producing a number of cytokines, including interleukin-5 (IL-5), IL-2, and gamma interferon (IFN-γ), as well as multifunctional CD4+ cytokine-producing T cells and memory T cells (38–41).
Expression of AMA-1 has been described in sporozoite and both early and late liver stages in addition to asexual blood stages (4, 42, 43). A role for AMA-1 in sporozoite invasion has been suggested (4), but it was recently demonstrated that while AMA-1 might mediate host cell recognition as well as parasite orientation and stabilization of hepatocyte binding, it is not essential for invasion and differentiation inside hepatocytes (44). The presence of AMA-1 in the sporozoite and liver stages suggests that it may be a potential target of preerythrocytic-stage immunity. However, although AMA-1 has been extensively studied as a candidate antigen for asexual erythrocytic malaria vaccines, information on its role in preerythrocytic immunity is scarce. There are numerous data sets showing that AMA-1 is recognized by antibodies from malaria-naive individuals immunized with radiation-attenuated sporozoites which do not develop into mature liver schizonts (45) as determined by enzyme-linked immunosorbent assay (ELISA), indirect fluorescent-antibody tests (IFATs) (J. Sacci, personal communication), and protein microarray studies (46). Those data show that AMA-1 is accessible to the immune system during early liver-stage development. However, to our knowledge, there are no reports demonstrating that AMA-1 vaccines can protect against Plasmodium sporozoite challenge in the absence of other Plasmodium antigens. Several studies in mice, nonhuman primates, and humans have investigated the protective capacity of multiantigen vaccines against sporozoite challenge, with no conclusive results. In the P. knowlesi nonhuman primate model, DNA prime/poxvirus boost immunization with a combination of four candidate vaccine antigens, i.e., circumsporozoite protein (CSP), sporozoite surface protein 2 (SSP2), AMA-1, and the 42-kDa fragment of merozoite surface protein 1 (MSP142), led to self-limited, low-level parasitemia in 60% to 80% of rhesus monkeys (47, 48). In humans, a virosome-based vaccine comprising peptides representing the P. falciparum CSP B cell repeat and the semiconserved loop I of domain III of AMA-1 induced very limited blood-stage protection (manifested primarily as a delay in development of parasitemia in 1 of 12 volunteers) (49). However, in both studies, protective effects could not explicitly be associated with defined immune responses to any one or all target antigens. Most recently, a two-component adenovirus-vectored vaccine consisting of one adenovector encoding the P. falciparum CSP protein and one encoding AMA-1 induced moderate T cell IFN-γ responses against both antigens but only low antibody responses, with no growth-inhibitory activity. However, the protective efficacy of the combination or of the AMA-1 component alone was not assessed (50).
Here, we report the first study to investigate the true potential of AMA-1 as a preerythrocytic-stage vaccine candidate. With this goal, we have employed two parallel strategies that target cellular immune responses against P. yoelii AMA-1 (PyAMA-1) using both a DNA prime/protein boost regimen and a T cell epitope/adjuvant immunization approach.
MATERIALS AND METHODS
Mice and parasites.
Specific-pathogen-free female BALB/c mice (Animal Resource Centre, Perth, Australia) were immunized at 6 weeks of age. The parasite strain used in all experiments was Plasmodium yoelii 17XNL. Cryopreserved sporozoites kindly provided by Stephen Hoffman (Sanaria Inc., Rockville, MD, USA) were used for sporozoite challenge. Parasitized red blood cells (pRBC) used for blood-stage challenge were obtained after one passage of frozen pRBC stock (derived from cryopreserved sporozoites at 14 days postinfection) through a BALB/c mouse. All studies were approved by the QIMR Animal Ethics Committee and were conducted in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (2004).
Parasite extract.
P. yoelii extract was generated from P. yoelii 17XNL grown in BALB/c mice to approximately 50% parasitemia. Blood was collected by cardiac puncture and diluted with 10 volumes of FCAB (flow cytometric assessment of blood) buffer (1× phosphate-buffered saline [PBS], 0.5% fetal calf serum [FCS], 2 mM EDTA) (51). The blood sample was centrifuged at 600 relative centrifugal force (RCF) for 10 min, and the cell pellet was resuspended in 0.5% (wt/vol) saponin (Sigma-Aldrich, Castle Hill, NSW, Australia) in FCAB buffer and incubated for 30 min at 37°C. The sample was then twice drawn through a 30-gauge needle, washed with 20 volumes of Milli-Q H2O, and centrifuged as described above. pRBC were fixed in FCAB fixation and lysis buffer (1× PBS, 4% [wt/vol] paraformaldehyde, and 0.0067% [wt/vol] saponin) for 10 min at 37°C. The extract was resuspended in FCAB buffer and stored at −20°C prior to use. All buffers were sterile filtered (0.2 μm) immediately before use.
Synthetic peptides.
Putative PyAMA-1 CD4+ and CD8+ T cell epitopes were predicted from the complete PyAMA-1 protein sequence (www.plasmoDB.org) (52, 53) using computerized major histocompatibility complex (MHC) binding prediction algorithms available from the Immune Epitope Data Base (www.IEDB.org) (54). Peptides were selected for MHC alleles expressed by BALB/c mice (MHC class I, H-2Kd and H-2Dd; MHC class II, IAd and IEd). A consensus algorithm approach was chosen to rank predicted peptides according to their scores from various prediction methods (ANN, SMM, and Comblib). The top scoring MHC class I (9-mer; n = 15) and MHC class II (15-mer; n = 5) nonoverlapping peptides predicted to bind with highest affinity were selected for study (see Table S1 in the supplemental material). Peptides were synthesized by Mimotopes Pty Ltd. (Clayton, VIC, Australia) at a purity of >80%, resuspended at 50 mg/ml in 100% dimethyl sulfoxide (DMSO), and stored at −80°C prior to use. A pool of all putative CD8+ and CD4+ T cell epitopes was used to immunize mice.
Plasmid DNA and recombinant protein immunogens.
P. yoelii 17XNL genomic DNA (gDNA) was extracted from pRBC of BALB/c mice 14 days after infection with P. yoelii 17XNL sporozoites. PyAMA-1 specific primers were designed to amplify the full-length gene based on the annotated protein sequence (www.plasmoDB.org) using Amplify 3.1 (http://engels.genetics.wisc.edu/amplify/; Bill Engels, University of Wisconsin) and OligoCalc (www.basic.northwestern.edu/biotools/oligocalc.html) software tools. Oligonucleotides were commercially synthesized by Sigma-Aldrich (Castle Hill, NSW, Australia). Full-length PyAMA-1 was cloned into pVR1020 (Vical, Inc., San Diego, CA), amplified in Escherichia coli DH5α, and purified using an endotoxin-free plasmid extraction kit (EndoFree Plasmid Giga kit; Qiagen Pty Ltd., Chadstone Centre, VIC, Australia).
For protein expression, a custom pIVEX-HIS/HA vector was created by modifying the pIVEX-HIS 3.2 vector (Roche Applied Science, Castle Hill, NSW, Australia) to express a C-terminal hemagglutinin (HA) tag (55). The cell-free protein expression system RTS 500 HY E. coli (5 Prime GmbH, Hamburg, Germany) was used to produce recombinant protein, which was then His purified using Cobalt Talon resin (Clontech Laboratories, Inc., Mountain View, CA). Briefly, Talon resin was equilibrated with wash buffer (20 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole) and incubated for 30 min at 4°C with the RTS product solution in wash buffer containing 8 M urea. The mixture was then transferred to an Econo-Pac chromatography column (Bio-Rad Laboratories, Gladesville, NSW, Australia), the endotoxin was removed with wash buffer containing 0.05% Triton X-114, and the purified protein was eluted with 150 mM imidazole. To exchange the elution buffer with PBS, the protein eluate was loaded onto a 4-ml 3K Amicon Ultra Filtration tube (Merck Millipore, Billerica, MA) and concentrated in three alternating spinning and washing steps. The protein concentration after the final centrifugation step was measured using a NanoDrop spectrometer (Thermo Fisher Scientific) with an A260/A280 ratio of 0.6 (A260/A280 ratios of <1.8 are indicative of low nucleic acid contamination, and a value of 0.6 is characteristic of pure protein). The endotoxin level of the purified protein was assessed by endpoint chromogenic Limulus amoebocyte lysate (LAL) assay (QCL-1000, Lonza Group, Basel, Switzerland) and ranged between 0.5 and 0.9 endotoxin units (EU)/ml.
Immunization and challenge.
BALB/c mice (n = 5) were immunized in two independent experiments either with two intramuscular doses (in the tibialis anterior muscle with the dose split between two legs) of 100 μg plasmid DNA and one intraperitoneal dose of 10 μg recombinant protein formulated 50:50 in Alhydrogel (Brenntag Biosector, Denmark) or with three subcutaneous doses of a pool of 20 synthetic peptides (29.5 μg/peptide/dose, 10% DMSO) formulated with 12 μg AbISCO100 (Isconova, Sweden) at three weekly intervals.
Mice administered an empty pVR1020 prime followed by a 50:50 alum–1× PBS boost were used as a control group for DNA/protein-vaccinated mice, and mice vaccinated with adjuvant AbISCO100 only were used as a control group for peptide/adjuvant-vaccinated mice. In addition, each experiment included a group of naive no-vaccine mice for immunogenicity assays and evaluation of protective capacity.
Two weeks after the last immunization, mice were challenged with either 103 cryopreserved sporozoites in 1% mouse serum or 103 pRBC in PBS in a volume of 200 μl intravenously in the tail vein. The sporozoite dose was based on counts conducted at vialing; the pRBC dose was determined immediately prior to infection. The process of cryopreservation is associated with a loss of sporozoite viability, and our challenge dose of 103 cryopreserved sporozoites corresponds to a challenge dose of approximately 140 freshly dissected sporozoites (56). We have previously established that 100% of mice inoculated with as few as 50 cryopreserved P. yoelii sporozoites developed blood-stage parasitemia (S. Schussek, submitted for publication). Infectivity controls (nonimmunized naive mice challenged with parasites in parallel to the test groups) were included in all experiments, and all developed blood-stage parasitemia from day 4 after sporozoite challenge (data not presented).
Assessment of liver-stage parasite burden via qRT-PCR.
Livers were harvested at 42 h after challenge and homogenized in 5 ml RNeasy lysis buffer (RLT; Qiagen Pty Ltd., Chadstone Centre, VIC, Australia) with 1% β-2-mercaptoethanol (Sigma-Aldrich Co LLC., St. Louis, MO, USA). RNA was extracted from 200 μl homogenate using the Qiagen RNeasy Minikit (Qiagen Pty Ltd.) according to the manufacturer's instructions. cDNA was synthesized from 2 μg RNA using Vilo reverse transcriptase (Life Technologies Pty Ltd., Applied Biosystems, Mulgrave, VIC, Australia) according to the manufacturer's instructions. cDNA synthesis reaction mixtures were assembled in 20 μl on ice and then incubated for 10 min at room temperature, followed by 2 h at 42°C. Reactions were terminated by heating the sample to 85°C for 5 min. cDNA samples were stored at −20°C. P. yoelii 18S cDNA was quantified using a newly developed and highly sensitive quantitative reverse transcription-PCR (qRT-PCR) assay (Schussek et al., submitted) modified from a previously described protocol (57) and using a custom-made TaqMan probe and primers (Applied Biosystems) and TaqMan Fast Advanced Master Mix (Applied Biosystems, Life Technologies Australia). The mouse β2-microglobulin gene was used as the housekeeping gene (Applied Biosystems kit, at 1/100) to normalize 18S cDNA threshold cycle (CT) values for analysis. Both P. yoelii 18S and β2-microglobulin cDNAs were quantified using a standard curve of cloned cDNA, and the ratio of P. yoelii 18S cDNA to total mouse cDNA (β2-microglobulin) was calculated and normalized against values for the no-vaccine control group to give the percentage of reduction of liver parasite burden (Microsoft Excel 2007, version 12; Microsoft Corporation, Redmond, WA, USA).
Assessment of blood-stage parasitemia via flow cytometric assessment of blood (FCAB assay).
The kinetics and burden of blood-stage infection were monitored daily from day 3 to day 14 and 3 times a week from day 14 to day 30 after either sporozoite or pRBC challenge, by flow cytometric assessment of the blood as described previously (51). Briefly, 3 μl blood was collected from the tip of the tail and diluted into 1 ml FCAB buffer. Then, 30 μl of diluted blood was transferred into a 96-well V-bottom plate (Sarstedt Australia Pty Ltd., Mawson Lakes, SA, Australia), and cells were fixed for 10 min at 37°C with 4% (wt/vol) paraformaldehyde and 0.0067% (wt/vol) saponin (Sigma-Aldrich Co., St. Louis, MO, USA). The parasite nucleic acid was stained with DAPI (4′,6′-diamidino-2-phenylindole) (1 μg/ml in FCAB buffer) for 30 min at 4°C, and the percentage of fluorescent cells was measured by flow cytometry on a high-throughput sampler (HTS)-equipped FACSCanto II (BD Biosciences, San Jose, CA). Data were analyzed using FlowJo software version 9.1 (Treestar Inc., Ashland, OR, USA) and displayed as comparative area under the curve (AUC): the percentage of parasitized RBCs was measured at multiple time points over the course of infection (daily from day 3 to day 14 after challenge and then twice a week until day 30 after challenge), and the curve determined by the percentage of pRBC over time was analyzed for immunized and control groups. The AUC of blood-stage parasitemia over time was calculated in order to take into consideration the kinetics of development of blood-stage parasitemia and subsequent recovery in immunized and control groups. In order to compare different experiments, the AUC values for immunized mice were normalized to the AUC values for no-vaccine controls of the same experiment that received the same challenge dose.
T cell assays.
Spleens and draining lymph nodes (DLNs) of immunized and unimmunized mice were harvested at the time of challenge (10 to 14 days after the third immunization) and restimulated with PyAMA-1 DNA or peptide pools using A20 cells (mouse B lymphocyte cell line, ATCC TIB-208) as antigen-presenting cells (APCs). Briefly, A20 cells were preincubated with 5 μg/ml PyAMA-1 peptide pool or transfected with PyAMA-1 plasmid DNA (5 μg DNA/5 × 106 cells) using the AMAXA Nucleofector system (kit V; Lonza Group, Basel, Switzerland) according to the manufacturer's instructions and then irradiated with approximately 16,666 cGy (Cs-137 irradiator). Splenocytes and lymph node cells were harvested by passing the organs through a 70-μm cell strainer, followed by RBC lysis. Then, 5 × 105 splenocytes or lymph node cells/well were coincubated with 1.5 × 105 antigen-presenting A20 cells/well at 37°C for 6 h (for intracellular cytokine staining [ICS]) or 48 h (for enzyme-linked immunosorbent spot [ELISpot] assay and cytokine bead array [CBA]). These restimulation times and conditions were previously optimized for the induction of cytokine responses using each of the above-mentioned assays. Mock-stimulated cells, coincubated with irradiated A20 cells not presenting any antigen, were included for each group of immunized mice.
(i) IFN-γ ELISpot assay.
The IFN-γ ELISpot assay was performed essentially as previously described (58, 59). A total of 5 × 105 splenocytes/well in Dulbecco's modified Eagle's medium/High Modified dry powder (KDMEM) (SAFC Global, St. Louis, MO) (6 mg/liter folic acid, 36 mg/liter l-asparagine, 116 mg/liter l-arginine, 2 g/liter sodium bicarbonate, 10 mM HEPES, 60 mg/liter penicillin, 100 mg/liter streptomycin), 10% FCS, 5 nM β-2-mercaptoethanol, and 1.5 mM l-glutamine were plated at 100 μl/well into 96-well MultiScreen filter plates (Millipore MAIPS4510 plates) precoated with 100 μg/ml anti-mouse IFN-γ monoclonal antibody (MAb) (BD Biosciences, San Jose, CA). Irradiated peptide-pulsed or antigen-transfected A20 APCs or mitogen control (concanavalin A) was added at 100 μl/well. After incubation at 37°C with 5% CO2 for 36 h, plates were washed 6 times with 1× PBS plus 0.05% Tween 20 (PBST) and incubated with 25 μg/ml biotinylated anti-mouse IFN-γ antibody (BD Biosciences, San Jose, CA) for 3 h at 37°C. Plates were again washed and incubated with 1 μg/ml streptavidin-horseradish peroxidase (HRP) (BD Biosciences, San Jose, CA) for 1 h at room temperature. Plates were then washed 3 times with PBST and 3 times with PBS, incubated with 3-amino-9-ethylcarbazole (AEC) substrate solution (BD Biosciences, San Jose, CA) for 5 min, flooded with tap water to stop the reaction, and dried overnight. Spot-forming cells (SFCs) were counted using the AID ELISpot reader (AID ELISpot reader classic; AID, San Diego, CA). Data were analyzed using Prism 4.02 software (GraphPad, San Diego, CA).
(ii) Cytokine bead array (CBA).
A total of 5 × 105 splenocytes and/or lymph node cells/well were restimulated with 1.5 × 105 A20 antigen-presenting cells (APCs)/well in a total volume of 200 μl in 96-well round-bottom plates. After incubation for 48 h at 37°C with 5% CO2, the cell supernatant was collected for comprehensive analysis of secreted cytokines (IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12, IL-13, IFN-γ, and tumor necrosis factor [TNF]) using the mouse cytometric bead array flex kit (BD Biosciences, San Jose, CA). Briefly, a mix of capture and detection beads for each cytokine was incubated with 5 μl sample for 2 h at room temperature. The fluorescence intensity of the detection beads was measured on a FACSArray instrument (BD Biosciences, San Jose, CA) and analyzed using FlowJo software.
(iii) Intracellular cytokine staining (ICS) and analysis of multifunctional T cell populations.
Splenocytes and/or lymph node cells were restimulated as described for CBA for 6 h and incubated with 0.1% Golgi Plug (BD Biosciences, San Jose, CA) from the start of the restimulation period at 37°C with 5% CO2 in 96-well V-shaped plates. Cells were spun at 600 RCF for 4 min, washed with FCAB buffer, and stained for surface molecules CD4 (mCD4-peridinin chlorophyll protein [PerCP]/Cy5; Invitrogen) and CD8 (mCD8-fluorescein isothiocyanate [FITC]; Biolegend) for 30 min at 4°C. Cells were then washed as before and fixed with 1% paraformaldehyde for 15 min at room temperature. Cells were washed again and stained for intracellular cytokines IL-2 (mIL-2–phycoerythrin [PE]; Biolegend), IFN-γ (mIFN-γ–allophycocyanin [APC]; Miltenyi Biotec), and TNF (mTNF-PE/Cy7; Bioscience) overnight at 4°C in the dark. Samples were acquired on an HTS-equipped FACSCanto II using FACS-DiVa software (BD Bioscience).
The percentage of IFN-γ-, IL-2-, or TNF-positive CD4+ and CD8+ splenocyte populations was calculated by subtracting the unstimulated background control value from the PyAMA-1-restimulated sample using Overton subtraction in the FlowJo Population Comparison tool (FlowJo software version 9.1; Treestar Inc.). To differentiate the percentages of cells positive for one, any two, or all three of the measured cytokines, Boolean gates (FlowJo software version 9.1; Treestar Inc.) were created for IFN-γ-, IL-2-, or TNF-producing CD4+ and CD8+ T cell populations. The background frequencies of cytokine-producing populations in unstimulated controls were subtracted using the Pestle program v1.6, and thresholding, visualization, and statistical analysis of data were performed using SPICE software v5.1 (http://exon.niaid.nih.gov/spice/) (60). The median fluorescence intensity (MFI) of the three cytokines in single-, double-, or triple-positive T cell populations relates to the amount of cytokine secreted from each of these populations. By multiplying the frequency of a multifunctional T cell population by the MFI of the individual cytokines for this population, the integrated MFI (iMFI) was calculated as an indicator of T cell functionality (61).
Indirect fluorescent-antibody test (IFAT) by flow cytometry.
The antiparasite antibody titer was determined by flow cytometry as described previously (62) (see Fig. S1 in the supplemental material). Briefly, sera of immunized and unimmunized mice were collected 2 weeks after each immunization and 21 days after challenge, and 30 μl of serum diluted in FCAB buffer (1/100 prechallenge and 1/300 postchallenge) was incubated with 6 μl of parasite extract for 20 min at room temperature. Antibodies binding to the P. yoelii parasite extract were stained with secondary goat anti-mouse IgG-DyLight 405 (Biolegend) or a mix of anti-mouse IgG1-APC (clone A85-1), anti-IgG2a–FITC (clone R19-15), anti-IgM–PE/Cy7 (clone R6-60.2), and anti-IgE–PE (clone RME-1) (all from BD Biosciences) for 15 min at 4°C. Following another wash, the samples were resuspended in 35 μl of FCAB buffer and analyzed on an HTS-equipped FACSCanto II. Postacquisition data analysis was performed with FlowJo software (see Fig. S1 in the supplemental material). All calculations were performed using Microsoft Excel.
Statistical analysis.
Geometric mean responses were calculated and the nonparametric, two-tailed Mann-Whitney test (Prism 4.02 software package; GraphPad, San Diego, CA) performed to compare PyAMA-1 immunization groups with control groups. The mean background cytokine response induced by mock stimulation with APC alone in each group was subtracted from the mean values for restimulated cells. To compare separate time points or cell populations, we analyzed the difference (calculated as a ratio) between two measurements of the same sample and evaluated the statistical significance of differences between immunized and control groups using the two-tailed Mann-Whitney test. Comparisons of multiple different stimulations (e.g., with individual peptides) were evaluated by one-way analysis of variance (ANOVA) followed by two-tailed Mann-Whitney test. Statistical analysis of multifunctional T cell populations was conducted using Pestle program v1.6 and SPICE software v5.1 (60). Significance was defined at the 5% level.
RESULTS
Protective capacity.
The primary outcome of the proposed study was to determine whether PyAMA-1-specific immune responses could reduce the liver-stage parasite burden or blood-stage parasitemia following sporozoite challenge, in order to evaluate AMA-1 as a target of sterile disease-preventing immune responses. To address this question, BALB/c mice were immunized with PyAMA-1 using DNA/protein or peptide/adjuvant approaches and challenged with 103 infectious P. yoelii 17XNL sporozoites or 103 parasitized red blood cells (pRBC). The parasite burdens in liver and blood were assessed by quantitative real-time PCR (qRT-PCR) or flow cytometric assessment of blood (FCAB), respectively, and compared to those in control mice.
(i) Liver-stage parasite burden following sporozoite challenge.
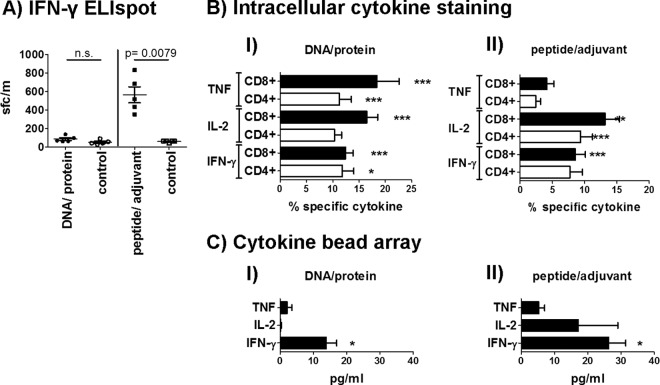
Immunization with either DNA/protein or pooled peptide epitopes in adjuvant significantly reduced parasite burden in the liver following sporozoite challenge, (Fig. 1A) as determined by qRT-PCR. The liver-stage parasite burden was reduced by 95.1% (mean; standard deviation [SD] = 2.9) in mice immunized with PyAMA-1 DNA/protein compared to no-vaccine controls (Fig. 1A) (P = 0.0195) and by 51.6% (mean; SD = 27.5) in mice immunized with PyAMA-1 peptide/adjuvant compared to the adjuvant-only controls (Fig. 1A) (P = 0.0119).
Fig 1.
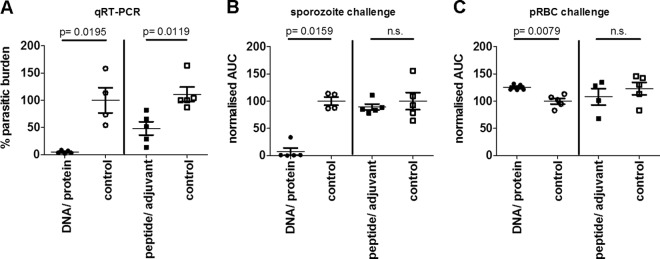
Protective capacity of PyAMA-1 plasmid DNA prime/protein boost immunization and peptide/adjuvant immunization. Mice (n = 5/group, representative of two independent experiments) were immunized 3 times at 3 weekly intervals with plasmid DNA encoding PyAMA-1 and boosted with PyAMA-1 protein or with PyAMA-1 pools of predicted PyAMA-1 T cell epitopes formulated in AbISCO adjuvant. Mice were challenged at 14 days after the last immunization with 1,000 P. yoelii sporozoites or 1,000 pRBC. Protection was assessed by quantitative RT-PCR (qRT-PCR) and FCAB assay. (A) qRT-PCR of Py18S rRNA in total liver RNA 42 h after sporozoite challenge, normalized against the parasite liver burden in infectivity control mice immunized with empty vector. (B) Parasitemia in the blood measured as area under the curve (AUC) after sporozoite challenge, normalized against the AUC for infected empty vector control-immunized mice. (C) Parasitemia in the blood (AUC) after blood-stage challenge, normalized against the AUC for infectivity control mice immunized with empty vector. Statistical analysis was conducted by a two-tailed Mann-Whitney test; significance was defined as a P value of < 0.05. n.s., not significant.
(ii) Blood-stage parasitemia following sporozoite or pRBC challenge.
In parallel groups, mice challenged with infectious sporozoites (preerythrocytic stage) or pRBC (blood stage) were monitored for development of blood-stage parasitemia via the FCAB assay (51). To investigate the effect of AMA-1 immunization on the entire course of infection, we calculated the AUC of blood-stage parasitemia over time and normalized AUC values of immunized mice to AUC values of control mice challenged at the same time.
Sterile protection against sporozoite challenge as indicated by the complete absence of blood-stage parasitemia was observed in 80% of mice immunized with PyAMA-1 DNA/protein (P = 0.0159). In the remaining 20%, blood-stage parasitemia was reduced by 65%, indicating partial protection against sporozoite challenge. However, there was no significant protection against the development of blood-stage parasitemia in mice immunized with the PyAMA-1 peptide pool (Fig. 1B).
Neither PyAMA-1 plasmid DNA/protein nor peptide/adjuvant immunization conferred protection against blood-stage challenge (Fig. 1C).
Our data demonstrate robust PyAMA-1-induced protection at the preerythrocytic stage following sporozoite challenge, as evidenced by a 98% reduction in liver-stage parasite burden and sterile immunity in 80% of mice (absence of parasitemia) following DNA/protein immunization. These data establish AMA-1 as a key target of preerythrocytic-stage immunity.
Immunological response.
To obtain information on the underlying immune responses mediating AMA-1-induced protection against sporozoite challenge, we analyzed cellular immune responses against the whole antigen or defined CD4+ or CD8+ T cell epitopes and measured antibody responses against whole parasite.
(i) Cellular responses.
Protection against preerythrocytic-stage malaria (directed at the parasite-infected hepatocyte) is thought to be mediated mainly by CD8+ T cells and Th1-type CD4+ T cells producing IFN-γ, IL-2, and TNF (13). Thus, we measured IFN-γ, IL-2, and TNF production by splenocytes from AMA-1-immunized mice via ELISpot assay, intracellular cytokine staining (ICS), and cytokine bead array (CBA). Recent studies suggest that cells expressing more than one cytokine have better effector function than single-cytokine-expressing cells and might correlate with protection against infections requiring T cells (38, 61, 63). Furthermore, T cells expressing more than one cytokine simultaneously might have a higher capacity to develop into memory cells (61). Thus, we also analyzed our ICS data in terms of simultaneous production of any two or all three Th1 cytokines, IFN-γ, IL-2, and TNF. Additionally, we investigated the extended cytokine profile induced by each immunization regime by analyzing the supernatants of splenocytes and lymph node cells after in vitro restimulation with PyAMA-1 for a panel of 10 cytokines (IFN-γ, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12p70, IL-13, IL-1β, and TNF) using CBA.
(a) Th1 type cytokines IFN-γ, IL-2, and TNF.
PyAMA-1 DNA/protein immunization increased the number of IFN-γ-expressing CD4+ and CD8+ splenocytes compared to those for no-vaccine controls as measured by ELISpot assay (2-fold [not significant]), ICS (CD4+, 2-fold [P = 0.0201]; CD8+, 7.8-fold [P < 0.0001]), and CBA (1.6-fold [P = 0.0159]) after restimulation with PyAMA-1 in vitro (Fig. 2). The percentage of IL-2-positive CD8+ T cells (4.6-fold [P < 0.0001]) as well as TNF-expressing CD4+ (7.1-fold [P < 0.0001]) and CD8+ (6.3-fold [P = 0.0002]) T cells was also increased in PyAMA-1 DNA/protein immunized mice compared to no-vaccine controls, as measured by ICS. Notably, Th1 cytokines were preferentially secreted by CD8+ T cells compared to CD4+ T cells for all three cytokines (IFN-γ, P = 0.0079; IL-2, P = 0.0079; TNF, P = 0.0119) (Fig. 2B; see Fig. S2 in the supplemental material). PyAMA-1 peptide immunization induced a significant 5-fold increase in IFN-γ spot-forming cells as measured by ELISpot assay (P = 0.0079) (Fig. 2A), significantly higher frequencies of IFN-γ expressing CD8+ T cells (6.1 [P < 0.0001) as measured by ICS, and a significant increase of IFN-γ secretion as detected by CBA in spleens (2.5-fold [P = 0.0119) but not lymph nodes of immunized mice compared to values for controls given adjuvant only (Fig. 2B and C). IL-2-expressing CD4+ (3.5-fold [P = 0.0005]) and CD8+ (3.7-fold [P = 0.002]) T cells were also increased in immunized compared to no-vaccine control mice in the ICS assay, and increased secretion of IL-2 from spleens (3.9-fold [P = 0.0120]) was detected by CBA. However, TNF was only marginally increased in ICS as well as CBA assays in both T cell populations and both tissues, respectively. As noted for DNA/protein immunization, there were significantly more IFN-γ-, IL-2-, or TNF-expressing CD8+ T cells than CD4+ T cells (IFN-γ, P = 0.0079; IL-2, P = 0.0119; TNF, P = 0.0117) (Fig. 3C; see Fig. S2 in the supplemental material) in spleen samples of AMA-1 peptide/adjuvant-immunized mice.
Fig 2.
IFN-γ, IL-2, and TNF responses at the time of challenge. (A) IFN-γ ELISpot assay of splenocytes restimulated in vitro with the PyAMA-1 peptide pool for 36 h. (B) Intracellular cytokine staining for IFN-γ, IL-2, and TNF for CD4+ (white bars) and CD8+ (black bars) cell populations after DNA/protein immunization (I) or peptide/adjuvant immunization (II) and restimulation for 6 h. Data were analyzed using FACS-DiVa software and the FlowJo population comparison tool. (C) CBA for IFN-γ, IL-2, and TNF secretion of splenocytes after restimulation with PyAMA-1 for 48 h. ELISpot data (SFC/million splenocytes) are presented as individual data points for 5 mice (representative of two independent experiments; n = 10). ICS (percent cytokine expression) and CBA (pg/ml secreted cytokine after correction for background in empty vector control groups) data are represented as the mean value (n = 10 mice, two independent experiments). Statistical significance compared to no-vaccine or adjuvant-only controls was determined by a two-tailed Mann-Whitney test (***, P < 0.0005; **, P < 0.005; *, P < 0.05).
Fig 3.
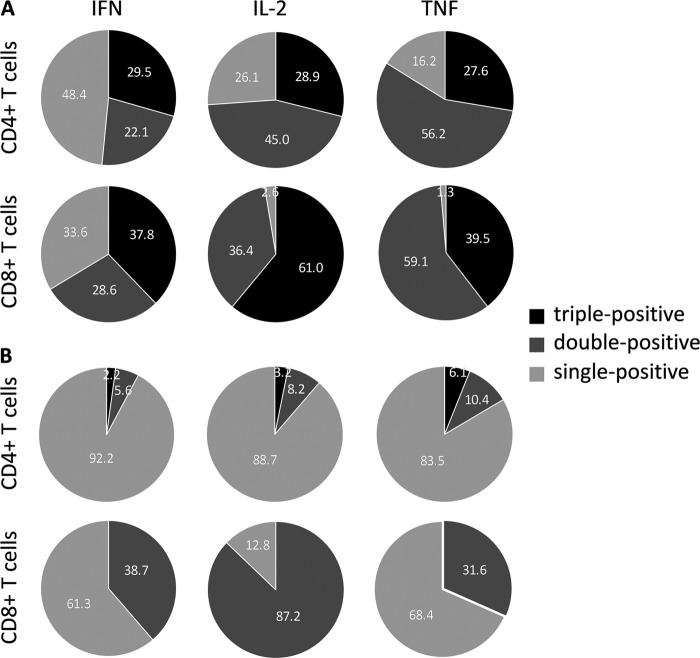
Multifunctional T cell responses induced by PyAMA-1 immunization. (A) The distribution of triple-cytokine-positive (red), double-cytokine-positive (orange), single-cytokine-positive (blue), and cytokine-negative (gray) T cell populations for PyAMA-1-immunized and control mice is presented as pie charts. Statistical evaluation of pie charts was done using a permutation test, comparing each distribution against the respective control group; significance was defined as a P value of < 0.05. (B to E) a comparison of splenic PyAMA-1-specific production of IFN-γ, TNF, and IL-2 in immunized (black) versus control (gray) mice in CD4+ T cells (B) and CD8+ T cells (C) of DNA/protein-immunized or CD4+ T cells (D) and CD8+ T cells (E) of peptide/adjuvant-immunized mice, assessed at the time of challenge as described in the legend to Fig. 1. Data are presented as relative frequency for each population after correction for background cytokine production in unstimulated cells, conducted using Pestle program v1.6 and SPICE software v5.1. The statistical significance of results for immunized versus control groups was determined using the Wilcoxon rank test (*, P < 0.05).
Overall these data show that both PyAMA-1 immunization strategies induced robust Th1 cytokine responses, although there was some inconsistency in the fine details of the response as noted by us (50) and others (64) previously. The antigen-specific IFN-γ response as detected by ELISpot assay and CBA was lower in PyAMA-1 DNA/protein-immunized mice than in peptide/adjuvant-immunized mice, but similar levels of IFN-γ were measured in splenocytes of both immunized groups by ICS. Both immunization strategies also induced antigen-specific TNF responses, although the TNF response was higher in DNA/protein-immunized mice than in peptide/adjuvant-immunized mice when measured by ICS (CD8+ T cells, P = 0.0071; CD4+ T cells, P = 0.0007). Furthermore, the robust antigen-specific IL-2 responses induced by either PyAMA-1 DNA/protein or PyAMA-1 peptide/adjuvant immunization detected by ICS were not measurable in the CBA. Consistent with a role for CD8+ T cells in preerythrocytic-stage immunity, PyAMA-1-specific CD8+ responses were more pronounced than CD4+ responses in both immunized groups (Fig. 2; see Fig. S2 in the supplemental material).
(b) Multifunctional T cells.
The relative distribution of the different CD4+ and CD8+ T cell populations expressing every possible combination of IFN-γ, IL-2, and TNF following PyAMA-1 immunization was significantly different from the relative distribution of those populations in no-vaccine control mice for DNA/protein (CD4+, P = 0.0176; CD8+, P = 0.0047) and peptide/adjuvant (CD4+, P = 0.0059; CD8+, P = 0.0068) immunization (Fig. 3A).
The number of IFN-γ/IL-2/TNF triple-positive T cells was significantly increased for CD4+ and CD8+ T cell populations extracted from spleens of PyAMA-1 DNA/protein-immunized mice compared to no-vaccine controls (CD4+, 20% [P = 0.018]; CD8+, 25% [P = 0.05]) (Fig. 3A and B). Additionally, the percentage of IFN-γ/IL-2 double-positive CD4+ and IL-2/TNF double-positive CD4+ and CD8+ T cells was increased in PyAMA-1 DNA/protein-immunized mice compared to no-vaccine controls (CD4+ IFN-γ/IL-2, 20% [P = 0.018]; CD4+ and CD8+ IL-2/TNF, 18% and 33%, respectively [P = 0.018]) (Fig. 3).
Consistent with the profile measured following DNA/protein immunization, multifunctional CD4+ T cell populations were also significantly higher in PyAMA-1 peptide/adjuvant-immunized mice than in adjuvant-only controls (triple positive, 10% [P = 0.018]; double-positive IFN-γ/IL-2 and IL-2/TNF, 14% and 17.5%, respectively [P = 0.018]) (Fig. 3C). However, and in contrast to the case for PyAMA-1 DNA/protein immunization, PyAMA-1 peptide/adjuvant immunization was very poor in inducing a multifunctional CD8+ T cell population (Fig. 3D); only the IL-2/TNF double-positive CD8+ T cell population was significantly greater in PyAMA-1 peptide/adjuvant-immunized mice than in adjuvant-only controls (33.5% [P = 0.018]).
The amount of cytokine expressed in multifunctional T cells was generally higher than that in single-cytokine-producing cells following PyAMA-1 DNA/protein immunization, as evidenced by the iMFI (Fig. 4A). Triple-positive CD4+ and CD8+ T cell populations accounted for 29.5% and 37.8%, respectively, of the total IFN-γ expression (Fig. 4A), and within the total populations of IL-2- and TNF-expressing CD8+ T cells, 61% and 39.5%, respectively, were triple positive (Fig. 4A). Similarly, IL-2 and TNF expression in double-positive CD4+ (45% of total IL-2 and 56.2% of total TNF) and double-positive CD8+ (36.4% of total IL-2 and 59.1% of total TNF) T cells was increased compared to that in single-cytokine-expressing T cell populations after PyAMA-1 DNA/protein immunization (Fig. 4A). However, and consistent with an increase in the frequency of IFN-γ-expressing CD4+ T cells compared to that in no-vaccine controls (17.5%; P = 0.018) (Fig. 3B), IFN-γ single-positive cells comprised approximately 48.4% and 33.6% of the CD4+ and CD8+ T cell populations, respectively (Fig. 4A).
Fig 4.
IFN-γ, IL-2, and TNF production induced by PyAMA-1 immunization. The contributions of different CD4+ and CD8+ T cell populations to total cytokine production are illustrated in pie charts based on the integrated median fluorescence intensity (iMFI), calculated by multiplying the frequency of cells producing a given cytokine by the magnitude of the cytokine response (measured as median fluorescence intensity). (A) iMFI of splenocytes after PyAMA-1 DNA/protein immunization; (B) iMFI of splenocytes after peptide/adjuvant immunization.
In contrast, following immunization with PyAMA-1 peptide/adjuvant, the highest contribution to IFN-γ and TNF expression was made by single-positive CD4+ T cells (IFN-γ, 92.2%; IL-2, 88.7%; TNF, 83.5%) and CD8+ T cells (IFN-γ, 61.3%; IL-2, 87.2%; TNF, 68.4%) rather than multifunctional cells (Fig. 4B). Very low levels of cytokines were expressed in triple-positive and double-positive CD4+ T cells (Fig. 4B). Consistent with the above-noted increase in the frequency of IL-2/TNF double-positive CD8+ T cells compared to that in adjuvant-only controls, 87.2% of the total IL-2 and 31.6% of the total TNF were produced by double-positive CD8+ T cells (Fig. 4B).
Overall, PyAMA-1 DNA/protein immunization, but not peptide/adjuvant immunization, induced robust multifunctional CD4+ and CD8+ T cell responses with a preference toward high triple-positive and IL-2/TNF double-positive CD8+ T cell responses.
(c) Other cytokine responses.
Using CBA, we measured the quantities of IL-12p70, IL-1β, IL-6, IL-13, IL-4, IL-5, and IL-10 secreted in the supernatant of splenocytes from PyAMA-1-immunized mice restimulated in vitro with full-length PyAMA-1 or a PyAMA-1 peptide pool for 48 h. To provide additional information on the PyAMA-1-specific cellular responses, we also profiled cytokine secretion by cells harvested from the draining lymph nodes.
Significant levels of IL-4 and IL-5 were detected in both splenocytes and lymph nodes of mice immunized with peptide/adjuvant (for IL-4, spleen P = 0.0347 and DLN P = 0.0317; for IL-5, spleen P = 0.0195 and DLN P = 0.0159) but not DNA/protein (Fig. 5). IL-4 secretion was significantly higher (23.2-fold; P < 0.0001) in the spleens than in the lymph nodes of PyAMA-1 peptide/adjuvant-immunized mice (Fig. 5B) and significantly higher in splenocytes of PyAMA-1 peptide/adjuvant- than DNA/protein-immunized mice (93-fold; P = 0.0001) (Fig. 5A). Significant levels of IL-12 and IL-13 were also detected in lymph nodes (IL-12, P = 0.0079; IL-13, P = 0.0286) but not spleens of peptide/adjuvant-immunized mice (Fig. 5B). All other measured cytokine responses were marginal.
Fig 5.
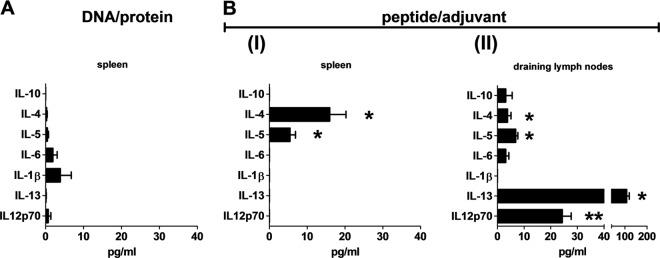
Cytokine bead array analysis of cytokine responses induced by PyAMA-1 immunization. (A) Splenocytes from mice immunized with PyAMA-1 DNA/protein restimulated in vitro with PyAMA-1 DNA-transfected A20 cells. (B) I, splenocytes from mice immunized with PyAMA-1 peptide pool/AbISCO100 adjuvant restimulated in vitro with PyAMA-1 peptide pool. II, lymph node cells from mice immunized with PyAMA-1 peptide pool/AbISCO100 adjuvant restimulated in vitro with PyAMA-1 peptide pool. Data are presented in pg/ml after correction for background cytokine production in empty vector or adjuvant-only controls. Statistical significance compared to controls was determined by a two-tailed Mann-Whitney test (**, P < 0.005; *, P < 0.05).
(ii) Identification of immunodominant epitopes.
The robust IFN-γ responses induced in our studies as reported above provided the opportunity to identify, for the first time, minimal CD4+ and CD8+ T cell epitopes on PyAMA-1. Understanding the IFN-γ response to the different immunization strategies could also allow us to identify differences in the immunodominant epitopes recognized. Indeed, we identified robust IFN-γ responses to five peptides of the pool of 20 putative T cell epitopes (15 CD8+ and 5 CD4+ peptides predicted to bind with high affinity to MHC class I or II molecules) in mice immunized with PyAMA-1 peptide/adjuvant (Fig. 6 and Table 1). Four of these five immunodominant PyAMA-1 epitopes are MHC class I specific (H-2Kd peptide I-1, H-2Dd peptide I-10, H-2Kd peptide I-3, and H-2Dd peptide I-6), and one is MHC class II specific (H-2 IAd peptide II-4), indicating a CD8+ T cell bias for the peptide immunization. A dominance hierarchy was noted, with two of the epitopes (H-2Kd peptide I-1 and H-2Dd peptide I-10) recognized by similar numbers of IFN-γ secreting cells as the complete peptide pool (Fig. 6).
Fig 6.
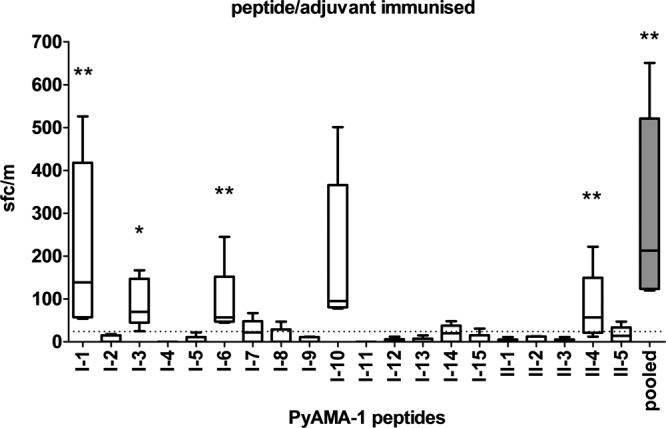
Immunodominant PyAMA-1 epitopes identified after peptide/adjuvant immunization. IFN-γ production by splenocytes harvested from mice immunized with peptide/adjuvant at 10 days after the third immunization and restimulated with individual peptides or PyAMA-1 peptide pool in vitro is shown. Data are presented as mean spot-forming cells (SFCs) for individual mice (n = 5), with error bars representing the total range of values. The dotted line represents the mean background reactivity of unstimulated cells. Statistical significance for the mean number of SFCs in restimulated cells compared to adjuvant-only controls was determined by one-way ANOVA and a two-tailed Mann-Whitney test (**, P < 0.01; *, P < 0.05).
Table 1.
PyAMA-1 immunodominant peptides and their conservation across rodent and primate malaria-causing species
| Peptide | MHC class | Sequence | Location (residues) | SFCs/million splenocytesa |
Conservation (%)b |
|||
|---|---|---|---|---|---|---|---|---|
| DNA/protein | Peptide | Rodent | Primate | Cross-species | ||||
| I-1 | I | YYIFILCSI | 5–13 | − | +++ | 81.5 | 64.8 | 54.0 |
| I-3 | I | QYIAENNDV | 455–463 | + | ++ | 77.8 | 72.2 | 58.7 |
| I-6 | I | EGPNQVISE | 23–31 | − | ++ | 70.4 | NA | 36.8 |
| I-10 | I | FTPEKIENY | 221–229 | − | +++ | 74.1 | 59.3 | 49.2 |
| II-4 | II | NDKNFIATTALSSTE | 366–380 | − | ++ | 87.5 | 76.0 | 71.1 |
| I-2 | I | SLCAKHTSL | 160–168 | + | − | 77.8 | 61.1 | 50.8 |
| I-7 | I | DKPVRSGGL | 118–126 | + | + | 66.7 | 74.1 | 52.2 |
| I-13 | I | AFPETDVHI | 127–135 | + | − | 66.7 | 55.6 | 52.4 |
| I-15 | I | GKGYNWGNY | 339–347 | + | − | 77.8 | 72.2 | 75.7 |
| AMA-1 | ++ | ++++ | 48.3 | 54.9 | 51.6 | |||
IFN-γ ELISpot response to AMA-1-expressing cells or AMA-1 peptide pool. +++, >100; ++, >50; +, >25.
Conservation was determined by CLUSTALW alignment of annotated protein sequences (www.plasmoDB.org) (52) for defined PyAMA-1 peptide sequences (class I or class II) or full-length AMA-1 protein. The analysis was split into conservation across all rodent species (rodent), conservation across all human and nonhuman primate species (primate), and conservation across all Plasmodium species (cross-species). NA, not applicable.
The IFN-γ response to individual peptides following DNA/protein immunization was much less robust than that following peptide/adjuvant immunization. Nevertheless, we were able to identify one H-2Kd binding peptide (I-2) and three H-2Dd binding peptides (I-7, I-13, and I-15) that were recognized by a 2-fold-higher number of cells compared to controls (Table 1; see Fig. S3 in the supplemental material); these responses were not significantly different from the background and did not overlap the epitopes identified following peptide immunization (Table 1). Additional studies are necessary to establish whether or not these peptides are also targets of PyAMA-1-specific CD4+ or CD8+ T cell responses.
Since polymorphism in parasite epitopes targeted by host immune responses is problematic for vaccine development (65), we next analyzed the polymorphism of the identified immunodominant epitopes. Surprisingly, given that AMA-1 is highly polymorphic (1), all five immunodominant PyAMA-1 epitopes identified in our studies were highly conserved (>66%) across rodent species (Table 1 and Fig. 7A and B), and two of the five epitopes (I-3 and II-4) showed high levels of conservation across all animal and human species (59 to 88%) (Fig. 7A and B and Table 1).
Fig 7.

Conservation of immunodominant AMA-1 epitopes across Plasmodium species. Immunodominant epitopes were defined by DNA/protein (A) or peptide/adjuvant (B) immunization. PY01580, P. yoelii; PBANKSA_0900, P. berghei; PCHAS_093100, P. chabaudi; PF11_0344, P. falciparum 3D7; PFIT_PF_0344, P. falciparum IT; PKH_093110, P. knowlesi; PVX_092275, P. vivax (www.plasmoDB.org) (52). Gray, conserved across two rodent Plasmodium species; dark turquoise, conserved across two human species; light turquoise, conserved across four animal and human species.
Two of the five newly identified epitopes (I-6 and I-10) overlap peptide sequences shown to be recognized by malaria-exposed adults in Kenya (66) (PL186 in 25% of peripheral blood mononuclear cell [PBMC] donors and PL192 in 18% of PBMC donors, respectively), and a third (I-3) overlaps with cryptic epitopes shown to be targeted by T helper cells associated with protective antibodies in P. chabaudi (35). One of the putative epitopes identified following DNA/protein immunization (I-2) was also identified in malaria-exposed adults in Kenya (PL190 in 10 to 12% of PBMC donors) (66), suggesting that this sequence does in fact represent a true T cell epitope.
(iii) Humoral responses.
Antibodies against sporozoite and merozoite proteins are known to inhibit parasite entry into host cells, and antibodies against parasite proteins expressed on the surface of infected hepatocytes or erythrocytes can mediate inhibition of parasite development (13, 15, 67, 68). To determine whether antibodies could potentially contribute to the observed reduction of blood-stage parasitemia after sporozoite challenge in PyAMA-1 DNA/protein-immunized mice, we measured the level of antibodies binding to whole blood-stage parasites in the sera of immunized and unimmunized mice before and 23 days after sporozoite challenge (see Fig. S1 in the supplemental material). The level of parasite-specific IgG antibody was significantly increased in mice immunized with PyAMA-1 DNA/protein (P = 0.0079) and peptide/adjuvant (P = 0.0079) compared to controls (Fig. 8).
Fig 8.
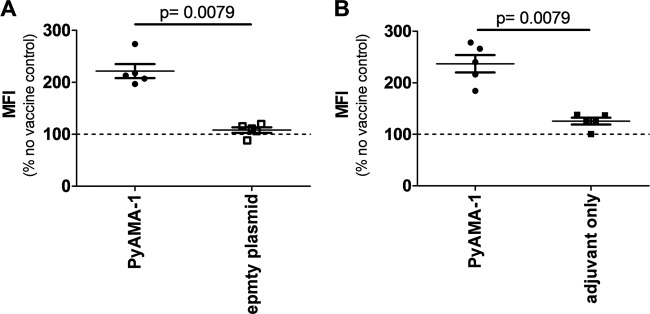
IgG recognition of whole Plasmodium parasite extract. Flow cytometric analysis of total IgG levels binding to P. yoelii blood-stage extract in the sera of DNA/protein (A)- or peptide/adjuvant (B)-immunized mice at the time of challenge is shown. Data are presented as median fluorescence index (MFI) correlating to the antibody titer. The dashed lines represent the unspecific IgG response measured in no-vaccine controls, which was used to normalize antibody titers of immunized and empty vector or adjuvant-only groups. Statistical significance was determined by a two-tailed Mann-Whitney test (P < 0.05).
Next, we analyzed IgG subtypes, measuring the levels of mIgG2a (correlating to human IgG1) and mIgG1 (correlating to human IgG2) in immunized mice before and after challenge (see Fig. S4 in the supplemental material). The ratio of mIgG2a to mIgG1 levels postchallenge was significantly (P = 0.0278) higher in DNA/protein-immunized mice than in no-vaccine controls (2.28 ± 0.4 compared to 0.46 ± 0.05), consistent with a bias toward a cytophilic antibody response. Neither IgM nor IgE was elevated by PyAMA-1 immunization compared to control treatment.
DISCUSSION
A number of studies in animal models (summarized in reference 1) confirm that AMA-1 is a target of antibody-mediated protective immunity against blood-stage Plasmodium parasites. AMA-1 has thus been considered a high-priority target antigen for a blood-stage malaria vaccine. However, AMA-1 is also expressed in Plasmodium sporozoites and liver stages (4, 42, 43) and may therefore also be a target of protective preerythrocytic-stage immune responses. A role for AMA-1 in the induction of protective immunity against preerythrocytic Plasmodium stages has not been demonstrated yet. Here, we performed the first evaluation of the immunogenicity and protective capacity of PyAMA-1 administered via DNA prime/protein boost or peptide/adjuvant immunization strategies against sporozoite challenge and established that AMA-1 is a target of sterile infection-blocking immune responses directed at the preerythrocytic parasite stage. As immunodominant epitopes for AMA-1 have not been previously identified, we utilized a whole-antigen immunization strategy incorporating immunization with PyAMA-1 plasmid DNA and recombinant protein in order to maximize immune responses to all possible AMA-1 epitopes; DNA/protein strategies have previously proved successful in inducing protection in animal models of malaria (69). The PyAMA-1 peptide/adjuvant strategy, on the other hand, was designed to focus immune responses on putative CD4+ and CD8+ T cell epitopes predicted to bind BALB/c MHC class I and class II molecules with high affinity. We elected to formulate the pool of predicted peptide epitopes with the adjuvant AbISCO100, a commercially available ISCOM-matrix formulation consisting of cholesterol, phospholipid, and a combined structure of two saponin fractions that has previously described to generate a balanced immune response with both Th1 and Th2 characteristics (70).
In independent experiments, sterile protection assessed as complete absence of blood-stage parasitemia was induced in 80% of mice immunized with PyAMA-1 DNA/protein (Fig. 1B). The liver-stage parasite burden, as assessed by quantitative RT-PCR, was reduced by 95.1% (mean; SD = 2.9) in mice immunized with PyAMA-1 DNA/protein compared to infected control groups. Immunization with the selected pool of putative PyAMA-1 T cell epitopes in adjuvant was less efficient and more variable but nonetheless resulted in a 51.6% reduction in liver-stage parasite burden (mean; SD = 27.5) (Fig. 1A). This difference in outcome is likely due to the presence of additional epitopes within the whole antigen sequence. An alternative explanation would be underlying differences in uptake and presentation of PyAMA-1 by antigen-presenting cells and subsequent priming of PyAMA-1-specific T cells (71). Additional studies evaluating additional minimal T cell epitopes, longer peptides, or different peptide formulations might improve a PyAMA-1 peptide-based immunization strategy.
In parallel to protection studies, we also evaluated the fine specificities of the PyAMA-1 DNA/protein- and peptide/adjuvant-induced immune responses. Although there are no established correlates of protective immunity against malaria, protection at the liver stage is thought to be mediated primarily by CD8+ T cells, with a role for CD4+ T cells, and production of Th1-type cytokines (13, 67). IFN-γ is routinely used as surrogate marker for preerythrocytic-stage immunity (72). Studies comparing malaria-specific cytokine profiles of protected and unprotected individuals and their capacity for inhibition of the development of early parasite stages confirmed the importance of IFN-γ for protection but also established TNF as mediator of inhibition of preerythrocytic development (73–78). In accordance with these findings, we found that levels of IFN-γ and TNF were significantly elevated in mice receiving the protective PyAMA-1 DNA/protein vaccine compared to unprotected mice (Fig. 2 and 5).
However, T cell cytokine responses induced by immunization are highly heterogeneous, and an imbalance of proinflammatory cytokines (such as IFN-γ and TNF) has been strongly associated with pathogenesis caused by infectious agents (28, 79–81). Also, differences in the types of cytokines produced by individual cells have profound implications for their capacity to mediate protective effector function and/or be sustained as memory cells (61). Thus, multifunctional T cells secreting more than one cytokine, especially IFN-γ, IL-2, and TNF, have recently been suggested as better correlated with protection than single-cytokine-producing cells (61, 82–84).
In our studies, the frequencies of triple-positive (IFN-γ/IL-2/TNF) CD4+ T cells and double-positive (TNF/IL-2) CD4+ and CD8+ T cells were significantly increased after DNA/protein immunization compared to those in controls (Fig. 3). Importantly, we observed that the CD8+ T cell response after PyAMA-1 DNA/protein immunization was significantly more robust than the CD4+ T cell response, with higher frequencies of multifunctional CD8+ T cells and higher magnitudes of cytokines secreted from triple- or double-positive CD8+ T cells than from CD4+ T cells (Fig. 3, Fig. 4). These findings are consistent with a primary role for CD8+ T cells in preerythrocytic immunity.
AMA-1-induced protection against blood-stage Plasmodium spp. in animal models has been associated with AMA-1-specific antibodies (1, 21, 23, 35). In our studies, immunization with PyAMA-1 DNA/protein did induce an antibody response to blood-stage parasites but failed to protect against P. yoelii 17XNL blood-stage challenge. Furthermore, levels of antiparasite IgG after immunization were only poorly boosted by homologous parasite challenge. Finally, subtyping studies identified a bias toward cytophilic antibodies but did not show a preferential induction of isotypes associated in humans with cellular immunity via ADCI (85). Taken together, our data support the concept that the robust PyAMA-1 DNA/protein-induced protection observed in our studies is directed at the preerythrocytic stages of the parasite and likely mediated by T cells rather than antibodies, although further studies are required to define the dependency of AMA-1-induced protection on specific T cell subsets versus antibodies.
Here, we also report the first identification of minimal CD8+ and CD4+ T cell epitopes on P. yoelii AMA-1. Specifically, we identified two immunodominant CD8+ T cell epitopes at residues 5 to 13 (YYIFILCSI) and residues 221 to 229 (FTPEKIENY) and two less dominant CD8+ T cell epitopes at residues 455 to 463 (QYIAENNDV) and residues 23 to 31 (EGPNQVISE), as well as an immunodominant CD4+ T cell epitope at residues 366 to 380 (NDKNFIATTALSSTE). The regions around YYIFILCSI and EGPNQVISE overlap two HLA-A-0201 epitopes, one HLA-A-2402 epitope, and two HLA_DRB5*0101 epitopes predicted for the P. falciparum AMA-1 protein. FTPEKIENY overlaps one HLA-A-1101 epitope predicted for the P. falciparum AMA-1 protein (D. L. Doolan and A. Sette, unpublished data). The crystal structure of the AMA-1 ectodomain shows a conserved central core and variable external loops formed by domains I, II, and III (86). The most polymorphic regions surround a hydrophobic groove containing cryptic and conserved epitopes (87, 88), but the well-studied inhibitory monoclonal antibody 1F9 maps to variable epitopes (89). Less is known regarding the T cell epitopes of AMA-1 and whether or not they fall in the region of highest variability. The synthetic peptides used in our study map to the prosequence as well as domains I, II, and III of AMA-1, spanning conserved as well as variable regions. Epitopes I-1 and I-6 map to the signal sequences and prodomain of AMA-1, whereas epitopes I-10, II-4, and I-3 map to domains I, II, and III, respectively. We establish that all of these newly identified epitopes are relatively conserved among Plasmodium strains, suggesting that the corresponding P. falciparum orthologs may represent immunodominant T cell targets in P. falciparum. Consistent with this, we noted that two of our newly identified immunodominant peptides overlap peptides previously described as targets of proliferative T cells in malaria-exposed adults in Kenya (66).
In summary, here we provide the first demonstration that AMA-1 is a target of sterile protective immunity against Plasmodium sporozoite challenge, validating the potential of AMA-1 as an effective target antigen for vaccines directed against preerythrocytic-stage malaria. We identified immunodominant CD8+ and CD4+ T cell epitopes derived from P. yoelii AMA-1 to facilitate preclinical development of such a vaccine, as well as other more basic research studies. We further established PyAMA-1 as potent inducer of Th1-type cellular immune responses, in particular IFN-γ, TNF, and IL-2, known to be beneficial in immunity against intracellular pathogens. Finally, we showed that AMA-1 stimulates multifunctional CD8+ T cells, which are implicated as more efficient than single-cytokine-secreting cells in protection against complex intracellular pathogens such as Plasmodium spp. These data validate the potential of AMA-1 as a promising target antigen for vaccines directed against preerythrocytic-stage malaria, as well as for inclusion in multistage malaria vaccines.
Supplementary Material
ACKNOWLEDGMENTS
S.S. is supported by an International Research Tuition Award from the University of Queensland and an award from the Australian Centre for Vaccine Development. D.L.D. is supported by a National Health and Medical Research Council (NHMRC) Principal Research Fellowship; support by a Pfizer Australia Senior Research Fellowship is also gratefully acknowledged. The research was supported by NHMRC and the Queensland Tropical Health Alliance.
We thank Stephen Hoffman and colleagues (Sanaria Inc., Rockville, MD, USA) for providing cryopreserved P. yoelii sporozoites, Grace Chojnowski and Paula Hall from the QIMR Flow Cytometry and Imaging Facility for assistance with flow cytometry, and Suzanne Cassidy and staff from the QIMR animal facility for animal husbandry.
Footnotes
Published ahead of print 8 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00544-13.
REFERENCES
- 1.Remarque EJ, Faber BW, Kocken CH, Thomas AW. 2008. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 24:74–84 [DOI] [PubMed] [Google Scholar]
- 2.Narum DL, Thomas AW. 1994. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 67:59–68 [DOI] [PubMed] [Google Scholar]
- 3.Thomas AW, Waters AP, Carr D. 1990. Analysis of variation in PF83, an erythrocytic merozoite vaccine candidate antigen of Plasmodium falciparum. Mol. Biochem. Parasitol. 42:285–287 [DOI] [PubMed] [Google Scholar]
- 4.Silvie O, Franetich JF, Charrin S, Mueller MS, Siau A, Bodescot M, Rubinstein E, Hannoun L, Charoenvit Y, Kocken CH, Thomas AW, Van Gemert GJ, Sauerwein RW, Blackman MJ, Anders RF, Pluschke G, Mazier D. 2004. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J. Biol. Chem. 279:9490–9496 [DOI] [PubMed] [Google Scholar]
- 5.Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. 2004. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 72:154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riglar DT, Richard D, Wilson DW, Boyle MJ, Dekiwadia C, Turnbull L, Angrisano F, Marapana DS, Rogers KL, Whitchurch CB, Beeson JG, Cowman AF, Ralph SA, Baum J. 2011. Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe 9:9–20 [DOI] [PubMed] [Google Scholar]
- 7.Tyler JS, Boothroyd JC. 2011. The C-terminus of Toxoplasma RON2 provides the crucial link between AMA1 and the host-associated invasion complex. PLoS Pathog. 7:e1001282. 10.1371/journal.ppat.1001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, Morlon-Guyot J, Dubremetz JF, Fauquenoy S, Tomavo S, Faber BW, Kocken CH, Thomas AW, Boulanger MJ, Bentley GA, Lebrun M. 2011. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 7:e1001276. 10.1371/journal.ppat.1001276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander DL, Mital J, Ward GE, Bradley P, Boothroyd JC. 2005. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 1:e17. 10.1371/journal.ppat.0010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos JM, Ferguson DJ, Blackman MJ, Soldati-Favre D. 2011. Intramembrane cleavage of AMA1 triggers Toxoplasma to switch from an invasive to a replicative mode. Science 331:473–477 [DOI] [PubMed] [Google Scholar]
- 11.Cowman AF, Tonkin CJ. 2011. Microbiology. A tail of division. Science 331:409–410 [DOI] [PubMed] [Google Scholar]
- 12.Olivieri A, Collins CR, Hackett F, Withers-Martinez C, Marshall J, Flynn HR, Skehel JM, Blackman MJ. 2011. Juxtamembrane shedding of Plasmodium falciparum AMA1 is sequence independent and essential, and helps evade invasion-inhibitory antibodies. PLoS Pathog. 7:e1002448. 10.1371/journal.ppat.1002448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doolan DL, Martinez-Alier N. 2006. Immune response to pre-erythrocytic stages of malaria parasites. Curr. Mol. Med. 6:169–185 [DOI] [PubMed] [Google Scholar]
- 14.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. 2008. Immunity to malaria: more questions than answers. Nat. Immunol. 9:725–732 [DOI] [PubMed] [Google Scholar]
- 15.Beeson JG, Osier FH, Engwerda CR. 2008. Recent insights into humoral and cellular immune responses against malaria. Trends Parasitol. 24:578–584 [DOI] [PubMed] [Google Scholar]
- 16.Douradinha B, Doolan DL. 2011. Harnessing immune responses against Plasmodium for rational vaccine design. Trends Parasitol. [DOI] [PubMed] [Google Scholar]
- 17.Offeddu V, Thathy V, Marsh K, Matuschewski K. 2012. Naturally acquired immune responses against Plasmodium falciparum sporozoites and liver infection. Int. J. Parasitol. 42:535–548 [DOI] [PubMed] [Google Scholar]
- 18.Deans JA, Knight AM, Jean WC, Waters AP, Cohen S, Mitchell GH. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10:535–552 [DOI] [PubMed] [Google Scholar]
- 19.Collins WE, Pye D, Crewther PE, Vandenberg KL, Galland GG, Sulzer AJ, Kemp DJ, Edwards SJ, Coppel RL, Sullivan JS, et al. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51:711–719 [DOI] [PubMed] [Google Scholar]
- 20.Crewther PE, Matthew ML, Flegg RH, Anders RF. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders RF, Crewther PE, Edwards S, Margetts M, Matthew ML, Pollock B, Pye D. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240–247 [DOI] [PubMed] [Google Scholar]
- 22.Kocken CH, Dubbeld MA, Van Der Wel A, Pronk JT, Waters AP, Langermans JA, Thomas AW. 1999. High-level expression of Plasmodium vivax apical membrane antigen 1 (AMA-1) in Pichia pastoris: strong immunogenicity in Macaca mulatta immunized with P. vivax AMA-1 and adjuvant SBAS2. Infect. Immun. 67:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narum DL, Ogun SA, Thomas AW, Holder AA. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stowers AW, Kennedy MC, Keegan BP, Saul A, Long CA, Miller LH. 2002. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect. Immun. 70:6961–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns JM, Jr, Flaherty PR, Romero MM, Weidanz WP. 2003. Immunization against Plasmodium chabaudi malaria using combined formulations of apical membrane antigen-1 and merozoite surface protein-1. Vaccine 21:1843–1852 [DOI] [PubMed] [Google Scholar]
- 26.Thomas AW, Trape JF, Rogier C, Goncalves A, Rosario VE, Narum DL. 1994. High prevalence of natural antibodies against Plasmodium falciparum 83-kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture-enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am. J. Trop. Med. Hyg. 51:730–740 [DOI] [PubMed] [Google Scholar]
- 27.Gray JC, Corran PH, Mangia E, Gaunt MW, Li Q, Tetteh KK, Polley SD, Conway DJ, Holder AA, Bacarese-Hamilton T, Riley EM, Crisanti A. 2007. Profiling the antibody immune response against blood stage malaria vaccine candidates. Clin. Chem. 53:1244–1253 [DOI] [PubMed] [Google Scholar]
- 28.Arnot DE, Cavanagh DR, Remarque EJ, Creasey AM, Sowa MP, Morgan WD, Holder AA, Longacre S, Thomas AW. 2008. Comparative testing of six antigen-based malaria vaccine candidates directed toward merozoite-stage Plasmodium falciparum. Clin. Vaccine Immunol. 15:1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullen GE, Ellis RD, Miura K, Malkin E, Nolan C, Hay M, Fay MP, Saul A, Zhu D, Rausch K, Moretz S, Zhou H, Long CA, Miller LH, Treanor J. 2008. Phase 1 trial of AMA1-C1/Alhydrogel plus CPG 7909: an asexual blood-stage vaccine for Plasmodium falciparum malaria. PLoS One 3:e2940. 10.1371/journal.pone.0002940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierce MA, Ellis RD, Martin LB, Malkin E, Tierney E, Miura K, Fay MP, Marjason J, Elliott SL, Mullen GE, Rausch K, Zhu D, Long CA, Miller LH. 2010. Phase 1 safety and immunogenicity trial of the Plasmodium falciparum blood-stage malaria vaccine AMA1-C1/ISA 720 in Australian adults. Vaccine 28:2236–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polhemus ME, Magill AJ, Cummings JF, Kester KE, Ockenhouse CF, Lanar DE, Dutta S, Barbosa A, Soisson L, Diggs CL, Robinson SA, Haynes JD, Stewart VA, Ware LA, Brando C, Krzych U, Bowden RA, Cohen JD, Dubois MC, Ofori-Anyinam O, De-Kock E, Ballou WR, Heppner DG., Jr 2007. Phase I dose escalation safety and immunogenicity trial of Plasmodium falciparum apical membrane protein (AMA-1) FMP2.1, adjuvanted with AS02A, in malaria-naive adults at the Walter Reed Army Institute of Research. Vaccine 25:4203–4212 [DOI] [PubMed] [Google Scholar]
- 32.Lyke KE, Daou M, Diarra I, Kone A, Kouriba B, Thera MA, Dutta S, Lanar DE, Heppner DG, Jr, Doumbo OK, Plowe CV, Sztein MB. 2009. Cell-mediated immunity elicited by the blood stage malaria vaccine apical membrane antigen 1 in Malian adults: results of a phase I randomized trial. Vaccine 27:2171–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagara I, Dicko A, Ellis RD, Fay MP, Diawara SI, Assadou MH, Sissoko MS, Kone M, Diallo AI, Saye R, Guindo MA, Kante O, Niambele MB, Miura K, Mullen GE, Pierce M, Martin LB, Dolo A, Diallo DA, Doumbo OK, Miller LH, Saul A. 2009. A randomized controlled phase 2 trial of the blood stage AMA1-C1/Alhydrogel malaria vaccine in children in Mali. Vaccine 27:3090–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Hodder AN, Yan H, Crewther PE, Anders RF, Good MF. 2000. CD4+ T cells acting independently of antibody contribute to protective immunity to Plasmodium chabaudi infection after apical membrane antigen 1 immunization. J. Immunol. 165:389–396 [DOI] [PubMed] [Google Scholar]
- 35.Amante FH, Crewther PE, Anders RF, Good MF. 1997. A cryptic T cell epitope on the apical membrane antigen 1 of Plasmodium chabaudi adami can prime for an anamnestic antibody response: implications for malaria vaccine design. J. Immunol. 159:5535–5544 [PubMed] [Google Scholar]
- 36.Brake DA, Long CA, Weidanz WP. 1988. Adoptive protection against Plasmodium chabaudi adami malaria in athymic nude mice by a cloned T cell line. J. Immunol. 140:1989–1993 [PubMed] [Google Scholar]
- 37.Biswas S, Spencer AJ, Forbes EK, Gilbert SC, Holder AA, Hill AV, Draper SJ. 2012. Recombinant viral-vectored vaccines expressing Plasmodium chabaudi AS apical membrane antigen 1: mechanisms of vaccine-induced blood-stage protection. J. Immunol. 188:5041–5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huaman MC, Mullen GE, Long CA, Mahanty S. 2009. Plasmodium falciparum apical membrane antigen 1 vaccine elicits multifunctional CD4 cytokine-producing and memory T cells. Vaccine 27:5239–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spring MD, Cummings JF, Ockenhouse CF, Dutta S, Reidler R, Angov E, Bergmann-Leitner E, Stewart VA, Bittner S, Juompan L, Kortepeter MG, Nielsen R, Krzych U, Tierney E, Ware LA, Dowler M, Hermsen CC, Sauerwein RW, de Vlas SJ, Ofori-Anyinam O, Lanar DE, Williams JL, Kester KE, Tucker K, Shi M, Malkin E, Long C, Diggs CL, Soisson L, Dubois MC, Ballou WR, Cohen J, Heppner DG., Jr 2009. Phase 1/2a study of the malaria vaccine candidate apical membrane antigen-1 (AMA-1) administered in adjuvant system AS01B or AS02A. PLoS One 4:e5254. 10.1371/journal.pone.0005254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malkin EM, Diemert DJ, McArthur JH, Perreault JR, Miles AP, Giersing BK, Mullen GE, Orcutt A, Muratova O, Awkal M, Zhou H, Wang J, Stowers A, Long CA, Mahanty S, Miller LH, Saul A, Durbin AP. 2005. Phase 1 clinical trial of apical membrane antigen 1: an asexual blood-stage vaccine for Plasmodium falciparum malaria. Infect. Immun. 73:3677–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thera MA, Doumbo OK, Coulibaly D, Diallo DA, Kone AK, Guindo AB, Traore K, Dicko A, Sagara I, Sissoko MS, Baby M, Sissoko M, Diarra I, Niangaly A, Dolo A, Daou M, Diawara SI, Heppner DG, Stewart VA, Angov E, Bergmann-Leitner ES, Lanar DE, Dutta S, Soisson L, Diggs CL, Leach A, Owusu A, Dubois MC, Cohen J, Nixon JN, Gregson A, Takala SL, Lyke KE, Plowe CV. 2008. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: results of a phase 1 randomized controlled trial. PLoS One 3:e1465. 10.1371/journal.pone.0001465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, Daly TM, Bergman LW, Kappe SH. 2008. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. U. S. A. 105:305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Ramachandran V, Kumar KA, Westenberger S, Refour P, Zhou B, Li F, Young JA, Chen K, Plouffe D, Henson K, Nussenzweig V, Carlton J, Vinetz JM, Duraisingh MT, Winzeler EA. 2008. Evidence-based annotation of the malaria parasite's genome using comparative expression profiling. PLoS One 3:e1570. 10.1371/journal.pone.0001570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giovannini D, Spath S, Lacroix C, Perazzi A, Bargieri D, Lagal V, Lebugle C, Combe A, Thiberge S, Baldacci P, Tardieux I, Menard R. 2011. Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa. Cell Host Microbe 10:591–602 [DOI] [PubMed] [Google Scholar]
- 45.Suhrbier A, Winger LA, Castellano E, Sinden RE. 1990. Survival and antigenic profile of irradiated malarial sporozoites in infected liver cells. Infect. Immun. 58:2834–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trieu A, Kayala MA, Burk C, Molina DM, Freilich DA, Richie TL, Baldi P, Felgner PL, Doolan DL. 2011. Sterile protective immunity to malaria is associated with a panel of novel P. falciparum antigens. Mol. Cell. Proteomics 10:M111.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamid MM, Remarque EJ, El Hassan IM, Hussain AA, Narum DL, Thomas AW, Kocken CH, Weiss WR, Faber BW. 2011. Malaria infection by sporozoite challenge induces high functional antibody titres against blood stage antigens after a DNA prime, poxvirus boost vaccination strategy in Rhesus macaques. Malar. J. 10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang G, Shi M, Conteh S, Richie N, Banania G, Geneshan H, Valencia A, Singh P, Aguiar J, Limbach K, Kamrud KI, Rayner J, Smith J, Bruder JT, King CR, Tsuboi T, Takeo S, Endo Y, Doolan DL, Richie TL, Weiss WR. 2009. Sterile protection against Plasmodium knowlesi in rhesus monkeys from a malaria vaccine: comparison of heterologous prime boost strategies. PLoS One 4:e6559. 10.1371/journal.pone.0006559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cech PG, Aebi T, Abdallah MS, Mpina M, Machunda EB, Westerfeld N, Stoffel SA, Zurbriggen R, Pluschke G, Tanner M, Daubenberger C, Genton B, Abdulla S. 2011. Virosome-formulated Plasmodium falciparum AMA-1 & CSP derived peptides as malaria vaccine: randomized phase 1b trial in semi-immune adults & children. PLoS One 6:e22273. 10.1371/journal.pone.0022273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sedegah M, Tamminga C, McGrath S, House B, Ganeshan H, Lejano J, Abot E, Banania GJ, Sayo R, Farooq F, Belmonte M, Manohar N, Richie NO, Wood C, Long CA, Regis D, Williams FT, Shi M, Chuang I, Spring M, Epstein JE, Mendoza-Silveiras J, Limbach K, Patterson NB, Bruder JT, Doolan DL, King CR, Soisson L, Diggs C, Carucci D, Dutta S, Hollingdale MR, Ockenhouse CF, Richie TL. 2011. Adenovirus 5-vectored P. falciparum vaccine expressing CSP and AMA1. Part A: safety and immunogenicity in seronegative adults. PLoS One 6:e24586. 10.1371/journal.pone.0024586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apte SH, Groves PL, Roddick JS, VPd Doolan HDL. 2011. High-throughput multi-parameter flow-cytometric analysis from micro-quantities of plasmodium-infected blood. Int. J. Parasitol. 41:1285–1294 [DOI] [PubMed] [Google Scholar]
- 52.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gairia B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr, Treatman C, Wang H. 2009. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37:D539–D543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, Peterson JD, Pop M, Kosack DS, Shumway MF, Bidwell SL, Shallom SJ, van Aken SE, Riedmuller SB, Feldblyum TV, Cho JK, Quackenbush J, Sedegah M, Shoaibi A, Cummings LM, Florens L, Yates JR, Raine JD, Sinden RE, Harris MA, Cunningham DA, Preiser PR, Bergman LW, Vaidya AB, van Lin LH, Janse CJ, Waters AP, Smith HO, White OR, Salzberg SL, Venter JC, Fraser CM, Hoffman SL, Gardner MJ, Carucci DJ. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512–519 [DOI] [PubMed] [Google Scholar]
- 54.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. 2010. The immune epitope database 2.0. Nucleic Acids Res. 38:D854–D862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardoso FC, Roddick JS, Groves P, Doolan DL. 2011. Evaluation of approaches to identify the targets of cellular immunity on a proteome-wide scale. PLoS One 6:e27666. 10.1371/journal.pone.0027666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roestenberg M, Bijker EM, Sim BK, Billingsley PF, James ER, Bastiaens GJ, Teirlinck AC, Scholzen A, Teelen K, Arens T, van der Ven AJ, Gunasekera A, Chakravarty S, Velmurugan S, Hermsen CC, Sauerwein RW, Hoffman SL. 2013. Controlled human malaria infections by intradermal injection of cryopreserved Plasmodium falciparum sporozoites. Am. J. Trop. Med. Hyg. 88:5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Witney AA, Doolan DL, Anthony RM, Weiss WR, Hoffman SL, Carucci DJ. 2001. Determining liver stage parasite burden by real time quantitative PCR as a method for evaluating pre-erythrocytic malaria vaccine efficacy. Mol. Biochem. Parasitol. 118:233–245 [DOI] [PubMed] [Google Scholar]
- 58.Czerkinsky C, Andersson G, Ekre HP, Nilsson LA, Klareskog L, Ouchterlony O. 1988. Reverse ELISPOT assay for clonal analysis of cytokine production. I. Enumeration of gamma-interferon-secreting cells. J. Immunol. Methods 110:29–36 [DOI] [PubMed] [Google Scholar]
- 59.Bruder JT, Stefaniak ME, Patterson NB, Chen P, Konovalova S, Limbach K, Campo JJ, Ettyreddy D, Li S, Dubovsky F, Richie TL, King CR, Long CA, Doolan DL. 2010. Adenovectors induce functional antibodies capable of potent inhibition of blood stage malaria parasite growth. Vaccine 28:3201–3210 [DOI] [PubMed] [Google Scholar]
- 60.Roederer M, Nozzi JL, Nason MX. 2011. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79:167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843–850 [DOI] [PubMed] [Google Scholar]
- 62.Apte SH, Groves PL, Skwarczynski M, Fujita Y, Chang C, Toth I, Doolan DL. 2012. Vaccination with lipid core peptides fails to induce epitope-specific T cell responses but confers non-specific protective immunity in a malaria model. PLoS One 7:e40928. 10.1371/journal.pone.0040928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaveh DA, Bachy VS, Hewinson RG, Hogarth PJ. 2011. Systemic BCG immunization induces persistent lung mucosal multifunctional CD4 T(EM) cells which expand following virulent mycobacterial challenge. PLoS One 6:e21566. 10.1371/journal.pone.0021566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flanagan KL, Lee EA, Gravenor MB, Reece WH, Urban BC, Doherty T, Bojang KA, Pinder M, Hill AV, Plebanski M. 2001. Unique T cell effector functions elicited by Plasmodium falciparum epitopes in malaria-exposed Africans tested by three T cell assays. J. Immunol. 167:4729–4737 [DOI] [PubMed] [Google Scholar]
- 65.Good MF, Doolan DL. 2010. Malaria vaccine design: immunological considerations. Immunity 33:555–566 [DOI] [PubMed] [Google Scholar]
- 66.Lal AA, Hughes MA, Oliveira DA, Nelson C, Bloland PB, Oloo AJ, Hawley WE, Hightower AW, Nahlen BL, Udhayakumar V. 1996. Identification of T-cell determinants in natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in an adult population exposed to malaria. Infect. Immun. 64:1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doolan DL, Dobano C, Baird JK. 2009. Acquired immunity to malaria. Clin. Microbiol. Rev. 22:13–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Druilhe P, Perignon JL. 1994. Mechanisms of defense against P. falciparum asexual blood stages in humans. Immunol. Lett. 41:115–120 [DOI] [PubMed] [Google Scholar]
- 69.Hoffman SL, Doolan DL. 2000. Can. malaria DNA vaccines on their own be as immunogenic and protective as prime-boost approaches to immunization? Dev. Biol. (Basel) 104:121–132 [PubMed] [Google Scholar]
- 70.Yu H, Jiang X, Shen C, Karunakaran KP, Jiang J, Rosin NL, Brunham RC. 2010. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect. Immun. 78:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donnelly JJ, Liu MA, Ulmer JB. 2000. Antigen presentation and DNA vaccines. Am. J. Respir. Crit. Care Med. 162:S190–193 [DOI] [PubMed] [Google Scholar]
- 72.Perlaza BL, Sauzet JP, Brahimi K, Benmohamed L, Druilhe P. 2011. Interferon-gamma, a valuable surrogate marker of Plasmodium falciparum pre-erythrocytic stages protective immunity. Malar. J. 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dunachie SJ, Berthoud T, Keating SM, Hill AV, Fletcher HA. 2010. MIG and the regulatory cytokines IL-10 and TGF-beta1 correlate with malaria vaccine immunogenicity and efficacy. PLoS One 5:e12557. 10.1371/journal.pone.0012557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng W, Wang QH, Liu YJ, Liu J, Feng H, Wu JJ, Cao YM. 2010. Distinct host-related dendritic cell responses during the early stage of Plasmodium yoelii infection in susceptible and resistant mice. Parasite Immunol. 32:324–334 [DOI] [PubMed] [Google Scholar]
- 75.Doolan DL, Hoffman SL. 2000. The complexity of protective immunity against liver-stage malaria. J. Immunol. 165:1453–1462 [DOI] [PubMed] [Google Scholar]
- 76.Schofield L, Villaquiran J, Ferreira A, Schellekens H, Nussenzweig R, Nussenzweig V. 1987. Gamma interferon, CD8+ T cells and antibodies required for immunity to malaria sporozoites. Nature 330:664–666 [DOI] [PubMed] [Google Scholar]
- 77.Bueno LL, Morais CG, Soares IS, Bouillet LE, Bruna-Romero O, Fontes CJ, Fujiwara RT, Braga EM. 2009. Plasmodium vivax recombinant vaccine candidate AMA-1 plays an important role in adaptive immune response eliciting differentiation of dendritic cells. Vaccine 27:5581–5588 [DOI] [PubMed] [Google Scholar]
- 78.Depinay N, Franetich JF, Gruner AC, Mauduit M, Chavatte JM, Luty AJ, van Gemert GJ, Sauerwein RW, Siksik JM, Hannoun L, Mazier D, Snounou G, Renia L. 2011. Inhibitory effect of TNF-alpha on malaria pre-erythrocytic stage development: influence of host hepatocyte/parasite combinations. PLoS One 6:e17464. 10.1371/journal.pone.0017464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Randall LM, Engwerda CR. 2010. TNF family members and malaria: old observations, new insights and future directions. Exp. Parasitol. 126:326–331 [DOI] [PubMed] [Google Scholar]
- 80.Armah H, Wired EK, Dodoo AK, Adjei AA, Tettey Y, Gyasi R. 2005. Cytokines and adhesion molecules expression in the brain in human cerebral malaria. Int. J. Environ. Res. Public Health 2:123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Claser C, Malleret B, Gun SY, Wong AY, Chang ZW, Teo P, See PC, Howland SW, Ginhoux F, Renia L. 2011. CD8+ T cells and IFN-gamma mediate the time-dependent accumulation of infected red blood cells in deep organs during experimental cerebral malaria. PLoS One 6:e18720. 10.1371/journal.pone.0018720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seder RA, Darrah PA, Roederer M. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8:247–258 [DOI] [PubMed] [Google Scholar]
- 83.Bogdan C, Moll H, Solbach W, Rollinghoff M. 1990. Tumor necrosis factor-alpha in combination with interferon-gamma, but not with interleukin 4 activates murine macrophages for elimination of Leishmania major amastigotes. Eur. J. Immunol. 20:1131–1135 [DOI] [PubMed] [Google Scholar]
- 84.Liew FY, Li Y, Millott S. 1990. Tumor necrosis factor-alpha synergizes with IFN-gamma in mediating killing of Leishmania major through the induction of nitric oxide. J. Immunol. 145:4306–4310 [PubMed] [Google Scholar]
- 85.Cortes A. 2008. Switching Plasmodium falciparum genes on and off for erythrocyte invasion. Trends Parasitol. 24:517–524 [DOI] [PubMed] [Google Scholar]
- 86.Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, Collins CR, Withers-Martinez C, Hackett F, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. 2005. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science 308:408–411 [DOI] [PubMed] [Google Scholar]
- 87.Bruder JT, Angov E, Limbach KJ, Richie TL. 2010. Molecular vaccines for malaria. Hum. Vaccin. 6:54–77 [DOI] [PubMed] [Google Scholar]
- 88.Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. 2005. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. U. S. A. 102:12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, Harris KS, Batchelor AH, Anders RF, Foley M. 2006. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect. Immun. 74:2628–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.