Abstract
Some species of the genus Mycoplasma code for the arginine deiminase pathway (ADI), which enables these bacteria to produce ATP from arginine by the successive reaction of three enzymes: arginine deiminase (ArcA), ornithine carbamoyltransferase (ArcB), and carbamate kinase (ArcC). It so far appears that independently isolated strains of Mycoplasma pneumoniae encode an almost identical truncated version of the ADI pathway in which the proteins ArcA and ArcB have lost their original enzymatic activities due to the deletion of significant regions of these proteins. To study the consequences of a functional ADI pathway, M. pneumoniae M129 was successfully transformed with the cloned functional arcA, arcB, and arcC genes from Mycoplasma fermentans. Enzymatic tests showed that while the M. pneumoniae ArcAB and ArcABC transformants possess functional arginine deiminase, ornithine carbamoyltransferase, and carbamate kinase, they were unable to grow on arginine as the sole energy source. Nevertheless, infection of a lung epithelial cell line, A549, with the M. pneumoniae transformants showed that almost 100% of the infected host cells were nonviable, while most of the lung cells infected with nontransformed M. pneumoniae were viable under the same experimental conditions.
INTRODUCTION
The mycoplasmas (class Mollicutes) form a large group of prokaryotic microorganisms that are divided into nine genera with over 200 species. They are distinguished from ordinary bacteria by their small size and minute genome (0.58 to 2.20 Mb) and the total lack of a cell wall (1, 2). Phylogenetically, the mycoplasmas are related to Gram-positive bacteria, from which they developed by genome reduction (3).
One criterion for classifying and characterizing mycoplasma species has been their energy source. For instance, glycolytic mycoplasmas generate energy from sugars by glycolysis, while some of the nonglycolytic mycoplasmas catabolize arginine by the arginine deiminase (ADI) pathway, consisting of three enzymes: arginine deiminase (ArcA), which hydrolyzes arginine to citrulline and ammonia; ornithine carbamoyltransferase (ArcB), which converts citrulline in the presence of phosphate to ornithine and carbamoylphosphate; and carbamate kinase (ArcC), which synthesizes ATP from carbamoylphosphate and ADP. For clarity and simplicity, throughout this article these protein and gene names are interchangeable.
The presence of glycolytic and/or ADI pathways in bacteria has been analyzed in the past by selective growth conditions and enzymatic assays. Barile and coworkers (4) tested 18 Mycoplasma species, of which 10, including Mycoplasma hominis, M. arthriditis, and M. fermentans, could convert arginine to ATP by the arginine deiminase pathway. Among the ADI-negative Mycoplasma species was M. pneumoniae M129, whose complete genome has been sequenced and annotated (5). Surprisingly, an operon that contains coding DNA sequences (CDSs) with significant similarities to all three enzymes of the ADI pathway and a hypothetical arginine transporter have been described for this organism (5, 6). Closer analyses of the proposed CDSs revealed that they do not code for enzymes with ArcA and ArcB activities. Due to frameshift mutations both, arcA and arcB were split, each encoding two partially overlapping CDSs. Mpn304 (238 amino acids) contains the N-terminal region and Mpn305 (198 amino acids) the C-terminal region of ArcA. Mpn306 (273 amino acids) corresponds to an ArcB without the first 70 N-terminal amino acids. These 70 amino acids were part of an 82-amino-acid-long CDS which was not considered in the list of annotated genes (5, 6). The third enzyme of the ADI pathway, carbamate kinase (Mpn307), seemed to be complete. The specificity of the permease (Mpn308) could not be unambiguously determined by comparison of sequence similarities to other permeases. In addition, a second CDS (Mpn560) with high similarity to functional arcA sequences was detected.
The phylogenetically most closely related species to M. pneumoniae within the genus Mycoplasma are M. gallisepticum, M. genitalium, M. alvi, M. imitans, M. pirum, M. testudinis, and M. amphoriforme (7, 8). Sequence analyses of the complete genomes of M. gallisepticum and M. genitalium showed that neither M. genitalium (9) nor M. gallisepticum (10) encoded functional enzymes or truncated CDSs of the ADI pathway. The conservation of one complete enzyme and two truncated enzymes of the ADI in M. pneumoniae strongly suggest that M. pneumoniae once encoded a functional ADI pathway (5). To test the consequences of a functional ADI pathway for the physiology of M. pneumoniae and its interaction with its host cells, M. pneumoniae was complemented by the three functional orthologous genes of the ADI pathway from M. fermentans (4, 11).
MATERIALS AND METHODS
Bacteria, cell line, and growth conditions.
M. fermentans JER and M. pneumoniae M129 (ATCC 29342) were used throughout the study. The organisms were grown for 48 to 72 h at 37°C in 150-cm2 tissue culture flasks containing 60 ml of a modified Hayflick medium consisting of 14.7 g/liter of PPLO broth (Difco), 10% freshly prepared yeast extract (DCL, Sutton, Surrey, Great Britain), 60 μM K2HPO4, 0.002% phenol red, 1,000 U/ml of penicillin G, and 0.05% thallium acetate. The medium for M. fermentans was supplemented with 5% (vol/vol) heat-inactivated horse serum (Biological Industries, Israel), whereas the medium for M. pneumoniae was supplemented with 20% heat-inactivated horse serum. The cultures were grown for 48 to 72 h at 37°C. The organisms were collected at the mid-exponential phase of growth by centrifugation at 12,000 × g for 20 min, washed twice, and resuspended in cold TN buffer containing 250 mM NaCl and10 mM Tris hydrochloride (pH 7.5). Total protein was determined and adjusted to a concentration of 0.5 to 1 mg/ml. Membrane and cytosolic preparations were obtained from intact M. pneumoniae cells by ultrasonic treatment as previously described (12). Membranes were separated from the supernatant by centrifugation in the cold at 37,000 × g for 30 min, washed three times, and resuspended in 10 mM Tris hydrochloride (pH 7.5). To obtain the cytosolic fraction, the supernatant was further centrifuged at 100,000 × g for 2 h to remove cell debris, membrane fragments, and ribosomes.
Plasmids were amplified in Escherichia coli DH5α cultured in Luria-Bertani broth. The cells were grown overnight at 37°C. Ampicillin (100 μg/ml) or kanamycin (25 μg/ml) was added to the medium to select for plasmid-containing clones.
The human lung carcinoma cell line A549 (ATCC CCL185) was maintained in F-12 medium. The medium was supplemented with 10% fetal bovine serum (FBS; Biological Industries, Israel), and the cultures were incubated at 37°C in 5% CO2 in tissue culture flasks or in 24-well polystyrene plates (Nunc, Roskilde, Denmark).
Cloning of arcA and arcB from M. fermentans.
The arcA (MFE_04160) and arcB (MFE_04150) genes were cloned from M. fermentans genomic DNA with a commercial PCR kit (Advantage 2 high-fidelity PCR kit; Clontech laboratories Inc., Mountain View, CA) with the forward primer Mf_ArcAB_F, which includes a BspHI restriction site, and the reverse primer Mf_ArcAB_R, which includes a BamHI restriction site (Table 1). The single 2.4-kbp PCR product containing the arcA and arcB genes (arcAB) was verified on a 1% agarose gel and purified with the Wizard SV gel and PCR cleanup system (Promega, Madison, WI).
Table 1.
List of primers used throughout this study
| Primer name | Primer sequencea | Restriction site introduced | Comments |
|---|---|---|---|
| Mf_ArcAB_F | ATGTTAATATTCATGAAAATTTATTTTATC | BspHI | Cloning of arcAB from M. fermentans |
| Mf_ArcAB_R | CTATTAGGATCCTTAGTAACCAATAGTTGC | BamHI | |
| pMT85_ArcAB_prom_F | GGACACACACTAGTACGGATCC | Verification of the insertion of arcAB into pMT85 by PCR and sequencing | |
| Verification of M. pneumoniae clones containing the inserted arcAB by PCR | |||
| pMT85_ArcAB_prom_R | ATCCTCTAGAGTTGCGGCC | ||
| pMT85_seq_L | TCAGTGAGCGAGGAAGCGGAAG | Determination of the point of integration of Tn4001 into M. pneumoniae genome |
Underlined nucleotides show the indicated restriction sites.
Ligation of ADI gene expression unit with arcAB genes and construction of pMT85:arcAB.
The gene expression unit in front of the arc operon of M. pneumoniae, consisting of the region 284 bp upstream of Mpn304 (13), was redesigned to include a NotI restriction site on the 5′ end and a BspHI restriction site on the 3′ end. Further, 16 bp from the 5′ end of the arcA gene sequence from M. fermentans, containing an internal BspHI site, was added. This modified expression unit was synthesized by GenScript (GenScript Inc., Piscataway, NJ), cloned into the vector pUC57 flanked by EcoRI restriction sites, propagated in E. coli DH5α under ampicillin selection, and purified with the DNA-spin plasmid DNA purification kit (iNtRON Biotechnology Inc., Gyeonggi-do, South Korea). The expression unit was excised by EcoRI digestion (Fermentas Inc., Burlington, Ontario, Canada), separated from the pUC57 vector on a 1.2% agarose gel, and purified (PureLink quick gel extraction kit; Invitrogen Corporation, Carlsbad, CA).
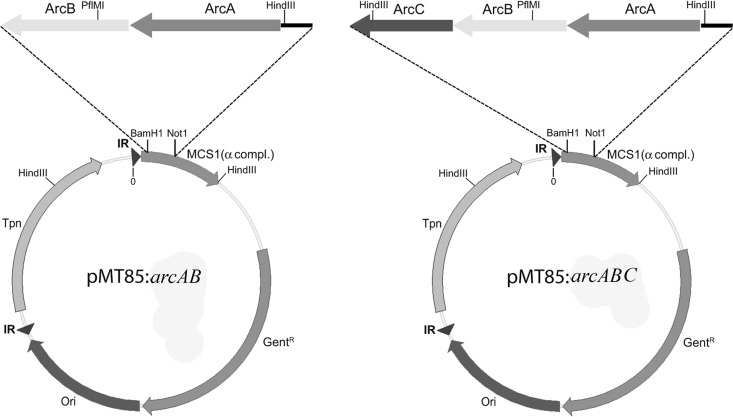
The purified expression unit and the arcAB PCR product were blunt ended with the Klenow fragment of DNA polymerase (New England BioLabs, Ipswich, MA), BspHI digested, and ligated with T4 DNA ligase (Fermentas Inc., Burlington, Ontario, Canada). The expression unit-arcAB construct (2.7 kbp) was inserted into the Tn4001 minitransposon that is part of the plasmid pMT85 (Fig. 1). The minitransposon is flanked by the two 26-bp-long inverted repeats (IR). It contains an origin of replication, a multiple-cloning site (MCS1) combined with a DNA fragment, which allows α complementation, and an aminoglycoside antibiotic resistance determinant as a selectable marker. It confers resistance to kanamycin on E. coli and to gentamicin on M. pneumoniae. Both the plasmid and the expression unit-arcAB construct were doubly digested with NotI and BamHI and ligated with T4 DNA ligase, yielding the plasmid pMT85:arcAB.
Fig 1.
Description of the plasmids pMT85:arcAB and pMT85:arcABC. These plasmids were constructed by inserting the arcA and arcB genes into the BamHI and NotI sites of the plasmid pMT85 (27), generating pMT85:arcAB, and by inserting the arcC gene into the PfiMI and BamHI sites of pMT85:arcAB, resulting in the plasmid pMT85:arcABC. For details, see Materials and Methods. The transposase (Tpn) is located outside the inverted repeats and will not be integrated into the genome of the M. pneumoniae transformants. The expression unit in front of arcA contains a HindIII site, which, together with the closest HindIII site on the genome, facilitates recloning of the genomic site of transposon integration.
Addition of the arcC gene to pMT85:arcAB.
A DNA sequence containing part of the arcB gene and the complete arcC gene from M. fermentans was amplified by PCR (1.9 kb), purified, and doubly digested with BamHI and PflMI. The plasmid pMT85:arcAB was doubly digested with the same enzymes. The linear vector pMT85:arcA (6.4 kbp) was purified by agarose gel electrophoresis. The arcBC PCR product and the linear pMT85:arcA were ligated with T4 ligase. E. coli DH5α was transformed with the ligation product and yielded the plasmid pMT85:arcABC (Fig. 1). This plasmid was digested with BamHI or HindIII to verify its length, and crucial DNA regions were sequenced. All the enzymes were purchased from Fermentas Inc., Burlington, Canada.
Transformation of competent E. coli DH5α.
Plasmids from 30 kanamycin-resistant E. coli clones were purified, BamHI digested, and analyzed on a 1% agarose gel. Plasmids with the expected size (7.5 kbp) were doubly digested with BamHI and NotI. The excised inserts were amplified by PCR and their identities confirmed by sequencing from both ends with the primers pMT85_ArcAB_prom_F and pMT85_ArcAB_prom_R. The primers were designed to align to the vector sequences flanking the insert ends (Table 1).
Transformation of M. pneumoniae.
Transformation of M. pneumoniae with pMT85:arcAB, pMT85:arcABC, and pMT85 was done by electroporation as described previously (14). After electroporation, the bacteria were allowed to recover in an antibiotic-free medium and then diluted in tissue culture flasks with a medium containing gentamicin (80 μg/ml) for the selection of transformants (14). The bacterial colonies were scraped off the tissue culture flasks, filtered through 0.45- and 0.2-μm-pore-size sterile polyvinylidene difluoride (PVDF) membrane Millex-HV filters (Millipore, Cork, Ireland), and plated on agar plates containing gentamicin (80 μg/ml). Colonies were excised from the agar plate, transferred to tubes with medium containing gentamicin (80 μg/ml), and incubated until the medium changed color from red to orange. These precultures were used to inoculate fresh medium (with gentamicin) in Nunclon 25-cm2 tissue culture flasks (Nunc, Roskilde, Denmark). The successful transformation of M. pneumoniae was evaluated and confirmed by PCR with the pMT85 sequencing primers.
The site of the transposon insertion was determined by HindIII restriction of the genomic DNA from transformants, religation of individual HindIII fragments under low DNA concentrations (about 5 μg/ml) to favor circle formation, and transformation of competent E. coli DH5α with the mixture of religated HindIII fragments (Fig. 1). Plasmids of kanamycin-resistant clones were isolated, prescreened by restriction analysis, and further analyzed by DNA sequencing to determine the transition between transposon and genomic DNA. A transformant of M. pneumoniae carrying the arcA and arcB genes from M. fermentans was named M. pneumoniae ArcAB, and M. pneumoniae ArcABC if it also contained the arcC gene from M. fermentans. Throughout this article, M. pneumoniae (M129; ATCC 29342) is referred to as the wild type (M. pneumoniae WT).
Enzymatic assays.
The activities of the enzymes of the arginine deiminase pathway—arginine deiminase (ADI; ArcA), which converts arginine to citrulline; ornithine carbamoyltransferase (OTC; ArcB), which converts citrulline to ornithine; and carbamate kinase (CK; ArcC), which synthesizes ATP from carbamoyl phosphate—were determined in the cytosolic fraction of cell lysates of M. pneumoniae WT and M. pneumoniae ArcAB. M. pneumoniae transformed only with pMT85 served as a control. To test for ADI and OTC activities, 1 mg of the cell lysate was incubated for up to 60 min at 37°C in TN buffer supplemented with 10 mM arginine. At various time intervals, samples were withdrawn and trichloroacetic acid (TCA) was added to a final concentration of 10% (wt/vol). The mixture was centrifuged at 12,000 × g for 3 min at 4°C, and the supernatants were collected for the colorimetric determination of citrulline (15) and ornithine (16). CK activity was determined by coupling ATP to glucose-6-phosphate via hexokinase, which, in turn, reduced NADP in the presence of excess glucose-6-phosphate dehydrogenase (17). The standard assay mixture (3 ml) consisted of 20 mM Tris-HCl (pH 8.3), 1 mM carbamoyl phosphate dilithium salt (Sigma), 5 mM ADP (Sigma), 30 mM MgCl2, 10 mM d-glucose, 1 mM NADP, 10 U of hexokinase, and 10 U of glucose-6-phosphate dehydrogenase. The enzymatic reaction was initiated by the addition of 0.1 ml of cell lysate. The samples were then incubated at 37°C for up to 5 min, and the change in absorbance at 340 nm was monitored continuously at 37°C. The amount of ATP produced is stoichiometrically equal to the amount of NADPH formed. CK activity was calculated by subtracting the values for controls from the values obtained for the complete reaction. Results of the synthesis of citrulline, ornithine, or ATP are presented in μmol/mg of cell protein. Assays were repeated at least three times, and results are presented as means ± standard deviations. When there were sufficient time points, the mean of each time was compared using the Wilcoxon matched-pairs test.
Uptake of l-arginine by M. pneumoniae.
Uptake of 3H-l-arginine by M. pneumoniae was tested in uptake mixtures (total volume of 1 ml) containing washed cells (1 mg of cell protein), 10 mM Tris-morpholineethanesulfonic acid (MES) buffer at various pH values, 250 mM NaCl, 1% bovine serum albumin (fraction V), and 200 μM l-arginine supplemented with 1.5 μCi of 3H-l-arginine (l-[2,3,4-3H]arginine-HCl; New England Nuclear, PerkinElmer Inc.). Chloramphenicol (100 μg/ml) was added to the uptake reaction mixture to prevent incorporation of the labeled arginine into protein, thus allowing the measurement of intracellular arginine. The cells were incubated in the uptake mixture for 5 min at 22°C before addition of the mixture of labeled and unlabeled l-arginine. Incubation was then continued for up to 5 min at 22°C. At various time intervals, 100-μl samples were withdrawn and the reaction was stopped by the addition of 5 ml of ice-cold 0.25 M NaCl. The samples were then passed through membrane glass filters (GF/C, 0.45 μm). The filters were washed with 20 ml of 0.25 M NaCl, air dried, transferred to scintillation vials, and counted in a Beckman scintillation counter. Results were presented as disintegrations per minute per mg of cell protein. Efflux of 3H-l-arginine from M. pneumoniae was examined in cells preloaded with the radioactive amino acid. The cells were loaded with 3H-l-arginine in the uptake mixture described for 5 min at 22°C. The cells were harvested by centrifugation, washed, and resuspended in the uptake mixture containing various amino acids except the labeled 3H-l-arginine. Samples were withdrawn at various time intervals, filtered, washed, and counted as described above.
Survival of A549 lung epithelial cells infected by M. pneumoniae.
Infection of A549 cells by M. pneumoniae was done in 24-well polystyrene plates (Nunc, Roskilde, Denmark), each well containing 5 × 105 A549 cells and 5 × 108 CFU of M. pneumoniae suspended in 1 ml of phosphate-buffered saline (PBS) supplemented with 10 mM CaCl2. The flasks were incubated for up to 24 h in a 5% CO2 atmosphere. The nonadhering mycoplasmas were removed, and the A549 cells were washed three times with 1 ml of PBS, trypsinized for 3 to 4 min, and resuspended in PBS. Viability was analyzed by the trypan blue method (18).
Analytical methods.
Protein was determined by the Bradford method (19) using bovine serum albumin as the standard. Genomic DNA from M. fermentans was extracted and purified with the AquaPure genomic DNA kit (Bio-Rad Laboratories, Hercules, CA). CDSs were predicted with YACOP (20), applying the CDS finders Glimmer, Critica, and Z-curve therein. The output was verified and edited manually by applying criteria such as the presence of a ribosome-binding site, GC frame plot analysis, and similarity to known protein-encoding sequences. Southern blots were carried out with a digoxigenin (DIG)-labeled probe prepared with the PCR DIG probe synthesis kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's recommendations with genomic DNA of M. fermentans as the template and forward and reverse primers Mf_ArcAB_F and Mf_ArcAB_R for the PCR (Table 1). Genomic DNA was extracted from clones of M. pneumoniae ArcAB and from the wild-type strain with the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) and digested with HindIII.
RESULTS
Comparative sequence analysis of mycoplasmal enzymes of the ADI pathway.
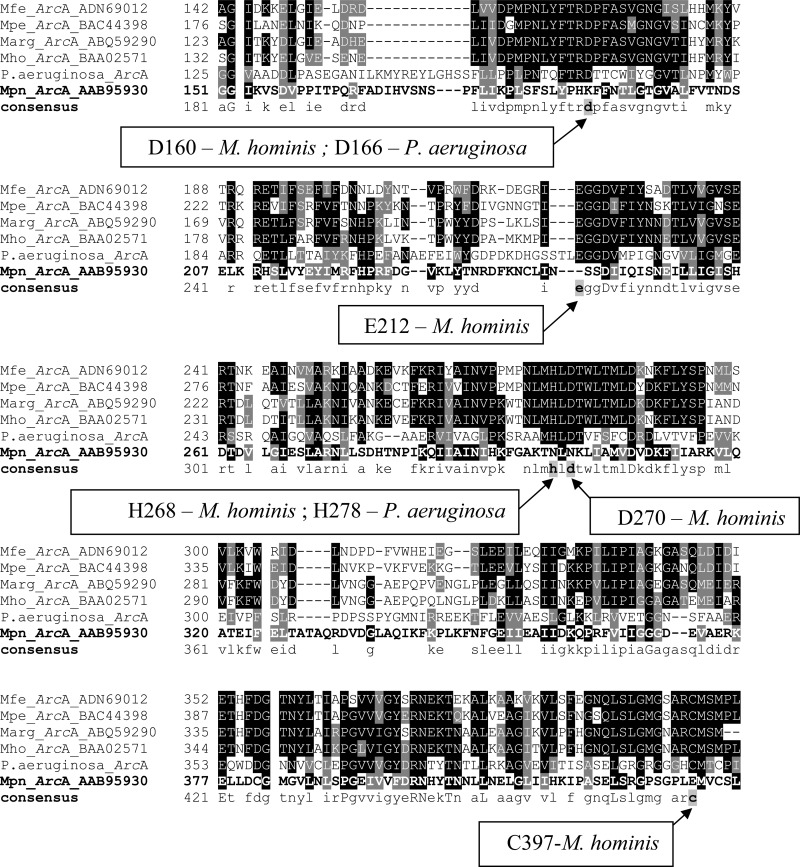
Multiple-sequence alignments of the M. pneumoniae proteins ArcA (Mpn304, Mpn305, and Mpn560), ArcB (Mpn306), and ArcC (Mpn307) with the functional orthologous proteins from M. fermentans (10), M. penetrans (21), M. arginini (22, 23), M. hominis (24), and Pseudomonas aeruginosa (25, 26) revealed that key amino acids located at specific protein positions and essential for enzymatic activity were not conserved in M. pneumoniae. It was obvious for MPN_304 and MPN_305, because these genes are truncated and encode either only the N-terminal region (MPN_304) or the C-terminal region (MPN_305) of ArcA, and also for MPN_306, which encodes only a truncated ArcB of which the first 70 N-terminal amino acids are missing (5). Mpn560 (ArcA), a second candidate for a functional ArcA protein of M. pneumoniae, has a size similar to that of the functional orthologous ArcA proteins from other bacteria, but several amino acids that were described as key amino acids in M. hominis (D160, D270, E212, H268, and C397) (24) were missing, suggesting that Mpn560 does not function as an arginine deiminase (Fig. 2). To confirm the results of the sequence comparison, the enzymatic activities of the arginine deiminase and ornithine carbamyl transferase were determined in M. pneumoniae WT and in insertion mutants (transformants) of M. pneumoniae that carried either arcA and arcB or, alternatively, the arcA, arcB, and arcC genes from M. fermentans integrated into their genomes.
Fig 2.
Multiple alignments of orthologous ArcA sequences. The predicted amino acid sequences (GenBank protein database) of the ArcA proteins from selected bacteria were aligned using ClustalW (47). Amino acid shading was done as described in Materials and Methods. Black represents conserved amino acids, and gray represents amino acids with side chains having similar chemical properties. The sequences are from M. fermentans (ADN69012) (Mfe), M. penetrans (BAC44398) (Mpe), M. arginini (ABQ59290) (Marg), M. hominis (BAA02571) (Mho), M. pneumoniae (AAB95930) (Mpn), Pseudomonas aeruginosa (P13981), and consensus sequence. Conserved amino acids in ArcA from M. hominis and P. aeruginosa essential for the enzymatic activity are framed by rectangles (25, 26), and their positions are indicated by arrows.
Construction of M. pneumoniae insertion mutants.
To construct M. pneumoniae ArcAB and M. pneumoniae ArcABC insertion mutants, M. pneumoniae WT cells were transformed with either pMT85:arcAB or pMT85:arcABC (Fig. 1) as described in Materials and Methods. Successful transformation was confirmed by PCR with pMT85-specific primers (27). The sites of insertion into the genome from five isolated clones named M. pneumoniae ArcAB1, M. pneumoniae ArcAB2, M. pneumoniae ArcAB3, M. pneumoniae ArcAB5, and M. pneumoniae ArcAB6 were determined by recloning and sequencing genomic DNA fragments that contained the transition between minitransposon and genomic DNA, the origin of replication, and the gentamicin and kanamycin resistance gene (Fig. 1). The recovered plasmids were named according to the transformants they were derived from, as follows: pMp arcAB1, pMp arcAB2, pMp arcAB3, pMp arcAB5, and pMp arcAB6. The clones M. pneumoniae ArcAB3, ArcAB5, and ArcAB6 showed the same site of genomic insertion, namely, after nucleotide position 440259 within the gene MPN_369. This gene encodes a putative lipoprotein, the function of which is unknown but nonessential according to results from a global transposon mutagenesis (28). The identical site of insertion indicates that these clones are probably descendants from one original transformant. The two other M. pneumoniae transformants had the transposon inserted after nucleotide position 324059 within the gene MPN_272 (M. pneumoniae ArcAB2), and the other one (M. pneumoniae ArcAB1) had it in the intergenic region between MPN_591 and MPN_592 after nucleotide position 713526. Mpn272 is a hypothetical protein and Mpn591 and Mpn592 are conserved hypothetical lipoproteins. Although the functions of these proteins are undefined, it seems unlikely that they influence the expression of any gene of the ADI pathway. The chromosomal site of insertion of pMT85:arcABC was not determined. This insertion mutant grew like the other insertion mutants; therefore, we did not determine its site of insertion and concluded that essential genes or genomic regions were not inactivated.
Enzymatic activities related to the arginine deiminase pathway and arginine uptake in M. pneumoniae WT and M. pneumoniae ArcAB.
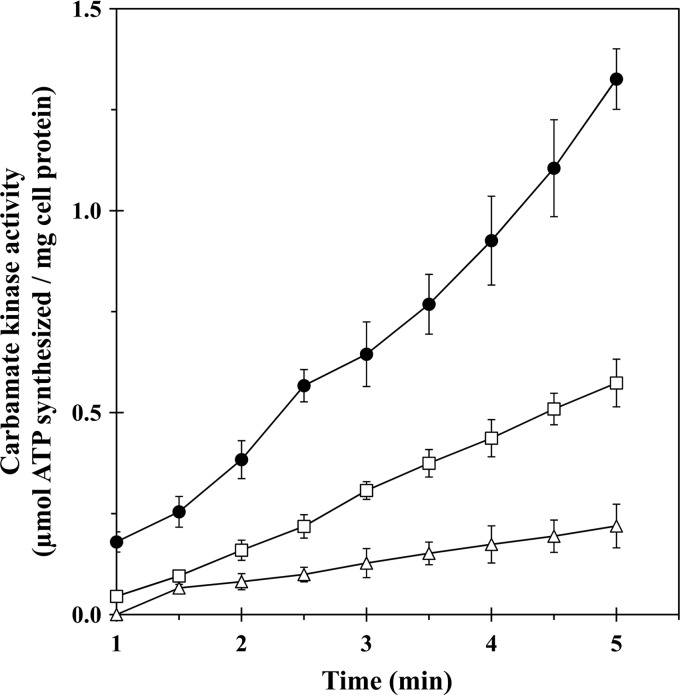
The expression of arcA, arcB, and arcC in M. pneumoniae WT and in a representative M. pneumoniae ArcAB transformant was tested by measuring the accumulation of citrulline and ornithine in M. pneumoniae grown in the presence of arginine and by determining carbamate kinase activity in cell lysates. The results showed that only the M. pneumoniae ArcAB transformant carrying the exogenous arcA and arcB genes was able to generate citrulline and ornithine, proving that the cloned arcA and arcB genes are expressed under the control of the expression unit of the ADI operon of M. pneumoniae and are functional (Table 2). Furthermore, the absence of enzymatic arginine deiminase activity in M. pneumoniae WT supports our notion that amino acids essential for an active arginine deiminase were also missing in Mpn560, as predicted from the sequence. The carbamate kinase activity of M. pneumoniae WT and of M. pneumoniae ArcABC was assessed by measuring ATP levels in cell lysates incubated with ADP and carbamoyl phosphate. The carbamate kinase activity was low in extracts of M. pneumoniae WT but high in extracts of M. pneumoniae ArcABC (Fig. 3). The carbamate kinase activity of the M. pneumoniae ArcABC transformant is significantly greater than the carbamate kinase activity of the M. pneumoniae WT (P < 0.002) as determined by an exact one-tail Wilcoxon test. Figure 3 also shows that the carbamate kinase activity in M. pneumoniae ArcABC was even higher than the activity in M. fermentans. This may be due to a gene dosage effect, since M. pneumoniae ArcABC expresses two active carbamate kinases. A systematic search of the M. pneumoniae genome database failed to identify a CDS homologous to ArcR (J. Kornspan, personal communication) that has been shown in several bacteria to govern expression of the ADI pathway in response to arginine (29).
Table 2.
l-Citrulline and l-ornithine synthesized by M. pneumoniae WT and M. pneumoniae ArcAB transformanta
| Organism | Incubation time (min) | Amt of citrulline synthesized (μmol/mg of cell protein) | Amt of ornithine synthesized (μmol/mg of cell protein) |
|---|---|---|---|
| M. pneumoniae WT | 30 | 0.11 ± 0.02 | 0.03 ± 0.01 |
| 60 | 0.09 ± 0.01 | 0.04 ± 0.01 | |
| M. pneumoniae ArcAB | 30 | 4.92 ± 0.52 | 1.06 ± 0.26 |
| 60 | 5.02 ± 0.69 | 1.73 ± 0.34 | |
| M. fermentans | 30 | 5.19 ± 0.63 | 1.08 ± 0.14 |
| 60 | 5.16 ± 0.61 | 1.83 ± 0.22 |
The biosynthesis of l-citrulline and l-ornithine was determined by colorimetric assays as described in Materials and Methods. M. fermentans served as a positive control. The results presented are means ± standard deviations of three independent experiments.
Fig 3.
Carbamate kinase activity by M. pneumoniae WT and M. pneumoniae ArcABC. Carbamate kinase activity of M. pneumoniae WT (triangles), M. pneumoniae ArcABC (circles), and M. fermentans (squares) was determined by following ATP synthesis from carbamoyl phosphate and ADP in a reaction mixture as described in Materials and Methods. Values are means ± standard deviations of results from three independent experiments.
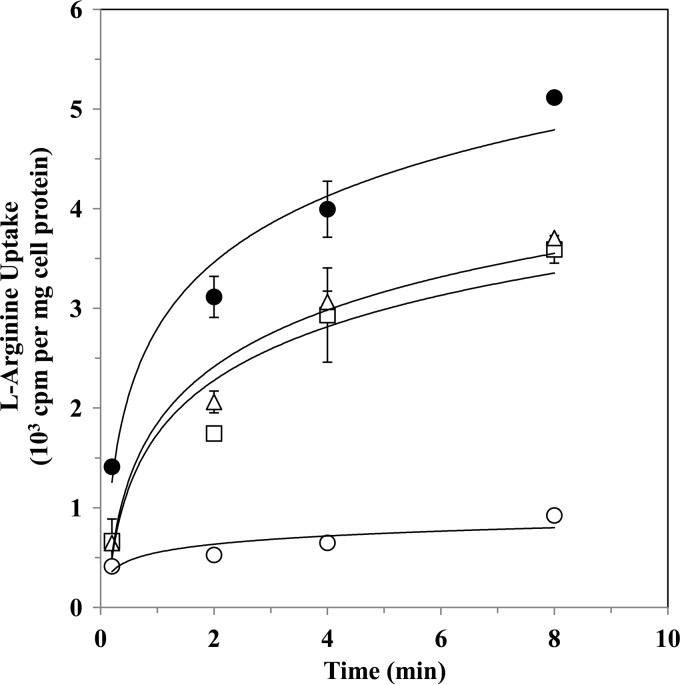
Arginine uptake is essential for M. pneumoniae, since this bacterium is unable to synthesize arginine. The uptake of 3H-labeled l-arginine by M. pneumoniae WT and the transformants was determined at 22°C, a temperature at which arginine metabolism is low or nonexistent (30). The results clearly show that the rate of uptake of 3H-labeled l-arginine by M. pneumoniae was much lower than that of M. fermentans (Fig. 4). Figure 4 also shows that l-arginine uptake by M. pneumoniae WT and that by M. pneumoniae ArcABC were not significantly different (P = 0.375), as determined by an exact two-tail Wilcoxon test. The uptake of 3H-labeled l-arginine by M. pneumoniae ArcAB was completely inhibited by unlabeled l-arginine but not by the d-isomer (data not shown). Uptake was markedly inhibited by l-canavanine and to a smaller extent by l-lysine but not by l-ornithine or l-citrulline (data not shown).
Fig 4.
Arginine transport by M. pneumoniae WT and M. pneumoniae ArcABC. Arginine transport was determined at 22°C as described in Materials and Methods. M. pneumoniae WT (triangles) and a representative clone of M. pneumoniae ArcABC transformants (squares) were used throughout the transport experiments. M. fermentans served as a positive control (filled circles), and transport of M. pneumoniae WT determined at 4°C served as a negative control (open circles). Values are means ± standard deviations of results from four independent experiments.
Growth of M. pneumoniae WT and M. pneumoniae ArcABC in the presence of arginine.
To assess the ability of M. pneumoniae WT and M. pneumoniae ArcAB to grow with arginine and glucose as energy sources and to make sure that an active carbamate kinase was present in transformed M. pneumoniae, the complete arc operon of M. fermentans was inserted into the genome of M. pneumoniae so that this transformant, M. pneumoniae ArcABC, contained all the three enzymes of the ADI pathway of M. fermentans.
When the sole energy source was arginine, the WT M. pneumoniae exhibited no growth and the transformed M. pneumoniae exhibited only very low growth according to protein synthesis measurement. Nonetheless, when both glucose and arginine were present, the growth of the transformed cells was enhanced, whereas growth of the wild type was similar to that in medium containing glucose alone (Table 3). These observations suggest that the energy generated by M. pneumoniae ArcABC grown in a medium with arginine as a sole energy source was not sufficient for regular growth.
Table 3.
Growth of M. pneumoniae WT and M. pneumoniae ArcABC in media supplemented with arginine and/or glucose
| Growth medium supplement(s) | Cell yield (μg of cell protein/100 ml of medium)a |
|
|---|---|---|
| WT | ArcABC transformant | |
| None | 5.0 ± 3.0 | 10.9 ± 3.5 |
| Arginine (20 mM) | 7.5 ± 3.0 | 109.0 ± 18.8 |
| Glucose (20 mM) | 442.2 ± 28.0 | 471.8 ± 35.0 |
| Glucose and arginine (20 mM each) | 521.4 ± 35.0 | 662.6 ± 38.2 |
The results presented are means ± standard deviations of four independent experiments done in triplicates.
Effects of M. pneumoniae WT and M. pneumoniae ArcABC on A549 lung epithelial cells.
Since arginine is essential for growth of eukaryotic cells, we analyzed the fate of A549 lung epithelial cells after infection with M. pneumoniae WT and M. pneumoniae ArcABC. The epithelial cells were infected at a multiplicity of infection of 1,000 CFU/cell, and the number of viable cells was determined 48 and 96 h postinfection by the trypan blue dye exclusion assay (Table 4). The results showed that almost all A549 cells, either noninfected or infected with M. pneumoniae WT, were viable after 96 h of incubation, but nearly 100% of the A549 cells infected with M. pneumoniae ArcABC were nonviable under the same experimental conditions.
Table 4.
Effects of M. pneumoniae WT and M. pneumoniae ArcABC on the viability of A549 cellsa
| Organism used to infect A549 cells | Duration of infection (h) | Total A549 cell count | Dead cells (%) |
|---|---|---|---|
| M. pneumoniae WT | 48 | (3.42 ± 0.75) × 105 | 1.46 ± 0.64 |
| 96 | (5.80 ± 0.74) × 105 | 2.01 ± 0.35 | |
| M. pneumoniae ArcABC | 48 | (1.83 ± 0.67) × 105 | 86.36 ± 10.77 |
| 96 | (2.28 ± 0.44) × 105 | 100.00 ± 0.00 | |
| None | 48 | (4.92 ± 0.42) × 105 | 0.34 ± 0.42 |
| 96 | (6.52 ± 0.46) × 105 | 1.53 ± 0.61 |
The results presented are means ± standard deviations of four independent experiments.
DISCUSSION
M. pneumoniae cannot utilize arginine as an energy source (4), although an operon with a high similarity to an arc operon was later discovered in its genome. A closer look at the sequence revealed that genes encoding the proposed arginine deiminase (Mpn304 and Mpn305) and the ornithine carbamoyl transferase (Mpn306) were truncated (5), while the carbamate kinase (Mpn307) gene seemed to be intact. A gene paralogous (the gene for Mpn560) to the truncated version of arcA had the expected size of a functional arcA gene, but the protein was inactive, as shown by enzymatic tests (Fig. 3). The inactivity is most probably due to several point mutations in all of the key positions (D160, D270, E212, H268, and C397) described previously as being required for enzymatic activity of ArcA from Mycoplasma hominis (24). Although the ADI pathway is not functional in M. pneumoniae M129, all the genes of the proposed arc operon (MPN_304-MPN_308), including the truncated ones, were transcribed (31, 32). The corresponding proteins, except for Mpn304, have also been identified (33, 34). This shows that the expression unit in front of the first gene (MPN_304) of the arc operon is functional. Mpn305, Mpn306, and Mpn307 were components of individual multiprotein complexes, as found in a genome-wide screen analyzing the proteome organization of M. pneumoniae (34). This suggests that these proteins have functions different from those in the ADI pathway. The functions of two of these complexes (complex 22 with MPN_305 and complex 79 with MPN_307) are unknown, and the third complex probably has a metabolic function. In addition to the pyruvate dehydrogenase complex (Pdh), it contains the chaperonin GroL and nine additional proteins, including Mpn306.
The described modifications (truncated versions) of the arcA and arcB genes are also conserved in M. pneumoniae FH (ATCC 15531), the prototype strain for serotype 2 (35) (GenBank accession no. P002077) and in an independently isolated M. pneumoniae strain from Japan (36, 37) (GenBank accession no. AP012303). They differ only by point mutations, strongly suggesting that the loss of the ADI pathway is a more general phenomenon in the species M. pneumoniae and not specific for the strain M129. It is very likely that the common ancestor of both these type strains once possessed a functional ADI pathway.
M. fermentans codes for a functional glycolysis pathway and for a functional ADI pathway. Although M. fermentans grows very poorly with arginine as a sole energy source, this bacterium shows that both pathways are expressed and coexist in one bacterium. Having more than a single energy-generating pathway enlarges the spectrum of substrates and should be beneficial for a bacterium. But this might not be true for M. pneumoniae, with its parasitic lifestyle. On the contrary, loss of the ADI pathway seems to be advantageous, since all three independently isolated and sequenced strains M129, FH, and 309 carry the truncated version of the genes arcA and arcB.
M. pneumoniae is tissue and host specific, and the many strains described in the literature have been mostly isolated from the respiratory tract of humans. The respiratory tract provides a rather constant environment vis-à-vis the composition of nutrients. Therefore, it is conceivable that enough sugars are available as energy sources, and arginine, which is essential for the bacterial protein biosynthesis but cannot be synthesized by M. pneumoniae itself, should not be “wasted” for inefficient ATP synthesis.
Arginine is most probably imported into M. pneumoniae by the protein Mpn308, a typical membrane transport protein with 12 predicted alpha helices forming transmembrane segments that is also homologous to positively charged amino acid permeases from other bacteria. The encoding gene, MPN_308, is also part of the arc operon. So far, no experimental data have been published connecting arginine import into M. pneumoniae with defined proteins and to determining the energy requirement of this process. Does the transporter function as uniporter, antiporter, or symporter? Among mollicutes, an arginine-ornithine exchange system has been reported only for spiroplasmas (30). Attempts have also failed to detect an arginine-ornithine exchange system for M. pneumoniae (A. Katzenell, unpublished data), although such a system has been identified in other bacteria, for example, Pseudomonas aeruginosa (38) and Streptococcus lactis (39). The energy dependence of arginine transport into M. pneumoniae would be an argument against the usefulness of the ADI pathway for M. pneumoniae, because the energy gained from this pathway would depend on the energy costs of the transport system (40).
Arginine deiminase inhibits growth of different cultured cell types, including vascular endothelial cells (41), melanoma cells (42), hepatocellular cells, the human colon cancer cell line DLD-1 (43), and human mammary adenocarcinoma cells (44). The inhibitory effect of native or recombinant arginine deiminase on the proliferation of tumor cells makes this enzyme a candidate for antitumor treatment (45). The main effects of arginine deiminase on these cells are depletion of arginine (46), inhibition of the cell cycle, and induction of apoptosis (41). Based on these findings, we assume that M. pneumoniae ArcABC transformants have very similar effects on A549 lung epithelial cells, and it is likely that the transformation of M. pneumoniae with an active ADI pathway increases its virulence for A549 cells.
The possibility that an active ADI pathway will increase the pathogenicity of M. pneumoniae merits further investigation in an animal model. It might also be worthwhile to investigate whether human patients with severe M. pneumoniae infection carry strains with an active ADI pathway.
ACKNOWLEDGMENT
The excellent help with statistical analyses of Norman B. Grover is acknowledged.
Footnotes
Published ahead of print 29 July 2013
REFERENCES
- 1.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rottem S. 2003. Interaction of mycoplasmas with host cells. Physiol. Rev. 83:417–432 [DOI] [PubMed] [Google Scholar]
- 3.Maniloff J. 2002. Phylogeny and evolution, p 31–43 In Razin S, Herrmann R. (ed), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 4.Barile MF, Schimke RT, Riggs DB. 1966. Presence of the arginine dihydrolase pathway in Mycoplasma. J. Bacteriol. 91:189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, Doerks T, Sanchez-Pulido L, Snel B, Suyama M, Yuan YP, Herrmann R, Bork P. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson K-E, Pettersson B. 2002. Taxonomy of Mollicutes, p 1–29 In Razin S, Herrmann R. (ed), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 8.Pitcher DG, Windsor D, Windsor H, Bradbury JM, Yavari C, Jensen JS, Ling C, Webster D. 2005. Mycoplasma amphoriforme sp. nov., isolated from a patient with chronic bronchopneumonia. Int. J. Syst. Evol. Microbiol. 55:2589–2594 [DOI] [PubMed] [Google Scholar]
- 9.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman RD, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb JF, Dougherty BA, Bott KF, Hu PC, Lucier TS, Peterson SN, Smith HO, Hutchison CA, III, Venter JC. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397–403 [DOI] [PubMed] [Google Scholar]
- 10.Papazisi L, Gorton TS, Kutish G, Markham PF, Browning GF, Nguyen DK, Swartzell S, Madan A, Mahairas G, Geary SJ. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149:2307–2316 [DOI] [PubMed] [Google Scholar]
- 11.Rechnitzer H, Brzuszkiewicz E, Strittmatter A, Liesegang H, Lysnyansky I, Daniel R, Gottschalk G, Rottem S. 2011. Genomic features and insights into the biology of Mycoplasma fermentans. Microbiology 157:760–773 [DOI] [PubMed] [Google Scholar]
- 12.Yavlovich A, Katzenell A, Tarshis M, Higazi AA, Rottem S. 2004. Mycoplasma fermentans binds to and invades HeLa cells: involvement of plasminogen and urokinase. Infect. Immun. 72:5004–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiner J, III, Herrmann R, Browning GF. 2000. Transcription in Mycoplasma pneumoniae. Nucleic Acids Res. 28:4488–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedreyda CT, Lee KK, Krause DC. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170–175 [DOI] [PubMed] [Google Scholar]
- 15.Knipp M, Vasak M. 2000. A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Anal. Biochem. 286:257–264 [DOI] [PubMed] [Google Scholar]
- 16.Chinard FP. 1952. Photometric estimation of proline and ornithine. J. Biol. Chem. 199:91–95 [PubMed] [Google Scholar]
- 17.Schimke RT, Berlin CM, Sweeney EW, Carroll WR. 1966. The generation of energy by the arginine dihydrolase pathway in Mycoplasma hominis 07. J. Biol. Chem. 241:2228–2236 [PubMed] [Google Scholar]
- 18.Louis KS, Siegel AC. 2011. Cell viability analysis using trypan blue: manual and automated methods. Methods Mol. Biol. 740:7–12 [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 20.Tech M, Merkl R. 2003. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 3:441–451 [PubMed] [Google Scholar]
- 21.Sasaki Y, Ishikawa J, Yamashita A, Oshima K, Kenri T, Furuya K, Yoshino C, Horino A, Shiba T, Sasaki T, Hattori M. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo K, Sone H, Yoshida H, Toida T, Kanatani K, Hong YM, Nishino N, Tanaka J. 1990. Cloning and sequence analysis of the arginine deiminase gene from Mycoplasma arginini. Mol. Gen. Genet. 221:81–86 [DOI] [PubMed] [Google Scholar]
- 23.Ohno T, Ando O, Sugimura K, Taniai M, Suzuki M, Fukuda S, Nagase Y, Yamamoto K, Azuma I. 1990. Cloning and nucleotide sequence of the gene encoding arginine deiminase of Mycoplasma arginini. Infect. Immun. 58:3788–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereyre S, Sirand-Pugnet P, Beven L, Charron A, Renaudin H, Barre A, Avenaud P, Jacob D, Couloux A, Barbe V, de Daruvar A, Blanchard A, Bebear C. 2009. Life on arginine for Mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 5:e1000677. 10.1371/journal.pgen.1000677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Xu D, Zhang L, Xie D, Guo H. 2007. Molecular dynamics and density functional studies of substrate binding and catalysis of arginine deiminase. J. Physiol. Chem. 111:3267–3273 [DOI] [PubMed] [Google Scholar]
- 26.Wei Y, Zhou H, Sun Y, He Y, Luo Y. 2007. Insight into the catalytic mechanism of arginine deiminase: functional studies on the crucial sites. Proteins 66:740–750 [DOI] [PubMed] [Google Scholar]
- 27.Zimmerman CU, Herrmann R. 2005. Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae. FEMS Microbiol. Lett. 253:315–321 [DOI] [PubMed] [Google Scholar]
- 28.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165–2169 [DOI] [PubMed] [Google Scholar]
- 29.Zúñiga M, Miralles Md Mdel C, Perez-Martinez G. 2002. The Product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl. Environ. Microbiol. 68:6051–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirazi I, Tarshis M, Rottem S. 1995. An arginine/ornithine exchange system in Spiroplasma melliferum. Microbiology 141:2323–2328 [Google Scholar]
- 31.Güell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, Rode M, Suyama M, Schmidt S, Gavin AC, Bork P, Serrano L. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326:1268–1271 [DOI] [PubMed] [Google Scholar]
- 32.Weiner J, III, Zimmerman CU, Gohlmann HW, Herrmann R. 2003. Transcription profiles of the bacterium Mycoplasma pneumoniae grown at different temperatures. Nucleic Acids Res. 31:6306–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, Kodira CD, Major J, Wang S, Wilkinson J, Nicol R, Nusbaum C, Birren B, Berg HC, Church GM. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kühner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castano-Diez D, Chen WH, Devos D, Guell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R, Herrmann R, Bottcher B, Frangakis AS, Russell RB, Serrano L, Bork P, Gavin AC. 2009. Proteome organization in a genome-reduced bacterium. Science 326:1235–1240 [DOI] [PubMed] [Google Scholar]
- 35.Krishnakumar R, Assad-Garcia N, Benders GA, Phan Q, Montague MG, Glass JI. 2010. Targeted chromosomal knockouts in Mycoplasma pneumoniae. Appl. Environ. Microbiol. 76:5297–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenri T, Horino A, Matsui M, Sasaki Y, Suzuki S, Narita M, Ohya H, Okazaki N, Shibayama K. 2012. Complete genome sequence of Mycoplasma pneumoniae type 2a strain 309, isolated in Japan. J. Bacteriol. 194:1253–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenri T, Taniguchi R, Sasaki Y, Okazaki N, Narita M, Izumikawa K, Umetsu M, Sasaki T. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 67:4557–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhoogt HJ, Smit H, Abee T, Gamper M, Driessen AJ, Haas D, Konings WN. 1992. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J. Bacteriol. 174:1568–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poolman B, Driessen AJ, Konings WN. 1987. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J. Bacteriol. 169:5597–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sjostrom KE, Chen KCS, Kenny GE. 1986. Detection of end products of the arginine dihydrolase pathway in both fermentative and nonfermentative Mycoplasma species by thin-layer chromatography. Int. J. Syst. Evol. Microbiol. 36:60–65 [Google Scholar]
- 41.Gong H, Zolzer F, von Recklinghausen G, Rossler J, Breit S, Havers W, Fotsis T, Schweigerer L. 1999. Arginine deiminase inhibits cell proliferation by arresting cell cycle and inducing apoptosis. Biochem. Biophys. Res. Commun. 261:10–14 [DOI] [PubMed] [Google Scholar]
- 42.Sugimura K, Ohno T, Kusuyama T, Azuma I. 1992. High sensitivity of human melanoma cell lines to the growth inhibitory activity of mycoplasmal arginine deiminase in vitro. Melanoma Res. 2:191–196 [DOI] [PubMed] [Google Scholar]
- 43.Kagemann G, Henrich B, Kuhn M, Kleinert H, Schnorr O. 2005. Impact of Mycoplasma hyorhinis infection on L-arginine metabolism: differential regulation of the human and murine iNOS gene. J. Biol. Chem. 386:1055–1063 [DOI] [PubMed] [Google Scholar]
- 44.Park H, Lee JB, Shim YJ, Shin YJ, Jeong SY, Oh J, Park GH, Lee KH, Min BH. 2008. Arginine deiminase enhances MCF-7 cell radiosensitivity by inducing changes in the expression of cell cycle-related proteins. Mol. Cells 25:305–311 [PubMed] [Google Scholar]
- 45.Kozai M, Sasamori E, Fujihara M, Yamashita T, Taira H, Harasawa R. 2009. Growth inhibition of human melanoma cells by a recombinant arginine deiminase expressed in Escherichia coli. J. Vet. Med. Sci. 71:1343–1347 [DOI] [PubMed] [Google Scholar]
- 46.Takaku H, Matsumoto M, Misawa S, Miyazaki K. 1995. Anti-tumor activity of arginine deiminase from Mycoplasma argini and its growth-inhibitory mechanism. Jpn. J. Cancer Res. 86:840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]