Abstract
Edwardsiella tarda is a Gram-negative bacterial pathogen with a broad host range that includes fish and humans. In this study, we examined the activity and function of the lysozyme inhibitor Ivy (named IvyEt) identified in the pathogenic E. tarda strain TX01. IvyEt possesses the Ivy signature motif CKPHDC in the form of 82CQPHNC87 and contains several highly conserved residues, including a tryptophan (W55). For the purpose of virulence analysis, an isogenic TX01 mutant, TXivy, was created. TXivy bears an in-frame deletion of the ivyEt gene. A live infection study in a turbot (Scophthalmus maximus) model showed that, compared to TX01, TXivy exhibited attenuated overall virulence, reduced tissue dissemination and colonization capacity, an impaired ability to replicate in host macrophages, and decreased resistance against the bactericidal effect of host serum. To facilitate functional analysis, recombinant IvyEt (rIvy) and three mutant proteins, i.e., rIvyW55A, rIvyC82S, and rIvyH85D, which bear Ala, Ser, and Asp substitutions at W55, C82, and H85, respectively, were prepared. In vitro studies showed that rIvy, rIvyW55A, and rIvyH85D were able to block the lytic effect of lysozyme on a Gram-positive bacterium, whereas rIvyC82S could not do so. Likewise, rIvy, but not rIvyC82S, inhibited the serum-facilitated killing effect of lysozyme on E. tarda. In vivo analysis showed that rIvy, but not rIvyC82S, restored the lost pathogenicity of TXivy and enhanced the infectivity of TX01. Together these results indicate that IvyEt is a lysozyme inhibitor and a virulence factor that depends on the conserved C82 for biological activity.
INTRODUCTION
Lysozymes are bactericidal proteins that exist in diverse organisms ranging from animals to bacteria. Lysozymes are classified into several different types, which include chicken-type (C-type), goose-type (g-type), and invertebrate-type (i-type) lysozymes (1). Lysozymes kill bacteria by breaking the 1,4-β-linkages between N-acetylmuramic acid and N-acetyl-d-glucosamine residues in peptidoglycans, a component of bacterial cell walls (2).
Bacteria, in particular, those of a pathogenic nature, have evolved various mechanisms to counteract the action of lysozymes. One of these mechanisms is production of lysozyme inhibitors. The first lysozyme inhibitor, Ivy (inhibitor of vertebrate lysozyme), was discovered in Escherichia coli in 2001 (3). Ivy is a periplasmic protein that is active mainly against C-type lysozymes. Subsequently, several different types of lysozyme inhibitors were identified exclusively in Gram-negative bacteria. These include PliC (periplasmic lysozyme inhibitor of C-type lysozyme)/MliC (membrane-associated lysozyme inhibitor of C-type lysozyme) (4), PliI (periplasmic inhibitor of I-type lysozyme) (5), and PliG (periplasmic inhibitor of G-type lysozyme) (6). Experimental evidence indicates that these inhibitors are able to confer lysozyme tolerance on Gram-negative bacteria when the outer membranes of the bacteria are permeabilized (4, 5, 7–9) and therefore contribute to evasion of the lysozyme-mediated immune response during infection of animal hosts (10, 11).
Ivy homologues have been identified in a large number of Gram-negative bacteria. Most of these proteins contain a strictly conserved CKPHDC motif, while a few contain a less conserved CKPHDC sequence (12). The structural organizations of Ivy alone and Ivy complexed with hen egg white lysozyme (HEWL), a C-type lysozyme, have been resolved. It appears that Ivy forms a homodimer, in which each monomer consists of a central β sheet made of five antiparallel β strands flanked by two short helices on one side and by an amphipathic helix on the other side (3, 12). In the Ivy-HEWL complex, the CKPHDC motif forms a loop that protrudes from Ivy and inserts into the active site of HEWL in a key-lock fashion, thus blocking the activity of the enzyme. Mutational analysis showed that of the conserved residues in CKPHDC, the His residue is essential to the activity of Ivy, whereas the disulfide linkage formed by the two cysteine residues is inessential (12).
Edwardsiella tarda is a Gram-negative bacterium and a pathogen for fish, birds, reptiles, and humans. In aquaculture, E. tarda is a severe fish pathogen and has caused heavy economic losses to many farmed fish species, including turbot (Scophthalmus maximus), flounder (Paralichthys olivaceus), tilapia (Oreochromis niloticus), and channel catfish (Ictalurus punctatus) (13, 14). Accumulating studies indicate that E. tarda possesses a large amount of virulence-associated factors/systems, notably, type III and type VI secretion systems, a quorum-sensing system, two-component systems, adhesin, invasin, and exoenzymes, which are required for optimal bacterial infection (15–18). As a facultative intracellular pathogen, E. tarda can grow extracellularly and inside fish phagocytes, though the intracellular replication mechanism is unclear (19, 20). In previous studies, we identified from E. tarda, via the in vivo-induced antigen technology (IVIAT) and a signal sequence trapping system, several in vivo-induced antigens and secreted proteins that are associated with pathogenicity, including the iron-cofactored superoxide dismutase, which inhibits the macrophage-mediated bactericidal effect, and the invasin Inv1 and the adhesin Eta1, both of which are involved in host infection (17–19, 21). In this report, we characterized another factor identified via IVIAT, i.e., Ivy (named IvyEt for E. tarda Ivy). We examined the activity of IvyEt, the potential role of IvyEt in host infection, and the dependence of IvyEt function on the conserved structural features of the protein. Our results revealed new insights into the biological properties of Ivy.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli BL21(DE3) was purchased from Tiangen (Beijing, China). E. coli S17-1λpir was purchased from Biomedal (Seville, Spain). E. tarda TX01 was isolated from diseased fish (22). The Gram-positive bacterium Micrococcus luteus was purchased from China General Microbiological Culture Collection Center, Beijing, China. Bacteria were cultured in Luria-Bertani (LB) broth at 37°C (for E. coli and M. luteus) or 28°C (for E. tarda). Where indicated, chloramphenicol was supplemented at a concentration of 30 μg/ml.
Fish used for infection study.
Clinically healthy turbot (Scophthalmus maximus) were purchased from a local fish farm and maintained at ∼22°C in aerated seawater. Fish were acclimatized in the laboratory for 2 weeks before experimental manipulation. The weight and size of the fish were 15.9 ± 0.6 g and 8.0 ± 0.3 cm, respectively. Fish were fed daily with commercial dry pellets (purchased from Shandong Sheng-suo Fish Feed Research Center, Shandong, China). Before the experiments, fish were randomly sampled for the examination of bacterial recovery from blood, liver, kidney, and spleen, and no bacteria were detected from the examined tissues of the sampled fish. For tissue collection, fish were euthanized with an overdose of MS-222 (tricaine methanesulfonate; Sigma, St. Louis, MO) as described previously (23). Experiments involving live animals were conducted in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals promulgated by the State Science and Technology Commission of Shandong Province. The animal study was approved by the Science and Research Department of the Institute of Oceanology, Chinese Academy of Sciences.
Sequence analysis.
ivyEt was initially cloned with the IVIAT technology as described previously (24). The sequence of IvyEt was analyzed using the BLAST program at the National Center for Biotechnology Information (NCBI) and the Expert Protein Analysis System. A domain search was performed with the conserved domain search program of NCBI. A signal peptide search was performed with the SignalP (v3.0) program. The theoretical molecular mass and theoretical isoelectric point were predicted using the EditSeq tool in the DNAStar software package (Madison, WI).
Plasmid and strain construction.
The primers used in this study are listed in Table 1. To construct pIvy, which expresses IvyEt, ivyEt was amplified by PCR with primers F1 and R1. The PCR product was ligated with the T-A cloning vector pBS-T (Tiangen, Beijing, China), and the recombinant plasmid was digested with EcoRV. The fragment containing ivyEt was retrieved and inserted into pET259 (25) at the SwaI site. The plasmid pIvyW55A, which expresses the mutant protein IvyW55A, was constructed by overlap extension PCR as follows. The first overlap PCR was performed with primers F1 and R2, the second overlap PCR was performed with primers F2 and R1, and the fusion PCR was performed with the primer pair F1/R1. The PCR product was ligated with pET259 as described above. Plasmids pIvyC82S and pIvyH85D, which express the mutant proteins IvyC82S and IvyH85D, respectively, were created by overlap extension PCR as described above. For the construction of pIvyC82S, the first and second overlap PCRs were performed with the primer pairs F1/R3 and F3/R1, respectively, and the fusion PCR was performed with the primer pair F1/R1. For the construction of pIvyH85D, the first and second overlap PCRs were performed with the primer pairs F1/R4 and F4/R1, respectively, and the fusion PCR was performed with the primer pair F1/R1. All PCR products were verified by sequence analysis.
Table 1.
Primers used in this study
| Primer | Sequence (5′ → 3′)a |
|---|---|
| F1 | GATATCGACGAGACTATTCCGCCCTCG (EcoRV) |
| R1 | GATATCTTTCCAATCCGGCTGCGAC (EcoRV) |
| F2 | CCGTCGGCGGTGCGCAG |
| R2 | GCACCGCCGACGGCAGATC |
| F3 | GCATGGTCAGCCAGCCGCACA |
| R3 | GGCTGGCTGACCATGCCGACC |
| F4 | GCCGGACAACTGCGGCAACC |
| R4 | CGCAGTTGTCCGGCTGGCA |
| F5 | GGATCCAGGGTTTTCGGTTTACT (BamHI) |
| R5 | CTATTTCCATAACGAGGGCGGAAT |
| F6 | CTCGTTATGGAAATAGCGCGC |
| R6 | GGATCCGTCAGCGATTTCACGA (BamHI) |
Underlined nucleotides are restriction sites of the enzymes indicated in the parentheses at the end of the sequence.
To construct the E. tarda mutant TXivy, in-frame deletion of a 360-bp segment (residues 88 to 447) of ivyEt was performed by overlap extension PCR as follows: the first overlap PCR was performed with primers F5 and R5, the second overlap PCR was performed with primers F6 and R6, and the fusion PCR was performed with the primer pair F5/R6. The PCR products were inserted into the suicide plasmid pDM4 (26) at the BglII site, resulting in pDMIvy. S17-1λpir was transformed with pDMIvy, and the transformants were conjugated with TX01 as described previously (22). The transconjugants were selected on LB agar plates supplemented with 10% sucrose. One of the colonies that were resistant to sucrose and sensitive to chloramphenicol (marker of pDM4) was analyzed by PCR, and the PCR products were subjected to DNA sequencing to confirm the in-frame deletion. This strain was named TXivy.
Purification of recombinant proteins.
E. coli BL21(DE3) was transformed separately with pIvy, pIvyW55A, pIvyC82S, and pIvyH85D. The transformants were cultured in LB medium at 37°C to mid-logarithmic phase, and expression of ivyEt was induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. After growing at 18°C for an additional 16 h, the cells were harvested by centrifugation, and His-tagged proteins were purified under native conditions using Ni-nitrilotriacetic acid agarose (Qiagen, Valencia, CA) as recommended by the manufacturer. The purified proteins were dialyzed for 24 h against phosphate-buffered saline (PBS) and treated with Triton X-114 to remove endotoxin, as reported previously (27). The proteins were concentrated with Amicon Ultra centrifugal filter devices (Millipore, Billerica, MA). The concentrated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized after staining with Coomassie brilliant blue R-250. The concentration of the proteins was determined using the Bradford method with bovine serum albumin as the standard.
Tissue dissemination and mortality analysis.
For tissue dissemination analysis, strains TXivy and TX01 were cultured in LB medium to an optical density at 600 nm (OD600) of 0.8. The cells were washed with PBS and resuspended in seawater. Turbot were randomly divided into four groups (n = 40) and immersed in seawater containing 1 × 108 CFU/ml TXivy or TX01 for 6 h. The fish were then moved to tanks containing fresh seawater and reared under normal conditions. At 0, 1, and 2 days after immersion infection, blood, kidney, liver, and spleen were aseptically taken from the fish (five per time point). The tissues were weighed and homogenized in a glass homogenizer containing PBS. The homogenates and blood were serially diluted and plated in triplicate on LB agar plates. The plates were incubated at 28°C for 48 h, and the colonies that appeared on the plates were enumerated. The genetic identity of the colonies was verified by PCR with the primers EsrAF2/EsrAR2 and ORF26F2/ORF26R3, which amplify the genes esrA and orf26, respectively, in TX01 and TXivy (28). Selected PCR products were subjected to sequence analysis. The remaining fish were monitored for mortality for 2 weeks. The experiment was performed three times.
Bacterial replication in macrophages.
Turbot head kidney (HK) macrophages were prepared as described previously (29). The macrophages were cultured in L-15 medium (Thermo Scientific HyClone, Beijing, China) in 96-well culture plates (∼105 cells/well). TX01 and TXivy suspensions in PBS were prepared as described above and added to macrophages (106 CFU/well). The cells were incubated at 25°C for 0.5 h and then washed three times with PBS. Fresh L-15 medium containing 100 μg/ml gentamicin (Thermo Scientific HyClone, Beijing, China) was added to the cells, and the cells were incubated at 25°C for 1 h to kill extracellular bacteria. The plates were then washed three times with PBS and incubated at 28°C for 1 h, 3 h, 5 h, and 7 h. After incubation, the plates were washed with PBS and the cells were lysed with 100 μl 1% Triton X-100. The cell lysate was serially diluted and plated in triplicate on LB agar plates. The plates were incubated at 28°C for 48 h, and the colonies that emerged on the plates were counted. The identities of the colonies were verified as described above. The experiment was performed three times.
Serum survival analysis.
Sera from three turbot were prepared as reported previously (19) and mixed together. TX01 and TXivy were cultured in LB medium to an OD600 of 0.8. The cells were washed with PBS and resuspended in PBS. Approximately 103 bacterial cells were mixed with or without (control) 50 μl turbot serum. After incubation with mild agitation at 30°C for 60 min, the mixture was serially diluted and plated in triplicate on LB agar plates. The plates were incubated at 28°C for 48 h. The colonies that appeared on the plates were enumerated. The survival rate was calculated as follows: (number of serum-treated cells/number of untreated control cells) × 100%. The experiment was performed three times.
Lysozyme inhibitor activity of rIvy.
M. luteus was cultured in LB medium to an OD600 of 0.8. The cells were washed with PBS and resuspended in PBS to an OD600 of 0.7. The bacterial suspension was mixed with or without (control) 5 μg/ml HEWL (Solarbio Science & Technology, Beijing, China) in the presence or absence of 7.5 μg/ml recombinant IvyEt (rIvy) or mutant protein rIvyW55A, rIvyC82S, or rIvyH85D. The mixture was incubated at 25°C, and cell density was monitored by measuring the absorbance at 600 nm at various time points. To examine the effect of rIvy on E. tarda survival in the presence of HEWL, TX01 and TXivy were cultured in LB medium to an OD600 of 0.8 and resuspended in PBS to 105 CFU/ml. Turbot serum was diluted five times in PBS. HEWL was suspended in PBS to 1 mg/ml. rIvy and rIvyC82S were suspended in PBS to 1.5 mg/ml. For the assay, a bacterial suspension was added to six 0.5-ml Eppendorf tubes (10 μl/tube), and the following solutions were then added separately to each tube: 90 μl PBS (control), 50 μl diluted serum, 10 μl HEWL, 50 μl diluted serum plus 10 μl HEWL, 50 μl diluted serum plus 10 μl HEWL and 10 μl rIvy, and 50 μl diluted serum plus 10 μl HEWL and 10 μl rIvyC82S. PBS was added to each tube to make the final volume to 100 μl. The tubes were incubated at 30°C for 1 h. After incubation, the mixture in each tube was diluted in LB medium and plated in triplicate on LB agar plates. The plates were incubated at 28°C for 48 h, and the colonies that emerged on the plates were counted. The identities of the colonies were verified as described above. The assays were performed three times.
In vivo effect of rIvy on TXivy and TX01 infection.
TX01 and TXivy were cultured in LB medium to an OD600 of 0.8. The cells were washed with PBS and resuspended in PBS to 1 × 107 CFU/ml. To examine the effect of rIvy on TXivy infection, turbot (average weight, 8.6 g) were randomly divided into four groups (five fish per group) and injected intraperitoneally with 100 μl TXivy, TX01, TXivy plus 50 μg rIvy, or TXivy plus 50 μg rIvyC82S. At 2 days postinfection, bacterial recoveries from blood, kidney, liver, and spleen were determined as described above. The effect of rIvy on TX01 infection was carried out in the same fashion. The experiments were performed three times.
Statistical analysis.
Except for the mortality assay, in which the log-rank test was used to compare the survival distributions of the fish, all other statistical analyses were performed with analysis of variance (ANOVA) of the SPSS (v15.0) package (SPSS Inc., Chicago, IL). In all cases, the significance level was defined as a P value of <0.05.
RESULTS
Sequence characterization of IvyEt.
The amino acid sequence of IvyEt is identical to the amino acid sequences of the inhibitor of the vertebrate lysozyme precursor of E. tarda FL6-60 and EIB202 (GenBank accession no. ADM41108.1 and ACY83906.1, respectively). IvyEt contains 151 residues and was predicted to have a molecular mass of 16.6 kDa. BLAST analysis showed that IvyEt shares the highest sequence identity (89%) with the Ivy of Edwardsiella ictaluri and moderate identities (25% to 46%) with the Ivy inhibitors of Serratia marcescens FGI94, Serratia proteamaculans 568, Yersinia pseudotuberculosis PB1/+, Yersinia enterocolitica subsp. palearctica Y11, Aeromonas hydrophila subsp. hydrophila ATCC 7966, Burkholderia pseudomallei MSHR346, and Escherichia coli K-12 (see Fig. S1 in the supplemental material). In silico analysis identified a putative signal peptide (residues 1 to 21) in IvyEt and a conserved Ivy domain (residues 32 to 149), with the latter containing the motif 82CQPHNC87. C82 and C87 are the only cysteine residues in IvyEt. In addition, IvyEt also contains several residues, i.e., A21, P53, W55, P65, and Y76, that are highly conserved among known Ivy lysozyme inhibitors.
Mutation of ivyEt and its effect on bacterial pathogenicity. (i) Effect on the capacity to disseminate in host tissues and induce host mortality.
To examine the biological importance of IvyEt, an ivyEt-defective E. tarda mutant, TXivy, was constructed by markerless in-frame deletion of a 360-bp internal segment of ivyEt. Compared to the wild-type strain TX01, TXivy exhibited a similar growth profile when cultured in LB medium (data not shown). To examine whether ivyEt mutation affected pathogenicity, turbot were inoculated with the same dose of TXivy or TX01 via immersion, and bacterial infection of blood, kidney, liver, and spleen was analyzed by bacterial recovery analysis. The results showed that at 0 days after immersion infection, the numbers of bacteria recovered from TXivy- and TX01-infected fish were comparable, while at 1 and 2 days postinfection, the numbers of bacterial cells recovered from all examined tissues of TXivy-infected fish were significantly less than those recovered from TX01-infected fish (Fig. 1). For TX01-infected fish, mortality began to occur at 3 days postinfection, and the accumulated mortality reached 80% by 6 days postinfection. For TXivy-infected fish, mortality began to occur at 5 days postinfection and stopped at 7 days postinfection, with an accumulated mortality of 30%. The survival rate of TXivy-infected fish (70%) was significantly (P < 0.01) higher than that (20%) of TX01-infected fish (Fig. 2).
Fig 1.
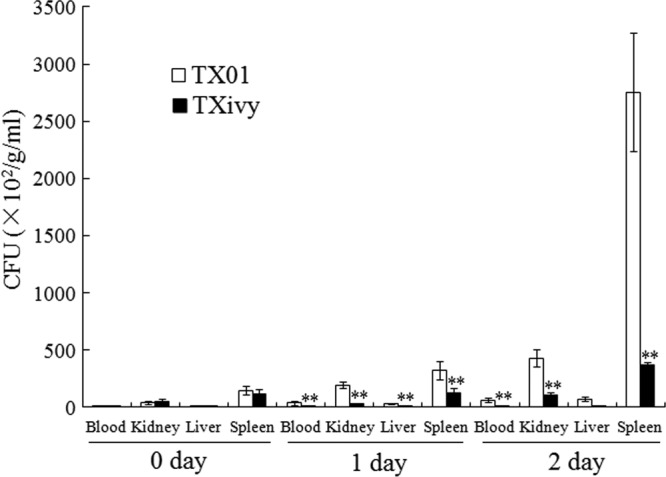
Dissemination and colonization of wild-type and mutant Edwardsiella tarda in fish tissues. Turbot were infected with E. tarda TX01 or TXivy via immersion. Bacterial recovery from blood, kidney, liver, and spleen was determined at 0, 1, and 2 days postinfection and is presented as the number of CFU per gram of tissue (kidney/liver/spleen) or per milliliter of blood. Data are the means of three independent experiments and presented as means ± SEMs. **, P < 0.01.
Fig 2.
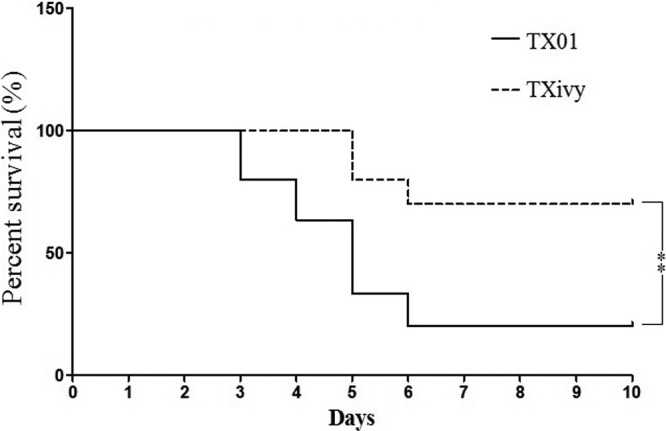
Survival distribution of infected fish. Turbot were infected with Edwardsiella tarda TX01 or TXivy via immersion. The fish were monitored daily for mortality and survival. Significance between the rates of survival of the two groups of fish was determined with the log-rank test. **, P < 0.01.
(ii) Effect on intracellular multiplication.
Replication in host phagocytes is a prominent virulence feature of E. tarda. To examine the potential effect of ivyEt mutation on this capacity of E. tarda, turbot HK macrophages were infected with TXivy or TX01, and intracellular bacterial recovery was determined at 1 h, 3 h, 5 h, and 7 h postinfection. The results showed that while the numbers of intracellular TX01 cells increased with time, the numbers of intracellular TXivy cells essentially remained unchanged (Fig. 3).
Fig 3.
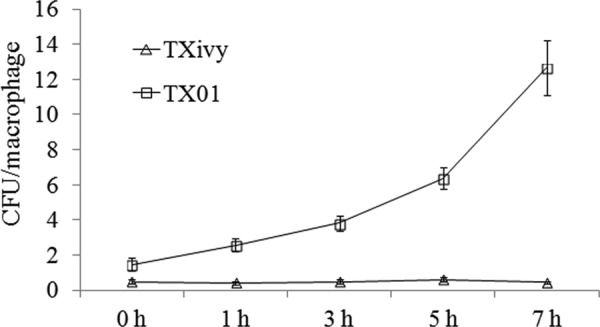
Replication of wild-type and mutant Edwardsiella tarda in HK macrophages. Turbot HK macrophages were infected with E. tarda TX01 or TXivy for 0.5 h. After removing the extracellular bacteria, the cells were incubated at 28°C, and intracellular bacterial recovery was determined at different time points. Data are the means of three independent experiments and presented as means ± SEMs.
(iii) Effect on survival in host serum.
E. tarda is known to resist the bactericidal effect of fish serum (17). To examine whether ivyEt mutation had any effect on serum resistance, TXivy and TX01 were incubated with turbot serum for 1 h, and survival of the bacteria was determined by plate count. The results showed that TXivy exhibited a relative survival rate of 47%, which was significantly (P < 0.01) lower than that (80%) of TX01.
In vitro activity of rIvy and its dependence on conserved residues. (i) Inhibition of lytic effect of lysozyme against a Gram-positive bacterium.
To examine the activity of IvyEt, rIvy was purified from E. coli as a His-tagged protein (see Fig. S2 in the supplemental material). Since, as shown above, W55, C82, and H85 are highly conserved among known Ivy lysozyme inhibitors (see Fig. S1 in the supplemental material), we also examined the functional importance of these residues by site-directed mutagenesis, which resulted in three mutants, rIvyW55A, rIvyC82S, and rIvyH85D, that bear Ala, Ser, and Asp substitutions at W55, C82, and H85, respectively. In the case of rIvyC82S, since serine is structurally the closest replacement to cysteine, the C82S mutation may not affect the structural feature of the mutant protein, and therefore, any alteration in the biological property of rIvyC82S would most likely be associated with the role of C82 in disulfide bond formation. Bacterial survival analysis showed that when the Gram-positive bacterium M. luteus was incubated with HEWL, the density of bacterial cells was severely reduced in a manner that depended on the incubation time; however, when rIvy was copresent with HEWL, no apparent reduction in bacterial density was observed at most of the time points examined (Fig. 4). Similarly, when rIvyW55A and rIvyH85D were copresent with HEWL, the bacterial density remained largely comparable to the densities of the control cells incubated in the absence of HEWL. In contrast, when rIvyC82S was copresent with HEWL, the density of bacterial cells was reduced in a fashion similar to that when the cells were incubated with HEWL alone.
Fig 4.
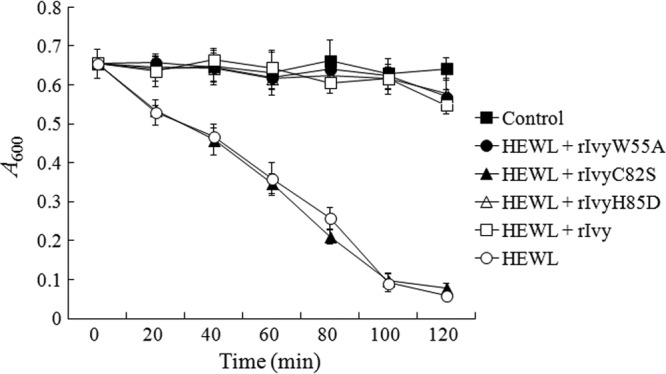
Lysozyme-inhibitory effect of wild-type and mutant Ivy. Micrococcus luteus was incubated in PBS alone (control) or in PBS containing HEWL in the presence or absence of rIvy, rIvyW55A, rIvyC82S, and rIvyH85D. Cell density was monitored by measuring the absorbance at 600 nm at various time points. Data are the means of three independent experiments and presented as means ± SEMs.
(ii) Inhibition of lytic effect of lysozyme against E. tarda.
Since Gram-negative bacteria are, in general, resistant to lysozyme, we examined the effect of serum-facilitated lysozyme hydrolysis of E. tarda, because serum complement can sensitize Gram-negative bacteria to lysozyme. For this purpose, E. tarda TX01 and TXivy were incubated with HEWL in the presence of diluted turbot serum. The results showed that under this condition, HEWL significantly reduced the survival rates of both TX01 and TXivy and that the survival rate of TXivy was significantly lower than that of TX01 (Fig. 5). When rIvy was copresent with HEWL, the survival rates of both TX01 and TXivy were comparable to those of the control cells incubated in the absence of HEWL. In contrast, the copresence of rIvyC82S with HEWL had no apparent effect on the survival of TX01 and TXivy.
Fig 5.
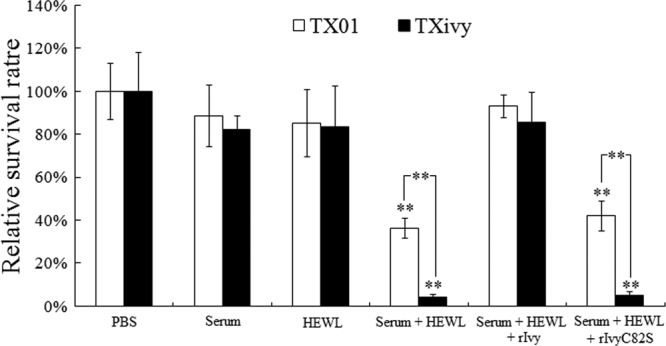
Effect of rIvy on serum-facilitated lysozyme killing of Edwardsiella tarda. E. tarda TX01 and TXivy were incubated with HEWL in the presence or absence of diluted fish serum plus rIvy or rIvyC82S. The control cells were incubated with PBS. The numbers of bacterial cells that survived were determined after the incubation. Data are the means of three independent experiments and presented as means ± SEMs. **, P < 0.01.
In vivo effect of rIvy. (i) Rescuing effect on infectivity of E. tarda TXivy.
Since rIvy exhibited an apparent lysozyme-inhibitory effect under in vitro conditions, we examined whether it could restore the lost infectivity of E. tarda TXivy. For this purpose, turbot were infected with the same dose of TX01, TXivy, TXivy plus rIvy, or TXivy plus rIvyC82S, and bacterial recoveries from tissues were determined at 2 days postinfection. The results showed that the presence of rIvy increased the bacterial recoveries of TXivy from blood, kidney, liver, and spleen to levels similar to those of TX01, whereas the presence of rIvyC82S had no significant effect on the levels of recovery of TXivy (Fig. 6). The experiment was performed three times with comparable results, and the numbers of CFU of one representative experiment are presented in Table S1 in the supplemental material.
Fig 6.
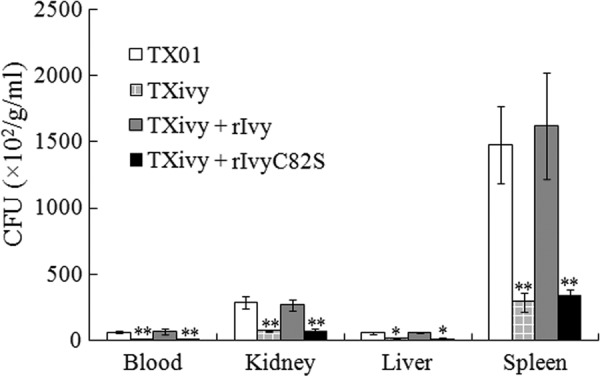
Effect of rIvy on the infectivity of Edwardsiella tarda TXivy. Turbot were infected with E. tarda TX01, TXivy, TXivy plus rIvy, or TXivy plus rIvyC82S. Bacterial recovery from blood, kidney, liver, and spleen was determined at 2 days postinfection and is presented as the number of CFU per gram of tissue (kidney/liver/spleen) or per milliliter of blood. Data are the means of three independent experiments and presented as means ± SEMs. **, P < 0.01; *, P < 0.05.
(ii) Augmenting effect on infectivity of E. tarda TX01.
With the results presented above, we further examined whether rIvy could enhance the infectivity of wild-type E. tarda TX01. For this purpose, turbot were infected with TX01 or TX01 plus rIvy. Subsequent bacterial recovery analysis showed that bacterial recoveries from the blood, kidney, liver, and spleen of the fish infected with TX01 plus rIvy were significantly higher than those from the fish infected with TX01 alone (Fig. 7).
Fig 7.
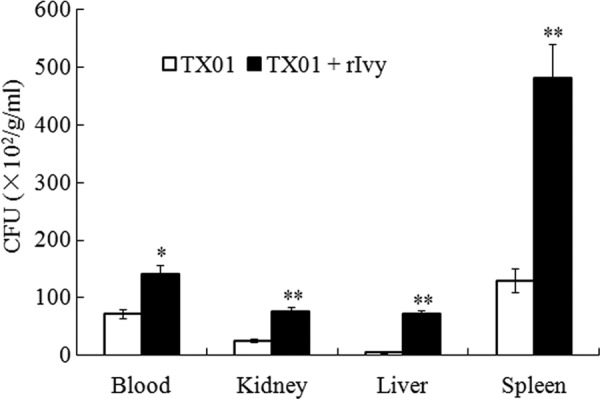
Effect of rIvy on the infectivity of Edwardsiella tarda TX01. Turbot were infected with E. tarda TX01 or TX01 plus rIvy. Bacterial recovery from blood, kidney, liver, and spleen was determined at 4 h postinfection and is presented as the number of CFU per gram of tissue (kidney/liver/spleen) or per milliliter of blood. Data are the means of three independent experiments and presented as means ± SEMs. **, P < 0.01; *, P < 0.05.
DISCUSSION
In this study, we examined the biological activity and function of E. tarda Ivy, IvyEt. IvyEt shares low to moderate sequence identities with known Ivy inhibitors. Unlike the majority of Ivy inhibitors, which exhibit strict sequence conservation in the CKPHDC motif, IvyEt has a variant version of CKPHDC, in which the lysine and the aspartate residues are replaced by glutamine and asparagine, respectively. The same replacement occurs in the Ivy of another species of Edwardsiella, E. ictaluri. Hence, CQPHNC may be a specific feature of the Edwardsiella genus. Similar sequence variations at CKPHDC have been observed in the Ivy of Gluconobacter oxydans (· CRPHDC) and Burkholderia sp. strain 383 (CKPHNC). Mutational analysis in the context of the three-dimensional structure of the E. coli Ivy-HEWL complex suggests that both CRPHDC and CKPHNC should still be able to have a proper interaction with lysozyme and cause lysozyme inhibition (12). In the case of IvyEt, we found that when M. luteus was incubated with HEWL in the presence of rIvy, bacterial survival was comparable to that in the absence of HEWL, suggesting that IvyEt is a functional lysozyme inhibitor that blocked the bactericidal activity of HEWL. Similar lysozyme inhibition effects were also observed with the mutants rIvy W55A and rIvyH85D, whereas no lysozyme inhibition was observed with the mutant rIvy C82S. These results indicate that C82, but not W55 and H85, is essential to the activity of IvyEt. These observations are in contrast to those reported for E. coli Ivy, which showed that H60 (equivalent to H85 of IvyEt) makes hydrogen bonds with D52 and E53 in the active site of HEWL, and as a result, mutation of H60 to a negatively charged amino acid dramatically reduces the activity of Ivy, whereas mutation of one of the Cys residues in CKPHDC has no significant impact (12). The difference in these observations favors the hypothesis that IvyEt, which shares low (25%) sequence identity with E. coli Ivy and has a variant CKPHDC motif, may utilize a lysozyme inhibition mechanism not exactly like that employed by E. coli Ivy. Since C82S mutation impairs the activity of IvyEt, the disulfide linkage between C82 and the only other cysteine residue (i.e., C87) is probably vital to the proper conformation of IvyEt. It is possible that the CQPHNC motif in IvyEt may still adopt a loop structure, which, however, differs from that assumed by the canonical CKPHDC sequence in the specific interactions between the residues of the loop and those in the active site of lysozyme.
Ivy, as well as other types of lysozyme inhibitors, is known to protect Gram-negative bacteria from lysozyme in the presence of the outer membrane permeabilizer lactoferrin (4–7). Other host factors, such as antimicrobial peptides, also enhance the lytic effect of lysozymes (30, 31). In our study, we found that when TX01 and TXivy were treated with HEWL in the presence of diluted serum, the survival rates of both strains were dramatically reduced, suggesting that serum components, probably complements and other innate immune factors, facilitate the bactericidal effect of HEWL. Compared to TX01, the survival rate of TXivy was significantly lower, which is consistent with the fact that TXivy is defective in the production of IvyEt and therefore more sensitive to lysozyme. When rIvy was copresent with HEWL, the survival rates of TX01 and TXivy were restored to those of the control cells, suggesting that rIvy inhibited the activity of HEWL. The observation that rIvyC82S had no apparent effect on HEWL confirmed the conclusion presented above that C82 is indispensable for the fundamental activity of IvyEt.
Recent reports showed that an mliC deletion in an E. coli mutant caused a strong reduction in serum resistance and in in vivo virulence, whereas ivy deletion had no effect on serum resistance and virulence (10); in contrast, for Yersinia pestis, which causes plague, mliC mutation did not affect lysozyme resistance and the development of plague, whereas ivy mutation impaired bacterial virulence (11). In our study, we found that, compared to the wild type, TXivy exhibited reduced dissemination and colonization in the tissues of turbot and caused significantly lower mortality in the host, suggesting that deletion of ivyEt attenuates the overall virulence of TXivy. Previous studies showed that E. tarda is an intracellular pathogen that can survive in host phagocytes (19, 20). In our study, we observed markedly reduced abilities of TXivy to replicate in turbot macrophages and serum, suggesting that IvyEt is likely to function against the lysozyme-mediated defense of both cellular and humoral innate immunity. In line with these observations, in vivo analysis showed that when rIvy was cointroduced into turbot with TXivy, it increased the tissue infection capacity of the mutant bacterium to a level comparable to that of the wild type, while when rIvy was cointroduced into turbot with TX01, it enhanced the tissue infection capacity of the bacterium to a significant extent. Taken together, these results indicate that IvyEt is a virulence factor that is involved in multiple aspects, including tissue dissemination and intracellular replication, of E. tarda infection.
In conclusion, we demonstrate in this study that IvyEt is a lysozyme inhibitor that possesses a noncanonical CKPHDC motif and that the biological activity of IvyEt is dependent on the conserved cysteine residues, rather than the histidine residue, of the CKPHDC motif. IvyEt confers protection on E. tarda against lysozyme lysis in the presence of serum and is required for optimal infection of the host. These results add new insights into the function of the Ivy type of lysozyme inhibitors.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (31025030 and 31202029), the Knowledge Innovation Program of the Chinese Academy of Sciences (KZCX2-EW-Q213 and KSCX2-EW-G-12B), and the Taishan Scholar Program of Shandong Province.
Footnotes
Published ahead of print 1 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00503-13.
REFERENCES
- 1.Callewaert L, Michiels CW. 2010. Lysozymes in the animal kingdom. J. Biosci. 35:127–160 [DOI] [PubMed] [Google Scholar]
- 2.Prager EM, Jollès P. 1996. Animal lysozymes c and g: an overview, p 9–31 In Jollès P. (ed), Lysozymes: model enzyme in biochemistry and biology, vol 75 Birkhäuser Basel Press, Basel, Switzerland [Google Scholar]
- 3.Monchois V, Abergel C, Sturgis J, Jeudy S, Claverie JM. 2001. Escherichia coli ykfE ORFan gene encodes a potent inhibitor of C-type lysozyme. J. Biol. Chem. 276:18437–18441 [DOI] [PubMed] [Google Scholar]
- 4.Callewaert L, Aertsen A, Deckers D, Vanoirbeek KG, Vanderkelen L, Van Herreweghe JM, Masschalck B, Nakimbugwe D, Robben J, Michiels CW. 2008. A new family of lysozyme inhibitors contributing to lysozyme tolerance in gram-negative bacteria. PLoS Pathog. 4:e1000019. 10.1371/journal.ppat.1000019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Herreweghe JM, Vanderkelen L, Callewaert L, Aertsen A, Compernolle G, Declerck PJ, Michiels CW. 2010. Lysozyme inhibitor conferring bacterial tolerance to invertebrate type lysozyme. Cell. Mol. Life Sci. 67:1177–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanderkelen L, Van Herreweghe JM, Vanoirbeek KGA, Baggerman G, Myrnes B, Declerck PJ, Nilsen IW, Michiels CW, Callewaert L. 2011. Identification of a bacterial inhibitor against g-type lysozyme. Cell. Mol. Life Sci. 68:1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deckers D, Masschalck B, Aertsen A, Callewaert L, Van Tiggelen CG, Atanassova M, Michiels CW. 2004. Periplasmic lysozyme inhibitor contributes to lysozyme resistance in Escherichia coli. Cell. Mol. Life Sci. 61:1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deckers D, Vanlint D, Callewaert L, Aertsen A, Michiels CW. 2008. Role of the lysozyme inhibitor Ivy in growth or survival of Escherichia coli and Pseudomonas aeruginosa bacteria in hen egg white and in human saliva and breast milk. Appl. Environ. Microbiol. 74:4434–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanderkelen L, Van Herreweghe JM, Callewaert L, Michiels CW. 2011. Goose-type lysozyme inhibitor (PliG) enhances survival of Escherichia coli in goose egg albumen. Appl. Environ. Microbiol. 77:4697–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanderkelen L, Ons E, Van Herreweghe JM, Callewaert L, Goddeeris BM, Michiels CW. 2012. Role of lysozyme inhibitors in the virulence of avian pathogenic Escherichia coli. PLoS One 7:e45954. 10.1371/journal.pone.0045954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derbise A, Pierre F, Merchez M, Pradel E, Laouami S, Ricard I, Sirard JC, Fritz J, Lemaître N, Akinbi H, Boneca IG, Sebbane F. 2013. Inheritance of the lysozyme inhibitor Ivy was an important evolutionary step by Yersinia pestis to avoid the host innate immune response. J. Infect. Dis. 207:1535–1543 [DOI] [PubMed] [Google Scholar]
- 12.Abergel C, Monchois V, Byrne D, Chenivesse S, Lembo F, Lazzaroni JC, Claverie JM. 2007. Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:6394–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohanty BR, Sahoo PK. 2007. Edwardsiellosis in fish: a brief review. J. Biosci. 32:1331–1344 [DOI] [PubMed] [Google Scholar]
- 14.Matsuyama T, Kamaishi T, Ooseko N, Kurohara K, Iida T. 2005. Pathogenicity of motile and non-motile Edwardsiella tarda to some marine fish. Fish Pathol. 40:133–135 [Google Scholar]
- 15.Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK. 2012. Edwardsiella tarda—virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 14:26–34 [DOI] [PubMed] [Google Scholar]
- 16.Park SB, Aoki T, Jung TS. 2012. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet. Res. 43:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Hu Y, Zheng W, Sun B, Wang C, Sun L. 2012. Inv1: an Edwardsiella tarda invasin and a protective immunogen that is required for host infection. Fish Shellfish Immunol. 32:586–592 [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Zheng W, Hu Y, Sun B, Sun L. 2012. Edwardsiella tarda Eta1: an in vivo-induced antigen that is involved in host infection. Infect. Immun. 80:2948–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng S, Zhang M, Sun L. 2010. The iron-cofactored superoxide dismutase of Edwardsiella tarda inhibits macrophage-mediated innate immune response. Fish Shellfish Immunol. 29:972–978 [DOI] [PubMed] [Google Scholar]
- 20.Srinivasa Rao PS, Lim TM, Leung KY. 2001. Opsonized virulent Edwardsiella tarda strains are able to adhere to and survive and replicate within fish phagocytes but fail to stimulate reactive oxygen intermediates. Infect. Immun. 69:5689–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao X, Dang W, Hu Y, Sun L. 2009. Identification and immunoprotective analysis of an in vivo-induced Edwardsiella tarda antigen. Fish Shellfish Immunol. 27:633–638 [DOI] [PubMed] [Google Scholar]
- 22.Sun K, Wang HL, Zhang M, Xiao ZZ, Sun L. 2009. Genetic mechanisms of multi-antimicrobial resistance in a pathogenic Edwardsiella tarda strain. Aquaculture 289:134–139 [Google Scholar]
- 23.Wang HR, Hu YH, Zhang WW, Sun L. 2009. Construction of an attenuated Pseudomonas fluorescens strain and evaluation of its potential as a cross-protective vaccine. Vaccine 27:4047–4055 [DOI] [PubMed] [Google Scholar]
- 24.Jiao XD, Zhang M, Hu YH, Sun L. 2009. Construction and evaluation of DNA vaccines encoding Edwardsiella tarda antigens. Vaccine 27:5195–5202 [DOI] [PubMed] [Google Scholar]
- 25.Hu YH, Zheng WJ, Sun L. 2010. Identification and molecular analysis of a ferritin subunit from red drum (Sciaenops ocellatus). Fish Shellfish Immunol. 28:678–686 [DOI] [PubMed] [Google Scholar]
- 26.Milton DL, O'Toole R, Hörstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aida Y, Pabst MJ. 1990. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods 132:191–195 [DOI] [PubMed] [Google Scholar]
- 28.Zheng W, Hu Y, Sun L. 2011. The two Dps of Edwardsiella tarda are involved in resistance against oxidative stress and host infection. Fish Shellfish Immunol. 31:985–992 [DOI] [PubMed] [Google Scholar]
- 29.Liu CS, Sun Y, Hu YH, Sun L. 2010. Identification and analysis of the immune effects of CpG motifs that protect Japanese flounder (Paralichthys olivaceus) against bacterial infection. Fish Shellfish Immun. 29:279–285 [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Niyonsaba F, Ushio H, Okuda DJ, Nagaoka I, Ikeda S, Okumura K, Ogawa H. 2005. Synergistic effect of antibacterial agents human beta-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J. Dermatol. Sci. 40:123–132 [DOI] [PubMed] [Google Scholar]
- 31.Hancock R, Scott M. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. U. S. A. 97:8856–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


