Abstract
Group G beta-hemolytic streptococcus (GGS) strains cause severe invasive infections, mostly in patients with comorbidities. GGS is known to possess virulence factors similar to those of its more virulent counterpart group A streptococcus (GAS). A streptococcal invasion locus, sil, was identified in GAS. sil encodes a competence-stimulating peptide named SilCR that activates bacterial quorum sensing and has the ability to attenuate virulence in GAS infections. We found that sil is present in most GGS strains (82%) but in only 25% of GAS strains, with a similar gene arrangement. GGS strains that contained sil expressed the SilCR peptide and secreted it into the growth medium. In a modified murine model of GGS soft tissue infection, GGS grown in the presence of SilCR caused a milder disease than GGS grown in the absence of SilCR. To further study the role of the peptide in bacterial virulence attenuation, we vaccinated mice with SilCR to produce specific anti-SilCR antibodies. Vaccinated mice developed a significantly more severe illness than nonvaccinated mice. Our results indicate that the sil locus is much more prevalent among the less virulent GGS strains than among GAS strains. GGS strains express and secrete SilCR, which has a role in attenuation of virulence in a murine model. We show that the SilCR peptide can protect mice from infection caused by GGS. Furthermore, vaccinated mice that produce specific anti-SilCR antibodies develop a significantly more severe infection. To our knowledge, this is a novel report demonstrating that specific antibodies against a bacterial component cause more severe infection by those bacteria.
INTRODUCTION
Large colony-forming group G beta-hemolytic streptococcus (GGS) was first isolated from humans in 1935, by Lancefield and Hare, from a case of puerperal sepsis (1). Streptococcus dysgalactiae subsp. equisimilis (GGS) strains are well known as commensals and pathogens in domestic animals. In humans, they may colonize the pharynx, skin, and gastrointestinal and female genital tracts (2). GGS causes a wide spectrum of human diseases, ranging from local uncomplicated infections in the pharynx and skin to life-threatening invasive diseases such as streptococcal toxic shock syndrome (STSS), bacteremia, and necrotizing fasciitis (NF) (3).
GGS and group A streptococcus (GAS) are phylogentically related and share virulence factors such as streptokinase, C5a peptidase, M protein, and certain exotoxin genes (4–9). Analysis of a GAS emm14 strain isolated from a patient with NF led us to identify and characterize the streptococcal invasion locus, sil, which is involved in GAS spreading into deeper tissues and is highly homologous to a regulon of Streptococcus pneumoniae involved in bacterial signaling (10). sil has six open reading frames (ORFs). silA and silB encode a putative two-component system, and silD and silE encode two putative ABC transporters. In the middle of sil there are two overlapping genes carried on different strands, silC and silCR. silC encodes a small peptide linked to virulence, starting with the com box promoter, while silCR encodes a putative competence-stimulating peptide (CSP). SilCR has the characteristics of a bacterial pheromone peptide (11) and includes the RKK motif in the C-terminal end and the Gly-Gly motif (double glycine leader) of the pre-CSP, a conserved cleavage site predicted to generate a 17-amino-acid (aa) mature peptide (10).
The exact distribution of sil in different GAS strains is unknown. However, only three of the 18 GAS strains which have currently been fully sequenced and for which data are available at the NCBI have the sil locus. These are MGAS8232 (emm18), MGAS10750 (emm1), and Alab49 (12–14). Although sil is highly homologous in these strains of GAS, several point mutations have been found. Whereas in the genomes of the emm18 and emm1 GAS strains, silCR possesses an ATG start codon (12, 13), in emm14 the start codon is eliminated due to a missense mutation, suggesting that the SilCR peptide may not be produced in GAS emm14 (10). In both the emm18 and emm1 strains, one of the predicted ATP transporter genes, silD, is truncated due to a point mutation (10). The secretion and processing of the SilCR peptide depends on intact transporter genes. The 41-aa SilCR precursor is exported through silDE and modified to the mature 17-aa active form. In the MGAS8232 strain, the silD gene is truncated and does not express the SilCR peptide. Thus, silCR may not be expressed.
We have shown that SilCR may play a role in regulating the ability of GAS to cause invasive infection. When synthetic SilCR (BioSight Ltd. Peptide Technologies, Israel) was added to GAS emm14 growth medium before injection into mice, it prevented mouse mortality (15).
In the current work, we studied whether the diminished virulence of GGS compared to GAS is correlated with sil in isolates obtained from patients with invasive disease. Most invasive GGS isolates contain sil in one of two types (sil, silS). Relative to GGS, invasive GAS isolates contain sil less frequently. We found that sil is present on the streptococcal chromosome in a single copy and that it is either entirely present or completely absent (silN).
Furthermore, we show that in GGS, SilCR is present on the bacterial surface and is also secreted into the growth medium and that the presence of sil does not necessarily mean that SilCR is expressed. In the modified murine soft tissue infection model, we show that like for GAS, adding synthetic SilCR to the growth medium of GGS before mice challenge attenuates virulence.
We developed a unique model in which mice were vaccinated with synthetic SilCR and produced specific anti-SilCR antibodies. Most interestingly, the vaccinated mice developed a significantly more severe infection than nonvaccinated mice. Histological sections of the necrotic wound showed numerous polymorphonuclear cells (PMNs) at the site of infection. We found that unlike GAS (15), GGS does not degrade interleukin-8 (IL-8), allowing PMN migration to the site of infection. We discuss several explanations for this phenomenon.
MATERIALS AND METHODS
Bacterial strains.
The GGS isolates used were previously described in a retrospective study of GGS bacteremia (3), where characteristics of the patients are described. The Hadassah-Hebrew University Medical Center clinical microbiology laboratory identified 100 isolates from blood of bacteremic patients as GGS by standard procedures, and the emm types of GGS isolates were determined as described. Seventy-nine isolates were used for this study (3). A total of 212 invasive GAS isolates were arbitrarily selected from a larger set of invasive GAS isolates which were described in a prospective study of invasive GAS infections (16–18). Of these, 10 were from patients with necrotizing fasciitis and 70 were isolated from blood cultures.
MGAS8232, an emm18 strain that has had its entire genome sequenced (13), was used as a control. JS95, an emm14 strain, was used as a negative control for SilCR expression. Typically, GGS and GAS were cultured in Todd-Hewitt broth supplemented with 0.2% (wt/vol) yeast extract (THY) in 37°C. Bacterial strains used for experiments are presented in Table 1.
Table 1.
Strains used in this study
DNA methodologies.
DNA for PCR was prepared as described previously (17). Primer pairs used for PCR are presented in Table 2. To determine the presence of an ATG start codon in silCR, we designed the primers Allel-A and Allel-G, consisting of the silCR 5′ sequence with either A or G at the 3′ end. Allel-A or Allel-G was used with SilB-f1.
Table 2.
Primers and peptides
| Primer or peptide | Description | Sequence |
|---|---|---|
| Primers | ||
| Primer1 | emm typing, forward | 5′-TATTCGCTTAGAAAATTAA-3′ |
| Primer2 | emm typing, reverse | 5′-GCAAGTTCTTCAGCTTGTTT-3′ |
| SilB-f1 | Detection of sil | 5′-GGAGTTGGTTTATCAAATGTCAG-3′ |
| SilD-r1 | Detection of sil | 5′-ATCTGCCACAAAGACTGATCAAG-3′ |
| PrtS 5′ f | Detection of protease | 5′-CGCAACATCTCACCAATAC-3′ |
| PrtS 5′ r | Detection of protease | 5′-CCCAGTCCTCATCAAAATC-3′ |
| prtS 3′ f | Detection of protease | 5′-GCATAACAGCCCGAGGAAGCAA-3′ |
| prtS 3′ r | Detection of protease | 5′-CATCTGCATCCCGCACAAGGTA-3′ |
| Allel-A | silCR start codon | 5′-AATTTTATTAGGAGATATTACAATA-3′ |
| Allel-G | silCR start codon | 5′-AATTTTATTAGGAGATATTACAATG-3′ |
| SilCR peptide | Synthetic | DIFKLVIDHISMKARKK |
Southern blotting of GGS for detection of sil.
Southern blotting of chromosomal DNA was performed according to standard procedures. Genomic DNA was purified according to protocol (19). Chromosomal DNA was digested with EcoRV, HincII, and BmgBI and placed on an agarose gel for electrophoresis (20). The gel was transferred to a nylon membrane and probed with the PCR product of silCR in GGS strain 3046, which was treated with the QIAquick PCR purification kit and labeled with digoxigenin (DIG) (nonradioactive technology to label nucleic acids).
Identification of SilCR on the bacterial surface.
The presence of SilCR on the bacterial surface was demonstrated by either horseradish peroxidase (HRP) dot blotting or immunofluorescence (IF) staining. The HRP dot blotting was performed by placing serial dilutions of an overnight culture on a nitrocellulose membrane and blocking with skim milk overnight. The membrane was then incubated with a chicken anti-SilCR antibody for 2 h, followed by incubation with an HRP-conjugated anti-chicken antibody. The synthetic SilCR peptide (1 μg/μl) was used as a positive control. A GAS strain that does not express SilCR (JS95) was used as a negative control. For detection of SilCR on the bacterial surface at different periods of the growth curve, we took bacteria from an overnight culture and grew them again for 30 min up to 4.5 h. At each time point, bacteria were taken for dot blot detection of SilCR. The same quantities of bacteria were calculated by optical density and taken for each period on the growth curve.
IF was performed using bacteria that were washed and diluted in 4% formaldehyde, washed again, and placed on a polylysine slide. After drying, the slide was place in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA) and 0.2% Triton X-100. The slide was then incubated with a rabbit anti-SilCR antibody. After incubation, goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) was added, and results were read under a fluorescence microscope.
Identification of SilCR secretion in the growth medium.
GGS strain 3046 (SilCR positive) and GAS strain JS95 (SilCR negative) were grown overnight in 100 ml Todd-Hewitt broth. The bacteria were filtered (0.2 μm); the remaining medium was lyophilized and resuspended in 4 ml PBS. Serial dilutions (200 μl) of this suspension were placed on a nitrocellulose transfer membrane using a slot blot unit and kept in skim milk overnight. The membrane was then incubated with an anti-SilCR antibody derived from chicken followed by a second anti-chicken–horseradish peroxidase (HRP) antibody. The synthetic SilCR peptide (1 μg/μl) was used as a positive control.
CY murine model of NF for GGS.
Female BALB/c mice, 3 to 4 weeks old and weighing ∼10 g, were used. The transiently compromised state in mice was achieved by one intraperitoneal (i.p.) injection of 200 mg/kg of the immunosuppressive agent cyclophosphamide (CY). Blood counts showed reduction of total white blood cells (WBC) and neutrophils. In the immunized mouse model, mice were given three doses of CY due to their increased weight and maturity. Four days after the last CY dose, mice were challenged subcutaneously with 108 CFU bacteria. For the SilCR virulence attenuation test we grew bacteria, before injection, in the presence of 100 μg/ml synthetic SilCR. Mice were monitored for their mobility, hair condition, wound severity, amount of bacteria in internal organs, and mortality. All animal experiments were approved by the Hadassah-Hebrew University Medical School Institutional Animal Care and Ethics Committee.
Vaccination of mice against SilCR.
Female BALB/c mice, 3 to 4 weeks old and weighing ∼10 g, were immunized with SilCR (100 μg/100 μl) with complete Freund's adjuvant and 10 days later were administered a SilCR booster (100 μg/100 μl) with incomplete Freund's adjuvant.
Detection of anti-SilCR antibodies in mouse blood.
Ten days after last vaccination, blood was examined for the presence of SilCR antibodies. Synthetic SilCR peptide (1 μg/μl) was placed on a nitrocellulose membrane and blocked with skim milk overnight. The membrane was incubated in serial dilutions of the mouse serum and incubated with a Goat anti-mouse HRP followed with chemiluminescent substrate.
Detection of bacteria in internal organs.
At 48 h after challenge with GGS (strain 3046), mice were sacrificed and the livers and spleens were harvested and minced in 1 ml PBS. Serial dilutions of the suspension were cultured for 24 h on blood agar; bacterial colonies were counted to determine the number of bacteria that spread to the internal organs.
IL-8 degradation test.
IL-8 degradation was performed twice and quantified by enzyme-linked immunosorbent assay (ELISA) using the Quantikine kit (R&D Systems, Minneapolis, MN, USA) as detailed previously (15). Briefly, bacteria were grown to log phase and washed, and the pellets were supplemented with fetal calf serum and incubated with 100 ng of IL-8 (recombinant human interleukin-8/CXCL8; Peprotech Asia).
Statistical analysis.
Descriptive statistics was performed using the SPSS statistical package release 12.0.2. The chi-square test or Fisher's exact test was used for differences in proportions. A two-sided P value of <0.05 was considered significant.
RESULTS
Presence of the sil locus in GGS.
Since the sil locus is highly conserved in at least 3 GAS strains of different emm types, we looked for its presence in GGS. sil was amplified in one of the GGS isolates (isolate 4674, emmG485), using the primers described in Materials and Methods. A product of the same size as in GAS was identified for silB, silE, and of silBCD. Since this suggested that GGS 4674 possesses a homologue of the GAS sil, the whole region was sequenced. Analysis of the sequence and the open reading frames revealed that the sil locus in GGS has a gene arrangement similar to that in GAS (data not shown).
Since it has been suggested that in GAS silCR has a role in regulating invasive infection, we further examined the sequence of silCR in GGS to determine whether it is of the ATG variety (in which silCR is potentially expressed) or the ATA variety (in which silCR is not expressed). The silCR nucleotide sequence of GGS isolate 4674 was found to be 100% homologous to that of GAS emm18 strain 8232. Thus, it possesses an ATG start codon. This may suggest that silCR can be efficiently translated in the GGS isolate. The nucleotide sequence of silD in GGS showed that there is no deletion or termination codon; thus, it possesses an intact silD ORF, similar to the case for JS95, suggesting that unlike in the emm18 strain, the SilD ABC transporter is produced in this isolate of GGS.
Prevalence of sil among GGS isolates.
To assess the distribution of sil in various GGS strains, we screened for its presence in 79 invasive isolates obtained from patients with bacteremia. In 65 isolates (82%), a product encompassing silBCD was amplified in one of two sizes: the first was similar to the size previously found in GAS (sil), and the second was shorter (silS) (GenBank accession number KF188416). As there was the possibility that the remaining 14 isolates harbor only part of the locus or that its genes are arranged differently, we further examined the silBCD-negative isolates for the presence of the specific genes in the locus, silB, silC, and silD, separately. By using seven different sets of primers (not shown), we found that none of the remaining isolates contained any of the sil genes. Thus, we concluded that GGS isolates either possess the locus in one of two forms (sil or silS) or completely lack it (silN).
Sequence variation of sil in GGS.
We compared the sequences of sil and silS. Whereas the ORFs of silB, silC, and silCR were conserved, there was an 87-bp deletion in the intergenic region immediately downstream of the stop codon of silB in silS. This sequence was deleted in silS and does not affect the expression of silB or silCR. Outside this deletion, the intergenic silBC region and the silC/CR coding sequence were completely conserved among sil and silS.
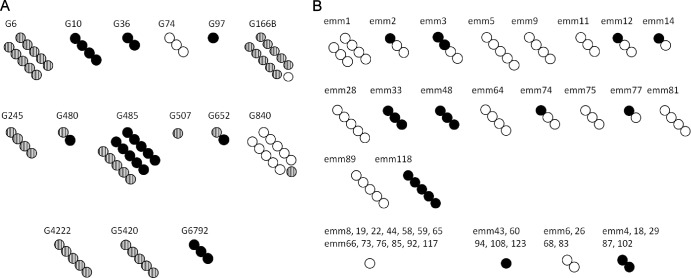
Of the 65 GGS isolates that possessed the sil locus, 43 (66%) had silS and 22 (34%) had sil. We found a high correlation between the presence of sil, its variety (sil/silS), and the emm type (Fig. 1A). Thus, most isolates of a certain emm type shared the presence or the absence of sil and the sil or silS pattern. For example, emmG10 and emmG6792 isolates tend to be sil (P = 0.011 and P = 0.035, respectively), emm6 and emm166B isolates tend to be silS (P = 0.044 and P = 0.012, respectively), and emmG74 and emmG840 isolates tend to be silN (P = 0.005 and P < 0.001, respectively).
Fig 1.
Distribution of emm type and sil in 79 GGS (A) and 105 GAS (B) isolates. Isolates are arranged according to emm type. The number of circles represents the number of isolates from the emm type designated above the circles. GGS strains are classified as sil (black circles), silS (striped circles), or silN (white circles), and GAS strains are classified as sil (black circles) or silN (white circles).
Presence of sil in GAS.
Since we found in GGS two varieties of sil, one different from that found in GAS, we screened our collection of invasive GAS isolates for sil types. Interestingly, silS was not found among any of 249 GAS isolates. sil was less frequent among GAS isolates than among GGS isolates (53 positive out of 212 isolates [25%] versus 65 of 79 [82%], respectively; P < 0.0001).
Similar to the situation in GGS, the presence or absence of sil in GAS was associated with isolate type as determined both by T and emm typing: increased sil presence was found in T types 3, 4, and 49 (P < 0.013 for all) and emm types 4, 33, 48, and 118 (P < 0.003 for all). In both T1 (22 isolates) and emm1 (17 isolates), sil was not found (P < 0.01) (Fig. 1B).
We found that in GAS the presence of sil was correlated with the source and type of infection from which the isolate was obtained; sil was present in 30.9% of isolates obtained from nonnecrotizing fasciitis invasive soft tissue infections without concomitant isolation from the blood (34 out 110 P = 0.008). Furthermore, a substantially lower proportion was found for isolates obtained from blood cultures (17.1%, 12 out of 70 isolates; P = 0.064). Thus, it is suggested that sil is present mainly in the less invasive GAS isolates. Whether sil actually has a separate role in the virulence of GAS is not clear. Different emm types are also associated with various degrees of virulence, and the relative contributions of sil, emm, and perhaps other factors in modulating virulence cannot as yet be determined.
We conclude that sil is more prevalent in GGS which is relatively less virulent than GAS and is also more prevalent in the less invasive isolates among GAS.
sil is detected in chromosomal DNA of GGS and GAS.
Using multiple restriction enzyme electrophoresis and hybridization with a silCR PCR product as probe, we found that sil was present in a single copy in both GGS strain 3046 and GAS strain JS95 and was absent in GGS strain 4750 (not shown). The sizes of the digestion products coincided with the expected sizes as calculated in the restriction sites in the sil locus.
Start codon of silCR in GGS.
We used an allele-specific PCR to examine whether the GGS isolates harbor an ATG or an ATA start codon in silCR (21). As controls, we used GAS strains 8232 (ATG) and JS95 (ATA). The Allel-G primer for the detection of ATG amplified sil only in GAS strain 8232 and not in JS95. The Allel-A primer for the detection of ATA gave the opposite results. These PCR results were in agreement with the sequence data. Using this approach, all the sil-containing GGS isolates (n = 65) were found to possess an ATG codon at the start of silCR, suggesting that they can express SilCR.
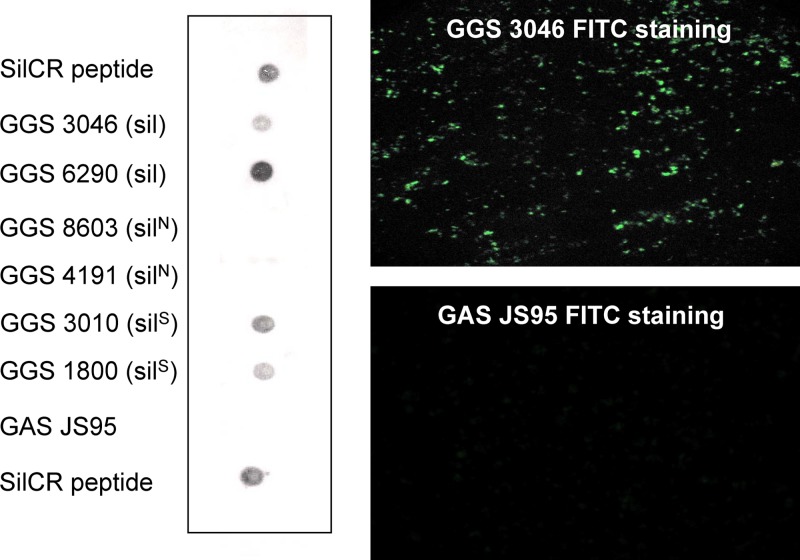
SilCR is present on the surface of GGS as detected by dot blotting and immunofluorescent staining.
SilCR was detected by immunofluorescence using a specific anti-SilCR antibody on the surfaces of GGS strains containing sil and silS but was not detected in GGS strains without sil. Similarly, SilCR was not detected in a GAS strain which does not have the SilCR start codon (Fig. 2).
Fig 2.
Dot blot (left) and immunofluorescent staining (right) demonstrating the presence or absence of the SilCR peptide on the surfaces of different GGS strains and GAS JS95.
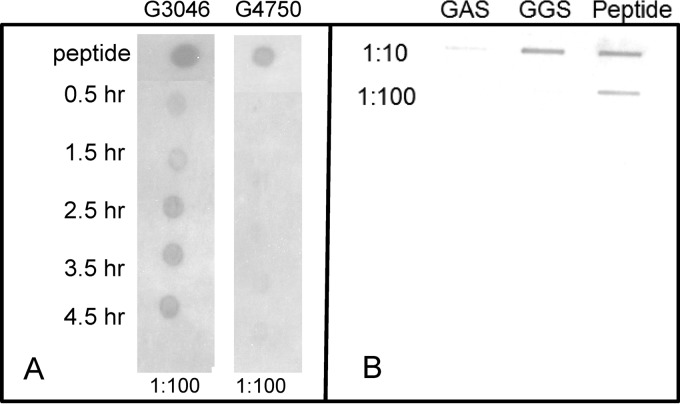
SilCR is expressed in correlation with the growth curve and secreted into the medium.
We found that the expression of SilCR on the surfaces of the bacteria is correlated with the growth curve progression (Fig. 3A). Similar to expression of the CSP family, the expression of the peptide peaks in the middle of the logarithmic growth-time curve expression.
Fig 3.
(A) Dot blot expression of SilCR in a sil strain (G3046) and a silN strain (G4750) during the growth curve. (B) Secretion of the SilCR peptide to the growth medium in GAS JS95 and GGS 3046.
Slot blotting performed on the growth medium during stationary phase detected that SilCR is secreted into the medium (Fig. 3B). Specifically, the sil-positive strain GGS 3046 secreted SilCR, whereas the sil-negative GAS strain JS95 did not.
SilCR protection against GGS challenge.
A modified model for NF (15) using cyclophosphamide was developed to adjust to the lesser virulence of GGS compared to GAS. Using this model, 6 mice challenged with GGS (strain 3046) grown in the presence of synthetic SilCR were protected from infection. Mortality rates declined (1/5 mice with SilCR versus 4/5 mice without SilCR), and the amount of bacteria in the internal organs was significantly reduced compared to that in 6 mice challenged with GGS grown in the absence of the peptide (data not shown). Each experiment was repeated three times.
Vaccinated mice.
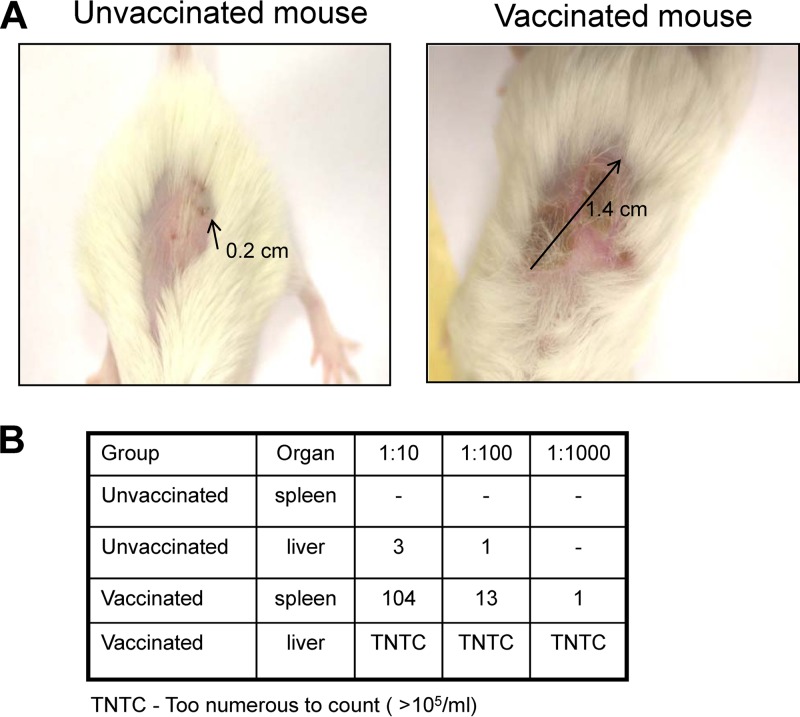
We vaccinated six mice with synthetic SilCR, resulting in the production of specific anti-SilCR antibodies (not shown). When vaccinated, six mice challenged with GGS (strain 3046) developed a significantly more severe infection than six nonvaccinated mice. This was shown mostly by a larger wound size at the site of injection (Fig. 4A) (P value for wound size difference between vaccinated and unvaccinated mice, <0.024) and a larger amount of bacteria spread to internal organs (Fig. 4B). This experiment was repeated twice (six mice in each group).
Fig 4.
Comparison between mice vaccinated against SilCR and unvaccinated mice in a murine model of necrotizing fasciitis. (A) Difference in wound size after bacterial challenge. (B) Bacterial counts in livers and spleens harvested 48 h after bacterial challenge.
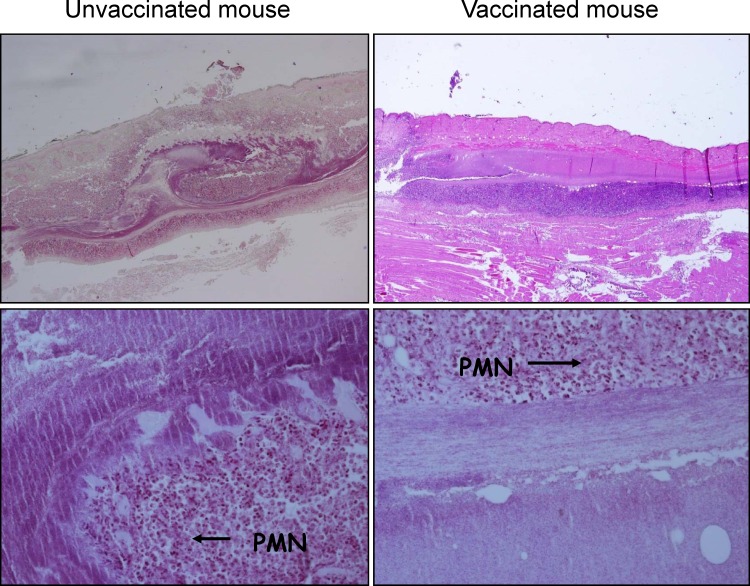
Histological sections of the wounds in the vaccinated mice showed greater damage and necrosis of skin tissue than in the nonvaccinated mice (Fig. 5). There was no difference in amounts of PMNs and bacteria at the site of infection between vaccinated and nonvaccinated mice.
Fig 5.
Histological sections of soft tissue infection site stained with hematoxylin and eosin. Abundant PMNs and bacteria are seen in unvaccinated and vaccinated mice. Greater tissue damage with necrosis of deep fascia is seen in the vaccinated mice.
IL-8 degradation test.
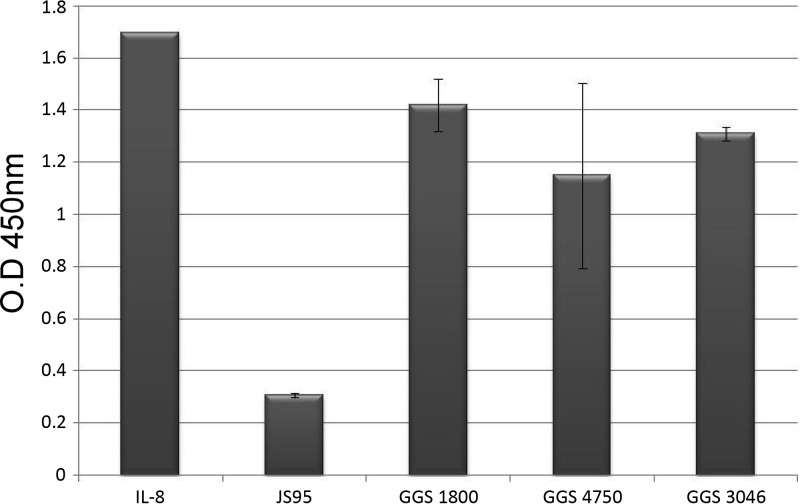
In GAS infection, PMN migration to the infection site is mediated by IL-8. In GAS JS95, which does not express SilCR, there is IL-8 degradation. The exogenous administration of SilCR to the medium prevents IL-8 degradation by a specific protease (ScpC) and allows PMNs to migrate to the infection site. To test whether a similar process is present in GGS, we performed an IL-8 degradation test to show whether GGS degrades IL-8 similarly to GAS (Fig. 6). Three strains of GGS (sil, silS, and silN) were tested and compared to the control GAS JS95 strain, which harbors the ScpC (PrtS) serine protease and degrades IL-8. We found that none of the GGS strains degraded IL-8 regardless of the presence or absence of sil in the genome.
Fig 6.
IL-8 degradation of GGS strains compared to GAS (JS95). None of the GGS strains degrade IL-8.
Detection of PrtS (ScpC) in GGS.
PCR using two different sets of primers (see Materials and Methods) was performed. All GGS isolates (n = 50) were negative for the gene (not shown). Taking these results together with those of the IL-8 degradation test, we suggest that GGS has no virulence factor that degrades IL-8.
DISCUSSION
We identified the sil locus in GGS. This locus has previously been shown to be involved in GAS virulence. sil of GGS is highly homologous to that found in GAS but is more prevalent. However, unlike GAS, all GGS isolates that possess sil harbor both intact silCR and silD and have one of two allele variants, sil or silS.
Transfer of alleles of housekeeping genes from GAS to GCS and GGS is known (22). The correlation between the presence of sil and the transposon-mediated tetracycline resistance is supportive of horizontal gene acquisition (unpublished data). Alleles of some housekeeping genes are diverged in GGS and in GAS (22), and virulence genes found in both bacteria, such as sagA and spegG, are even less homologous (79% and 87%, respectively) (23, 24). In light of this information, the tremendous conservation of the sil is remarkable, as exemplified by high homology of the ORF of silC in different emm types of GGS and GAS. The silBC intergenic region is also 100% homologous (except for the deletion in silS), even though noncoding sequences are usually less conserved than ORFs.
In this study, we found that the sil locus is present in a single copy in GGS isolates that are positive for sil by PCR and is not present in strains negative for sil. Thus, the decreased virulence of GGS relative to GAS is not due to multiple copies of the silCR gene. We further describe that the presence of the sil locus in the form of sil or silS correlates to the T/emm type of GGS and that the silS form is more prevalent than sil.
Our findings are that in GAS, the presence of sil correlates to bacteria isolated from nonnecrotizing fasciitis soft tissue infections, whereas isolation from blood correlates with the lack of sil. In GGS, the less virulent streptococcus, there is a high prevalence of sil. Furthermore, sil is not present in any of the GAS emm1 isolates we examined. This type for years has been considered the leading type associated with invasive disease. Similar findings were described by Bidet et al. (25), where none of 19 emm1 isolates had sil. SilCR producers are less virulent, and SilCR nonproducers are more virulent. The ability to produce SilCR is determined by the presence of the ATG rather than the ATA start codon in silCR. The ATG variety was found in all GGS isolates we examined, supporting this hypothesis. This is similar to the finding by Belotserkovsky et al., where the GGS isolates they examined (five isolates) contained ATG (26).
We found (10) in the past that the sil locus is a determinant of invasion (thus its name, streptococcal invasion locus). It is now becoming clear that the presence of sil by itself is not sufficient to determine virulence. Rather, the bacterial ability to produce SilCR is the important factor influencing virulence.
We identified SilCR on the surfaces of GGS strains positive for sil but not on those negative for sil. This finding was true for strains which carry the sil as well as the silS variant. The 87-bp deletion of silS does not affect the expression of SilCR. We did not identify SilCR on the surface of GAS which has sil but is defective in the silCR start codon (strain JS95). Thus, sil not only is present in the genomes of these strains; it also is expressed on the surface, correlated with the growth progress in the logarithmic phase, and secreted into the growth medium.
SilCR belongs to a group of short peptides (CSPs) with the same structure and a role in signal transduction through bacterial sensing systems. It is known (11) that the expression of these peptides correlates with the growth phase, which is thus in agreement with our findings.
In Gram-positive bacteria, quorum sensing involves a complex of a competence-stimulating peptide, two-component systems, and ABC transporters. The species specificity of this sort of quorum sensing is allowed by the exquisite specificity of amino acid sequences of the pheromone peptides (27). It has been shown that competence-stimulating peptides are secreted and function as signals for two-component signal transduction systems (28). The sil locus is a typical peptide/two-component-type circuit of Gram-positive bacteria. In our current work, we provided proof that SilCR is expressed and secreted in all GGS strains containing sil and therefore fits the paradigm of quorum-sensing CSPs.
GAS and GGS strains often coreside in the same niche and may provide a mechanism of virulence regulation via the same pheromone peptide capable of sensing the peptide SilCR, which means that SilCR can be sensed across these streptococcal species (26). We speculate that through such communication, the relatively milder bacterium can control the more pathogenic bacterium and confine it to a more benign, carrier-state phenotype.
Many common virulence factors are present both in GAS and in GGS. These include streptolysin S, M protein, streptokinase, C5a peptidase, and others. However, GGS is a relatively rare cause of severe invasive disease, and when it does cause such disease, it is almost always in a compromised host. This suggests that other factors in GAS, which are not present in GGS, account for its higher invasive potential and virulence. It has been suggested that the paucity of the GAS pyrogenic exotoxins in GGS may explain why, among the beta-hemolytic streptococci that cause bacteremia and necrotizing fasciitis, GGS is associated with the lowest mortality (23). The sil paradigm may suggest that it is not only that GGS lacks GAS determinants that make the latter more virulent but also that it is regulatory circuits that restrain virulence mechanisms and thus contain GGS infections within a more benign form.
Taking the results together, we conclude that the GGS strains may be less virulent because they more often contain sil, express SilCR, and secrete it, while the more virulent GAS strains tend to either lack sil or contain sil but lack the ability to express SilCR due to a defective start codon.
Mice vaccinated against SilCR and challenged with GGS developed a much more severe disease than nonvaccinated mice. The traditional concept is that antibodies generated against bacterial components protect the host against disease. In this study, the opposite was shown. Antibodies directed against a bacterial peptide increased the severity of the disease. This result supports the assumption that the SilCR peptide reduces virulence and protects against severe disease.
sil was first discovered as a virulence factor by transposon insertion into the silC gene. This caused a reduction of virulence compared to that of the wild-type bacteria. Streptococcal strains that contain sil and produce the SilCR peptide reduce virulence by suppression of silC transcription (29). We hypothesize that in the vaccinated mice, neutralization of SilCR by specific antibodies allows silC expression, which increases virulence.
In histological sections of the necrotizing fasciitis area, numerous PMNs migrating to the site of infection were seen in both vaccinated and unvaccinated mice. This is in contrast to what was observed with GAS (15), where PMNs were not seen at the site of severe infection. The ScpC protease has a significant role in virulence of invasive GAS, where it degrades IL-8 and eliminates the chemotactic activity of PMNs to the site of infection.
We assumed that GGS strains do not possess a serine protease such as ScpC as GAS does. We showed that indeed the scpC gene is not found in any of the 50 GGS strains we examined. Furthermore, there was no IL-8 degradation in the strains we tested. This explains the presence of PMNs at the site of infection and the lesser virulence of GGS.
Despite the finding that in both vaccinated and unvaccinated mice there was PMN migration to the infection site, we found a significant difference in disease severity in the two groups. This finding may be partially explained by the virulence capacity of SilC and the ability of SilCR to suppress it, as seen previously (15, 29). Other, as-yet-undefined reasons for this may exist.
The common perception is that bacterial pathogenicity genes are often found as discrete islands in the chromosomes and extrachromosomal elements of pathogenic species but are absent from nonpathogenic members of the same genus or species. In contrast to the conception that there are factors that promote bacterial virulence, the idea of bacterial factors that negate pathogenicity is rather novel.
We have previously shown that the SilCR peptide prevents the development of a fulminant, lethal streptococcal infection in the mouse model of soft tissue infection (15). We have now discovered that in GGS infection, host antibodies against SilCR increase disease severity. Such a phenomenon has not been described in the past and needs further studies in other bacteria.
ACKNOWLEDGMENTS
None of the authors has a commercial or other association that might pose a conflict of interest.
This study was funded by a grant from the Chief Scientist of the Israel Ministry of Health to A.E.M.
Footnotes
Published ahead of print 26 August 2013
REFERENCES
- 1.Lancefield RC, Hare R. 1935. The serological differentiation of pathogenic and non-pathogenic strains of hemolytic streptococci from parturient women. J. Exp. Med. 61:335–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vartian C, Lerner PI, Shlaes DM, Gopalakrishna KV. 1985. Infections due to Lancefield group G streptococci. Medicine (Baltimore) 64:75–88 [DOI] [PubMed] [Google Scholar]
- 3.Cohen-Poradosu R, Jaffe J, Lavi D, Grisariu-Greenzaid S, Nir-Paz R, Valinsky L, Dan-Goor M, Block C, Beall B, Moses AE. 2004. Group G streptococcal bacteremia in Jerusalem. Emerg. Infect. Dis. 10:1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisno AL, Craven DE, McCabe WR. 1987. M proteins of group G streptococci isolated from bacteremic human infections. Infect. Immun. 55:753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleary PP, Peterson J, Chen C, Nelson C. 1991. Virulent human strains of group G streptococci express a C5a peptidase enzyme similar to that produced by group A streptococci. Infect. Immun. 59:2305–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Igwe EI, Shewmaker PL, Facklam RR, Farley MM, van Beneden C, Beall B. 2003. Identification of superantigen genes speM, ssa, and smeZ in invasive strains of beta-hemolytic group C and G streptococci recovered from humans. FEMS Microbiol. Lett. 229:259–264 [DOI] [PubMed] [Google Scholar]
- 7.Jones KF, Fischetti VA. 1987. Biological and immunochemical identity of M protein on group G streptococci with M protein on group A streptococci. Infect. Immun. 55:502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalia A, Bessen DE. 2003. Presence of streptococcal pyrogenic exotoxin A and C genes in human isolates of group G streptococci. FEMS Microbiol. Lett. 219:291–295 [DOI] [PubMed] [Google Scholar]
- 9.Simpson WJ, Robbins JC, Cleary PP. 1987. Evidence for group A-related M protein genes in human but not animal-associated group G streptococcal pathogens. Microb. Pathog. 3:339–350 [DOI] [PubMed] [Google Scholar]
- 10.Hidalgo-Grass C, Ravins M, Dan-Goor M, Jaffe J, Moses AE, Hanski E. 2002. A locus of group A Streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46:87–99 [DOI] [PubMed] [Google Scholar]
- 11.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 103:7059–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, Sturdevant DE, Ricklefs SM, Porcella SF, Parkins LD, Beres SB, Campbell DS, Smith TM, Zhang Q, Kapur V, Daly JA, Veasy LG, Musser JM. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. U. S. A. 99:4668–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessen DE, Kumar N, Hall GS, Riley DR, Luo F, Lizano S, Ford CN, McShan WM, Nguyen SV, Dunning Hotopp JC, Tettelin H. 2011. Whole-genome association study on tissue tropism phenotypes in group A Streptococcus. J. Bacteriol. 193:6651–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidalgo-Grass C, Dan-Goor M, Maly A, Eran Y, Kwinn LA, Nizet V, Ravins M, Jaffe J, Peyser A, Moses AE, Hanski E. 2004. Effect of a bacterial pheromone peptide on host chemokine degradation in group A streptococcal necrotising soft-tissue infections. Lancet 363:696–703 [DOI] [PubMed] [Google Scholar]
- 16.Moses AE, Goldberg S, Korenman Z, Ravins M, Hanski E, Shapiro M. 2002. Invasive group a streptococcal infections, Israel. Emerg. Infect. Dis. 8:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moses AE, Hidalgo-Grass C, Dan-Goor M, Jaffe J, Shetzigovsky I, Ravins M, Korenman Z, Cohen-Poradosu R, Nir-Paz R. 2003. emm typing of M nontypeable invasive group A streptococcal isolates in Israel. J. Clin. Microbiol. 41:4655–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nir-Paz R, Block C, Shasha D, Korenman Z, Gorodnitzky Z, Jaffe J, Ron M, Michael-Gayego A, Cohen-Poradosu R, Shapiro M, Moses AE. 2006. Macrolide, lincosamide and tetracycline susceptibility and emm characterisation of invasive Streptococcus pyogenes isolates in Israel. Int. J. Antimicrob. Agents 28:313–319 [DOI] [PubMed] [Google Scholar]
- 19.Caparon MG, Scott JR. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556–586 [DOI] [PubMed] [Google Scholar]
- 20.Southern EM. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 21.Wu DY, Ugozzoli L, Pal BK, Wallace RB. 1989. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc. Natl. Acad. Sci. U. S. A. 86:2757–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalia A, Enright MC, Spratt BG, Bessen DE. 2001. Directional gene movement from human-pathogenic to commensal-like streptococci. Infect. Immun. 69:4858–4869 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Humar D, Datta V, Bast DJ, Beall B, De Azavedo JC, Nizet V. 2002. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet 359:124–129 [DOI] [PubMed] [Google Scholar]
- 24.Sachse S, Seidel P, Gerlach D, Gunther E, Rodel J, Straube E, Schmidt KH. 2002. Superantigen-like gene(s) in human pathogenic Streptococcus dysgalactiae, subsp equisimilis: genomic localisation of the gene encoding streptococcal pyrogenic exotoxin G (speG(dys)). FEMS Immunol. Med. Microbiol. 34:159–167 [DOI] [PubMed] [Google Scholar]
- 25.Bidet P, Courroux C, Salgueiro C, Carol A, Mariani-Kurkdjian P, Bonacorsi S, Bingen E. 2007. Molecular epidemiology of the sil streptococcal invasive locus in group A streptococci causing invasive infections in French children. J. Clin. Microbiol. 45:2002–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belotserkovsky I, Baruch M, Peer A, Dov E, Ravins M, Mishalian I, Persky M, Smith Y, Hanski E. 2009. Functional analysis of the quorum-sensing streptococcal invasion locus (sil). PLoS Pathog. 5:e1000651. 10.1371/journal.ppat.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Federle MJ, Bassler BL. 2003. Interspecies communication in bacteria. J. Clin. Invest. 112:1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleerebezem M, Quadri LE, Kuipers OP, de Vos WM. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895–904 [DOI] [PubMed] [Google Scholar]
- 29.Eran Y, Getter Y, Baruch M, Belotserkovsky I, Padalon G, Mishalian I, Podbielski A, Kreikemeyer B, Hanski E. 2007. Transcriptional regulation of the sil locus by the SilCR signalling peptide and its implications on group A streptococcus virulence. Mol. Microbiol. 63:1209–1222 [DOI] [PubMed] [Google Scholar]