Abstract
Cattle are naturally infected with Salmonella enterica serotype Typhimurium and exhibit pathological features of enteric salmonellosis that closely resemble those in humans. Cattle are the most relevant model of gastrointestinal disease resulting from nontyphoidal Salmonella infection in an animal with an intact microbiota. We utilized this model to screen a library of targeted single-gene deletion mutants to identify novel genes of Salmonella Typhimurium required for survival during enteric infection. Fifty-four candidate mutants were strongly selected, including numerous mutations in genes known to be important for gastrointestinal survival of salmonellae. Three genes with previously unproven phenotypes in gastrointestinal infection were tested in bovine ligated ileal loops. Two of these mutants, STM3602 and STM3846, recapitulated the phenotype observed in the mutant pool. Complementation experiments successfully reversed the observed phenotypes, directly linking these genes to the colonization defects of the corresponding mutant strains. STM3602 encodes a putative transcriptional regulator that may be involved in phosphonate utilization, and STM3846 encodes a retron reverse transcriptase that produces a unique RNA-DNA hybrid molecule called multicopy single-stranded DNA. The genes identified in this study represent an exciting new class of virulence determinants for further mechanistic study to elucidate the strategies employed by Salmonella to survive within the small intestines of cattle.
INTRODUCTION
Nontyphoidal salmonellae (NTS) are the leading cause of bacterial food-borne gastroenteritis in humans worldwide (1, 2) and are responsible for hundreds of millions of cases of gastroenteritis and bacteremia annually (3). In humans, gastrointestinal disease caused by NTS is characterized by neutrophilic infiltrates within the ileum and symptoms of inflammatory diarrhea (4).
Cattle are naturally susceptible to infection with NTS and develop inflammatory diarrhea histologically characterized by neutrophilic inflammation (5, 6). Cattle either clear the organism after resolution of disease or become persistently infected and continually shed Salmonella enterica in their feces (7). Approximately 30% of human cases of enteric salmonellosis originate from bovine sources (8). Therefore, knowledge of factors important for survival of Salmonella within the gastrointestinal tracts of cattle allows not only extrapolation to human disease but also the opportunity for creation of new strategies to reduce bovine colonization and thus reduce the contamination to the food supply and environment. Additionally, the use of calves as a model organism makes discoveries in this model directly applicable to farm animal populations.
Although there are many tractable animal models of salmonellosis, the majority of screening and the development of mechanistic understanding of NTS infection have historically been done in small animal models that do not naturally develop inflammatory diarrhea upon infection with NTS. These models include mice of the BALB/c, C57B6, 129SvJ, and CBA/J lineages (9–11). To more closely resemble human disease, mice can be treated with antimicrobial agents to eliminate natural microflora prior to infection with Salmonella. These pretreated animals do develop neutrophilic inflammation (commonly known as the murine colitis model) (12–15). The murine colitis model is attractive because it requires minimal technical expertise and allows study of host factors involved in Salmonella pathogenesis through use of widely available immunological reagents and genetically altered mice. However, the lack of an intact microbiota precludes full evaluation of the strategies used by Salmonella to survive in the complex microbial ecosystem of the gastrointestinal tract. Thus, the use of an animal that is a natural host of Salmonella is optimal to understand the biology of Salmonella during infection.
The current animal model with intact microbiota that most closely resembles gastrointestinal salmonellosis in humans in both clinical presentation and histopathology is the calf model of infection (5, 16, 17). This model, although expensive and complex to use, has become very useful for identification of bacterial factors necessary for NTS to thrive in the complex environment of the gastrointestinal tract (18–21). Bovine ligated ileal loops have been used to elucidate the absolute requirement of the type III secretion system (TTSS) and effectors encoded by genes on the Salmonella pathogenicity island 1 (SPI-1) for development of neutrophilic enteritis (6, 17, 21, 22). They have also been used to study the importance of flagella for virulence (18) and to understand the mechanism by which Salmonella employs the host inflammatory response to gain a survival advantage by the use of tetrathionate as a terminal electron acceptor (19). An additional benefit of the calf model is that it reliably replicates enteric salmonellosis in cattle, a population that contributes to the maintenance of Salmonella in the food supply and environment, allowing for development of novel preharvest interventions for this important zoonotic pathogen (5, 11, 23–25). However, because of its complexity, this model has not previously been used in an unbiased approach to study novel virulence factors.
Ligated ileal loops in calves provide a unique environment for the study of Salmonella pathogenesis where virulence factors necessary for establishing early infection may be identified in the presence of intact microbiota. We previously constructed a library of targeted single-gene deletion (SGD) mutants of Salmonella enterica serotype Typhimurium that we used to discover novel genes required for survival during systemic infection in BALB/c mice (26). In the work described here, we used this library of targeted single-gene deletions in combination with the calf ligated ileal loop model to identify novel genes used by Salmonella during enteric infection of a natural host. Using this strategy, we identified 54 mutant genes under selection. Over 20 of these genes have not previously been described as under selection in this model. We tested three mutants (ΔSTM3602, ΔSTM3846, and ΔSTM4602 mutants) and confirmed two in individual competitive infections, in addition to testing and confirming ΔphoP and ΔphoQ mutants (for a total of five mutants tested). Complementation in trans restored the ability of the STM3602 and STM3846 mutants to colonize ligated ileal loops. The genes we reveal here to have roles in colonization represent an exciting group for further study to elucidate the mechanisms that Salmonella species use to survive within and cause disease in the complex environment of the small intestine of cattle.
MATERIALS AND METHODS
Ethics statement.
The Texas A&M University Institutional Animal Care and Use Committee approved all animal experiments, and all experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (27) and USDA Animal Welfare Regulations. Texas A&M has AAALAC-accredited animal facilities.
Bacterial strains.
All bacterial strains are isogenic derivatives of virulent Salmonella enterica serotype Typhimurium ATCC 14028. The SGD mutant library was constructed as previously described (26). All bacteria were grown in Luria-Bertani (LB) broth or LB agar supplemented with kanamycin (50 mg/liter), nalidixic acid (50 mg/liter), carbenicillin (100 mg/liter), or streptomycin (100 mg/liter) where appropriate.
Construction of complementing plasmids.
PCR products were generated by colony PCR using Pfu polymerase (Agilent Technologies). To obtain a 1.1-kb PCR product for STM3602, we used an annealing temperature of 45°C for 5 cycles, 58°C for a further 25 cycles, and the following primers: 3602forward (5′-GTTGAATTCTTCCGCCTCGATCATTTC-3′) and 3602reverse (5′-GTCAAGCTTTCATACGGTAAACCGTATTTTATC-3′). To clone STM3846, a PCR using an annealing temperature of 45°C for 5 cycles, and 57°C for a further 25 cycles, generated a 1.7-kb product using the following primers: 3846forward (5′-GTCGAATTCAAGTCTCATCCTCTGTTGTAATCTATC-3′) and 3846reverse (5′-GTCAAGCTTTATCTACAGCGTTCTGTCTGC-3′). The appropriate length of the PCR products was ensured by agarose gel electrophoresis. A poly(A) tail was added to the 3′ end of the product using Taq polymerase (New England BioLabs) for 9 min at 72°C. PCR products were then ligated into pCR2.1 (TOPO TA cloning; Invitrogen) and transformed into chemically competent One Shot Escherichia coli (Invitrogen) using heat shock, following the manufacturer's instructions. Plasmids were isolated using the Qiagen miniprep kit (Qiagen), and the insert was removed by digestion with EcoRI (New England BioLabs). The insert was isolated from the plasmid backbone by agarose gel electrophoresis and gel purified using a QIAquick gel purification kit (Qiagen). The insert was then ligated into EcoRI-digested and gel-purified pWSK29 (28). Ligations were performed overnight at 14°C, using T4 DNA ligase (New England BioLabs). Ligation reactions were transformed into chemically competent E. coli XL1-Blue (pSTM3846) or Mach One E. coli (pSTM3602; Invitrogen). Transformants were obtained by selection on LB agar supplemented with carbenicillin and were streaked twice to single colonies. Plasmids were isolated using the Qiagen miniprep kit (Qiagen), and correct inserts were verified by restriction digestion of plasmids using BamHI or BstXI (New England BioLabs; pSTM3846 and pSTM3602, respectively). The desired sequence was confirmed by sequencing. Complementing plasmids were transformed into chemically competent S. Typhimurium LB5000 (restriction negative, modification positive) (29), and transformants were obtained by selection on LB with carbenicillin. Plasmids were then isolated as described above and transformed into ΔSTM3846 and ΔSTM3602 mutants using heat shock or electroporation, respectively. Mutants bearing complementing plasmids were purified by streaking twice for single colonies prior to use in competitive infection experiments.
Calves and ligated ileal loop surgery.
Angus cross calves were obtained from a breeding herd at the Veterinary Medical Park at Texas A&M University. A total of 12 calves were used in this study, 3 for screening the mutant library and 9 for competitive infection and complementation analysis. Calves were separated from the dam at 1 day of age, and adequate passive transfer was estimated by measurement of serum total protein. Calves were housed in an AAALAC-approved barn, fed milk replacer twice daily, and provided with water and grass hay. Selective fecal cultures were performed at least once weekly to ensure calves remained negative for Salmonella spp. (6, 30).
At 3 to 6 weeks of age, calves were anesthetized for ligated ileal loop surgery as previously described (6, 16). A detailed description of the surgical procedure is available in the supplemental material. Briefly, calves were placed in left lateral recumbency, and a right flank incision was made. Twenty-four to thirty-eight 4- to 6-cm loops were tied within the ileum within grossly visible Peyer's patches, leaving 1-cm spacers between adjacent loops. Loops were infected individually with 3 ml of LB containing approximately 109 Salmonella Typhimurium. The intestine was returned to the abdomen, the incision was closed, and the calves were monitored under inhalant anesthesia for the duration of the experiment. At 12 h postinfection, the incision was opened, and each loop was individually excised. Calves were euthanized by barbiturate overdose (pentobarbital) administered intravenously.
SGD pool preparation, inoculation, and recovery from ligated ileal loops.
The pool of ∼1,000 SGD mutants prepared and described previously (26) was grown overnight at 37°C with agitation in LB supplemented with kanamycin. Overnight cultures were subcultured 1:100 into LB with kanamycin and incubated for 3 h at 37°C with agitation. The cultures were washed twice in sterile LB broth, and the concentration of organisms was adjusted to 109 CFU in 3 ml LB. A wild-type strain marked with streptomycin resistance in a neutral location, HA697 (ΔphoN::strep) (31; H. J. Yang, L. Bogomolnaya, T. Endicott-Yazdani, M. M. Reynolds, S. Porwollik, M. McClelland, and H. Andrews-Polymenis, unpublished data), was added at a ratio of 1:500 (HA697/total inoculum) to measure the random loss of mutants in the pool. Eight ligated ileal loops were inoculated with the SGD library in three calves. Inoculum titers were determined by serial dilution and plating. Following excision of the infected loops, intestinal fluid, mucus, and tissue samples were harvested and processed separately. Fluid volume, which is correlated with inflammatory response (21), was calculated by weighing each loop before and after removal of fluid. Data from only those loops with fluid accumulation similar to a wild-type (WT)-infected control loop were used. Mucus was gently scraped from the epithelial surface and diluted in 3 ml phosphate-buffered saline (PBS). The remaining tissue was diluted in 5 ml PBS. These specimens were subsequently homogenized, serially diluted in PBS, and plated for enumeration of CFU. The remaining homogenates were grown to stationary phase in LB supplemented with kanamycin and washed in PBS prior to extraction of total DNA.
Mutants with an observed phenotype during the library screen were selected for competitive infection experiments against the wild-type strain, HA420 (ATCC 14028 Nalr). Mutations were moved into a clean genetic background using P22 transduction (32). Bacterial strains used for competitive infection experiments are listed in Table 1. Mutant strains and the isogenic WT were grown in LB with kanamycin and nalidixic acid (mutant) or with nalidixic acid alone (WT) as described above. The inoculum was prepared by mixing SGD mutant and WT at a 1:1 ratio. Ligated ileal loops from 3 to 8 calves were infected with the prepared inoculum, and the WT-to-mutant ratio of the inoculum was determined by serial dilutions and plating. Intestinal fluid, mucus, and tissue samples were processed as described above, and the WT-to-mutant ratio was determined by differential plating. The competitive index (CI) was determined by dividing the output ratio of WT to mutant by the inoculum ratio.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description or relevant characteristica | Source or reference |
|---|---|---|
| Bacterial strains | ||
| 14028s | Wild type | ATCC |
| HA420 | 14028s; spontaneous Nalr | 31 |
| ΔphoP mutant | ΔphoP::kan; Kanr | 26 |
| ΔphoQ mutant | ΔphoQ::kan; Kanr | 26 |
| ΔSTM3602 mutant | ΔSTM3602::kan; Kanr | 26 |
| ΔSTM3846 mutant | ΔSTM3846::kan; Kanr | 26 |
| ΔSTM4206 mutant | ΔSTM4206::kan; Kanr | 26 |
| HA697 | ΔphoN::strep; Strr | Yang et al., unpublished |
| HA1501 | HA420 ΔphoQ::kan; Nalr Kanr | This study |
| HA1473 | HA420 ΔSTM3602::kan; Nalr Kanr | This study |
| HA1444 | HA420 ΔSTM3846::kan; Nalr Kanr | This study |
| HA1482 | HA420 ΔSTM4206::kan; Nalr Kanr | This study |
| HA1474 | HA1473 carrying pSTM3602 | This study |
| HA1446 | HA1444 carrying pSTM3846 | This study |
| Plasmids | ||
| pWSK29 | Cloning vector; Ampr | 28 |
| pSTM3602 | pWSK29::STM3602; Ampr | This study |
| pSTM3846 | pWSK29::STM3846; Ampr | This study |
Nalr, nalidixic acid resistant; Kanr, kanamycin resistant; Strr, streptomycin resistant; Ampr, ampicillin resistant.
Microarray analysis.
The protocol used to prepare transcripts from input and output pools for microarray analysis was essentially as previously described (26). Briefly, total DNA of input or output mutant pools was sonicated, poly(A) tails were added to the DNA fragments, and PCR was amplified with a primer targeting the shared portion of each mutant and a primer including oligo(dT) at the 3′ end (26). PCR products were subjected to reverse transcription from a T7 RNA polymerase promoter located inside each mutant and a mixture of nucleoside triphosphates (NTPs) that included a fluorescently labeled UTP. The RNA was purified using the RNeasy minikit (Qiagen), and approximately 4 μg of labeled RNA was hybridized to a NimbleGen tiling array of 387,000 50-mer oligonucleotides at 42°C for 16 h. The arrays were washed according to the manufacturer's protocol and scanned using a GenePix 4000B laser scanner (Molecular Devices, Sunnyvale, CA) at 5-μm resolution. Data were uploaded into WebArrayDB (33–35), and data were analyzed for peak height in the DNA directly downstream of each mutant location. The relative signal of each mutant was compared to the relative signal in a corresponding array of the same library prior to selection. All large changes in mutant representation were manually inspected and converted into a numerical score between −1 (strongly underrepresented in the output pool) and 1 (overrepresented in the output pool).
Data analysis.
The SGD library was screened in a total of eight loops: four loops in one calf and two loops each in two further calves. The mean score for mutants in each calf was determined by calculating the mean score from multiple loops. The overall score for each mutant was the mean of data from all loops in the three calves. Mutants that were not represented in the input pool in all of the loops were excluded from further analysis. The intercalf variation was defined as the absolute value of the standard deviation of the mean scores from each calf. Mutants under selection in our screen of the SGD library were defined as those mutants with scores outside the 90% confidence interval of the mean scores and with an intercalf variation less than the calculated mean score.
For competitive infection experiments, the competitive index was defined as the ratio of WT to mutant in output normalized to the input ratio. Statistical significance was determined using Student's two-tailed t test with significance set at P < 0.05.
RESULTS
Screen for mutants under selection during enteritis in calves.
In order to assess the fluctuation of mutant representation in the pool, we added strain HA697 (ΔphoN::strep), a derivative of the wild type marked with a streptomycin resistance cassette in a neutral location, to the input pool (31; Yang et al., unpublished). In the input pool, strain HA697 was present in a ratio relative to the total inoculum that approximated the representation of each individual mutant in the pool. By enumerating the representation of this mutant in the output pool relative to the total recovery of the pool, we observed less than 1.4-fold fluctuation of HA697 in intestinal tissue samples compared to the input pool (Fig. 1). As HA697 was inoculated into and recovered from loops in approximately the same proportion relative to the total pool, there appears to be only minimal random loss of mutants occurring during the incubation of our pool in ligated ileal loops.
Fig 1.
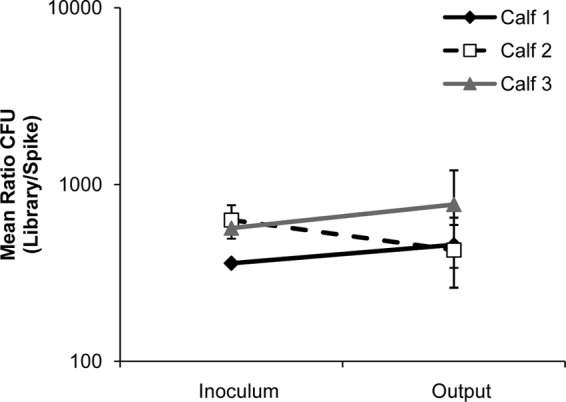
Mutant representation in the pool remains stable. The “input” pool of kanamycin-marked mutants was spiked with strain HA697 (ΔphoN::strep) at a ratio of 1:500 (HA697/total pool). Comparison of the representation of this mutant in the inoculum to the representation of this mutant in the output pools indicated less than 1.4-fold change in HA697 representation over the duration of the incubation of the pool in ligated ileal loops.
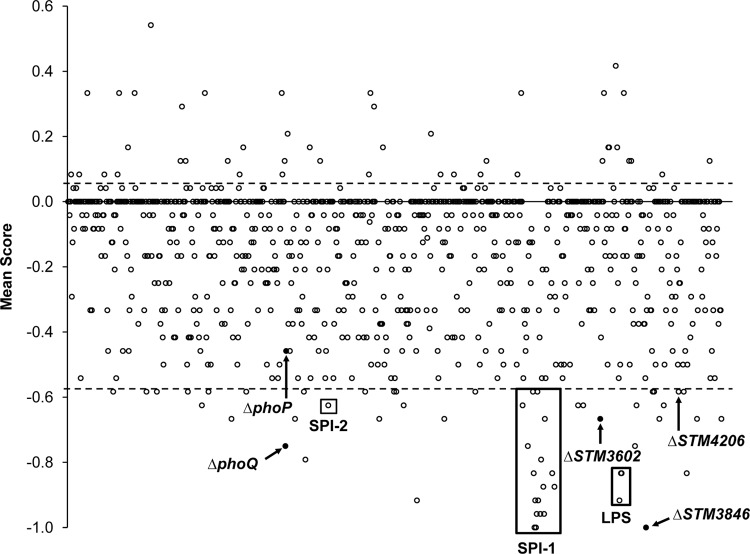
In order to identify candidate mutant genes under selection in ligated ileal loops, both the input pool and the output pool were used to prepare labeled transcripts unique to each mutant. Representation of mutants in the input pool versus the output pool was performed by analysis of labeled transcripts using a NimbleGen tiling array. The resulting data are presented both in Table S1 in the supplemental material and by genome position of each deleted gene represented in the SGD library in Fig. 2. We identified 54 mutant genes under selection in our screen with a mean score outside a 90% confidence interval and with an intercalf variation smaller than the mean (Table 2). We chose to exclude mutant genes with high intercalf variation because we found numerous mutants with a strong phenotype in only a single calf. These mutants had a mean score outside the 90% confidence interval but are considered outliers and not reported here.
Fig 2.
Loss of targeted mutant genes from the pool after inoculation into ligated ileal loops shown by genomic position. Eight ligated ileal loops from three different calves were infected, collected, and analyzed. The scores from multiple loops in a single calf were averaged, and then an overall mean score was generated from all three animals. The x axis represents mutant genes by genomic position with the mean score plotted on the y axis. The horizontal dotted lines indicate the 90% confidence interval. Mutants under selection were outside the 90% confidence interval with an intercalf standard deviation less than the absolute value of the given score. Labeled mutants are in known virulence factors, and those that were confirmed by individual competitive infection are shown in black.
Table 2.
Mutant genes under selection
| Pathogenicity group or cluster of orthologous groups (COG) | Locus tag | Gene name |
|---|---|---|
| Salmonella pathogenicity island 1 | STM2867a | hilC |
| STM2875b,c | hilD | |
| STM2883b,c | sipD | |
| STM2884b,c | sipC | |
| STM2885c | sipB | |
| STM2886a,b,c | sicA | |
| STM2887a,c | spaS | |
| STM2888a,b,c | spaR | |
| STM2889c | spaQ | |
| STM2892c | invJ | |
| STM2893a,c | invI | |
| STM2896c | invA | |
| STM2897b,c | invE | |
| STM2898 | invG | |
| Salmonella pathogenicity island 2 | STM1441a,b | ssaK |
| Salmonella pathogenicity island 4 | STM4261b,c | |
| Salmonella pathogenicity island 6 | STM0296 | |
| Cell envelope biogenesis | STM0719a | |
| STM1737 | tonB | |
| STM3719a,b,c | rfaB | |
| STM3722a,b,c | rfaG | |
| STM3723a,c | rfaQ | |
| Cell motility and secretion | STM3975a,c | tatC |
| DNA replication, recombination, and repair | STM3846a | rrtT |
| Metabolism | STM0522c | allP |
| STM1636 | ||
| STM2437 | ||
| STM3781 | ||
| Posttranslational modification | STM3342c | sspA |
| STM4067 | ||
| Signal transduction | STM0398 | phoR |
| STM1230a,c | phoQ | |
| STM1947a | sirA or uvrY | |
| STM2958a,c | barA | |
| Transcriptional regulation | STM0031 | |
| STM0552a,c | fimW | |
| STM1588c | yncC | |
| STM3245c | tdcA | |
| STM3602b | ||
| STM4417a,c | iolR | |
| Unknown | STM0278 | |
| STM0285c | ||
| STM1258a | ||
| STM1329 | ||
| STM1331 | ||
| STM1785 | ||
| STM1861 | ||
| STM2209 | ||
| STM3026 | ||
| STM3954 | yigG | |
| STM4030a | ||
| STM4206c | ||
| STM4302 | ||
| STM4596c |
This mutant was under selection during a library screen in systemic infection of BALB/c mice (26).
This mutant was identified during a screen of a transposon library in oral infection of a single calf (53).
This mutant was identified during a screen of a transposon library in oral infection of a single calf (52).
Among the mutants with reduced fitness, 14 mutations were in genes located in Salmonella pathogenicity island 1 (SPI-1), and three were in genes needed for lipopolysaccharide (LPS) biosynthesis, all known to be important virulence determinants of Salmonella during enteric infection (21, 36–40). Numerous genes previously identified as virulence factors in animal hosts were also identified in our screen: ssaK, a gene within SPI-2 encoding a portion of the TTSS apparatus (41, 42); phoQ, the sensor in the two-component regulatory system phoPQ responsible for regulation of virulence genes (43, 44); barA and sirA, the sensor and regulator in a two-component regulatory system that regulates SPI-1 (45–47); tatC, a sec-independent transport protein responsible for resistance to bile salts (48); and tonB, a transport protein necessary for iron acquisition in the intestine (49).
We have already used this library to screen for mutant genes under selection during systemic infection of BALB/c mice (26). We found 19 mutants to be under selection in both models (Table 2). Not surprisingly, of the genes not previously implicated to be important in enteric disease, only seven mutants were under selection in both models. These results confirm the necessity of different genes of Salmonella for survival in different niches during infection and show that our library is useful for identification of new virulence factors in different animal models.
Thirty-one mutant genes under selection were not previously proven to be essential for colonization of the bovine host. Of these 31 genes, six encode transcriptional regulators, four encode proteins involved in metabolism, two each encode proteins involved in protein modification and cell envelope biogenesis, and one each encodes a protein involved in DNA modification, cell motility, and secretion. Sixteen of our new mutants under selection have unknown function or are not assigned a group based on clusters of orthologous group assignments (50, 51). Ten of these genes had predicted phenotypes in screening of a library of transposon mutants during oral infection of a single calf, but no further characterization was performed to validate the results of either screen (52, 53).
Confirmation of fitness defects of candidate mutants in calves.
We chose four mutants for confirmation by individual competitive infections with a derivative of the isogenic parental wild-type strain, ATCC 14028. These mutants, the ΔphoQ, ΔSTM3602, ΔSTM3846, and ΔSTM4206 mutants, were transduced to a clean genetic background by P22 transduction and tested in competitive infections in ligated ileal loops in at least three animals.
A ΔphoP mutant, although it was just outside the stringent cutoff of our screen, was also studied in competitive infection experiments because of its known function in resistance against host-derived antimicrobial peptides. We determined that the ΔphoP mutant has a statistically significant survival disadvantage relative to the wild-type organism in ligated ileal loops in calves, as expected (Fig. 3). We also confirmed that the ΔphoQ mutant, a candidate mutant from our screen, has statistically significant survival defects in this model (Fig. 3).
Fig 3.
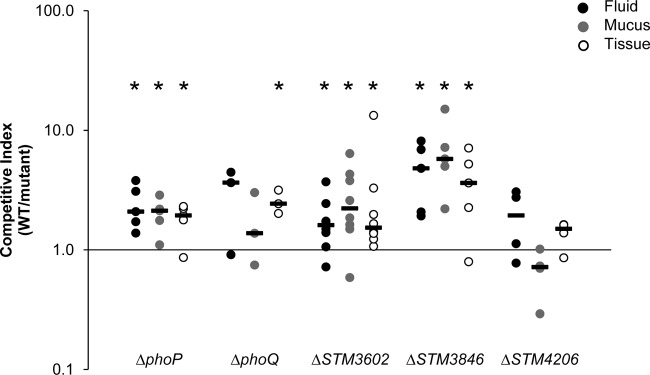
Individual competitive infection experiments confirm requirement for three genes during infection of ligated ileal loops. Ligated ileal loops were infected with a 1:1 mixture of the WT and an isogenic mutant, either the ΔphoP (n = 5), ΔphoQ (HA1501; n = 3), ΔSTM3602 (HA1473; n = 8), ΔSTM3846 (HA1444; n = 5), or ΔSTM4206 (HA1482; n = 4) mutant, for 12 h. Infected loops were harvested, and the fluid contents and mucus layer and intestinal tissue samples were processed for enumeration of bacteria and determination of competitive index. Each symbol represents the value for an individual ligated ileal loop. The short solid black bars are the median values. Asterisks indicate statistically significant differences between the ratio of the mutant and wild type in the inoculum and that ratio in the material collected at the termination of the infection (“output” ratio). A significant difference (P < 0.05) in the output-to-input ratio was determined using a two-tailed Student t test.
Using competitive infections, we confirmed that mutants with deletions of STM3602 and STM3846, genes not previously linked to virulence during enteritis, colonize poorly during competitive infections in ligated ileal loops (Fig. 3). It was surprising to us that the ΔSTM3846 mutant is more severely affected in the calf intestine than the phoP and phoQ mutant genes, genes with a previously defined role in pathogenesis that confer only very modest phenotypes in the calf model (Fig. 3).
We also attempted to confirm the phenotype of a mutant with a deletion in STM4206 during competitive infection in four calves. Although we were unable to confirm the phenotype of this mutant, the calf model has high variability between loops and between genetically nonidentical animals. Thus, we cannot exclude the possibility that the predicted phenotype of the ΔSTM4206 mutant could be confirmed if competitive infection experiments are performed in additional animals. To summarize, we were able to confirm the phenotypes of three (STM3602, STM3846, and phoQ) of four candidate mutant genes that met the stringent inclusion criteria of our screen in ligated ileal loops in calves.
Complementation analysis.
In order to link the observed phenotypes to disrupted genes definitively, we chose to complement the two mutants with confirmed phenotypes in trans and retest these complemented mutants during infection of ligated ileal loops in calves. We cloned STM3602, which encodes a putative transcriptional regulator, and STM3846, which encodes a putative reverse transcriptase, onto a stable, low-copy-number plasmid and transformed these constructs into the corresponding deletion mutants. Complemented deletion mutants were tested in competitive infections with the isogenic wild-type organism in ligated ileal loops. In both cases, the fitness defect of the deletion mutant was reversed by placement of the corresponding gene in trans (Fig. 4).
Fig 4.
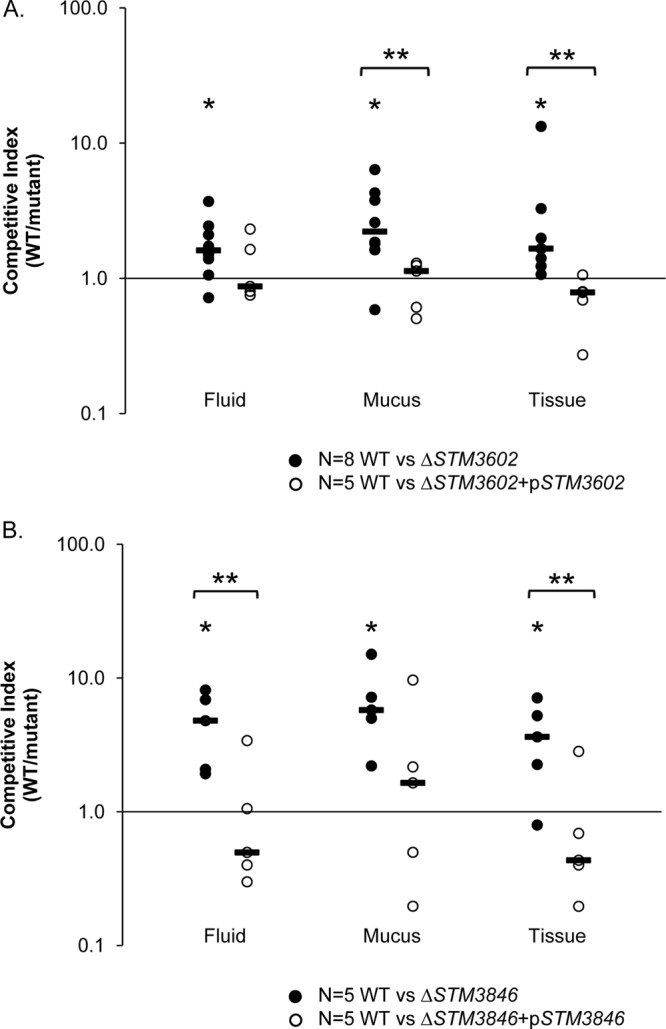
Complementation in trans reverses the observed phenotypes of ΔSTM3602 and ΔSTM3846 mutants. Ligated ileal loops from five calves were inoculated with the WT and the targeted deletion mutant complemented with an intact copy of the corresponding open reading frame in trans (open symbols). Data from the competitive infection experiments between the WT and targeted deletion mutant (Fig. 3, closed symbols), and statistical significance allows direct comparison to complementation data. (A) Competitive index for the wild type versus the complemented ΔSTM3602 mutant. (B) Competitive index for the wild type versus the complemented ΔSTM3846 mutant. In all panels, short solid black bars indicate the median values. Two asterisks indicate statistically significant differences between the ratio of the mutant and wild type in the inoculum and the ratio in the material collected at the termination of the infection (“output” ratio) (P < 0.05) between groups as determined by a two-tailed Student t test.
The genomic contexts of STM3602 and STM3846 are shown in Fig. 5 (54). STM3602 is located between STM3601, encoding a putative phosphosugar isomerase, and treF, encoding a trehalase. STM3602 encodes a putative GntR family regulator and shares a conserved domain with phnF, encoding a regulator of phosphonate utilization (55). STM3846 encodes a reverse transcriptase that catalyzes the formation of an RNA-DNA hybrid molecule called multicopy single-stranded DNA, or msDNA (56–58).
Fig 5.
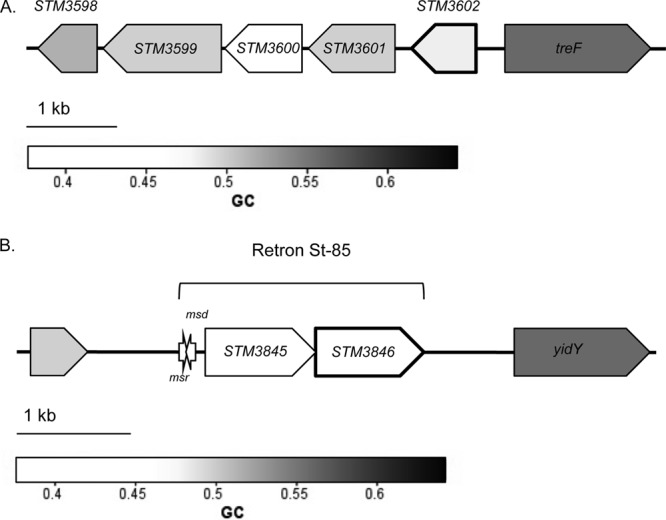
Schematic representation of the genomic regions containing STM3602, STM3846, and retron St-85. (A) Schematic diagram of the region surrounding STM3602. (B) Schematic diagram of the region surrounding STM3846. This gene is located on a 2.13-kb retron at position 4,051,144 on the chromosome between STM3844, a pseudogene encoding an integrase with a truncation at amino acid residue 164, and yidY, encoding a putative multidrug efflux system protein. The orientations of the msr and msd genes are indicated by the arrows. This figure was constructed using coliBASE (86) and data from the study by Ahmed and Shimamoto (56).
DISCUSSION
We used a highly relevant model of enteric salmonellosis, bovine ligated ileal loops, to identify mutants under selection from our library of targeted deletion mutants in Salmonella Typhimurium (26). Our work is the first example of a screen of a mutant library in ligated ileal loops in calves, a technically challenging model that is highly relevant to human enteric salmonellosis. In addition, this work is the first to confirm predicted phenotypes in the bovine model.
The ligated ileal loop model is ideal for screening of a library of mutants to identify the early strategies utilized by Salmonella to survive within the small intestinal environment. Using this model, the “input” pool is administered directly into the ileum, reducing the random loss of mutants traveling through the upper gastrointestinal tract and ensuring that all mutants arrive at the small intestine at the same time. The short duration of infection allows reliable invasion of the epithelium and development of a robust host neutrophilic inflammatory response (6, 16). Thus, we are able to study the early factors responsible for survival of Salmonella within the lumen of the intestine and those factors necessary for creation of and survival during the host inflammatory response. In addition, this model provides an opportunity to dissect the genetic strategies required for survival in the different microenvironments within the small intestine—intestinal fluid, mucus, and tissue layers.
In this study, we evaluated the representation of mutants in the output pool isolated from intestinal tissue from 3 separate calves, because this site might include all mutants under selection in the intestinal luminal and mucus layers in addition to those mutants defective for invasion or survival within epithelial cells. We show that mutants in our pool experience a very low level of random fluctuation during infection of loops, removing a significant barrier to screening in this model. We screened our mutant library in eight loops in three calves, the first screen in any bovine model to use multiple calves to identify candidate mutant genes under selection. We repeated our screen in numerous animals because each of our outbred animals may respond to infection differently, thus placing different selection pressures on our library. To develop a list of candidate mutants for further study that had the strongest phenotypes and the highest probability of being true positives, we used stringent criteria to define a mutant as under selection, and this may have excluded some candidate mutants with relevant phenotypes from further analysis. One example of a mutant we know to be under selection in competitive infection that did not meet our defining criteria for significance in our library screen is the ΔphoP mutant.
Only two other screens have been performed in calves (52, 53). In both cases, randomly generated transposon libraries were screened after oral infection, and phenotypes were assigned to mutants from infection of a single calf. No further characterization of mutant phenotypes was performed in the bovine model. Despite the excellent genome coverage obtained by transposon mutagenesis, such studies provide only a list of candidate genes needed in the bovine host. They lack an estimated true-positive rate of discovery, making it difficult to determine how many of the candidate genes one would expect to have a relevant biological effect in the bovine host. This drawback is a critical roadblock to the design of future studies evaluating the importance of candidate Salmonella genes in the bovine host, a model that requires specialized housing, technical expertise, and great expense compared with conventional small animal models of disease.
Fourteen of the 54 genes under strong selection in our screen were located within SPI-1, and a single gene was located within SPI-2. Among these genes were regulators of expression of SPI-1 (hilC and hilD) and genes encoding portions of the TTSS-1 apparatus (sipD, sipC, sipB, sicA, spaS, spaR, spaQ, invJ, invI, invA, invE, and invG) (59). The requirement of the TTSS-1 and associated effectors for invasion of epithelial cells and creation of a host neutrophilic inflammatory response has been previously described using bovine models of enteric disease (21, 36–38) and has been replicated in the murine colitis model (13, 60). We also predict a phenotype for a single gene (ssaK) encoding a portion of the TTSS-2 apparatus. The TTSS-2 and associated effectors are necessary for virulence during systemic disease (61) and for induction of an inflammatory response in the intestine (36, 62). However, it is possible that 12 h of infection was not long enough to show a more pronounced phenotype for the remainder of the SPI-2 genes. These data show that our screen appropriately identifies virulence factors known to be important in both bovine and murine models of enteric salmonellosis.
Both sirA and barA mutants have predicted phenotypes in the calf model. These genes comprise a two-component regulatory system that senses short-chain fatty acids within the intestine, causing activation of invasion gene expression via hilA, the master regulator of SPI-1 (46, 47, 63). Strains with mutations in each of these genes have reduced virulence during oral infection of BALB/c mice (47) but have not previously been proven to have a role during enteric infection in the bovine host.
In order to survive within the gastrointestinal tract, bacteria have mechanisms to resist antimicrobial peptides produced by the host. Within the small intestine, numerous antimicrobial peptides are constitutively produced by Paneth cells and are concentrated in the mucus covering the mucosa (64–66). One response of the mucosa to proinflammatory cytokines released as a result of Salmonella infection is to increase the production of defensins (67). Polymorphonuclear cells also contain numerous classes of antimicrobial peptides within cytoplasmic granules (68, 69). PhoP and PhoQ comprise a two-component regulatory system that responds to antimicrobial peptides to regulate genes for LPS biosynthesis and virulence (43, 70–73).
Therefore, we tested the phenotypes of ΔphoQ and ΔphoP mutants in competitive infection, even though the latter gene did not meet the stringent cutoff of our screen. We confirmed the predicted phenotype of our ΔphoQ mutant in intestinal tissue and found that a ΔphoP mutant also has a phenotype in bovine ligated ileal loops (Fig. 3). The ΔphoQ mutant was tested in competitive infection in only three calves, and the lack of an observed phenotype in intestinal mucus, the location with the greatest concentration of antimicrobial peptides, may be due to the small number of calves used in the study. The phenotypes we observed for each of these mutants in ligated ileal loops were mild but statistically significant (CI of ∼2). These mild phenotypes are likely due to the short duration of infection or the variable production of antimicrobial peptides as a result of the variation in ages of calves used in this study (3 to 6 weeks). Recent reports indicate that 3-week-old Holstein-Friesian calves may not constitutively express much β-defensin in the gastrointestinal tract but that this expression increases with age (74). However, it is not known whether antimicrobial peptide production in intestinal tissue occurs in response to bacterial infections in calves of this age. Our data are the first to directly support the roles of the phoPQ regulatory system during survival of Salmonella in the inflamed intestinal tract.
STM3602 encodes a putative transcriptional regulator (54), and we show that this gene is necessary for survival in fluid, mucus, and tissue in ligated ileal loops (Fig. 3 and 4A). STM3602 was predicted to be under selection in a signature-tagged mutagenesis screen of transposon mutants during oral infection of a calf (53), but the predicted phenotype was never confirmed. This gene belongs to the GntR (gluconate operon repressor) family of regulators (75) and shares conserved domains with phnF (phosphonate utilization, E value of 1.29e−68) (76, 77), the regulator of the phosphonate utilization operon in E. coli (55).
Phosphonates are stable carbon-phosphorus bonds produced by bacteria and some marine invertebrates as a means of storage of phosphate (78, 79). Salmonella Typhimurium has a complete operon containing two genes for metabolism of phosphonate (phnVUTSRWX, STM0426 to STM0432, GC content 56 to 60%) that is activated by inorganic phosphate during periods of phosphate starvation (55) and an additional locus involved in phosphonate metabolism (phnOBA, STM4287 to STM4289, GC content 49 to 55%) (80). STM3602 is located at a different chromosomal site (Fig. 5A) and has a much lower GC content (49.3%) than the phnVUTSRWX operon. Whether STM3602 is involved in regulating phosphonate metabolism and whether this is related to the phenotype we observe during enteric infection is not yet clear. STM3602 is a very interesting bacterial regulatory protein that merits further study to elucidate its precise function during enteric infection.
The second deletion mutant that we studied in this work, ΔSTM3846, is deleted for a putative reverse transcriptase (54). This gene is encoded on a bacterial retroelement termed a retron (56–58, 81, 82). Bacterial retrons may be both horizontally and vertically acquired and produce a small multicopy single-stranded DNA molecule called msDNA, a unique RNA-DNA hybrid (58, 81, 82). STM3846 is carried on the St-85 retron (Fig. 5b) containing two open reading frames (STM3845 and STM3846) and a small segment of DNA upstream of these open reading frames that encodes the primer and template (msr and msd) used by the reverse transcriptase to produce the msDNA (56). Bacterial reverse transcriptases produce msDNA by using a leader RNA encoded by msr to prime the reaction and produce a 2′,5′ phosphodiester linkage between an RNA (encoded by msr) and DNA (encoded by msd) molecule (83).
The msDNA produced by STM3846 is 85 bp in length, has a predicted stem-loop structure with no mismatched base pairs in the stem (56), and may have lost the RNA template (84). The STM3846 reverse transcriptase is present in the genomes of all 19 S. Typhimurium isolates that we have sequenced (M. McClelland and P. Desai, unpublished data). No role for the St-85 retron has been established despite several previous studies of genes in this region (84, 85). Furthermore, other enterovirulent Gram-negative organisms, including Vibrio spp. and virulent E. coli, produce msDNAs (56), yet no phenotypes have been identified for mutants unable to produce any of these msDNAs. We are the first to unambiguously show a phenotype for a mutant lacking a bacterial reverse transcriptase, and this phenotype is for virulence in a highly relevant model of disease.
In the work we report here, we have used a library of targeted single-gene deletion mutants to identify novel colonization and virulence determinants of Salmonella Typhimurium during infection of bovine ileal loops, a technically challenging model highly relevant to human gastrointestinal salmonellosis. The bovine ileal loop model has not previously been used for unbiased screening of Salmonella mutants, although it closely replicates early events of enteric salmonellosis in humans. We identified more than 30 genes not previously proven to be important for survival of Salmonella in this model, and we confirmed 3 of these mutants individually in competitive infections. Complementation analysis linked the observed phenotypes directly to the disrupted genes for mutations in a putative regulator (STM3602) and a reverse transcriptase (STM3846). We show that the reverse transcriptase encoded by STM3846 is essential for virulence, and we show the first phenotype of any kind for a bacterial reverse transcriptase gene located on a retron. Finally, we report an exciting group of genes for further study to elucidate the mechanisms utilized by Salmonella for survival in the complex niche of the host small intestine during the inflammatory response.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fred Long and Xiao-Qin Xia for help with data analysis methods and Clay Ashley and Destiny Taylor for assistance with experiments involving calves.
M.M., P.C., and S.P. were supported in part by NIH grants R21AI083964, R01AI083646, R56AI077645, and R01AI075093, AFRI CSREES grant 2009-03579, and BARD Fund grant IS-4267-09. This work was supported by NIH/NIAID R21AI083964-01 awarded to H.A.-P., and J.R.E. was supported by the NIH-NCRR 1T32RR0312291 and is currently supported by the Office of Research Infrastructure Programs/OD NIH grant 8T32OD011083. We also acknowledge the Texas A&M University (TAMU) College of Veterinary Medicine & Biomedical Sciences (CVMBS) Office of the Dean and the Department of Large Animal Clinical Sciences for additional funding to J.R.E.
Footnotes
Published ahead of print 9 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00874-13.
REFERENCES
- 1.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV, Emerging Infections Program FoodNet Working Group 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 3 8(Suppl 3):S127–S134 [DOI] [PubMed] [Google Scholar]
- 3.Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32:263–269 [DOI] [PubMed] [Google Scholar]
- 4.Blaser MJ, Newman LS. 1982. A review of human salmonellosis. I. Infective dose. Rev. Infect. Dis. 4:1096–1106 [DOI] [PubMed] [Google Scholar]
- 5.Costa LF, Paixão TA, Tsolis RM, Bäumler AJ, Santos RL. 2012. Salmonellosis in cattle: advantages of being an experimental model. Res. Vet. Sci. 93:1–6 [DOI] [PubMed] [Google Scholar]
- 6.Santos RL, Zhang S, Tsolis RM, Bäumler AJ, Adams LG. 2002. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet. Pathol. 39:200–215 [DOI] [PubMed] [Google Scholar]
- 7.Cobbold RN, Rice DH, Davis MA, Besser TE, Hancock DD. 2006. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J. Am. Vet. Med. Assoc. 228:585–591 [DOI] [PubMed] [Google Scholar]
- 8.Guo C, Hoekstra RM, Schroeder CM, Pires SM, Ong KL, Hartnett E, Naugle A, Harman J, Bennett P, Cieslak P, Scallan E, Rose B, Holt KG, Kissler B, Mbandi E, Roodsari R, Angulo FJ, Cole D. 2011. Application of Bayesian techniques to model the burden of human salmonellosis attributable to U.S. food commodities at the point of processing: adaptation of a Danish model. Foodborne Pathog. Dis. 8:509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller I, Maskell D, Hormaeche C, Johnson K, Pickard D, Dougan G. 1989. Isolation of orally attenuated Salmonella typhimurium following TnphoA mutagenesis. Infect. Immun. 57:2758–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H-J, Choy H. 2010. Identification of genes that are dispensable for animal infection by Salmonella typhimurium. J. Microbiol. 48:399–403 [DOI] [PubMed] [Google Scholar]
- 11.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Bäumler AJ. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335–1344 [DOI] [PubMed] [Google Scholar]
- 12.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hapfelmeier S, Hardt WD. 2005. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol. 13:497–503 [DOI] [PubMed] [Google Scholar]
- 14.Kaiser P, Diard M, Stecher B, Hardt WD. 2012. The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen's virulence factors, and the host's mucosal immune response. Immunol. Rev. 245:56–83 [DOI] [PubMed] [Google Scholar]
- 15.Stecher B, Paesold G, Barthel M, Kremer M, Jantsch J, Stallmach T, Heikenwalder M, Hardt WD. 2006. Chronic Salmonella enterica serovar Typhimurium-induced colitis and cholangitis in streptomycin-pretreated Nramp1+/+ mice. Infect. Immun. 74:5047–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost AJ, Bland AP, Wallis TS. 1997. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet. Pathol. 34:369–386 [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Bäumler AJ. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter SE, Thiennimitr P, Nuccio SP, Haneda T, Winter MG, Wilson RP, Russell JM, Henry T, Tran QT, Lawhon SD, Gomez G, Bevins CL, Russmann H, Monack DM, Adams LG, Baumler AJ. 2009. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype typhimurium infection. Infect. Immun. 77:1904–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran QT, Gomez G, Khare S, Lawhon SD, Raffatellu M, Bäumler AJ, Ajithdoss D, Dhavala S, Adams LG. 2010. The Salmonella enterica serotype Typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infect. Immun. 78:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Santos RL, Tsolis RM, Stender S, Hardt W-D, Bäumler AJ, Adams LG. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawhon SD, Khare S, Rossetti CA, Everts RE, Galindo CL, Luciano SA, Figueiredo JF, Nunes JES, Gull T, Davidson GS, Drake KL, Garner HR, Lewin HA, Bäumler AJ, Adams LG. 2011. Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica serovar Typhimurium: a systems biology analysis approach. PLoS One 6:e26869. 10.1371/journal.pone.0026869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsolis RM, Kingsley RA, Townsend SM, Ficht TA, Adams LG, Baumler AJ. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261–274 [PubMed] [Google Scholar]
- 24.Tsolis RM, Xavier MN, Santos RL, Baumler AJ. 2011. How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect. Immun. 79:1806–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wray C, Sojka WJ. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139–143 [PubMed] [Google Scholar]
- 26.Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. 10.1371/journal.ppat.1000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 28.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
- 29.Bullas LR, Ryu JI. 1983. Salmonella typhimurium LT2 strains which are r− m+ for all three chromosomally located systems of DNA restriction and modification. J. Bacteriol. 156:471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Adams LG, Nunes J, Khare S, Tsolis RM, Baumler AJ. 2003. Secreted effector proteins of Salmonella enterica serotype typhimurium elicit host-specific chemokine profiles in animal models of typhoid fever and enterocolitis. Infect. Immun. 71:4795–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogomolnaya LM, Santiviago CA, Yang HJ, Baumler AJ, Andrews-Polymenis HL. 2008. ‘Form variation' of the O12 antigen is critical for persistence of Salmonella Typhimurium in the murine intestine. Mol. Microbiol. 70:1105–1119 [DOI] [PubMed] [Google Scholar]
- 32.Sternberg NL, Maurer R. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204:18–43 [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, McClelland M, Xia XQ. 2009. Analyzing microarray data using WebArray. Cold Spring Harb. Protoc. 2009:pdb.prot5260. 10.1101/pdb.prot5260 [DOI] [PubMed] [Google Scholar]
- 34.Xia XQ, McClelland M, Porwollik S, Song W, Cong X, Wang Y. 2009. WebArrayDB: cross-platform microarray data analysis and public data repository. Bioinformatics 25:2425–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia X, McClelland M, Wang Y. 2005. WebArray: an online platform for microarray data analysis. BMC Bioinformatics 6:306. 10.1186/1471-2105-6-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. 2005. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect. Immun. 73:7161–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, Tran QT, Lawhon S, Khare S, Adams LG, Bäumler AJ. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velge P, Wiedemann A, Rosselin M, Abed N, Boumart Z, Chaussé AM, Grépinet O, Namdari F, Roche SM, Rossignol A, Virlogeux-Payant I. 2012. Multiplicity of Salmonella entry mechanisms, a new paradigm for Salmonella pathogenesis. MicrobiologyOpen 1:243–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilg K, Endt K, Misselwitz B, Stecher B, Aebi M, Hardt WD. 2009. O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect. Immun. 77:2568–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt WD. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 174:1675–1685 [DOI] [PubMed] [Google Scholar]
- 42.Muller AJ, Kaiser P, Dittmar KE, Weber TC, Haueter S, Endt K, Songhet P, Zellweger C, Kremer M, Fehling HJ, Hardt WD. 2012. Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe 11:19–32 [DOI] [PubMed] [Google Scholar]
- 43.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 [DOI] [PubMed] [Google Scholar]
- 44.Miller SI, Mekalanos JJ, Pulkkinen WS. 1990. Salmonella vaccines with mutations in the phoP virulence regulon. Res. Microbiol. 141:817–821 [DOI] [PubMed] [Google Scholar]
- 45.Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971–982 [DOI] [PubMed] [Google Scholar]
- 46.Altier C, Suyemoto M, Ruiz AI, Burnham KD, Maurer R. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635–646 [DOI] [PubMed] [Google Scholar]
- 47.Lawhon SD, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451–1464 [DOI] [PubMed] [Google Scholar]
- 48.Reynolds MM, Bogomolnaya L, Guo J, Aldrich L, Bokhari D, Santiviago CA, McClelland M, Andrews-Polymenis H. 2011. Abrogation of the twin arginine transport system in Salmonella enterica serovar Typhimurium leads to colonization defects during infection. PLoS One 6:e15800. 10.1371/journal.pone.0015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsolis RM, Bäumler AJ, Heffron F, Stojiljkovic I. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64:4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 51.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. 2013. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 9:e1003456. 10.1371/journal.pgen.1003456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, Barrow PA, Maskell DJ, Wallis TS. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994–1010 [DOI] [PubMed] [Google Scholar]
- 54.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 55.Jiang W, Metcalf WW, Lee KS, Wanner BL. 1995. Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J. Bacteriol. 177:6411–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed AM, Shimamoto T. 2003. msDNA-St85, a multicopy single-stranded DNA isolated from Salmonella enterica serovar Typhimurium LT2 with the genomic analysis of its retron. FEMS Microbiol. Lett. 224:291–297 [DOI] [PubMed] [Google Scholar]
- 57.Inouye S, Inouye M. 1993. The retron: a bacterial retroelement required for the synthesis of msDNA. Curr. Opin. Genet. Dev. 3:713–718 [DOI] [PubMed] [Google Scholar]
- 58.Inouye S, Inouye M. 1995. Structure, function, and evolution of bacterial reverse transcriptase. Virus Genes 11:81–94 [DOI] [PubMed] [Google Scholar]
- 59.Ong SY, Ng FL, Badai SS, Yuryev A, Alam M. 2010. Analysis and construction of pathogenicity island regulatory pathways in Salmonella enterica serovar Typhi. J. Integr. Bioinform. 7:145. 10.2390/biecoll-jib-2010-145 [DOI] [PubMed] [Google Scholar]
- 60.Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Figueira R, Holden DW. 2012. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158:1147–1161 [DOI] [PubMed] [Google Scholar]
- 62.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 73:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altier C. 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 43:85–92 [PubMed] [Google Scholar]
- 64.McGuckin MA, Linden SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9:265–278 [DOI] [PubMed] [Google Scholar]
- 65.Bevins CL, Salzman NH. 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9:356–368 [DOI] [PubMed] [Google Scholar]
- 66.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, Andersson M. 2008. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 57:764–771 [DOI] [PubMed] [Google Scholar]
- 67.Muniz LR, Knosp C, Yeretssian G. 2012. Intestinal antimicrobial peptides during homeostasis, infection and disease. Front. Immunol. 3:310. 10.3389/fimmu.2012.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kościuczuk EM, Lisowski P, Jarczak J, Strzałkowska N, Jóźwik A, Horbańczuk AJ, Krzyżewski J, Zwierzchowski L, Bagnicka E. 2012. Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 39:10957–10970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hager M, Cowland JB, Borregaard N. 2010. Neutrophil granules in health and disease. J. Intern. Med. 268:25–34 [DOI] [PubMed] [Google Scholar]
- 70.Miller SI, Pulkkinen WS, Selsted ME, Mekalanos JJ. 1990. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium. Infect. Immun. 58:3706–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richards SM, Strandberg KL, Conroy M, Gunn JS. 2012. Cationic antimicrobial peptides serve as activation signals for the Salmonella Typhimurium PhoPQ and PmrAB regulons in vitro and in vivo. Front. Cell. Infect. Microbiol. 2:102. 10.3389/fcimb.2012.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shprung T, Peleg A, Rosenfeld Y, Trieu-Cuot P, Shai Y. 2012. Effect of PhoP-PhoQ activation by broad repertoire of antimicrobial peptides on bacterial resistance. J. Biol. Chem. 287:4544–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219–230 [DOI] [PubMed] [Google Scholar]
- 74.Malmuthuge N, Li M, Fries P, Griebel PJ, Guan LL. 2012. Regional and age dependent changes in gene expression of Toll-like receptors and key antimicrobial defence molecules throughout the gastrointestinal tract of dairy calves. Vet. Immunol. Immunopathol. 146:18–26 [DOI] [PubMed] [Google Scholar]
- 75.Haydon DJ, Guest JR. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 79:291–295 [DOI] [PubMed] [Google Scholar]
- 76.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39(Suppl 1):D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41(Database issue):D348–D352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinn JP, Kulakova AN, Cooley NA, McGrath JW. 2007. New ways to break an old bond: the bacterial carbon–phosphorus hydrolases and their role in biogeochemical phosphorus cycling. Environ. Microbiol. 9:2392–2400 [DOI] [PubMed] [Google Scholar]
- 79.Villarreal-Chiu JF, Quinn JP, McGrath JW. 2012. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front. Microbiol. 3:19. 10.3389/fmicb.2012.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Errey JC, Blanchard JS. 2006. Functional annotation and kinetic characterization of PhnO from Salmonella enterica. Biochemistry 45:3033–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lampson BC, Inouye M, Inouye S. 2005. Retrons, msDNA, and the bacterial genome. Cytogenet. Genome Res. 110:491–499 [DOI] [PubMed] [Google Scholar]
- 82.Rice SA, Lampson BC. 1995. Bacterial reverse transcriptase and msDNA. Virus Genes 11:95–104 [DOI] [PubMed] [Google Scholar]
- 83.Hsu MY, Inouye S, Inouye M. 1989. Structural requirements of the RNA precursor for the biosynthesis of the branched RNA-linked multicopy single-stranded DNA of Myxococcus xanthus. J. Biol. Chem. 264:6214–6219 [PubMed] [Google Scholar]
- 84.Matiasovicova J, Faldynova M, Pravcova M, Karpiskova R, Kolackova I, Damborsky J, Rychlik I. 2003. Retron reverse transcriptase rrtT is ubiquitous in strains of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 223:281–286 [DOI] [PubMed] [Google Scholar]
- 85.Pilousova L, Matiasovicova J, Sisak F, Havlickova H, Rychlik I. 2005. Retron reverse transcriptase (rrtT) can be lost in multidrug resistant Salmonella enterica serovar Typhimurium DT 104 strains and influences virulence for mice. Vet. Microbiol. 111:191–197 [DOI] [PubMed] [Google Scholar]
- 86.Chaudhuri RR, Khan AM, Pallen MJ. 2004. coliBASE: an online database for Escherichia coli, Shigella and Salmonella comparative genomics. Nucleic Acids Res. 32(Database issue):D296–D299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.