Abstract
The human pathogen enterohemorrhagic Escherichia coli (EHEC) O157:H7 colonizes the rectoanal junction (RAJ) in cattle, its natural reservoir. Colonization at the RAJ poses a serious risk for fecal shedding and contamination of the environment. We previously demonstrated that EHEC senses acyl-homoserine lactones (AHLs) produced by the microbiota in the rumen to activate the gad acid resistance genes necessary for survival through the acidic stomachs in cattle and to repress the locus of enterocyte effacement (LEE) genes important for colonization of the RAJ, but unnecessary in the rumen. Devoid of AHLs, the RAJ is the prominent site of colonization of EHEC in cattle. To determine if the presence of AHLs in the RAJ could repress colonization at this site, we engineered EHEC to express the Yersinia enterocolitica AHL synthase gene yenI, which constitutively produces AHLs, to mimic a constant exposure of AHLs in the environment. The yenI+ EHEC produces oxo-C6-homoserine lactone (oxo-C6-HSL) and had a significant reduction in LEE expression, effector protein secretion, and attaching and effacing (A/E) lesion formation in vitro compared to the wild type (WT). The yenI+ EHEC also activated expression of the gad genes. To assess whether AHL production, which decreases LEE expression, would decrease RAJ colonization by EHEC, cattle were challenged at the RAJ with WT or yenI+ EHEC. Although the yenI+ EHEC colonized the RAJ with efficiency equal to that of the WT, there was a trend for the cattle to shed the WT strain longer than the yenI+ EHEC.
INTRODUCTION
Enterohemorrhagic Escherichia coli serotype O157:H7 (EHEC) is a human pathogen that causes complications that range from abdominal cramps and bloody diarrhea to the life-threatening sequelae known as hemolytic uremic syndrome (1–3). Although EHEC colonizes the large intestine and causes disease in humans, EHEC is a member of the transient normal bovine microbial flora and naturally colonizes the rectoanal junction (RAJ) mucosa of cattle and is then subsequently shed into the environment with the animal's feces (4). To colonize the host, EHEC forms attaching and effacing (A/E) lesions on epithelial cells (5). These lesions are characterized by the effacement of the epithelium's microvilli, the intimate attachment of bacteria to the epithelial cells, and the rearrangement of the host actin cytoskeleton to form a pedestal-like structure cupping the individual bacterium (6, 7). The majority of the genes required to form A/E lesions are encoded within a chromosomal pathogenicity island known as the locus of enterocyte effacement (LEE) (8, 9). The LEE is comprised of 41 genes, most of them organized in five major operons (9). These genes encode transcriptional regulators (10, 11), a type III secretion system (12), the adhesin intimin (13), and its receptor Tir (13), as well as several effector proteins (14–18).
A complex network of proteins and genes has been shown to regulate the LEE, including H-NS (19), GadX (20), Per (11), EtrA and EivF (21), QseA (22), SdiA (23), CpxR (24), LexA (25), Pch (26), Hha (27), and Ler (11, 28, 29). Ler, encoded by the first gene in the LEE1 operon, is the transcriptional master regulator of the other LEE genes (11, 28, 29). The nucleoid-associated protein H-NS silences the LEE; however, Ler antagonizes H-NS to overcome silencing and to activate the LEE (19). Recently, a member of the LuxR protein family, the transcription factor SdiA, was shown to modulate transcription of the LEE by directly repressing the expression of ler (30, 31).
The first LuxR-I quorum sensing (QS) system was described in Vibrio fischeri (32). LuxI is a synthase, while LuxR is a cognate transcription factor. The LuxI synthase produces small chemical signaling molecules known as acyl-homoserine lactones (AHLs) that diffuse freely out of the bacteria into the environment. Once an external threshold concentration is reached, AHLs diffuse back into the cells and bind to their cognate cytoplasmic LuxR transcription factor. Ligand binding initiates an increase in LuxR protein stability and also promotes LuxR protein oligomerization (33). The AHL-LuxR complex then binds to target promoters and regulates their expression (33). For example, the LuxR-I system of V. fischeri activates the production of light by inducing the expression of genes important for bioluminescence (32). Since this initial discovery, homologs of the LuxR-I system have been found in over 50 bacterial species, including the human pathogens Yersinia enterocolitica and Pseudomonas aeruginosa and the plant pathogens Erwinia carotovora and Agrobacterium tumefaciens (33). A majority of these species encode both a synthase and a transcriptional regulator, but interestingly, a subset of species encode only the LuxR homologs but not their cognate LuxI synthases. The LuxR homolog SdiA present in E. coli and Salmonella spp. is an example of such “orphan” LuxR proteins.
SdiA has been shown to be involved in interspecies communication, as evidenced by the fact that SdiA is able to detect and respond to AHLs produced by other bacteria (30, 34–36). Dyszel and colleagues demonstrated that SdiA from Salmonella enterica serovar Typhimurium detects AHLs produced by the pathogen Yersinia enterocolitica in mice. To mimic a constant interaction with Y. enterocolitica, Dyszel et al. constructed an S. Typhimurium strain to constitutively express the Y. enterocolitica gene yenI, which encodes an AHL synthase. SdiA-dependent genes activated by AHLs conferred a fitness advantage in S. Typhimurium organisms carrying yenI and sdiA compared to those carrying yenI but not sdiA, implicating the importance of SdiA and AHLs in competition within a niche.
Additionally, recent evidence from studies in cattle indicates that EHEC may sense AHLs in an SdiA-dependent manner in order to discern the appropriate niche for bacterial colonization. An investigation of the bovine digestive tract determined that AHLs are present only in the rumen (30, 37). EHEC activates the gad acid resistance genes in response to AHLs, likely as a mechanism to safely passage through the cattle's acidic environment (30). Conversely, rumen-derived AHLs repress the LEE (23, 30). The rationale for this inhibition is that rumen colonization is unfavorable, and the absence of AHLs in the RAJ, the prominent site of colonization, alleviates SdiA-AHL mediated-repression of the LEE, thus promoting colonization at the RAJ. Ruminants known as “supershedders” shed high numbers of EHEC in their feces over a prolonged period, and EHEC strains isolated from supershedders more intimately colonize the RAJ (38). Epidemiologic studies have shown that supershedders account for approximately 95% of all EHEC shed into the environment (39–41). Since most humans become infected with EHEC either by ingestion of food products contaminated by infected animals (42–45) or through direct contact with infected animals (46–49), understanding how EHEC promotes intimate colonization at the RAJ in its natural host is crucial for development of preventive strategies to decrease EHEC shedding into the environment and consequent transmission to humans.
In this study, we explore the role of AHLs in colonization of the RAJ by EHEC by constructing an AHL-producing EHEC strain to mimic constant exposure to AHLs in the environment. Our data provide evidence that continuous exposure of EHEC to AHLs decreases A/E lesion formation on epithelial cells in vitro but does not prohibit colonization of EHEC at the RAJ in cattle. These data suggest that EHEC colonization at the RAJ is complex, with multiple factors contributing to efficient colonization.
MATERIALS AND METHODS
Strains and plasmids.
All strains and plasmids used in this study are listed in Table 1. Unless otherwise stated, E. coli strains were grown aerobically in Luria-Bertani (LB) broth at 37°C and 250 rpm. Where indicated, strains were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Invitrogen). Medium was supplemented where necessary with the selective antibiotics streptomycin, kanamycin, ampicillin, and chloramphenicol, which were added to a final concentration of 100, 50, 100, or 30 μg/ml, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| 86-24 | Wild-type Stx2+ EHEC strain (serotype O157:H7) | 67 |
| YNN01 | 86-24 Δstx2a | This study |
| YNN02 | YNN01 λPR | This study |
| YNN03 | YNN01λPR-yenI | This study |
| Plasmids | ||
| pKD3 | λ-Red template plasmid | 51 |
| pKD46 | λ-Red helper plasmid (recombinase) | 51 |
| pCP20 | λRed resolvase plasmid | 51 |
| pJLD1600 | λPR-yenI-FRT-kan-FRT template plasmid | 36 |
| pJLD500 | λPR-FRT-kan-FRT vector template plasmid | 36 |
Recombinant DNA methods.
Methods used for PCR amplification, plasmid purification, and transformations followed standard protocols as previously described (50). Oligonucleotide primers (Table 2) were designed by using Primer Express v1.5 (Applied Biosystems). Δstx2a, yenI+ Δstx2a, and vector+ Δstx2a strains were constructed using λ Red mutagenesis (51). Briefly, to generate the Δstx2a mutant in a wild-type EHEC strain (strain 86-24), PCR product was amplified using Phusion polymerase (Invitrogen), primers stx2AλRed F and stx2AλRed R, and pKD3 plasmid as the template. PCR product was digested with DpnI to remove the template DNA and then gel purified (Qiagen). Wild-type EHEC strains transformed with the helper plasmid pKD46 were prepared for electroporation and transformed with the resulting stx2a PCR product. Colonies were screened for ampicillin sensitivity and chloramphenicol resistance. The chloramphenicol cassette was removed from Δstx2a deletion candidates with the resolvase plasmid pCP20. Final verification was performed by PCR amplification and sequencing to yield the resolved Δstx2a EHEC strain (YNN01). yenI and empty vector were chromosomally integrated into the YNN01 background to yield yenI+ Δstx2a and vector+ Δstx2a EHEC strains using the same λ Red mutagenesis method. Primers yenIλRed F and yenIλRed R were used to amplify the λPR-yenI-FRT-kan-FRT or λPR-FRT-kan-FRT cassettes from pJLD1600 and pJD500 (36), respectively, and the sequences homologous to the lacI integration site in the chromosome of Δstx2a EHEC.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| stx2AλRed F | CTTTTTTATATCTGCGCCGGGTCTGGTGCTGATTACTTCAGCCAAAAGGAACACCTGTATGTGTAGGCTGGAGCTGCTTCG |
| stx2AλRed R | CATTAACAGAAGCTAATGCAAATAAAACCGCCATAAACATCTTCTTCATGCTTAACTCCTCATATGAATATCCTCCTTAG |
| yenIλRed F | AGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATCTGTAATCATAGTCATGATACGACTCACTATAGGGCG |
| yenIλRed R | AAAACCTTTCGCGGTATGGCATGATAGCGCCCGGAAGAGAGTCAATTCAGGGTGGTGAATGTTGACCATGATTACGCCAAGC |
| stx2AUP | CTGCATTATGCGTTGTTAGCTCAG |
| stx2ADown | ATCCGCCGCCATTGCATTAAC |
| lacZF | TGCAAGGCGATTAAGTTGGGTAACG |
| IRlacIR | TGTGACCTGGCGTCAGCATTTTAAATCT |
Western blotting.
Wild-type and yenI+ strains were grown in high-glucose DMEM at 37°C in the presence of either 10 μM oxo-C6-HSL (synthesized in the Flack's laboratory) or an equivalent amount of dimethyl sulfoxide (DMSO) to an optical density at 600 nm (OD600) of 1.0, and secreted proteins were prepared as previously described (12). Protein samples were electrophoresed in sodium dodecyl sulfate–12% polyacrylamide gels. Samples were subjected to Western blotting as described previously (50). Blots were probed with rabbit polyclonal antisera to EspA and EspB (Cocalico Biologicals) and visualized with enhanced chemiluminescence (Bio-Rad).
FAS assay.
To assess A/E lesion formation, fluorescent actin staining (FAS) assays were performed as previously described (52). Briefly, HeLa cells were grown on coverslips in low-glucose DMEM supplemented with 10% fetal bovine serum (FBS) (Invitrogen) at 37°C in 5% CO2 and were infected with a 1:100 dilution of overnight static bacterial cultures for 6 h. The coverslips were washed, fixed, and permeabilized with 0.2% Triton X. Fluorescein isothiocyanate (FITC)-labeled phalloidin (Sigma-Aldrich) was used to visualize actin accumulation, and propidium iodide was added to stain bacteria and HeLa nuclei.
RNA extraction.
Overnight cultures grown aerobically at 37°C in LB were diluted at 1:100 into high-glucose DMEM in the presence or absence of 10 μM oxo-C6-HSL and grown in triplicate to late exponential growth phase (OD600 = 1.0). Oxo-C6-HSL was dissolved in DMSO at 10 mM concentration and added directly to DMEM at 1:1,000 dilutions. For samples assessed without exogenous signals, the respective concentration of DMSO was used to ensure that the solvent did not alter gene expression. RNA from these biological replicates was extracted using TRIzol (Invitrogen) and the RiboPure Bacteria RNA isolation kit (Ambion) according to the manufacturer's instructions.
RT-qPCR.
The primers used for the real-time PCR assays were designed by using Primer Express v1.5 (Applied Biosystems) (Table 2). The amplification efficiency and template specificity of each of the primers were validated, and reaction mixtures were prepared as previously described (53). Quantitative real-time reverse transcriptase PCR (RT-qPCR) was performed in a one-step reaction using an ABI 7500 sequence detection system (Applied Biosystems). Using the ABI sequence Detection 1.2 software (Applied Biosystems), data were collected and normalized to endogenous levels of rpoZ. Data were analyzed by using the comparative critical threshold cycle (CT) method and presented as fold changes compared to WT levels. Error bars represent the standard deviations of the ΔΔCT values. Statistical significance was determined by Student's t test, and a P value of ≤0.05 was considered significant.
AHL detection.
AHL extraction and detection were performed as previously described (54). Briefly, wild-type, yenI+, and vector control strains were grown in high-glucose DMEM to late exponential phase. AHLs were extracted three times with ethyl acetate and concentrated to 50 μl. For analytical thin-layer chromatography (TLC), 1 μl of concentrated extracts from wild-type and vector strains or 10 μl of concentrated extract diluted at 1:100 from yenI+ strains was applied to C18 reverse-phase TLC plates (200-μm layer; Whatman). The chromatograms were developed with 70% methanol, dried, and overlaid with a culture of the Agrobacterium tumefaciens traI lacZ (55) indicator strain and 60 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for 16 h at 30°C. For positive control, 1 μl of 10 μM 3-oxo-C6-HSL was also included.
Cattle experiments.
Two groups of five mature Holstein steers were housed in quarantined facilities. All personnel followed strict biosafety procedures, and all procedures were approved by the Institutional Animal Care and Use and Biosafety Committees. O157 cultures were adjusted to the desired bacterial concentration by dilution in phosphate-buffered saline and confirmed by viable plate count. Cattle received a single rectal application of 109 CFU of wild-type or yenI+ O157, as previously described (56). O157 was cultured from rectoanal junction mucosa swabs as previously described (39) on the days indicated in Results.
RESULTS
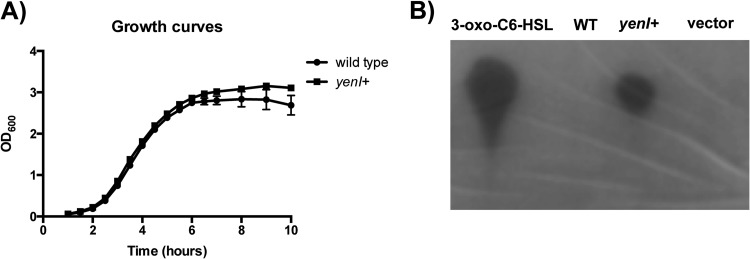
The canonical LuxR/LuxI-type quorum-sensing system encodes a transcription factor (LuxR) and an associated AHL synthase (LuxI). Several examples exist, however, whereby a LuxR regulator lacks an associated AHL synthase. In E. coli, for example, the LuxR regulator SdiA functions to regulate several important virulence and colonization genes yet lacks a cognate synthase. The EHEC homolog of SdiA senses AHLs synthesized by other bacteria rather than self-produced AHLs to modulate virulence gene expression (30). Previously, we demonstrated that an EHEC lacking sdiA colonizes the RAJ of cattle with reduced efficiency compared to EHEC carrying sdiA (30), suggesting that SdiA plays an important role to promote colonization at the RAJ. Additionally, we demonstrated that AHLs repress the LEE pathogenicity island in vitro. An absence of AHLs promotes alleviation of SdiA-AHL-mediated repression of the LEE and likely contributes to successful colonization of EHEC at the RAJ. To explore the effect of AHLs on EHEC virulence and colonization, we engineered an EHEC strain to constitutively express the Yersinia enterocolitica AHL synthase YenI to mimic a constant exposure of AHLs. The U.S. Department of Agriculture (USDA) prohibits the use of Shiga toxin-producing E. coli in cattle experiments; therefore, the stx2a gene was deleted from both wild-type and yenI+ EHEC strains. Initial studies confirmed that the chromosomal integration of yenI in EHEC did not affect growth (Fig. 1A). Additionally, we confirmed that the yenI+ EHEC strain also produced endogenous AHLs as detected by the thin-layer chromatography comparisons with the wild type and the vector control (Fig. 1B).
Fig 1.
yenI+ EHEC produces AHLs. (A) Growth curves of wild-type and yenI+ strains grown in triplicate in high-glucose DMEM at 37°C. Optical density at 600 nm (OD600) was measured at the indicated times. (B) yenI+ EHEC produces endogenous AHLs, as shown by TLC of AHLs extracted from wild-type, yenI+, and vector+ strains; 3-oxo-C6-HSL was used as a positive control.
Endogenous AHLs repress the transcription of the LEE.
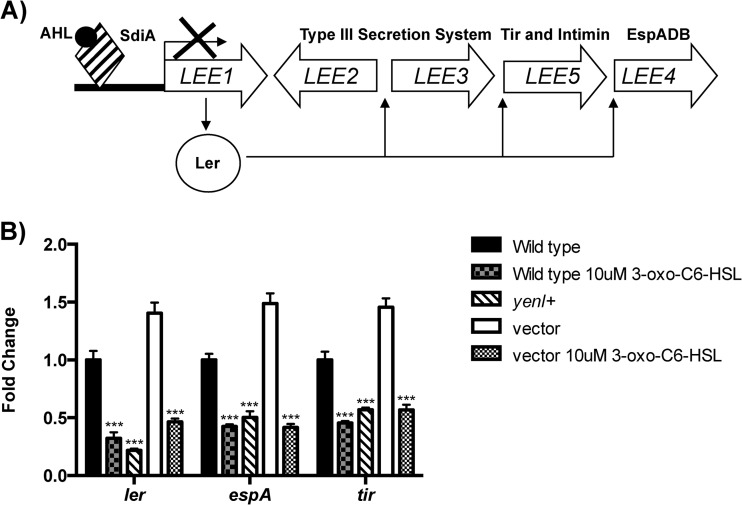
In response to exogenous AHLs, SdiA represses transcription of the LEE genes by directly binding to the promoter and repressing the master transcriptional regulator ler (30) (Fig. 2A). Consistent with our previous results, addition of exogenous AHLs reduced the expression of the LEE. The endogenous production of AHLs in the yenI+ EHEC strain decreased expression of ler, espA, and tir (all of which are LEE genes) to a level similar to the one obtained with the addition of exogenous signal. Transcriptional repression of the LEE in the yenI+ strain was dependent on endogenous AHLs, since chromosomal integration of the empty vector control had no effect on transcription of the LEE in the absence of exogenous AHL signal.
Fig 2.
LEE regulation by AHLs. (A) Schematic of SdiA-AHL complex regulation of the LEE genes. SdiA complexed with AHLs binds to the LEE1 promoter to repress its expression. LEE1 encodes the Ler transcription activator of the LEE genes; hence, SdiA inhibition of LEE1 inhibits transcription of all LEE genes in a cascade fashion. (B) RT-qPCR analyses of ler (LEE1), espA (LLE4), and tir (LEE5) transcription in the wild type, wild type with exogenous AHLs, yenI+ strain (produces AHLs endogenously), wild type with vector (vector used to insert yenI into the EHEC chromosome, no AHLs, negative control), and wild type with vector plus endogenous AHLs. The mRNA levels are graphed as fold changes compared to WT levels. Statistical significance was determined by Student's t test; ***, P ≤ 0.001.
yenI+ EHEC forms fewer A/E lesions.
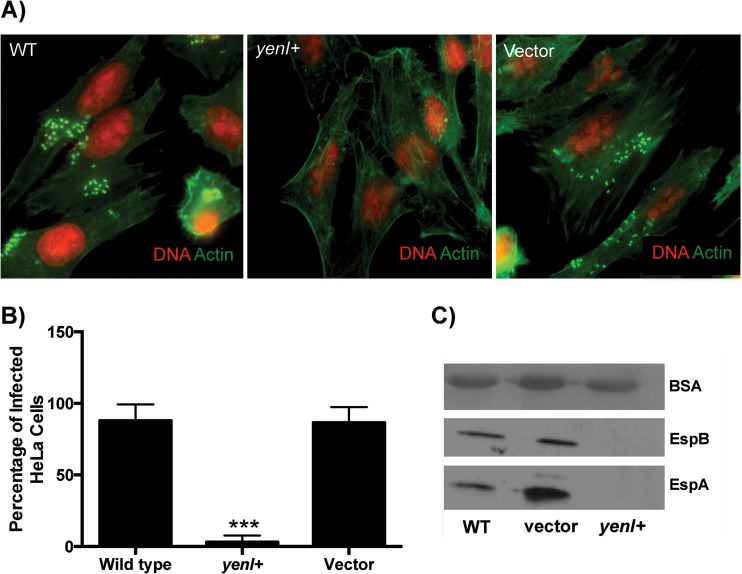
Since endogenous AHLs reduced expression of the LEE, we next used fluorescent actin staining (FAS) to investigate whether yenI+ EHEC has a reduced ability to form A/E lesions on epithelial cells compared to the wild type. HeLa epithelial cells were infected for 6 h with wild-type, yenI+, or vector-only EHEC strains. Wild-type and vector control EHEC formed A/E lesions on 88% and 87% of the HeLa cells, respectively (Fig. 3). In contrast, yenI+ produced significantly fewer A/E lesions, infecting only 3% of the HeLa cells. These results suggest that endogenous AHLs decrease the ability of EHEC to form A/E lesions. Congruently with the decrease in LEE expression (Fig. 2) and A/E lesion formation, the yenI+ strain was unable to secrete the EspA and EspB LEE type three secreted proteins (Fig. 3C).
Fig 3.
AHLs decrease A/E lesion formation by EHEC. (A) Fluorescent actin staining (FAS) assays. HeLa cells were infected with wild-type, yenI+, and vector control strains for 6 h. Actin was stained in green with FITC-phalloidin, and HeLa cell nuclei and bacteria were stained in red with propidium iodide. A/E lesion formations were visualized as bright green cupping the red bacteria. (B) Quantification of infected cells or cells with A/E lesions. (C) Western blots of the secreted proteins of wild-type, vector, and yenI+ strains probed with antiserum against EspB and EspA; bovine serum albumin (BSA) was used as a loading control. Statistical significance was determined by Student's t test; ***, P ≤ 0.001.
Endogenously produced AHLs activate gad gene expression.
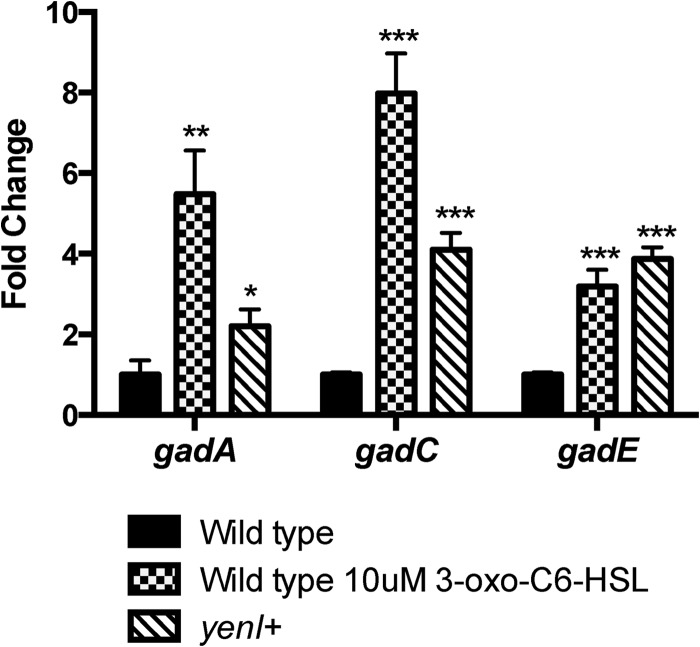
Although AHLs repress LEE gene expression, they activate expression of the gad acid resistance genes (30). Here, again, we show that transcription of the gadA, gadC, and gadE genes is significantly increased by both exogenous and endogenous (yenI+) AHLs. These data combined suggest that the yenI+ EHEC behaves as if it is constantly in the presence of this signal.
Wild-type and yenI+ EHEC organisms colonize the RAJ mucosa of cattle similarly.
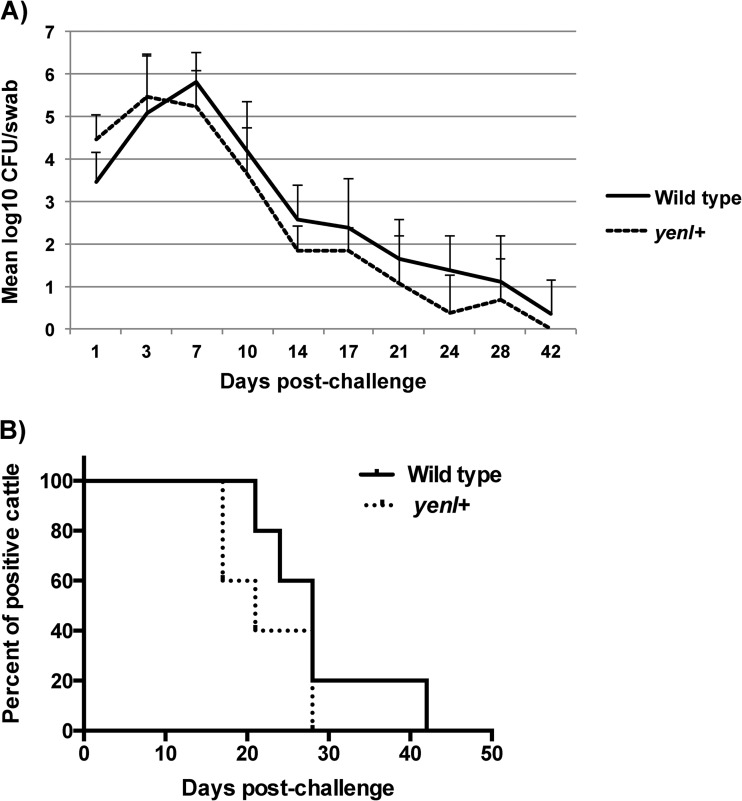
Our in vitro studies suggest that endogenous AHL production decreases LEE expression and consequently reduces A/E lesion formation on HeLa cells. Next, we investigated if yenI+ EHEC colonizes the host in vivo less efficiently than does the wild type. Five steers were challenged with a single rectal application of wild-type or yenI+ EHEC. Wild-type or yenI+ EHEC was cultured on day 0 prior to challenge and then on days 1, 3, 7, 10, 14, 17, 21, 24, 28, and 42 postchallenge. All animals were O157 culture negative prior to challenge, and all animals were culture positive for at least 14 days postchallenge. The wild-type and yenI+ EHEC colonized the bovine RAJ similarly, although the wild-type tended to persist slightly longer than the yenI+ EHEC (see Fig. 5).
Fig 5.
Cattle carriage of EHEC after challenge. The rectoanal junction (RAJ) mucosas of 10 Holstein steers were dosed with a single application of 109 CFU of either wild-type or yenI+ EHEC. The challenge strains were cultured from RAJ mucosal swabs on the indicated postchallenge days. (A) Mean log CFU of wild-type or yenI+ strain per swab from cattle throughout the experiment. (B) Percentage of animals (inoculated with either wild-type or yenI+ strain) culture positive with EHEC postchallenge throughout the period of 56 days.
DISCUSSION
EHEC can colonize multiple hosts. In order to survive host defenses and successfully colonize specific niches, EHEC must rely on environmental cues to modulate appropriate gene expression. Gram-negative bacteria use the chemical signal acyl-homoserine lactones to monitor their own population density and mount appropriate responses (33). EHEC can hijack this bacterial cell-to-cell communication by sensing AHLs produced by other bacteria in the environment to regulate their genes for survival and colonization in its natural reservoir, cattle (30). To establish colonization in cattle at the RAJ (4), EHEC must survive the acidic environment of the stomachs and activate A/E lesion formation at the RAJ. Construction of a type III secretion system for injection of bacterial effector proteins into the host cells to form A/E lesions requires a lot of energy resources; therefore, expression of the LEE genes that are required for A/E lesions has to be tightly regulated to prevent waste of energy in the wrong gastrointestinal compartment. Previous findings suggest that activation of acid resistance genes by AHLs synthesized by other bacteria present in the rumen of cattle prepares EHEC to survive the acidic environment of cattle stomachs while decreasing the LEE, since the rumen is not a preferable niche for colonization (30). Interestingly, the RAJ contains no AHLs (30, 37). The absence of AHLs in the RAJ alleviates AHL-mediated repression of the LEE to allow for A/E lesion formation on the mucosal epithelial cells at the RAJ, favoring colonization at the RAJ. These data suggest that AHLs produced by the bacterial population in cattle guide EHEC to survive and establish a niche in an appropriate environment once inside the cattle.
If AHLs provide cues to EHEC that the rumen is an unfavorable environment for colonization, then an interesting question to explore is whether AHLs present in the RAJ would decrease bacterial colonization of EHEC at the RAJ and decrease subsequent shedding of EHEC into the environment. We explored this idea by creating an EHEC strain that contains the LuxI-like synthase gene yenI from Y. enterocolitica to imitate the constant exposure to AHLs that occurs in the environment. yenI+ EHEC produced endogenous AHLs that significantly decreased expression of the LEE genes while increasing expression of the gad acid resistance genes (Fig. 4). Lowered transcriptional expression of the LEE also resulted in a significantly decreased production of secreted proteins and, consequently, A/E lesion formation on HeLa cells (Fig. 3). Although the in vitro data are consistent with our previous data demonstrating the importance of AHLs in the downregulation of the LEE, yenI+ EHEC colonizes the RAJ similarly to the wild type, albeit there is a trend that animals infected with yenI+ EHEC shed EHEC for a shorter time than those infected with WT (Fig. 5). Diffusion of AHLs into the large environment of the RAJ rather than an enclosed system in vitro may account for the lack of AHL-mediated repression of EHEC colonization of this site.
Fig 4.
AHLs increase gad gene expression in EHEC. RT-qPCR analyses of gadA, gadC, and gadE transcription in the wild type, in the wild-type strain with exogenous AHLs, and in the yenI+ strain (produces endogenous AHLs). The mRNA levels are graphed as fold changes compared to WT levels. Statistical significance was determined by Student's t test; **, P ≤ 0.01; ***, P ≤ 0.001.
The data also suggest that unidentified signals sensed by SdiA are contributing to EHEC colonization at the RAJ, as supported by our previous findings that the wild-type EHEC outcompeted the mutant lacking sdiA in colonization at the RAJ (30). Since AHLs are not naturally present in the RAJ, it is also possible that there are enzymes or factors expressed by the epithelial cells in the RAJ that can readily degrade AHLs. For example, a class of AHL-degrading enzymes found in mammals called paraoxonases (PONs) has been shown to degrade AHLs and inhibit the quorum-sensing regulation of bacteria (57). PON2 has the highest enzymatic capacity to degrade AHLs compared to other PONs (58, 59). Interestingly, various tissues, including epithelial cells of the gastrointestinal tract, where EHEC colonizes humans and AHLs are absent (60), express PON2 (61). This suggests that EHEC utilizes other available signals such as epinephrine or norepinephrine (62–64) to modulate the LEE genes to promote colonization in the human gut. Paraoxonases are also found in cattle (65), and bovine serum has been shown to degrade AHLs (59). This infers that either paraoxonases or other similar AHL-degrading enzymes could degrade the AHLs produced by yenI+ strains in the RAJ and, as a result, EHEC is utilizing other, more-abundant signals to promote colonization.
Other non-LEE factors may also be important for colonization of EHEC at the RAJ. For example, curli and other fimbriae have been implicated as important for colonization of EHEC in cattle (66). This demonstrates how EHEC can utilize an array of complex mechanisms and signals to regulate genes required for colonization in cattle. Further identification of new potential signals and elucidation of the mechanisms used by EHEC to colonize its natural host will help develop better preventive strategies to reduce EHEC colonization at the RAJ and consequent shedding of EHEC into the environment and transmission to humans.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI077613, the Burroughs Wellcome Fund (V.S.), and the Robert A. Welch Foundation (GL625910) (J.R.F.). This work was supported, in part, by the Idaho Agriculture Experiment Station (C.J.H.) and by NIH grants P20-RR16454 (NCRR) (C.J.H.) and P20-GM103408 (NIGMS) (C.J.H.).
We thank Lonie Austin for technical assistance and animal handling and thank Brian Ahmer for plasmids PJLD1600 and pJLD500.
Footnotes
Published ahead of print 26 August 2013
REFERENCES
- 1.Karmali MA, Steele BT, Petric M, Lim C. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619–620 [DOI] [PubMed] [Google Scholar]
- 2.Karmali MA. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60–98 [DOI] [PubMed] [Google Scholar]
- 4.Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DG, Gally DL. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knutton S, Baldini MM, Kaper JB, McNeish AS. 1987. Role of plasmid-encoded adherence factors in adhesion of enteropathogenic Escherichia coli to HEp-2 cells. Infect. Immun. 55:78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng YK, Lai LC, McNamara BP, Donnenberg MS, Kaper JB. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4 [DOI] [PubMed] [Google Scholar]
- 10.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296–306 [DOI] [PubMed] [Google Scholar]
- 12.Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U. S. A. 92:7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott SJ, Krejany EO, Mellies JL, Robins-Browne RM, Sasakawa C, Kaper JB. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanack KJ, Crawford JA, Tatsuno I, Karmali MA, Kaper JB. 2005. SepZ/EspZ is secreted and translocated into HeLa cells by the enteropathogenic Escherichia coli type III secretion system. Infect. Immun. 73:4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny B, Jepson M. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579–590 [DOI] [PubMed] [Google Scholar]
- 17.McNamara BP, Donnenberg MS. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71–78 [DOI] [PubMed] [Google Scholar]
- 18.Tu X, Nisan I, Yona C, Hanski E, Rosenshine I. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595–606 [DOI] [PubMed] [Google Scholar]
- 19.Bustamante VH, Santana FJ, Calva E, Puente JL. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664–678 [DOI] [PubMed] [Google Scholar]
- 20.Shin S, Castanie-Cornet MP, Foster JW, Crawford JA, Brinkley C, Kaper JB. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133–1150 [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Chaudhuri RR, Constantinidou C, Hobman JL, Patel MD, Jones AC, Sarti D, Roe AJ, Vlisidou I, Shaw RK, Falciani F, Stevens MP, Gally DL, Knutton S, Frankel G, Penn CW, Pallen MJ. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp FC, Sperandio V. 2007. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 75:2432–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanamaru K, Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805–816 [DOI] [PubMed] [Google Scholar]
- 24.Macritchie DM, Ward JD, Nevesinjac AZ, Raivio TL. 2008. Activation of the Cpx envelope stress response down-regulates expression of several locus of enterocyte effacement-encoded genes in enteropathogenic Escherichia coli. Infect. Immun. 76:1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellies JL, Haack KR, Galligan DC. 2007. SOS regulation of the type III secretion system of enteropathogenic Escherichia coli. J. Bacteriol. 189:2863–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyoda S, Watanabe H. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150:2357–2571 [DOI] [PubMed] [Google Scholar]
- 27.Sharma VK, Zuerner RL. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott SJ, Sperandio V, Giron JA, Shin S, Mellies JL, Wainwright L, Hutcheson SW, McDaniel TK, Kaper JB. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sperandio V, Mellies JL, Delahay RM, Frankel G, Crawford JA, Nguyen W, Kaper JB. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781–793 [DOI] [PubMed] [Google Scholar]
- 30.Hughes DT, Terekhova DA, Liou L, Hovde CJ, Sahl JW, Patankar AV, Gonzalez JE, Edrington TS, Rasko DA, Sperandio V. 2010. Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U. S. A. 107:9831–9836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuqua C, Parsek MR, Greenberg EP. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439–468 [DOI] [PubMed] [Google Scholar]
- 34.Ahmer BM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F. 1998. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180:1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith JN, Dyszel JL, Soares JA, Ellermeier CD, Altier C, Lawhon SD, Adams LG, Konjufca V, Curtiss R, III, Slauch JM, Ahmer BM. 2008. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826. 10.1371/journal.pone.0002826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dyszel JL, Smith JN, Lucas DE, Soares JA, Swearingen MC, Vross MA, Young GM, Ahmer BM. 2010. Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice. J. Bacteriol. 192:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erickson DL, Nsereko VL, Morgavi DP, Selinger LB, Rode LM, Beauchemin KA. 2002. Evidence of quorum sensing in the rumen ecosystem: detection of N-acyl homoserine lactone autoinducers in ruminal contents. Can. J. Microbiol. 48:374–378 [DOI] [PubMed] [Google Scholar]
- 38.Cobbold RN, Hancock DD, Rice DH, Berg J, Stilborn R, Hovde CJ, Besser TE. 2007. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 73:1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice DH, Sheng HQ, Wynia SA, Hovde CJ. 2003. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J. Clin. Microbiol. 41:4924–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omisakin F, MacRae M, Ogden ID, Strachan NJ. 2003. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 69:2444–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chase-Topping ME, McKendrick IJ, Pearce MC, MacDonald P, Matthews L, Halliday J, Allison L, Fenlon D, Low JC, Gunn G, Woolhouse ME. 2007. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. J. Clin. Microbiol. 45:1594–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armstrong GL, Hollingsworth J, Morris JG., Jr 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29–51 [DOI] [PubMed] [Google Scholar]
- 43.Cody SH, Glynn MK, Farrar JA, Cairns KL, Griffin PM, Kobayashi J, Fyfe M, Hoffman R, King AS, Lewis JH, Swaminathan B, Bryant RG, Vugia DJ. 1999. An outbreak of Escherichia coli O157:H7 infection from unpasteurized commercial apple juice. Ann. Intern. Med. 130:202–209 [DOI] [PubMed] [Google Scholar]
- 44.Hilborn ED, Mermin JH, Mshar PA, Hadler JL, Voetsch A, Wojtkunski C, Swartz M, Mshar R, Lambert-Fair MA, Farrar JA, Glynn MK, Slutsker L. 1999. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Intern. Med. 159:1758–1764 [DOI] [PubMed] [Google Scholar]
- 45.Olsen SJ, Miller G, Breuer T, Kennedy M, Higgins C, Walford J, McKee G, Fox K, Bibb W, Mead P. 2002. A waterborne outbreak of Escherichia coli O157:H7 infections and hemolytic uremic syndrome: implications for rural water systems. Emerg. Infect. Dis. 8:370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spika JS, Parsons JE, Nordenberg D, Wells JG, Gunn RA, Blake PA. 1986. Hemolytic uremic syndrome and diarrhea associated with Escherichia coli O157:H7 in a day care center. J. Pediatr. 109:287–291 [DOI] [PubMed] [Google Scholar]
- 47.Carter AO, Borczyk AA, Carlson JA, Harvey B, Hockin JC, Karmali MA, Krishnan C, Korn DA, Lior H. 1987. A severe outbreak of Escherichia coli O157:H7–associated hemorrhagic colitis in a nursing home. N. Engl. J. Med. 317:1496–1500 [DOI] [PubMed] [Google Scholar]
- 48.Rowe PC, Orrbine E, Lior H, Wells GA, McLaine PN. 1993. Diarrhoea in close contacts as a risk factor for childhood haemolytic uraemic syndrome. The CPKDRC co-investigators. Epidemiol. Infect. 110:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walters M, Sperandio V. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74:5445–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Jr, Rinehart KL, Farrand SK. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U. S. A. 94:6036–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuqua C, Winans SC. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheng H, Davis MA, Knecht HJ, Hovde CJ. 2004. Rectal administration of Escherichia coli O157:H7: novel model for colonization of ruminants. Appl. Environ. Microbiol. 70:4588–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camps J, Pujol I, Ballester F, Joven J, Simo JM. 2011. Paraoxonases as potential antibiofilm agents: their relationship with quorum-sensing signals in Gram-negative bacteria. Antimicrob. Agents Chemother. 55:1325–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Draganov DI, Teiber JF, Speelman A, Osawa Y, Sunahara R, La Du BN. 2005. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 46:1239–1247 [DOI] [PubMed] [Google Scholar]
- 59.Yang F, Wang LH, Wang J, Dong YH, Hu JY, Zhang LH. 2005. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS Lett. 579:3713–3717 [DOI] [PubMed] [Google Scholar]
- 60.Swearingen MC, Sabag-Daigle A, Ahmer BM. 2013. Are there acyl-homoserine lactones within mammalian intestines? J. Bacteriol. 195:173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mackness B, Beltran-Debon R, Aragones G, Joven J, Camps J, Mackness M. 2010. Human tissue distribution of paraoxonases 1 and 2 mRNA. IUBMB Life 62:480–482 [DOI] [PubMed] [Google Scholar]
- 62.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 103:10420–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reading NC, Rasko DA, Torres AG, Sperandio V. 2009. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:5889–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100:8951–8956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyamoto T, Takahashi Y, Oohashi T, Sato K, Oikawa S. 2005. Bovine paraoxonase 1 activities in serum and distribution in lipoproteins. J. Vet. Med. Sci. 67:243–248 [DOI] [PubMed] [Google Scholar]
- 66.Farfan MJ, Torres AG. 2012. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 80:903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705–712 [DOI] [PubMed] [Google Scholar]