Abstract
Most bacterial pathogens require iron to grow and colonize host tissues. The Gram-negative bacterium Salmonella enterica serovar Typhimurium causes a natural systemic infection of mice that models acute and chronic human typhoid fever. S. Typhimurium resides in tissues within cells of the monocyte lineage, which limit pathogen access to iron, a mechanism of nutritional immunity. The primary ferric iron import system encoded by Salmonella is the siderophore ABC transporter FepBDGC. The Fep system has a known role in acute infection, but it is unclear whether ferric iron uptake or the ferric iron binding siderophores enterobactin and salmochelin are required for persistent infection. We defined the role of the Fep iron transporter and siderophores in the replication of Salmonella in macrophages and in mice that develop acute followed by persistent infections. Replication of wild-type and iron transporter mutant Salmonella strains was quantified in cultured macrophages, fecal pellets, and host tissues in mixed- and single-infection experiments. We show that deletion of fepB attenuated Salmonella replication and colonization within macrophages and mice. Additionally, the genes required to produce and transport enterobactin and salmochelin across the outer membrane receptors, fepA and iroN, are needed for colonization of all tissues examined. However, salmochelin appears to be more important than enterobactin in the colonization of the spleen and liver, both sites of dissemination. Thus, the FepBDGC ferric iron transporter and the siderophores enterobactin and salmochelin are required by Salmonella to evade nutritional immunity in macrophages and cause persistent infection in mice.
INTRODUCTION
In humans, Salmonella enterica serovars Typhi and Paratyphi A, B, and C cause typhoid fever, a systemic infection with mortality rates exceeding 15% if left untreated (1, 2). S. enterica serovar Typhimurium causes murine typhoid in mice, a disease that models both the acute and chronic stages of human typhoidal infection. Upon ingestion, Salmonella bacteria pass through the stomach and traverse the intestinal epithelial barrier to colonize Peyer's patches, mesenteric lymph nodes, spleen, and liver (3, 4). Salmonella bacteria reside within cells of the monocyte lineage, typically macrophages that contain the bacteria within a specialized membrane-bound compartment called a Salmonella-containing vesicle (5, 6).
Iron is an essential nutrient for animals. However, iron must be tightly regulated, as it can both accept and donate electrons, leading to the generation of damaging free radicals (7). Dietary ferric iron (Fe3+) is reduced to ferrous iron (Fe2+) by a ferrireductase (DcytB) on the luminal side of intestinal epithelial cells (8, 9). Ferrous iron is then taken up by enterocytes via DMT1/Nramp2/Slc11A2, exported into the blood via ferroportin 1, and oxidized to ferric iron by hephaestin on the basolateral membrane (10). Blood ferric iron is captured by transferrin, a high-affinity iron binding protein (Kd [dissociation constant] = 1022 M−1) (11). In tissues, cells endocytose transferrin, and ferric iron is released upon endosome acidification. The released ferric iron is then reduced (12) and transported to the cytosol (13). Cytosolic iron is sequestered by ferritin, which accounts for about 16% of iron stores in the body (14).
A major host defense against infection is nutritional immunity, the sequestration of essential molecules, including metals, to prevent pathogen outgrowth (15). Sequestration of iron is an effective antimicrobial defense because iron is a cofactor required for crucial processes, including energy production and DNA replication. For instance, during acute or chronic immune activation, ferritin-bound iron accumulates in macrophages, a process that withholds iron from extracellular pathogens and is characteristic of the anemia of chronic disease (14, 16). In contrast, macrophages infected with Salmonella increase the export of iron, and the spleen and liver of Salmonella-infected mice do not accumulate iron (17–19). Decreased iron concentrations in macrophages limit Salmonella replication (20, 21), highlighting the importance of delineating how Salmonella acquires iron under such starved conditions.
Salmonella acquires ferric iron by secreting the catecholate siderophores enterobactin and salmochelin. Once bound to iron, enterobactin and salmochelin transit the bacterial outer membrane receptors FepA and IroN, respectively, in a TonB-dependent manner (22). Enterobactin and salmochelin then bind to FepB in the periplasm for import through FepDGC (23). During infection, macrophages respond to gamma interferon (IFN-γ) by increasing the secretion of lipocalin-2, a siderophore-capturing protein (20, 24–27). Lipocalin-2 binds enterobactin but not salmochelin, a glycosylated derivative of enterobactin that gives Salmonella a selective growth advantage in the intestine (26).
Whether Salmonella requires access to ferric iron has been examined in mice that lack functional Nramp1/Slc11a1, a ferrous iron transporter in the membrane of the phagosome. Nramp1-deficient mice have dysregulated iron metabolism and are exquisitely sensitive to microbial pathogens that reside within vesicles, including Salmonella, Mycobacterium tuberculosis, and Leishmania species. Salmonella strains lacking fepA and iroN have no apparent phenotype in Nramp1−/− mice in single and mixed infections (23, 28) or in survival assays upon intragastric or intraperitoneal inoculation compared with the wild type (29). Together, these studies indicate that enterobactin and salmochelin uptake via FepA and/or IroN is not essential for Salmonella infection of Nramp1−/− mice.
Nramp1+/+ mice, such as the Sv129S6 strain, survive acute infection and develop chronic infection. In these animals, the synthesis and secretion of salmochelin are required for Salmonella virulence upon intraperitoneal inoculation, as measured by survival assays (26, 30). Intraperitoneal inoculation allows bacteria to bypass the gastrointestinal tract and directly access the spleen (31). However, Bearson et al. demonstrated that upon intranasal inoculation of piglets, there was no colonization defect of a triple mutant lacking fepA, iroN, and cirA compared to the wild-type strain (32). CirA is a third outer membrane receptor for siderophore-mediated iron uptake via FepBDGC (27, 28, 33). However, CirA does not directly bind enterobactin or salmochelin but rather captures catecholate breakdown products containing ferric iron. In the work presented here, we establish the requirement of Salmonella for FepB and the enterobactin and salmochelin binding proteins FepA and IroN for gastric and deep-tissue colonization of Nramp1+/+ mice upon orogastric inoculation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Typhimurium wild-type strain SL1344 (34) and mutant derivatives were grown overnight at 37°C with aeration in Luria-Bertani (LB) medium prior to infections. Antibiotics were used at the following concentrations: streptomycin at 30 μg/ml and kanamycin at 30 μg/ml.
Deletion mutants in genes involved in iron uptake and metabolism were constructed by using lambda Red methodology (35, 36). ΔiroN, ΔfepA, and ΔentC mutants of background strain 14028 were generated previously by using a similar methodology (36), and the ΔfepB mutant was generated in this study by using primers fepB fwd (5′-GCGCTAACCTAAGAGTAAAACGTCGCTCTGTCAACTGTGTAGGCTGGAGCTGCTTC-3′) and fepB rev (5′-AATCGGTCTGGTCAGTCGGATAAGACTCCGATAAGGCATATGAATATCCTCCTTAG-3′). PCR confirmed the insertion of the cassette by using site-specific and internal kanamycin primers. P22 phage lysates were prepared from each mutant strain and used to transduce each mutation into wild-type strain SL1344.
Restoration of fepB was achieved by P22 phage transduction of a wild-type copy into the fepB-deleted strain. To enrich for colonies in which fepB was restored to the wild type, P22-exposed bacteria were first incubated in M9 minimal medium for 6 h. During this time, the wild-type strain reaches an optical density at 600 nm (OD600) of ∼0.5, while the ΔfepB strain is still in the lag phase. Cells were diluted and plated onto streptomycin-containing LB plates. Colonies were replica plated onto kanamycin-containing LB plates, and those that grew on streptomycin but not kanamycin were tested and confirmed for restoration of a wild-type fepB copy by PCR.
Measurement of growth in vitro.
Cultures grown overnight were diluted to an OD600 of 0.01 in 200 μl of LB or M9 minimal medium (M9) with the vehicle control, ferric chloride (Sigma), or 2,2′-dipyridyl (Sigma). Bacteria were grown in 96-well plates with shaking in a Synergy2 plate reader (BioTek) at 37°C for 17 h. At 20-min intervals, the OD600 was recorded. Addition of 25 to 400 μM deferasirox in broth is not practical because the presence of deferasirox enhances Salmonella growth under these conditions for unknown reasons (data not shown).
Murine infection.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (37), and all protocols were approved by the University of Colorado Institutional Committees for Biosafety and for Animal Care and Use. For competitive infection studies, 7-week-old male and female 129SvEvTac (Nramp1+/+) mice (Taconic Laboratories) bred in-house were fasted for 2 h prior to orogastric inoculation with a total of 2 × 109 CFU of a 1:1 mixture of streptomycin-resistant wild-type (1 × 109 CFU) and kanamycin-resistant mutant (1 × 109 CFU) bacteria in 100 μl phosphate-buffered saline (PBS), as verified by plating for CFU on selective LB agar. For single-infection experiments, mice were fasted for 2 h and orogastrically inoculated with 1 × 109 bacteria in 100 μl PBS. The infectious dose was verified by plating for CFU on selective LB agar. During the period in which poor grooming may be observed, mice were monitored twice daily. Cages containing “scruffy” mice were supplied with food and water in small dishes on the floor of the cage to alleviate suffering from dehydration and malnourishment. Two weeks after inoculation, infected animals were euthanized by CO2 asphyxiation, followed by cervical dislocation. Spleen, liver, mesenteric lymph nodes, Peyer's patches, and cecum were collected; homogenized in 1 ml PBS; and then serially diluted for plating to enumerate CFU. Competitive indexes (CIs) were calculated as follows: (CFUwild type/CFUmutant) output/(CFUwild type/CFUmutant) input.
Gentamicin protection assays.
RAW264.7 cells stably transfected with the pHbA-1-neo expression plasmid containing the full-length Slc11a1 cDNA (25) were seeded at 1.5 × 105 cells per well in poly-l-lysine-coated 24-well tissue culture plates. Cells were activated with 20 ng/ml lipopolysaccharide (LPS) (S. enterica serovar Typhimurium LPS; Sigma-Aldrich) and 20 U/ml IFN-γ (PeproTech) for 18 h. Bacteria opsonized with normal mouse serum (Sigma) were added to macrophages at a multiplicity of infection of 10:1 (bacteria to macrophage). After 30 min, free and loosely adherent bacteria were removed by washing with PBS twice, and cells were incubated for a further 1.5 h at 37°C in fresh medium supplemented with gentamicin (100 μg/ml) to kill extracellular bacteria. Medium was then exchanged for medium supplemented with 10 μg/ml gentamicin to inhibit extracellular bacterial growth. At 2, 18, and 24 h, wells were washed twice with prewarmed PBS, incubated with 1% Triton X-100 for 5 min, and lysed, and serial dilutions were plated to enumerate CFU.
Statistics.
P values were calculated with GraphPad Prism 5 (GraphPad Software Inc.) and considered significant if the P value was <0.05. For nonparametric data, Wilcoxon signed-rank or Mann-Whitney tests were used. Otherwise, one-sample t tests or Student's t tests were used.
RESULTS
Salmonella fepB ferric iron transporter mutants colonize mice poorly in competitive infections.
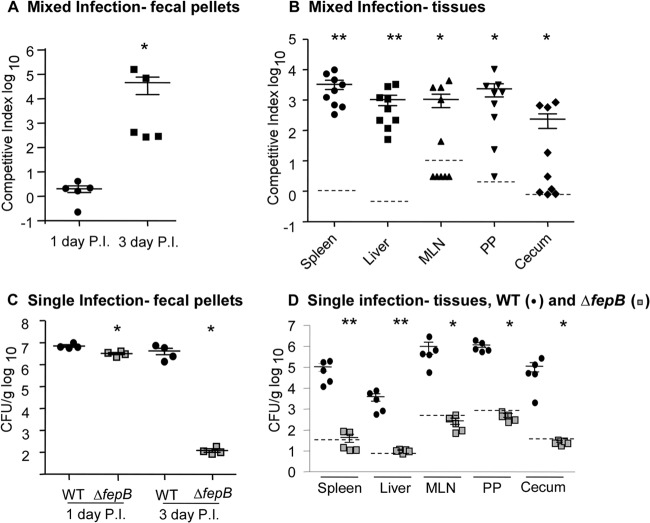
We constructed a mutant Salmonella Typhimurium SL1344 strain with a deletion of fepB, encoding a protein crucial for ferric iron uptake. To establish whether FepB is needed for Salmonella survival and growth in tissue, mixed-infection experiments were performed in Sv129S6 (Nramp1+/+) mice. These mice become chronically infected with Salmonella and can be used to monitor infection long term (38). We evaluated the number of wild-type organisms and the number of fepB mutant organisms in feces, a close proxy for the number of organisms in the cecum (39), at 1 and 3 days postinfection. At 1 day postinfection, equivalent numbers of wild-type and ΔfepB mutant bacteria were recovered from fecal pellets, indicating that the ΔfepB mutant colonizes the intestine well initially (Fig. 1A). However, by 3 days postinfection, the ΔfepB mutant had 100- to 10,000-fold-decreased colonization relative to the wild-type organism (Fig. 1A).
Fig 1.
Salmonella requires the FepB ferric iron transporter for persistent infection of Sv129S6 (Nramp1+/+) mice. (A and B) Mice were orogastrically inoculated with a 1:1 mixture of wild-type (WT) and ΔfepB strains. (A) One and 3 days postinfection (P.I.), fecal samples were collected and homogenized to determine bacterial loads. ∗, P = 0.003 compared with the null hypothesis. (B) At 2 weeks, mice were sacrificed, and the spleen, liver, mesenteric lymph nodes (MLN), Peyer's patches (PP), and cecum were immediately homogenized and plated to determine bacterial loads. Dashed lines indicate the limit of detection based on average tissue weight. Each symbol represents one mouse. ∗, P < 0.05; ∗∗, P < 0.00001 compared with the null hypothesis. (C and D) Mice were orogastrically inoculated with the wild-type or ΔfepB strain. (C) One and 3 days postinfection, fecal samples were collected and homogenized to determine bacterial loads in the intestine. ∗, P = 0.03 versus the wild type. (D) After 2 weeks, mice were sacrificed, and the spleen, liver, mesenteric lymph nodes, Peyer's patches, and cecum were immediately homogenized and plated to determine bacterial loads. Each symbol represents one mouse (n = 5). ∗, P < 0.05; ∗∗, P < 0.001 versus the wild type for each organ. Dashed lines indicate the limit of detection based on average tissue weight.
At 2 weeks postinfection, mice were euthanized, and tissues were collected to enumerate CFU. In gut-associated tissues, including the cecum, Peyer's patches, and mesenteric lymph nodes, the numbers of ΔfepB strain bacteria were 100-fold reduced relative to the numbers of wild-type organisms (Fig. 1B). In the spleen and liver, the phenotype was more severe, as ΔfepB colonies were recovered 1,000 times less frequently than wild-type colonies. These data suggest that ferric iron transport by the Fep system is required for tissue colonization in the context of coinfection with wild-type bacteria.
Salmonella requires the FepB ferric iron transporter for persistent infection of mice.
We used single-infection experiments in Sv129S6 (Nramp1+/+) mice to establish whether the ferric iron transporter mutant colonizes tissues normally in the absence of the wild-type organism. At 1 day postinfection, the ΔfepB mutant was recovered at slightly but significantly lower numbers from fecal pellets of mice infected with the ΔfepB mutant than from mice infected with the wild-type organism (Fig. 1C). Within 3 days postinfection, the difference in recovery from fecal pellets was >10,000-fold, with mice infected with the ΔfepB mutant exhibiting a strong colonization defect. The large difference in relative strain recovery at 1 and 3 days suggests that the ΔfepB mutant initially colonizes the intestine well. At 2 weeks postinfection, there were 103 to 106 CFU per gram of wild-type Salmonella bacteria in each tissue examined, while tissues of mice infected with the ΔfepB mutant lacked detectable colonization (Fig. 1D). These results, in combination with the mixed-infection experiments, clearly demonstrate that Salmonella infection of Nramp1+/+ mice requires the FepB ferric iron transporter in all tissues examined.
Salmonella requires the FepB ferric iron transporter for replication in macrophages.
During persistent infection, S. Typhimurium resides within macrophages (5, 38). To establish whether FepB is required for Salmonella replication in macrophages, macrophage-like Nramp1+/+ tissue culture cells were individually infected with equivalent numbers of wild-type or ΔfepB mutant Salmonella bacteria. Infected cells were lysed, and CFU were enumerated over a time course (Fig. 2A). The wild-type strain replicated during the first 18 to 24 h, while CFU recovered for the ΔfepB mutant strain remained similar throughout the experiment. Expression of fepB on a plasmid is toxic to Escherichia coli (40) and Salmonella (data not shown). Therefore, to confirm that fepB is needed for replication in macrophages, wild-type fepB was restored in the fepB-deleted strain by phage transduction. Replication of the restored strain in macrophages was comparable to that of the wild-type strain. Macrophages were also treated with 50 μM deferasirox, a ferric iron-specific chelator, over a time course of infection. At each time point examined, replication of the wild type in the presence of deferasirox was significantly reduced compared to the replication of bacteria in the presence of the vehicle alone (Fig. 2B), similar to results obtained by Nairz et al. (41). These results demonstrate that ferric iron transport via FepB is required for replication of Salmonella in macrophages.
Fig 2.
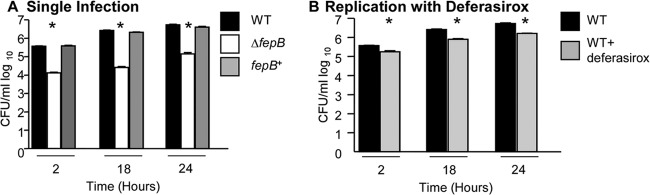
Salmonella requires ferric iron (fepB) for growth in macrophages. (A) Cell culture macrophages (Nramp1+) were infected with the wild-type, ΔfepB, or fepB-restored (fepB+) strain at a multiplicity of infection of 10. At 2, 18, and 24 h postinfection, macrophages were lysed, serially diluted, and plated to determine numbers of CFU/ml. Error bars indicate standard errors of the means (n ≥ 3 experiments). ∗, P < 0.05 versus the wild type. (B) Cell culture macrophages (Nramp1+) were infected with the wild type in the absence or presence of 50 μM deferasirox at a multiplicity of infection of 10. At 2, 18, and 24 h postinfection, macrophages were lysed, serially diluted, and plated to determine numbers of CFU/ml. Error bars indicate standard errors of the means (n ≥ 3 experiments). ∗, P < 0.05 versus the wild type.
Salmonella can utilize ferric iron via FepB as its sole iron source.
Next, we determined whether ferric iron is sufficient to support Salmonella growth in broth. Control experiments demonstrated no significant growth differences between the ΔfepB mutant and the wild-type strain in nutrient-rich medium (LB medium) at 37°C over 16 h (Fig. 3A). The addition of the iron chelator 2,2′-dipyridyl, which has a similar binding affinity for ferrous and ferric iron (42), prevented the growth of both strains similarly. In M9 minimal medium without added iron, the wild-type strain grew significantly better than the ΔfepB mutant strain (Fig. 3B). Treatment with 100 but not 50 μM ferric chloride significantly increased growth of the wild-type strain. Growth of the ΔfepB mutant strain was not improved by the addition of ferric chloride. Chelation of iron with 2,2′-dipyridyl in M9 medium reduced the growth of both wild-type Salmonella and the ΔfepB strain, as observed in LB medium. Collectively, these observations suggest that Salmonella can utilize ferric iron acquired via the catecholate transporter FepB as its sole iron source.
Fig 3.
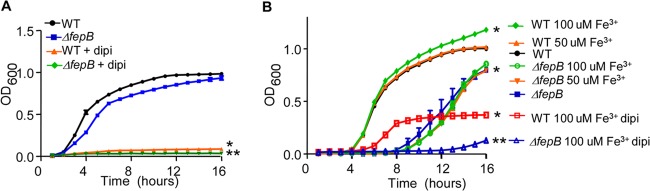
Salmonella can grow with ferric iron as the sole iron source. (A) The indicated strains were grown in LB medium supplemented with 200 μM iron chelator 2,2′-dipyridyl (dipi). The optical density at 600 nm was monitored for 16 h. Error bars indicate standard deviations (n ≥ 3 experiments). ∗, P < 0.05 versus the wild type; ∗∗, P < 0.05 versus the ΔfepB mutant. (B) The indicated strains were grown in M9 minimal medium supplemented with ferric chloride (Fe3+) and/or 200 μM dipyridyl. The optical density at 600 nm was monitored for 16 h. Error bars indicate standard deviations (n ≥ 3 experiments). ∗, P < 0.05 versus the wild type; ∗∗, P < 0.05 versus the ΔfepB mutant.
Salmonella requires Fe3+ binding siderophore uptake via FepA and IroN in mice.
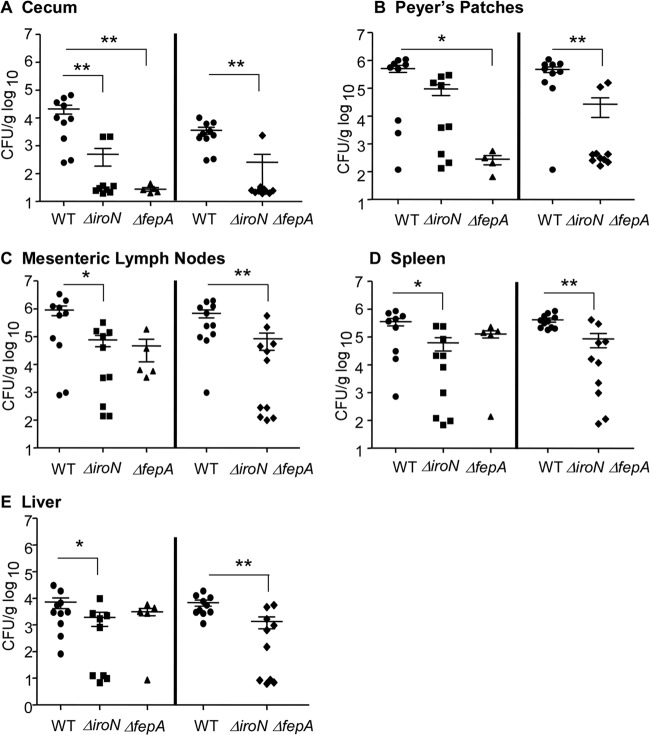
Salmonella acquires ferric iron by producing and secreting the siderophores enterobactin and salmochelin, which, once bound to iron, are transported through the bacterial outer membrane receptor FepA (enterobactin) or IroN (enterobactin or salmochelin) and bind FepB in the periplasm. To determine whether ferric iron uptake via one or both of these siderophores is necessary for persistent infection, mice were orogastrically inoculated with the individual wild-type, ΔfepA, or ΔiroN strain or with a ΔiroN ΔfepA double mutant strain. After 2 weeks, mice were sacrificed, and tissue colonization was determined. Tissue colonization by ΔfepA and ΔiroN single and double mutants was significantly reduced compared to that by the wild-type strain in the cecum (Fig. 4A). The ΔfepA mutant also had a significant colonization defect in Peyer's patches compared to wild-type Salmonella (Fig. 4B). In contrast, the ΔiroN mutant strain colonized the mesenteric lymph nodes, spleen, and liver poorly (Fig. 4C and D). Deletion of both iroN and fepA resulted in a severe colonization defect in the mesenteric lymph nodes, spleen, and liver compared to wild-type bacteria. These results suggest that both FepA and IroN are utilized in all tissues but that there may be a greater requirement for IroN in deep tissues during Salmonella infection of Nramp1+/+ mice.
Fig 4.
Salmonella requires enterobactin and salmochelin during persistent infection of mice. In separate experiments (left and right), mice were orogastrically inoculated with the wild-type, ΔiroN, ΔfepA, or ΔiroN ΔfepA strain. After 2 weeks, mice were sacrificed, and the cecum (A), Peyer's patches (B), mesenteric lymph nodes (C), spleen (D), and liver (E) were immediately homogenized and plated to determine bacterial loads. Each symbol represents one mouse (n = 5 to 10). ∗, P < 0.05 versus the wild type; ∗∗, P < 0.003 versus the wild type.
fepA or iroN is required for growth in macrophages.
To establish whether fepA and/or iroN is required for Salmonella replication in macrophages, macrophage-like Nramp1+/+ tissue culture cells were singly infected with the Salmonella wild-type, ΔfepA, ΔiroN, or ΔiroN ΔfepA mutant strain. Over a time course of infection, CFU enumeration indicated that deletion of ΔfepA or ΔiroN did not significantly affect the ability of Salmonella to replicate in macrophages compared to wild-type bacteria. However, deletion of both ΔiroN and ΔfepA prevented Salmonella replication within macrophages at between 2 and 24 h postinfection (Fig. 5). These results demonstrate that Salmonella requires either iroN (salmochelin or enterobactin outer membrane uptake receptor) or fepA (enterobactin outer membrane uptake receptor) for replication in macrophages.
Fig 5.
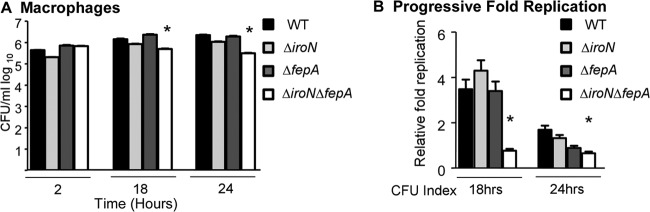
Salmonella requires either fepA or iroN for growth in macrophages. Cell culture macrophages (Nramp1+) were infected with the wild-type, ΔiroN, ΔfepA, or ΔiroN ΔfepA strain at a multiplicity of infection of 10. At 2, 18, and 24 h postinfection, macrophages were lysed, serially diluted, and plated to determine numbers of CFU/ml. In panel B, the CFU index is defined as the number of CFU at 18 h relative to the number of CFU at 2 h (18 h) and the number of CFU at 24 h relative to the number of CFU at 18 h (24 h). Error bars indicate standard deviations (n ≥ 3 experiments). ∗, P < 0.05 versus the wild type.
Salmonella requires siderophore synthesis in mice.
Bacteria overproduce siderophores in strain backgrounds lacking siderophore import genes such as iroN and fepA (28). Excessive siderophore production could result in local iron starvation and thus poor tissue colonization in vivo. To determine whether the in vivo growth defects observed for the ΔiroN and ΔfepA strains reflected enhanced siderophore production, a strain unable to import or produce enterobactin and salmochelin (ΔiroN ΔfepA ΔentC) was generated. Mice orogastrically inoculated with individual wild-type or ΔiroN ΔfepA ΔentC bacteria were sacrificed at 2 weeks postinfection, and tissue colonization was determined (Fig. 6). Tissue colonization by the triple mutant strain was significantly reduced compared to that of the wild-type strain in the cecum, Peyer's patches, mesenteric lymph nodes, and spleen, although the difference in liver colonization was not significant. These observations indicate that the reduced tissue colonization observed for ΔfepA and ΔiroN mutants is not dependent upon siderophore overproduction.
Fig 6.
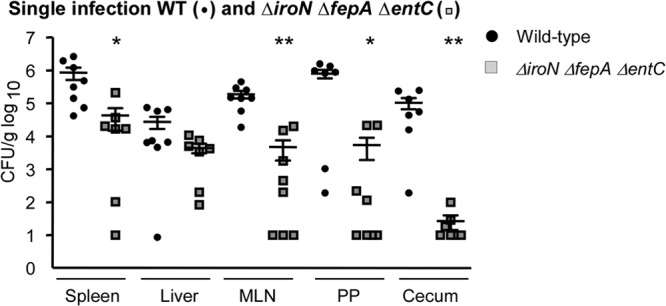
Salmonella requires siderophore synthesis in mice. Mice were orogastrically inoculated with the wild-type or ΔiroN ΔfepA ΔentC strain. After 2 weeks, mice were sacrificed, and the spleen, liver, mesenteric lymph nodes, Peyer's patches, and cecum were immediately homogenized and plated to determine bacterial loads. Each symbol represents one mouse (n ≥ 7). ∗, P < 0.006 versus the wild type for each organ; ∗∗, P < 0.0006 versus the wild type for each organ.
DISCUSSION
Salmonella encodes transporters for both ferric and ferrous iron. Ferrous iron is taken up by FeoB (43) and by divalent cation transporters (MntH and SitA) (44, 45). FeoB and SitA have known roles in BALB/c Nramp1−/− mice during acute infection (43, 44). Ferric iron is acquired upon the import of iron-bound siderophores through FepA and IroN and siderophore capture in the periplasm by FepB (23). Previous work established that fepA and iroN are dispensable for infection of BALB/c Nramp1−/− mice (23, 28, 29). We demonstrate that siderophore capture and import via fepB are required for Salmonella survival and replication within macrophages as well as for colonization of mice upon orogastric inoculation. In addition, the ability to produce and/or take up either the enterobactin or salmochelin siderophores is needed for Salmonella colonization of both gastric and deep tissues in Sv129S6 Nramp1+/+ mice. Our observations are consistent with those of Gorbacheva et al., who found that an S. Typhi mutant strain lacking the ability to synthesize enterobactin (and salmochelin) failed to replicate in human macrophages (46). The growth defect observed in strains lacking fepB was more severe than that in strains unable to produce or import either siderophore. This may reflect that FepBDGC also takes up derivatives of catecholates that are imported by CirA (27, 28, 33). Overall, the data suggest that S. Typhimurium requires ferric iron during persistent infection of mice. Moreover, the results from infection of mice with single mutant strains indicate that enterobactin acquisition through FepA is important for colonization of the cecum and Peyer's patches and that enterobactin or salmochelin import via IroN is important for colonization of all tissues examined. Our studies thus provide an experimental foundation to make hypotheses about the temporal-spatial requirements of siderophores during different stages of infection.
Mice that lack hemochromatosis gene alleles (Hfe) have increased resistance to Salmonella infection. This is due to decreased levels of macrophage iron via an enhanced production of ferroportin (47) as well as increased iron sequestration via augmented lipocalin-2 production, which scavenges enterobactin (25). These results are in concordance with our current studies indicating a preferential uptake of ferric iron via IroN and FepA during infection. Because hemochromatosis studies have been performed in Nramp1−/− mice, it will be of interest to determine if similar mechanisms occur in Nramp1+/+ mice, which tightly regulate iron stores and mimic the acute-to-chronic progression of human typhoid fever.
Patients with β-thalassemia and thus ferric iron overload, including within macrophages, have increased susceptibility to bacterial infection. Mice with experimentally induced iron overload have significantly enhanced Salmonella growth rates in all tissues examined (48, 49). Dysregulation of hepcidin, a peptide that normally limits excess iron absorption, leads to iron overload and increased susceptibility to Salmonella infection (50). The mechanism by which iron overload increases susceptibility to Salmonella infection remains to be fully resolved, but the results of previous studies and the current work indicate that iron overload may provide abundant stores of ferric iron for bacterial utilization.
There are at least two pathways by which Salmonella may access ferric iron in macrophages. First, ferric iron is transported to different tissues by the host protein transferrin, which binds cognate receptors on cells, including macrophages (51). Host transferrin receptor expression is increased in response to many intracellular pathogens, such as Francisella tularensis, Mycobacterium tuberculosis, and Ehrlichia (52–54). Upon acidification of transferrin-containing endosomes, ferric iron is released from transferrin and may be accessed by these pathogens. In Nramp1−/− macrophages, Salmonella does not require transferrin expression for successful intracellular survival (52). However, Nramp1+ macrophages limit Salmonella access to iron (20, 25). Therefore, Salmonella may, like other pathogens that live within vesicles, intercept transferrin-containing vesicles to obtain ferric iron. Ferric iron released from transferrin can alternatively be transported to the cytosol and stored within ferritin. Some cytosolic microbes, such as Neisseria meningitidis and Listeria monocytogenes, utilize ferritin as an iron source through mechanisms that directly degrade ferritin pools in the cytosol (55, 56). Since Salmonella is typically an intravesicular pathogen in macrophages, it may not be able to directly access cytosolic ferritin. However, iron starvation and cellular stress induce autophagy of ferritin, followed be ferritin degradation in vesicles (57–59). Salmonella has been demonstrated to induce autophagy in macrophages (60, 61) and can also direct vesicular trafficking events (62). Thus, one hypothesis is that Salmonella facilitates fusion of the vesicle where it resides with ferritin-containing vesicles and then uses enterobactin or salmochelin to remove Fe3+ from the cytosolic storage protein ferritin. The addition of exogenous iron-loaded ferritin to Nramp1−/− macrophage-like cells increases Salmonella survival, consistent with this hypothesis (63).
Together, our studies combined with others highlight the particular importance of ferric iron in both nutritional immunity and Salmonella survival in the host. Salmonella has successfully evolved ferric iron-specific siderophores, enterobactin and salmochelin, that may be especially suited to take advantage of intracellular ferric iron storage pools. The current work also indicates that Salmonella may utilize siderophores differentially based upon tissue localization. Modulation of host iron regulators or the preferentially utilized Salmonella iron uptake systems has the potential to become pivotal in the development of new pharmacological approaches to control and/or prevent disease.
ACKNOWLEDGMENTS
We thank all the members of the Detweiler laboratory, M. Carolina Pilonieta, Erin McDonald, Eugenia Silva-Herzog, Christopher English, Garth Kreitz, and Angelika Krivenko, for helpful discussions and technical help over the course of this project.
This work was supported by NIH grants 1F32AI094766 (T.A.N.), AI072492, and AI095395 (C.S.D.).
We declare that there are no conflicts of interest.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 19 August 2013
REFERENCES
- 1.Miller SI, Pegues DA. 2009. Salmonella species, including Salmonella typhi, p 2887–2904 In Mandell GL, Bennett JE, Dolin R.(ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 7th ed. Churchill Livingstone, Philadelphia, PA [Google Scholar]
- 2.Ohl ME, Miller SI. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259–274 [DOI] [PubMed] [Google Scholar]
- 3.Baumler AJ, Tsolis RM, Heffron F. 1996. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64:1862–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones BD, Ghori N, Falkow S. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nix RN, Altschuler SE, Henson PM, Detweiler CS. 2007. Hemophagocytic macrophages harbor Salmonella enterica during persistent infection. PLoS Pathog. 3:e193. 10.1371/journal.ppat.0030193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva-Herzog E, Detweiler CS. 2010. Salmonella enterica replication in hemophagocytic macrophages requires two type three secretion systems. Infect. Immun. 78:3369–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenton HJH. 1894. Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 65:899–911 [Google Scholar]
- 8.Latunde-Dada GO, Van der Westhuizen J, Vulpe CD, Anderson GJ, Simpson RJ, McKie AT. 2002. Molecular and functional roles of duodenal cytochrome B (Dcytb) in iron metabolism. Blood Cells Mol. Dis. 29:356–360 [DOI] [PubMed] [Google Scholar]
- 9.McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, Peters TJ, Raja KB, Shirali S, Hediger MA, Farzaneh F, Simpson RJ. 2001. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 291:1755–1759 [DOI] [PubMed] [Google Scholar]
- 10.Vulpe CD, Kuo YM, Murphy TL, Cowley L, Askwith C, Libina N, Gitschier J, Anderson GJ. 1999. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 21:195–199 [DOI] [PubMed] [Google Scholar]
- 11.Aisen P, Listowsky I. 1980. Iron transport and storage proteins. Annu. Rev. Biochem. 49:357–393 [DOI] [PubMed] [Google Scholar]
- 12.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. 2005. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat. Genet. 37:1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su MA, Trenor CC, Fleming JC, Fleming MD, Andrews NC. 1998. The G185R mutation disrupts function of the iron transporter Nramp2. Blood 92:2157–2163 [PubMed] [Google Scholar]
- 14.Gkouvatsos K, Papanikolaou G, Pantopoulos K. 2012. Regulation of iron transport and the role of transferrin. Biochim. Biophys. Acta 1820:188–202 [DOI] [PubMed] [Google Scholar]
- 15.Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14:218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss G, Goodnough LT. 2005. Anemia of chronic disease. N. Engl. J. Med. 352:1011–1023 [DOI] [PubMed] [Google Scholar]
- 17.Brown DE, Libby S, Moreland SM, McCoy MW, Brabb T, Stepanek A, Fang FC, Detweiler CS. 27 February 2013. Salmonella enterica causes more severe inflammatory disease in C57/BL6 Nramp1G169 mice than Sv129S6 mice. Vet. Pathol. [Epub ahead of print.] 10.1177/0300985813478213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nairz M, Fritsche G, Crouch ML, Barton HC, Fang FC, Weiss G. 2009. Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell. Microbiol. 11:1365–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown DE, McCoy MW, Pilonieta MC, Nix RN, Detweiler CS. 2010. Chronic murine typhoid fever is a natural model of secondary hemophagocytic lymphohistiocytosis. PLoS One 5:e9441. 10.1371/journal.pone.0009441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nairz M, Fritsche G, Brunner P, Talasz H, Hantke K, Weiss G. 2008. Interferon-gamma limits the availability of iron for intramacrophage Salmonella typhimurium. Eur. J. Immunol. 38:1923–1936 [DOI] [PubMed] [Google Scholar]
- 21.Chlosta S, Fishman DS, Harrington L, Johnson EE, Knutson MD, Wessling-Resnick M, Cherayil BJ. 2006. The iron efflux protein ferroportin regulates the intracellular growth of Salmonella enterica. Infect. Immun. 74:3065–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skare JT, Ahmer BMM, Seachord CL, Darveau RP, Postle K. 1993. Energy transduction between membranes: TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302–16308 [PubMed] [Google Scholar]
- 23.Rabsch W, Voigt W, Reissbrodt R, Tsolis RM, Bäumler AJ. 1999. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J. Bacteriol. 181:3610–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917–921 [DOI] [PubMed] [Google Scholar]
- 25.Fritsche G, Nairz M, Libby SJ, Fang FC, Weiss G. 2012. Slc11a1 (Nramp1) impairs growth of Salmonella enterica serovar Typhimurium in macrophages via stimulation of lipocalin-2 expression. J. Leukoc. Biol. 92:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hantke K, Nicholson G, Rabsch W, Winkelmann G. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A. 100:3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabsch W, Methner U, Voigt W, Tschäpe H, Reissbrodt R, Williams PH. 2003. Role of receptor proteins for enterobactin and 2,3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 71:6953–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams PH, Rabsch W, Methner U, Voigt W, Tschäpe H, Reissbrodt R. 2006. Catecholate receptor proteins in Salmonella enterica: role in virulence and implications for vaccine development. Vaccine 24:3840–3844 [DOI] [PubMed] [Google Scholar]
- 30.Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. 2008. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 67:971–983 [DOI] [PubMed] [Google Scholar]
- 31.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400–403 [DOI] [PubMed] [Google Scholar]
- 32.Bearson BL, Bearson SM, Uthe JJ, Dowd SE, Houghton JO, Lee I, Toscano MJ, Lay DC., Jr 2008. Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect. 10:807–816 [DOI] [PubMed] [Google Scholar]
- 33.Hantke K. 1990. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol. Lett. 67:5–8 [DOI] [PubMed] [Google Scholar]
- 34.Merritt FF, Smith BP, Reina-Guerra M, Habasha F, Johnson E. 1984. Relationship of cutaneous delayed hypersensitivity to protection from challenge exposure with Salmonella typhimurium in calves. Am. J. Vet. Res. 45:1081–1085 [PubMed] [Google Scholar]
- 35.Datsenko KA, Wanner B. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. 10.1371/journal.ppat.1000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 38.Monack DM, Bouley DM, Falkow S. 2004. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J. Exp. Med. 199:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivula CP, Bogomolnaya LM, Andrews-Polymenis HL. 2008. A comparison of cecal colonization of Salmonella enterica serotype Typhimurium in white leghorn chicks and Salmonella-resistant mice. BMC Microbiol. 8:182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 41.Nairz M, Schleicher U, Schroll A, Sonnweber T, Theurl I, Ludwiczek S, Talasz H, Brandacher G, Moser PL, Muckenthaler MU, Fang FC, Bogdan C, Weiss G. 2013. Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J. Exp. Med. 210:855–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou T, Ma Y, Kong X, Hider RC. 2002. Design of iron chelators with therapeutic application. Coord. Chem. Rev. 232:151–171 [DOI] [PubMed] [Google Scholar]
- 43.Tsolis RM, Bäumler AJ, Heffron F, Stojiljkovic I. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella Typhimurium in the mouse. Infect. Immun. 64:4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer E, Bergevin I, Malo D, Gros P, Cellier MF. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janakiraman A, Slauch JM. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146–1155 [DOI] [PubMed] [Google Scholar]
- 46.Gorbacheva VY, Faundez G, Godfrey HP, Cabello FC. 2001. Restricted growth of ent(−) and tonB mutants of Salmonella enterica serovar Typhi in human Mono Mac 6 monocytic cells. FEMS Microbiol. Lett. 196:7–11 [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Johnson EE, Shi HN, Walker WA, Wessling-Resnick M, Cherayil BJ. 2008. Attenuated inflammatory responses in hemochromatosis reveal a role for iron in the regulation of macrophage cytokine translation. J. Immunol. 181:2723–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caroline L, Kozinn PJ. 1969. Infection and iron overload in thalassemia. Ann. N. Y. Acad. Sci. 165:148–155 [DOI] [PubMed] [Google Scholar]
- 49.Sawatzki G, Hoffmann FA, Kubanek B. 1983. Acute iron overload in mice: pathogenesis of Salmonella typhimurium infection. Infect. Immun. 39:659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuki KE, Eva MM, Richer E, Chung D, Paquet M, Cellier M, Canonne-Hergaux F, Vaulont S, Vidal SM, Malo D. 2013. Suppression of hepcidin expression and iron overload mediate Salmonella susceptibility in ankyrin 1 ENU-induced mutant. PLoS One 8:e55331. 10.1371/journal.pone.0055331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theurl I, Fritsche G, Ludwiczek S, Garimorth K, Bellmann-Weiler R, Weiss G. 2005. The macrophage: a cellular factory at the interphase between iron and immunity for the control of infections. Biometals. 18:359–367 [DOI] [PubMed] [Google Scholar]
- 52.Pan X, Tamilselvam B, Hansen EJ, Daefler S. 2010. Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens. BMC Microbiol. 10:64. 10.1186/1471-2180-10-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnewall RE, Ohashi N, Rikihisa Y. 1999. Ehrlichia chaffeensis and E. sennetsu, but not the human granulocytic ehrlichiosis agent, colocalize with transferrin receptor and up-regulate transferrin receptor mRNA by activating iron-responsive protein 1. Infect. Immun. 67:2258–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clemens DL, Horwitz M. 1996. The Mycobacterium tuberculosis phagosome interacts with early endosomes and is accessible to exogenously administered transferrin. J. Exp. Med. 184:1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larson JA, Howie HL, So M. 2004. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol. Microbiol. 53:807–820 [DOI] [PubMed] [Google Scholar]
- 56.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946–953 [DOI] [PubMed] [Google Scholar]
- 57.Konijn AM, Glickstein H, Vaisman B, Meyron-Holtz EG, Slotki IN, Cabantchik ZI. 1999. The cellular labile iron pool and intracellular ferritin in K562 cells. Blood 94:2128–2134 [PubMed] [Google Scholar]
- 58.Roberts S, Bomford A. 1988. Ferritin iron kinetics and protein turnover in K562 cells. J. Biol. Chem. 263:19181–19187 [PubMed] [Google Scholar]
- 59.Ollinger K, Roberg K. 1997. Nutrient deprivation of cultured rat hepatocytes increases the desferrioxamine-available iron pool and augments the sensitivity to hydrogen peroxide. J. Biol. Chem. 272:23707–23711 [DOI] [PubMed] [Google Scholar]
- 60.Mesquita FS, Thomas M, Sachse M, Santos AJ, Figueira R, Holden DW. 2012. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 8:e1002743. 10.1371/journal.ppat.1002743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hernandez LD, Pypaert M, Flavell RA, Galán JE. 2003. A Salmonella protein causes macrophage cell death by inducing autophagy. J. Cell Biol. 163:1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knodler LA, Steele-Mortimer O. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587–599 [DOI] [PubMed] [Google Scholar]
- 63.Nairz M, Theurl I, Ludwiczek S, Theurl M, Mair SM, Fritsche G, Weiss G. 2007. The co-ordinated regulation of iron homeostasis in murine macrophages limits the availability of iron for intracellular Salmonella typhimurium. Cell. Microbiol. 9:2126–2140 [DOI] [PubMed] [Google Scholar]