Abstract
Yersinia pestis carries homologues of the toxin complex (Tc) family proteins, which were first identified in other Gram-negative bacteria as having potent insecticidal activity. The Y. pestis Tc proteins are neither toxic to fleas nor essential for survival of the bacterium in the flea, even though tc gene expression is highly upregulated and much more of the Tc proteins YitA and YipA are produced in the flea than when Y. pestis is grown in vitro. We show that Tc+ and Tc− Y. pestis strains are transmitted equivalently from coinfected fleas, further demonstrating that the Tc proteins have no discernible role, either positive or negative, in transmission by the flea vector. Tc proteins did, however, confer Y. pestis with increased resistance to killing by polymorphonuclear leukocytes (PMNs). Resistance to killing was not the result of decreased PMN viability or increased intracellular survival but instead correlated with a Tc protein-dependent resistance to phagocytosis that was independent of the type III secretion system (T3SS). Correspondingly, we did not detect T3SS-dependent secretion of the native Tc proteins YitA and YipA or the translocation of YitA– or YipA–β-lactamase fusion proteins into CHO-K1 (CHO) cells or human PMNs. Thus, although highly produced by Y. pestis within the flea and related to insecticidal toxins, the Tc proteins do not affect interaction with the flea or transmission. Rather, the Y. pestis Tc proteins inhibit phagocytosis by mouse PMNs, independent of the T3SS, and may be important for subverting the mammalian innate immune response immediately following transmission from the flea.
INTRODUCTION
Toxin complex (Tc) proteins were first identified as potent insecticidal toxins in the roundworm symbiont and insect pathogen Photorhabdus luminescens (1–4). Subsequently, homologues of the P. luminescens Tc genes were identified in Yersinia pestis. Y. pestis is the causative agent of bubonic plague and utilizes fleas as a vector for transmission to the mammalian host. Fleas become infected with Y. pestis following ingestion of an infected blood meal, and the bacteria survive and grow as a biofilm within the flea digestive tract. Y. pestis biofilm can adhere to the flea proventriculus (PV), a valve that connects the esophagus to the midgut (MG), and can cause blockage of blood flow to the midgut. Proventricular biofilm formation and blockage greatly enhance the transmission of Y. pestis to the mammalian host (5). Several Y. pestis genes are specifically upregulated in the flea compared to in vitro culture at the same temperature (6, 7). Notable among these are the Tc genes, which are upregulated 6- to 50-fold in the flea (7). The Y. pestis Tc proteins are encoded within a single chromosomal locus and are termed YitA (TcaA-like), YitB (TcaB-like), YitC (TcaC-like), YipA, and YipB (both TccC-like) (8, 9). Y. pestis Tc proteins are maximally produced within infected fleas maintained at 21°C (7, 8) and minimally produced in vitro only during growth at <26°C (7–11). Upstream of the Tc locus is the LysR-like regulator gene yitR, which encodes a positive regulator of Tc gene expression (8, 9). Deletion of yitR results in greatly decreased production of Tc proteins (9), whereas overexpression of yitR increases the production of Tc proteins in vitro (8).
Unlike the insecticidal Tc proteins of P. luminescens, Y. pestis Tc proteins are not toxic to Manduca sexta larvae or active against Spodoptera frugiperda (Sf9) insect cells (12). Furthermore, although the Y. pestis Tc proteins are highly upregulated during infection of the rat flea, Xenopsylla cheopis (7, 8), they are not toxic to fleas (13), nor are they required for infection of or survival within the flea digestive tract or for blockage of the proventriculus (8). However, Y. pestis Tc proteins show some cytotoxicity to NIH 3T3 mouse fibroblast cells (12), and the Tc genes (yitA, -B, and -C) are upregulated when Y. pestis is within J774A.1 macrophages (14). Y. pestis isolated from infected fleas has increased resistance to uptake and killing by human and murine polymorphonuclear leukocytes (PMNs) (15, 16) and murine macrophages (7). Conspicuously, Y. pestis lacking YitR is no longer resistant to phagocytosis by murine macrophages (7).
A previous study suggested that the Y. pestis Tc proteins may be secreted through the type III secretion system (T3SS), which is encoded on the Y. pestis pCD1 virulence plasmid and essential for secretion of the effector Yop proteins (9). That study also reported that Bordetella pertussis adenylate cyclase CyaA constructs fused with the 100 or 250 N-terminal amino acids of the Tc protein YipB were translocated into Sf9 cells, as well as mammalian RAW macrophages and HeLa cells, and that translocation was dependent on the presence of the T3SS (9).
Thus, although the Y. pestis Tc proteins are maximally produced in the flea, they are neither toxic to fleas nor required to produce a transmissible infection. Instead, the Tc proteins may be secreted by the T3SS and act to subvert the initial immune response of the mammalian host following transmission of Y. pestis from the flea. In particular, they may act to limit killing by PMNs which are rapidly recruited to sites of Y. pestis infection (17). We examined these possibilities in this study.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are listed in Table 1. All experiments were performed with highly attenuated Y. pestis strains which lack either the pCD1 (Lcr) virulence plasmid encoding the T3SS and effector Yop proteins (KIM6+), the pigmentation (Pgm) locus (KIM5), or both (KIM6 and KIM10). Where indicated, strains were transformed with pCD1Δ1234, which encodes a functional T3SS but not the Yop virulence factors (18), and/or the low-copy-number plasmid pWKS130::yitR (7) or the high-copy-number plasmid pCR-XL-TOPO::yitR (8) or pUC19::yitR to increase YitA and YipA synthesis during growth in vitro (8). Plasmid pUC19::yitR was constructed by PCR amplifying the YitR open reading frame flanked by ∼300 bp of upstream and downstream sequence into pUC19 (19) by using the primers 5′-AGTTGAGCTCGTCTGCATTGATTATTTGACC-3′ and 5′-AGTTTCTAGAGATCGTTGCGTAGCTGTGTTGC-3′, digesting the PCR product with the restriction enzymes SacI and XbaI (sites indicated by the underlined text), and ligating the purified fragment into pUC19. Unless stated otherwise, Y. pestis was grown in brain heart infusion (BHI) broth at 21°C overnight from frozen stocks and subcultured into fresh BHI broth at 21°C prior to incubation at 21°C or 37°C. Prior to each assay, bacteria were washed in Dulbecco's phosphate-buffered saline (DPBS; Life Technologies, Grand Island, NY), resuspended in RPMI 1640 medium with 10 mM HEPES (RPMI medium; Life Technologies), enumerated by Petroff-Hauser counts, and adjusted to the desired concentration. Where appropriate, kanamycin (30 μg/ml), carbenicillin (100 μg/ml), or chloramphenicol (10 μg/ml) was added to the broth cultures at the indicated final concentration. All Y. pestis strains used in this study are exempt from CDC select agent rules and were approved for use under biosafety level 2 conditions by the RML NIAID Biosafety Committee.
Table 1.
Y. pestis strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| KIM6+ | Tc genes present, pCD1 negative, pgm positive, pPCP1 positive | 51 |
| KIM6 | Tc genes present, pCD1 negative, pgm negative, pPCP1 positive | 51 |
| KIM10 | Tc genes present, pCD1 negative, pgm negative, pPCP1 negative | 9 |
| KIM5 | Tc genes present, pCD1 positive, pgm negative, pPCP1 positive | 52 |
| KIM6+ ΔyitA-yipB | Deletion of the Tc locus from yitA to yipB, yitR still present | 8 |
| KIM6+ ΔyitR | Deletion of the regulator YitR | 7 |
| KIM6+ YitA–β-lactamase | YitA::β-lactamase translational fusion | 8 |
| KIM6+ YipA–β-lactamase | YipA::β-lactamase translational fusion | 8 |
| KIM5 YitA–β-lactamase | YitA::β-lactamase translational fusion | 8 |
| KIM5 YipA–β-lactamase | YipA::β-lactamase translational fusion | 8 |
| KIM6 YitA–β-lactamase | YitA::β-lactamase translational fusion | 8 |
| KIM6 YipA–β-lactamase | YipA::β-lactamase translational fusion | 8 |
| Plasmids | ||
| pCR-XL-TOPO::yitR | yitR expressed under native promoter control from a high-copy-number plasmid (Kan, Zeo) | 8 |
| pUC19::yitR | yitR expressed under native promoter control from a high-copy-number plasmid (Amp) | This study |
| pWKS130::yitR | yitR expressed under native promoter control from a low-copy-number plasmid (Kan) | 7 |
| pCD1Δ1234 | Plasmid encoding the genes required for the T3SS, with no effector Yops (Cam) | 18 |
| pCD1::kan | pCD1ΔyadA::kan (Kan) | 53 |
| pMM83 | Plasmid pHSG576 encoding YopM–β-lactamase under control of its native promoter (Cam) | 32 |
| pMM85 | Plasmid pHSG576 encoding YopE–β-lactamase under control of its native promoter (Cam) | 32 |
| pAcGFP1 | Constitutively produces GFP (Amp) | Clontech |
| pmCherry | Constitutively expresses mCherry (Amp) | Clontech |
Antibiotic resistances, where present, are noted in parentheses. Kan, kanamycin; Amp, ampicillin; Cam, chloramphenicol; Zeo, zeocin.
Flea infections and transmission efficiency.
Y. pestis strains used for flea infections were grown overnight at 28°C without aeration. The following day, bacteria were subcultured into fresh BHI broth and grown overnight at 37°C without aeration. Cultures were centrifuged, and the bacterial pellet was resuspended in 1 ml phosphate-buffered saline (PBS) and added to 5 ml of fresh heparinized mouse blood in a previously described artificial feeding chamber (20, 21).
For coinfections, X. cheopis fleas were allowed to feed on blood containing approximately equal numbers of Y. pestis KIM6+ containing pAcGFP1 (Clontech Laboratories, Mountain View, CA) and KIM6+ ΔyitA-yipB containing pmCherry (Clontech) in 5 ml of fresh heparinized mouse blood. Alternatively, KIM6+(pmCherry) and KIM6+ ΔyitA-yipB(pAcGFP1) were used. The initial blood meals contained a combined total of 3.0 × 109 CFU/ml of KIM6+ and KIM6+ ΔyitA-yipB for a high infectious dose or 5.8 × 107 CFU/ml for a low infectious dose. Fleas that had taken an infectious blood meal were maintained at 21°C and fed twice weekly on normal uninfected mice for 37 days. Immediately after each feeding, fleas were checked by microscopy for blockage of the proventriculus as previously described (20, 21). Every 2 to 7 days postinfection, 5 to 10 flea digestive tracts were dissected from infected fleas. The proventriculus (PV) was separated from the midgut (MG) and triturated, serially diluted, and plated on blood agar plates. The numbers of KIM6+ and KIM6+ ΔyitA-yipB cells in the PV and MG for each flea at each time were determined by counting and scoring red and green fluorescent colonies under a UV lamp. Fourteen days after the initial infection, the digestive tracts of coinfected fleas with a blocked proventriculus were dissected and imaged by fluorescence microscopy. Images were obtained using a Photometrics CoolSnap HQ (Tucson, AZ) digital camera, and images were artificially colored and combined using MetaMorph software, version 7.5.6.0 (Molecular Devices, Sunnyvale, CA).
Transmission efficiency of Y. pestis KIM6+ and KIM6+ ΔyitA-yipB from coinfected fleas was determined 14, 21, and 28 days after infection by allowing groups of ∼100 fleas to feed on an artificial feeding chamber loaded with sterile mouse blood. After a 2-h feeding period, the blood in the feeding chamber was collected, and the interior of the skin through which the fleas had fed was thoroughly washed with PBS. All of the collected blood and pooled washes were plated on blood agar containing carbenicillin. The total numbers of transmitted KIM6+ and KIM6+ ΔyitA-yipB cells were determined by counting numbers of red and green CFU under a UV lamp. Each transmission experiment was performed in duplicate on days 14, 21, and 28, with at least two independent flea infections.
To obtain flea-derived bacteria, fleas were fed on mouse blood containing ∼6 × 109 CFU/ml of either Tc+ or Tc− Y. pestis. Fleas that had taken an infectious blood meal were maintained at 21°C and fed twice weekly on normal uninfected mice. Bacteria were subsequently recovered from the fleas 2 to 4 weeks after infection as previously described (15).
Purification of murine PMNs.
Murine PMNs were isolated from pooled bone marrow from 8- to 12-week-old RML Swiss-Webster mice as described previously (15, 22). Bone marrow was flushed from the femurs and tibias of 3 or 4 RML mice and pooled. Following Percoll and Histopaque 1119 density gradient purification, PMNs were resuspended in RPMI medium and enumerated by hemocytometer counts. PMN viability was 95 to 99% as determined by trypan blue dye exclusion (22). PMN purity was estimated to be 70 to 85% by hemocytometer direct counts and by white blood cell differential counts.
Survival of Y. pestis after incubation with murine PMNs.
Viability of Y. pestis following exposure to PMNs was determined as described previously (23), with the following modifications. PMNs (1 × 106) suspended in RPMI medium were added to a 96-well cell culture plate that had been precoated with 20% mouse serum (Innovative Research, Novi, MI) for 30 min at 37°C. For experiments comparing bacteria isolated from fleas to bacteria grown in vitro, bacteria grown in BHI were triturated with an equivalent number of uninfected flea midguts. Trituration was performed for 5 s in lysing matrix H tubes (MP Biomedicals, Solon, OH), using a FastPrep FP120 device (Qbiogene, Inc., Carlsbad, CA) with speed setting 4.0. Following trituration, bacteria were washed with PBS and resuspended in RPMI medium. Bacteria isolated from flea midguts or grown in BHI and triturated with uninfected flea midguts were added (2:1 bacterium-to-PMN ratio). Plates were centrifuged at 400 × g for 5 min at 4°C to synchronize phagocytosis and were incubated at 37°C and 5% CO2. At different times after infection, PMNs were lysed with 0.01% Triton X-100 (EMD Chemicals, Gibbstown, NJ) in DPBS for 5 min on ice, followed by trituration for 5 s (24). Dilutions of the lysed suspension were plated on BHI agar, and CFU were enumerated after incubation for 3 days at 28°C. To determine the total number (both within PMNs and extracellular) of viable Y. pestis cells, replicate samples were processed immediately to determine the initial number of bacteria loaded (T0), or after 30, 60, or 180 min of incubation with PMNs. The percent survival (intracellular and extracellular) was calculated as follows: % survival = [CFUT(x)/CFUT0] × 100.
To determine the survival of only intracellular Y. pestis, samples were prepared as described above, except that 45 min after the initial infection, gentamicin was added (25 μg/ml) for 15 min to eliminate remaining extracellular bacteria. Medium containing gentamicin was removed from the wells by aspiration, cells were washed twice with RPMI medium, fresh medium without gentamicin was added, and plates were incubated at 37°C and 5% CO2. Samples were processed immediately after gentamicin treatment to determine the initial number of viable intracellular bacteria [T(I, gent)] and the number after an additional 60 min or 180 min of incubation [T(x, gent)]. Percent intracellular survival was calculated as follows: % survival = [CFUT(x, gent)/CFUT(I, gent)] × 100. Experiments were performed in triplicate and repeated at least three times.
Murine PMN phagocytosis assay.
Phagocytosis of bacteria by murine bone marrow-derived PMNs was determined by fluorescence microscopy as described previously (7, 15, 23). One hundred microliters of a chilled suspension containing 5 × 105 PMNs in RPMI medium was added to acid-washed glass coverslips in individual wells of a 24-well cell culture plate, a chilled suspension of green fluorescent protein (GFP)-expressing Y. pestis(pAcGFP1) in RPMI medium was added (1:2 PMN-to-bacterium ratio), and the plates were centrifuged for 7 min at 400 × g to synchronize phagocytosis. Following centrifugation, plates were incubated at 37°C and 5% CO2 for 1 h before fixation with 4% paraformaldehyde. Extracellular bacteria were labeled with a hyperimmune rabbit anti-Y. pestis polyclonal antibody (16) and then stained with goat anti-rabbit IgG conjugated to Alexa Fluor 568 (Life Technologies). Intracellular bacteria fluoresced only green (GFP), whereas extracellular bacteria fluoresced red (Alexa Fluor 568) and green. A minimum of 100 PMNs from random microscopic fields were counted for each experiment. Percent phagocytosis was calculated by dividing the number of intracellular bacteria by the total number of PMN-associated bacteria (intracellular plus extracellular) and multiplying the result by 100.
PMN membrane integrity and LDH release assay.
Cell lysis was quantified by measuring released lactate dehydrogenase (LDH) activity by use of a Cytotoxicity Detection Plus kit (Roche Applied Science, Indianapolis, IN). Cell membrane integrity was assessed using a LIVE/DEAD viability/cytotoxicity kit for mammalian cells (Life Technologies). For both assays, Y. pestis was grown overnight at 21°C in BHI, washed, and resuspended in RPMI medium. Y. pestis (5 × 105 organisms) was added to murine PMNs (1 × 105) in wells of 96-well cell culture plates, centrifuged for 7 min at 400 × g, and incubated at 37°C and 5% CO2 for 1 or 6 h prior to analysis. For membrane integrity, ethidium homodimer 1 (EthD-1; 4.5 mM final concentration) was added at the indicated time points and allowed to incubate for 30 min at room temperature. Fluorescence was measured using a microplate fluorometer (Synergy Mx; Biotek Instruments). The percent LDH release and the percent membrane integrity were calculated using the following formula: % LDH release = [(experimental release − T0 background)/(maximum release − T0 background)] × 100. The maximum release represents the value for detergent-lysed cells. Experiments were repeated at least three times in triplicate.
Secretion of Tc proteins into culture supernatants.
Y. pestis was grown overnight at 21°C in 100 ml of TMH medium with 2.5 mM CaCl2 (25). Cultures were centrifuged at 3,200 × g, washed with fresh TMH medium, and resuspended in 20 ml of TMH medium. Culture density was normalized to an optical density at 600 nm (OD600) of 1.2, and 5 ml of the normalized suspension was added to 250 ml of TMH medium, with or without CaCl2, and subsequently incubated at either 21°C or 37°C for an additional 4 h. Samples were then centrifuged at 10,000 × g at 4°C for 15 min, and the culture supernatants were removed, filtered through a 0.2-μm filter, and concentrated using centrifugal filter devices with a 10-kDa molecular mass cutoff (Centricon Plus; Millipore, Billerica, MA). Concentrated supernatants were separated by SDS-PAGE in lanes of precast 4 to 15% polyacrylamide gels (Criterion TGX; Bio-Rad, Hercules, CA). Samples were then transferred to 0.2-μm nitrocellulose membranes for Western blot analysis. YitA and YipA were detected using anti-YitA and anti-YipA sera (8). Proteins fused with β-lactamase were detected using a rabbit anti-β-lactamase polyclonal antibody (Millipore). Mouse antiserum against the intracellular protein GroEL (Enzo Life Sciences, Farmingdale, NY) was used to assess cellular lysis. Goat anti-rabbit IgG or goat anti-mouse IgG conjugated to alkaline phosphatase (Life Technologies) and 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NBT) liquid substrate (Sigma-Aldrich, St. Louis, MO) were used to visualize protein bands.
Alternatively, as a more sensitive way to detect secreted proteins, culture supernatants from Y. pestis strains expressing YitA–β-lactamase, YipA–β-lactamase, YopE–β-lactamase, or YopM–β-lactamase were collected and concentrated as described above and then incubated with the β-lactamase substrate fluorocillin (Life Technologies). Fifty microliters of concentrated culture supernatant or bacterial lysate, prepared by resuspending ∼109 bacteria (from the same cultures from which supernatants were derived) in 1 ml PBS and sonicating the suspension on ice for 30 s at power setting 5 and duty cycle setting 25 (model XL2015 sonicator; Heat Systems, Farmingdale, NY), was added to wells of a 96-well cell culture plate along with 50 μl of 10 μM fluorocillin in PBS. Samples were incubated for 4 h at 37°C, and fluorescence was measured in a microplate fluorometer (Synergy Mx; Biotek Instruments, Winooski, VT) at 495 nm (excitation) and 525 nm (emission).
Translocation of Tc–β-lactamase fusion proteins into human PMNs and CHO cells.
Heparinized human blood was obtained from healthy volunteers in accordance with a protocol approved by the Institutional Review Board for Human Subjects of the NIAID. PMNs were purified as described previously and used immediately (26). CHO-K1 (CHO) cells were maintained in tissue culture-treated plastic flasks in RPMI 1640 with 10% fetal bovine serum (FBS) and antibiotic-antimycotic (100 units/ml of penicillin, 100 μg/ml of streptomycin, and 0.25 μg/ml of amphotericin B [Fungizone]; Life Technologies) at 37°C and 5% CO2. Twenty-four hours prior to use, CHO cells were detached with 0.05% trypsin-EDTA (Life Technologies), washed three times, resuspended in antibiotic-antimycotic-free RPMI 1640 with 10% FBS, enumerated by hemocytometer counts, and seeded into wells of 24-well cell culture plates (2.5 × 105/well).
To infect isolated human PMNs and CHO cells, bacteria grown in BHI and maintained at 21°C or transferred to 37°C for 2 h were added at a 1:10 or 1:40 (cell-to-bacterium) ratio to isolated human PMNs or CHO cells (∼2.5 × 105 cells/well) in wells of 24-well glass-bottom (Greiner Bio-One, Monroe, NC) or polystyrene cell culture plates. For experiments using human PMNs, plates were first coated with autologous normal human serum (NHS; 20% NHS for polystyrene plates and 50% NHS for glass plates) for 30 min at 37°C. Plates were centrifuged at 400 × g for 5 min at 4°C to synchronize phagocytosis and then incubated at 37°C and 5% CO2 for 2 or 4 h. Wells were gently washed, and 0.4 ml of 1 μM CCF2-AM (Life Technologies) was added for 1 h at room temperature. Cells were washed and immediately imaged or collected for analysis by flow cytometry. Image acquisition and analysis were performed on a Nikon Eclipse TE2000 epifluorescence microscope using a Plan Fluor 40× objective. Images were captured using the following emission and excitation filter combinations (Chroma Technology, Rockingham, VT): D360/40x and D460/50m for detection of cleaved CCF2-AM and D360/40x and S535/40m for detection of uncleaved CCF2-AM. A CoolSnap HQ2 digital camera (Photometrics) was used to capture images for analysis using MetaMorph software, version 7.7.9.0 (Molecular Devices). Samples for flow cytometry were analyzed with an LSRII SORP cytometer (BD Biosciences, San Jose, CA), using a 405-nm laser. Cleaved CCF2-AM was detected with a 450-nm filter (blue), and uncleaved CCF2-AM was detected with a 525-nm filter (green). Analysis and gating were performed using FlowJo software, version 10.0.4 (Tree Star, Ashland, OR). Cells that had not been loaded with CCF2-AM and CCF2-AM-loaded cells that were not infected or were infected with Y. pestis lacking any β-lactamase were used as controls to establish the negative gate. Results are displayed as ratios of cleaved to intact CCF2-AM.
Statistics.
Data were analyzed by paired Student's t test or by one-way analysis of variance (ANOVA) with the Bonferroni posttest, using GraphPad Prism, version 5.04 (GraphPad Software, San Diego, CA).
RESULTS
Tc proteins do not confer a selective advantage in the flea vector.
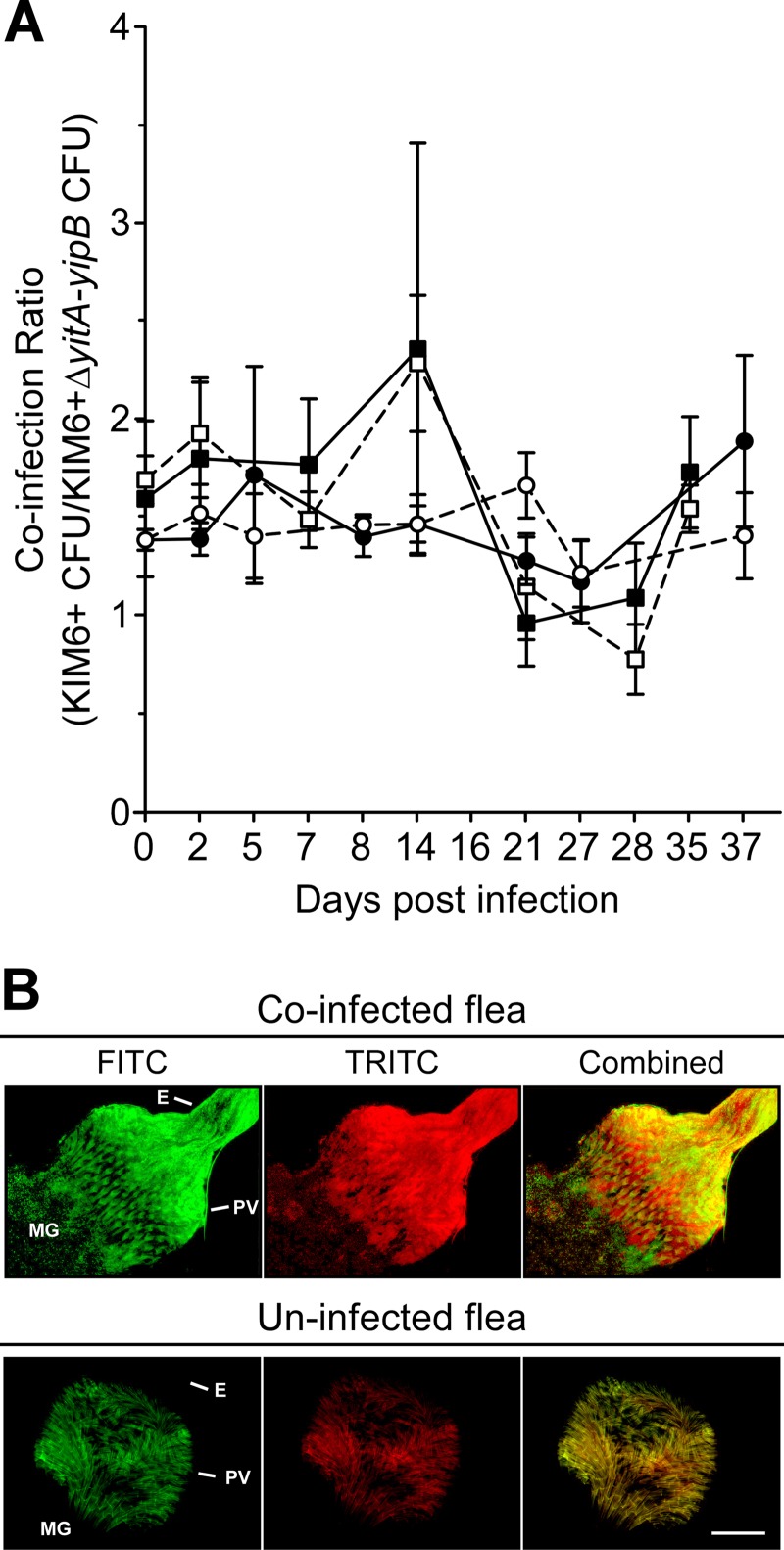
Y. pestis mutant strains that do not make Tc proteins are able to infect and block fleas as well as the parental strains (8), so we evaluated whether the Tc proteins provide a competitive advantage for survival within the flea or for transmission by flea bite. At different times after infection with approximately equal numbers of T3SS+ Y. pestis KIM6+(pCD1Δ1234) and KIM6+ ΔyitA-yipB(pCD1Δ1234), the digestive tracts of infected fleas were dissected, and the PV and MG were separated and analyzed independently. Regardless of whether the initial infectious dose was high or low, or the region of the flea digestive tract examined, coinfected fleas maintained a consistent ratio of KIM6+(pCD1Δ1234) to KIM6+ ΔyitA-yipB(pCD1Δ1234) cells that was comparable to the ratio in the infectious blood meal (Fig. 1A). The KIM6+ parent and the ΔyitA-yipB mutant colocalized in both the PV and MG and did not appear to aggregate independently (Fig. 1B and data not shown). Altering which Y. pestis strain carried which fluorescent protein-expressing plasmid had no effect on the results (data not shown). Thus, even though the Tc proteins are highly produced in the flea gut (8), the Tc+ strain did not have a competitive advantage over the Tc− strain.
Fig 1.
Tc+ and Tc− Y. pestis cells survive equally and colocalize within X. cheopis fleas. Fleas were fed on blood containing a combined total of ∼5.75 × 107 (squares) or 3.0 × 109 (circles) CFU of KIM6+ and KIM6+ ΔyitA-yipB per ml of blood. On average, fleas that fed on a blood meal containing the low infectious dose initially contained ∼1.41 × 104 CFU of KIM6+ and ∼8.40 × 103 CFU of KIM6+ ΔyitA-yipB, and fleas that fed on a blood meal containing the high infectious dose initially contained ∼1.93 × 106 CFU of KIM6+ and ∼1.39 × 106 CFU of KIM6+ ΔyitA-yipB. (A) The mean (± standard error) ratios of KIM6+ to KIM6+ ΔyitA-yipB in the flea proventriculus (solid symbols with solid lines) and midgut (open symbols with dashed lines) were determined for 5 to 10 fleas on the indicated days after infection. (B) Colocalization of KIM6+(pAcGFP1) and KIM6+ ΔyitA-yipB(pmCherry) in the flea digestive tract. The proventriculus and upper midgut regions of digestive tracts dissected from coinfected and uninfected fleas were imaged by fluorescence microscopy, using a fluorescein isothiocyanate (FITC) filter for GFP (green) and a tetramethylrhodamine isothiocyanate (TRITC) filter for mCherry (red). Images were artificially colored and merged (combined). Images of representative uninfected and coinfected fleas containing masses of colocalized Tc+ and Tc− bacteria in the proventriculus (PV; the proventricular spines autofluoresce in both the TRITC and FITC channels), esophagus (E), and upper midgut (MG) are shown. Bar, 50 μm.
To determine if Y. pestis expressing the Tc proteins is somehow more transmissible during a flea bite, coinfected fleas were allowed to feed on sterile blood in an artificial feeding chamber 14, 21, and 28 days after infection. The total number of CFU recovered from the blood after feeding varied among experiments. This is attributable to inherent variation in the number of blocked, transmission-competent fleas on a given day and in the large range for the number of CFU a blocked flea transmits (27). Nonetheless, fleas initially coinfected with either the high dose or low dose transmitted an average ratio of KIM6+(pCD1Δ1234) to KIM6+ ΔyitA-yipB(pCD1Δ1234) after each feeding that closely matched the initial infectious blood meal ratio (Table 2). Thus, the Tc proteins are not essential or notably advantageous for transmission by flea bite.
Table 2.
Transmission of KIM6+(pCD1Δ1234) and KIM6+ ΔyitA-yipB(pCD1Δ1234) from coinfected fleas in two independent experiments
| Combined no. of CFU/ml in blood meala | Initial ratiob | Replicate | Strainc | No. of CFU transmittedd on day: |
Total no. of CFU transmitted | Transmission ratioe | ||
|---|---|---|---|---|---|---|---|---|
| 14 | 21 | 28 | ||||||
| 3.00e9 | 1.39 | 1 | Tc+ | 0 | 1,218 | 102 | 1,320 | 1.43 |
| 1 | Tc− | 2 | 598 | 73 | 673 | |||
| 2 | Tc+ | 5 | 1,344 | 335 | 1,684 | |||
| 2 | Tc− | 3 | 1,085 | 334 | 1,422 | |||
| 5.75e7 | 1.65 | 1 | Tc+ | 129 | 113 | 14,040 | 14,282 | 1.56 |
| 1 | Tc− | 75 | 52 | 8,940 | 9,067 | |||
| 2 | Tc+ | 15 | 0 | 262 | 277 | |||
| 2 | Tc− | 9 | 0 | 232 | 241 | |||
Total number of CFU/ml of KIM6+(pCD1Δ1234) and KIM6+ ΔyitA-yipB(pCD1Δ1234) in the infectious blood meal.
Ratio of KIM6+(pCD1Δ1234) CFU to KIM6+ ΔyitA-yipB(pCD1Δ1234) CFU in the infectious blood meal.
Tc+, KIM6+(pCD1Δ1234); Tc−, KIM6+ ΔyitA-yipB(pCD1Δ1234).
Number of CFU of each strain transmitted in two independent flea feedings of two different groups of fleas for each experiment.
Ratio of cumulative number of Tc+ CFU to cumulative number of Tc− CFU transmitted.
Tc proteins are responsible for flea-derived Y. pestis resistance to killing by murine PMNs.
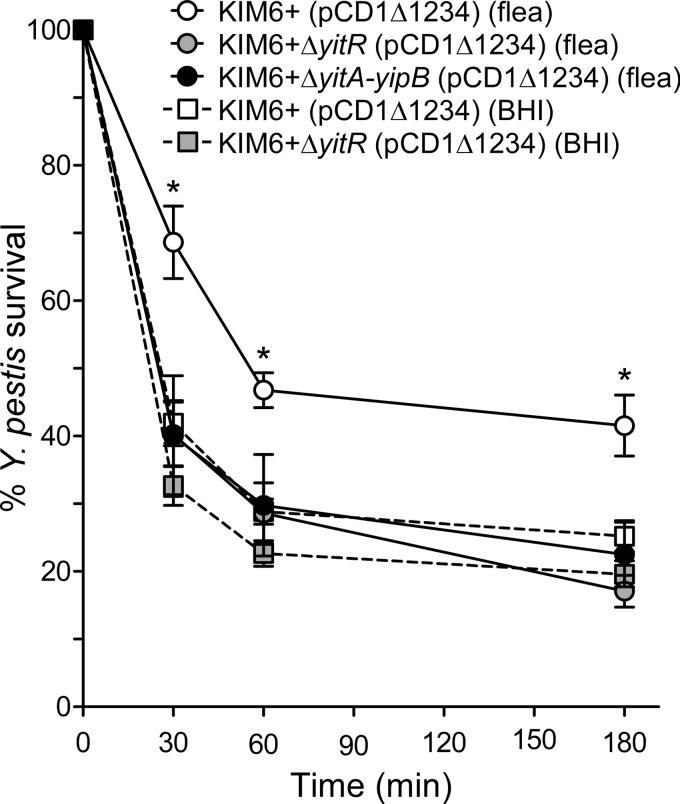
Y. pestis isolated from fleas is more resistant to killing by human (16) and murine (15) PMNs than Y. pestis grown in vitro. Because the Tc proteins are produced much more highly in the flea than in vitro (7, 15), we tested whether they play an important role in the flea-specific resistance phenotype. After 30 min of incubation with PMNs, Y. pestis KIM6+(pCD1Δ1234) isolated from fleas had an ∼27% higher survival rate than bacteria grown in BHI at the same temperature (68.6% ± 5.3% [mean ± standard error of the mean {SEM}] compared to 41.9% ± 3.3%) (Fig. 2). After 60 min and 180 min of incubation, KIM6+(pCD1Δ1234) isolated from fleas showed ∼18% and ∼16%, respectively, greater survival than the same strain grown in BHI (Fig. 2). When production of the Tc proteins was prevented, by deleting either the transcriptional activator gene yitR or the entire Tc locus (ΔyitA-yipB), bacteria isolated from fleas were as susceptible to killing by PMNs as bacteria grown in vitro (Fig. 2), indicating that the Tc proteins are essential for flea-derived Y. pestis resistance to killing by PMNs.
Fig 2.
Y. pestis Tc proteins confer resistance to killing by murine PMNs. Y. pestis strains isolated from flea midguts (circles) or grown in BHI at 21°C (squares) were incubated with murine PMNs, and numbers of Y. pestis CFU were determined 30, 60, and 180 min after infection. Results are presented as percentages of the initial CFU. The means and standard errors for at least three experiments are indicated. Data were analyzed by one-way ANOVA with the Bonferroni posttest. *, P < 0.01 [KIM6+(pCD1Δ1234) isolated from fleas versus all other sample types].
Tc proteins do not disrupt membrane integrity of murine PMNs.
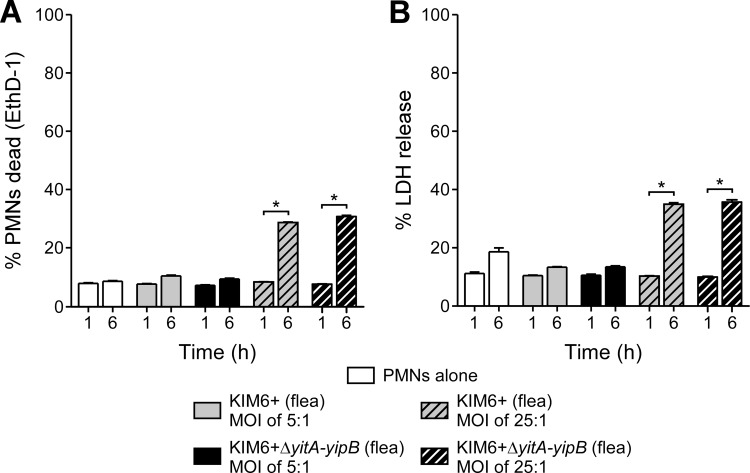
To determine if the resistance of flea-derived Y. pestis to PMNs was due to disruption of PMN integrity or viability by the Tc proteins, we evaluated PMN membrane integrity and cytotoxicity following infection with Tc+ or Tc− Y. pestis isolated from fleas. Murine PMNs were incubated with KIM6+(pCD1Δ1234) and KIM6+ ΔyitA-yipB(pCD1Δ1234) for 1 h or 6 h at a multiplicity of infection (MOI) of 5 or 25 bacteria per PMN. Y. pestis isolated from fleas and expressing the Tc proteins did not alter PMN membrane integrity (Fig. 3A) or cause cell cytotoxicity (Fig. 3B) to a greater extent than that with Y. pestis KIM6+ ΔyitA-yipB(pCD1Δ1234). After 6 h of incubation at an MOI of 25:1, there were significant increases in EthD-1 staining and LDH release compared to those of uninfected PMNs; however, these were not dependent on the presence of the Tc proteins (Fig. 3A and B). Thus, under our assay conditions, the Tc proteins do not notably decrease PMN membrane integrity or cause cell lysis.
Fig 3.
Y. pestis Tc proteins do not disrupt PMN membrane integrity. The membrane integrity of infected PMNs was evaluated by EthD-1 staining (A) and release of intracellular LDH into the medium (B). PMNs were incubated with Y. pestis strains for 1 h or 6 h at an MOI (bacteria/PMN) of 5 (solid bars) or 25 (hatched bars). All strains contained pCD1Δ1234 (T3SS+). The means and standard errors for at least three experiments are indicated. Data were analyzed by paired Student's t test or by one-way ANOVA with the Bonferroni posttest. *, P < 0.001.
Y. pestis Tc proteins do not enhance survival within murine PMNs.
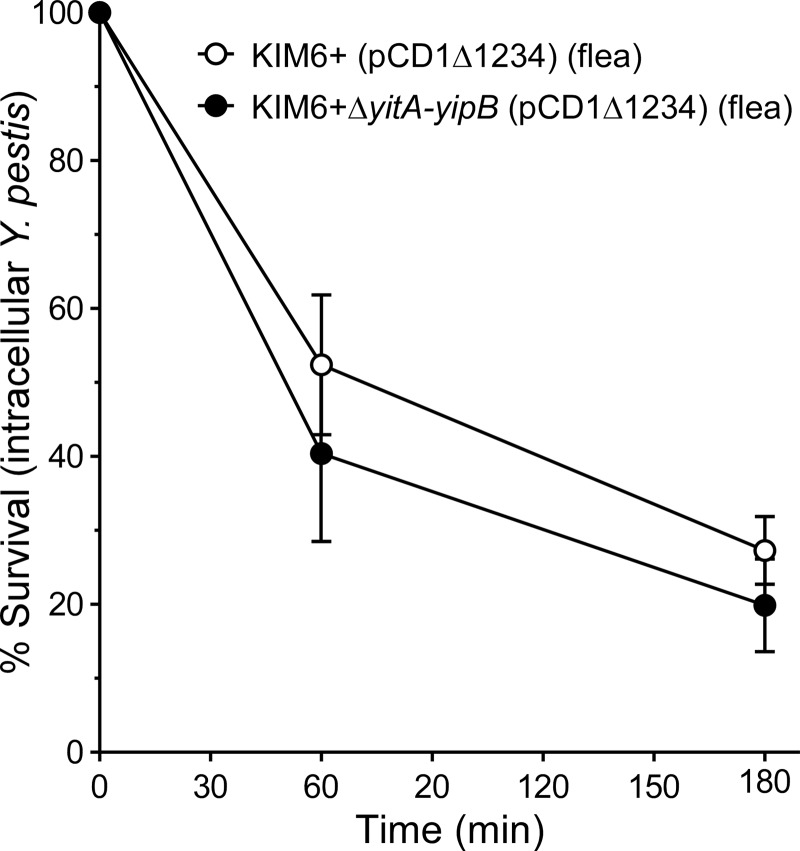
Since the Tc proteins did not alter PMN viability (Fig. 3), we evaluated whether the observed resistance to killing by PMNs (Fig. 2) was attributable to increased intracellular survival of Tc+ Y. pestis. PMNs were incubated with Y. pestis for 45 min to allow for phagocytosis and then treated with gentamicin and washed to remove any remaining extracellular bacteria. The survival of the intracellular population was then determined after an additional 60 and 180 min of incubation. The survival of intracellular KIM6+(pCD1Δ1234) and KIM6+ ΔyitA-yipB(pCD1Δ1234) isolated from fleas was equivalent (Fig. 4). On average, after 60 min, 52.4% ± 9.4% (mean ± SEM) of the initial intracellular population of KIM6+(pCD1Δ1234) cells and 40.4% ± 11.9% of that of KIM6+ ΔyitA-yipB(pCD1Δ1234) cells remained viable. After 180 min, 27.3% ± 4.6% and 19.9% ± 6.2% of the initial intracellular populations were viable. Thus, the rates at which Y. pestis cells were killed after being phagocytosed by PMNs were similar regardless of the Tc proteins.
Fig 4.
Tc proteins do not inhibit killing of intracellular Y. pestis by PMNs. Y. pestis KIM6+(pCD1Δ1234) (open circles) or KIM6+ ΔyitA-yipB(pCD1Δ1234) (solid circles) recovered from infected fleas was used to infect PMNs. After 45 min, gentamicin was added to eliminate remaining extracellular bacteria. Medium containing gentamicin was then removed, and fresh medium without gentamicin was added (T0 sample). Additional samples were collected after 60 and 180 min of incubation, and the number of recovered CFU was compared to that of the T0 sample. The means and standard errors for at least three experiments are indicated. Data were analyzed by paired Student's t test. No significant differences between the strains were observed.
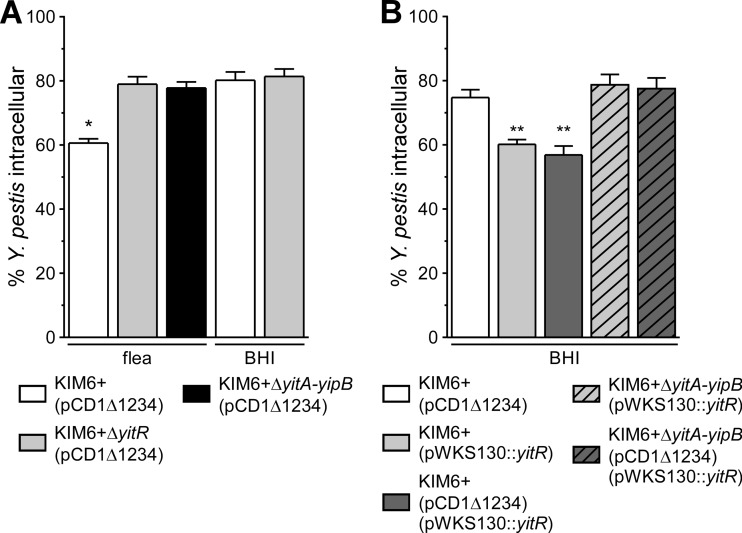
Y. pestis Tc proteins confer resistance to phagocytosis by murine PMNs.
We next investigated whether Y. pestis producing Tc proteins is more resistant to phagocytosis by murine PMNs. Only 60.6% ± 1.3% (mean ± SEM) of Y. pestis KIM6+(pCD1Δ1234) cells isolated from fleas were phagocytosed by murine PMNs after 1 h, in contrast to KIM6+ ΔyitR(pCD1Δ1234) or KIM6+ ΔyitA-yipB(pCD1Δ1234), which were phagocytosed to significantly greater extents (79% ± 2.3% and 77.8% ± 1.9% intracellular, respectively) (Fig. 5A). Y. pestis KIM6+(pCD1Δ1234) grown in BHI and minimally expressing the Tc proteins (8) was phagocytosed similarly to KIM6+ ΔyitR(pCD1Δ1234) and KIM6+ ΔyitA-yipB(pCD1Δ1234) that were either isolated from fleas or grown in BHI (Fig. 5A), demonstrating that Tc protein expression is responsible for the increased resistance of flea-derived Y. pestis to phagocytosis by PMNs.
Fig 5.
Y. pestis Tc proteins are essential for T3SS-independent resistance to phagocytosis by PMNs. (A) Phagocytosis of Tc+ or Tc− (ΔyitR or ΔyitA-yipB) Y. pestis KIM6+ isolated from fleas or from stationary-phase BHI cultures grown at 21°C. All 3 strains contained pCD1Δ1234 (T3SS+). (B) Phagocytosis of in vitro-grown Y. pestis KIM6+ (minimally expressing the Tc proteins) or Tc+ KIM6+ overexpressing YitR (pWKS130::yitR) as a means to elicit expression of the Tc proteins. Phagocytosis of KIM6+ strains containing pCD1Δ1234, which encodes the T3SS, as well as that of strains lacking the T3SS, is shown. The means and standard errors for at least three experiments are indicated. Data were analyzed by one-way ANOVA with the Bonferroni posttest. *, P < 0.001 compared to all other samples; **, P < 0.002 compared to parental KIM6+ or Tc− KIM6+. No significant differences were observed between Y. pestis pairs with and without the pCD1Δ1234 plasmid.
Overexpression of YitR increases expression of the Tc proteins when Y. pestis is grown in BHI broth (8, 9). We investigated whether increasing the expression of the Tc proteins would confer Y. pestis grown in broth with a resistance to phagocytosis similar to levels of resistance seen when Y. pestis is isolated from fleas. Y. pestis containing the pWKS130::yitR plasmid overexpresses YitR and produces much more of the Tc proteins in vitro (8). KIM6+(pCD1Δ1234) containing pWKS130::yitR was more resistant to phagocytosis by PMNs (with only 56.9% ± 2.8% [mean ± SEM] of the bacteria being phagocytosed within 1 h) than the parental KIM6+(pCD1Δ1234) strain, which was 74.8% ± 2.4% intracellular (Fig. 5B). When the yitR plasmid was transformed into the ΔyitA-yipB mutant, no increased resistance to phagocytosis was observed (Fig. 5B), verifying that the resistance to phagocytosis was due to the Tc proteins and not the expression plasmid. Furthermore, we examined whether the Tc protein-dependent inhibition of phagocytosis by PMNs required the T3SS. Y. pestis KIM6+(pWKS130::yitR), which lacks a T3SS and overexpresses the Tc proteins, was as resistant to phagocytosis by PMNs as the T3SS-positive strain KIM6+(pCD1Δ1234)(pWKS130::yitR) (Fig. 5B). Therefore, the Tc proteins do not require the pCD1-encoded T3SS for the observed resistance to phagocytosis by PMNs.
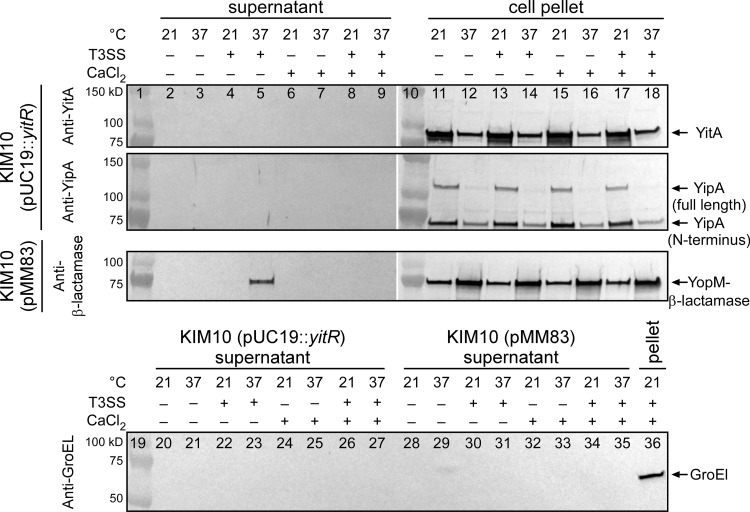
Y. pestis Tc proteins YitA and YipA are not secreted via the T3SS.
Y. pestis Tc proteins are produced predominately during growth at temperatures of <26°C (8, 9), whereas the T3SS is maximally expressed at 37°C (28). Nonetheless, Gendlina et al. indicated that the Tc proteins may be secreted via the T3SS (9). To further investigate this, we determined if the Y. pestis Tc proteins YitA and YipA are secreted into the culture supernatant during growth under conditions known to be optimal for T3SS-dependent secretion of effector Yop proteins (9, 25, 29, 30). Y. pestis strain KIM10 was used preferentially because it lacks the Pla protease, which is implicated in the degradation of secreted effector proteins (31), although KIM5 and KIM6 variants were also tested. Y. pestis KIM10 containing the plasmid pMM83 (32), which expresses YopM–β-lactamase, was used as a control for T3SS-dependent secretion. Strains were transformed with pCR-XL-TOPO::yitR or pUC19::yitR to increase the expression of the Tc proteins under liquid culture conditions. To verify that cellular lysis was not the reason for the presence of proteins in the supernatant, the level of the intracellular protein GroEL was also monitored. As expected, we were able to detect T3SS-dependent secretion of YopM–β-lactamase in the culture supernatant after incubation of Y. pestis at 37°C for ∼2 h in medium lacking CaCl2 (Fig. 6). Importantly, YopM–β-lactamase was detectable in the culture supernatant when GroEL was not (Fig. 6, lanes 28 to 35). However, despite comparably high levels of YitA and YipA production (Fig. 6, lanes 11 to 18), we never detected T3SS-dependent secretion of either Tc protein (Fig. 6, lanes 2 to 9). The same results were obtained using both KIM5 and KIM6 overexpressing YitR or KIM10 and KIM6 transformed with pCD1Δ1234 and overexpressing YitR (data not shown).
Fig 6.
T3SS-dependent secretion of the Y. pestis effector protein YopM but not the Tc proteins YitA and YipA. Y. pestis KIM10(pUC19::yitR) and Y. pestis(pMM83) (expresses YopM–β-lactamase) with or without pCD1::kan (T3SS+ or T3SS−) were grown in TMH overnight at 21°C, normalized, and then subcultured into TMH with or without CaCl2 and incubated at either 21°C or 37°C for an additional 4 h. Concentrated culture supernatants were analyzed by Western blotting using anti-YitA, anti-YipA, and anti-β-lactamase antibodies. Anti-GroEL was used to evaluate cellular lysis. Results are representative of three separate experiments.
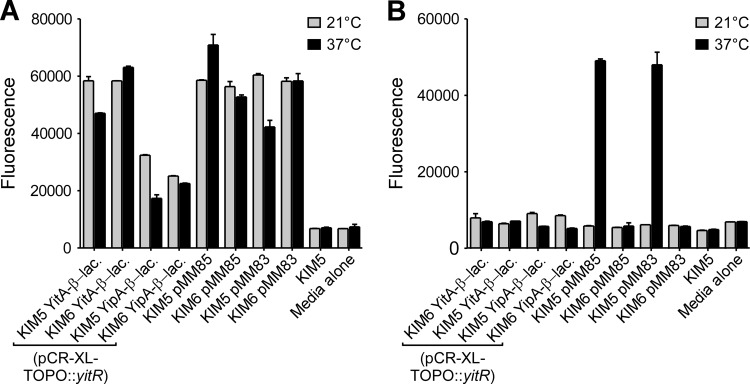
To ensure that our method of detection was sensitive enough to detect low levels of protein in the supernatant, we also measured β-lactamase activity in the culture supernatants of KIM5 YitA– or YipA–β-lactamase (pCR-XL-TOPO::yitR) and KIM6 YitA– or YipA–β-lactamase (pCR-XL-TOPO::yitR) strains in assays using the β-lactamase substrate fluorocillin. KIM5(pMM83) or KIM5(pMM85) and KIM6(pMM83) or KIM6(pMM85), expressing YopM–β-lactamase or YopE–β-lactamase, respectively, were used as positive controls for T3SS-dependent protein secretion. As before, although YitA–β-lactamase and, to a lesser extent, YipA–β-lactamase were detected in whole-cell lysates (Fig. 7A), β-lactamase activity was not detected in the culture supernatants under any growth conditions (Fig. 7B). Conversely, both YopE–β-lactamase and YopM–β-lactamase were detected only in the culture supernatants of Y. pestis with a T3SS (KIM5) grown at 37°C (Fig. 7B). Thus, under conditions where YopE and YopM are secreted via the T3SS, YitA and YipA were not detected.
Fig 7.
Detection of T3SS-dependent secretion of YopE– and YopM–β-lactamase but not YitA– or YipA–β-lactamase in culture supernatants. Y. pestis(pCR-XL-TOPO::yitR) strains with (KIM5) or without (KIM6) the pCD1 virulence plasmid encoding the T3SS and engineered to express YitA–, YipA–, YopM– (pMM83), or YopE–β-lactamase (pMM85) were grown in TMH overnight at 21°C, normalized, and then subcultured in TMH (without CaCl2) and incubated at 21°C (gray columns) or 37°C (black columns) for an additional 4 h. Bacterial lysates (A) and culture supernatants (B) were incubated with fluorocillin for 4 h, and the fluorescence of each sample was measured. The means and standard errors for three experiments are indicated. Y. pestis KIM5 lacking β-lactamase fusion proteins and sterile TMH medium were used as background controls.
Y. pestis YitA– and YipA–β-lactamase fusion proteins are not translocated into CHO-K1 cells or human PMNs via the T3SS.
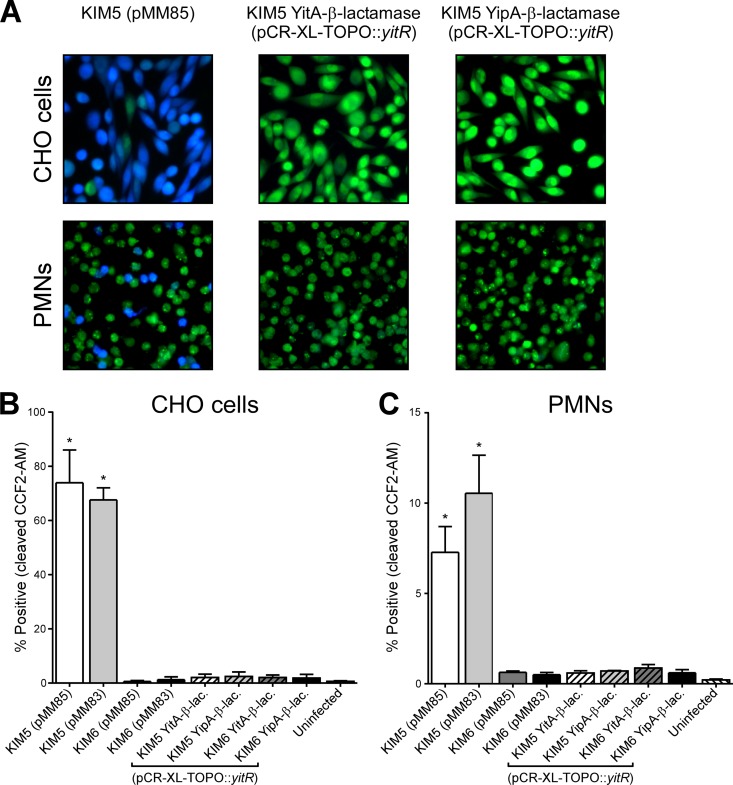
The N terminus of the Y. pestis Tc protein YipB can reportedly direct translocation of CyaA via the T3SS into insect and mammalian cells (9). Since the N terminus of YipA is ∼96% homologous to that of YipB (9), we utilized our YitA–β-lactamase- and YipA–β-lactamase-expressing strains to determine if either of these full-length proteins is translocated into either CHO cells or human PMNs. Y. pestis KIM5 and KIM6 containing either plasmid pMM85 or pMM83 were used as positive controls for T3SS-dependent translocation of YopE–β-lactamase or YopM–β-lactamase, respectively (32). Y. pestis KIM5, KIM6, and KIM6(pCD1Δ1234) were transformed with pCR-XL-TOPO::yitR to increase the expression of the Tc proteins. Y. pestis was grown overnight at 21°C and either maintained at 21°C or transferred to 37°C for ∼2 h prior to infection of CHO cells or PMNs. When the bacteria were maintained at 21°C, we did not detect translocation of YopE–, YopM–, YitA–, or YipA–β-lactamase into CHO cells or PMNs (data not shown). When the cells had been transferred to 37°C for 2 h, T3SS-dependent translocation of YopE–β-lactamase and YopM–β-lactamase into CHO cells was readily detected by microscopy (Fig. 8A) and flow cytometry (Fig. 8B). T3SS-dependent translocation of YopE–β-lactamase and YopM–β-lactamase into human PMNs was also detected (Fig. 8A and C), although not to the same extent as that with CHO cells (7.3 to 10.5% compared to 62.6 to 74.0%). No translocation of YopE–β-lactamase or YopM–β-lactamase was detected above background levels when Y. pestis did not have the T3SS (KIM6) (Fig. 8B and C and data not shown). In contrast, we did not detect any T3SS-dependent or -independent translocation of YitA– or YipA–β-lactamase into CHO cells (Fig. 8A and B) or human PMNs (Fig. 8A and C) with any of our strains. Our results indicate that under our assay conditions, native full-length YitA and YipA or YitA– and YipA–β-lactamase are neither secreted into culture supernatant via the T3SS nor translocated into CHO cells or PMNs.
Fig 8.
YopM– and YopE–β-lactamase are translocated into CHO cells and PMNs via the T3SS, but YitA– and YipA–β-lactamase are not. (A) Representative images of CHO cells and human PMNs that were loaded with the fluorescent β-lactamase substrate CCF2-AM and incubated with Y. pestis KIM5(pMM85), expressing YopE–β-lactamase, KIM5(pCR-XL-TOPO::yitR), expressing YitA–β-lactamase, or KIM5(pCR-XL-TOPO::yitR), expressing YipA–β-lactamase. Intact CCF2-AM fluoresces green, and β-lactamase-cleaved CCF2-AM fluoresces blue. The cumulative flow cytometry results for three separate experiments are shown as the percentages of CHO cells (B) and PMNs (C) that contained cleaved CCF2-AM following incubation with Y. pestis with (KIM5) or without (KIM6) the T3SS-encoding pCD1 and expressing either YitA–, YipA–, YopE–, or YopM–β-lactamase. Uninfected cells or cells incubated with KIM5 lacking β-lactamase fusion proteins were used as negative controls. The means and standard errors are indicated. Data were analyzed by one-way ANOVA with the Bonferroni posttest. *, P < 0.05 [KIM5(pMM85) and KIM5(pMM83) compared to the other strains tested].
DISCUSSION
Y. pestis tc genes are highly expressed, and the Tc proteins YitA and YipA have been shown to be produced while Y. pestis is in the flea vector (7, 8). However, the Tc proteins are not toxic to fleas (7, 8, 13) or required to infect the flea or establish blockage of the flea proventriculus (8), which is necessary for late-stage transmission of Y. pestis to the mammalian host (5, 16). In this study, we show that fleas coinfected with Tc+ and Tc− Y. pestis maintain a consistent ratio of parental to mutant Y. pestis in the midgut and proventriculus. Furthermore, the ΔyitA-yipB mutant forms a biofilm within the proventriculus and colocalizes with Tc+ Y. pestis. This finding affirms that neither strain has a competitive advantage or is selected for in the flea environment. Consistent with these data, parental and mutant Y. pestis strains were transmitted from coinfected fleas at approximately the same efficiency, further indicating that both are equally capable of transmission to the mammalian host. Thus, the Y. pestis Tc proteins have no detectable role for survival within or biofilm-dependent transmission from the rat flea X. cheopis.
Y. pestis cells isolated from fleas (and expressing Tc proteins) are, however, more resistant to killing by human (16) and murine (15) PMNs. Flea-derived Y. pestis is also resistant to phagocytosis by murine macrophages (7). In this study, we show that the Tc proteins are responsible for the flea-specific increased resistance of Y. pestis to killing by murine PMNs. The Tc protein-dependent resistance to killing was not a result of increased survival of Y. pestis within PMNs or PMN cytotoxicity induced by the Tc proteins. Rather, resistance of flea-derived Y. pestis to killing by PMNs correlated with a Tc protein-dependent resistance to phagocytosis. Accordingly, overexpression of the Tc proteins conferred culture-grown Y. pestis with a T3SS-independent resistance to phagocytosis and killing comparable to that of flea-derived bacteria. Our results indicate that although the Y. pestis Tc proteins are not insecticidal, they are active against PMNs. This is in agreement with previous studies which have shown that Y. pestis and Yersinia pseudotuberculosis Tc proteins are not toxic to Sf9 insect cells, M. sexta larvae, or X. cheopis (12, 13) but are active against mammalian NIH 3T3 and Caco-2 cells (12). Additionally, Yersinia enterocolitica Tc protein homologues are only mildly insecticidal and nematocidal when fed orally (33–35) but are important in colonization of the gastrointestinal tract of mice (36). In contrast, the Tc proteins of P. luminescens, the closely related Xenorhabdus nematophilus, and Yersinia entomophaga (isolated from a diseased Costelytra zealandica larva) are all highly insecticidal (3, 34, 37, 38). Therefore, whereas the Tc proteins of P. luminescens, X. nematophilus, and Y. entomophaga are potent insecticidal toxins, the Tc proteins of Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica appear to have evolved to be active primarily against mammalian rather than insect cells.
Although the Tc proteins conferred Y. pestis with a resistance to phagocytosis by PMNs, the mechanism by which the Tc proteins act to hinder phagocytosis is still unclear. Tc proteins of Y. pestis (9), Y. pseudotuberculosis (12, 39), P. luminescens (1, 40), Y. entomophaga (37), and X. nematophilus (41) have been shown to form large-molecular-weight protein complexes. In general, Tc protein complexes are composed of class A (TcA; Y. pestis YitA and YitB), class B (TcB; Y. pestis YitC), and class C (TcC; Y. pestis YipA and YipB) Tc proteins (9, 37, 41–46). Recent studies on class A Tc proteins from P. luminescens (46), X. nematophilus (41), and Y. entomophaga (43) indicate that class A (TcA) proteins form a multimeric transmembrane pore which interacts with a complex composed of a class B and a class C Tc protein. This holotoxin is theorized to bind an as yet unknown receptor on the target cell surface, which induces endocytosis of the Tc protein complex. Acidification of the endosome is thought to trigger penetration of the membrane by the pore and translocation of the B-C complex into the cytoplasm of the cell (45, 46), although actual translocation of the B-C complex has not yet been observed (46). Once in the cell, B-C Tc protein complexes are able to directly modify the actin of the target cell (4, 12, 44) and can prevent phagocytosis by affected cells (44).
How well the less-insecticidal Y. pestis Tc proteins fit the model described above is unknown. The Y. pestis Tc proteins YipA and YipB share amino acid similarity with the class C TcC proteins of P. luminescens, and YipA also contains a putative protein tyrosine phosphatase (PTP) domain similar to that of Y. pestis YopH, a T3SS-secreted effector protein able to alter host cell signaling pathways and disrupt phagocytosis by host cells (47). Therefore, either YipA or YipB (in conjunction with YitA, -B, and -C) may gain access to the PMN cytosol and hinder phagocytosis of Y. pestis by the affected PMN. However, how individual Tc proteins or Tc protein complexes are released from the bacteria in order to access target host cells has not been described. Theoretically, the Tc proteins could interact with host cells following their active secretion from bacteria or after their direct translocation into the host cell (T3SS dependent or T3SS independent). They could also be released from lysed bacteria or exert their mode of action from the surfaces of the bacteria following direct contact with the host cell.
To determine if Y. pestis Tc proteins are actively secreted as a means to access host cells and whether their secretion is T3SS dependent or T3SS independent, we tested whether full-length YitA or YipA is secreted into culture supernatants under conditions permissive and nonpermissive for T3SS-dependent secretion. Although we could readily detect T3SS-dependent secretion of YopE– and YopM–β-lactamase, we were unable, under assay conditions where bacterial lysis was not evident (based on the absence of the intracellular bacterial chaperonin GroEL in the culture supernatants), to detect any secretion of either native YitA or YipA by Western blotting or YitA– or YipA–β-lactamase fusion proteins by the more sensitive fluorocillin enzymatic assay. Our results are in agreement with the findings of Bresolin et al., which indicated that Y. enterocolitica Tc proteins were not present in the culture supernatant after growth of bacteria at 10°C or 30°C (34). Therefore, our data suggest that unlike the Tc proteins of P. luminescens and Y. entomophaga, which are readily detected in culture supernatant (1, 37, 40, 43, 48), the Tc proteins (particularly YitA and YipA) of Y. pestis are not secreted. However, under bacterial growth conditions similar to ours, Gendlina et al. previously showed that the N-terminal 100 amino acids of YitA and YipA were able to direct T3SS-dependent secretion of an epitope tag into culture supernatants (9). Since we did not detect secretion of full-length YitA or YipA, the additional amino acids of native YitA and YipA may interfere with or preclude T3SS-dependent secretion. This could be due to incorporation of full-length YitA or YipA proteins into Tc-ABC protein complexes (9). Prevention of T3SS-dependent Tc protein secretion or translocation due to formation of protein complexes is consistent with the findings of Gendlina et al., which showed that a CyaA construct consisting of the N-terminal 100 amino acids of YipB was translocated into Sf9 cells more efficiently than a construct with the N-terminal 250 amino acids of YipB (9).
In contrast to our results, Gendlina et al. did detect full-length YitB in culture supernatants (9), and Hares et al. reported that the Y. pseudotuberculosis B-C Tc protein complex (TcaC-TccC; homologous to Y. pestis YitB-YipA or -B) was detectable in culture supernatants (12). We did not evaluate secretion of YitB, and these data suggest that unlike YitA and YipA, YitB is secreted by Y. pestis. However, a control for bacterial lysis was not included in either study, so it is possible that YitB was released only following bacterial cell lysis rather than being actively secreted. For example, in some of our experiments, both YitA and YipA were detectable in culture supernatants, but only when GroEL was also detected (data not shown).
We also examined whether β-lactamase fusions of the Tc proteins YitA and YipA are translocated into phagocytic (PMNs) and nonphagocytic (CHO) cells via the T3SS or by an alternative pathway. Although we were consistently able to detect T3SS-dependent translocation of the control proteins YopE–β-lactamase and YopM–β-lactamase into CHO cells and human PMNs, we were unable to detect translocation of YitA–β-lactamase or YipA–β-lactamase under any conditions. Together with the lack of evidence for secretion into culture supernatants, our data indicate that the Tc proteins YitA and YipA are neither actively secreted nor translocated into PMNs or CHO cells via the T3SS. Note that since YipA is processed and detected at ∼106 kDa as a full-length protein, and at ∼73 kDa for the N-terminal portion (8), it was only possible to detect translocation of the full-length YipA–β-lactamase and the YipA C terminus–β-lactamase, which contains the putative PTP domain (8). Nonetheless, T3SS-dependent secretion of the N-terminal portion of YipA was not detected in culture supernatants (Fig. 6), suggesting that it is also not translocated into host cells by the T3SS. Additionally, it is unknown whether the other Tc proteins (YitB, YitC, or YipB) are translocated into host cells. The N terminus of YipB (a class C Tc protein) was previously shown to be able to direct translocation of Cya (9), suggesting that YipB rather than YipA is translocated and plays an essential role in Tc protein-dependent resistance to phagocytosis by PMNs, plausibly by disrupting their actin cytoskeleton as reported with NIH 3T3 cells (12). However, our results also show that Tc protein-dependent inhibition of phagocytosis by PMNs does not require the T3SS (Fig. 5), indicating that the mechanism of action for the Tc proteins is independent of the T3SS.
A third possibility is that direct contact of the bacterial membrane-associated Tc proteins with PMNs, without secretion or translocation, is sufficient to inhibit phagocytosis. Both YitA and YipA are present in the outer membrane fraction of Y. pestis, and YitA is outer surface exposed (8). Since GroEL has been associated with outer membrane vesicles (OMVs) (37, 49) and YitA and YipA were detectable in culture supernatants that also contained GroEL, the Tc proteins might also be released from the bacteria in association with OMVs prior to or in conjunction with bacterial lysis. In support of this, Y. entomophaga Tc proteins were associated with GroEL in culture supernatants, and transmission electron microscopy images indicated the presence of OMVs that contained pyramidal structures which were absent in a Tc− strain (37). The production of OMVs by Y. pestis is not well described, although small vesicles which were either closely associated with or possibly blebbing from Y. pestis within macrophage vacuoles have been observed (50). Further studies are necessary to determine the conditions under which Y. pestis may release OMVs, whether Tc proteins are associated with them, and the mechanisms by which the Y. pestis Tc proteins inhibit uptake by PMNs.
In conclusion, although the Tc proteins are highly expressed by Y. pestis while in the flea vector, the Tc proteins appear to have no role in flea infection or transmission to the mammalian host. In contrast, we show that the Tc proteins play a role in resistance to phagocytosis and killing by PMNs. Tc protein-dependent resistance to phagocytosis did not require the T3SS. In keeping with this, the Tc proteins YitA and YipA did not appear to be secreted or translocated into CHO cells or PMNs via the T3SS. Rather, our data indicate that Tc proteins are present on the surface of Y. pestis, where they may have a role in limiting uptake by phagocytes immediately following transmission from the flea. This would provide Y. pestis transmitted from fleas with an initial defense against the innate immune response prior to upregulation of the T3SS and effector Yop proteins.
ACKNOWLEDGMENTS
We thank Carleen Collins and Doris LaRock (University of Washington) for providing strain KIM6+ ΔyitA-yipB. We thank Melanie Marketon and Rebecca Dewoody (Indiana University) for plasmids pMM83 and pMM85. We thank Adeline Whitney, Kevin Braughton, Scott Kobayashi, and Frank DeLeo for providing human PMNs and Kelsi Sandoz and Bob Heinzen for providing CHO-K1 cells. We thank Levi O'Loughlin (Washington State University), Rebecca Dewoody, Chris Bosio, Jeffrey Shannon, and Iman Chouikha for critical reviews of the manuscript.
This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print 19 August 2013
REFERENCES
- 1.Bowen D, Rocheleau TA, Blackburn M, Andreev O, Golubeva E, Bhartia R, Ffrench-Constant RH. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129–2132 [DOI] [PubMed] [Google Scholar]
- 2.Waterfield NR, Bowen DJ, Fetherston JD, Perry RD, Ffrench-Constant RH. 2001. The tc genes of Photorhabdus: a growing family. Trends Microbiol. 9:185–191 [DOI] [PubMed] [Google Scholar]
- 3.Waterfield NR, Dowling A, Sharma S, Daborn PJ, Potter U, Ffrench-Constant RH. 2001. Oral toxicity of Photorhabdus luminescens W14 toxin complexes in Escherichia coli. Appl. Environ. Microbiol. 67:5017–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterfield NR, Hares MC, Yang G, Dowling A, Ffrench-Constant RH. 2005. Potentiation and cellular phenotypes of the insecticidal toxin complexes of Photorhabdus bacteria. Cell. Microbiol. 7:373–382 [DOI] [PubMed] [Google Scholar]
- 5.Hinnebusch BJ. 2012. Biofilm-dependent and biofilm-independent mechanisms of transmission of Yersinia pestis by fleas. Adv. Exp. Med. Biol. 954:237–243 [DOI] [PubMed] [Google Scholar]
- 6.Vadyvaloo V, Jarrett C, Sturdevant D, Sebbane F, Hinnebusch BJ. 2007. Analysis of Yersinia pestis gene expression in the flea vector. Adv. Exp. Med. Biol. 603:192–200 [DOI] [PubMed] [Google Scholar]
- 7.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog. 6:e1000783. 10.1371/journal.ppat.1000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spinner JL, Jarrett CO, Larock DL, Miller SI, Collins CM, Hinnebusch BJ. 2012. Yersinia pestis insecticidal-like toxin complex (Tc) family proteins: characterization of expression, subcellular localization, and potential role in infection of the flea vector. BMC Microbiol. 12:296. 10.1186/1471-2180-12-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gendlina I, Held KG, Bartra SS, Gallis BM, Doneanu CE, Goodlett DR, Plano GV, Collins CM. 2007. Identification and type III-dependent secretion of the Yersinia pestis insecticidal-like proteins. Mol. Microbiol. 64:1214–1227 [DOI] [PubMed] [Google Scholar]
- 10.Motin VL, Georgescu AM, Fitch JP, Gu PP, Nelson DO, Mabery SL, Garnham JB, Sokhansanj BA, Ott LL, Coleman MA, Elliott JM, Kegelmeyer LM, Wyrobek AJ, Slezak TR, Brubaker RR, Garcia E. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186:6298–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. 2006. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. U. S. A. 103:11766–11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hares MC, Hinchliffe SJ, Strong PC, Eleftherianos I, Dowling AJ, Ffrench-Constant RH, Waterfield NR. 2008. The Yersinia pseudotuberculosis and Yersinia pestis toxin complex is active against cultured mammalian cells. Microbiology 154:3503–3517 [DOI] [PubMed] [Google Scholar]
- 13.Erickson DL, Waterfield NR, Vadyvaloo V, Long D, Fischer ER, Ffrench-Constant RH, Hinnebusch BJ. 2007. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: implications for the evolution of vector-borne transmission of plague. Cell. Microbiol. 9:2658–2666 [DOI] [PubMed] [Google Scholar]
- 14.Fukuto HS, Svetlanov A, Palmer LE, Karzai AW, Bliska JB. 2010. Global gene expression profiling of Yersinia pestis replicating inside macrophages reveals the roles of a putative stress-induced operon in regulating type III secretion and intracellular cell division. Infect. Immun. 78:3700–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spinner JL, Hinnebusch BJ. 2012. The life stage of Yersinia pestis in the flea vector confers increased resistance to phagocytosis and killing by murine polymorphonuclear leukocytes. Adv. Exp. Med. Biol. 954:159–163 [DOI] [PubMed] [Google Scholar]
- 16.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, Hinnebusch BJ. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783–792 [DOI] [PubMed] [Google Scholar]
- 17.Bosio CF, Jarrett CO, Gardner D, Hinnebusch BJ. 2012. Kinetics of innate immune response to Yersinia pestis after intradermal infection in a mouse model. Infect. Immun. 80:4034–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartra SS, Jackson MW, Ross JA, Plano GV. 2006. Calcium-regulated type III secretion of Yop proteins by an Escherichia coli hha mutant carrying a Yersinia pestis pCD1 virulence plasmid. Infect. Immun. 74:1381–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 20.Hinnebusch BJ, Fischer ER, Schwan TG. 1998. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Infect. Dis. 178:1406–1415 [DOI] [PubMed] [Google Scholar]
- 21.Hinnebusch BJ, Perry RD, Schwan TG. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367–370 [DOI] [PubMed] [Google Scholar]
- 22.Siemsen DW, Schepetkin IA, Kirpotina LN, Lei B, Quinn MT. 2007. Neutrophil isolation from nonhuman species. Methods Mol. Biol. 412:21–34 [DOI] [PubMed] [Google Scholar]
- 23.Spinner JL, Cundiff JA, Kobayashi SD. 2008. Yersinia pestis type III secretion system-dependent inhibition of human polymorphonuclear leukocyte function. Infect. Immun. 76:3754–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pujol C, Bliska JB. 2003. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect. Immun. 71:5892–5899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straley SC, Bowmer WS. 1986. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect. Immun. 51:445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi SD, Voyich JM, Buhl CL, Stahl RM, DeLeo FR. 2002. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc. Natl. Acad. Sci. U. S. A. 99:6901–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. 2005. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J. Infect. Dis. 191:1907–1912 [DOI] [PubMed] [Google Scholar]
- 28.Plano GV, Straley SC. 1995. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J. Bacteriol. 177:3843–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehigh RJ, Sample AK, Brubaker RR. 1989. Expression of the low calcium response in Yersinia pestis. Microb. Pathog. 6:203–217 [DOI] [PubMed] [Google Scholar]
- 30.Mehigh RJ, Brubaker RR. 1993. Major stable peptides of Yersinia pestis synthesized during the low-calcium response. Infect. Immun. 61:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sodeinde OA, Sample AK, Brubaker RR, Goguen JD. 1988. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect. Immun. 56:2749–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marketon MM, DePaolo RW, DeBord KL, Jabri B, Schneewind O. 2005. Plague bacteria target immune cells during infection. Science 309:1739–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchs TM, Bresolin G, Marcinowski L, Schachtner J, Scherer S. 2008. Insecticidal genes of Yersinia spp.: taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol. 8:214. 10.1186/1471-2180-8-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bresolin G, Morgan JA, Ilgen D, Scherer S, Fuchs TM. 2006. Low temperature-induced insecticidal activity of Yersinia enterocolitica. Mol. Microbiol. 59:503–512 [DOI] [PubMed] [Google Scholar]
- 35.Spanier B, Starke M, Higel F, Scherer S, Fuchs TM. 2010. Yersinia enterocolitica infection and tcaA-dependent killing of Caenorhabditis elegans. Appl. Environ. Microbiol. 76:6277–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tennant SM, Skinner NA, Joe A, Robins-Browne RM. 2005. Homologues of insecticidal toxin complex genes in Yersinia enterocolitica biotype 1A and their contribution to virulence. Infect. Immun. 73:6860–6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurst MR, Jones SA, Binglin T, Harper LA, Jackson TA, Glare TR. 2011. The main virulence determinant of Yersinia entomophaga MH96 is a broad-host-range toxin complex active against insects. J. Bacteriol. 193:1966–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurst MR, Becher SA, Young SD, Nelson TL, Glare TR. 2011. Yersinia entomophaga sp. nov., isolated from the New Zealand grass grub Costelytra zealandica. Int. J. Syst. Evol. Microbiol. 61:844–849 [DOI] [PubMed] [Google Scholar]
- 39.Pinheiro VB, Ellar DJ. 2007. Expression and insecticidal activity of Yersinia pseudotuberculosis and Photorhabdus luminescens toxin complex proteins. Cell. Microbiol. 9:2372–2380 [DOI] [PubMed] [Google Scholar]
- 40.Bowen DJ, Ensign JC. 1998. Purification and characterization of a high molecular weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl. Environ. Microbiol. 64:3029–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheets JJ, Hey TD, Fencil KJ, Burton SL, Ni W, Lang AE, Benz R, Aktories K. 2011. Insecticidal toxin complex proteins from Xenorhabdus nematophilus: structure and pore formation. J. Biol. Chem. 286:22742–22749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ffrench-Constant R, Waterfield N. 2006. An ABC guide to the bacterial toxin complexes. Adv. Appl. Microbiol. 58:169–183 [PubMed] [Google Scholar]
- 43.Landsberg MJ, Jones SA, Rothnagel R, Busby JN, Marshall SD, Simpson RM, Lott JS, Hankamer B, Hurst MR. 2011. 3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity. Proc. Natl. Acad. Sci. U. S. A. 108:20544–20549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, Sheets JJ, Mannherz HG, Aktories K. 2010. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science 327:1139–1142 [DOI] [PubMed] [Google Scholar]
- 45.Lang AE, Schmidt G, Sheets JJ, Aktories K. 2011. Targeting of the actin cytoskeleton by insecticidal toxins from Photorhabdus luminescens. Naunyn Schmiedebergs Arch. Pharmacol. 383:227–235 [DOI] [PubMed] [Google Scholar]
- 46.Gatsogiannis C, Lang AE, Meusch D, Pfaumann V, Hofnagel O, Benz R, Aktories K, Raunser S. 2013. A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature 495:520–523 [DOI] [PubMed] [Google Scholar]
- 47.Viboud GI, Bliska JB. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59:69–89 [DOI] [PubMed] [Google Scholar]
- 48.Blackburn M, Golubeva E, Bowen D, Ffrench-Constant RH. 1998. A novel insecticidal toxin from Photorhabdus luminescens, toxin complex a (Tca), and its histopathological effects on the midgut of Manduca sexta. Appl. Environ. Microbiol. 64:3036–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi MC, Sharma A, Kant S, Birah A, Gupta GP, Khan SR, Bhatnagar R, Banerjee N. 2008. An insecticidal GroEL protein with chitin binding activity from Xenorhabdus nematophila. J. Biol. Chem. 283:28287–28296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straley SC, Harmon PA. 1984. Yersinia pestis grows within phagolysosomes in mouse peritoneal macrophages. Infect. Immun. 45:655–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Une T, Brubaker RR. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43:895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perry RD, Fetherston JD. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun YC, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, Darby C, Hinnebusch BJ. 2011. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6:e19267. 10.1371/journal.pone.0019267 [DOI] [PMC free article] [PubMed] [Google Scholar]