Abstract
Rad23a and Rad23b proteins are linked to nucleotide excision DNA repair (NER) via association with the DNA damage recognition protein xeroderma pigmentosum group C (XPC) are and known to be implicated in protein turnover by the 26S proteasome. Rad23b-null mice are NER proficient, likely due to the redundant function of the Rad23b paralogue, Rad23a. However, Rad23b-null midgestation embryos are anemic, and most embryos die before birth. Using an unbiased proteomics approach, we found that the majority of Rad23b-interacting partners are associated with the ubiquitin-proteasome system (UPS). We tested the requirement for Rad23b-dependent UPS activity in cellular proliferation and more specifically in the process of erythropoiesis. In cultured fibroblasts derived from embryos lacking Rad23b, proliferation rates were reduced. In fetal livers of Rad23b-null embryos, we observed reduced proliferation, accumulation of early erythroid progenitors, and a block during erythroid maturation. In primary wild-type (WT) erythroid cells, knockdown of Rad23b or chemical inhibition of the proteasome reduced survival and differentiation capability. Finally, the defects linked to Rad23b loss specifically affected fetal definitive erythropoiesis and stress erythropoiesis in adult mice. Together, these data indicate a previously unappreciated requirement for Rad23b and the UPS in regulation of proliferation in different cell types.
INTRODUCTION
Mammalian orthologues of the yeast RAD23 gene, Rad23a and Rad23b, play a role in two distinct processes, nucleotide excision DNA repair (NER) and the ubiquitin-proteasome system (UPS) (1–3). The role of Rad23a and Rad23b in these processes has been inferred from characterization of knockout mice, as well as extrapolations from yeast studies (4–7).
NER is a genome maintenance pathway responsible for repairing a wide variety of DNA bulky lesions, including those induced by sunlight (8). Rad23p was first identified in yeast after mutant screening for UV sensitivity and was shown to be implicated in NER (4). In mammals, a trimeric complex consisting of xeroderma pigmentosum group C (XPC), CEN2, and either RAD23A or RAD23B (9) recognizes distortions in the DNA helix caused by bulky lesions. After damage recognition, other NER factors are recruited to complete the cut-and-patch-type repair (10, 11). The role of Rad23b in NER is to stabilize Xpc and to facilitate effective loading of Xpc on DNA lesions (5).
The UPS performs controlled protein degradation by the orchestrated targeting, ubiquitin labeling, and degradation of proteins in such diverse processes as signal transduction, cell cycle control, apoptosis, antigen presentation, and DNA repair (12–17). Ubiquitylation of proteins occurs by the sequential action of an ubiquitin activation enzyme (E1), a conjugation enzyme (E2), a ligase (E3), and in some cases a polyubiquitin chain conjugation enzyme (E4). The Rad23 family proteins function in the UPS to bind and shuttle polyubiquitylated proteins to the 26S proteasome where actual degradation occurs, although it has also been reported to protect proteins from degradation (1, 2). Rad23 proteins harbor two ubiquitin-binding (UBA) domains capable of binding size-restricted polyubiquitin chains of 4 to 6 moieties (18, 19) and an amino-terminal ubiquitin-like (UBL) domain that interacts with the 26S proteasome. In yeast, substrates targeted to the proteasome for degradation by Rad23 include the G1 cyclin-Cdk inhibitor Far1p, the S Cdk inhibitor Sic1p, the delta-9 fatty acid desaturase Ole1p, the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase isozyme Hmg2p, and unsaturated fatty acid synthesis transcription factor Spt23p (21–23). Rad23 activity is partially redundant with the polyubiquitin-binding proteosomal subunit Rpn10p and the UBL- and UBA-containing Dsk2p (22, 24, 25). Proteasomes isolated from rad23Δ yeast have greatly reduced proteasome activity toward synthetic polyubiquitylated substrates, while 20S peptidase activity per se is not affected (23).
Although Rad23a-null mice are born normally (5), Rad23b-null mice are born at sub-Mendelian ratios (approximately 10% of expected) (6). Surviving mice suffer from facial dysmorphologies and exhibit male sterility. Rad23b-null midgestation embryos are anemic. This condition develops from 12.5 days postcoitum (dpc) onwards, is still present at 15.5 dpc, and might partly account for the embryonic lethality observed. However, the few surviving Rad23b-null mice do not suffer from anemia (6), suggesting a transient defect. Rad23b-null mice with only one Rad23a allele display a more severe anemia and die at 14.5 dpc; Rad23a/Rad23b double-knockout (KO) mice are the most severely affected and die at 8.5 dpc (5). Although embryonic fibroblasts derived from double-KO animals are deficient in NER (5), Rad23a or Rad23b single-KO fibroblasts are NER proficient (5, 6). Taken together, these data suggest that Rad23a and Rad23b have overlapping functions in NER but that Rad23a can only partially complement Rad23b in non-NER-related functions, including fetal erythropoiesis.
Erythropoiesis is the process by which erythroid cells are formed. There are two consecutive waves of erythropoiesis in mammals: primitive and definitive (26). In the mouse, primitive erythropoiesis starts in the yolk sac at 7.5 dpc. Primitive erythroid cells are released nucleated to the embryonic bloodstream, where they divide and eventually enucleate from 8.0 to 15.0 dpc (26). Definitive erythropoiesis starts around 11 dpc in the fetal liver and moves to the spleen and later to the bone marrow, which remain the adult sites of hemato/erythropoeisis. Proliferation and terminal differentiation are tightly linked in definitive erythropoiesis. Proerythroblasts, which are committed erythroid progenitors, proceed through four differentiation cell divisions, followed by enucleation and finally maturation of reticulocytes into erythrocytes (27). This process requires orchestrated protein synthesis and degradation in order to produce the enormous amount of hemoglobin (HCB) relative to the total protein content of the cell. The product of definitive erythropoiesis, the enucleated erythrocyte, appears in the blood at 11.0 to 12.0 dpc and gradually replaces primitive erythroid cells by 16.0 dpc.
Here, we tested the hypothesis that Rad23b deficiency in mammals would cause a malfunction in proteasomal degradation due to the poor recognition/delivery of ubiquitylated proteins to the proteasome, resulting in the observed midgestation anemia. Although the anemia is likely to contribute to the high rate of embryonic lethality, it is not the only process that is affected by Rad23b deficiency. Since Rad23 targets in yeast include cell cycle regulatory proteins (21), we anticipated that Rad23b deficiency in mammals would also impact general cellular proliferation. To test this hypothesis, we used an unbiased proteomics approach to identify interacting partners of Rad23b and functional assays in Rad23b-null embryos and cells to test the effects of Rad23b depletion on cellular proliferation and erythropoiesis.
MATERIALS AND METHODS
Mice.
Rad23b-null mice were generated previously and crossed into the C57BL/6 background (6). Matings were set with heterozygous Rad23b-null mice, and embryos were collected at 11.5 dpc to 13.5 dpc. Embryonic blood and fetal liver single-cell suspensions were prepared as described previously (28), and cell counts were determined in an electronic cell counter (CASY-1; Schärfe Systems).
In vivo transplantation and stress erythropoiesis assay.
Wild-type (WT) or Rad23b-null embryos at 12.5 dpc were isolated, fetal livers were dissected and dissociated as previously described (29), and PCR genotyping was performed on a piece of tail. Intravenous injection of total fetal liver cells (from male WT or Rad23b-null embryos) into irradiated adult WT female recipients (C57BL/6) was performed as previously described (30). Recipient mice received a split dose of 9 Gy of gamma irradiation (137Cs source) prior to injection of 0.1 or 0.5 embryo equivalents (ee) of fetal liver cells together with 2 × 105 spleen cells (recipient background) to promote short-term survival. Blood obtained at 1 month posttransplantation was used for semiquantitative PCR against a donor cell marker (Ymt) to calculate the percentage of donor contribution relative to a standard curve of DNA control dilutions (from 0 to 100%). Recipients with chimerism of greater than 50% were used for further analysis. Phenylhydrazine (PHZ) treatment was performed as described previously (31). All animal experiments were approved by the animal ethics and welfare committee of Erasmus MC.
Cell culture.
Fetal liver-derived single-cell suspensions were cultured either in hanging drops or in suspension as described previously (31, 32), in the presence of 0.5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) (Invitrogen) or 5 μM MG132 (proteasome inhibitor; Sigma) when specified. The relative cell division was calculated by dividing the CFSE mean fluorescence intensity (MFI) at the beginning of the culture by the CFSE MFI at collection. Burst-forming unit erythroid (BFUe) colony assays were performed as previously described (31–33).
The mouse I/11 proerythroblast cell line was cultured as described previously (32). For proliferation assays, I/11 cells were cultured in the presence of 5 μM, 0.5 μM, or 0.05 μM MG132 (proteasome inhibitor; Sigma) or 5 μM, 0.5 μM, or 0.05 μM PS341 (proteasome inhibitor; BioVision) when specified and collected at days 1, 2, and 3 after treatment. For differentiation assays, I/11 cells were treated with the same concentrations of proteasome inhibitors at day 0 (start of the differentiation culture) and at day 1 or at day 2 after inducing differentiation and were analyzed every 24 h until day 3 of differentiation.
Mouse embryonic fibroblasts (MEFs) were generated as described previously (34). For serial passaging, cultures were trypsinized at ∼80% confluence, reseeded at a density of 3 × 105 cells/10-cm dish, and maintained at either 3% or 20% oxygen tension. For low- versus high-seeding-density experiments, cells were seeded at an initial density of 2.5 ×105 (low) or 3.75 × 105 (high) cells/10-cm dish and cultured in a 3%O2–5%CO2 incubator. Acute-sensitivity assays were performed by seeding 40,000 cells per well of a six-well plate in duplicate and then treating them the next day with UV light, gamma irradiation, or heat. Cells were trypsinized and counted in a Coulter counter before the cells in the untreated control wells reached confluence.
Generation of Rad23b-YFP-FLAG embryonic stem cells and culture.
Bacterial artificial chromosome (BAC) clone RP23-302N23, spanning 200 kb of genomic mouse C57BL/6 sequence including the entire Rad23b locus, was ordered from the BACPAC Resource Center (BPRC) at Children's Hospital Oakland Research Institute (Oakland, CA, USA). A yellow fluorescent protein (YFP)-FLAG-LoxP cassette was inserted into the stop codon of the Rad23b gene using RecA-mediated recombination in Escherichia coli (35). A kanamycin/neomycin gene driven by the dual bacterial/mammalian gb2/Pgk promoter and flanked by LoxP sites was inserted in the SacB gene of the BAC vector using λ Red-mediated recombination (36). Two Rad23b-YFP-FLAG 129 embryonic stem (ES) cell clones, F11 and B11, were selected for further experiments based on construct integrity (as analyzed by Southern blotting and fluorescent in situ hybridization [FISH]) and YFP expression (as measured by flow cytometry). Both had a correct karyotype and 2 and 3 tandem copies, respectively, of the transgene. ES cells were cultured as described previously (5, 6).
Flow cytometry.
Flow cytometry analysis was performed on fresh single-cell suspensions. Antibodies used were annexin V-fluorescein isothiocyanate (FITC), cKit-phycoerythrin (PE), cKit-PECy7, CD9-allophycocyanin (APC), CD71-FITC, and Ter119-PE (all from BD Biosciences). Intracellular staining of Gata1 was done using the CytoFix/CytoPerm kit (BD Biosciences) with the antibodies Gata1 N6 (Santa Cruz) and anti-rat–FITC (Dako). 7-Aminoactinomycin D (7AAD), propidium iodide, or Hoechst-33342 (Molecular Probes, Invitrogen) was used to distinguish dead cells. For prospective isolation of erythroid cells during differentiation, 13.5-dpc fetal liver cells were stained with cKit, CD71, and Ter119 and sorted in the following populations: early (Ter119− CD117+ CD71+), maturing (CD117− CD71+ Ter119+), and mature (CD117− CD71− Ter119+). Circulating blood cells at 13.5 dpc were stained with Ter119 and CD9, and enucleated primitive cells were gated as Ter119+ CD9+ SSClow. Cell cycle analysis was done by staining 5 × 106 cells fixed in 75% ethanol (EtOH) with 8 μg propidium iodide plus 80 μg RNase in 400 μl 0.1% Triton X-100–phosphate-buffered saline (PBS). Cell sorting was performed in a FACSAria I, and measurements were performed in a FACScan (Becton, Dickinson Immunocytometry Systems). Data analysis was done using FlowJo software (version 9).
Western blotting.
Equal numbers of cells (4 × 107 cells/ml) were lysed with 2× Laemmli buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue, and 0.125 M Tris HCl, pH 6.8). Antibodies included those against Gata1 N6 (Santa Cruz) and mono- and polyubiquitinylated proteins (FK2 Enzo Life Sciences) and secondary antibodies conjugated to horseradish peroxidase (Dako). Blots were developed with enhanced chemiluminescence (ECL) as described by the manufacturer (GE Healthcare).
Immunoprecipitation and mass spectrometry.
ES cells expressing Rad23b-YFP-FLAG were lysed in radioimmunoprecipitation assay plus (RIPA+) buffer (50 mM Tris [pH 8], 1 mM EDTA, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 10% glycerol, protease inhibitors, 0.1 mM dithiothreitol [DTT]) on ice. Cleared extracts were incubated with anti-FLAG beads (M2; Sigma A2220) for 3 h at 4°C, washed in RIPA+ buffer, and eluted with 3×FLAG peptide (Sigma F4799) for 15 min on ice. Eluates were analyzed by Western blotting and Coomassie blue and silver staining and concentrated by trichloroacetic acid (TCA) precipitation if necessary before analysis by mass spectrometry (MS) as previously described (37). I/11 and fetal liver cells were lysed in RIPA+ buffer, and endogenous Rad23b and binding partners were coimmunoprecipitated using Rad23b antibody (Abcam) and protein A-magnetic beads (Dynabeads; Invitrogen) following the manufacturer's guidelines.
Nanoflow liquid chromatography-tandem MS (LC-MS/MS) was performed on an 1100 series capillary LC system (Agilent Technologies) coupled to an LTQ-Orbitrap mass spectrometer (Thermo). Peak lists were automatically created from raw data files using the Mascot Distiller software (version 2.1; MatrixScience). The Mascot search algorithm (version 2.2; MatrixScience) was used for searching against the NCBInr database (release NCBInr 20070812). The Mascot score cutoff value for a positive protein hit was set to 60. Individual peptide MS/MS spectra with Mascot scores below 40 were checked manually and either interpreted as valid identifications or discarded. Typical contaminants, also present in immunopurifications using beads coated with preimmune serum or antibodies directed against irrelevant proteins, were omitted. Data were filtered, compared, and analyzed with Ingenuity Pathway Analysis (Ingenuity Systems) in order to obtain a list of Rad23b-interacting partners common to both clones. This list was compared with two previously published sets of proteasome-interacting proteins (38, 39) to assess the percentage of known proteasome-interacting proteins among the Rad23b-interacting protein list. A graphic depiction of interacting partners was obtained with the Pathway tool from Ingenuity Pathway Analysis and edited with Adobe Illustrator CS4. Gene ontology (GO) classification was performed using Ensembl/Biomart. A similar procedure was followed to analyze Rad23b immunoprecipitations from erythroid cells.
Downregulation of Rad23b and Ubc in I/11 cells.
The mouse I/11 proerythroblast cell line was generated previously, and the cells were cultured as described previously (32). TRC short hairpin RNA (shRNA) library (Sigma, St. Louis, MO, USA) clones TRCN0000127119 (R1), TRCN0000127120 (R2), TRCN0000127121 (R3), TRCN0000127122 (R4), and TRCN0000127123 (R5) were used for knockdown of Rad23b, and TRCN0000098655 (U1), TRCN0000098656 (U2), TRCN0000098657 (U3), TRCN0000098658 (U4), and TRCN0000098659 (U5) were used for knockdown of Ubc. Lentivirus production was done as described previously (40). I/11 cells were transduced, and at 2 days after transduction the cells were grown in the presence of 1 μg/ml puromycin for 4 days. Cultures with verified Rad23b or Ubc downregulation were used for analysis.
qPCR.
Total RNA (1 μg) was converted to cDNA using SuperScript II reverse transcriptase according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). Expression levels of mRNAs were analyzed by quantitative real-time PCR (qPCR) with LightCycler SYBR green I PCR master mix on a LightCycler carousel-based system (both from Roche Applied Science). All reactions were performed in duplicate. Gene expression levels were calculated with the 2−ΔΔCT method (41). Target gene expression was normalized to Gapdh/Hprt expression. Primers used were as follows: Rad23b, 5′-ATGGCAACACTGGATAATGGC-3′ and 5′-TGTGAAGCAGCAACGATGAC-3′; Rad23a, 5′-GGAACCTGACGAGACGGTAA-3′ and 5′-TGCCAGCATAGATGAGTTTCTG-3′; Xpc, 5′-GGAGGATGATGAAGCGTTTC-3′ and 5′-GCTGTCGGCAGATGCTATT-3′; Ubc, 5′-AGGTCAAACAGGAAGACAGACGTA-3′ and 5′-TCACACCCAAGAACAAGCACA-3′; Uba52, 5′-ATTGAGCCATCCCTTCGTC-3′ and 5′-CACTTCTTCTTGCGGCAGT-3′; Ccnb1, 5′-ACCAGAGGTGGAACTTGCT-3′ and 5′-CGGGCTTGGAGAGGGATTAT-3′; Ccnb2, 5′-ACACCAGTTCCCAAATCCG-3′ and 5′-AAGAAGTGAAGTGGCAAAGGTC-3′; Gapdh, 5′-CCTGCCAAGTATGATGACAT-3′ and 5′-GTCCTCAGTGTAGCCCAAG-3′; and Hprt, 5′-AGCCTAAGATGAGCGCAAGT-3′ and 5′-ATGGCCACAGGACTAGAACA-3′.
Statistical analysis.
Two-tailed t tests were performed as indicated.
Proteasome activity.
Briefly, fluorescence-activated cell sorter (FACS)-sorted cells were washed with PBS, resuspended in 50 mM Tris HCl, and lysed by sonication for 5 s. Protein lysates were incubated with the substrate Suc–Leu–Val–Tyr–7-amino-4-methylcoumarin (AMC) (Boston Biochem), and AMC liberated from the substrate by proteasome activity was determined by measuring AMC fluorescence (Glomax Multi Promega) using a filter set with excitation at 365 nm and emission at 410 to 460 nm.
RESULTS
Rad23-null embryos are anemic as a result of impaired definitive erythropoiesis.
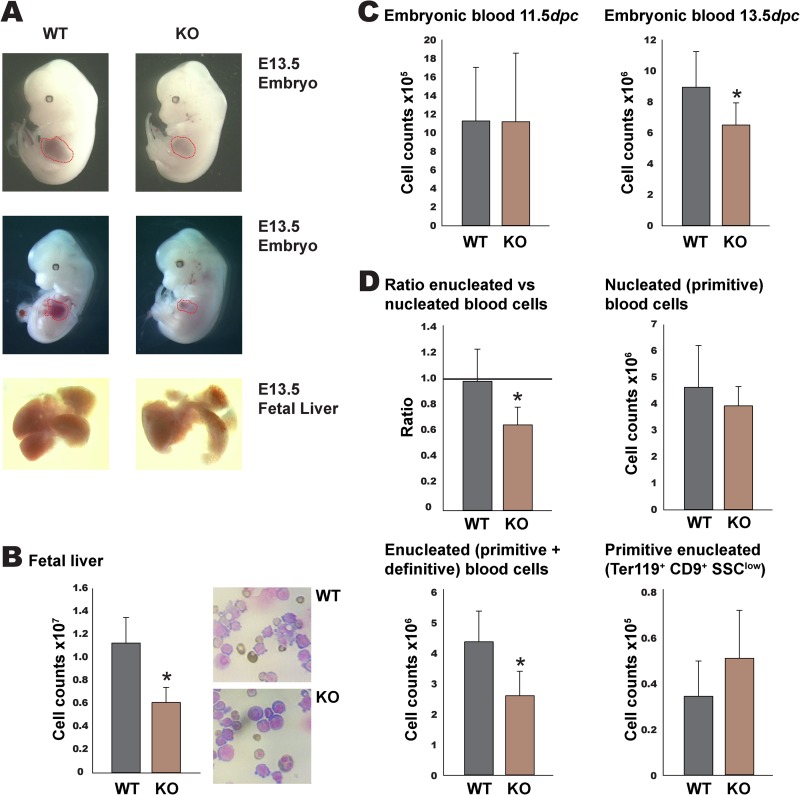
Embryos from heterozygous Rad23b+/− crosses were collected at 11.5 dpc and 13.5 dpc for analysis. Rad23b-null embryos within their yolk sacs were paler (data not shown) and had a smaller fetal liver than WT littermates at 13.5 dpc, while their overall size was slightly smaller but within the normal range (Fig. 1A). We observed reduced cell counts in the source of definitive erythropoiesis, i.e., the fetal liver (Fig. 1B), and cytospin preparations of fetal liver cell suspensions showed that mature erythroid cells are underrepresented in Rad23b-null fetal livers (Fig. 1B). The anemia status was confirmed by embryonic blood cell counts, which were consistently reduced in Rad23b-null embryos relative to WT littermates at 13.5 dpc while being normal at 11.5 dpc (Fig. 1C).
Fig 1.
Rad23b-null embryos at 13.5 dpc display anemia. (A) Representative pictures of WT and Rad23b-null (KO) embryos (upper two rows) with fetal livers surrounded by red lines and of fetal livers (lower row). (B) Fetal liver total cell count. WT, n = 14; KO, n = 12. Cytospins of fetal liver cell suspensions are depicted. Note the reduction of maturing cells in the KO sample. (C) Embryonic blood total cell count at 11.5 dpc (WT, n = 8; KO, n = 7) and 13.5 dpc (WT, n = 10; KO, n = 8). (D) Analysis of 13.5-dpc embryonic blood. Upper left, ratio of enucleated to nucleated erythroid cells. Upper right, absolute number of nucleated blood cells. Lower left, absolute number of enucleated blood cells. Lower right, absolute number of primitive enucleated blood cells, as calculated using flow cytometry. SSC, side scatter. Asterisks indicate statistical significance.
As embryonic blood at 13.5 dpc contains products of both primitive and definitive erythropoiesis, we next looked at the ratio of nucleated to enucleated erythrocytes in Rad23b-null embryos and littermate controls (Fig. 1D). While the numbers of nucleated and enucleated erythrocytes were equal in WT embryos (i.e., average ratio of 1), the ratio of enucleated to nucleated erythrocytes was significantly lowered in Rad23b-null embryos due to a significant reduction in the absolute number of enucleated blood cells (Fig. 1D). Since in normal embryos at this stage, more than 80% of the enucleated cells account for definitive erythroid cells (26, 42, 43), we conclude that the reduction in the ratio of enucleated to nucleated cells is accounted for mainly by the absence of enucleated definitive cells. In order to rule out a possible defect in the enucleation of primitive erythrocytes, circulating red blood cells were stained with Ter119 (present in all red blood cells) and CD9 (specific for primitive red cells). Differential exclusion of enucleated primitive (Ter119+ CD9+ SSClow) cells revealed normal enucleation of primitive cells in Rad23b KO mice (Fig. 1D). Together, these data suggest that the anemia in Rad23b-null embryos is caused by a specific defect in definitive erythropoiesis.
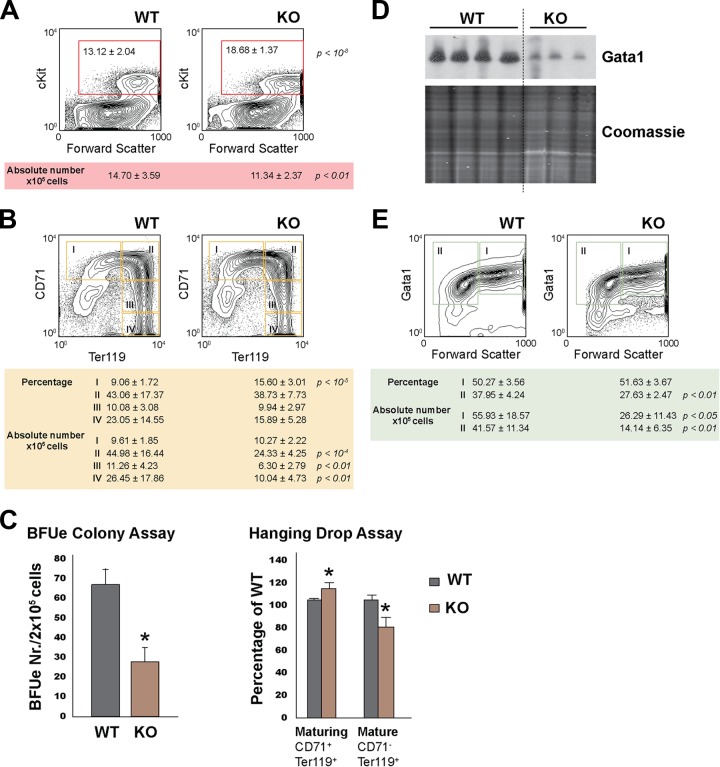
We next analyzed the erythroid compartment in the fetal liver by flow cytometry. Staining with 7AAD revealed equal percentages of live cells for the different genotypes (data not shown). Staining fetal livers with cKit, a marker for hematopoietic stem cells, early myeloid and erythroid progenitors, megakaryocytes, and mast cells, showed a significant relative increase in the cKit+ live cell population in Rad23b-null fetal livers, while the absolute numbers were slightly reduced (Fig. 2A). This suggests that colonization of fetal livers by progenitor cells is not the main problem affecting definitive erythropoiesis, but a blockade in subsequent developmental steps might be.
Fig 2.
Defective definitive erythropoiesis in Rad23b-null embryos. (A) Representative flow cytometric analysis of cKit expression in fetal livers of WT and Rad23b-null (KO) embryos. The percentage of live cKit+ cells is depicted in the contour plots of representative WT and KO livers. Absolute numbers (average and standard deviation [SD] for WT [n = 14] and KO [n = 12]) are indicated in the table. (B) Flow cytometric analysis of CD71 and Ter119 expression in fetal livers of WT and KO embryos. Populations I to IV represent subsequent stages during erythroid maturation (44). The percentage of live cells and the absolute numbers are indicated in the table as average and SD (WT, n = 14; KO, n = 12). (C) Bar graphs representing burst-forming unit erythroid (BFUe) colony assays and hanging-drop erythroid differentiation culture. Asterisks indicate statistical significance. (D) Western blot analysis of Gata1 expression in fetal liver cells. The same number of cells was used per lane. Coomassie blue staining is shown as loading control. (E) Flow cytometric analysis of intracellular Gata1 expression in fetal livers of WT and KO embryos. Populations I (FSChigh Gata1high) and II (FSClow Gata1low) represent subsequent stages during erythroid maturation. The percentage of live cells and the absolute numbers are indicated in the table as average and SD (WT, n = 14; KO, n = 12).
Therefore, we set out to analyze the committed erythroid lineage in fetal livers by staining with CD71 (transferrin receptor, a marker for early erythroid progenitors) and Ter119 (a marker for late erythroid cells). We classified erythroid populations I to IV in order of maturation as described by Socolovsky et al. (44). Rad23b-null fetal livers had increased relative numbers of erythroid progenitor cells (I). This was counterbalanced by a decrease in more mature cells (IV) compared to those in WT littermates (Fig. 2B). Absolute numbers revealed a normal contribution of early erythroid committed progenitors (I) and a decrease in more mature cell types (II to IV) in Rad23b-null fetal livers. This is consistent with a blockade during erythroid differentiation. To test this possibility directly, we performed burst-forming unit erythroid (BFUe) colony assays. BFUe colonies from Rad23b-null fetal livers were significantly reduced by 50% relative to those from WT controls (Fig. 2C). Although their overall size was not significantly different, a subset of Rad23b-null colonies was less compact than WT (data not shown). To test the potential of later committed progenitors, we performed hanging-drop assays and observed a reduction in the differentiation capacity of cultured cells, as measured by the accumulation of intermediate maturing cells, and reduction of mature cells (Fig. 2C). Furthermore, protein levels of the transcription factor Gata1 were lowered (Fig. 2D), mainly due to a reduction in the number of Gata1+ cells as measured by flow cytometry (Fig. 2E). Collectively, these data are in concordance with the notion that in Rad23b-null embryos, hematopoietic progenitors colonize fetal livers normally but are unable to generate sufficient numbers of committed and late erythroid cells, resulting in anemia.
Characterization of the Rad23b interactome in ES cells and erythroid cells.
In order to elucidate non-NER related functions of Rad23b, we performed an unbiased screen for Rad23b-interacting partners. To this end, we generated embryonic stem (ES) cells expressing a transgenic Rad23b-YFP-FLAG fusion protein and performed FLAG affinity chromatography on RIPA extracts derived from two independent Rad23b-YFP-FLAG-expressing clones or WT ES cells lacking the transgene. Bound proteins, eluted by FLAG peptide, were identified by mass spectrometry. The common Rad23b interactome, defined by proteins enriched at least 4-fold in Mascot score in eluted material from both transgenic clones compared to WT ES cells, is depicted in Fig. S1 in the supplemental material. Based on Ingenuity Pathway Analysis and Ensembl/Biomart GO term classification of the Rad23b-interacting protein list, proteins were classified as either known proteasome core constituents, known proteasome targets, or not previously described to interact with the proteasome. GO term classification and Mascot scores for each entry are indicated in Table S1 in the supplemental material.
In addition to the known interactions with Rad23a and Xpc, we confirmed interactions with members of the UPS. In fact, UPS members or related proteins and known targets accounted for 60% of the Rad23b interactome. Supporting this observation, Ingenuity Pathway Analysis reveals a predominance of the ubiquitin-proteasome pathway (see Table S2 in the supplemental material). Additional literature searches allowed classification of some of the remaining entries, which are marked with a reference number linked to Table S3 in the supplemental material. These findings strongly suggest that a role of Rad23b in the UPS rather than DNA repair may be crucial for understanding the phenotype of Rad23b-null mice.
We therefore pursued identification of binding partners of Rad23b in erythroid cells, using an antibody suitable for immunoprecipitation of the endogenous protein followed by mass spectrometry analysis, in both I/11 cells under proliferation conditions and fetal liver cells at 13.5 dpc. I/11 cells are an immortalized erythroid cell line with the capacity to differentiate and enucleate into mature erythrocytes upon stimulation with erythropoietin (Epo) (32). We detected proteasome subunit members that have been shown to directly interact with Rad23b (Psmcs and Psmds) (see Fig. S2 in the supplemental material). In erythroid cells, UPS members or related proteins and known targets accounted for 70% of the Rad23b interactome. Additional data from literature searches, GO term classification, and Ingenuity Pathway Analysis are shown in Tables S3 to S5 in the supplemental material. In contrast to the case for the interactome of Rad23b in ES cells, we coimmunoprecipitated with Rad23b a significant number of cell cycle regulators (i.e., Cdks). This led us to investigate cell proliferation in Rad23b-null cells.
Rad23b-null cells show a proliferative defect.
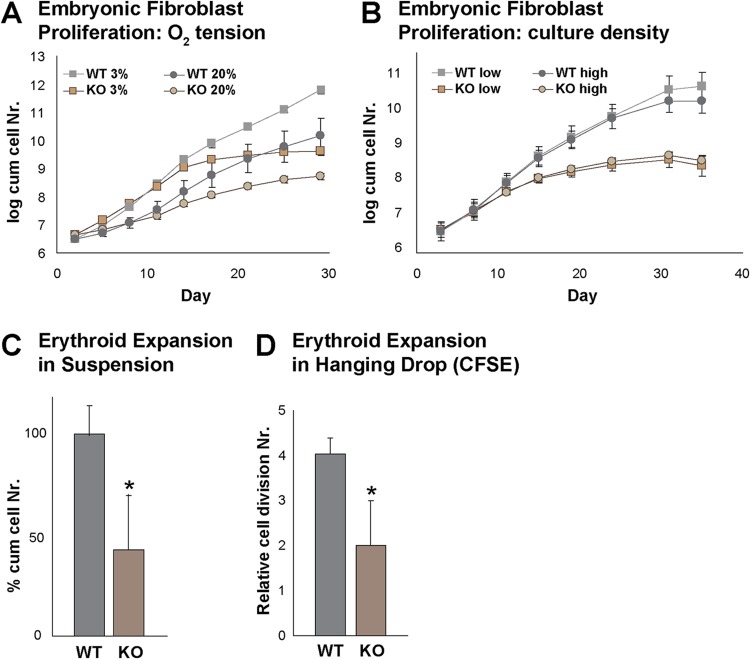
The interaction of Rad23b with cell cycle regulators in erythroid cells and the notion that yeast Rad23 substrates include G1 and S cyclin-dependent kinase inhibitors (21) pointed to a possible defect in cell cycle control in Rad23b-null cells. To test this, we generated mouse embryonic fibroblasts (MEFs) from 13.5-dpc embryos and maintained them in log-phase growth by serial passaging. We observed a growth defect in multiple independent Rad23b-null MEF cultures relative to growth of WT controls (Fig. 3A). MEFs cultured at physiologic oxygen tension have reduced oxidative stress and greater proliferative capacity than those cultured at atmospheric oxygen tension (45). To exclude oxidative stress hypersensitivity related to DNA repair deficiency of Rad23b-deficient MEFs being causative of the proliferation defect, we cultured cells at 3% oxygen. Despite an improved proliferative capacity at 3% versus 20% oxygen, the difference in proliferation between WT and Rad23b-null MEFs was similar at both oxygen tensions (Fig. 3A). Cell density in vitro also influences growth rate; therefore, we repeated the serial proliferation assay at 3% oxygen tension with two different seeding densities at each passage. Under these conditions, the growth defect of Rad23b-null MEFs was density independent (Fig. 3B). Finally, we tested the sensitivity of Rad23b-null cells to acute genotoxic stress and heat shock. Consistent with previous reports (5, 6), cells were not overtly sensitive to UV-C light, nor were they hypersensitive to gamma irradiation, heat shock, or proteasome inhibition (data not shown) (6). This suggests that the proliferation defect of Rad23b-null cells is independent of genotoxic or heat shock responses and supports the notion that UPS function is critical to the phenotype of Rad23b-null MEFs.
Fig 3.
Proliferation defect in Rad23b-null cells. (A) Proliferation of WT and KO embryonic fibroblasts at 3% and 20% O2 tension. (B) Proliferation of WT and KO embryonic fibroblasts plated at low and high density upon serial passaging at 3% O2 tension. (C) Expansion of WT and KO fetal liver erythroid cells cultured under conditions that favor the proliferation of early erythroid progenitors, preventing their differentiation. (D) Hanging-drop CFSE erythroid differentiation culture showing relative proliferation differences between WT and KO erythroid cells. Asterisks indicate statistical significance.
Since erythropoiesis is tightly coupled to cell proliferation, we investigated whether the impaired differentiation we observed in the Rad23b-null embryos could be due to a defect in proliferation. Expansion of fetal liver progenitor cells in suspension cultures under conditions favoring the proliferation of undifferentiated erythroid cells (32) was consistent with a diminished proliferation rate of Rad23b-null cells (Fig. 3C). To assess whether this is due to a reduced cell division rate, we performed hanging-drop differentiation cultures of fetal liver cells previously stained with CFSE and measured by flow cytometry the CFSE mean fluorescence intensity (MFI) at the onset and 2 days after starting the cultures. This revealed that at day 2, the Rad23b-null cells still displayed high levels of CFSE, consistent with the notion that the reduction in cell number is due to a defect in proliferation (Fig. 3D).
Rad23b and Ubc knockdown erythroid cells show a proliferative defect.
To further substantiate these data, we tested the effects of Rad23b depletion in erythroid cells using lentiviral shRNA-mediated knockdown in I/11 cells (32). We used several Rad23b shRNA targeting constructs and a scrambled nontargeting shRNA construct (Scr) as control.
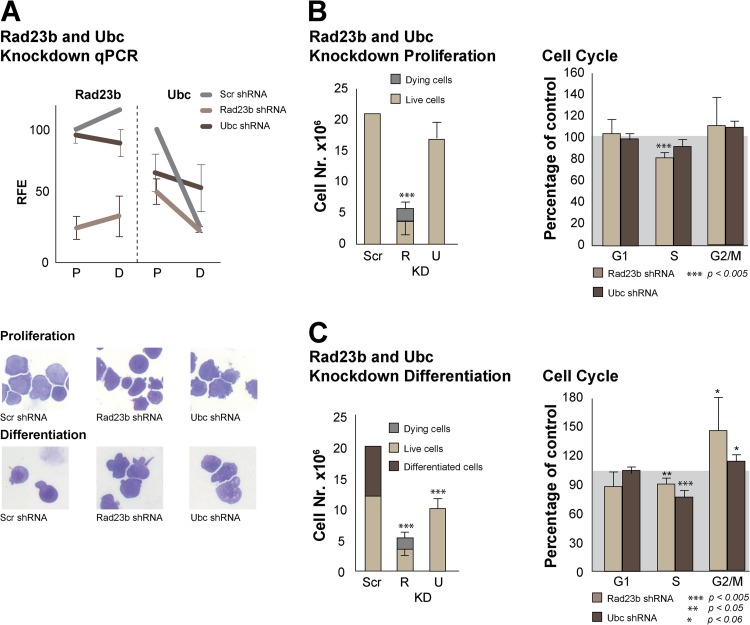
After transduction with shRNA-expressing lentivirus and 4 days of puromycin selection, qPCR revealed an average 70% reduction of Rad23b expression levels (Fig. 4A). Equal numbers of these cells were seeded and subjected to either proliferation or differentiation conditions for 2 days. Knockdown of Rad23b by either Rad23b shRNA targeting construct resulted in markedly reduced cell numbers under both proliferation and differentiation conditions (Fig. 4B and C). Importantly, under differentiation conditions we failed to observe differentiating cells in Rad23b knockdown samples as opposed to Scr control cells.
Fig 4.
Intrinsic proliferation defect in Rad23b knockdown erythroid cells. (A) The relative fold enrichment (RFE) of cDNA expression as calculated by qPCR is shown for Rad23b and Ubc. Several shRNA constructs were used to knock down Rad23b or Ubc, and control cells were transduced with a scrambled shRNA lentivirus construct (Scr). P, proliferation conditions; D, differentiation conditions. Cytospins of knockdown cells under proliferation or differentiation conditions are depicted in the lower panels. (B) Cell number (left) and cell cycle analysis (right) after 2 days under proliferation conditions, starting with 5 × 106 cells. Rad23 knockdown (R KD) or Ubc knockdown (U KD) cells fail to proliferate and/or die compared to the cells transduced with a scrambled shRNA lentivirus construct (Scr). Dead cells were scored as small cells (<4 μm) in the CASY cell counter, and percentages were confirmed by 7AAD staining and flow cytometry (not shown). The average and SD are depicted for the 4 Rad23b shRNA lentivirus-transduced cells. (C) Cell number (left) and cell cycle analysis (right) after 2 days under differentiation conditions, starting with 5 × 106 cells. Rad23 knockdown (R KD) or Ubc knockdown (U KD) cells fail to proliferate and/or die compared to the cells transduced with a scrambled shRNA lentivirus construct (Scr). Dead cells were scored as small cells (<4 μm) in the CASY cell counter, and percentages were confirmed by 7AAD staining and flow cytometry (not shown, 7AAD+). Differentiating cells were scored as smaller than 7 μm in the CASY cell counter, and percentages were confirmed by flow cytometry (not shown, 7AAD− FSClow). The average and SD are depicted for the Rad23b or Ubc shRNA lentivirus-transduced cells.
Cell cycle distribution analysis of propidium iodide-stained cells by flow cytometry indicated a significant reduction of Rad23b knockdown cells in S phase and a trend toward accumulation in G2 under proliferation conditions (Fig. 4B). Both of these effects were significant under differentiation conditions (Fig. 4C). This result is concordant with previous studies, in which a delay in G2/M cell cycle stage has been observed in Rad23 yeast mutants (22, 46). Since terminally differentiating cells undergo G1 arrest prior to enucleation (47), the increase in G2-arrested cells may provide, in part, an explanation for the observed inefficient terminal erythroid differentiation.
It has been previously reported that loss of Ubc, an inducible polymeric form of ubiquitin, results in fetal liver growth retardation and preterm mortality in mice, similar to the phenotype of Rad23b-null mice (48). Ubc-null cells share the proliferative defects and G2/M arrest that we observed in Rad23-null cells (48). Therefore, we measured Ubc expression levels in Rad23 knockdown cells. Interestingly, we found that these were reduced (Fig. 4A), and we performed Ubc knockdown experiments. We were able to knock down Ubc levels to 50% of that of the Scr control (Fig. 4A). We observed a proliferative defect that was more obvious under differentiation conditions (Fig. 4B and C), upon which Ubc knockdown cells also failed to differentiate. These data are substantiated by the aberrant morphology of Rad23b or Ubc knockdown cultured cells as depicted in Fig. 4A (lower panels). Under differentiation conditions, Rad23b and Ubc knockdown cells fail to undergo differentiation, as can be deduced from the larger size of the knockdown cells than of Scr control cells.
In conclusion, these data show that the proliferation/differentiation defect observed in erythroid cells from Rad23b-null mice is cell intrinsic. Furthermore, although there are similarities in the phenotype when comparing shRNA targeting of Rad23b and Ubc in cultured erythroid cells, their functions appear not to overlap completely.
UPS requirement at an early stage of erythroid differentiation.
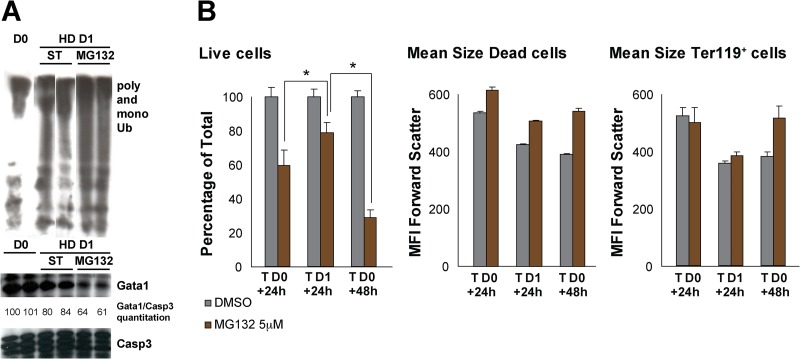
To further test the requirement for proteasome function in erythroid differentiation, we measured the effects of proteasome inhibition on differentiation of WT fetal liver erythroid cells using MG132, a peptide that saturates the proteasome machinery. MG132 was added to hanging-drop cultures at day 0 or day 1, and cultures were analyzed at day 1 or day 2. Western blot analyses revealed an increase in mono- and polyubiquitinylated proteins and a decrease in Gata1 in the presence of MG132 within 1 day of culture (Fig. 5A). Cultures treated with MG132 at the onset of the hanging-drop cultures had 60% live cells after 1 day (T D0 + 24 h) and only 30% live cells after 2 days (T D0 + 48 h) (Fig. 5B). We noted that dead cells were larger in MG132-treated cultures than in standard (ST) cultures (Fig. 5B). Among the still-living cells, the cell size was not decreased to the same extent after 2 days of culture as in untreated cultures (Fig. 5B). These data are consistent with a requirement for proteasome function in erythroid cell survival and differentiation.
Fig 5.
Proteasome activity is required early during erythroid differentiation. (A) Western blot analysis of poly- and monoubiquitylated proteins and Gata1 expression in 13.5-dpc fetal liver at the day of collection (D0) or at day 1 after hanging-drop culture (HD D1) under standard (ST) conditions or in the presence of the proteasome inhibitor MG132. Loading was normalized by cell number. (B) Flow cytometry analysis of standard (ST) or MG132-treated hanging-drop cultures at the onset of cultures (T D0) and analyzed at day 1 or 2 of culture (+24 h or + 48 h) or treated after 1 day in hanging-drop culture (T D1) and analyzed a day later (+24 h). The percentage of live cells, the mean size of dead (7AAD+) cells, and the mean size of Ter119+ cells are depicted. *, P < 0.005. Values are normalized to those for untreated cultures.
Interestingly, the percentage of live cells was not reduced to the same extent when MG132 was added later during differentiation; i.e., there were 79% live cells in cultures treated for 24 h after 1 day differentiating in hanging drops (TD1 + 24 h) versus 60% live cells in cultures treated for 24 h at the onset of setting the hanging drops (T D0 + 24 h) (Fig. 5B). This suggests that once cells start differentiating, they are less sensitive to proteasome inhibition.
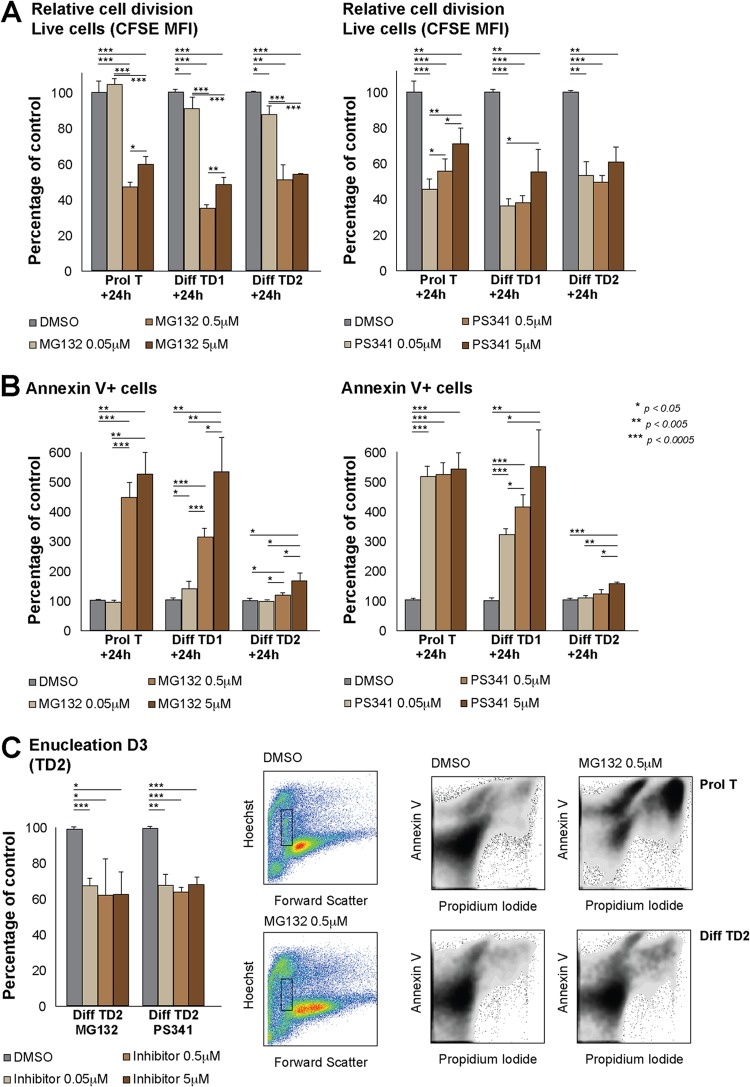
These results led us to test the effects of proteasome inhibition in I/11 cells, which are a homogeneous erythroid population and which when induced to differentiate do so in a synchronous manner, as opposed to the mixed fetal liver composition. We therefore treated I/11 cells with either MG132 or PS341 proteasome inhibitor for 24 h at the indicated differentiation stages and concentrations. We always measured a reduction in cell number at the time of collection in treated cells (data not shown). When we performed the treatments in I/11 cells previously labeled with CFSE, we were able to measure proliferation in living cells (Hoechst negative) at the time of collection (Fig. 6A). Independent of the differentiation stage, we observed a proliferative defect in I/11 cells treated with either of the two inhibitors. Interestingly, when measuring the percentage of annexin V-positive cells in order to assess whether cell death or apoptosis contributes to the reduced cell number, we observed that almost all cells were annexin V positive when treated under proliferating conditions. The percentage of annexin V-positive cells gradually decreased and was almost not detectable when cells were treated later during differentiation, as if they were resistant to apoptosis induction (Fig. 6B). Still, and as previously described (20), proteasome inhibition at this late stage had an impact on enucleation, as shown in Fig. 6C.
Fig 6.
Effects of proteasome inhibitors on erythropoiesis depend on differentiation status. (A) Relative cell proliferation as measured by CFSE mean fluorescence intensity (MFI) after culturing I/11 cells for 24 h in the presence of MG132 or PS341 proteasome inhibitor at the concentrations and culture stages indicated. (B) Percentage of annexin V-positive cells after culturing I/11 cells for 24 h in the presence of MG132 or PS341 proteasome inhibitor at the concentrations and culture stages indicated. Representative density plots are depicted below. (C) Percentage of enucleation, as measured by flow cytometry (Hoechst intermediate, forward scatter small) after culturing I/11 cells for 24 h in the presence of MG132 or PS341 proteasome inhibitor at the indicated concentrations on day 2 of differentiation (analysis was done on day 3). The results of one representative experiment out of three are depicted in the dot plots.
This implies that proteasome activity is essential for cell cycle regulation of erythroid cells during differentiation and that its inhibition compromises cell survival specifically at early stages, while it compromises enucleation at later stages.
Proteasome activity decreases toward the end of erythroid maturation.
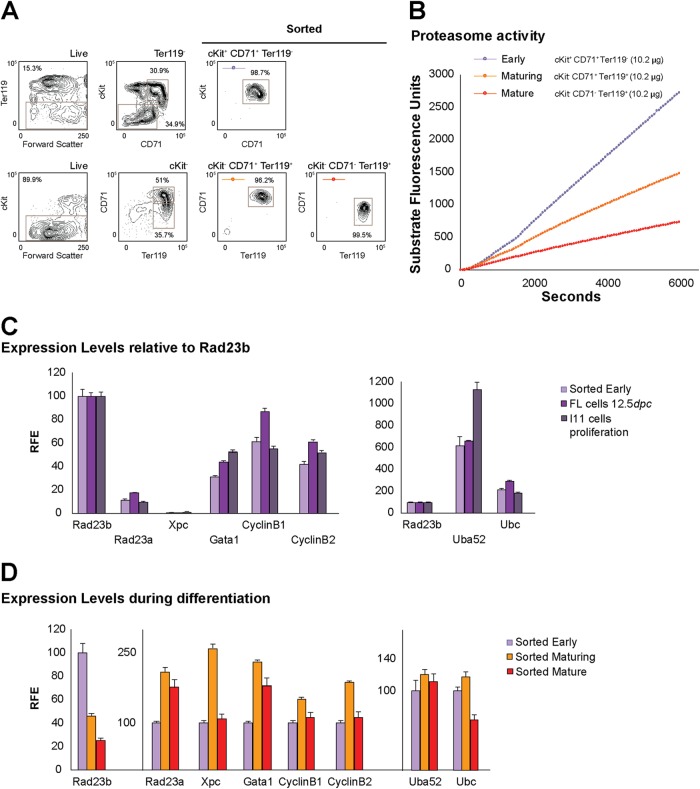
These data led to the prediction that proteasome activity is highest in early erythroid progenitors and diminishes progressively in more mature stages. To test this, we measured proteasome activity using a fluorescent substrate-based technique in prospectively isolated WT fetal liver erythroid progenitors (Fig. 7A and B). Earlier progenitors sorted as cKit+ CD71+ Ter119− had the highest proteasome activity, followed by maturing progenitors (cKit− CD71+ Ter119+) and more mature erythroid cells (cKit− CD71− Ter119+), which displayed the lowest proteasome activity (Fig. 7A and B). Notably, this marked reduction in proteasome activity is accompanied by a gradual reduction in Rad23b expression (Fig. 7D), which is in line with the notion that Rad23b- and UPS-related activities lose their relevance toward the end of erythroid differentiation. We also observed that Rad23b transcripts are expressed to a much higher level than Xpc and Rad23a transcripts (Fig. 7C), again supporting a prominent role for Rad23b outside NER.
Fig 7.
Proteasome activity is required early during erythroid differentiation. (A) Flow cytometry gating strategy to sort prospectively erythroid cells at different stages of maturation from fetal livers. The order from immature to more mature is as follows: early, cKit+ CD71+ Ter119−; maturing, cKit− CD71+ Ter119+; mature, cKit− CD71− Ter119+. Respective contour plots showing the purity of sorted populations are depicted. (B) Proteasome activity of representative prospectively isolated erythroid populations. (C) mRNA expression levels as measured by qPCR of Rad23b and other transcripts in early FACS-sorted erythroid cells, 12.5-dpc fetal liver cells, and proliferating I/11 cells. Relative fold enrichment (RFE) is depicted, setting Rad23b to 100. (D) mRNA expression levels as measured by qPCR of Rad23b and other transcripts measured by qPCR during erythroid differentiation in FACS-sorted erythroid populations. Relative fold enrichment (RFE) is depicted, setting the early cell population to 100 for each transcript. Representative analyses are shown.
These data overall suggest that the proteasome or UPS is required for erythroid differentiation and that it exerts its main actions (i.e., those required for cell survival) in committed early erythroid progenitors.
Rad23b loss impairs stress erythropoiesis in adult bone marrow chimeric mice.
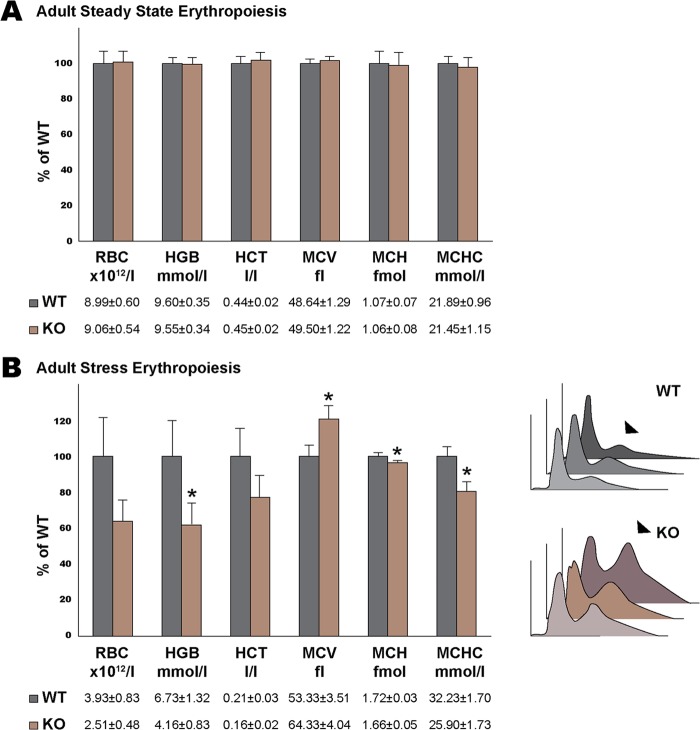
While most Rad23b-null mice die before birth, those that survive to adulthood do not present with anemia or other hematological defects (6), suggesting a developmental specificity to the observed erythropoietic defect. Fetal erythropoiesis occurs in a relatively hypoxic intrauterine environment. It is thus considered an example of stress erythropoiesis, which is different from steady-state erythropoiesis occurring in bone marrow of adult animals. These considerations prompted us to investigate Rad23b loss in adult erythropoiesis under both steady-state and stress erythropoietic conditions. To this end, we injected WT or Rad23b-null fetal liver cells into lethally irradiated mice (here referred to as WT or Rad23b-null chimeras, respectively). Mice with an engraftment of 50% or higher were selected for further analysis. Six weeks after transplantation, no significant differences were found between Rad23b-null and WT chimeras with respect to standard hematological parameters (Fig. 8A). In sharp contrast, mice treated with the hemolytic agent PHZ to induce stress erythropoiesis had a significantly reduced hemoglobin (HGB) content in blood, a lower mean cell hemoglobin (MCH), a larger mean cell volume (MCV), a lower mean corpuscular hemoglobin concentration (MCHC), and a higher proportion of reticulocytes in circulation (Fig. 8B). Enlarged spleens were observed in both Rad23b-null and WT chimeras (data not shown). Taken together, these data suggest that while the initiation of extramedullary erythropoiesis per se was unaffected, Rad23b-null chimeras had an inefficient stress response and delayed erythroid recovery compared to WT chimeras.
Fig 8.
Rad23b-null chimeric mice show a delayed stress erythropoiesis response. (A) Blood hematological parameters of Rad23b-null and WT chimeric mice 6 weeks after transplantation measured on a Vet ABC hematocytocounter. (B) Blood hematological parameters of Rad23b-null and WT chimeric mice 8 weeks after transplantation and 5 days after hemolysis induction with PHZ. Asterisks indicate statistical significance. RBC, red blood cell; HGB, hemoglobin; HCT, hematocrit; MCV, mean cell volume; MCH, mean corpuscular hemoglobin; MCHC, mean cellular hemoglobin concentration. Histogram overlays on the right show the scatter of RBC, with arrowheads pointing to reticulocyte peaks.
DISCUSSION
Here we showed that Rad23b loss affects proteasome function rather than DNA repair, resulting in proliferation defects in at least two different types of highly proliferating cells, MEFs and erythroid cells. Indeed, Rad23b-null embryos are anemic, and most of them die before birth. The few surviving mice suffer from bone dysmorphologies, sterility, and growth retardation, although without hematological defects (6). These features are likely not derived from the known function of Rad23b in DNA repair (7), as null cells are NER proficient (5) and not sensitive to oxidative DNA damage, suggesting that other repair mechanisms are not compromised by Rad23b absence (5, 6). The NER function of Rad23b appears to be redundant with that of Rad23a, as only double-KO (Rad23a−/−/Rad23b−/−) fibroblasts exhibit NER deficiency (5). Rad23 proteins are essential for the stability of Xpc (5) and for efficient binding of Xpc to DNA lesions (49). It is known, however, that only a fraction of Rad23b is bound to Xpc, suggesting that the non-Xpc-associated pool of Rad23b functions in other pathways (50).
Several previous studies have shown that another important function of Rad23 is its role as shuttling factor in the UPS pathway (51). Here, proteomic profiling of Rad23b-interacting proteins, purified under native conditions by affinity chromatography of full-length FLAG-tagged Rad23b expressed at physiological levels, revealed that 40 to 60% of the Rad23b interactome is related to the UPS system. This corroborates the importance of the UPS system in Rad23b function and confirms previous studies based on overexpression of the ubiquitin binding domain of human Rad23a or Rad23b (38, 39). Immunoprecipitation of endogenous Rad23b protein in erythroid cells confirmed the interaction with the proteasome and revealed a number of interacting proteins with functions in cell cycle regulation that, if altered in abundance or localization, could potentially explain the observed defects in erythroid differentiation and more generally in fibroblast proliferation. Therefore, we aimed to understand how the role of Rad23b in protein turnover could impact the anemia observed in Rad23b-null mice.
While the cause of death from 12.5 dpc onward in Rad23b-null mice is not known, anemia is a highly penetrant phenotype that can result in embryonic lethality. We thus characterized the erythropoietic compartments of 11.5-dpc and 13.5-dpc embryos and concluded that Rad23b loss specifically affects definitive erythropoiesis during the fetal stages, most likely through defects in proteosomal function. Inhibition of the proteasome in erythroid cultures resulted in a similar block in differentiation and increase in cell death, most notably at early stages of erythropoiesis. Concordant with results for fetal livers from Rad23b-null mice, Gata1 protein levels in remaining live cells were significantly reduced upon proteasome inhibition in this cell system. By means of lentivirus-mediated shRNA knockdown and generation of chimeric mice, we also showed that the defect generated by loss of Rad23b is cell intrinsic to erythroid cells and not dependent on the environment of a Rad23b-deficient embryo.
Because rare Rad23b-null mice that survive into adulthood do not present with anemia, we tested the effects of Rad23b deficiency on erythropoiesis in adult chimeras. Similarly to Rad23b-null mice, Rad23b-null bone marrow chimeras displayed no defects in hematological parameters. However, because erythropoiesis in fetal liver is a form of stress erythropoiesis, we also tested the effects of Rad23b deficiency after induction of stress erythropoiesis by treatment of chimeras with a hemolytic agent. Consistent with an essential role of Rad23b specifically in stress erythropoiesis, a number of hematological defects were seen after treatment with a hemolytic agent.
The expression levels of Ubc, which has been described as a “stress”-dependent ubiquitin as opposed to Uba, is induced during early erythropoiesis, possibly due to an increased demand for UPS activity. Ubc mRNA levels decrease through terminal differentiation of erythroid cells. Interestingly, Ubc-null mice have a phenotype similar to that of Rad23b-null mice; i.e., they die after 12.5 dpc, suffer from anemia and retarded fetal liver growth, and have cell proliferative defects due to delayed G2/M transition (48). Here, we observed significant downregulation of Ubc expression levels in Rad23b knockdown erythroid cells. Upon Ubc knockdown in I/11 erythroid cells, the cell numbers were reduced due to a cell cycle defect, in a manner comparable to that for Rad23b knockdown I/11 cells.
In further support of a link between proteasome activity and erythropoeisis, we note the involvement of UPS-related proteins in hereditary human diseases with perturbations in erythroid compartments. For example, defects in core components of a complex with E3 ubiquitin ligase activity give rise to Fanconi anemia (52, 53). It has been recently proposed that the proteasome machinery is required to maintain the balance of alpha- and beta-globin chains within a maturing erythroid cell and that protein quality control is crucial in the erythropoietic process (54). Additionally, anemia, thrombocytopenia, and neutropenia are common secondary effects caused by proteasome inhibitors applied to cancer patients (55). Of note, G2/M cell cycle arrest is also caused by the proteasome inhibitor bortezomib (Velcade) in endothelial cells (56).
Proteasome activity is believed to be required for enucleation of erythroid cells (20). Although we could confirm that inhibition of the proteasome at late stages of differentiation compromises enucleation, our data suggest an additional and more stringent requirement for proteasome activity in early committed erythroid progenitors, i.e., cKit+ CD71+ Ter119−, prior to enucleation. In support of this, we observed that inhibition of proteasome activity later during differentiation had less of an impact on erythrocyte viability than when inhibition was under proliferation conditions, while cell division was reduced at all stages. It has been suggested that the proteolytic activity is greater in immature erythroid cells and that it decreases with maturation (54, 57). Furthermore, we demonstrated reduced proteasome activity and lower levels of Rad23b in the later stages of erythroid differentiation in prospectively isolated WT fetal liver cells. We hypothesize that Rad23b deficiency results in impairment in the delivery of ubiquitylated substrates to the UPS or in their protection from the UPS, which has a profound effect on highly proliferative tissues. Taken together, our data support an increased demand for proteasome activity during the early phase of erythroid maturation, followed by a gradual decrease in later stages of maturation.
Our data support a cooperative role between Rad23b and the proteasome in proliferation, with Rad23b loss leading to delayed G2/M transition and subsequent cell death in differentiating erythroid cells. These data include reduced erythroid proliferation under combined proliferation/differentiation conditions or isolated proliferation conditions in Rad23b-null erythroid progenitors, reduced proliferation of Rad23b-null MEFs, and reduced proliferation/survival in erythroid cells depleted of Rad23b. Currently, we do not know whether Rad23b coordinates the degradation of a specific set of proteins or whether it cooperates with the proteasome in a broader manner and how its loss and/or proteasome saturation could affect transcription of ubiquitins. Boutet et al. showed the specific requirement of Rad23b for Pax3 degradation in muscle (58), favoring the notion of Rad23b specificity. Our mass spectrometry analysis of Rad23b-interacting proteins in two different lineages, erythroid and embryonic stem cells, revealed an interaction with core proteasome components, as well as with known proteasome degradation targets representing 40% and 20% of the Rad23b interactome in erythroid and embryonic stem cell lineages, respectively. This may indicate a specific role for Rad23b in shuttling these proteins to the proteasome. However, in DM2 muscular dystrophy, Rusconi et al. linked downregulation of Rad23b with a decrease in ubiquitylated proteins (59), which could be interpreted as an additional role for Rad23b in protecting some proteins for degradation or a general reduction in ubiquitin proteins due to a negative feedback upon proteasome saturation. In erythroid lineages specifically, we found cell cycle regulators interacting with Rad23b that could, if differentially regulated in the absence of Rad23b, explain proliferation defects observed in null and knockdown cells of different lineages. However, the functional relevance of these interactions must still be tested, as proliferation/cell cycle defects might be either direct effects on stability/instability of cell cycle regulators or indirect effects of Rad23b, for example, on transcriptional differentiation programs (60), that may or may not be a result of perturbations in protein degradation. Future studies will be required to identify which, if any, of these proteins are altered as a function of Rad23b deficiency. Indeed, due to the predicted transient interaction of Rad23b with substrates targeted for proteasomal degradation, the current strategy may not be well suited to identify such proteins. Nonetheless, these results strongly suggest that interaction with the UPS and regulation of cell cycle progression are a key function of Rad23b in mammals. The exact consequences of Rad23b loss in different tissues that are subject to proliferative stress remain an intriguing topic that deserves further attention, especially in light of the successful usage of proteasome inhibitors as drug targets.
Supplementary Material
ACKNOWLEDGMENTS
We thank Raymond Poot for technical advice regarding FLAG affinity chromatography.
This work was supported by grants from the Dutch Genomics Initiative (NGI) and The Netherlands Organization for Scientific Research (NWO) to S.P. (700.55.006, DN 82-294), L.G. (863.09.012), and J.A.M. (917.96.120).
S.B. designed and performed experiments and wrote the paper; A.F.T. and W.T. performed experiments; I.M.D.C., D.I.K., T.C., R.V.D.L., J.A.D., E.P.M., F.P.V.A., J.A.M., T.V.G., and A.M. performed experiments; C.R. designed and performed experiments; S.P. and W.V. designed experiments and wrote the paper; and J.R.M. and L.G. designed and performed experiments and wrote the paper.
Footnotes
Published ahead of print 29 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.05772-11.
REFERENCES
- 1.Dantuma NP, Heinen C, Hoogstraten D. 2009. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst.) 8:449–460 [DOI] [PubMed] [Google Scholar]
- 2.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. 1999. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem. 274:28019–28025 [DOI] [PubMed] [Google Scholar]
- 3.van der Spek PJ, Visser CE, Hanaoka F, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers JH. 1996. Cloning, comparative mapping, and RNA expression of the mouse homologues of the Saccharomyces cerevisiae nucleotide excision repair gene RAD23. Genomics 31:20–27 [DOI] [PubMed] [Google Scholar]
- 4.Miller RD, Prakash L, Prakash S. 1982. Defective excision of pyrimidine dimers and interstrand DNA crosslinks in rad7 and rad23 mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 188:235–239 [DOI] [PubMed] [Google Scholar]
- 5.Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH. 2003. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 17:1630–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng JM, Vrieling H, Sugasawa K, Ooms MP, Grootegoed JA, Vreeburg JT, Visser P, Beems RB, Gorgels TG, Hanaoka F, Hoeijmakers JH, van der Horst GT. 2002. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol. Cell Biol. 22:1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugasawa K, Ng JM, Masutani C, Maekawa T, Uchida A, van der Spek PJ, Eker AP, Rademakers S, Visser C, Aboussekhra A, Wood RD, Hanaoka F, Bootsma D, Hoeijmakers JH. 1997. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell Biol. 17:6924–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugasawa K. 2010. Regulation of damage recognition in mammalian global genomic nucleotide excision repair. Mutat. Res. 685:29–37 [DOI] [PubMed] [Google Scholar]
- 9.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276:18665–18672 [DOI] [PubMed] [Google Scholar]
- 10.Volker M, Mone MJ, Karmakar P, HAvan Schul W, Vermeulen W, Hoeijmakers JH, DRvan van Zeeland AA, Mullenders LH. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213–224 [DOI] [PubMed] [Google Scholar]
- 11.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. 2000. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 275:9870–9875 [DOI] [PubMed] [Google Scholar]
- 12.Bader M, Steller H. 2009. Regulation of cell death by the ubiquitin-proteasome system. Curr. Opin. Cell Biol. 21:878–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciechanover A, Finley D, Varshavsky A. 1984. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell 37:57–66 [DOI] [PubMed] [Google Scholar]
- 14.Fasanaro P, Capogrossi MC, Martelli F. 2010. Regulation of the endothelial cell cycle by the ubiquitin-proteasome system. Cardiovasc. Res. 85:272–280 [DOI] [PubMed] [Google Scholar]
- 15.Finley D, Ciechanover A, Varshavsky A. 2004. Ubiquitin as a central cellular regulator. Cell 116:S29–32, 2 [DOI] [PubMed] [Google Scholar]
- 16.Groettrup M, Kirk CJ, Basler M. 2010. Proteasomes in immune cells: more than peptide producers? Nat. Rev. Immunol. 10:73–78 [DOI] [PubMed] [Google Scholar]
- 17.Rinetti GV, Schweizer FE. 2010. Ubiquitination acutely regulates presynaptic neurotransmitter release in mammalian neurons. J. Neurosci. 30:3157–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raasi S, Orlov I, Fleming KG, Pickart CM. 2004. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J. Mol. Biol. 341:1367–1379 [DOI] [PubMed] [Google Scholar]
- 19.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120:73–84 [DOI] [PubMed] [Google Scholar]
- 20.Chen CY, Pajak L, Tamburlin J, Bofinger D, Koury ST. 2002. The effect of proteasome inhibitors on mammalian erythroid terminal differentiation. Exp. Hematol. 30:634–639 [DOI] [PubMed] [Google Scholar]
- 21.Cocklin RR, Heyen JW, Larry TR, Tyers M, Goebl MG. 2011. New insight into the role of the Cdc34 ubiquitin-conjugating enzyme in cell cycle regulation via Ace2 and Sic1. Genetics 187:701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambertson D, Chen L, Madura K. 1999. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma R, Oania R, Graumann J, Deshaies RJ. 2004. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118:99–110 [DOI] [PubMed] [Google Scholar]
- 24.Biggins S, Ivanovska I, Rose MD. 1996. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 133:1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeki Y, Saitoh A, Toh-e A, Yokosawa H. 2002. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 293:986–992 [DOI] [PubMed] [Google Scholar]
- 26.Palis J. 2008. Ontogeny of erythropoiesis. Curr. Opin. Hematol. 15:155–161 [DOI] [PubMed] [Google Scholar]
- 27.Allen TD, Testa NG. 1991. Cellular interactions in erythroblastic islands in long-term bone marrow cultures, as studied by time-lapse video. Blood Cells 17:29–38 [PubMed] [Google Scholar]
- 28.Gutierrez L, Lindeboom F, Langeveld A, Grosveld F, Philipsen S, Whyatt D. 2004. Homotypic signalling regulates Gata1 activity in the erythroblastic island. Development 131:3183–3193 [DOI] [PubMed] [Google Scholar]
- 29.Robin C, Dzierzak E. 2005. Hematopoietic stem cell enrichment from the AGM region of the mouse embryo. Methods Mol. Med. 105:257–272 [DOI] [PubMed] [Google Scholar]
- 30.Robin C, Ottersbach K, Durand C, Peeters M, Vanes L, Tybulewicz V, Dzierzak E. 2006. An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev. Cell 11:171–180 [DOI] [PubMed] [Google Scholar]
- 31.Gutierrez L, Tsukamoto S, Suzuki M, Yamamoto-Mukai H, Yamamoto M, Philipsen S, Ohneda K. 2008. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood 111:4375–4385 [DOI] [PubMed] [Google Scholar]
- 32.Dolznig H, Boulme F, Stangl K, Deiner EM, Mikulits W, Beug H, Mullner EW. 2001. Establishment of normal, terminally differentiating mouse erythroid progenitors: molecular characterization by cDNA arrays. FASEB J. 15:1442–1444 [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez L, Lindeboom F, Ferreira R, Drissen R, Grosveld F, Whyatt D, Philipsen S. 2005. A hanging drop culture method to study terminal erythroid differentiation. Exp. Hematol. 33:1083–1091 [DOI] [PubMed] [Google Scholar]
- 34.van de Ven M, Andressoo JO, Holcomb VB, von Lindern M, Jong WM, De Zeeuw CI, Suh Y, Hasty P, Hoeijmakers JH, van der Horst GT, Mitchell JR. 2006. Adaptive stress response in segmental progeria resembles long-lived dwarfism and calorie restriction in mice. PLoS Genet. 2:e192. 10.1371/journal.pgen.0020192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imam AM, Patrinos GP, KMde Bottardi S, Janssens RJ, Katsantoni E, Wai AW, Sherratt DJ, Grosveld FG. 2000. Modification of human beta-globin locus PAC clones by homologous recombination in Escherichia coli. Nucleic Acids Res. 28:E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci.U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466–469 [DOI] [PubMed] [Google Scholar]
- 38.Besche HC, Haas W, Gygi SP, Goldberg AL. 2009. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48:2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scanlon TC, Gottlieb B, Durcan TM, Fon EA, Beitel LK, Trifiro MA. 2009. Isolation of human proteasomes and putative proteasome-interacting proteins using a novel affinity chromatography method. Exp. Cell Res. 315:176–189 [DOI] [PubMed] [Google Scholar]
- 40.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, Cassar W, Galdies R, IWvan Ozgur Z, Gillemans N, Hou J, Bugeja M, Grosveld FG, LMvon Felice AE, Patrinos GP, Philipsen S. 2010. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 42:801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 42.Kingsley PD, Malik J, Fantauzzo KA, Palis J. 2004. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood 104:19–25 [DOI] [PubMed] [Google Scholar]
- 43.Palis J, Segel GB. 1998. Developmental biology of erythropoiesis. Blood Rev. 12:106–114 [DOI] [PubMed] [Google Scholar]
- 44.Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. 2001. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood 98:3261–3273 [DOI] [PubMed] [Google Scholar]
- 45.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elder RT, Song XQ, Chen M, Hopkins KM, Lieberman HB, Zhao Y. 2002. Involvement of rhp23, a Schizosaccharomyces pombe homolog of the human HHR23A and Saccharomyces cerevisiae RAD23 nucleotide excision repair genes, in cell cycle control and protein ubiquitination. Nucleic Acids Res. 30:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsieh FF, Barnett LA, Green WF, Freedman K, Matushansky I, Skoultchi AI, Kelley LL. 2000. Cell cycle exit during terminal erythroid differentiation is associated with accumulation of p27(Kip1) and inactivation of cdk2 kinase. Blood 96:2746–2754 [PubMed] [Google Scholar]
- 48.Ryu KY, Maehr R, Gilchrist CA, Long MA, Bouley DM, Mueller B, Ploegh HL, Kopito RR. 2007. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 26:2693–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergink S, Toussaint W, Luijsterburg MS, Dinant C, Alekseev S, Hoeijmakers JH, Dantuma NP, Houtsmuller AB, Vermeulen W. 2012. Recognition of DNA damage by XPC coincides with disruption of the XPC-RAD23 complex. J. Cell Biol. 196:681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Spek PJ, Eker A, Rademakers S, Visser C, Sugasawa K, Masutani C, Hanaoka F, Bootsma D, Hoeijmakers JH. 1996. XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 24:2551–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Madura K. 2006. Evidence for distinct functions for human DNA repair factors hHR23A and hHR23B. FEBS Lett. 580:3401–3408 [DOI] [PubMed] [Google Scholar]
- 52.Jiang YH, Beaudet AL. 2004. Human disorders of ubiquitination and proteasomal degradation. Curr. Opin. Pediatr. 16:419–426 [DOI] [PubMed] [Google Scholar]
- 53.Marteijn JA, Jansen JH, van der Reijden BA. 2006. Ubiquitylation in normal and malignant hematopoiesis: novel therapeutic targets. Leukemia 20:1511–1518 [DOI] [PubMed] [Google Scholar]
- 54.Khandros E, Weiss MJ. 2010. Protein quality control during erythropoiesis and hemoglobin synthesis. Hematol. Oncol. Clin. North Am. 24:1071–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D, Lonial S, Goldschmidt H, Reece D, San-Miguel JF, Blade J, Boccadoro M, Cavenagh J, Dalton WS, Boral AL, Esseltine DL, Porter JB, Schenkein D, Anderson KC. 2005. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N. Engl. J. Med. 352:2487–2498 [DOI] [PubMed] [Google Scholar]
- 56.Tamura D, Arao T, Tanaka K, Kaneda H, Matsumoto K, Kudo K, Aomatsu K, Fujita Y, Watanabe T, Saijo N, Kotani Y, Nishimura Y, Nishio K. 2010. Bortezomib potentially inhibits cellular growth of vascular endothelial cells through suppression of G2/M transition. Cancer Sci. 101:1403–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanash SM, Rucknagel DL. 1978. Proteolytic activity in erythrocyte precursors. Proc. Natl. Acad. Sci. U. S. A. 75:3427–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. 2007. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 130:349–362 [DOI] [PubMed] [Google Scholar]
- 59.Rusconi F, Mancinelli E, Colombo G, Cardani R, Da RL, Bongarzone I, Meola G, Zippel R. 2010. Proteome profile in myotonic dystrophy type 2 myotubes reveals dysfunction in protein processing and mitochondrial pathways. Neurobiol. Dis. 38:273–280 [DOI] [PubMed] [Google Scholar]
- 60.Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. 2011. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 147:120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.