Abstract
Neurite growth requires neurite extension and retraction, which are associated with protein degradation. Autophagy is a conserved bulk degradation pathway that regulates several cellular processes. However, little is known about autophagic regulation during early neurite growth. In this study, we investigated whether autophagy was involved in early neurite growth and how it regulated neurite growth in primary cortical neurons. Components of autophagy were expressed and autophagy was activated during early neurite growth. Interestingly, inhibition of autophagy by atg7 small interfering RNA (siRNA) caused elongation of axons, while activation of autophagy by rapamycin suppressed axon growth. Surprisingly, inhibition of autophagy reduced the protein level of RhoA. Moreover, expression of RhoA suppressed axon overelongation mediated by autophagy inhibition, whereas inhibition of the RhoA signaling pathway by Y-27632 recovered rapamycin-mediated suppression of axon growth. Interestingly, hnRNP-Q1, which negatively regulates RhoA, accumulated in autophagy-deficient neurons, while its protein level was reduced by autophagy activation. Overall, our study suggests that autophagy negatively regulates axon extension via the RhoA-ROCK pathway by regulating hnRNP-Q1 in primary cortical neurons. Therefore, autophagy might serve as a fine-tuning mechanism to regulate early axon extension.
INTRODUCTION
Neurons have highly polarized structures, including axons and dendrites, which are important to their specialized functions. Accordingly, their proper development is crucial to the formation of appropriate neuronal connections and function (1–3). Neurons extend excessive neuritic projections into target regions, after which orphan neurites are pruned (1, 4). During early growth and active developmental refinement of axons and dendrites, new protein synthesis and protein degradation are important to the formation of a functional neural network (5–8). Interestingly, pruning during neurite development is morphologically similar to neurite degeneration following nerve injury (9). The ubiquitin-proteasome pathway (UPS) is a protein degradation pathway involved in neurite retraction, synaptic elimination during neurite and synaptic development, and neurite regeneration after injury (10–12). In addition to UPS, autophagy is a conserved bulk lysosomal-degradation pathway that is involved in cell survival, differentiation, development, and homeostasis (13, 14). There are three types of autophagic pathways, microautophagy, chaperon-mediated autophagy, and macroautophagy (referred to as autophagy), and they have been extensively investigated in organisms ranging from yeast to humans using genetic and biochemical approaches (15, 16). Although all cell types have autophagic pathways, cells comprising different tissues have different activities and mechanisms for regulation of autophagy (17). Once autophagy is activated, cytoplasmic proteins or organelles are sequestered by double- or multi-membrane-bound autophagosomes, after which they are fused with lysosomes for final degradation (18).
There is a growing body of evidence that autophagy is constitutively active in healthy neurons (17, 19, 20). Mice with specific deletion of atg5 or atg7, which are essential genes for autophagosome formation (21, 22), from the central nervous system (CNS) show accumulation of ubiquitin-positive inclusions, neurodegeneration, and motor dysfunction, indicating that constitutive neuronal autophagy regulates cellular homeostasis (21, 22). Moreover, Purkinje neuron-specific autophagy-deficient mice exhibit axon swelling, indicating that autophagy regulates axon homeostasis (23). According to a recent report, the autophagic pathway can also regulate presynaptic structure and function in dopaminergic neurons, indicating that it is involved in synaptic plasticity (24).
Autophagy in neurons is also associated with the pathology of several neurodegenerative diseases, including Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), and frontotemporal dementia (FTD) (25–28). The disruption of autophagic flux contributes to disease progression, indicating that proper regulation of autophagy is essential to neuronal-cell survival (29). Indeed, massive accumulation of autophagic vacuoles, including autophagosomes and autolysosomes, is also observed in axon regions and cytoplasm in many neurodegenerative diseases (19, 28). Moreover, autophagy is induced and activated to reduce neuronal cytotoxicity and protect neurons under various stress conditions, such as protein accumulation, hypoxic ischemia, or excitotoxic stimuli (28, 30).
Despite increasing studies of autophagy in mature neurons, the exact role of autophagy during early neurite growth is largely unknown in vitro and in vivo. Autophagosomes have been reported to accumulate in developing neurons during brain development (31). Lysosomal activity has also been associated with axon pruning in vivo (32). A recent study showed that autophagosomes exist in both axon and somatodendritic regions and move dynamically along the axon in cultured neurons, indicating their possible roles in early neurite growth and morphological plasticity (28, 33–35). Despite some indications of involvement of the autophagic pathway in neurite formation and growth, little is known about the regulation and exact function of the autophagic pathway during early axon or dendritic growth of postmitotic neurons.
In this study, we investigated the roles of the autophagic pathway in early neurite growth in cultured cortical neurons, a well-characterized model for investigating early neurite growth in a time-dependent fashion (36, 37). We showed that autophagy-related genes were indeed expressed and mTOR was inhibited during early neurite growth and that autophagy was activated during this stage in cultured cortical neurons. Inhibition of autophagy by atg7 small interfering RNA (siRNA) led to elongation of the axon, while activation of autophagy by rapamycin reduced early axon growth. We also showed that autophagy negatively regulates early axon elongation through the hnRNP-Q1–RhoA–Rho-associated protein kinase (ROCK) signaling pathway. Based on these findings, we propose that autophagy acts as a fine-tuning mechanism to regulate axon morphological plasticity by controlling axon extension during early neurite growth in vitro.
MATERIALS AND METHODS
Molecular cloning.
Human cDNA of wild-type RhoA or the constitutive active form (RhoAG14V) was obtained from Addgene (Cambridge, MA). For mammalian expression, each RhoA cDNA was generated using specific primers (sense, 5′-CGCCAAGCTTATGGCTGCCATCCGGAAG-3′; antisense, 5′-CGACCTCGAGTCACAAGACAAGGCACCC-3′) and then subcloned into the BamHI-HindIII site of a 3×Flag CMV7.1 vector (Sigma, St. Louis, MO). For hnRNPQ-1 siRNA or atg7 siRNA, synthetic siRNA against mouse and rat hnRNP-Q or ATG7 was generated as previously described (38, 39). Specifically, the siRNAs were as follows: hnRNP-Q1, 5′GCAAGCAGCAAAGAAUCAAAUGUAU-3′ and 5′-AUACAUUUGAUUCUUUGCUGCUUGC-3′; control, 5′-CCUACGCCACCAAUUUCGU(dTdT)-3′ and 5′-ACGAAAUUGGUGGCGUAGG(dTdT)-3′; si-atg7, 5′-GCAUCAUCUUUGAAGUGAA-3′ and 5′-UUCACUUCAAAGAUGAUGC-3′ (Bioneer, Munpyeongseo-ro, South Korea).
Cell cultures, transfection, and immunocytochemistry.
Cortical neurons from embryonic day 17 (E17) or E18 Sprague-Dawley rats (Samtako, Gyeonggi-do, South Korea) were cultured as previously described (26, 40). Each DNA plasmid was transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or calcium phosphate reagents (Clontech, Mountain View, CA) into cortical neurons at 1 or 3 days in vitro (DIV1 or DIV3) according to the manufacturer's protocol. For siRNA transfection, each siRNA (75 to 90 pmol/μl/well) aliquot was incubated in cell culture medium using RNAi Max reagents (Invitrogen), followed by the addition of trypsinized neurons or mouse embryonic fibroblasts (MEFs) according to the manufacturer's protocol.
For activation of autophagy, 10 nM rapamycin (Sigma) or dimethyl sulfoxide (DMSO) was incubated at DIV1 for 12 to 14 h in cultured cortical neurons. For inhibition of ROCK under autophagy activation, 50 μM Y-27632 (Sigma) was added to cultured cortical neurons 1 h after rapamycin treatment for 12 to 14 h. To inhibit autophagy, atg7 siRNA with or without siRNA-resistant human atg7–c-myc was transfected into cultured cortical neurons at DIV1, and the neurite length was measured 2 days later. For the rescue experiment, wild-type (WT) RhoA (RhoAWT) or the constitutive active form of RhoA (RhoaG14V) was transfected with either scrambled siRNA or atg7 siRNA at DIV1, and neurons were fixed with 4% paraformaldehyde (PFA) for 10 min 2 days later. For immunostaining, transfected neurons were permeabilized with 0.1% Triton X-100 for 5 min and then blocked with 3% bovine serum albumin (BSA) for 1 h at room temperature. After blocking, cells were incubated with anti-c-myc (Thermo Science, Waltham, MA 1:100) for 1 h and then incubated with anti-Cy3 or anti-DyLight 488 secondary antibodies (Jackson Laboratory, Bar Harbor, ME) for 1 h at room temperature. For inhibition of autophagy by pharmacological methods, 3-methyladenine (3-MA) (10 mM) was applied to cultured cortical neurons at DIV2 for 14 to 16 h.
Quantification of neurite length.
Anti-Tau1 (Millipore, Billerica, MA; 1:200)-positive and anti-microtubule-associated protein 2 (MAP2) (Millipore; 1:200)-positive neurites were used for measurement of the axon or dendrite length, respectively. Neuronal images were then obtained using an LSM 510 confocal laser scanning microscope (LSM700; Carl Zeiss, Oberkochen, Germany). Neurite length was measured and quantified using the Image J, Adobe Photoshop CS3, and GraphPad Prism5 programs. The lengths of 20 to 30 neurons in each experimental group were measured and quantified from 3 to 5 independent experiments. Statistical analyses (one-way analysis of variance [ANOVA] and Student's t test) were conducted using the GraphPad Prism5 program.
Western blotting.
Total cell lysates from cultured cortical neurons from DIV1 to DIV5 or MEFs from wild-type or atg5 knockout (atg5−/−) mice (41) were prepared in lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 10% sodium dodecyl sulfate [SDS], and protease inhibitor). Cell lysates (30 to 50 μg protein) were analyzed by Western blotting with anti-LC3 antibody (Novus Biologicals, Littleton, CO; 1:5,000), anti-ATG7 antibody (Santa Cruz Biotech, Santa Cruz, CA; 1:1,000), anti-phosphorylated p70S6 kinase antibody or anti-p70S6 kinase antibody (Cell Signaling, Danvers, MA; 1:1,000), anti-RhoA antibody (Santa Cruz Biotech; 1:1,000), anti-hnRNP-Q1 (Abcam, Cambridge, MA; 1:1,000), and horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary antibody (1:10,000 to 1:20,000).
Semiquantitative RT-PCR analysis.
Total RNA was extracted from neurons or MEFs using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RT-PCR experiments were carried out with cDNA generated from 1 μg of total RNA using the Superscript III first-strand synthesis system (Invitrogen). The RT-PCR exponential phase was determined based on 22 to 30 cycles to allow semiquantitative comparisons of cDNA obtained from the same reactions with nTaq DNA Polymerase (Enzynomics, Daejeon, South Korea). The primers were as follows: rat hnRNP-Q1, sense, 5′-GTAAAGTGACAGAGGGTCTC-3′, and antisense, 5′-TCCATAGCCTTGACAGCACC-3′; mouse hnRNP-Q1, sense, 5′-TGGTGCTGTCAAGGCTATGG-3′, and antisense, 5′-CATATCCACCACGATAGTTAT-3′. RT-PCR was conducted by subjecting the samples to cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 15 s.
RESULTS
Autophagy is activated during early neurite growth of cultured cortical neurons.
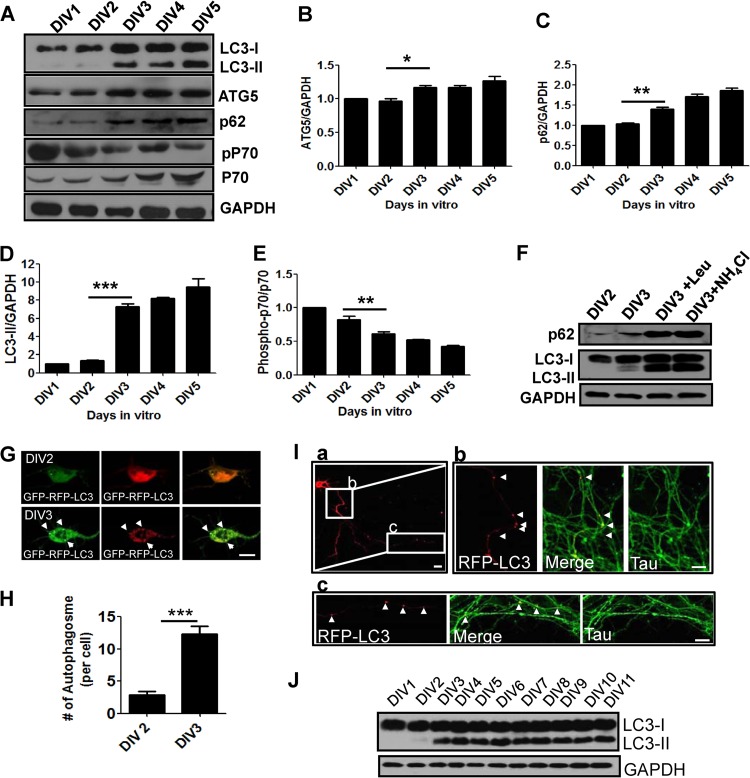
To investigate whether autophagy is involved in early neurite growth in mammalian neurons, we first examined the expression of ATG5, p62, and LC3, which are key components required for autophagy (21, 42, 43). Interestingly, the levels of ATG5, p62, and LC3-II were significantly elevated between DIV2 and DIV3 and sustained during early neurite growth (DIV1 to DIV5), indicating that autophagic components are indeed expressed in the recapitulated culture system of early neurite growth (Fig. 1A to C and F).
Fig 1.
Autophagy-related genes are expressed and autophagy is activated during early neurite growth in cultured cortical neurons. (A to E) Western blot (A) and quantification plots of bands (B to E) of cultured cortical neurons from DIV1 to DIV5 showing increased ATG5, LC3-II, and p62 and decreased phospho-p70 (pP70) S6 kinase/p70 S6 kinase ratios (indicative of mTOR inhibition), indicating autophagy induction. GAPDH was used as a loading control and a normalizing band for quantification. The phosphorylated p70 S6 kinase band was normalized to that of p70 S6 kinase. The values are the means and standard errors of the mean (SEM) of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (F) Western blot of cultured cortical neurons at DIV2 and DIV3 and in the presence of ammonium chloride (NH4Cl) or leupeptin (Leup) at DIV3 revealing increased LC3 and p62 levels. (G to I) Representative images (G) and quantification (H) of RFP-GFP-positive autophagosomes of cultured cortical neurons at DIV2 and DIV3 expressing GFP-RFP-LC3 or RFP-LC3 showing accumulation of autophagosomes (puncta) in cell bodies (G) and axons (I, a) at DIV3. The values are the means and SEM of three independent replicates. Student's t test: ***, P < 0.001. RFP-LC3-positive autophagosomes were localized to the proximal (I, b) and distal (I, c) regions of axons labeled with Tau. The arrowheads indicate LC3-positive autophagosomes. Scale bars, 20 μm (G and I, a) and 5 μm (I, b and c). (J) Western blot analysis of LC3 using cultured developing cortical neurons (from DIV1 to DIV11).
To further characterize autophagy during early neurite growth, we checked the level of LC3-II, a marker of autophagosomes that is processed from LC3-I and specifically localized to autophagosomes (43). The LC3-II level was significantly elevated at DIV3, after which it remained high (Fig. 1A and D). Increases in LC3-II could be due to the activation of autophagy or inhibition of the maturation or degradation of autophagosomes (44). To determine autophagic flux during early neurite growth, we investigated LC3 and p62 turnover by Western blotting in the presence of an inhibitor of lysosomal protease, leupeptin or ammonium chloride (NH4Cl), which inhibits acidification in lysosomes (44, 45). The levels of LC3-II and p62 increased in the presence of an inhibitor of lysosome protease or a lysosome-trophic reagent, indicating that LC3-II and p62 were actively degraded in lysosomes during early neurite growth in cultured cortical neurons (Fig. 1F).
Consistent with our Western blot analysis, red fluorescent protein (RFP)-green fluorescent protein (GFP)-LC3-positive autophagosomes accumulated in cell bodies (Fig. 1G and H), and RFP-LC3-positive autophagic vacuoles were localized to proximal or distal regions of axons at DIV3 (Fig. 1I, a, b, and c). Notably, the increase in LC3-II was sustained until DIV11, at which time the neuronal morphology was almost mature, suggesting an active role of autophagy in neuronal morphogenesis and function (Fig. 1J).
We further examined the mTOR pathway, a negative regulator of autophagy induction, by checking the phosphorylation of p70S6 kinase (46). Phosphorylation of p70 was significantly reduced from DIV3 onward, suggesting that inhibition of mTOR may be associated with activation of autophagy during the early stages of neurite growth in culture, even though inhibition of mTOR affects the autophagy-independent pathway (47) (Fig. 1A and E). Taken together, these data indicate that components of autophagy are expressed and autophagy is activated during early neurite growth in cultured neurons.
Autophagy suppression by atg7 siRNA induces axon elongation.
To elucidate the functional significance of autophagy activation during early neurite growth, we inhibited the autophagy pathway by transfecting atg7 siRNA into cultured cortical neurons (DIV1). atg7 siRNA indeed reduced the ATG7 level in differentiated cortical neurons (Fig. 2A; see Fig. 4E). atg7 siRNA also reduced the LC3-II level and the number of GFP-LC3-positive autophagosomes, confirming the reduction of autophagy (Fig. 2C). To determine if autophagy regulates early neurite growth, atg7 siRNA and GFP were cotransfected into cortical neurons at DIV1. Two days posttransfection, total neurite length was increased significantly in response to the knockdown of ATG7 (Fig. 2D and E). Neuronal viability was not affected by atg7 siRNA for 2 to 3 days posttransfection (Fig. 2G) (40). To rule out any off-target effects of atg7 siRNA, siRNA-resistant human atg7–c-myc was cotransfected with siRNA at DIV1. siRNA-resistant ATG7 rescued increased neurite length (Fig. 2D and E). Moreover, inhibition of autophagy by 3-MA, a general inhibitor of autophagy, also increased total neurite length, suggesting the possible roles of autophagy in neurite growth (Fig. 2H and I). These results show that autophagy controls neurite extension during the early stages of neurite growth.
Fig 2.
Autophagy inhibition by atg7 siRNA leads to axon elongation during early neurite growth. (A to C) Western blot (A), quantification of the Western blot (LC3-II and ATG7) (B), and representative images of GFP-LC3-positive autophagosomes (indicated by arrows) (C) of cultured cortical neurons at DIV2 and DIV3 transfected with atg7 siRNA showing a decrease in ATG7 and LC3-II levels (A and B) and autophagosomes in cell bodies (C) compared to controls (CTL) transfected with scrambled siRNA, indicating inhibition of autophagy. Neurons rescued by cotransfection of siRNA-resistant human atg7–c-myc (atg7–c-myc*) and atg7 siRNA (Rescue) showed no noticeable decrease in ATG7, LC3-II, or autophagosomes, demonstrating the specificity of siRNA treatment. GAPDH was used as a loading control. (D) Composite images of neurite morphology were generated from 2 or 3 separate images obtained from the same neuron to track the total neurite length. (D and E) Representative images of neurite growth (D) and a quantification plot of neurite length (E) of cultured cortical neurons at DIV1 to DIV3 transfected with atg7 siRNA showing a significant increase in total neurites and axons but not in dendrite length, whereas atg7–c-myc* (inset labeled with c-myc antibody) showed no significant change in total neurite, axon, and dendrite length. Each siRNA was cotransfected with GFP to determine the neurite morphology. CTL, control neurons transfected with scrambled siRNA. The values are shown as means ± SEM of three independent experiments. One-way ANOVA and Tukey's multiple-comparison test; *, P < 0.05; **, P < 0.01; ns, not significant. Scale bar, 20 μm. (F) Scrambled siRNA (CTL) or atg7 siRNA was transfected into cultured cortical neurons to examine axon morphology at DIV1 to DIV3 in developing neurons, and the number of axon tips was determined. The values are shown as means and SEM. Student's t test; ns, not significant. (G) Scrambled siRNA (CTL) or atg7 siRNA was transfected into cultured cortical neurons at DIV1. Cell viability was measured by counting surviving neurons at 24 h and 48 h after transfection. The values are shown as means and SEM. Student's t test; ns, not significant. (H and I) Cultured neurons were transfected with GFP to visualize cell morphology at DIV2. Cells were treated with 3-MA (10 mM) at DIV2 for 12 h. Shown are representative images of neurite growth (H) and a quantification plot of neurite length (I) of cultured cortical neurons at DIV2 and DIV3. (H) Composite images of neurite morphology were generated from 2 separate images obtained from the same neuron to track the total neurite length of atg7 siRNA-transfected neurons. The values are shown as means and SEM. Student's t test; ***, P < 0.001. Scale bar, 20 μm.
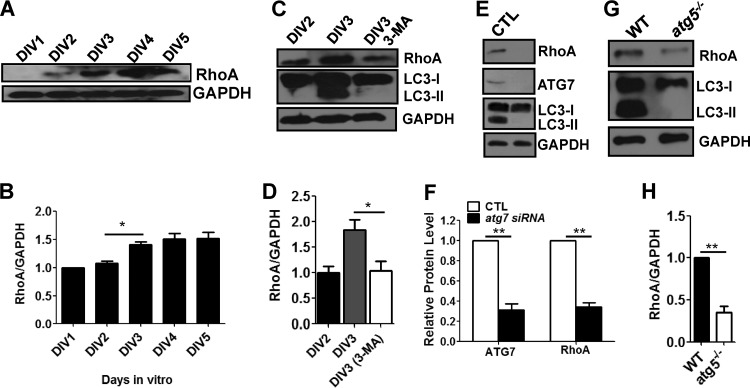
Fig 4.
RhoA is downregulated by autophagy inhibition and upregulated during early neurite growth. (A and B) Western blot (A) and quantification plots of bands (B) of cultured cortical neurons from DIV1 to DIV5 showing increased RhoA levels. GAPDH was used as a loading control and a normalizing band for quantification. The values are means and SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; *, P < 0.05. (C and D) Western blot (C) and quantification plots of bands (D) of cultured cortical neurons at DIV2 and DIV3 treated with 10 mM 3-MA showing a significant decrease in RhoA and LC3-II levels compared to DMSO (vehicle)-treated controls (CTL), indicating that inhibition of autophagy downregulates RhoA. GAPDH was used as a loading control. The values are the means and SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; *, P < 0.05. (E and F) Western blot (E) and quantification plots of bands (F) of cultured cortical neurons at DIV2 and DIV3 transfected with atg7 siRNA showing significant decreases in RhoA, ATG7, and LC3-II relative to those transfected with scrambled siRNA (CTL), indicating that inhibition of autophagy downregulates RhoA. GAPDH was used as a loading control. The values are the means and SEM of normalized band intensities from three independent replicates. Student's t test; **, P < 0.01. (G and H) Western blot (G) and quantification plots of bands (H) of atg5−/− MEFs showing a significant decrease in RhoA and LC3-II levels compared to wild-type MEFs (WT), indicating that inhibition of autophagy downregulates RhoA. GAPDH was used as a loading control. The values are the means and SEM of three independent replicates. Student's t test; **, P < 0.01.
To examine whether autophagy affects axon and/or dendritic growth, axon and dendritic lengths were measured by immunolabeling of the axon marker Tau1 or the dendritic marker MAP2 (data not shown). Axon length was significantly increased in response to the knockdown of ATG7, while no significant difference in dendrite length was observed (Fig. 2E). Moreover, axon branching was not affected by reduction of ATG7 (Fig. 2F). Taken together, these results show that reduction of autophagy by knockdown of ATG7 causes abnormal overelongation of axons during the early stages of neurite growth.
Autophagy induction by rapamycin suppresses axon growth during early neurite growth.
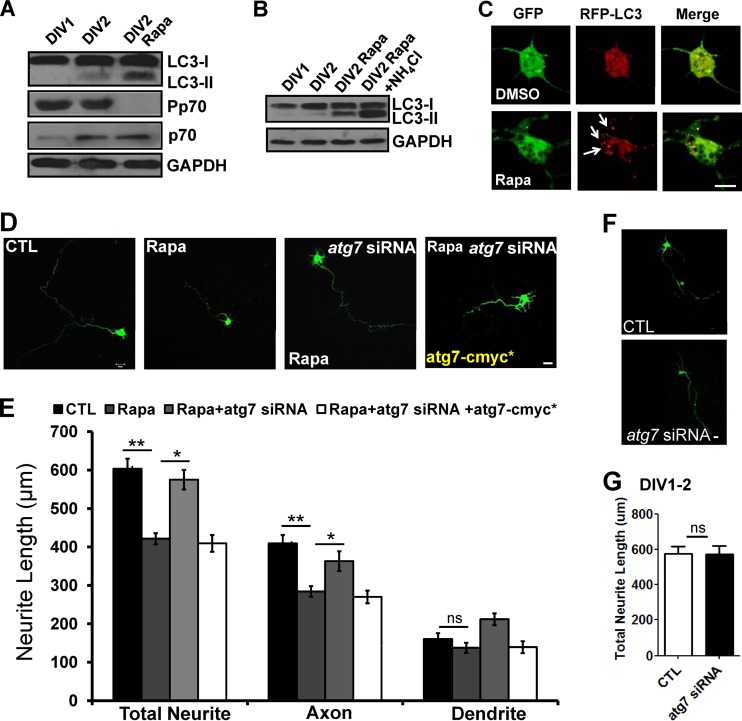
To further confirm the role of autophagy during early neurite growth, we pharmacologically induced the autophagic pathway with rapamycin during DIV1 and DIV2, a stage in which autophagosomes were almost undetectable in developing cultured neurons. Rapamycin treatment increased the level of LC3-II (Fig. 3A), which was further increased in the presence of ammonium chloride (Fig. 3B) and caused accumulation of RFP-LC3-positive autophagosomes (Fig. 3C). Rapamycin treatment also decreased the phosphorylation of p70S6 kinase, a serine/threonine kinase that is a downstream target of mTORC1 and is considered a negative regulator of autophagy (46) (Fig. 3A). Interestingly, neurons treated with rapamycin showed significant reduction in axon length (Fig. 3D and E). To rule out non-autophagy-related effects of rapamycin on neurite growth, we inhibited the autophagic pathway with atg7 siRNA under rapamycin treatment. Suppression of autophagy by atg7 siRNA rescued the rapamycin-mediated axon reduction phenotype, confirming that rapamycin-mediated autophagy suppresses axon extension during the early stages of neurite growth in primary neurons (Fig. 3D and E). However, atg7 siRNA itself had no significant effects on neurite length at DIV1 to DIV2 (Fig. 3F and G). This might have been due to low expression of autophagic machinery or to autophagy having little effect on neurite growth at DIV1 and DIV2 (Fig. 1A and B and 3D to G). Taken together, these results show that autophagy negatively regulates early axon extension in cultured cortical neurons.
Fig 3.
Autophagy activation by rapamycin causes suppression of axon growth during early neurite growth. (A to E) Western blots (A and B) in the absence (A) or presence (B) of ammonium chloride (10 mM), representative images of RFP-LC3-positive autophagosomes (indicated by arrows) (C), neurite length (D), and quantification plot of neurite length (E) of cultured cortical neurons at DIV1 and DIV2 treated with 10 nM rapamycin (Rapa) at DIV1 showing increased LC3 (more robust for LC3-II) and a decreased phospho-p70 S6 kinase/p70 S6 kinase ratio (A), accumulation of autophagosomes (C), and significant reduction in total neurite and axon but not dendrite length (D and E) at DIV2 compared to vehicle-treated control (DMSO), indicating that rapamycin induced autophagy. GAPDH was used as a loading control. Transfection with atg7 siRNA followed by rapamycin treatment led to significant improvement of total neurite and axon length, whereas atg7–c-myc* showed no improvement in total neurite and axon length. Each siRNA was cotransfected with GFP to determine the neurite morphology. CTL, control neurons were transfected with scrambled siRNA. The values are shown as means ± SEM of three independent experiments. One-way ANOVA and Tukey's multiple-comparison test; *, P < 0.05; **, P < 0.01; ns, not significant. Scale bar, 20 μm. (F and G) Scrambled siRNA (CTL) or atg7 siRNA was transfected into cultured cortical neurons at DIV1. The total neurite length was quantified 24 h after transfection. Shown are representative images of neurite growth (F) and a quantification plot of the neurite length (G) of cultured cortical neurons at DIV1 and DIV2. The values are shown as means and SEM. Student's t test; ns, not significant. Scale bar, 20 μm.
RhoA is downregulated by inhibition of autophagy, while it is upregulated during autophagy activation in developing neurons.
Since autophagy negatively controls axon growth during the early stages of neurite growth, it is important to determine how it affects early axon growth. Therefore, we attempted to identify the mechanistic link between autophagy and axon growth. To accomplish this, we examined RhoA, a small GTPase belonging to the Rho family that is a well-characterized modulator of axon morphology (48). RhoA has been shown to play a role in various aspects of axon development, such as axon initiation, guidance, elongation, and branching (49). RhoA has also been shown to negatively regulate neurite outgrowth and extension (50). Therefore, we investigated whether RhoA was involved in autophagy-associated axon growth in cultured developing neurons.
We reasoned that if RhoA modulates axon growth via autophagy, its expression pattern might be correlated with that of autophagy-related gene expression during early axon growth. Interestingly, the RhoA protein level was elevated between DIV2 and DIV3, indicating that the timing of upregulation of RhoA coincides with that of autophagy-related gene expression (Fig. 1 and 4A and B). We next investigated the RhoA level upon knockdown of ATG7 by siRNA or 3-MA during the early neurite growth of cultured cortical neurons (DIV1 to DIV3). Exposure to 3-MA or knockdown of ATG7 at DIV1 to DIV3 significantly reduced the RhoA level, indicating that inhibition or reduction of autophagy downregulates RhoA (Fig. 4C to F). To further confirm whether RhoA is affected by autophagy, we examined RhoA levels in the MEFs of autophagy-deficient atg5−/− mice (41) and found that they were reduced (Fig. 4G and H). Taken together, these data strongly suggest that autophagy affects RhoA levels.
Ectopic expression of RhoA rescues the axon elongation phenotype by autophagy inhibition during early neurite growth.
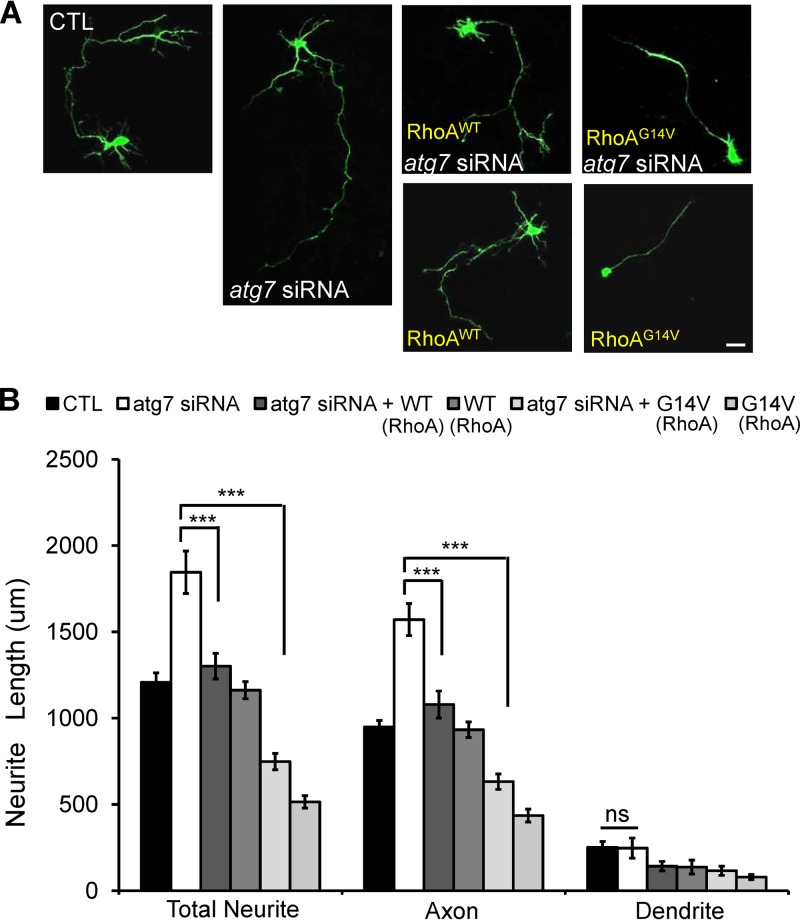
To examine whether RhoA is responsible for early axon growth affected by autophagy, we investigated the effects of ectopic expression of RhoAWT and RhoAG14V on axon overelongation in atg7 siRNA-mediated autophagy-deficient neurons. To accomplish this, Flag-RhoAWT or Flag-RhoAG14V (51) was cotransfected with atg7 siRNA or scrambled siRNA into cortical neurons at DIV1. The expression of RhoAWT and RhoAG14V was then confirmed by immunostaining using anti-Flag antibody (data not shown). Forced expression of RhoAWT and RhoAG14V in atg7 siRNA-transfected neurons significantly rescued the axon elongation defect (Fig. 5A and B), while ectopic expression of RhoAWT alone did not affect axon length. However, ectopic expression of RhoAG14V reduced axon length, indicating that expression and sustained activity of RhoA enhance suppression of axon growth (Fig. 5A and B). These data suggest that RhoA is involved in autophagy-mediated axon growth.
Fig 5.
RhoAWT or RhoAG14V rescues abnormal axon extension caused by autophagy inhibition. (A and B) Representative images of neurite growth (A) and quantification plot of neurite length (B) of cultured cortical neurons at DIV1 to DIV3 transfected with atg7 siRNA showing significant increases in total neurite and axon length but not in dendrite length compared to transfection with scrambled siRNA. Shown is cotransfection of atg7 siRNA with wild-type (Flag-RhoAWT [WT]) or constitutively active (Flag-RhoAG14V [G14V]) RhoA. RhoA significantly recovered the axon overextension phenotype, indicating that ectopic expression of RhoA recovers the deficient autophagy-mediated reduction of RhoA. Transfection with Flag-RhoAWT alone did not show a neurite phenotype; however, transfection with constitutively active RhoAG14V alone showed a remarkable reduction in neurite length compared to the control. Each siRNA was cotransfected with GFP to determine the neurite morphology. CTL, control neurons transfected with scrambled siRNA. The values are shown as the means ± SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; ***, P < 0.001; ns, not significant. Scale bar, 20 μm.
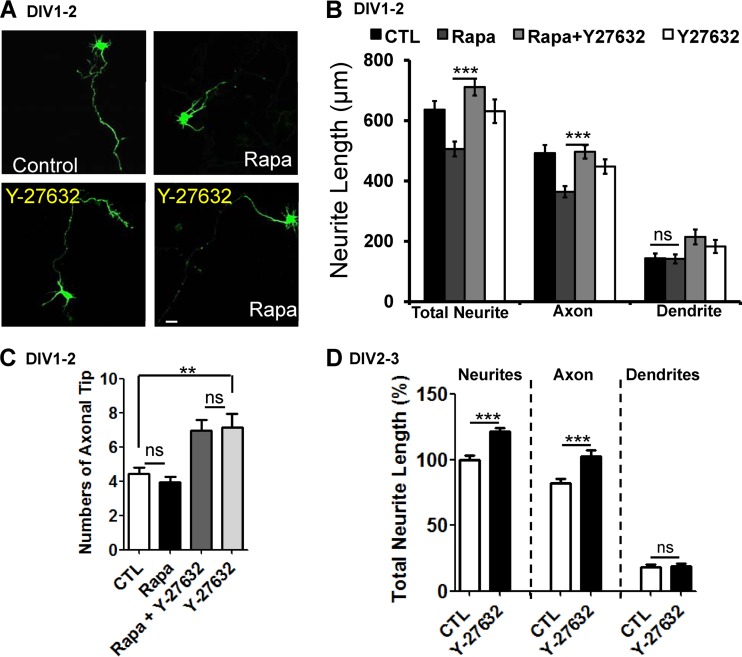
Inhibition of ROCK ameliorates suppressed axon growth by autophagy activation during early neurite growth.
We next examined the effects of inhibiting endogenous RhoA activity on suppressed axon growth in autophagy-activated neurons. RhoA signaling is blocked by Y-27632, an inhibitor of ROCK, which is a serine threonine kinase and a major downstream effector of RhoA (52). Interestingly, Y-27632 treatment significantly recovered the rapamycin-mediated suppression of axon growth (Fig. 6A and B). Y-27632 treatment alone slightly increased axon branching (Fig. 6C) but did not have a remarkable effect on axon elongation during DIV1 and DIV2 (Fig. 6A and B). However, Y-27632 treatment alone at DIV2 and DIV3 (during the stage in which autophagy-related gene expression is increased and activated) slightly increased axon length, indicating that autophagy activation affects RhoA-ROCK signaling during early axon growth (Fig. 6D). Overall, these results suggest that RhoA-ROCK activity regulates autophagy-controlled growth of axons in cultured developing cortical neurons.
Fig 6.
Inhibition of ROCK by Y-27632 recovers the suppression of axon growth induced by rapamycin. (A and B) Representative images of neurite growth (A) and quantification plot of neurite length (B) of cultured cortical neurons at DIV2 treated with 10 nM rapamycin showing a significant decrease in total neurite and axon length but not in dendrite length compared to vehicle (DMSO) treatment. Cotreatment with 10 nM rapamycin and 50 μM Y-27632, an inhibitor of ROCK, significantly rescued the rapamycin phenotype, indicating that inhibition of the ROCK pathway mitigates the reduction in axon length due to rapamycin-mediated activation of autophagy. Treatment with Y-27632 alone did not show any dramatic changes in the neurite phenotype. Each siRNA was cotransfected with GFP to determine the neurite morphology. CTL, control neurons treated with DMSO. The values are shown as means ± SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; ***, P < 0.001; ns, not significant. Scale bar, 20 μm. (C) Cortical neurons were treated with either rapamycin (10 mM) or Y-27632 (50 μm) or both at DIV1 for 14 to 16 h. The number of axon tips was determined in neurons treated with rapamycin at DIV2. The values are shown as means and SEM. One-way ANOVA and Tukey's multiple-comparison test; **, P < 0.01; ns, not significant. (D) Cortical neurons transfected with GFP were treated with 50 μM Y-27632 at DIV2 and DIV3. The total neurite length was quantified, and the axon length was found to have increased significantly. The values are means and SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; ***, P < 0.001.
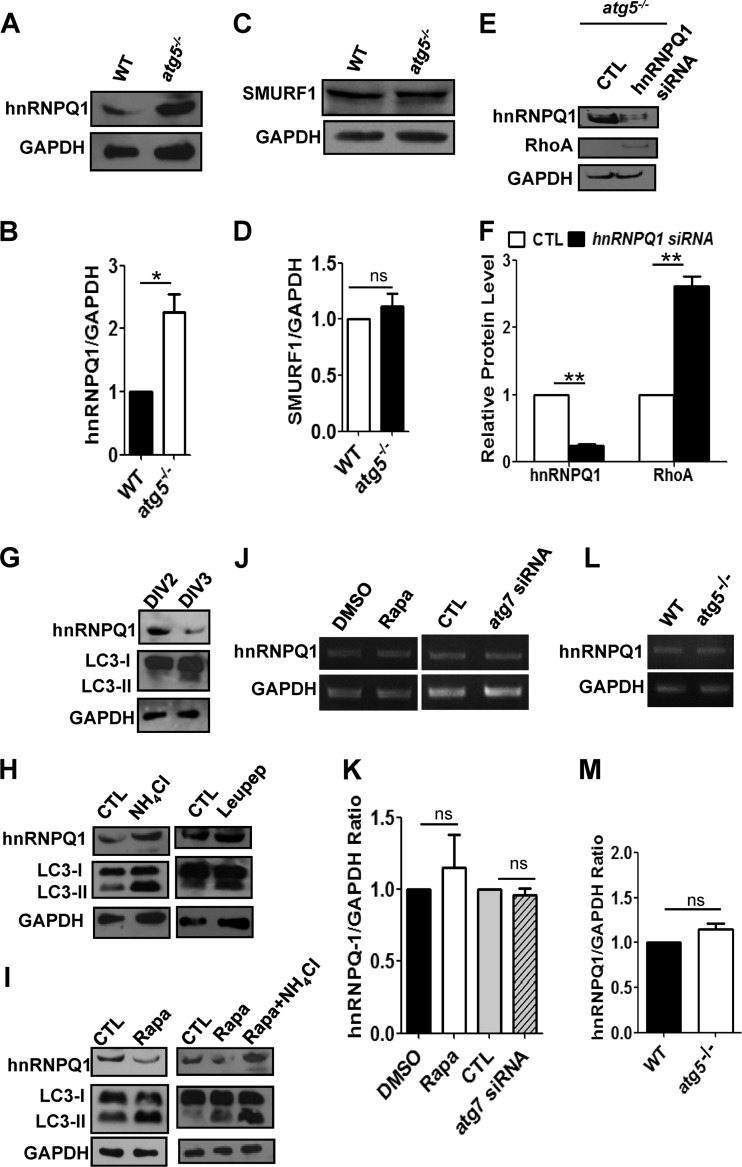
hnRNP-Q1, which negatively regulates the level of RhoA, is accumulated in autophagy-deficient cells, while its protein level was reduced by autophagy activation.
To determine how autophagy affects RhoA levels, we investigated whether the autophagy pathway could degrade negative regulators of RhoA expression. hnRNP-Q1 and SMAD-specific E3 ubiquitin protein ligase 1 (SMURF1) have been shown to negatively regulate RhoA, leading to changes in neuronal morphogenesis and neurite outgrowth (53, 54). Therefore, we examined whether hnRNP-Q1 and SMURF1 levels were affected by autophagy. Indeed, the levels of hnRNP-Q1 but not SMURF1 were significantly elevated in atg5−/− MEFs compared to wild-type MEFs (Fig. 7A to D). We next examined whether hnRNP-Q1 regulates the RhoA level in atg5−/− MEFs. Knockdown of hnRNP-Q1 by siRNA reduced the hnRNP-Q1 level and increased the RhoA level in atg5−/− MEFs, indicating that hnRNP-Q1 negatively regulates the RhoA level (Fig. 7E and F).
Fig 7.
The protein level of hnRNP-Q1, but not its mRNA level, is regulated by autophagy during early neurite growth. (A and B) Western blot (A) and quantification plots of bands (B) of atg5−/− MEFs showing an increase in hnRNP-Q1 levels, indicating that autophagy negatively regulates hnRNP-Q1. GAPDH was used as a loading control and a normalizing band for quantification. The values are means and SEM of three independent replicates. Student's t test; *, P < 0.05. (C) Western blot analysis of SMURF1 using wild-type or atg5−/− MEFs. GAPDH was used as a loading control. (D) Levels of SMURF1 in wild-type and atg5−/− MEFs. The values are the means and SEM of three independent replicates. ns, not significant. (E and F) Western blot (E) and quantification plots of bands (F) of atg5−/− MEFs transfected with hnRNP-Q1 siRNA showing a significant decrease in hnRNP-Q1 levels (F) and a significant increase in RhoA levels (F) relative to control scrambled siRNA, indicating that hnRNP-Q1 negatively regulates RhoA. GAPDH was used as a loading control. The values are the means and SEM of three independent replicates. Student's t test; **, P < 0.01. (G) Western blot of cultured cortical neurons at DIV2 and DIV3 showing a remarkable decrease in hnRNP-Q1 and increase in LC3-II at DIV3 compared to DIV2, indicating that activation of autophagy coincides with a reduction in hnRNP-Q1 during early neurite development. GAPDH was used as a loading control. (H) Western blot of cultured cortical neurons at DIV2 and DIV3 treated with ammonium chloride (NH4Cl) and leupeptin (Leupep) to block lysosomal degradation showing increased hnRNP-Q1 and LC3-II levels relative to the vehicle control (CTL). (I) Western blot of cultured cortical neurons at DIV2 and DIV3 treated with rapamycin (Rapa) showing a decrease in hnRNP-Q1 and an increase in LC3-II in the presence or absence of NH4Cl. GAPDH was used as a loading control. (J and K) Representative gel images of semiquantitative RT-PCR (J) and densitometric quantification of band intensities (K) illustrating the relative levels of hnRNP-Q1 mRNA in cultured neurons either treated with 10 nM rapamycin or DMSO (CTL) or transfected with atg7 siRNA or scrambled siRNA (CTL). There was no significant difference in the mRNA levels of hnRNP-Q1 between CTL (DMSO or scrambled siRNA) and neurons regulated by autophagy. The relative gene expression of hnRNP-Q1 mRNA was normalized to that of GAPDH. The values are the means and SEM of three independent replicates. Student's t test; ns, not significant. (L and M) Representative gel images of semiquantitative RT-PCR (L) and densitometric quantification of band intensities (M) illustrating the relative levels of hnRNP-Q1 mRNA in wild-type and atg5−/− MEFs. There was no significant difference in the mRNA levels of hnRNP-Q1 between wild-type and atg5−/− MEFs. The relative gene expression of hnRNP-Q1 mRNA was normalized to that of GAPDH. The values are the means and SEM of three independent replicates. Student's t test; ns, not significant.
Next, we examined whether the protein level of hnRNP-Q1 was regulated during early axon growth (DIV2 and DIV3). We found that the protein level of hnRNP-Q1 decreased during this period (Fig. 7G). Furthermore, inhibition of lysosomal degradation by leupeptin or NH4Cl accumulated hnRNP-Q1 during early axon growth (DIV2 and DIV3), while autophagy induction by rapamycin reduced the hnRNP-Q1 level, indicating that activation of autophagy regulates the hnRNP-Q1 level (Fig. 7H and I).
To check the possibility that autophagy regulates gene expression of hnRNP-Q1 mRNA during early neurite growth, we performed semiquantitative RT-PCR using neurons in which autophagy was activated by rapamycin or inhibited by atg7 siRNA. The mRNA level of hnRNP-Q1 was not affected by autophagy activation or inhibition during early neurite growth in neurons (Fig. 7J and K). Furthermore, there was no significant difference in the mRNA level of hnRNP-Q1 between wild-type and atg5−/− MEFs (Fig. 7L and M). Taken together, these results suggest that hnRNP-Q1 was likely degraded via the autophagy pathway during early neurite growth in cultured neurons.
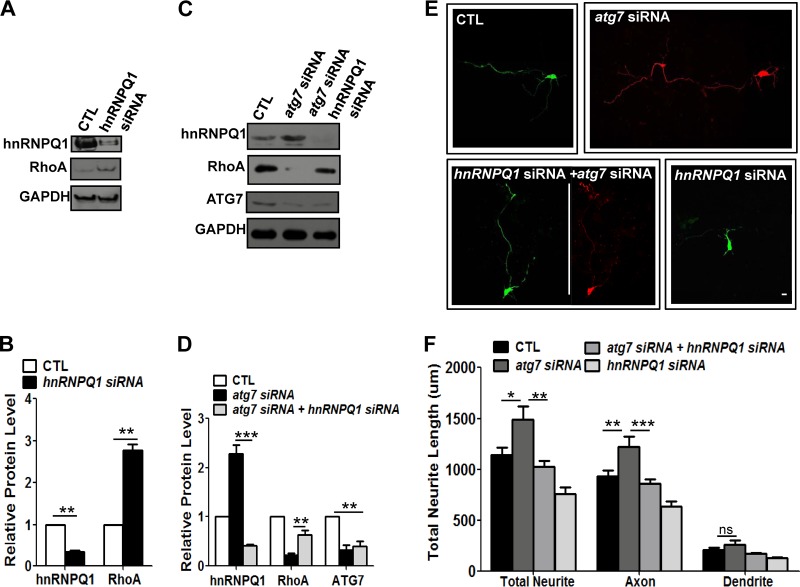
Reduction of hnRNP-Q1 by hnRNP-Q1 siRNA in autophagy-deficient neurons rescues the RhoA level and suppresses axon elongation.
To determine if the RhoA level was affected by hnRNP-Q1 in cultured cortical neurons, we reduced the hnRNP-Q1 level during autophagy activation at DIV2 and DIV3 using siRNA in neurons. Reduction of hnRNP-Q1 increased the RhoA level, indicating that hnRNP-Q1 negatively controls the level of RhoA in cultured cortical neurons (Fig. 8A and B). Moreover, siRNA-mediated double knockdown of hnRNP-Q1 and ATG7 in neurons increased the RhoA level that had been reduced by autophagy deficiency (Fig. 8C and D). These results suggest that hnRNP-Q1, which is reduced by autophagy activation, negatively regulates the RhoA level in cultured cortical neurons.
Fig 8.
hnRNP-Q1 regulated by autophagy negatively regulates RhoA and affects early axon growth in cultured neurons. (A and B) Western blot (A) and quantification plots of bands (B) of cultured cortical neurons at DIV1 to DIV3 transfected with hnRNP-Q1 siRNA showed a significant decrease in hnRNP-Q1 levels (B) and a significant increase in RhoA levels (B) compared to the control (scrambled siRNA), indicating that hnRNP-Q1 negatively regulates RhoA in cortical neurons. GAPDH was used as a loading control and a normalizing band for quantification. The values are the means and SEM of three independent replicates. Student's t test; **, P < 0.01. (C and D) Western blot (C) and quantification plots of bands (D) of cultured cortical neurons at DIV2 and DIV3 with cotransfection of atg7 and hnRNP-Q1 siRNA showing recovery of the autophagy deficiency-mediated reduction in RhoA levels. GAPDH was used as a loading control. The values are the means and SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; **, P < 0.01; ***, P < 0.001. (E and F) Composite images of neurite morphology (E) were generated from 2 or 3 separate images obtained from the same neuron to track the total neurite length of atg7 siRNA-transfected neurons. Representative images of neurite growth (E) and a quantification plot of the neurite length (F) of cultured cortical neurons at DIV1 to DIV3 transfected with atg7 siRNA, hnRNP-Q1 siRNA, or both show that cotransfection of atg7 siRNA (in pSICOR-RFP) with hnRNP-Q1 siRNA (in pSUPER-GFP) significantly recued the axon overextension phenotype, indicating that reduction of hnRNP-Q1 recovers the level of RhoA and suppresses abnormal axon elongation. Transfection with hnRNP-Q1 siRNA alone showed a slight reduction in neurite length compared to the control. Transfection with hnRNP-Q1 or atg7 siRNA and neurite morphology was determined by expression of GFP or RFP, respectively, or both. CTL, control neurons transfected with scrambled siRNA. The values are shown as the means and SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. Scale bar, 20 μm.
Finally, to determine whether increased hnRNP-Q1 in autophagy-deficient neurons is indeed associated with the axon elongation phenotype, hnRNP-Q1 siRNA was transfected into atg7 knockdown neurons at DIV1 to DIV3. Interestingly, axon elongation caused by atg7 siRNA was suppressed by hnRNP-Q1 siRNA, although reduction of hnRNP-Q1 itself suppressed axon growth partially via increased RhoA, as shown in Fig. 8A, B, E, and F. Taken together, these data indicate that hnRNP-Q1 regulated by autophagy could regulate the RhoA level and early axon growth.
DISCUSSION
Metabolic homeostasis in developing neurons, which are nondividing cells, is especially important for remodeling of their polarized structures, axons and dendrites. Structural plasticity during developmental processes requires proper protein or organelle degradation. Among degradation pathways in neurons, autophagy is the only bulk lysosomal-degradation pathway that regulates cytosolic components, including proteins, RNA, lipids, and organelles (14). The autophagy pathway is essential to development, differentiation, homeostasis, longevity, tumor biology, cell death, and immunity, as well as to stress-associated conditions (55, 56). Therefore, dysfunction of autophagy causes various human diseases, including cardiovascular diseases, infectious diseases, cancer, and neurodegenerative diseases (17, 55).
Autophagy is highly active in mature neurons, indicating that neuronal homeostasis is important for their specialized functions and survival. Despite increased focus on the role of autophagy in physiology and pathology in mature neurons, little is known about the roles of autophagy in early neurite growth in postmitotic neurons. Indeed, it is challenging to track neurite growth in vivo using current methods and models due to their rapid development and the complexity of brain development. Therefore, in this study, we used cultured postmitotic neurons, which comprise a well-characterized model that represents the sequence of morphological events in early axon or dendrite growth during neurite development in vivo (36, 37). To study autophagy, we used siRNA-mediated knockdown and MEFs from knockout animals instead of atg knockout mice, because complete knockout of autophagy causes extensive neurodegeneration in the CNS, raising the possibility of overlooking subtle phenotypes (21, 22, 57). Using these approaches in cultured mammalian cortical neurons, we showed for the first time that autophagy during early axon growth negatively regulates hnRNP-Q1 levels, which negatively controls RhoA levels and in turn negatively regulates axon extension. Autophagosomes are abundant in the distal region of axons and the proximal region of dendrites, indicating their possible roles in these structures (30, 33, 58). Our finding that autophagy was activated and mTOR inhibition was inhibited during early neurite growth supports the possibility that autophagy plays a role in early neurite growth. One of the pathways involved in mTOR inhibition during early axon growth in vitro is the phosphoinositide 3-kinase (PI3K)–Akt–tuberous sclerosis complex (TSC) signaling pathway (59, 60). TSC1 and TSC2 play important roles in restricting cell size and cell growth (61, 62). The TSC1-TSC2 complex has been shown to inhibit mTOR signaling (63, 64), and TSC has been reported to be localized in axons in developing cultured neurons, while Tsc1-Tsc2 acts upstream of the mTOR-mediated axonal growth-regulated pathway (65). Interestingly, overexpression of Tsc1/2 or rapamycin in cultured hippocampal neurons suppresses axon growth (65). Therefore, expression of Tsc1-Tsc2 might be increased during early axon growth, resulting in inactivation of mTOR and leading to suppression of axon growth.
Moreover, in our study, the stage of expression of autophagy-related genes coincides with that of axonal growth in culture. Indeed, studies have suggested an involvement of autophagy in axon development. Hollenbeck (66) demonstrated that autophagosomes were present within growth cones and the distal region of axons, indicating that their dynamic retrograde transport for degradation might affect axon growth and maintenance. Neurite outgrowth has also been reported to be induced by bafilomycin A1 as an inhibitor of vacuolar H+-ATPase, which is an inhibitor of lysosomal degradation in PC12 cells. Accordingly, these findings indicate that the autophagic pathway may also play a role in neurite outgrowth (67). Despite these findings, autophagic regulation of axon extension during neural development needs to be confirmed in vivo. Recent genetic studies have indicated a role of autophagy in axon growth in vivo. Pro-opimelanocortin (POMC) neurons showed active autophagic vacuoles in their projections, while loss of autophagy in POMC neurons perturbed axon growth in vivo (68). A murine study using Unc51.1/Ulk1, which is also required for autophagy induction, revealed that inhibition of Ulk1 suppresses neurite outgrowth (69), which is inconsistent with our finding that autophagy deficiency causes axon elongation. However, Ulk1/ATG1 might regulate neurite outgrowth through an autophagy-independent pathway that controls membrane vesicle trafficking, as previously reported (70, 71). Although there are some differences in their effects on axon growth, these might be due to differences in neuronal-cell types, the level of reduced autophagic activity, or the time or duration of autophagy deficiency. Another study demonstrated that lysosomal activity and autophagic vacuoles were associated with axonal pruning in developing nervous systems in mice (32). These data support our finding that autophagy in postmitotic neurons is involved in regulation of early axon growth.
In addition to the role of autophagy in early axon growth, we also provide evidence of the underlying mechanism through which autophagy regulates early axon growth. Our results suggest that autophagy regulates axon growth via upregulation of the RhoA-ROCK pathway in developing neurons. RhoA and its downstream effector, ROCK, are well-known negative regulators of the early stages of axon extension in cultured neurons (72–74). Indeed, forced expression of the wild-type or constitutive active form of RhoA in autophagy-deficient neurons rescued the axon elongation phenotype caused by autophagy reduction. Furthermore, inhibition of the ROCK pathway under autophagy induction recovered reduced axon growth. Taken together, these findings indicate that RhoA plays a key role in autophagy-associated suppression of axon elongation during the early stages of axonal growth.
It is unclear how autophagy, a predominantly degradative pathway, positively regulates RhoA levels. One possibility is that autophagy directly or indirectly degrades a negative regulator of RhoA expression or signaling. Our study showed that hnRNP-Q1 could be one of the substrates degraded by autophagy during early axon growth. Xing et al. demonstrated that hnRNP-Q1 represses RhoA expression at the posttranscriptional level and that knockdown of hnRNP-Q1 causes the phenotype associated with elevated RhoA levels (54). hnRNP-Q1 has been characterized as an NS1-associating protein 1 (Nsap1), and its expression in the CNS has been shown to be developmentally regulated (75–77). It is interesting that the level of hnRNP-Q1 is also regulated by the autophagic pathway during early neurite development. Therefore, it appears that autophagy activation during axon extension downregulates hnRNP-Q1, which subsequently leads to RhoA upregulation, thereby regulating axon elongation. Another possibility is that a transcriptional repressor of RhoA is a substrate for autophagic degradation, since autophagy may regulate RhoA at the transcriptional level. It will be interesting to identify additional autophagy substrates responsible for this regulation at the transcriptional or posttranscriptional level.
A number of studies have suggested that impairment of autophagy in mature neurons is primarily linked to axonopathy and results in neurodegeneration, indicating that axonal homeostasis and maintenance of axon structure are highly dependent on constitutive neuronal autophagy (78, 79). A recent report indicated that inhibition of the lysosomal pathway selectively impairs axonal retrograde transport of autophagosomes containing degradative organelles, which causes Alzheimer's-like axonal dystrophy (58). This discrepancy in the effect of autophagy deficiency on the axon phenotype could be due in part to differences in the level of autophagy reduction between studies. Complete knockout of autophagy is associated with axon degeneration, while reduction (or transient impairment) of autophagy might be involved in refinement of axon length during early neurite growth. Overall, accumulating evidence strongly suggests the importance of autophagy in axonal physiology and pathology.
In conclusion, although autophagy is reportedly important for survival in mature neurons, our findings suggest that it plays a role in the early morphological plasticity of neurite growth in cultured cortical neurons. We propose that autophagy might be a potential cellular and molecular mechanism underlying axon extension and degeneration during early axonal growth. Autophagy could also be a fine regulatory mechanism of axon extension and degeneration during early neurite growth in mammalian neurons. Acute defects in autophagy during early neurite development may cause impairment of axon growth and result in synaptic malfunction before severe neurodegeneration. Thus, manipulation of the autophagic pathway may represent an important therapeutic strategy for promotion of axonal growth or regeneration in neurodevelopmental disorders, as well as neurodegenerative diseases with axonopathy.
ACKNOWLEDGMENTS
We thank Noboru Mizushima for the generous gift of MEFs from atg5 knockout mice. We also thank Eun Kyung Cho (Chungnam University) and Dong Hyong Cho (Kyunghee University) for generously providing reagents for autophagy.
This work was supported by the Basic Science Research Program through the NRF (2010-0010824 and 2011-0022813).
J.-A.L. designed the experiments; J.-A.L., B.-K.B., D.-J.J., and S.T.A. analyzed the data; J.-A.L. and S.T.A. wrote the manuscript; and B.-K.B, M.-H.J., and H.-H.R. performed most of the experiments.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 5 August 2013
REFERENCES
- 1.Low LK, Cheng HJ. 2006. Axon pruning: an essential step underlying the developmental plasticity of neuronal connections. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1531–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo L, O'Leary DD. 2005. Axon retraction and degeneration in development and disease. Annu. Rev. Neurosci. 28:127–156 [DOI] [PubMed] [Google Scholar]
- 3.Saxena S, Caroni P. 2007. Mechanisms of axon degeneration: from development to disease. Prog. Neurobiol. 83:174–191 [DOI] [PubMed] [Google Scholar]
- 4.Murakami F, Song WJ, Katsumaru H. 1992. Plasticity of neuronal connections in developing brains of mammals. Neurosci. Res. 15:235–253 [DOI] [PubMed] [Google Scholar]
- 5.Eberwine J. 2001. Molecular biology of axons: “a turning point…”. Neuron 32:959–960 [DOI] [PubMed] [Google Scholar]
- 6.Gumy LF, Tan CL, Fawcett JW. 2010. The role of local protein synthesis and degradation in axon regeneration. Exp. Neurol. 223:28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klimaschewski L, Hausott B, Ingorokva S, Pfaller K. 2006. Constitutively expressed catalytic proteasomal subunits are up-regulated during neuronal differentiation and required for axon initiation, elongation and maintenance. J. Neurochem. 96:1708–1717 [DOI] [PubMed] [Google Scholar]
- 8.Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. 2005. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J. Neurosci. 25:331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raff MC, Whitmore AV, Finn JT. 2002. Axonal self-destruction and neurodegeneration. Science 296:868–871 [DOI] [PubMed] [Google Scholar]
- 10.Bingol B, Schuman EM. 2005. Synaptic protein degradation by the ubiquitin proteasome system. Curr. Opin. Neurobiol. 15:536–541 [DOI] [PubMed] [Google Scholar]
- 11.Korhonen L, Lindholm D. 2004. The ubiquitin proteasome system in synaptic and axonal degeneration: a new twist to an old cycle. J. Cell Biol. 165:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K. 2003. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr. Biol. 13:899–910 [DOI] [PubMed] [Google Scholar]
- 13.Kundu M, Thompson CB. 2008. Autophagy: basic principles and relevance to disease. Annu. Rev. Pathol. 3:427–455 [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N, Komatsu M. 2011. Autophagy: renovation of cells and tissues. Cell 147:728–741 [DOI] [PubMed] [Google Scholar]
- 15.Klionsky DJ. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Klionsky DJ. 2010. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12:814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JA. 2012. Neuronal autophagy: a housekeeper or a fighter in neuronal cell survival? Exp. Neurobiol. 21:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Klionsky DJ. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 [DOI] [PubMed] [Google Scholar]
- 19.Nixon RA. 2005. Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiol. Aging 26:373–382 [DOI] [PubMed] [Google Scholar]
- 20.Xilouri M, Stefanis L. 2010. Autophagy in the central nervous system: implications for neurodegenerative disorders. CNS Neurol. Disord. Drug Targets 9:701–719 [DOI] [PubMed] [Google Scholar]
- 21.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889 [DOI] [PubMed] [Google Scholar]
- 22.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884 [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. 2007. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U. S. A. 104:14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez D, Torres CA, Setlik W, Cebrian C, Mosharov EV, Tang G, Cheng HC, Kholodilov N, Yarygina O, Burke RE, Gershon M, Sulzer D. 2012. Regulation of presynaptic neurotransmission by macroautophagy. Neuron 74:277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boland B, Nixon RA. 2006. Neuronal macroautophagy: from development to degeneration. Mol. Aspects Med. 27:503–519 [DOI] [PubMed] [Google Scholar]
- 26.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. 2007. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17:1561–1567 [DOI] [PubMed] [Google Scholar]
- 27.Marino G, Madeo F, Kroemer G. 2011. Autophagy for tissue homeostasis and neuroprotection. Curr. Opin. Cell Biol. 23:198–206 [DOI] [PubMed] [Google Scholar]
- 28.Yue Z, Friedman L, Komatsu M, Tanaka K. 2009. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim. Biophys. Acta 1793:1496–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu CT. 2006. Autophagic stress in neuronal injury and disease. J. Neuropathol. Exp. Neurol. 65:423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, Stefanis L, Tolkovsky A. 2005. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 1:11–22 [DOI] [PubMed] [Google Scholar]
- 31.Nixon RA, Cataldo AM. 2006. Lysosomal system pathways: genes to neurodegeneration in Alzheimer's disease. J. Alzheimers Dis. 9:277–289 [DOI] [PubMed] [Google Scholar]
- 32.Song JW, Misgeld T, Kang H, Knecht S, Lu J, Cao Y, Cotman SL, Bishop DL, Lichtman JW. 2008. Lysosomal activity associated with developmental axon pruning. J. Neurosci. 28:8993–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. 2008. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 28:6926–6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maday S, Wallace KE, Holzbaur EL. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dotti CG, Sullivan CA, Banker GA. 1988. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8:1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goslin K, Banker G. 1989. Experimental observations on the development of polarity by hippocampal neurons in culture. J. Cell Biol. 108:1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadasivan S, Zhang Z, Larner SF, Liu MC, Zheng W, Kobeissy FH, Hayes RL, Wang KK. 2010. Acute NMDA toxicity in cultured rat cerebellar granule neurons is accompanied by autophagy induction and late onset autophagic cell death phenotype. BMC Neurosci. 11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng Y, Yang X, Wang J, Fan J, Kong Q, Yu X. 2012. Aristolochic acid I induced autophagy extenuates cell apoptosis via ERK 1/2 pathway in renal tubular epithelial cells. PLoS One 7:e30312. 10.1371/journal.pone.0030312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JA, Gao FB. 2009. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J. Neurosci. 29:8506–8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036 [DOI] [PubMed] [Google Scholar]
- 42.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Banhegyi G, Bartholomew CR, Bassham DC, Bast RC, Jr, Batoko H, Bay BH, Beau I, Bechet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8:445–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang C. 2010. Negative regulation of autophagy. Cell Death Differ. 17:1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H, Huang S. 2010. The complexes of mammalian target of rapamycin. Curr. Protein Pept. Sci. 11:409–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polleux F, Snider W. 2010. Initiating and growing an axon. Cold Spring Harb. Perspect. Biol. 2:a001925. 10.1101/cshperspect.a001925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall A, Lalli G. 2010. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2:a001818. 10.1101/cshperspect.a001818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryan B, Cai Y, Wrighton K, Wu G, Feng XH, Liu M. 2005. Ubiquitination of RhoA by Smurf1 promotes neurite outgrowth. FEBS Lett. 579:1015–1019 [DOI] [PubMed] [Google Scholar]
- 51.Amano M, Chihara K, Nakamura N, Fukata Y, Yano T, Shibata M, Ikebe M, Kaibuchi K. 1998. Myosin II activation promotes neurite retraction during the action of Rho and Rho-kinase. Genes Cells 3:177–188 [DOI] [PubMed] [Google Scholar]
- 52.Hahmann C, Schroeter T. 2010. Rho-kinase inhibitors as therapeutics: from pan inhibition to isoform selectivity. Cell Mol. Life Sci. 67:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen DW, Pouliot LM, Gillet JP, Ma W, Johnson AC, Hall MD, Gottesman MM. 2012. The transcription factor GCF2 is an upstream repressor of the small GTPAse RhoA, regulating membrane protein trafficking, sensitivity to doxorubicin, and resistance to cisplatin. Mol. Pharm. 9:1822–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xing L, Yao X, Williams KR, Bassell GJ. 2012. Negative regulation of RhoA translation and signaling by hnRNP-Q1 affects cellular morphogenesis. Mol. Biol. Cell 23:1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubinsztein DC. 2012. Autophagy—alias self-eating—appetite and ageing. EMBO Rep. 13:173–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z. 2012. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J. Neurosci. 32:7585–7593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee S, Sato Y, Nixon RA. 2011. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J. Neurosci. 31:7817–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang H, Guo W, Liang X, Rao Y. 2005. Both the establishment and the maintenance of neuronal polarity require active mechanisms: critical roles of GSK-3beta and its upstream regulators. Cell 120:123–135 [DOI] [PubMed] [Google Scholar]
- 60.Shi SH, Jan LY, Jan YN. 2003. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112:63–75 [DOI] [PubMed] [Google Scholar]
- 61.Kwiatkowski DJ, Manning BD. 2005. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum. Mol. Genet. 14(Spec. no. 2):R251–R258 [DOI] [PubMed] [Google Scholar]
- 62.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. U. S. A. 99:13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13:1259–1268 [DOI] [PubMed] [Google Scholar]
- 64.Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ. 2003. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Invest. 112:1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi YJ, Di Nardo A, Kramvis I, Meikle L, Kwiatkowski DJ, Sahin M, He X. 2008. Tuberous sclerosis complex proteins control axon formation. Genes Dev. 22:2485–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hollenbeck PJ. 1993. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell. Biol. 121:305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura H, Ohkuma S. 1991. Induction of neurite outgrowth of PC12 cells by an inhibitor of vacuolar H(+)-ATPase, bafilomycin A1. FEBS Lett. 294:51–55 [DOI] [PubMed] [Google Scholar]
- 68.Coupe B, Ishii Y, Dietrich MO, Komatsu M, Horvath TL, Bouret SG. 2012. Loss of autophagy in pro-opiomelanocortin neurons perturbs axon growth and causes metabolic dysregulation. Cell Metab. 15:247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. 1999. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24:833–846 [DOI] [PubMed] [Google Scholar]
- 70.Mochizuki H, Toda H, Ando M, Kurusu M, Tomoda T, Furukubo-Tokunaga K. 2011. Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PLoS One 6:e19632. 10.1371/journal.pone.0019632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toda H, Mochizuki H, Flores R, III, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, Tomoda T. 2008. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 22:3292–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Auer M, Hausott B, Klimaschewski L. 2011. Rho GTPases as regulators of morphological neuroplasticity. Ann. Anat. 193:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Govek EE, Newey SE, Van Aelst L. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49 [DOI] [PubMed] [Google Scholar]
- 74.Luo L. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1:173–180 [DOI] [PubMed] [Google Scholar]
- 75.Bannai H, Fukatsu K, Mizutani A, Natsume T, Iemura S, Ikegami T, Inoue T, Mikoshiba K. 2004. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 279:53427–53434 [DOI] [PubMed] [Google Scholar]
- 76.Harris CE, Boden RA, Astell CR. 1999. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol. 73:72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, Sendtner M. 2002. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum. Mol. Genet. 11:93–105 [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez-Muela N, Boya P. 2012. Axonal damage, autophagy and neuronal survival. Autophagy 8:286–288 [DOI] [PubMed] [Google Scholar]
- 79.Yue Z, Wang QJ, Komatsu M. 2008. Neuronal autophagy: going the distance to the axon. Autophagy 4:94–96 [DOI] [PMC free article] [PubMed] [Google Scholar]