Abstract
The eukaryotic reverse transcriptase, telomerase, adds tandem telomeric repeats to chromosome ends to promote genome stability. The fully assembled telomerase holoenzyme contains a ribonucleoprotein (RNP) catalytic core and additional proteins that modulate the ability of the RNP catalytic core to elongate telomeres. Electron microscopy (EM) structures of Tetrahymena telomerase holoenzyme revealed a central location of the relatively uncharacterized p50 subunit. Here we have investigated the biochemical and structural basis for p50 function. We have shown that the p50-bound RNP catalytic core has a relatively low rate of tandem repeat synthesis but high processivity of repeat addition, indicative of high stability of enzyme-product interaction. The rate of tandem repeat synthesis is enhanced by p50-dependent recruitment of the holoenzyme single-stranded DNA binding subunit, Teb1. An N-terminal p50 domain is sufficient to stimulate tandem repeat synthesis and bridge the RNP catalytic core, Teb1, and the p75 subunit of the holoenzyme subcomplex p75/p19/p45. In cells, the N-terminal p50 domain assembles a complete holoenzyme that is functional for telomere maintenance, albeit at shortened telomere lengths. Also, in EM structures of holoenzymes, only the N-terminal domain of p50 is visible. Our findings provide new insights about subunit and domain interactions and functions within the Tetrahymena telomerase holoenzyme.
INTRODUCTION
In most eukaryotic nuclei, chromosomes are capped with an array of tandem simple-sequence DNA repeats. These telomeric repeats, along with bound proteins, create an end-protective structure that distinguishes authentic chromosome ends from unintended double-stranded DNA breaks (1–3). Telomere integrity is compromised by the attrition of repeats as an inevitable consequence of genome replication and the many steps of chromosome end processing (4–6). Replication-linked telomere shortening is progressive, leading eventually to one or more telomeres of a length insufficient for end-protective function. Critically short telomeres induce genome instability, proliferative senescence, and tissue renewal failures in human disease (7, 8).
The telomerase ribonucleoprotein (RNP) can compensate for telomere erosion, using a region within its integral RNA component to template the synthesis of new repeats. Telomerase catalytic activity can be reconstituted from the biologically cofolded subunits of a telomerase RNP catalytic core, including telomerase reverse transcriptase (TERT), the template-containing telomerase RNA (TER), and proteins that fold and stabilize TER to promote TERT-TER interaction (9, 10). Beyond the RNP catalytic core, additional telomerase holoenzyme proteins are required for repeat synthesis at telomeres in vivo (11, 12). Some holoenzyme proteins assemble with telomerase RNP only in the DNA synthesis phase of the cell cycle (13, 14). Differing models have been offered to explain telomerase holoenzyme protein functions at a molecular level, and accordingly, many open questions remain to be addressed.
Telomere-rich ciliated protozoa are experimentally favorable organisms for insights about telomerase (15). As a ciliate model organism, Tetrahymena thermophila has numerous advantages, including large-scale culture growth, a sequenced genome, and methods for genetic manipulation (16). These features enabled endogenously assembled telomerase holoenzyme purification to homogeneity and comprehensive subunit identification at a molecular level (17–19). The Tetrahymena telomerase holoenzyme purified from cell extract contains 8 subunits, each of which is telomerase specific and essential for telomere maintenance: TERT, TER, the telomerase assembly factor p65, and 5 additional proteins that give p65-TER-TERT (the RNP catalytic core) an ability to elongate telomeres in vivo (17–20). These additional subunits are the holoenzyme single-stranded telomeric-repeat DNA binding protein Teb1; one copy each of p75, p45, and p19 assembled as a subcomplex designated 7-1-4; and p50 (19, 21). Of these subunits, only Teb1 has domains readily detectable by sequence homology and fold prediction. The 4 oligonucleotide/oligosaccharide-binding (OB)-fold-domain architecture of Teb1 is paralogous to that of the large subunit of the general single-stranded DNA binding factor replication protein A, with 3 of the predicted Teb1 OB-fold domains confirmed by high-resolution structures (19, 22).
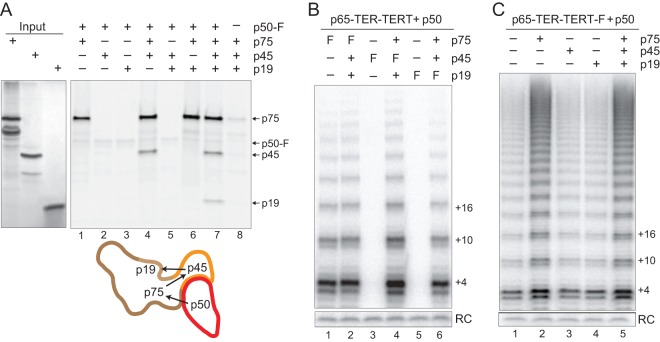
Functional associations of recombinant Tetrahymena telomerase RNP catalytic core, Teb1, and 7-1-4 require p50 (21). The p50 subunit alone stimulates RNP catalytic core activity, and p50 is required for additional activity stimulation by Teb1 or 7-1-4 (Fig. 1A, left). The physical arrangement of these Tetrahymena telomerase holoenzyme subunits was defined by difference imaging, comparing electron microscopy (EM) two-dimensional (2D) image class averages and/or 3D reconstructions of telomerase complexes purified from cell extracts and labeled with Fab antibodies (21). Individual subunits or subunit combinations were fused to tandem protein A domains (ZZ tag) and/or 3 consecutive FLAG peptides (F tag). Taking advantage of the F tag still present after 2-step affinity purification, binding of anti-FLAG Fab antibody fragment was used to localize individual subunits within the holoenzyme. C-terminal tagging of p65, TERT, p75, p19, or Teb1 preserved telomerase function for each of these subunits in vivo. C-terminally tagged p50 also retained biological function but purified a very low holoenzyme yield. Therefore, N-terminally tagged F-p50 telomerase was used to map the location of p50. N-terminal tagging of p50 disrupted holoenzyme association of the Teb1 C-terminal OB-fold domain (Teb1C), which tethers the central Teb1 high-affinity DNA binding domains (Teb1A and Teb1B) to the holoenzyme complex (21, 23). By using holoenzyme subcomplexes in addition to differential subunit tagging, a model for holoenzyme architecture was developed that pinpoints p50 as the central hub of telomerase holoenzyme subcomplex coordination (Fig. 1A, right).
Fig 1.
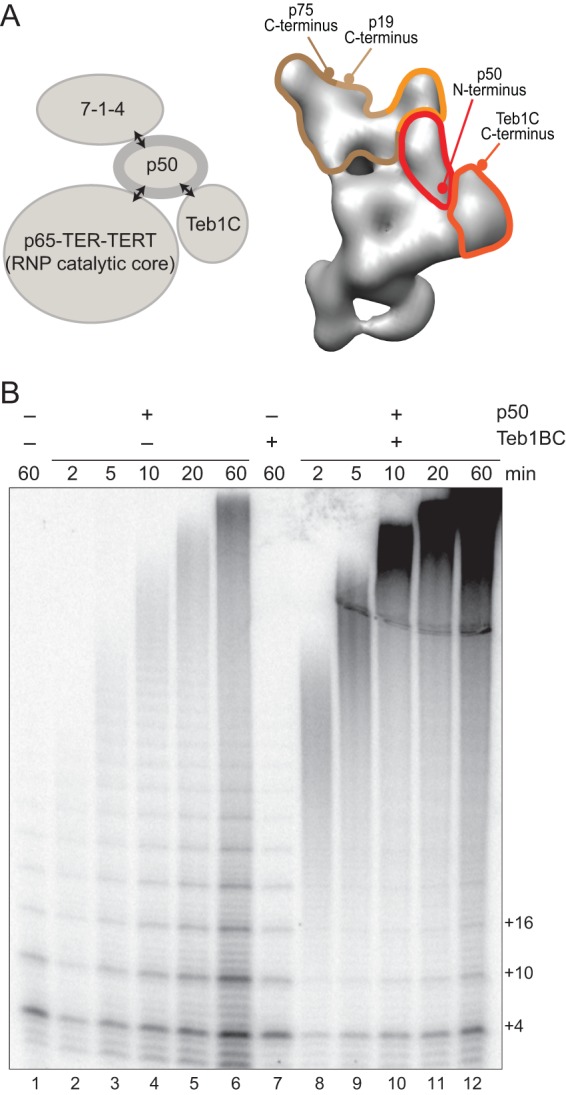
Stimulation of catalytic activity by Teb1 involves an increase in the rate of product synthesis. (A) At left, the schematic summarizes the functional dependence of holoenzyme subcomplexes. At right, a 3D reconstruction of Teb1-F holoenzyme is shown with outlined regions assigned to 7-1-4 (brown, beige, and orange edges for p75, p19, and p45, respectively), p50, and Teb1 (21). The protein termini of these subunits mapped by Fab labeling are indicated. (B) Activity assays were performed for the RNP catalytic core alone (lane 1) or the RNP catalytic core with or without p50 and/or Teb1BC.
Despite the critical role of p50 in Tetrahymena telomerase holoenzyme assembly and repeat synthesis processivity, little is known about this protein at a molecular level. The p50, Teb1, and p65 proteins are particularly sensitive to partial proteolysis in cell extract (19, 21). For Teb1 and p65, partial proteolysis releases N-terminal domains of unknown function from the C-terminal domains that mediate holoenzyme assembly and catalytic activation. Unlike Teb1 and p65, p50 does not have a known multidomain architecture or even any domains predicted by sequence analysis. Here we have investigated the biochemical, biological, and structural roles of N-terminal and C-terminal regions of p50. We identify an N-terminal ∼30-kDa domain of p50 that is necessary and sufficient for physical and functional association of the RNP catalytic core, Teb1, and 7-1-4 both in vitro and in vivo. Our results suggest that the p50 C-terminal region has a regulatory role. Overall, our findings bring new insight into the mechanism of processive repeat synthesis, the specificity of subunit interactions, and the overall architectural organization of Tetrahymena telomerase holoenzyme.
MATERIALS AND METHODS
Telomerase reconstitutions in vitro.
We used synthetic open reading frames for TERT, p75, p65, p50, p45, and p19 expression in rabbit reticulocyte lysate (RRL) and for Teb1BC expression in Escherichia coli. TER was gel purified following in vitro transcription by T7 RNA polymerase. Subunit expression, reconstitution, and affinity purification from RRL protein synthesis reactions was done according to previously optimized protocols (21, 23). Unless otherwise indicated, complexes bound to anti-FLAG antibody resin were washed into T2MG (20 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol [DTT]). More extensive washing was performed prior to SDS-PAGE analysis of physical interactions using T2MG with 0.1% Igepal CA-630. Peptide elutions were performed for subunit interaction assays and 2-step affinity purification using 150 to 200 ng/μl peptide.
Detection of protein, RNA, DNA, and telomerase activity.
Protein expression in RRL was monitored by [35S]methionine ([35S]Met) incorporation. TER detection was done by blot hybridization using an oligonucleotide 32P end labeled by T4 polynucleotide kinase. Genomic DNA was purified, digested, and hybridized for Southern blot analysis as described previously (24), using a sequence 3′ flanking the targeted region to detect differentially sized wild-type and recombinant chromosomes. Telomeric restriction fragments were detected following denaturing gel electrophoresis of HindIII-digested genomic DNA using a 5′-end-labeled oligonucleotide probe complementary to the subtelomeric region of the palindromic chromosome encoding ribosomal RNAs (20). The HindIII-cleaved telomeric restriction fragment has ∼350 bp of nontelomeric sequence as well as a variable number of telomeric repeats. Activity assays used a standard Tetrahymena telomerase reaction buffer containing 50 mM Tris-acetate, pH 8.0, 2 mM MgCl2, and 5 mM β-mercaptoethanol or 2 mM DTT or 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl). Product synthesis reactions additionally contained 3 μM [α-32P]dGTP, 200 μM dTTP, 100 to 200 nM primer (GT2G3)3, and 50 to 200 nM Teb1BC and were performed for 10 min at room temperature unless indicated otherwise. A 5′-labeled oligonucleotide DNA recovery control was added to telomerase products before precipitation.
Tetrahymena strain construction and enzyme purification from cell extracts.
Flanking genomic regions were used for targeting the endogenous TAP50 locus to express synthetic open reading frames of full-length or truncated p50, with the F and ZZ tag modules separated by a tobacco etch virus protease cleavage site (19). Selection was performed using the bsr2 cassette (24). Cell extract production and affinity purification were done as optimized previously (19, 21) using T2EG50 buffer (20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 10% glycerol, 50 mM NaCl, and 2 mM DTT or 1 mM TCEP-HCl). Prior to an activity assay, the enzyme was washed into T2MG with 50 mM NaCl and 0.1% Igepal CA-630. For EM samples, the 2-step antibody affinity purification (21) was followed by microscale gel filtration over a Sephadex 200 PC 3.2/30 column in 20 mM HEPES-NaOH, pH 8.0, 50 mM NaCl, 1 mM MgCl2, and 1 mM TCEP-HCl. Fab labeling was done as described previously (21).
Electron microscopy and image analysis.
Methods used were largely as described previously (21). Briefly, purified holoenzyme was subjected to negative staining using uranyl formate (0.8%) on glow-discharged grids coated with carbon film. Micrographs were recorded on a Tietz F415MP 16-megapixel charge-coupled-device (CCD) camera at magnification of ×68,027 in an FEI Tecnai F20 electron microscope operated at 200 kV. This study used 19,325 particles of p50N30-F telomerase and 15,427 particles of Fab-labeled p50N30-F telomerase for 2D image classification and 5,593 particles of p50N30-F telomerase for 3D reconstruction by the random-conical-tilt (RCT) method (25).
RESULTS
Holoenzyme protein interactions increase the rate of tandem repeat synthesis.
The p50 subunit occupies a central position among the Tetrahymena telomerase holoenzyme subunits mapped by EM (21). Consistent with its location, p50 is required for stimulation of RNP catalytic core activity by Teb1 or 7-1-4 (21). Compared to the profile of product synthesis by the reconstituted RNP catalytic core alone, product lengths increased with the addition of p50 and increased even more with the addition of both p50 and Teb1 (21). We investigated the mechanism for these increases in product length using a time course of direct telomeric primer elongation with radiolabeled dGTP and dTTP (Fig. 1B). RNP catalytic core containing TERT C-terminally tagged with a triple FLAG epitope (TERT-F) was assembled in RRL and then combined with separately RRL-expressed p50. Importantly, C-terminal tagging of Tetrahymena TERT does not impede telomerase holoenzyme function in vivo (17). Telomerase complexes were removed from other RRL reaction components by affinity purification with anti-FLAG antibody resin. Direct primer extension assays of purified complexes were then performed with or without the addition of bacterially expressed Teb1BC, which is the C-terminal half of Teb1 sufficient to confer holoenzyme-like high repeat addition processivity (RAP) (23).
Given sufficient reaction time, the Tetrahymena telomerase RNP catalytic core assembled with p50 gave substantial high-RAP activity (Fig. 1B, lanes 2 to 6), much greater than that of the RNP catalytic core alone (lane 1). As reported previously (21), supplementing the p50-bound RNP catalytic core with Teb1 dramatically increased the overall level of catalytic activity and high-RAP product synthesis (Fig. 1B, lanes 8 to 12). Notably, unlike the RNP catalytic core alone (26), and with or without Teb1, the p50-bound RNP catalytic core continued to elongate high-RAP products for more than 60 min without reaching an equilibrium distribution of product lengths. We conclude that p50 association with the RNP catalytic core is sufficient to confer a high stability of enzyme-product interaction even without Teb1.
Curiously, addition of Teb1 increased the rate of tandem repeat synthesis: products were longer at each time point in the reaction with Teb1 than in the reaction without Teb1 (Fig. 1B; for example, compare lanes 2 and 3 to lanes 8 and 9). In addition to increasing the rate of high-RAP product synthesis, Teb1 also enhanced productive association of p50 with the RNP catalytic core: the RNP catalytic core low-RAP product synthesis was almost completely converted to high-RAP product synthesis in the presence but not absence of Teb1 (Fig. 1B; compare products +4, +10, and +16 in lanes 2 to 6 versus lanes 8 to 12). This suggests that Teb1 stabilizes a p50 conformation favorable for its assembly with or activation of the RNP catalytic core.
The p75 subunit of 7-1-4 interacts with p50.
In addition to bridging the RNP catalytic core and Teb1, p50 also bridges the RNP catalytic core to 7-1-4. Endogenously expressed p75, p45, and p19 form a remarkably tight subcomplex that is resistant to dissociation by micrococcal nuclease treatment of p45-F holoenzyme, a treatment that degrades TER and releases p65, TERT, Teb1, and a large fraction of p50 from intact 7-1-4 (19, 21, 23). The 7-1-4 subcomplex is positionally dynamic relative to the rest of the holoenzyme, hinging as a unit around p50, which suggests a role for this subcomplex in holoenzyme coordination with unknown additional factors involved in telomere synthesis (21). Addition of p75, p45, and p19 coexpressed in RRL stimulated product synthesis by the p50-bound RNP catalytic core with or without Teb1 (21). However, unlike Teb1, 7-1-4 increased the overall level of RNP catalytic activity without increasing product length.
To investigate whether the stimulatory influence of 7-1-4 correlates with direct physical interaction of 7-1-4 subunits and p50, we combined C-terminally triple-FLAG-tagged but not radiolabeled p50-F with [35S]Met-labeled but untagged p75, p45, and/or p19, each synthesized independently in RRL reactions. This radiolabeling strategy resolved the potential overlap in migration of p45 and p50 expression products. Protein-protein interactions were assessed by the amount of radiolabeled protein that copurified with p50-F using anti-FLAG antibody resin. As a negative control, we verified that untagged p75, p45, and p19 did not bind anti-FLAG antibody resin in the absence of p50-F (Fig. 2A, lane 8). As expected, the combination of all 3 of the 7-1-4 proteins allowed all 3 to associate with p50 (Fig. 2A, lane 7). For the 7-1-4 subunits individually, only p75 could copurify with p50 (Fig. 2A, lanes 1 to 3). The interaction of p45 with p50 was dependent on the presence of p75, and interaction of p19 with p50 was dependent on the presence of both p75 and p45 (Fig. 2A, lanes 4 to 7). The simplest explanation for these findings would be that p75 binds to p50, and then p45 can bind to p75, and finally, p19 can bind to p45 (Fig. 2A, schematic). However, it remains possible that each 7-1-4 subunit physically contacts each other subunit and/or p50 in a manner dependent on conformational changes induced by subunit interactions.
Fig 2.
The p75 subunit of 7-1-4 interacts directly with p50. (A) Unlabeled p50-F was combined with RRL synthesis reactions of [35S]Met-labeled p75, p45, and/or p19, and the mixture was incubated with anti-FLAG antibody resin. Bound proteins were eluted and analyzed by SDS-PAGE. Trace radiolabeling of p50-F derived from protein synthesis after RRL mixing. Below the gel image, using the subunit outlining from Fig. 1A, arrows indicate assembly dependence. (B and C) Activity assays are shown for telomerase complexes recovered by anti-FLAG antibody purification of the catalytic core, p50, and the indicated 7-1-4 subunit(s). In panel B, “F” indicates the F-tagged subunit. “RC” is the recovery control added to telomerase products before precipitation. Quantification and normalization for relative methionine content give a molar ratio for p75/p45/p19 of 1.0:0.6:0.2, consistent with the stepwise rather than exclusively cooperative assembly of 7-1-4 by p75 interaction with p45 and subsequent recruitment of p19.
To confirm that p75 associates with p50 in the context of a catalytically active enzyme, we used p75, p45, or p19 tagged at the protein C terminus, which preserves subunit function in vivo (19). Each subunit was expressed as an F-tagged protein and combined individually or as 7-1-4 with a p50-bound RNP catalytic core. Telomerase complexes assembled on an F-tagged 7-1-4 subunit were then recovered by affinity purification using anti-FLAG antibody resin. In the presence or absence of untagged p45 and p19, p75-F copurified telomerase activity (Fig. 2B, lanes 1 and 2). In contrast, p45-F and p19-F copurified telomerase activity only in the presence of other untagged 7-1-4 subunits (Fig. 2B, lanes 4 and 6). We also tested whether p75 alone could recapitulate the 7-1-4 stimulation of overall telomeric-repeat product synthesis (21). Addition of p75 to the p50-bound RNP catalytic core did induce the same increase in telomerase activity as addition of 7-1-4, whereas addition of p45 or p19 alone did not (Fig. 2C).
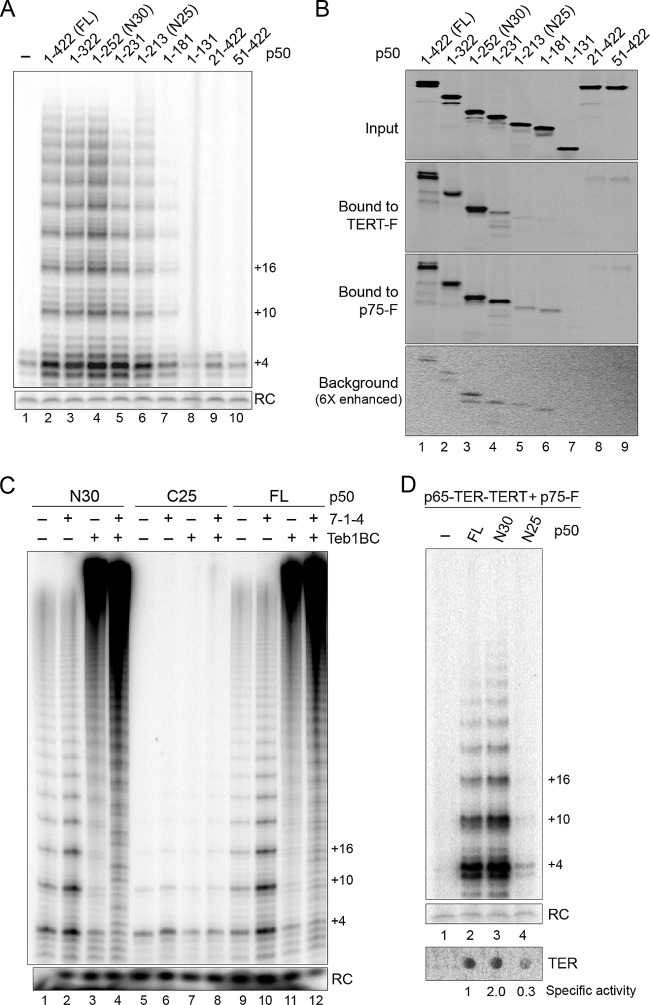
An N-terminal domain mediates p50 biochemical roles in telomerase activation.
We next investigated whether the activity stimulation and subunit bridging functions of p50 could be mapped to specific regions of the protein. The RNP catalytic core assembled on TERT-F was combined with full-length or various p50 N- or C-terminal truncations. Full-length p50 stimulated the overall level and RAP of product synthesis (Fig. 3A, compare lanes 1 and 2). The same result was observed for C-terminally truncated p50 polypeptides lacking up to half of the protein primary sequence (Fig. 3A, lanes 3 to 6). No activity stimulation was detected in assays of N-terminally truncated p50 polypeptides, including modest truncations of only 20 to 50 amino acids (Fig. 3A, lanes 9 and 10). The presence of a structurally autonomous p50 N-terminal domain is supported by partial proteolysis of endogenously expressed p50 during purifications from Tetrahymena cell extract (see, e.g., Fig. S1 in the supplemental material), which generates an ∼30-kDa fragment of p50 that would roughly correspond to amino acids 1 to 252 (here designated p50N30). The C-terminal 170 amino acids of p50 that are absent in p50N30 are not required for processive repeat synthesis (Fig. 3A, lane 4), and even the slightly shorter p50N25 (amino acids 1 to 213) could stimulate catalytic activity, although to a lesser extent (Fig. 3A, lane 6).
Fig 3.
An N-terminal domain of p50 reconstitutes holoenzyme interactions and catalytic activity in vitro. (A) Full-length (FL) and truncated p50 polypeptides were combined with RNP catalytic core containing TERT-F, and complexes purified by anti-FLAG antibody resin were assayed for primer extension activity. (B) The same untagged p50 polypeptides were assayed for physical interaction with RNP catalytic core containing TERT-F or for interaction with p75-F in the presence of untagged RNP catalytic core. The bottom panel is shown with radiolabeled protein signal intensity increased about 6 times relative to that of the other panels to allow visualization of the weak background of nonspecific binding. (C) Activity assays were performed for 20 min using complexes assembled on RNP catalytic core containing TERT-F. (D) Activity assays were performed for complexes purified using p75-F that harbored the indicated untagged p50 polypeptide, with spot blot hybridization for TER shown in the lower panel. Relative specific activity was calculated as the quantified ratio of product intensity to TER, normalizing full-length p50 to 1. Triplicate independent replicates of protein expression, RNP assembly, purification, and activity assay were performed on separate days, with a specific activity increase for p50N30 relative to that of full-length p50 of 2.1 ± 0.38 standard error of the mean.
To complement the definition of p50 domain requirements for biochemical activity described above, we determined which regions of p50 are required for physical interaction with the RNP catalytic core and p75. We [35S]Met labeled the variously truncated p50 polypeptides (Fig. 3B, Input) and assayed for copurification with unlabeled TERT-F RNP catalytic core or unlabeled, untagged TERT RNP catalytic core and p75-F. The TERT-F RNP catalytic core and p75-F copurified full-length p50 and p50 C-terminal truncations up to p50N30 with similar recovery (Fig. 3B, lanes 1 to 3). Any additional truncation reduced p50 association with the RNP catalytic core (Fig. 3B, lane 4), and truncation to p50N25 reduced p50 association with p75 (lane 5). By physical association assay, p50N25 interaction with the RNP catalytic core was reduced more than expected by its copurification of RNP catalytic activity (Fig. 3A, lane 6, and B, lane 5). This difference is likely due to the higher stringency of washes used in the physical association assay (see Materials and Methods), which was necessary to remove nonspecific background protein association with the purification resin.
Additional tests of p50N30 biochemical activities did not detect any loss of function in comparison to results for full-length p50. For example, we compared the abilities of full-length p50 and p50N30 to confer activity stimulation by 7-1-4 and Teb1. For all combinations of telomerase holoenzyme subcomplexes, the profiles of product synthesis were indistinguishable for enzymes with p50N30 or full-length p50 (Fig. 3C, compare lanes 1 to 4 and 9 to 12). In comparison, the C-terminal half of p50 (C25, amino acids 214 to 422) did not stimulate RNP catalytic core activity with or without 7-1-4 and/or Teb1 (Fig. 3C, lanes 5 to 8). Also, no difference from full-length p50 was detected in the p50N30 specificity of activity stimulation by individual 7-1-4 subunits (see Fig. S2 in the supplemental material). In these and additional biochemical assays, no loss of function was detected as a consequence of p50 C-terminal truncation to p50N30.
Curiously, in the course of experiments above, we noticed that the overall level of catalytic activity was often higher for enzymes reconstituted using the same volume of RRL expression reaction for p50N30 versus full-length p50. This difference could arise from an expression or folding advantage of the smaller p50N30 protein, or it could reflect the removal of an inhibitory C-terminal domain of p50. To address this distinction, we devised a strategy to compare the specific activity of telomerase complexes containing full-length p50 or p50N30. Because the two polypeptides have different structural environments at their C-terminal ends and because N-terminal tagging of p50 is disruptive for Teb1 assembly into holoenzyme (21), it was necessary to use untagged versions of the p50 proteins. This was accomplished by capturing p50-bound telomerase complexes using p75-F and anti-FLAG antibody resin, which also eliminated any activity contribution from the RNP catalytic core without p50. We normalized the amount of purified RNP using parallel aliquots of the p75-F complexes to detect catalytic activity (Fig. 3D, top panel) and TER (lower panel). RNP with p50N30 reproducibly had about twice the specific activity of RNP with full-length p50 (Fig. 3D, lanes 2 and 3). Only a low level of activity was detected for RNP assembled by p50N25 (Fig. 3D, lane 4), consistent with reduced stability of p50N25 interaction with p75 and the RNP catalytic core.
The N-terminal domain of p50 supports physiological telomerase holoenzyme assembly and function.
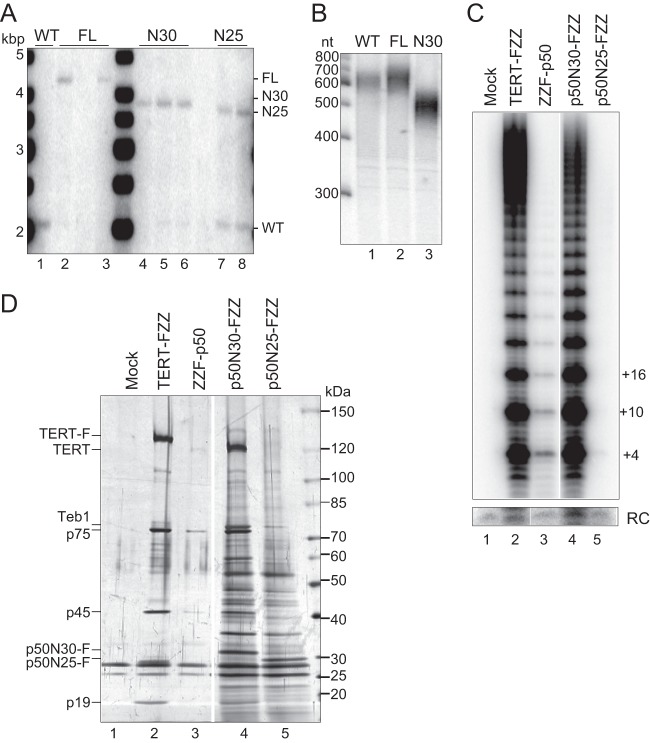
Because the p50 C-terminal ∼20 kDa was unnecessary for recombinant telomerase holoenzyme assembly or catalytic activity in vitro, we investigated whether this region was required for telomere maintenance in vivo. We targeted the TAP50 locus for replacement with C-terminally tagged full-length p50, p50N30, or p50N25. In each targeting construct, we used the synthetic open reading frame of the p50 RRL expression vector and the FZZ tag developed for holoenzyme affinity purification from Tetrahymena cell extracts (see Materials and Methods). Integration of the targeting vector initially replaced only a few of the 45 expressed macronuclear copies of TAP50 with the modified open reading frame and selectable marker cassette. A standard protocol of additional selection, clonal cell line isolation, and release from selection was used to discriminate whether the recombinant chromosome can genetically substitute for the wild-type chromosome (27).
We performed genomic DNA Southern blot hybridization to distinguish wild-type and recombinant chromosomes. As expected from previous results, which introduced the FZZ tag in fusion with the endogenous rather than the synthetic p50 open reading frame (19), the newly generated p50-FZZ expression construct could completely replace the wild-type TAP50 locus (Fig. 4A, lanes 1 to 3; the p50-FZZ cell line analyzed in lane 2 was used for subsequent studies). Wild-type chromosomes were also replaced by chromosomes with the p50N30-FZZ expression construct (Fig. 4A, lanes 4 to 6; the p50N30-FZZ cell line in lane 4 was used for subsequent studies). The silent micronuclear gene locus not targeted for replacement gives rise to a low wild-type TAP50 locus signal that does not recover to higher copy number after release from selection. In contrast, all cell lines expressing p50N25-FZZ had a high copy number of wild-type chromosomes (Fig. 4A, lanes 7 and 8; the p50N25-FZZ cell line in lane 7 was used for subsequent studies). This abundance of wild-type chromosomes indicates that p50N25 is not able to provide genetically essential p50 function. Southern blot results were confirmed using reverse transcription-PCR (RT-PCR) to verify the absence (in p50-FZZ and p50N30-FZZ cell lines) or presence (in p50N25-FZZ cell lines) of wild-type TAP50 mRNA (data not shown).
Fig 4.
An N-terminal domain of p50 reconstitutes holoenzyme assembly in vivo. Cells or cell extracts had no tagged protein (WT) or a C-terminally tagged p50 polypeptide with recombinant open reading frame (full-length, N30, or N25, as indicated). (A) Genomic DNA Southern blots were performed to assess wild-type versus recombinant chromosome content. (B) Telomeric restriction fragment length was assayed after denaturing gel electrophoresis by hybridization with an oligonucleotide probe complementary to the subtelomeric region of the palindromic chromosome encoding rRNA. (C and D) Two-step affinity purifications from equivalent amounts of cell extract were assayed for telomerase catalytic activity (C) or protein composition (D). The right and left sides of each panel were cropped and aligned from the same gel with the same intensity settings.
Although p50N30 retained the genetically essential functions of full-length p50, telomeres were shorter in cells expressing p50N30-FZZ than in those expressing untagged p50 (Fig. 4B, compare lanes 1 and 3). In contrast, telomeres were longer than those of the wild type in both the p50-FZZ cell line created here by integration of a complete p50-FZZ synthetic open reading frame (Fig. 4B, lane 2) and the p50-FZZ strain created previously (19) by integration of just the FZZ tag (see Fig. S3 in the supplemental material). Because p50 knockdown and overexpression both shorten telomere length (19), the increase in telomere length resulting from p50 C-terminal tagging is unlikely to reflect a simple change in protein abundance. Instead, we suggest that it reflects tag interference with an inhibitory activity of the p50 C-terminal domain (see Discussion).
Single-step affinity purification of p50N30-FZZ or p50N25-FZZ from Tetrahymena cell extracts showed that both proteins could coenrich holoenzyme catalytic activity (see Fig. S4 in the supplemental material). More stringent 2-step affinity purification coenriched activity in association with p50N30-F but not p50N25-F (Fig. 4C), consistent with weaker p50N25 interaction with the RNP catalytic core (Fig. 3B). Only p50N30-FZZ purification recovered associated proteins with the expected SDS-PAGE profile of telomerase holoenzyme subunits (Fig. 4D). Consistent with complete replacement of endogenous p50 by p50N30-FZZ, the yield of p50N30-F telomerase was comparable to the yield of TERT-F telomerase and was more than the yield of N-terminally tagged F-p50 telomerase (Fig. 4D), which has reduced high-RAP catalytic activity (Fig. 4C) and does not provide genetically essential p50 function (21). Of note, under standard purification conditions, p50N30-FZZ and p50N25-FZZ coenriched a heterogenous background of proteins not present in purifications of TERT-FZZ (Fig. 4D; see also Fig. 5C).
Fig 5.
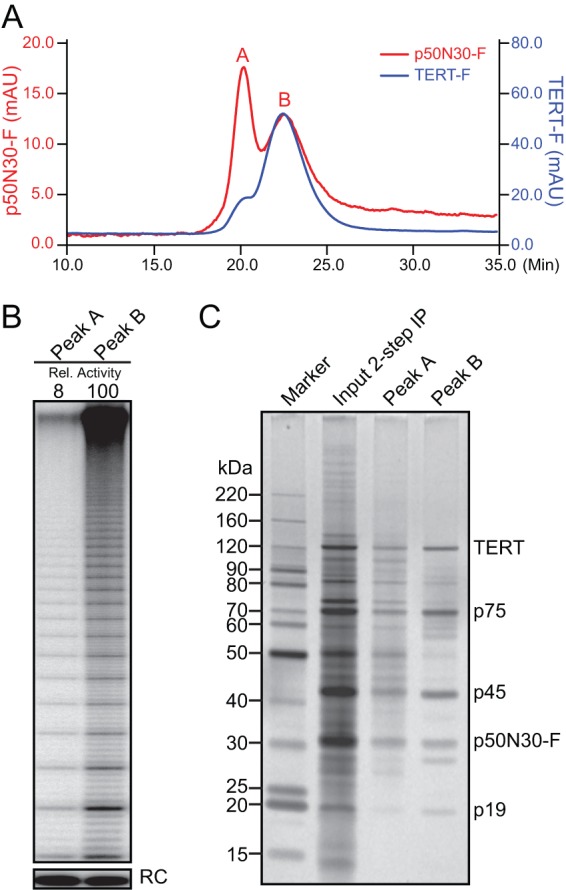
Purification of p50N30-F telomerase for EM. (A) Microscale size exclusion purification of p50N30-F (red) and TERT-F (blue) after 2-step affinity purification. (B and C) Activity assay (B) or SDS-PAGE and silver staining (C) of p50N30-F telomerase purification by the size exclusion chromatography shown in panel A. In panel B, relative (Rel.) activity is total product synthesis quantified for the assays shown, which used peak fractions normalized by absorbance at 280 nm. Note that activity detected in peak A could derive from overlap with peak B.
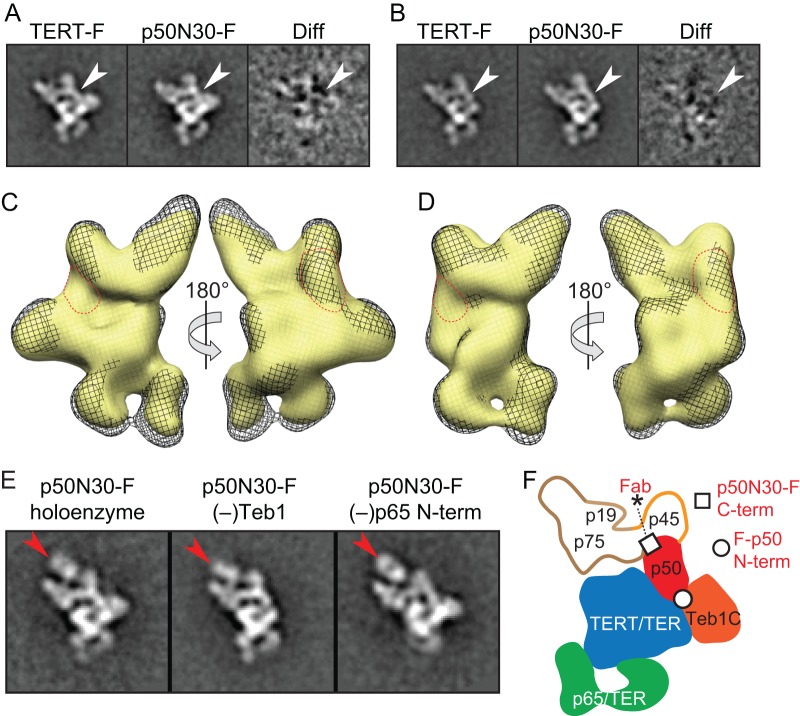
Deletion of the p50 C-terminal domain does not change the holoenzyme structure determined by EM.
We exploited the genetically stable substitution of untagged p50 by p50N30-FZZ (Fig. 4A) and the high yield of holoenzyme obtained from p50N30-FZZ 2-step affinity purification (Fig. 4D) to investigate whether the holoenzyme structure determined by EM previously (21) encompasses the p50 C-terminal domain as well as the N-terminal domain located by Fab binding to F-p50. Gel filtration was required after p50N30-FZZ 2-step affinity purification (Fig. 5A) to remove an interfering background of large particles that, based on EM and image analysis, were heterogeneous in size and did not contain consistent structural features (see Fig. S5 in the supplemental material). The large particles that eluted first had low if any telomerase activity compared to the peak of fractionated telomerase holoenzyme (Fig. 5A and B). Only a small fraction of aggregate-sized particles was detected for telomerase holoenzyme purified using TERT-FZZ (Fig. 5A). SDS-PAGE analysis showed a background of polypeptides in the aggregate-size p50N30-F peak A complexes in addition to the expected telomerase holoenzyme subunits, which were selectively enriched in the peak B samples used for additional studies (Fig. 5C). Truncation of full-length p50 by partial proteolysis during purification yields an SDS-PAGE profile of holoenzyme subunits with apparently substoichiometric p50 (19). In comparison, minimal extract proteolysis of genetically truncated p50N30 allowed its visualization by silver staining after SDS-PAGE at stoichiometry more comparable to that of other holoenzyme subunits (Fig. 5C, Peak B), consistent with the central location of p50 in the holoenzyme structure determined by EM (21).
Difference images of EM class averages generated from p50N30-F telomerase versus TERT-F telomerase (21) showed no additional density in the complete holoenzyme (Fig. 6A) or holoenzyme lacking Teb1 (Fig. 6B). Comparison of 3D EM reconstructions of p50N30-F telomerase to TERT-F telomerase from our previous study (21) showed no difference significant for the 26-Å resolution of the structures (Fig. 6C and D; see also Fig. S6 in the supplemental material). Comparison of 2D class averages and 3D RCT reconstructions of the complete holoenzyme (Fig. 6A and C) to the small subset (∼5%) of p50N30-F particles lacking Teb1 (Fig. 6B and D) allowed the boundary between p50N30 and Teb1 to be more readily visualized. Overall, the structures suggest that telomerase enzymes containing endogenous p50 are purified without retaining the p50 C-terminal region due to proteolysis and/or that the C-terminal region of p50 is positionally flexible relative to its N-terminal domain.
Fig 6.
The p50 C-terminal region occupies an unknown location relative to other holoenzyme proteins. (A and B) Class averages of TERT-F, p50N30-F, and difference maps for holoenzyme (A) or particles lacking Teb1 (B). White arrows indicate the p50 location. (C and D) Comparison of RCT 3D reconstructions of TERT-F (black mesh) and p50N30-F (yellow surface) for holoenzyme (C) or particles lacking Teb1 (D), with p50 outlined (red dashes). The structure of p50N30-F telomerase with or without Teb1 was reconstructed using 1,162 or 945 particles, respectively. (E) Class averages of Fab-labeled p50N30-F telomerase. Red arrows indicate Fab density. (F) Model for subunit placement, highlighting the previously defined location of the p50 N terminus (circle) (21) and the proposed location of the p50N30 C terminus (square) based on the location of the single Fab-bound paratope (asterisk). Box side length of class averages is 350 Å in panels A, B, and E.
We used anti-FLAG Fab labeling of p50N30-F telomerase complexes to define the physical location of the p50N30 C terminus (residue 252 of full-length p50) within the holoenzyme. Our F tag can bind up to 3 Fabs, as shown in previous labeling experiments (21). Intriguingly, only a single Fab is seen in the class averages of Fab-labeled p50N30-F telomerase (Fig. 6E), with its paratope (Fig. 6F, asterisk) proximal to p45/p75 and ∼40 Å from the apparent boundary of p50. Assuming that only the most terminal of the 3 FLAG epitopes was accessible for Fab binding and that each FLAG epitope extends a maximum of 20 Å based on peptide length, the C terminus of p50N30 is likely to be near the p50 boundary with p45 and/or p75 (Fig. 6F, square). However, because a maximum of 1 Fab bound to p50N30-F holoenzyme, it was not possible to determine the location of the p50N30 C terminus as precisely as the locations of tagged protein termini with more than 1 Fab bound.
DISCUSSION
EM structures of Tetrahymena telomerase holoenzyme revealed a central location of the p50 subunit (21). Combined with findings from holoenzyme reconstitution in vitro (21), p50 was proposed as the structural and functional hub for coordination of the RNP catalytic core, 7-1-4, and Teb1. Here we have biochemically and structurally characterized the roles of p50 in recombinant and endogenously assembled telomerase holoenzymes. We have shown that a p50 N-terminal domain supports all of the previously identified functions of p50, including physical bridging of other holoenyzme subunits and stimulation of processive repeat synthesis. The p50-bound RNP catalytic core has an unexpectedly slow elongation rate of tandem repeat synthesis but high stability of product interaction and RAP. Although direct p50-DNA interaction could account for the p50-mediated increase in RAP, we suggest that p50 confers high RAP by inducing a conformational change in the RNP catalytic core. In the holoenzyme subunit model based on EM structures (21), p50 is close to the TERT N-terminal (TEN) domain, which is crucial for conferring RAP to the otherwise single-repeat synthesis activity of the remaining TERT domains (28).
Addition of Teb1 to the p50-bound RNP catalytic core dramatically increased the elongation rate for tandem repeat synthesis. At least under the conditions typical for telomerase assays in vitro, product synthesis is rate limited by dissociation of the thermodynamically favorable product-template hybrid (12, 29). Therefore, we suggest a model in which Teb1C favors hybrid dissociation after synthesis to the template 5′ end, disfavors template-product reassociation in the postsynthesis register (product base paired to the template 5′ end), and/or favors template-product reassociation for the next repeat synthesis (product base-paired to the template 3′ end). In parallel, direct protein-DNA interactions mediated by Teb1A and Teb1B would enhance retention of the single-stranded DNA product. Given the closely triangulated arrangement of the TERT TEN domain, p50, and Teb1C (21), Teb1C could act by allosteric regulation or by direct DNA or RNA contact. A direct nucleic acid binding activity of Teb1C and/or p50 could escape detection in biochemical assays of the isolated subunits if holoenzyme protein-protein interactions are necessary for folding or positioning the contact surface. Future studies of the physical path of DNA would be aided by a reconstitution system that recapitulated the high physiological stability of Teb1 interaction with the remainder of the holoenzyme, which is compromised in the current reconstitutions (21, 23).
In the structure of Tetrahymena telomerase holoenzyme determined by EM, we could distinguish the general locations but not the individual boundaries of p75, p45, and p19, since these subunits were always present as a complete 7-1-4 subcomplex. By differential tagging and individual expression of the 7-1-4 subunits described above, we could separate the requirements of each for assembly with p50 in a manner that was not possible to do using endogenously assembled telomerase complexes. We have shown that the interaction of RRL-expressed p75 and p50 is specific and independent of p45 or p19. In addition, p75 alone influenced the activity of the p50-bound RNP catalytic core to an extent similar to that for the entire 7-1-4 complex. From EM structures, both p75 and p45 were modeled to contact p50 (21). The biochemical results here confirm a direct interaction of p75 with p50 and leave open the possibility that p45 also contacts p50 in a p75-dependent manner. The EM structure locations of the p19 and p75 C termini place these 2 subunits adjacent to each other (21). Because we show that p45 promotes the association of p19 with p75, p45 likely extends farther toward p19 than previously modeled. The biochemical stability of the 7-1-4 subcomplex predicts an extensive network of subunit interactions, only some of which we may be able to determine using proteins expressed in RRL.
Because all of the known biochemical functions of p50 are mediated by the p50 N-terminal domain, the function of the C-terminal half of p50 is uncertain. This region of p50 is not detectable in the EM structures of telomerase holoenzyme and has a relatively small effect on telomerase function in vitro or in vivo. One tantalizing speculation is that this domain is regulatory. Negative regulation is suggested by the higher specific activity of telomerase reconstituted with p50N30 than that with full-length p50. Negative regulation is also consistent with telomere elongation in cells expressing p50-FZZ versus untagged p50, because C-terminal tagging could interfere with p50 C-terminal domain function. However, caveats to interpretation for all of the biochemical and biological assays of p50 C-terminal domain function preclude a firm conclusion about its regulatory role. Considering results from holoenzyme reconstitution in vitro, the higher specific activity of telomerase RNP with p50N30 than with full-length p50 could derive from an impact of heterologous expression on domain folding. Unfortunately, in vitro activity comparison of purified holoenzymes endogenously assembled with p50 versus p50N30 is not reliable due to the heterogeneity arising from partial proteolysis of full-length p50. Finally, considering telomere length as an in vivo readout of holoenzyme activity, the shortened telomeres observed in p50N30-FZZ cells could arise from a difference in holoenzyme function but also could be secondary to p50N30-FZZ aggregation or functional interference from its C-terminal tag. Taken together, our studies establish that p50 has an N-terminal domain hub for holoenzyme assembly and activation and also a structurally uncharacterized C-terminal domain with potential regulatory function.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants NIH GM54198 to K.C., NSF MCB1022379 and NIH GM48123 to J.F., and NIH GM071940 and AI069015 to Z.H.Z., an NSF predoctoral fellowship to H.U., and a Ruth L. Kirschstein NRSA postdoctoral fellowship, GM101874, to E.J.M. We acknowledge the use of instruments at the Electron Imaging Center for NanoMachines, supported by NIH (1S10RR23057; to Z.H.Z.) and CNSI at UCLA.
Footnotes
Published ahead of print 5 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00792-13.
REFERENCES
- 1.Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301–334 [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan RJ, Karlseder J. 2010. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11:171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart JA, Chaiken MF, Wang F, Price CM. 2012. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 730:12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sampathi S, Chai W. 2011. Telomere replication: poised but puzzling. J. Cell Mol. Med. 15:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu P, Takai H, de Lange T. 2012. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150:39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeiffer V, Lingner J. 2013. Replication of telomeres and the regulation of telomerase. Cold Spring Harb. Perspect. Biol. 5:a010405. 10.1101/cshperspect.a010405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shay JW, Wright WE. 2011. Role of telomeres and telomerase in cancer. Semin. Cancer Biol. 21:349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armanios M, Blackburn EH. 2012. The telomere syndromes. Nat. Rev. Genet. 13:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hengesbach M, Akiyama BM, Stone MD. 2011. Single-molecule analysis of telomerase structure and function. Curr. Opin. Chem. Biol. 15:845–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackburn EH, Collins K. 2011. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 3:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan ED, Collins K. 2012. Biogenesis of telomerase ribonucleoproteins. RNA 18:1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podlevsky JD, Chen JJ. 2012. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutat. Res. 730:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Londono-Vallejo JA, Wellinger RJ. 2012. Telomeres and telomerase dance to the rhythm of the cell cycle. Trends Biochem. Sci. 37:391–399 [DOI] [PubMed] [Google Scholar]
- 14.Nandakumar J, Cech TR. 2013. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14:69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackburn EH, Greider CW, Szostak JW. 2006. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 12:1133–1138 [DOI] [PubMed] [Google Scholar]
- 16.Collins K. (ed). 2012. Methods in Cell Biology, vol. 109. Tetrahymena thermophila. Elsevier Inc., Waltham, MA: [DOI] [PubMed] [Google Scholar]
- 17.Witkin KL, Collins K. 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 18:1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witkin KL, Prathapam R, Collins K. 2007. Positive and negative regulation of Tetrahymena telomerase holoenzyme. Mol. Cell. Biol. 27:2074–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min B, Collins K. 2009. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell 36:609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller MC, Collins K. 2000. The Tetrahymena p80/p95 complex is required for proper telomere length maintenance and micronuclear genome stability. Mol. Cell 6:827–837 [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. 2013. The architecture of Tetrahymena telomerase holoenzyme. Nature 496:187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M. 2011. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc. Natl. Acad. Sci. U. S. A. 108:20357–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min B, Collins K. 2010. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J. Biol. Chem. 285:16434–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couvillion MT, Collins K. 2012. Biochemical approaches including the design and use of strains expressing epitope-tagged proteins. Methods Cell Biol. 109:347–355 [DOI] [PubMed] [Google Scholar]
- 25.Radermacher M, Wagenknecht T, Verschoor A, Frank J. 1987. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146:113–136 [DOI] [PubMed] [Google Scholar]
- 26.Hardy CD, Schultz CS, Collins K. 2001. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276:4863–4871 [DOI] [PubMed] [Google Scholar]
- 27.Chalker DL. 2012. Transformation and strain engineering of Tetrahymena. Methods Cell Biol. 109:327–345 [DOI] [PubMed] [Google Scholar]
- 28.Robart AR, Collins K. 2011. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol. Cell 42:308–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins K. 2011. Single-stranded DNA repeat synthesis by telomerase. Curr. Opin. Chem. Biol. 15:643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.