Abstract
SLC5A8 is a putative tumor suppressor that is inactivated in more than 10 different types of cancer, but neither the oncogenic signaling responsible for SLC5A8 inactivation nor the functional relevance of SLC5A8 loss to tumor growth has been elucidated. Here, we identify oncogenic HRAS (HRASG12V) as a potent mediator of SLC5A8 silencing in human nontransformed normal mammary epithelial cell lines and in mouse mammary tumors through DNMT1. Further, we demonstrate that loss of Slc5a8 increases cancer-initiating stem cell formation and promotes mammary tumorigenesis and lung metastasis in an HRAS-driven murine model of mammary tumors. Mammary-gland-specific overexpression of Slc5a8 (mouse mammary tumor virus-Slc5a8 transgenic mice), as well as induction of endogenous Slc5a8 in mice with inhibitors of DNA methylation, protects against HRAS-driven mammary tumors. Collectively, our results provide the tumor-suppressive role of SLC5A8 and identify the oncogenic HRAS as a mediator of tumor-associated silencing of this tumor suppressor in mammary glands. These findings suggest that pharmacological approaches to reactivate SLC5A8 expression in tumor cells have potential as a novel therapeutic strategy for breast cancer treatment.
INTRODUCTION
SLC5A8 is a sodium-coupled transporter for short-chain fatty acids (acetate, propionate, and butyrate), monocarboxylates (lactate, pyruvate, and β-hydroxybutyrate), and the B-complex vitamin nicotinate (1–5). SLC5A8 was first identified as a potential tumor suppressor in the colon (6); since then, the transporter has been shown to be silenced in cancers of many other organs, including stomach, brain, thyroid, lung, breast, prostate, pancreas, head and neck, lymphocytes, and kidney (7, 8). The tumor suppressor function of SLC5A8 is mainly associated with inhibition of histone deacetylases (HDACs) in tumor cells (9). Butyrate, one of the substrates of SLC5A8, is a well-known HDAC inhibitor that induces differentiation in normal epithelial cells but causes apoptosis in cancer cells (10–13). The tumor-selective sensitization of the cells to apoptosis by butyrate involves the tumor cell-specific induction of the death receptor pathway or activation of the proapoptotic protein Bim (14–17). Butyrate is generated at high concentrations in the colonic lumen by bacterial fermentation of dietary fiber, and SLC5A8 is expressed in the lumen-facing apical membrane of colonic epithelial cells, mediating the entry of butyrate into the cells (18, 19). This provides a molecular mechanism for the transporter's role as a tumor suppressor in the colon. However, SLC5A8 is silenced in tumors of various noncolonic tissues in which butyrate is not relevant under physiologic conditions. Attempts in our laboratory to address this conundrum led to the discovery that pyruvate, an endogenous metabolite and also a substrate for SLC5A8, is a potent inhibitor of HDACs and an inducer of tumor cell-specific apoptosis (11, 13). Further, SLC5A8 is a transcriptional target of C/EBPδ and p53 in the kidney, as well as in mammary epithelium (20). All these findings explain not only why SLC5A8 is silenced in many tumors but also why tumor cells effectively convert pyruvate into lactate. Lactate is also a substrate for SLC5A8, but it does not inhibit HDACs. In order to avoid the entry of the HDAC inhibitors pyruvate and butyrate, tumor cells purposely silence SLC5A8 to escape from cell death.
SLC5A8 inactivation in cancer occurs via hypermethylation of the SLC5A8 promoter (6). However, the molecular mechanisms responsible for this hypermethylation are not known. It has been shown that increased DNA methyltransferase (DNMT) activity is an early event in carcinogen-initiated lung tumorigenesis, and this phenomenon has also been demonstrated in several other tumors, cancer cell lines, and mouse tumor models (21–24). DNA hypermethylation is a hallmark of cancer (25, 26). DNA methylation is catalyzed by DNMTs; in mammals, there are at least three DNMT isoforms (DNMT1, DNMT3a, and DNMT3b). DNMT1 is responsible for maintaining the DNA methylation pattern during embryonic development and cell division (27, 28). Further, DNMT1 deregulation has been proposed to play a critical role in cellular transformation; forced expression of DNMT1 in nontransformed cells leads to cellular transformation (29), whereas DNMT1 knockdown protects mice from cancer (30).
Several oncogenic signaling pathways, especially RAS/RAF/MAPK signaling, lead to activation of DNMT1 through transcriptional and posttranscriptional control (31–34). Stable expression of HRASG12V induces transcription of DNMT1 through an AP-1 site in the promoter region (35). Further, RAS-induced DNMT1 activation is a prerequisite for fos-mediated cellular transformation (36). These observations suggest that oncogenic HRAS plays a prominent role in DNMT1 activation and subsequent cellular transformation. Oncogenic transformation arises from accumulation of both genetic and epigenetic alterations that result in activation of oncogenes and inactivation of tumor suppressor genes. Of the many oncogenes activated in human cancers, RAS is one of the most extensively studied. Although the incidence of mutations in RAS is very low in human breast cancer, over 50% of human breast carcinomas express elevated levels of normal HRAS protein (37, 38). High levels of HRAS protein have also been observed in hyperplasias from patients who subsequently develop breast cancer (39). Since the silencing of SLC5A8 in tumors occurs via promoter hypermethylation, we hypothesized that there could be a functional link among RAS-induced cellular transformation, DNMT1 activation, and SLC5A8 inactivation in breast cancer and that this signaling pathway could be a very early event in mammary tumor development. Here, we provide evidence in support of this hypothesis.
MATERIALS AND METHODS
Cell lines, plasmids, and transfection.
The human nontransformed normal mammary epithelial cell lines HMEC and MCF10A were grown in complete mammary epithelial cell growth medium (MEGM; obtained from Lonza Inc., Walkersville, MD). HBL100 cells were grown in Dulbecco's modified Eagle's medium (DMEM)–F-12 containing 10% fetal bovine serum (FBS). MCF7 cells were grown in DMEM containing 10% FBS. T47D, ZR75.1, MB231, MB453, and MB468 cells were grown in RPMI medium containing 10% FBS with antibiotics at 37°C in the presence of 5% CO2. RNA prepared from these cells was used for expression analyses. Nuclear extracts were prepared from the cells for use in measurement of DNMT activity. Genomic DNA prepared from the cells was used for methylation-specific (MS)-PCR analysis.
MCF10A cells and the series of oncogenic cell lines derived from this cell line, which represent nontransformed normal (MCF10A1 [M1]), transformed premalignant (MCF10AT1k.cl2 [M2]), ductal carcinoma in situ (DCIS) (MCF10CA1h [M3]), and invasive breast cancer (MCF10CA1a.cl1 [M4]) cells, were kindly provided by Fred Miller and Steven Santner at the Barbara Ann Karmanos Cancer Institute, Detroit, MI. M1 and M2 cells were grown in DMEM–F-12 containing 5% horse serum, insulin (10 μg/ml), epidermal growth factor (EGF) (20 ng/ml), hydrocortisone (0.5 μg/ml), and cholera toxin (100 ng/ml). M3 and M4 cells were grown in DMEM–F-12 containing 5% horse serum with antibiotics at 37°C in the presence of 5% CO2. RNA prepared from these cells was used for microarray and gene expression analyses. Nuclear extracts were prepared from these cells and used for measurement of DNMT activity. Genomic DNA prepared from the cells was used for MS-PCR analysis.
The HCT116 cell line, which is positive for DNMTs (DNMT+/+), and the isogenic cell lines with homologous deletion of DNMT1 (DNMT1−/−), DNMT3b (DNMT3b−/−), and DNMT1 plus DNMT3b (double knockout [DKO]), were kindly provided by Bert Vogelstein at the Johns Hopkins University, Baltimore, MD. DNMT+/+, DNMT3b−/−, and DKO cells were grown in McCoy 5A medium with 10% FBS, and DNMT1−/− cells were grown in McCoy 5A medium with 10% FBS, 0.1 mg/ml hygromycin B, and antibiotics at 37°C in the presence of 5% CO2.
The expression constructs pBaBe-Puro, HRASG12V-pBaBe-Puro, cytomegalovirus-long terminal repeat (CMV-LTR), c-Myc, pcDNA3.1, STAT3C, C/EBPβ, Bcl-2, pCMV-neo, and E2F1 were transfected into MCF10A cells using FuGene 6 according to the manufacturer's instructions (Roche Applied Science). After 48 h of transfection, cells were harvested and processed for RNA and protein extraction for gene expression analysis. We also extracted nuclear fractions from these cells for DNMT assay. DNA from the cells was used for MS-PCR analysis.
Generation of HRASG12V stable cell lines.
Transfection of M1 cells with activated HRAS (HRASG12V) and selection from xenografted tumors yielded the M2 cell line; this cell gives rise to premalignant lesions with the potential for neoplastic progression (40). M3 and M4 cells were derived from occasional carcinomas arising from xenografts of MCF10AT1k.cl2. M3 cells give rise to predominantly well-differentiated carcinomas in xenografts, while M4 cells give rise to relatively undifferentiated carcinomas and also metastasize to the lung upon injection into the tail vein (41). In the present study, we refer to these cell lines as M1, M2, M3, and M4.
Generation of the SLC5A8-pLVX stable cell line.
SLC5A8 cDNA was subcloned into the pLVX-TetOn-advanced vector (Clontech). Recombinant lentivirus was produced by cotransfection of pLVX-TetOn vector or SLC5A8-pLVX-TetOn with helper plasmids (Viral Power Lentiviral Expression System; Invitrogen) into HEK-293FT human embryonic kidney cells using Lipofectamine 2000 transfection reagent. M4-SLC5A8-TetOn cells were generated using virus produced from the SLC5A8-pLVX-TetOn plasmid in the metastatic breast cancer cell line M4.
Generation of MMTV-Hras/Slc5a8−/− transgenic mice.
Slc5a8+/− mice (42) were backcrossed for at least 7 generations into an FVB/N genetic background and then intercrossed to generate all three Slc5a8 genotypic mice. Genotyping was performed with appropriate primers (see Table S2 in the supplemental material). A mouse mammary tumor virus (MMTV) Hras transgenic (Tg) mouse was purchased from Jackson Laboratory (stock number 004363), and the presence of the MMTV-Hras transgene was confirmed by genotype analysis (see Table S2 in the supplemental material). To generate MMTV-Hras-Tg mice with three different Slc5a8 genetic backgrounds (Slc5a8+/+, Slc5a8+/−, and Slc5a8−/−), Slc5a8−/− mice were bred with MMTV-Hras-Tg mice, and the resulting MMTV-Hras/Slc5a8+/− mice were crossed again to generate MMTV-Hras/Slc5a8−/− mice.
Generation of the MMTV-Slc5a8-Tg mouse.
Mouse Slc5a8 cDNA for the coding sequence was amplified using the original mouse pSPORT1-mSlc5a8 plasmid (3) as the template and then subcloned into pMMTV-SV40-BSSK (Addgene plasmid 1824). The resulting plasmid, pMMTV-SLC5A8-BSSK, consists of 2.3 kb of the MMTV LTR and 600 bp of the 5′ untranslated region (UTR) of c-Hras fused to a 5.34-kb Slc5a8-EcoRI/XbaI fragment containing the Slc5a8 coding region, 3′ UTR, and polyadenylation signal. This plasmid was used to generate the mammary-gland-specific overexpression of SLC5A8. The transgenic mouse was generated by standard techniques at the Mouse ES Cell and Transgenesis Core Facility, Georgia Health Sciences University. The MMTV-Slc5a8-Tg mouse was genotyped and screened to confirm the overexpression of SLC5A8 in the mammary gland.
Cloning of the DNMT1 promoter.
The human DNMT1 promoter-EGFP and DNMT1 promoter-Luc constructs were generated by cloning the 1.2-kb DNMT1 promoter (obtained by PCR using human genomic DNA as a template) into the pGEM-T Easy vector and then subcloned using the SalI/NcoI site into pTurboGFP-PRL (Evrogen, Russia) and using the SacI/NcoI site into pGL3-Luc (Promega, Madison WI) vectors. The primers used for PCR were 5′-GGCCCAATCATGGCTCATTGCAG-3′ (sense) and 5′-CCATGGCGGAGGCTTCAGCAG-3′ (antisense). For the transactivation assays, MCF10A cells (2 × 105) were seeded in 35-mm tissue culture dishes and allowed to grow in MEGM for 24 h. The effector and reporter plasmids were transfected using FuGene 6. The enhanced green fluorescent protein (EGFP) expression was monitored by epifluorescence after 36 h posttransfection under the fluorescence microscope. With the luciferase reporter, cells were collected after 36 h posttransfection, and the lysates were used for the measurement of luciferase activity. The activity was normalized to β-galactosidase (β-Gal) levels and compared with vector-transfected cells.
Knockdown of HRAS.
To inactivate HRAS, we used retrovirus-mediated expression of small interfering RNAs (siRNAs) that specifically target HRASG12V (H1/siRNA) and the wild-type HRAS gene (H2/siRNA), kindly provided by Jinson Liu at the University of Texas M. D. Anderson Cancer Center. We transfected M4 and MB231 cells with H1/HRAS siRNA to inactivate oncogenic HRAS (HRASG12V) and H2/HRAS siRNA to inactivate wild-type HRAS, and this served as a control siRNA (43). Seventy-two hours postinfection, cells were collected for gene expression analysis. For cell cycle analysis, cells were infected with H1/siRNA and H2/siRNA for 72 h and then treated with 1 mM butyrate and pyruvate for 48 h. For the positive control, cells were treated with 1 μM tryptic soy agar (TSA) for 48 h. After the treatment, cells were collected and processed for cell cycle analysis.
Knockdown of C/EBPβ.
To knock down C/EBPβ, an HRAS nuclear effector, we transfected M4 and MB231 cells with scrambled siRNA and C/EBPβ siRNA, obtained from Santa Cruz Biotechnology, according to the manufacturer's instructions. Seventy-two hours posttransfection, cells were collected for gene expression analysis. For cell cycle analysis, cells were infected with scrambled and C/EBPβ siRNAs for 72 h and then treated with 1 mM butyrate and pyruvate for 48 h. For the positive control, cells were treated with 1 μM TSA for 48 h. After the treatment, cells were collected and processed for cell cycle analysis.
5-AzaDC treatment.
For 5-aza-2-deoxycytidine (5-AzaDC) (a pan-DNMT inhibitor) treatment, MCF10A, HBL100, MCF7, MB231, M1, and M4 cells (0.5 × 106) were seeded in 10-cm cell culture dishes. After 24 h, the cells were treated with 5-AzaDC (Sigma) at 2 μg/ml for 24 h on days 2, 4, and 6. The medium was changed 24 h after addition of the 5-AzaDC (i.e., on days 3, 5, and 7). Cells were collected on day 8 and processed for gene expression analysis. For cell cycle analysis, cells were treated with 5-AzaDC for 72 h and then treated with 1 mM butyrate for 48 h. For the positive control, cells were treated with 1 μM TSA for 48 h. After the treatment, cells were collected and processed for cell cycle analysis.
Procainamide treatment.
For procainamide (P-NH2) (a DNMT1-specific inhibitor) treatment, MCF10A, HBL100, MCF7, and MB231 cells (2 × 106) were seeded in 10-cm cell culture dishes. After 24 h, the cells were treated with P-NH2 (Sigma) at 25 μg/ml for 72 h. The medium was changed 24 h after addition of the P-NH2, and cells were collected and processed for gene expression analysis. For cell cycle analysis, cells were treated with P-NH2 for 72 h and then treated with 1 mM butyrate for 48 h. For the positive control, cells were treated with 1 μM TSA for 48 h. After the treatment, cells were collected and processed for cell cycle analysis.
U0126 treatment.
M4 and MB231 cells were treated with 5 μM U0126, a MEK/extracellular signal-regulated kinase (ERK) inhibitor obtained from EMD Biosciences, for 48 h, and then cells were collected and processed for gene expression analysis. For cell cycle analysis, cells were treated with U0126 for 48 h and then treated with 1 mM butyrate and pyruvate for another 48 h. For the positive control, cells were treated with 1 μM TSA for 48 h.
Sodium bisulfate treatment and MS-PCR.
DNA samples from human nontransformed normal mammary epithelial cells, human breast cancer cells, mouse normal mammary tissues, and mouse tumor tissues were used for sodium bisulfite treatment to convert unmethylated cytosine to thymidine using the EpiTect 96 Bisulfite DNA methylation assay kit (Qiagen) following the manufacturer's instructions. The primers used to amplify the methylated allele were AS-meth-442-459s (5′-TCGAACGTATTTCGAGGC-3′) and AS-meth-550as (5′-ACAACGAATCGATTT TCCG-3′). We used MyoD1 as an internal control, and the primers used for MyoD1 were 5′-CCAACTCCAAATCCCCTCTCTAT-3′ (forward) and 5′-TGATTAATTTAGATTGGGTTTAGAGAAGGA-3′ (reverse).The PCR parameters were 31 cycles of 95°C for 45 s, 56°C for 45 s, 72°C for 45 s, and 72°C for 10 min, and 4°C to cool. The amplified PCR products were resolved in 2% agarose gels, stained with ethidium bromide, and photographed.
Generation of mammospheres and tumorospheres.
For the mammosphere and tumorosphere cultures, sorted cells were seeded in a 96-well culture plate coated with Matrigel at a concentration of 1,000 cells per well in DMEM–F-12 supplemented with B-27, recombinant human EGF (rhEGF), recombinant human basic fibroblast growth factor (rhbFGF), heparin, and penicillin/streptomycin. For suspension culture, cells were seeded on ultra-low-adherence plates (Corning) at 20,000 cells/ml in DMEM–F-12 containing B-27 supplement (Invitrogen), rhEGF (Invitrogen), rhbFGF (Invitrogen), heparin (Sigma), and penicillin/streptomycin.
Ras kinase assay.
The presence of GTP-bound Ras was determined by using an active Ras pulldown and detection kit (Pierce, Rockford, IL). Briefly, cells and tissues were lysed with lysis buffer (25 mM Tris-HCl, pH 7.2, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, and 5% glycerol), and the lysates (500 μg of protein) were incubated with glutathione S-transferase (GST)-Raf1-RBD and the glutathione resin for 1 h at 4°C in a spin column. Then, the spin column was washed three times with lysis buffer and eluted with 2× SDS sample buffer. The eluted samples were separated by SDS-PAGE, transferred, and probed with anti-Ras antibody.
Immunoblot analysis.
For immunoblot (IB) analysis, cell lysates were prepared by sonication in cell lysis buffer with protease inhibitors. Protein samples were fractionated on SDS-PAGE gels and transferred to Protran nitrocellulose membranes (Whatman GmbH). The membranes were blocked with 5% nonfat dry milk and exposed to primary antibody at 4°C overnight, followed by treatment with appropriate secondary antibody conjugated to horseradish peroxidase at room temperature for 1 h, and developed with an enhanced chemiluminescence SuperSignal Western system. In this study, we used anti-SLC5A8 (APR44110; Aviva System Biology), anti-MCT1 (AB3428B; Millipore), anti-HRAS (Sc-520; Santa Cruz Biotechnology), anti-β-actin (Sc-47778; Santa Cruz Biotechnology), anti-E-cadherin (610181, BD Bioscience), and anti-green fluorescent protein (anti-GFP) (G6539; Sigma) antibodies. We used all primary antibodies at 1:1,000 dilution and secondary antibodies at 1:5,000 dilution.
ChIP assays.
MCF10A cells were transfected with expression constructs of HRASG12V, c-Myc, STAT3C, C/EBPβ, Bcl-2, and E2F1. Chromatin immunoprecipitation (ChIP) assays were carried out using a ChIP assay kit (Millipore), human DNMT1-specific antibody, and mouse IgG antibodies. After ChIP, the protein-DNA complexes were eluted from the beads, and the cross-linking was reversed. The DNA present in the immunoprecipitates was purified from the eluted solution and used for PCR amplification of the SLC5A8 promoter using SLC5A8 promoter-specific primers (see Table S2 in the supplemental material).
RT-PCR.
Human SLC5A8, DNMT1, DNMT3a, and DNMT3b and mouse Slc5a8, Dnmt1, Dnmt3a, and Dnmt3b expression was determined by reverse transcription (RT)-PCR. Total RNA was isolated from MCF10A cells transfected with various oncogenes and from mammary gland and tumor tissues. RNA was reverse transcribed using the GeneAmp RNA PCR kit (Applied Biosystems), and PCR was performed on a Veriti thermocycler (Applied Biosystems) using specific primers (see Table S3 in the supplemental material). Representative images of triplicate experiments are shown.
Real-time PCR analysis.
Real-time PCR analysis was carried out in an Applied Biosystems Step One Plus instrument using the power SYBR green PCR master mix (Applied Biosystems, United Kingdom) according to the manufacturer's instructions. The primer sequences used for the real-time PCR analysis are listed in Table S3 in the supplemental material.
Cell cycle analysis.
Cells were fixed in 50% ethanol; treated with 0.1% sodium citrate, 1 mg/ml RNase A, and 50 μg/ml propidium iodide; and subjected to fluorescence-activated cell sorting (FACS) (Becton, Dickinson) analysis.
Measurement of HDAC activity.
A commercially available kit (Biovision, Mountain View, CA, USA) was used to determine HDAC activity according to the manufacturer's instructions. Cell lysates from human nontransformed normal mammary epithelial cells, breast cancer cells, M1 to M4 cells, and nontransformed normal mouse mammary gland and tumor tissues were prepared and used as the source of HDAC activity.
Measurement of DNMT activity.
A commercially available kit (Epigentek, Farmingdale, NY, USA) was used to determine DNMT enzymatic activity according to the manufacturer's instructions. Nuclear fractions from human nontransformed normal mammary epithelial cells, breast cancer cells, M1 to M4 cells, and nontransformed normal mouse mammary gland and tumor tissues were prepared, using the commercially available kit (Epigentek, Farmingdale, NY, USA), and used as the source of DNMT activity.
Mouse xenograft studies.
Female athymic nude mice (6 to 8 weeks old) were purchased from Taconic, Hudson, NY). The animals were housed in a pathogen-free isolation facility with a light/dark cycle of 12 h/12 h and fed with rodent chow and water ad libitum. We included 6 mice in each group. To test the tumor-forming potential of M1, M2, M3, and M4 cells, 1 × 107 cells in 100 μl phosphate-buffered saline (PBS) were injected subcutaneously into the mammary fat pad. Tumor incidence was monitored for another 8 weeks, and tumor volume was quantified by measuring the length and width of the tumor every 7 days for 8 weeks using calipers. We repeated the experiment two times using 6 mice in each group in each experiment.
To analyze the SLC5A8 substrates butyrate, pyruvate, and propionate, which inhibit HDACs, and acetate, lactate, and nicotinate, which do not inhibit HDACs, in tumor growth, we implanted M4-SLC5A8-pLVX stable cells (1 × 107) in 100 μl PBS subcutaneously in the mammary fat pad. To induce SLC5A8 expression, doxycycline (2 mg/ml) in drinking water was given on the same day as tumor implantation. Similarly, butyrate, pyruvate, propionate, acetate, lactate, and nicotinate (2 mg/ml) were given in drinking water 48 h after tumor implantation. Tumor size was measured periodically with calipers, and tumor volume was calculated using the following formula: (width2 × length)/2. These experiments were approved by the Georgia Health Sciences University (GHSU) Institutional Animal Care and Use Committee (IACUC).
5-Azacytidine and butyrate implantation.
Twelve- to 14-week-old MMTV-Hras/Slc5a8+/+-Tg and MMTV-Hras/Slc5a8−/−-Tg mice were anesthetized, and 3-mm pellets containing either 5-azacytidine (0.5 mg; 90-day release) or sodium butyrate (10 mg; 90-day release), obtained from Innovative Research of America, were implanted either alone or in combination subcutaneously in each animal's back. The pellets provided continuous release of 5-azacytidine and butyrate for 90 days. The tumor incidence, time of tumor onset, number of tumors in each mouse, tumor volume, and animal survival time were monitored.
Collection of tissue samples from mice.
Mice were killed by CO2 asphyxiation, followed by cervical dislocation. For studies using frozen sections, normal mammary tissues and mammary tumor and lung metastatic tumor tissues were harvested and oriented in Tissue-Tek O. C. T. compound. After slow freezing, the cryosections were prepared and mounted on slides in the GHSU Histology and Pathology Core. The sections were stained with hematoxylin and eosin (H&E) and used for morphometric studies. Additional cryosections were used for immunohistochemical studies. To isolate RNA, DNA, and protein samples, tissue samples were immediately processed or snap-frozen in liquid nitrogen.
Single-cell preparation.
Normal inguinal mammary glands were obtained from 12- to 16-week-old FVB/N female mice and tumors were obtained from an MMTV-Hras-Tg mouse. Aseptic technique was maintained during adult mammary gland isolation and dissections for single-cell preparations. Adult mammary glands and tumors were digested for 8 to 10 h at 37°C in DMEM–F-12 containing 10% FBS, 300 U ml−1 collagenase (Sigma), and 100 U ml−1 hyaluronidase (Sigma). Mammary organoids resulting from overnight digestion were centrifuged and treated with ammonium chloride solution (Stem Cell Technology) and then treated with trypsin, followed by dispase (Stem Cell Technology) and DNase (Worthington), with washes and resuspension after each treatment using Hank's balanced salt solution (HBSS) supplemented with 2% FBS. Single cells were collected by passing them through a 40-μm nylon filter (BD Biosciences).
Stem cell medium.
M1 and M2 cells were grown in DMEM–F-12 supplemented with B-27, 0.1% methylcellulose, heparin (Sigma), rhEGF, rhbFGF (Invitrogen), and penicillin/streptomycin.
Flow cytometry procedures.
Single cells were stained with the following antibodies. Anti-CD49f (Stem Cell Technology) and anti-CD24 (Stem Cell Technology) antibodies were used to label the mammary stem cell and progenitor populations. For human cell lines, anti-CD49F (BD Bioscience) and anti-CD24 (BD Bioscience) antibodies were used. Lineage-negative (Lin−) cocktail was comprised of allophycocyanin (APC) anti-CD31 (BD Bioscience), APC anti-CD45 (BD Bioscience), and Ter119 (ebioscience). To assess viability, cells were stained with 1 mg/ml propidium iodide (Sigma) for 10 min. FACS analysis and cell sorting were performed using FACS LSRII, a MoFlo cell sorter, and FlowJo software. The purity of sorted cell populations was routinely >90%.
Immunofluorescence.
Cells were washed with PBS and fixed for 15 min in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences), washed three times in PBS, and permeabilized in 0.1% Triton X-100 for 10 min. The slides were washed again, blocked for 1 h in 10% goat serum (Invitrogen) at room temperature, and then incubated with primary antibodies overnight at 4°C, followed by secondary antibodies for 1 h. All sections were imaged using a confocal microscope or a Zeiss LSM 710 laser scanning confocal microscope. The antibodies used were anti-SLC5A8 (Aviva Systems Biology; 1:250), anti-CD49f (R&D; 1:500), and anti-DNMT1 (Cell Signaling; 1:500).
Histological sections.
Normal mammary glands and mammary tumor and lung tumor tissues were harvested, fixed, stained with H&E, and visualized at ×40 and ×63 magnifications.
Measurement of SLC5A8 transport function.
The transport function of SLC5A8 was monitored by Na+-coupled nicotinate uptake as described in our previous report (5). Uptake of [14C]nicotinate (20 μmol/liter) for 15 min was measured in the presence of NaCl and N-methyl-d-glucamine (NMDG)–Cl in monolayer cell cultures.
Measurement of MCT transport function.
The transport function of MCT was monitored by Na+-free uptake buffer containing 25 mM HEPES-Tris (pH 7.5), 140 mM N-methyl-d-glucamine, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, and 5 mM glucose, and transport activity was monitored with a 2-min incubation period. Uptake of [3H]pyruvic acid using 26.25 nM per assay (0.5 μCi/well) in these cells was measured as described previously (3, 5).
Northern blot analysis.
Ten micrograms of RNA from each sample was fractionated and analyzed by standard Northern blotting techniques. 32P-radiolabeled DNA probes for mouse keratin 18 (K18) and cyclophilin loading controls were prepared from isolated cDNA clones.
Institutional compliance.
All experimental procedures involving animals adhered to the National Institutes of Health Principles of Laboratory Animal Care (44) and were approved by the Georgia Health Sciences University IACUC and Biosafety Committee.
Statistical analyses.
Statistical analysis was done using one-way analysis of variance (ANOVA), followed by a Bonferroni multiple-comparison test. The software used was Graph Pad Prism, version 5.0. A P value of <0.05 was considered statistically significant.
RESULTS
HRAS inactivates SLC5A8 expression.
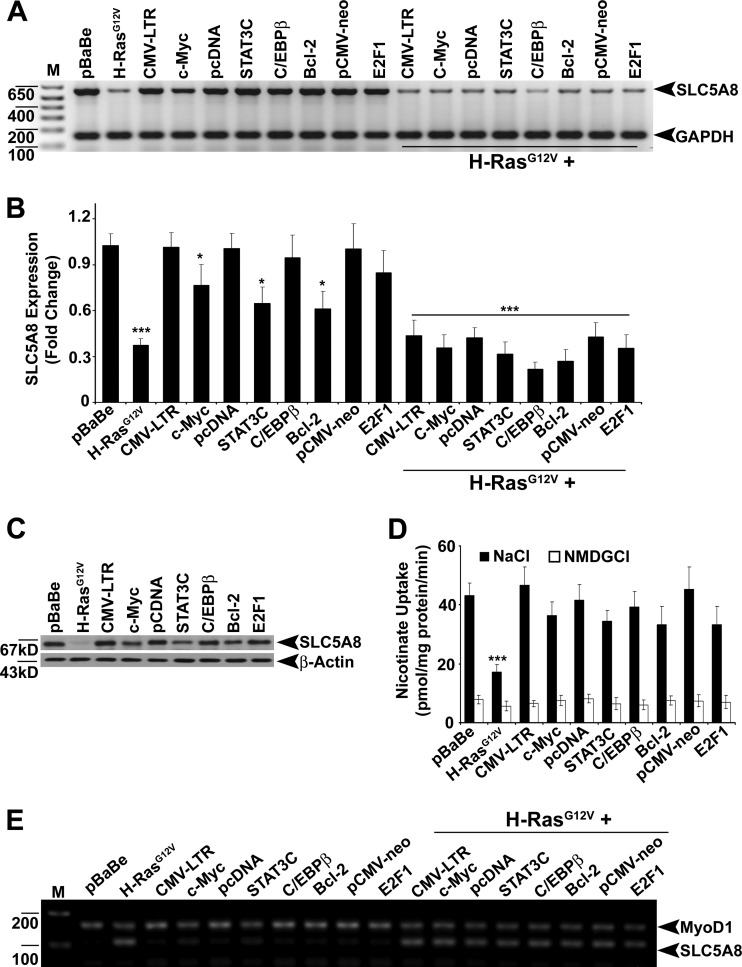
To explore the oncogenic signaling responsible for SLC5A8 inactivation in breast cancer, we expressed the HRAS (HRASG12V), c-Myc, constitutively active STAT3 (STAT3C), HRAS nuclear effector C/EBPβ, antiapoptotic protein Bcl-2, and cell cycle regulator E2F1 oncogenes in MCF10A cells, a human normal nontransformed mammary epithelial cell line, and analyzed SLC5A8 expression. Ectopic expression of the HRAS oncogene, either alone or in combination with other oncogenes, significantly decreased SLC5A8 expression and function (Fig. 1A to D). c-Myc, STAT3C, and Bcl-2 by themselves decreased the levels of SLC5A8 mRNA, but the magnitude of the effect was much smaller than that observed with HRAS. Since SLC5A8 inactivation in cancers is associated with DNA methylation of the SLC5A8 gene promoter, we analyzed the methylation status of the SLC5A8 promoter by methylation-specific PCR. Only HRAS was able to induce methylation of the SLC5A8 promoter (Fig. 1E). RAS was relatively inactive in normal nontransformed mammary epithelial cell lines, whereas it was activated in cancer cell lines (see Fig. S1A in the supplemental material). There was a reciprocal correlation between the RAS activity status and SLC5A8 expression in these cell lines (see Fig. S1B in the supplemental material). The HRAS-induced silencing of SLC5A8 was associated with methylation of the SLC5A8 promoter (see Fig. S1C in the supplemental material).
Fig 1.
HRASG12V inactivates SLC5A8 expression. (A and B) RT-PCR and real-time PCR analyses demonstrating that SLC5A8 transcript expression is inactivated in MCF10A cells by ectopic expression of HRASG12V. The data are means and standard errors of the mean (SEM). *, P < 0.05; ***, P < 0.001 by t test. (C) Western blot analysis showing downregulation of SLC5A8 protein in MCF10A cells by ectopic expression of HRASG12V. (D) [14C]nicotinate uptake demonstrating HRASG12V-induced decrease in SLC5A8 functional activity in MCF10A cells. The data are means ± SEM. ***, P < 0.001 by t test. (E) MS-PCR analysis providing evidence for HRASG12V-induced SLC5A8 methylation in MCF10A cells. MyoD1 was used as an internal control. Lanes M, 1-kb plus DNA ladder.
HRASG12V transactivates DNMT1 expression and binds to the SLC5A8 promoter.
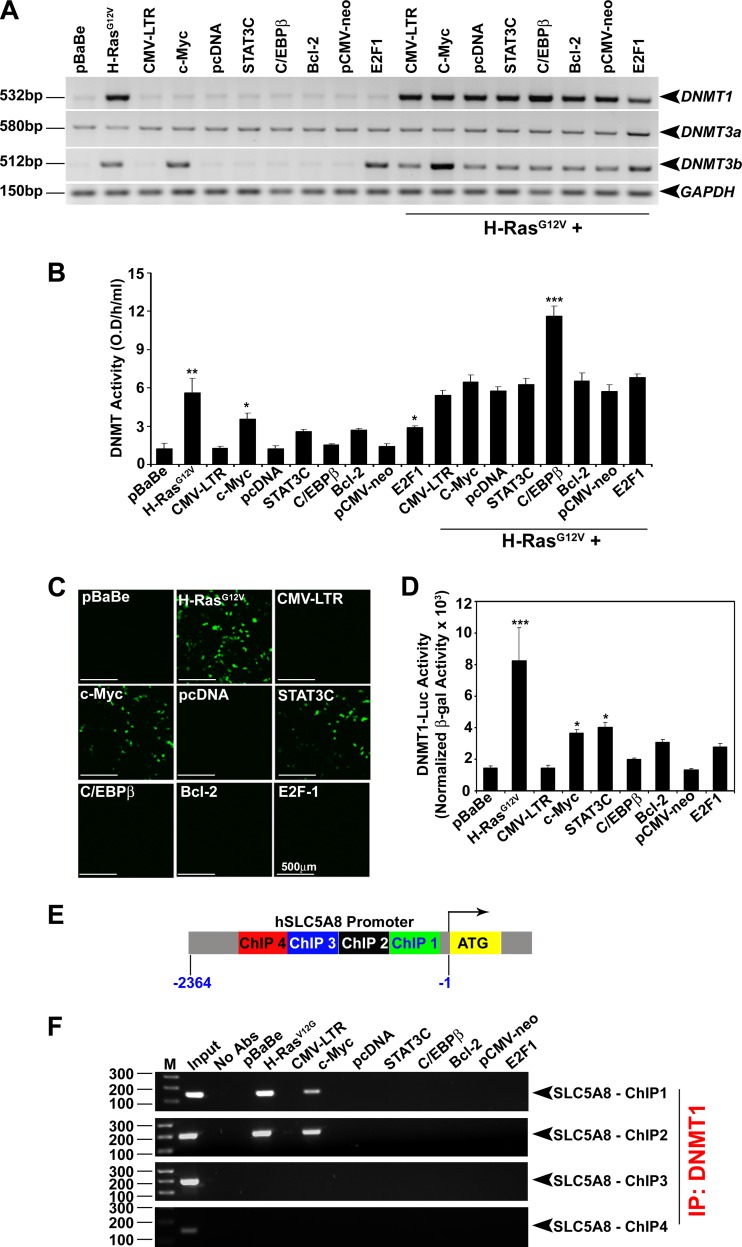
To examine the role of DNMTs in HRAS-induced SLC5A8 silencing in breast cancer cells, we analyzed the expression profiles of DNMTs (DNMT1, -3a, and -3b) and RAS activity status in nontransformed normal and cancer cell lines. In nontransformed normal cells, RAS activity was low, as was the expression/activity of DNMTs. In cancer cells, RAS activity was high, and this was accompanied by high expression levels of DNMT1 and DNMT3b and high DNMT activity (see Fig. S1D and E in the supplemental material). To explore the influence of HRAS on DNMT expression directly, we monitored the expression levels of DNMTs in MCF10A cells with and without ectopic expression of HRAS and other oncogenes. Oncogenic HRAS induced DNMT1 and DNMT3b but did not have any effect on DNMT3a (Fig. 2A and B). c-Myc and E2F1 did not affect DNMT1 and DNMT3a but induced DNMT3b. These results demonstrate that HRAS-induced DNMT1 and DNMT3b may play a role in SLC5A8 inactivation in breast cancer. We had already shown, using DNMT1+/+, DNMT1−/−, DNMT3b−/−, and DNMT1−/−/DNMT3b−/− (DKO) isogenic HCT116 colon cancer cells, that DNMT1 controls SLC5A8 expression (13). Here, we confirmed these findings with SLC5A8 protein and activity data (see Fig. S2A and B in the supplemental material). To determine if this was also true with breast cancer cells, we treated normal nontransformed mammary epithelial cell lines (MCF10A and HBL100) and breast cancer cell lines (MCF7 and MB231) with 5-AzaDC, a pan-DNMT inhibitor, and P-NH2, a DNMT1-specific inhibitor, and analyzed SLC5A8 expression. Both 5-AzaDC and P-NH2 reactivated SLC5A8 expression in breast cancer cells but not in nontransformed normal mammary epithelial cells (see Fig. S2C in the supplemental material). These results show that HRAS-induced DNMT1 expression specifically inactivates SLC5A8 in breast cancer cells.
Fig 2.
HRASG12V transactivates DNMT1 expression and forms a complex with the SLC5A8 promoter. (A) RT-PCR analysis showing HRASG12V-induced DNMT1 and DNMT3b transcript expression in MCF10A cells. (B) HRASG12V-induced increase in DNMT enzymatic activity in MCF10A cells. The data are means and SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test. O. D., optical density. (C and D) Ectopic expression of HRASG12V in MCF10A cells transactivates the DNMT1 promoter; reporters, EGFP (C) and luciferase (D). The data are means and SEM. *, P < 0.05; ***, P < 0.001 by t test. (E) Locations of ChIP primers designed from the human SLC5A8 promoter. (F) ChIP-PCR analysis showing that HRAS-induced DNMT1 binds to the SLC5A8 promoter.
The human DNMT1 promoter has several AP-1 and SP1 binding sequences, where oncogenic HRAS presumably binds and transactivates transcription. This was supported by our findings that oncogenic HRAS expression induced DNMT1 promoter-driven EGFP fluorescence (Fig. 2C) and luciferase activity (Fig. 2D). c-Myc and STAT3C expression also induced DNMT1 promoter activity to some extent (Fig. 2C and D), but the relative contribution to SLC5A8 inactivation was minor (Fig. 1A to E). We then wanted to determine whether oncogenic HRAS-induced DNMT1 directly binds to the SLC5A8 promoter to inactivate SLC5A8 gene transcription, and therefore, we performed ChIP assays of the human SLC5A8 gene promoter from bp −2400 to +1 relative to the SLC5A8 gene transcription initiation site. For this, we designed four PCR primers in the SLC5A8 promoter, two from the CG-rich CpG island and two away from the CpG island. We expressed vector controls, HRAS, c-Myc, STAT3C, C/EBPβ, Bcl-2, and E2F1 in MCF10A cells; performed chromatin immunoprecipitation with antibody specific for human DNMT1; and examined the presence of the SLC5A8 promoter in the immune complex by ChIP-PCR analysis. As shown in Fig. 2F, HRAS and DNMT1 bind to the SLC5A8 promoter specifically in the CG-rich CpG island. c-Myc and DNMT1 also bind to the SLC5A8 promoter in the CG-rich CpG island and inactivate its gene transcription to some extent. Although STAT3C and E2F1 induced DNMT1 activity to a certain extent (Fig. 2D), they were unable to bind to the SLC5A8 promoter and inactivate its gene transcription. These results demonstrate that oncogenic HRAS-induced DNMT1 binds to the SLC5A8 promoter and thereby inactivates SLC5A8 gene transcription in mammary epithelial cells.
SLC5A8 inactivation is an early event in oncogenic HRAS-induced cellular transformation.
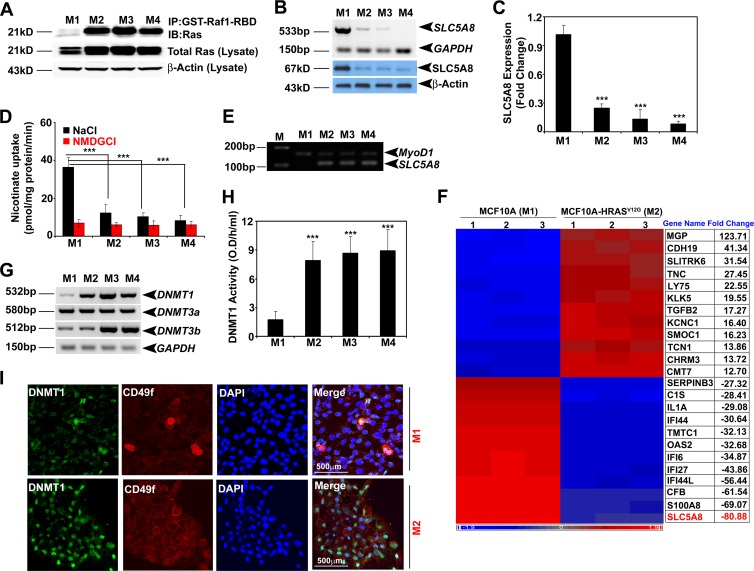
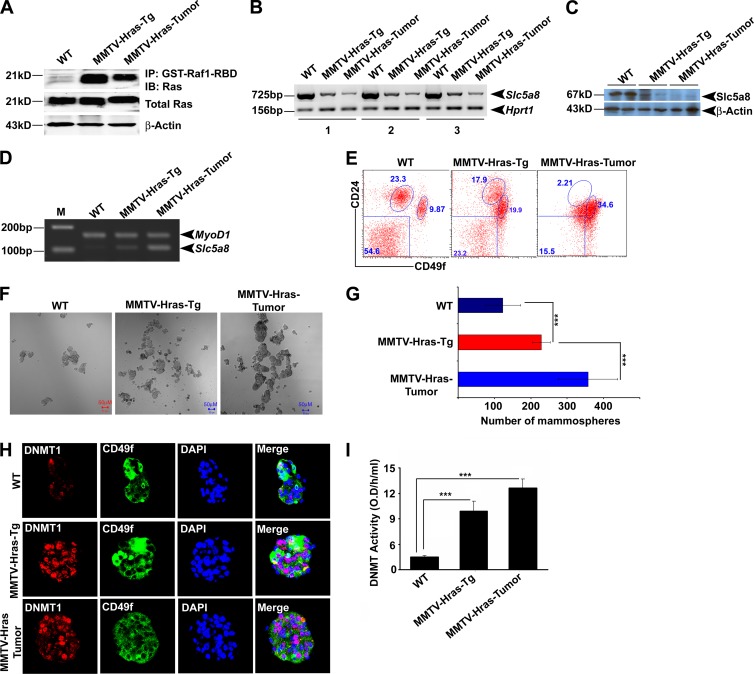
We analyzed the expression of SLC5A8 in a series of MCF10A-derived and HRAS-transformed cell lines: nontransformed normal (M1), HRAS-transformed premalignant (M2), HRAS-transformed malignant (M3), and HRAS-transformed metastatic and invasive (M4) cells (45). We found high RAS activity (Fig. 3A) and correspondingly low expression and activity of SLC5A8 in M2, M3, and M4 cells (Fig. 3B to D), as well as SLC5A8 promoter methylation in M2, M3, and M4 cells, but not in M1 cells (Fig. 3E). We then monitored the tumorigenic potential of M1, M2, M3, and M4 cells in BALB/c nude mice. M1 cells did not form tumors, whereas M2 cells were able to form tumors in 83% of the mice and M3 and M4 cells were able to form tumors in 100% of the mice (see Table S1 in the supplemental material). We compared the transcriptomes in these four cell lines by microarray analysis: M1 versus M2, M1 versus M3, M1 versus M4, M2 versus M3, and M3 versus M4. There were marked differences in gene expression between nontransformed normal cells and transformed cells (Fig. 3F; see Fig. S3A to D in the supplemental material). Interestingly, SLC5A8 expression was reduced drastically even in the premalignant M2 cell line compared to the nontransformed normal M1 cell line. The expression level stayed low in the malignant M3 cell line and the metastatic M4 cell line. We also analyzed the expression and activity of DNMTs. HRAS-induced cellular transformation was specifically associated with induction of DNMT1 even in the premalignant M2 cell line (Fig. 3G and H). There was no significant change in the expression of DNMT3a and DNMT3b at this early stage. DNMT3b was induced in M3 and M4 cells. In addition, we analyzed whether there is functional correlation among HRAS-induced cellular transformation, DNMT1 expression, and mammary stem cell numbers in the M1 and M2 cell lines using CD49f, a mammary stem cell marker. The expression of both DNMT1 and CD49f was significantly increased in M2 cells compared to M1 cells (Fig. 3I; see Fig. S4A and B in the supplemental material). Similarly, DNMT activity was positively associated with the CD49f level in M1, M2, M3, and M4 cells (Fig. 3H; see Fig. S4A and B in the supplemental material).
Fig 3.
SLC5A8 inactivation is an early event in oncogenic HRAS-induced cellular transformation. (A) RAS kinase assay demonstrating RAS activation in M2, M3, and M4 cells. (B) RT-PCR and Western blot analyses showing that SLC5A8 mRNA and protein expression is silenced in M2, M3, and M4 cells. (C) Real-time PCR analysis demonstrating SLC5A8 inactivation in M2, M3, and M4 cells. The data are means and SEM. ***, P < 0.001 by t test. (D) [C14]nicotinate uptake demonstrating that SLC5A8 functional activity is significantly decreased in M2, M3, and M4 cells. The data are means ± SEM. ***, P < 0.001 by t test. (E) MS-PCR demonstrating SLC5A8 methylation in M2, M3, and M4 cells. MyoD1 was used as an internal control. (F) Microarray heat map showing differential gene expression between MCF10A cells and HRASG12V-expressing MCF10A cells. The SLC5A8 gene transcript is downregulated ∼80-fold in M2 cells compared to M1 cells. 1, 2, and 3 indicate triplicates of each sample. (G) RT-PCR analysis showing DNMT1, DNMT3a, and DNMT3b transcript expression in M1, M2, M3, and M4 cells. (H) DNMT enzymatic activity showing that constitutive expression of HRAS in MCF10A cells increased DNMT activity. The data are means and SEM. ***, P < 0.001 by t test. (I) Immunofluorescence data showing that DNMT1 expression and CD49f expression are induced in M2 cells compared to M1 cells. DAPI (4′,6-diamidino-2-phenylindole) was used to label nuclei.
Involvement of the RAS/RAF/ERK signaling pathway in HRAS-induced silencing of SLC5A8.
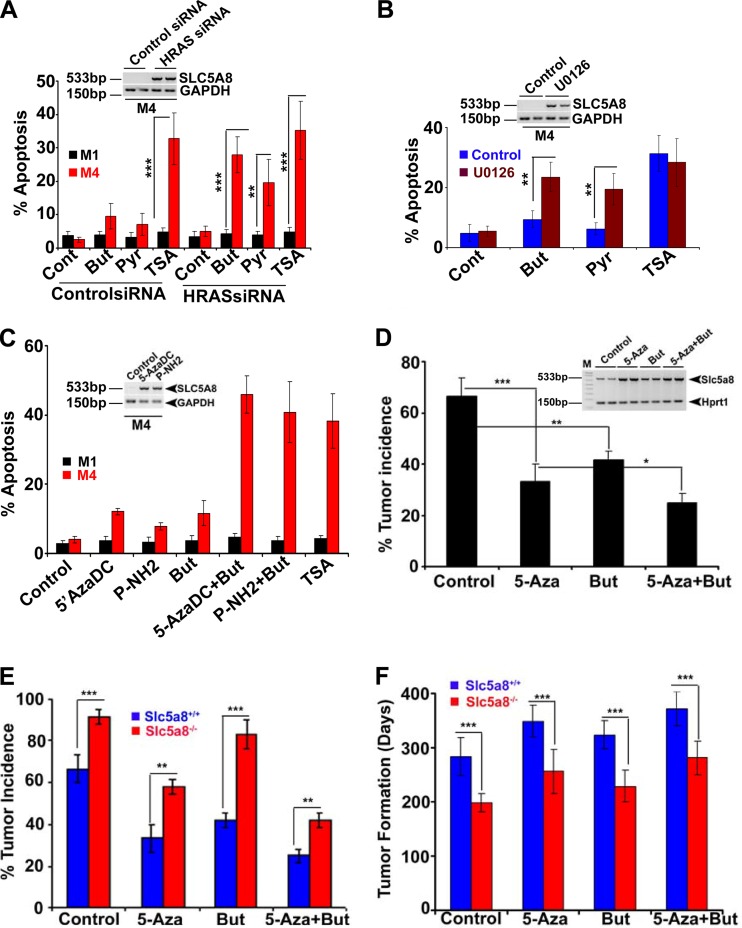
HRAS is involved in a broad spectrum of cellular processes, including cell proliferation and oncogenesis, by regulating various signaling pathways, such as RAF/MEK/ERK, RalGDS/Ral/RBP1, and phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR. To identify the signaling pathway that mediates HRAS-induced silencing of SLC5A8, we expressed different HRAS-activating mutants that are known to control different signaling pathways: HRASG12V E37G, which allows binding and activation of Ral-GEF but not Raf or PI3K; HRASG12V Y40G, which allows binding and activation of PI3K but not Raf or Ral-GEF; and HRASG12V T35S, which allows binding and activation of Raf but not Ral-GEF or PI3K in MCF10A cells. We used similar amounts of HRAS mutant plasmids for the transfection and used the pEGFP plasmid to monitor the transfection efficiency. To control the different HRAS mutant expression levels, we used the same amount of protein in each mutant and also analyzed HRAS and GFP protein expression (see Fig. S5A in the supplemental material). Then, we analyzed SLC5A8 expression, the methylation status of SLC5A8, and DNMT activity. Neither HRASG12V E37G nor HRASG12V Y40G was able to silence SLC5A8 expression, induce DNMT1 expression, or increase SLC5A8 promoter methylation. On the other hand, HRASG12V T35S was able to elicit all three of those effects (see Fig. S5B to D in the supplemental material). These results suggest that HRAS-induced silencing of SLC5A8 occurs through activation of RAF/MEK/ERK signaling rather than through RalGDS/Ral/RBP1 and PI3K/AKT/mTOR signaling. This observation was also confirmed by inhibition of M4 cells, which constitutively express active HRASG12V, with MEK inhibitor (U0126) that reactivates SLC5A8 expression and function (see Fig. 7B; see Fig. S14E and F in the supplemental material).
Fig 7.
Reactivation of endogenous Slc5a8 inhibits mammary tumorigenesis and reduces tumor cell survival. (A) RT-PCR and FACS analysis showing that siRNA-induced silencing of HRAS in M4 cells reactivates endogenous SLC5A8 gene expression (inset) and induces apoptosis in the presence of butyrate or pyruvate (HDAC inhibitors and substrates for SLC5A8; 1 mM). TSA (1 μM) was used as a positive control for HDAC inhibition. The data are means ± SEM from three independent experiments. (B) RT-PCR and FACS analysis showing that blockade of the ERK signaling pathway by U0126 reactivates endogenous SLC5A8 gene expression (inset) and induces apoptosis in the presence of butyrate (But) or pyruvate (Pyr). The data are means ± SEM from three independent experiments. (C) RT-PCR and FACS analysis showing that inactivation of DNMTs by 5-AzaDC and of DNMT1 by P-NH2 reactivates endogenous SLC5A8 gene expression (inset) and induces apoptosis in the presence of butyrate. The data are means ± SEM from three independent experiments. (D) Tumor incidence in MMTV-Hras-Tg mice without any treatment and in MMTV-Hras-Tg mice treated with either 5-Aza or butyrate, independently or together, by implantation of slow-release pellets. The data are means and SEM (number of mice in each group, 12). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test. (Inset) RT-PCR analysis showing the reactivation of Slc5a8 gene expression in mammary glands of mice treated with 5-azacytidine, butyrate, or 5-azacytidine plus butyrate. (E and F) Tumor incidence (E) and age of mice at the time of tumor onset (F) in MMTV-Hras/Slc5a8+/+ transgenic mice and MMTV-Hras/Slc5a8−/− double-transgenic mice under conditions of no treatment (control) or treatment with 5-azacytidine, treatment with butyrate, or treatment with 5-azacytidine plus butyrate via implantation of slow-release pellets. The data are means ± SEM (number of mice in each group, 12). **, P < 0.01; ***, P < 0.001 by t test.
Slc5a8 inactivation is an early event in MMTV-Hras-driven murine breast cancer.
To translate our in vitro findings with cell lines to in vivo circumstances, we analyzed Hras expression in normal mammary glands, mammary glands from 5-month-old MMTV-Hras-Tg mice that were premalignant with no visible tumors, and breast tumors collected from 10- to 12-month-old MMTV-Hras-Tg mice. RAS was activated in premalignant and mammary-tumor tissues (Fig. 4A). In conjunction, Slc5a8 expression was significantly decreased in premalignant mammary tissue and decreased further in mammary-tumor tissues (Fig. 4B and C). The decrease in Slc5a8 expression followed the increased methylation status of the Slc5a8 promoter (Fig. 4D). To test whether the epithelial contents in normal, premalignant, and tumor tissues were comparable, we analyzed the epithelial cell markers E-cadherin and K18. We found that E-cadherin expression was also lower in premalignant and tumor tissues than in the normal mammary gland (see Fig. S6 in the supplemental material). However, keratin 18 expression levels were comparable in all three tissues (see Fig. S6).
Fig 4.
Slc5a8 inactivation is an early event in MMTV-Hras-driven mammary tumorigenesis. (A) RAS kinase assay demonstrating RAS activation in premalignant mammary tissue and in tumor tissue derived from MMTV-Hras-Tg mice. WT, wild type. (B and C) RT-PCR and Western analyses demonstrating that Slc5a8 gene transcript and SLC5A8 protein expression are downregulated in premalignant mammary tissue and in tumor tissue derived from MMTV-Hras-Tg mice. (D) MS-PCR analysis demonstrating Slc5a8 methylation in premalignant mammary tissue and in tumor tissue derived from MMTV-Hras-Tg mice. (E) Representative images of FACS data showing induction of cancer-initiating stem cells (Lin− CD49fhigh CD24low) in premalignant mammary tissue and in tumor tissue derived from MMTV-Hras-Tg mice. (F) Representative images of mammospheres (wild-type and premalignant mammary glands) and tumorospheres (tumor tissue) demonstrating increased numbers of mammospheres and tumorospheres in premalignant and tumor tissues. (G) Quantification of mammospheres and tumorospheres in premalignant and tumor tissue. The data are means ± SEM. ***, P < 0.001 by t test. (H) Representative immunofluorescence data showing induced expression of DNMT1 and CD48f in premalignant and tumor tissues. DAPI was used to label nuclei. (I) DNMT enzymatic activity showing increased DNMT activity in premalignant mammary and tumor tissues of MMTV-Hras-Tg mice. The data are means and SEM. ***, P < 0.001 by t test.
Further, a longstanding question in cancer research has been whether cancer arises through mutations in stem cells or whether differentiated cells undergoing malignant transformation reacquire stem cell characteristics through a process of dedifferentiation (46, 47). Studies have shown that promoter DNA methylation of the repressed genes, such as SLC5A8, could lock into stem cell phenotypes and initiate abnormal clonal expansion and cancer (48). To determine whether there is a functional link between loss of Slc5a8 expression and formation of mammary stem cells, mammary progenitor cells, and cancer stem cells, we collected mammary glands from healthy mice and premalignant glands and tumor tissues from MMTV-Hras-Tg mice. From these tissues, we isolated mammary stem cells and progenitor cells using specific cell surface markers. The normal tissue and premalignant tissue had three distinct cell populations: fibroblasts and stromal cells (Lin− CD49f−/CD24−), progenitor cells (Lin− CD49flow/CD24high), and stem cells (Lin− CD49fhigh/CD24low). However, the tumor tissue had only one population, which was almost exclusively stem cells (Lin− CD49fhigh/CD24low). There was a gradual increase in the number of stem cells from normal to premalignant to tumor (9.87% versus 19.9% versus 34.6%) (Fig. 4E). We also analyzed Slc5a8 expression in stromal, luminal, and myoepithelial or basal cell types from the normal mouse mammary gland. Slc5a8 is mainly expressed in basal and relatively less in luminal but not in stromal cell types (see Fig. S7A and B in the supplemental material). Interestingly, Slc5a8 expression was significantly lower in cancer stem cells than in the normal basal cell type (see Fig. S7C). Further, to correlate the expression of Slc5a8 with that of DNMT1 in stem cells, we generated mammospheres (wild type and premalignant) and tumorospheres (tumor tissue). The mammospheres from stem cells of normal mammary glands were smaller and fewer than those from stem cells of premalignant glands. The values were even larger for the tumorospheres from stem cells of tumor tissues (Fig. 4F and G). The expression of Dnmt1 and CD49f was very low or undetectable in mammospheres generated from the wild-type mammary gland. However, their expression was significantly induced in mammospheres generated from premalignant mammary glands. The increase was even greater in tumorospheres developed from tumor tissues (Fig. 4H and I). These observations suggest that Hras-induced mammary tumor formation leads to Dnmt1 activation and subsequent Slc5a8 inactivation. This is associated with increased cancer-initiating stem cell formation.
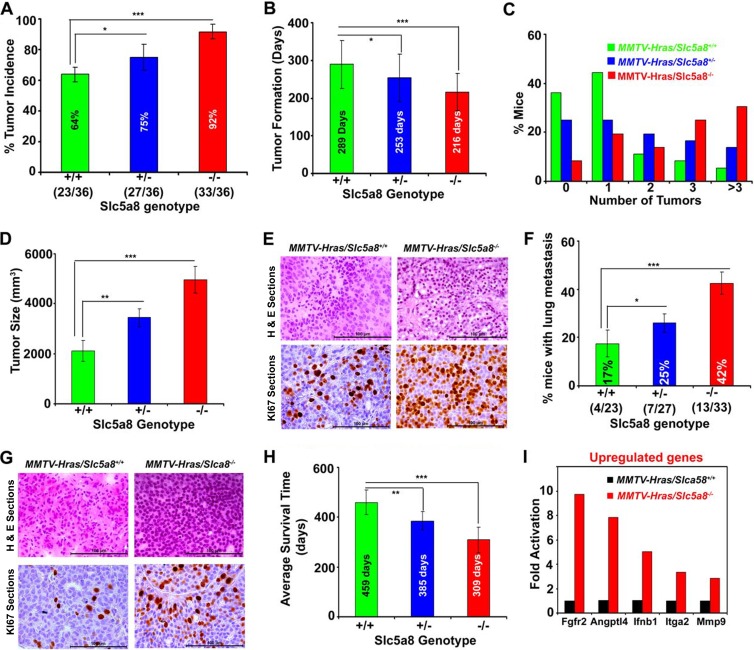
Deletion of Slc5a8 in mice is associated with early onset of mammary tumor formation, accelerated lung metastasis, and decreased overall survival.
If SLC5A8 functions in vivo as a tumor suppressor, absence of the transporter should promote breast cancer. We tested this in a mouse model of MMTV-Hras-driven mammary tumorigenesis by comparing the incidence, progression, and lung metastasis of breast cancer between MMTV-Hras/Slc5a8+/+ mice and MMTV-Hras/Slc5a8−/− mice. The incidence of tumors was 64% in MMTV-Hras/Slc5a8+/+ mice, 75% in MMTV-Hras/Slc5a8+/− mice, and 92% in MMTV-Hras/Slc5a8−/− mice (Fig. 5A), indicating that deletion of Slc5a8 increased the incidence of tumors in a dose-dependent manner in MMTV-Hras-Tg mice. The deletion of Slc5a8 also significantly decreased the age of the animals at which tumors began to appear and increased the number of tumors per animal and the tumor size (Fig. 5B to D). Tumors in MMTV-Hras/Slc5a8−/− mice were aggressive; histopathology and Ki-67 immunostaining suggested that these tumors were highly invasive ductal adenocarcinomas (Fig. 5E; see Fig. S8A and B in the supplemental material). There were also marked differences in cancer stem cell populations in tumors depending on the Slc5a8 genotype. The population of cancer stem cells (Lin− CD49fhigh/CD24low) increased from 17% in MMTV-Hras/Slc5a8+/+ mice to 35% in MMTV-Hras/Slc5a8+/− mice and to 48% in MMTV-Hras/Slc5a8−/− mice (see Fig. S9 in the supplemental material). Tumors in MMTV-Hras/Slc5a8+/+ mice metastasized to the lung occasionally, but the incidence of lung metastasis increased significantly with Slc5a8 deletion (Fig. 5F). The time needed for lung metastasis decreased, and the multiplicity of metastatic nodules increased dramatically with Slc5a8 deletion (see Fig. S10A and B in the supplemental material). Histopathology and Ki-67 immunostaining of the nodules indicated that the metastasized tumors were more aggressive in MMTV-Hras/Slc5a8−/− mice than in MMTV-Hras/Slc5a8+/+ mice (Fig. 5G). These changes in MMTV-Hras/Slc5a8−/− mice were accompanied by substantially decreased survival time (Fig. 5H).
Fig 5.
Deletion of Slc5a8 in mice is associated with early onset of mammary tumorigenesis, accelerated lung metastasis, and decreased overall survival. (A to H) Tumor incidence (A), age of animals at tumor onset (B), number of tumors in each mouse (C), tumor size (D), H&E and Ki-67 staining of tumors (E), lung metastasis (F), H&E and Ki-67 staining of metastatic lung tumors (G), and average survival time (H) in MMTV-Hras/Slc5a8+/+ mice, MMTV-Hras/Slc5a8+/− mice, and MMTV-Hras/Slc5a8−/− mice. The data are means ± SEM (number of mice in each group, 36). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test. (I) Upregulation of specific genes in MMTV-Hras/Slc5a8−/− mouse tumors compared to tumors from MMTV-Hras/Slc5a8+/+ mice (number of mice in each group, 3).
To investigate the molecular mechanisms underlying the aggressiveness of mammary tumors associated with Slc5a8 deletion, we subjected tumor tissue RNA samples from MMTV-Hras/Slc5a8+/+ mice and MMTV-Hras/Slc5a8−/− mice to cancer pathway finder real-time PCR array analysis. The five most upregulated and the five most downregulated genes are shown in Fig. 5I (see Fig. S11 in the supplemental material). The upregulated genes included the prometastatic genes Fgfr2, Angptl4, and Mmp9, and the downregulated genes included the anti-proliferative gene Cdkn2a.
Carcinogen-induced mammary tumors are associated with HRAS activation and subsequent SLC5A8 inactivation.
We evaluated the interplay among HRAS, DNMT1, and SLC5A8 in a carcinogen-induced murine mammary-tumor model to corroborate our findings in MMTV-Hras mice. We used DMBA [7,12-dimethylbenz(a)anthracene] to induce mammary tumors in mice. DMBA-induced mammary tumors have been shown to result in a gain-of-function mutation in HRAS (49, 50). We found decreased expression of Slc5a8 in DMBA-induced tumors (see Fig. S12A and B in the supplemental material). We then compared the carcinogenic potentials of DMBA in wild-type mice and Slc5a8−/− mice. The incidence of carcinogen-induced tumors was significantly higher in Slc5a8−/− mice than in wild-type mice (see Fig. S12C in the supplemental material). DNMT activity was higher in tumors than in normal mammary glands, but the difference was increased even further with Slc5a8 deletion (see Fig. S12D in the supplemental material).
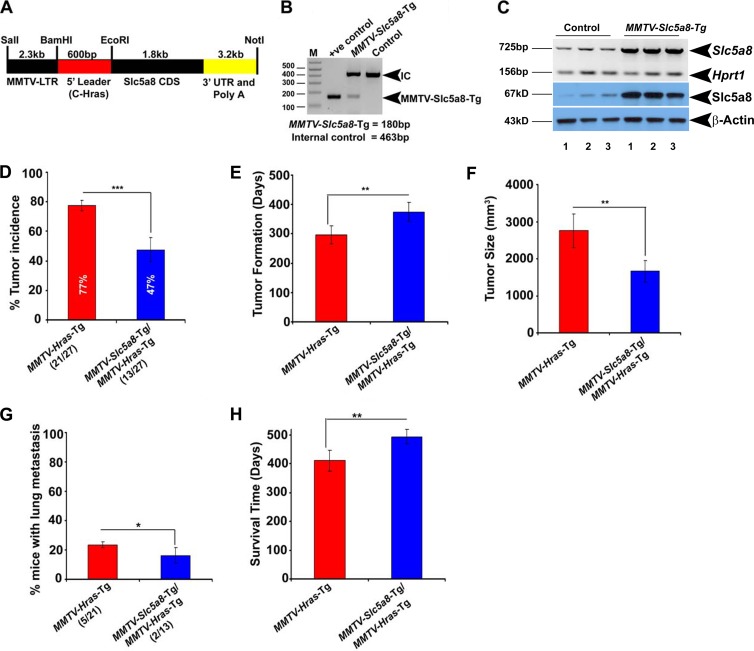
Overexpression of SLC5A8 in mammary glands protects against Hras-driven tumorigenesis.
The tumor-suppressive function of SLC5A8 in mammary glands would be further supported if overexpression of the transporter specifically in mammary glands protects against breast cancer. To examine this, we generated the MMTV-Slc5a8 transgenic mouse for mammary-gland-specific overexpression of SLC5A8 (Fig. 6A and B). Since the transgene is under the control of the MMTV promoter, its expression is not silenced in tumors. The specific expression of the transgene in mammary glands was confirmed at the mRNA and protein levels (Fig. 6C; see Fig. S13A in the supplemental material). We then used this mouse line to generate MMTV-Hras/MMTV-Slc5a8 double-transgenic mice, as confirmed by PCR (see Fig. S13B in the supplemental material). We compared tumor incidence, time to tumor onset, tumor size, lung metastasis, and average survival time between MMTV-Hras mice and MMTV-Hras/MMTV-Slc5a8 mice. As shown in Fig. 6D to H, mammary-gland-specific overexpression of SLC5A8 provided significant protection against Hras-induced mammary tumorigenesis, delayed the time of tumor onset, decreased the tumor size and lung metastasis, and increased the survival time.
Fig 6.
Mammary-gland-specific expression of SLC5A8 in mice protects against MMTV-Hras-driven mammary tumorigenesis. (A) Diagrammatic representation of the construct for generation of MMTV-Slc5a8-Tg mice. CDS, coding sequence. (B) Genotype analysis confirming the expression of the transgene. IC, internal control. (C) RT-PCR and Western blot analyses demonstrating overexpression of the Slc5a8 gene transcript and protein in mammary glands from MMTV-Slc5a8-Tg mice. (D to H) Tumor incidence (D), age of mice at tumor onset (E), tumor size (F), lung metastasis (G), and average survival time (H) in MMTV-Hras-Tg mice and MMTV-Hras/Slc5a8 double-transgenic mice. The data are means ± SEM (number of mice in each group, 27). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by t test.
Blockade of tumor-associated silencing of endogenous Slc5a8 decreases mammary-tumor growth and progression.
SLC5A8-dependent tumor suppression is related to its ability to transport the HDAC inhibitors butyrate and pyruvate into cells. This was confirmed by reactivation of SLC5A8 in the metastatic breast cancer cell lines M4 and MB231 by siRNA-induced silencing of HRAS and reduced cell survival in the presence of its substrates butyrate and pyruvate (Fig. 7A; see Fig. S14A in the supplemental material). Silencing of HRAS-induced SLC5A8 expression in these cells and subsequent exposure of the cells to butyrate or pyruvate caused apoptosis. Similar results were obtained if SLC5A8 was induced with an inhibitor of RAF/MEK/ERK signaling (Fig. 7B; see Fig. S14B in the supplemental material) or by siRNA-induced silencing of C/EBPβ (see Fig. S14C in the supplemental material). In these experiments, we used TSA, which causes HDAC inhibition independently of SLC5A8, as a positive control.
Since tumor-associated silencing of SLC5A8 occurs through DNA methylation, we reactivated SLC5A8 in M4 and MB231 cells with 5-AzaDC, a pan-DNMT inhibitor, and P-NH2, a DNMT1-specific inhibitor, and examined the influence of butyrate and pyruvate on cell survival. Again, exposure of the cells to these HDAC inhibitors following reactivation of SLC5A8 promoted apoptosis (Fig. 7C; see Fig. S14D in the supplemental material). To test whether the reactivation of SLC5A8 by HRAS siRNA, C/EBPβ siRNA, MEK, and DNMT inhibitors is functional in M4 and MB231 cells, we performed nicotinate uptake assays (see Fig. S14E and F in the supplemental material). Further, we confirmed these findings in vivo using MMTV-Hras transgenic mice. We implanted 90-day slow-release 5-azacytidine (5-Aza) and butyrate tablets, either alone or in combination, in 12-week-old MMTV-Hras mice and then monitored the tumor incidence and time to tumor onset. We also harvested tumor tissues from these mice and analyzed Slc5a8 expression and measured DNMT and HDAC activities. As shown in Fig. 7D, treatment with 5-Aza decreased tumor incidence, and the combination of 5-Aza and butyrate decreased it even further. The same was true for tumor incidence and the time required for tumor onset (Fig. 7E and F). As expected, the DNMT activity was less in tumor tissues from 5-Aza-treated mice than in tumor tissues from untreated mice, and HDAC activity was less in tumor tissues from 5-Aza–butyrate-treated mice than in tumor tissues from untreated mice (see Fig. S15 in the supplemental material). Interestingly, 5-Aza and butyrate implantation itself reactivated SLC5A8 expression significantly, and this induction was further accelerated by the combination of 5-Aza plus butyrate (Fig. 7D, inset; see Fig. S16A and B in the supplemental material). Since 5-Aza is expected to induce not only SLC5A8 but also other genes, we also performed these experiments in MMTV-Hras/Slc5a8+/+ mice and MMTV-Hras/Slc5a8−/− mice in parallel. The effects of 5-Aza and 5-Aza plus butyrate were significantly blunted in MMTV-Hras/Slc5a8−/− mice compared to MMTV-Hras/Slc5a8+/+ mice (Fig. 7E and F), indicating the relevance of reactivated Slc5a8 to the observed findings. We also analyzed Slc5a8 expression in MMTV-Hras/Slc5a8+/+ mice and MMTV-Hras/Slc5a8−/− mice that were treated with 5-Aza, butyrate, and a combination of the two. As shown in Fig. S16C and D in the supplemental material, 5-Aza, butyrate, and the combination reactivated Slc5a8 only in MMTV-Hras/Slc5a8+/+ mice but not in MMTV-Hras/Slc5a8−/− mice. Interestingly, butyrate treatment itself significantly reduced tumor incidence and onset of tumor formation in wild-type mice but not in Slc5a8-null mice, suggesting that functional Slc5a8, even a residual amount, is required to transport butyrate in the tumor tissue. Further, treatment of breast cancer cell lines with butyrate alone modestly induced apoptosis, and the combination of 5-Aza plus butyrate significantly enhanced the effect (Fig. 7A), while butyrate transplantation itself in MMTV-Hras-Tg mice significantly reduced tumor incidence and onset (Fig. 1D to F), suggesting that persistent butyrate exposure could relieve SLC5A8 from the suppressor complex and reactivate its expression or that butyrate itself induces SLC5A8 transcription (Fig. 7D; see Fig. S16 in the supplemental material). However, this observation needs to be confirmed by detailed analysis.
SLC5A8-transported butyrate and pyruvate, but not lactate and nicotinate, induce apoptosis and suppress tumor growth.
To demonstrate that the tumor suppressor function of SLC5A8 is mainly associated with its transportable substrates butyrate, pyruvate, and propionate, which inhibit HDACs, but not due to acetate, lactate, and nicotinate, which do not inhibit HDACs, we made a TetOn system-regulated lentivirus-mediated stable cell line in the metastatic breast cancer cell line M4. SLC5A8 expression and function were confirmed in the presence and absence of doxycycline (2 μg/ml) for 24 h (see Fig. S17A to C in the supplemental material). Then, we treated these cells or not with the SLC5A8 substrates butyrate, pyruvate, and propionate, which inhibit HDACs, and acetate, lactate, and nicotinate, which do not inhibit HDACs, and measured apoptosis. Butyrate, pyruvate, and propionate induced significant apoptosis in SCL5A8-induced M4 cells, but acetate, lactate, and nicotinate were unable to induce apoptosis in these cells (see Fig. S17D in the supplemental material). To analyze the tumor suppressor function of SLC5A8, we made a mouse xenograft using an M4-SLC5A8-pLVX stable cell line. SLC5A8 expression was induced with doxycycline (2 mg/ml in drinking water) and then treated with butyrate, pyruvate, propionate, acetate, lactate, and nicotinate (2 mg/ml) in drinking water. As shown in Fig. S17E to G in the supplemental material, butyrate, pyruvate, and propionate efficiently reduced tumor growth, but there were no significant changes in acetate-, lactate-, and nicotinate-treated animals (see Fig. S17H to J in the supplemental material). These results confirmed that SLC5A8-induced tumor suppression is mainly associated with its ability to transport the HDAC inhibitors butyrate, pyruvate, and propionate but not acetate, lactate, and nicotinate.
Constitutively active HRAS expression induces MCT1 expression and function in normal nontransformed mammary epithelial cell lines.
A recent study has shown that the monocarboxylate transporter MCT1, which also transports butyrate, pyruvate, and lactate in a sodium-independent manner, is silenced in some breast cancer cell lines (51). Because both SLC5A8 and MCT1 transport monocarboxylates, we wanted to analyze whether MCT1 also plays a role in HRAS-induced cellular transformation and associated mammary-tumor formation. First, we analyzed MCT1 expression and function in M1, M2, M3, and M4 cells. MCT1 expression and function were higher in the HRAS-induced transformed cell line (M2) than in M1 cells and further increased in M3 and M4 cells (see Fig. S18A and B in the supplemental material). Further, we also tested the expression and functional status of MCT1 in M1 and M4 cells treated with DNMT inhibitors (5-AzaDC and P-NH2) and a MEK inhibitor (U0126). As shown in Fig. S18C and D in the supplemental material, DNMT inhibitors did not affect either MCT1expression or function. However, the MEK inhibitor significantly reduced MCT1 expression and function, suggesting that HRAS-induced MCT1 expression does not occur through HRAS-induced DNA methylation. We also confirmed the involvement of HRAS in the activation of MCT1 expression and function using HRAS siRNA (see Fig. S18E and F in the supplemental material). Finally, we analyzed Mct1 expression in normal and tumor tissues obtained from DMBA-induced mammary tumors, as well as MMTV-HRAS-Tg mouse mammary tumors. The expression of MCT1 was lower in normal mammary tissues, but there was a significant increase in mammary-tumor tissues (see Fig. S18G in the supplemental material). All these results suggest that MCT1 and SLC5A8 are reciprocally regulated by HRAS in the mammary epithelium and that constitutively active HRAS silences SLC5A8 but activates MCT1.
DISCUSSION
SLC5A8 is a candidate tumor suppressor whose inactivation is a common event in many cancers. Even though tumor-associated silencing of SLC5A8 has been shown to occur primarily through DNA methylation, little is known about the signaling pathways that underlie this phenomenon. Here, we have shown for the first time that oncogenic HRAS is responsible for this process. Oncogenic RAS proteins are commonly detected in many cancers, suggesting that RAS plays a key role in tumor development (52, 53). Although mutations of RAS genes are rare in breast cancer, >50% of breast carcinomas express elevated levels of HRAS protein (37–39, 54). Activated RAS plays an important role in epigenetic inactivation of several tumor suppressor genes, and there is evidence for functional cross talk between oncogenic HRAS and DNMT1 (33, 55). Here, we have shown that the candidate tumor suppressor SLC5A8 is a downstream target for HRAS/DNMT1 and that the RAS/RAF/MAPK pathway is involved in the process. The microarray data also confirmed that SLC5A8 is the most abundantly downregulated gene in HRAS-transformed premalignant, malignant, and metastatic breast cancer cells. SLC5A8 inactivation in papillary thyroid carcinoma is associated with BRAF mutation (56). BRAF, a member of the RAF family of serine/threonine kinases, mediates cellular responses to growth signals through the RAS/RAF/MAPK signaling pathway (57, 58), and BRAF-activating mutations have been reported in more than 70% of cancers (59, 60). Since there is a positive correlation between BRAF-activating mutations and DNA hypermethylation in cancer (61), we speculate that activated BRAF may silence SLC5A8 in cancer through DNA hypermethylation. We have shown in the present study that Slc5a8 is silenced in murine mammary tumors, either induced by Hras or caused by a carcinogen. The silencing of Slc5a8 in MMTV-Hras mouse tumors is associated with an increase in tumor-initiating stem cells (Lin− CD49fhigh CD24low), suggesting that prevention of cancer stem cell formation is one of the mechanisms underlying the tumor-suppressive function of SLC5A8.
Even though several studies have shown tumor-associated silencing of SLC5A8 in a variety of cancers and have demonstrated the tumor-suppressive function of the transporter in vitro with cancer cell lines, there has been no documentation of this transporter functioning as a tumor suppressor in vivo. Here, we have provided unequivocal evidence for its tumor-suppressive role in vivo using multiple animal models and experimental approaches. Deletion of Slc5a8 in the MMTV-Hras transgenic-mouse model of mammary tumors increases tumor incidence, accelerates the onset of tumors, increases the size and number of tumors, and promotes lung metastasis. Similarly, Slc5a8−/− mice are more susceptible to carcinogen-induced mammary tumors. Overexpression of SLC5A8 in mammary glands decreases tumor incidence and progression and also lung metastasis. These data provide strong evidence that SLC5A8 functions as a tumor suppressor in vivo. A recent study (42) attempted to examine the role of SLC5A8 in tumor progression in vivo using Slc5a8−/− mice. The investigators focused on colon cancer because SLC5A8 was first identified as a putative tumor suppressor in the colon (6). With the ApcMin/+ mouse as a model of intestinal/colon cancer, the investigators found no difference in colon cancer incidence and progression between wild-type mice and Slc5a8−/− mice, leading to the conclusion that SLC5A8 does not function as a tumor suppressor in vivo. Our present studies with breast cancer have yielded a totally opposite conclusion. The differences in the concentrations of butyrate in the colon versus mammary glands might offer a rational explanation for the discrepancy between the two studies. Butyrate is present in millimolar concentrations in the colon as a result of bacterial fermentation of dietary fiber. Since SLC5A8 transports butyrate with high affinity, the transporter is obligatory for butyrate entry into cells only at micromolar concentrations; butyrate at millimolar concentrations can enter the cells by diffusion without the transporter. This is not the case in mammary glands, in which pyruvate rather than butyrate is the metabolite that is relevant to SLC5A8-dependent HDAC inhibition. Pyruvate is present in circulation only at micromolar concentrations, at which the transporter is obligatory for the entry of the metabolite into cells.
If SLC5A8 is silenced in breast cancer, how can the transporter be exploited for cancer treatment? Our present studies address this issue. Silencing of SLC5A8 in tumors occurs via DNA methylation, and inhibitors of DNA methylation induce reactivation of SLC5A8. We have shown in the present study that treatment of mice with inhibitors of DNA methylation induces SLC5A8 expression in breast cancer cell lines and in mouse models of breast cancer. This reactivation of the gene is associated with tumor suppression. These data show that pharmacological strategies to reactivate SLC5A8 in tumors might have potential in cancer treatment. 5-Azacytidine is in clinical use for treatment of certain cancers. This DNA methylation inhibitor might also have potential in the treatment of breast cancer. Our previous studies have shown that SLC5A8 is silenced in all subtypes of breast cancer: estrogen receptor (ER) positive, ER negative, and triple negative (11). This means that pharmacological reactivation of this tumor suppressor may have potential for treatment of breast cancer irrespective of its ER status.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bert Vogelstein for providing DNMT1 knockdown isogenic HCT116 cell lines. We also thank Fred Miller and Steven Santner for providing MCF10A-derived cell lines.
This work was supported by grants from the National Institutes of Health (R01 CA131402) and the Department of Defense (BC074289) and by a GHSU Intramural Pilot Study grant and GHSU startup funds (M.T.).
S.E., V.G., and M.T. designed the research; S.E., S.R., and M.T. performed expression analysis, DNMT and HDAC assays, FACS analysis, and animal experiments; R.P. performed experiments related to stem cells, mammospheres, and tumorospheres; S.E., P.D.P., P.V.S., S.B.S., and M.T. performed siRNA, promoter, and ChIP assays; S.A., S.R.S., and E.B. performed uptake assays; L.H. performed microarray analysis; T.B. generated Slc5a8−/− mice; and S.E., V.G., and M.T. wrote the manuscript.
We declare no conflict of interest.
Footnotes
Published ahead of print 5 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01702-12.
REFERENCES
- 1.Coady MJ, Chang MH, Charron FM, Plata C, Wallendorff B, Sah JF, Markowitz SD, Romero MF, Lapointe JY. 2004. The human tumor suppressor gene SLC5A8 expresses a Na+-monocarboxylate cotransporter. J. Physiol. 557:719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyauchi S, Gopal E, Fei YJ, Ganapathy V. 2004. Functional identification of SLC5A8, a tumor suppressor down-regulated in colon cancer, as a Na+-coupled transporter for short-chain fatty acids. J. Biol. Chem. 279:13293–13296 [DOI] [PubMed] [Google Scholar]
- 3.Gopal E, Fei YJ, Sugawara M, Miyauchi S, Zhuang L, Martin P, Smith SB, Prasad PD, Ganapathy V. 2004. Expression of slc5a8 in kidney and its role in Na+-coupled transport of lactate. J. Biol. Chem. 279:44522–44532 [DOI] [PubMed] [Google Scholar]
- 4.Martin PM, Gopal E, Ananth S, Zhuang L, Itagaki S, Prasad BM, Smith SB, Prasad PD, Ganapathy V. 2006. Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ketone bodies in the brain. J. Neurochem. 98:279–288 [DOI] [PubMed] [Google Scholar]
- 5.Gopal E, Fei YJ, Miyauchi S, Zhuang L, Prasad PD, Ganapathy V. 2005. Sodium-coupled and electrogenic transport of B-complex vitamin nicotinic acid by slc5a8, a member of the Na/glucose co-transporter gene family. Biochem. J. 388:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Kasturi L, Lutterbaugh J, Rerko RM, Casey G, Issa JP, Willis J, Willson JK, Plass C, Markowitz SD. 2003. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc. Natl. Acad. Sci. U. S. A. 100:8412–8417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganapathy V, Thangaraju M, Gopal E, Itagaki S, Miyauchi S, Prasad PD. 2008. Sodium-coupled monocarboxylate transporters in normal tissues and in cancer. AAPS J. 10:193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganapathy V, Thangaraju M, Prasad PD. 2009. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol. Ther. 121:29–40 [DOI] [PubMed] [Google Scholar]
- 9.Gupta N, Martin PM, Prasad PD, Ganapathy V. 2006. SLC5A8 (SMCT1)-mediated transport of butyrate forms the basis for the tumor suppressive function of the transporter. Life Sci. 78:2419–2425 [DOI] [PubMed] [Google Scholar]
- 10.Davie JR. 2003. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133(Suppl. 7):2485S–2493S [DOI] [PubMed] [Google Scholar]
- 11.Thangaraju M, Gopal E, Martin PM, Ananth S, Smith SB, Prasad PD, Sterneck E, Ganapathy V. 2006. SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res. 66:11560–11564 [DOI] [PubMed] [Google Scholar]
- 12.Thangaraju M, Cresci G, Itagaki S, Mellinger J, Browning DD, Berger FG, Prasad PD, Ganapathy V. 2008. Sodium-coupled transport of the short-chain fatty acid butyrate by SLC5A8 and its relevance to colon cancer. J. Gastrointest. Surg. 12:1773–1782 [DOI] [PubMed] [Google Scholar]
- 13.Thangaraju M, Carswell KN, Prasad PD, Ganapathy V. 2009. Colon cancer cells maintain low levels of pyruvate to avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem. J. 417:379–389 [DOI] [PubMed] [Google Scholar]
- 14.Insinga A, Monestrioli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, Altucci L, Nervi C, Minucci S, Pelicci PG. 2005. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat. Med. 11:71–76 [DOI] [PubMed] [Google Scholar]
- 15.Nakata S, Yoshida T, Horinaka M, Shiraishi T, Wakada M, Sakai T. 2004. Histone deacetylase inhibitors upregulate death receptor 5/TRAIL-R2 and sensitize apoptosis induced by TRAIL/APO2-L in human malignant tumor cells. Oncogene 23:6261–6271 [DOI] [PubMed] [Google Scholar]
- 16.Nebbioso A, Clarke N, Voltz E, Germain E, Ambrosino C, Bontempo P, Alvarez R, Schiavone EM, Ferrara F, Bresciani F, Weisz A, de Lera AR, Gronemeyer H, Altucci L. 2005. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat. Med. 11:77–84 [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Tan J, Zhang L, Jiang X, Liu ET, Yu Q. 2005. Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc. Natl. Acad. Sci. U. S. A. 102:16090–16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopal E, Miyauchi S, Martin PM, Ananth S, Roon P, Smith SB, Ganapathy V. 2007. Transport of nicotinate and structurally related compounds by human SMCT1 (SLC5A8) and its relevance to drug transport in the mammalian intestinal tract. Pharm. Res. 24:575–584 [DOI] [PubMed] [Google Scholar]
- 19.Manning TS, Gibson GR. 2004. Microbial-gut interactions in health and disease. Prebiotics. Best Pract. Res. Clin. Gastroenterol. 18:287–298 [DOI] [PubMed] [Google Scholar]
- 20.Thangaraju M, Ananth S, Martin PM, Roon P, Smith SB, Sterneck E, Prasad PE, Ganapathy V. 2006. c/ebpdelta null mouse as a model for the double knock-out of slc5a8 and slc5a12 in kidney. J. Biol. Chem. 281:26769–26773 [DOI] [PubMed] [Google Scholar]
- 21.Belinsky SA, Nikula KJ, Baylin SB, Issa J-P. 1996. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc. Natl. Acad. Sci. U. S. A. 93:4045–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Deiry WS, Nelkin BD, Celano P, Yen RW, Falco JP, Hamilton SR, Baylin SB. 1991. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc. Natl. Acad. Sci. U. S. A. 88:3470–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PA, Baylin SB. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415–428 [DOI] [PubMed] [Google Scholar]
- 24.Kautiainen TL, Jones PA. 1986. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J. Biol. Chem. 261:1594–1598 [PubMed] [Google Scholar]
- 25.Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 27.Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferases gene results in embryonic lethality. Cell 69:915–926 [DOI] [PubMed] [Google Scholar]
- 28.Szyf M. 1991. DNA methylation patterns: an additional level of information? Biochem. Cell Biol. 69:764–767 [DOI] [PubMed] [Google Scholar]
- 29.Wu J, Issa JP, Herman J, Bassett DE, Jr, Nelkin BD, Baylin SB. 1993. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc. Natl. Acad. Sci. U. S. A. 90:8891–8895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81:197–205 [DOI] [PubMed] [Google Scholar]
- 31.Bigey P, Ramchandani S, Theberg JM, Araujo FD, Szyf M. 2000. Transcriptional regulation of the human DNA methyltransferase (dnmt1) gene. Gene 242:407–418 [DOI] [PubMed] [Google Scholar]
- 32.Detich N, Ramchandani S, Szyf M. 2001. A conserved 3′-untranslated element mediates growth regulation of DNA methyltransferase 1 and inhibits its transforming activity. J. Biol. Chem. 276:24881–24890 [DOI] [PubMed] [Google Scholar]
- 33.MacLeod AR, Rouleau J, Szyf M. 1995. Regulation of DNA methylation by the Ras signaling pathway. J. Biol. Chem. 70:11327–11337 [DOI] [PubMed] [Google Scholar]
- 34.Slack A, Cervoni N, Pinard M, Szyf M. 1999. DNA methyltransferase is a downstream effector of cellular transformation triggered by simian virus 40 large T antigen. J. Biol. Chem. 274:10105–10112 [DOI] [PubMed] [Google Scholar]
- 35.Rouleau J, Macleod AR, Szyf M. 1995. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J. Biol. Chem. 270:1595–1601 [DOI] [PubMed] [Google Scholar]
- 36.Bakin AV, Curran T. 1999. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science 283:387–390 [DOI] [PubMed] [Google Scholar]
- 37.Hand PH, Vilasi V, Thor A, Ohuchi N, Schlom J. 1987. Quantitation of Harvey ras p21 enhanced expression in human breast and colon carcinomas. J. Natl. Cancer Inst. 79:59–65 [PubMed] [Google Scholar]
- 38.Spandidos DA, Agnantis NJ. 1984. Human malignant tumors of the breast, as compared to their respective normal tissue, have elevated expression of the Harvey ras oncogene. Anticancer Res. 4:269–272 [PubMed] [Google Scholar]
- 39.Ohuchi N, Thor A, Page DL, Hand PH, Halter SA, Schlom J. 1986. Expression of the 21,000 molecular weight ras protein in a spectrum of benign and malignant human mammary tissues. Cancer Res. 46:2511–2519 [PubMed] [Google Scholar]
- 40.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. 1996. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 148:313–319 [PMC free article] [PubMed] [Google Scholar]
- 41.Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. 2001. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 65:101–110 [DOI] [PubMed] [Google Scholar]
- 42.Frank H, Groger N, Diener M, Becker C, Braun T, Boettger T. 2008. Lactaturia and loss of sodium-dependent lactate uptake in the colon of SLC5A8-deficient mice. J. Biol. Chem. 283:24729–24737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang G, Thompson JA, Fang B, Liu J. 2003. Silencing of H-ras gene expression by retrovirus-mediated siRNA decrease transformation efficiently and tumor growth in a model of human ovarian cancer. Oncogene 22:5694–5701 [DOI] [PubMed] [Google Scholar]
- 44.National Institutes of Health 1985. Principles of laboratory animal care. National Institutes of Health publication 85-23. National Institutes of Health, Bethesda, MD [Google Scholar]
- 45.Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakfield LM. 2003. TGF-beta switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J. Clin. Invest. 112:1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houghton J, Morozov A, Smirnova I, Wang TC. 2007. Stem cells and cancer. Semin. Cancer Biol. 17:191–203 [DOI] [PubMed] [Google Scholar]
- 47.Passegue E. 2006. Cancer biology: a game of subversion. Nature 442:754–755 [DOI] [PubMed] [Google Scholar]
- 48.Schuebel K, Chen W, Baylin SB. 2006. CIMPle origin for promoter hypermethylation in colorectal cancer? Nat. Genet. 38:738–740 [DOI] [PubMed] [Google Scholar]
- 49.Dandekar S, Sukumar S, Zarbl H, Young LJ, Cardiff RD. 1986. Specific activation of the cellular Harvey-ras oncogene in dimethylbenzanthracene-induced mouse mammary tumors. Mol. Cell. Biol. 6:4104–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zarbl H, Sukumar S, Arthur AV, Martin-Zanca D, Barbacid M. 1985. Direct mutagenesis of Ha-ras-1 oncogenes by N-nitroso-N-methylurea during initiation of mammary carcinogenesis in rats. Nature 315:382–385 [DOI] [PubMed] [Google Scholar]
- 51.Birsoy K, Wang T, Possemato R, Yilmaz OH, Koch CE, Chen WW, Hutchins AW, Gultekin Y, Peterson TR, Carette JE, Brummelkamp TR, Clish CB, Sabatini DM. 2013. MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat. Genet. 45:104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bardeesy N, DePinho RA. 2002. Pancreatic cancer biology and genetics. Nat. Rev. Cancer 2:897–909 [DOI] [PubMed] [Google Scholar]
- 53.Bos JL. 1989. Ras oncogenes in human cancer: a review. Cancer Res. 49:4682–4689 [PubMed] [Google Scholar]
- 54.De Biasi F, Del Sal G, Hand PH. 1989. Evidence of enhancement of the ras oncogene protein product (p21) in a spectrum of human tumors. Int. J. Cancer 43:431–435 [DOI] [PubMed] [Google Scholar]
- 55.Lund P, Weisshaupt K, Mikesha T, Jammas D, Chen X, Kuban RJ, Ungethum U, Krapfenbauer U, Herzel HP, Schafer R. 2006. Oncogenic H-Ras suppresses clusterin expression through promoter hypermethylation. Oncogene 25:4890–4903 [DOI] [PubMed] [Google Scholar]
- 56.Porra V, Ferraro-Peyret C, Durand C, Selmi-Ruby S, Giroud H, Berger-Dutrieux N, Decaussin M, Peix JL, Bournaud C, Orgiazzi J, Borson-Chazot F, Dante R, Rousset B. 2005. Silencing of the tumor suppressor gene SLC5A8 is associated with BRAF mutations in classical papillary thyroid carcinomas. J. Clin. Endocrinol. Metab. 90:3028–3035 [DOI] [PubMed] [Google Scholar]
- 57.Baccarini M. 2002. An old kinase on a new path: Raf and apoptosis. Cell Death Differ. 9:783–785 [DOI] [PubMed] [Google Scholar]
- 58.Peyssonnaux C, Eychene A. 2001. The Raf/MEK/ERK pathway: new concepts of activation. Biol. Cell 93:53–62 [DOI] [PubMed] [Google Scholar]
- 59.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949–954 [DOI] [PubMed] [Google Scholar]
- 60.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. 2002. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 418:934. [DOI] [PubMed] [Google Scholar]
- 61.Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, Jass JR, Tanaka N, Matsubara N. 2004. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J. Clin. Oncol. 22:4584–4594 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.