COMMENTARY
Eukaryotic cells are constantly bombarded with a plethora of extracellular and intracellular stresses that they must quickly respond to in order to survive (1). These stresses can come in the form of changes in temperature, nutrient availability, osmotic changes, and DNA-damaging events (extracellular), as well as oxidative stress from normal metabolism and replicative/transcriptional DNA damage (intracellular). In order to respond to a wide range of stresses, cells must be able to rapidly translate a stress response signal into a specific transcriptional program (2). While there are some common themes that underlie the general “environmental stress response” (ESR), the transcriptional programs each stressor initiates are unique and tailored to deal with each specific type of stress (3). The exact mechanisms underlying the cellular response to a particular stress are poorly defined and continue to be an exciting area of active research.
A question of particular significance is how the cell is able to modulate the chromatin environment surrounding the genes necessary to respond to specific stresses encountered by the cell. This is a complex problem, as many genes may need to be quickly, and precisely, up- or downregulated in response to each particular type of stress (3). In order to achieve this level of control, the cell must utilize one or more signaling cascades activated by the stressor to alter the chromatin landscape precisely at many genomic loci by recruiting a host of chromatin-modifying enzymes (4). It is likely that the posttranslational modifications (PTMs) created by these enzymes form a chromatin signature, or code, that can help to initiate the ESR through recruitment of chromatin effector proteins that dock on these histone PTMs (5–8). In this issue of Molecular and Cellular Biology, using Saccharomyces cerevisiae, Baker and colleagues (9) explore a connection between a complex containing the histone deacetylase Rpd3 and the ESR pathway, thereby increasing our understanding of how cell signaling and chromatin come together to regulate the cellular response to stress. As described below, these studies break new ground and give rise to many fascinating new questions that await future discoveries.
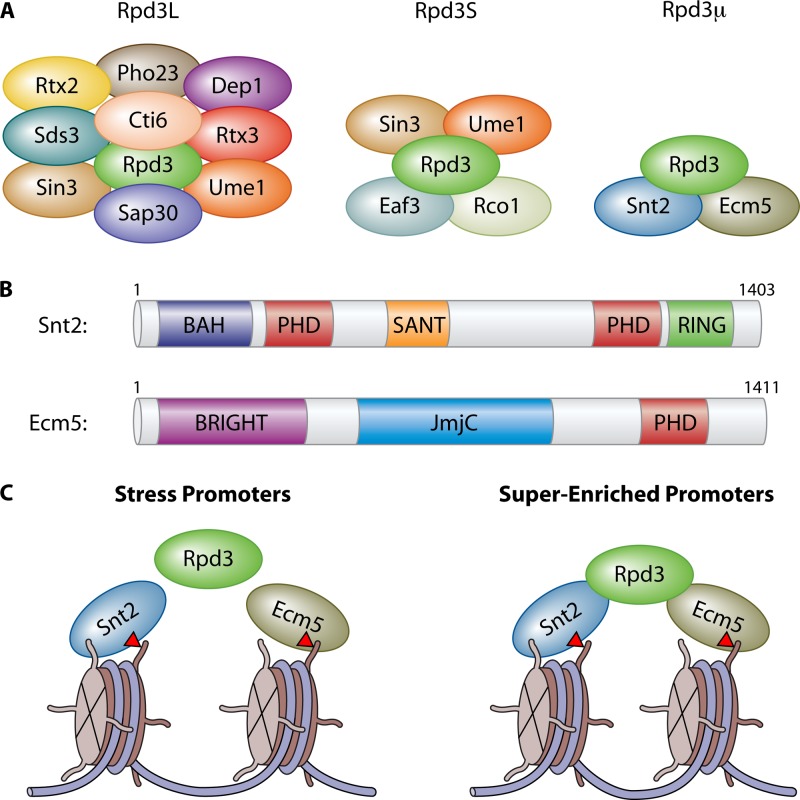
In this report (9), Baker et al. demonstrate that the histone deacetylase Rpd3 forms a third complex, which we call herein Rpd3 “micro” (Rpd3μ), given that it is the smallest of the three Rpd3 complexes identified thus far (Fig. 1A). This result is in agreement with a previous finding (10). Rpd3 has been associated with two other complexes, Rpd3L and Rpd3S (11), each with unique functions. Rpd3L has been shown to be involved in the response to heat stress and ribosome biogenesis (12, 13), while Rpd3S functions to maintain chromatin integrity and repress cryptic transcription within gene bodies (14). Using liquid chromatography-mass spectrometry (LC-MS), the authors demonstrated that Rpd3 physically associates with Snt2 and Ecm5 to form the Rpd3μ complex. Interestingly, Snt2 and Ecm5 each contain recognizable histone interaction motifs, including plant homeodomain (PHD) fingers and bromo-adjacent homology (BAH) domains (Fig. 1B), strongly implicating them in chromatin interaction. Intriguingly, Ecm5 also contains a putative histone demethylase domain (Fig. 1B)—however, whether this protein contains demethylase activity remains unknown.
Fig 1.
(A) Cartoon schematic illustrating the Rpd3L, Rpd3S, and Rpd3μ complexes. (B) The domain architecture of Snt2 and Ecm5 reveals multiple, putative chromatin and DNA binding domains. (C) Rpd3μ localizes to two distinct types of promoters. Upon oxidative stress, all three members of the Rpd3μ complex are recruited to oxidative stress response promoters, though Rpd3 recruitment is not dependent on Snt2 or Ecm5. The superenriched promoters are constitutively bound by Snt2 and Ecm5, and recruitment of Rpd3 is dependent upon both Snt2 and Ecm5. Recent work suggests that Ecm5 may bind histone H3 lysine 36 trimethylation (red triangles).
An initial clue that Rpd3μ might be fundamental to the stress response pathway was provided by a prior study linking Snt2 to osmotic stress (15). To further investigate this possibility, the authors created strains lacking Rpd3, Snt2, and Ecm5, both singly and in pairs. Interestingly, while strains lacking Snt2 or Rpd3 showed resistance to hydrogen peroxide (H2O2), cells lacking Ecm5 were highly sensitive. This suggests that while both Ecm5 and Snt2 are present in the same complex, they may play opposing functions in the response to oxidative stress. This result was also seen when these strains were treated with rapamycin, an inhibitor of the TOR (target of rapamycin) pathway that simulates nitrogen starvation. Further arguing for opposing roles, the authors observed opposing changes in gene expression when either Snt2 or Ecm5 was deleted. Whether the opposing functions between Snt2 and Ecm5 are driven through DNA sequence recognition, the epigenetic landscape, posttranslational modification of Ecm5 or Snt2, protein-protein interactions with specific transcription factors, or yet another mechanism remains to be elucidated.
To gain further insight into where Rpd3μ is found across the genome, the authors performed genome-wide localization studies. All three members of Rpd3μ were found to colocalize primarily at promoter regions. Strikingly, the authors discovered two distinct sets of promoters: a set that recruited Snt2 and Ecm5 only upon treatment with H2O2 and a set of “superenriched” promoters that had Snt2 and Ecm5 constitutively bound (Fig. 1C). Interestingly, Rpd3 recruitment did not require Ecm5 and Snt2 at the H2O2-responsive promoters but was necessary for Rpd3 to localize at the superenriched promoters, suggesting a fundamental difference in how Rpd3 is targeted to these genes. Whether Snt2 and Ecm5 function independently of Rpd3 at H2O2-responsive promoters remains an intriguing question. Additionally, a Pdr1/Pdr3 sequence motif was identified; this motif overlapped with the 20 most highly H2O2-enriched Snt2/Ecm5-bound promoters, whereas other Snt2/Ecm5 target genes are regulated by Rap1 and Ste12. These observations suggest a possible mechanism for the positive or negative gene expression changes observed at individual genes when bound by Rpd3μ and may help to explain how the cell finely tunes its response to individual types of stress at discrete genomic sites.
This new study from Baker et al. provides an important step forward in understanding how stress response signals work together with chromatin to execute specific transcriptional programs. Nevertheless, many exciting questions remain. For example, what role does Rpd3-mediated histone deacetylation play in activation or repression of the genes found to be regulated? What are the roles of the multiple conserved chromatin and DNA binding domains in Snt2 and Ecm5? Do these domains function to target Rpd3μ to specific chromatin PTM signatures and/or DNA motifs? We note that the PHD domain of Ecm5 has been shown to interact with histone H3 lysine 36 methylation in vitro (16), which could contribute to the small amount of gene body localization of Rpd3μ. Furthermore, what defines how Rpd3μ is targeted to the superenriched promoters versus the H2O2-responsive promoters? Could this be mediated through specific interactions Rpd3μ has with other transcription factors or through binding to specific DNA sequences? Perhaps Rpd3μ is also posttranslationally modified in one or more of its subunits to control its localization and/or function, which may explain how Snt2 and Ecm5 can exhibit distinct functions. Any combination of these elements could be working together to fine-tune gene expression. Baker and colleagues have provided critical insight into how cells modulate their transcriptional programs to respond to stress. We conclude by asking whether Rpd3μ will be limited to oxidative stress and nitrogen starvation or will Rpd3μ prove to be crucial in responding to a wide variety of cellular stresses? Future studies addressing these questions will surely lead to many exciting discoveries.
ACKNOWLEDGMENTS
We thank Scott Rothbart, Deepak Jha, and Lindsay Rizzardi for critical reading the manuscript and apologize to our colleagues whose work we were not able to cite due to space limitations.
Work in the Strahl lab is funded by NIH grant R01 GM068088.
Footnotes
Published ahead of print 12 August 2013
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Morano KA, Grant CM, Moye-Rowley WS. 2012. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190:1157–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Nadal E, Ammerer G, Posas F. 2011. Controlling gene expression in response to stress. Nat. Rev. Genet. 12:833–845 [DOI] [PubMed] [Google Scholar]
- 3.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res. 21:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner KE, Allis CD, Strahl BD. 2011. Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JS, Smith E, Shilatifard A. 2010. The language of histone crosstalk. Cell 142:682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith E, Shilatifard A. 2010. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell 40:689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 9.Baker LA, Ueberheide BM, Dewell S, Chait BT, Zheng D, Allis CD. 2013. The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxide-mediated oxidative stress. Mol. Cell. Biol. 33:3735–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shevchenko A, Roguev A, Schaft D, Buchanan L, Habermann B, Sakalar C, Thomas H, Krogan NJ, Shevchenko A, Stewart AF. 2008. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 9:R167. 10.1186/gb-2008-9-11-r167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592 [DOI] [PubMed] [Google Scholar]
- 12.Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL, Loewith R. 2011. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 30:3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz-Roig C, Vieitez C, Posas F, de Nadal E. 2010. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 76:1049–1062 [DOI] [PubMed] [Google Scholar]
- 14.Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13:115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, Martin DE, Tresch A, Cramer P. 2011. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 7:458. 10.1038/msb.2010.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282:2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]