Abstract
Nucleomethylin (NML), a novel nucleolar protein, is important for mediating the assembly of the energy-dependent nucleolar silencing complex (eNoSC), which also contains SirT1 and SUV39H1. eNoSC represses rRNA transcription during nutrient deprivation, thus reducing energy expenditure and improving cell survival. We found that NML is an RNA binding protein that copurifies with 5S, 5.8S, and 28S rRNA. The SirT1 and RNA binding regions on NML showed partial overlap, and the NML-SirT1 interaction was competitively inhibited by rRNA. Nutrient deprivation triggered downregulation of rRNA transcription, reduced the level of NML-associated rRNA, and stimulated NML-SirT1 binding. Assembly of eNoSC facilitated repression of pre-rRNA transcription. These results suggest that nascent rRNA generates a positive-feedback signal by suppressing the assembly of eNoSC and protecting active ribosomal DNA units from heterochromatin formation. This RNA-mediated mechanism enables the eNoSC to amplify the effects of upstream nutrient-responsive regulators.
INTRODUCTION
Ribosome biogenesis is a major biosynthetic and energy-consuming process. rRNA synthesis accounts for >50% of cellular transcriptional activity and must be tightly coupled to nutrient availability and growth signaling (1). Synthesis of rRNA by RNA polymerase I (Pol I) and Pol III is a rate-limiting step in ribosome biogenesis. RNA Pol I produces the common 45S precursor of mature 5.8S, 18S, and 28S rRNA, while RNA Pol III produces the precursor of 5S rRNA. The 45S pre-rRNA is transcribed from ∼200 copies of ribosomal DNA (rDNA) genes distributed as clusters on 5 different human chromosomes and organizes the formation of nucleoli (2). The human 5S rDNA gene cluster is separately localized to the chromosomal 1q42 to 1q44 region (3, 4) but spatially associates with the nucleolar periphery (5, 6). Nucleolar pre-rRNA transcriptional output is determined by the rate of transcription from each rDNA copy and the fraction of rDNA in a transcriptionally competent state.
Numerous growth and stress signals regulate Pol I activity and rRNA transcription (7–9). Nutrients and growth factors control rRNA synthesis via mTOR (1). Transcription of pre-rRNA requires basal factors TIF-1A, SL1, and UBF to form a transcriptional initiation complex with Pol I. Formation of this complex is regulated by mTOR through S6K1 kinase-mediated phosphorylation of TIF-1A and UBF, which promotes the recruitment of Pol I to rDNA promoters (2). UBF is also targeted by the Rb, ARF, and p53 tumor suppressor proteins. mTOR also controls the activity of Pol III and the transcription of 5S rRNA and tRNAs (1).
Nearly 50% of nucleolar rDNA repeats are present as heterochromatin in growing cells (10). Maintenance of the heterochromatin state is important for maintaining the stability of rDNA repeats. It has been shown that the nucleolar remodeling complex (NoRC) is important for switching rDNA between the silent and the active state. NoRC is an SNF2h-containing chromatin remodeling complex that recruits DNA methyltransferase and histone deacetylase to the promoter to trigger heterochromatin formation and silencing (11). The NoRC complex also contains a noncoding RNA (ncRNA), which is derived from a Pol I-dependent transcript from the rDNA intergenic spacer and is called pRNA (12, 13). The pRNA is processed into short fragments, one of which is bound by the RNA binding protein TIP5 in NoRC (14). Elimination of pRNA by the antisense approach or disruption of the RNA binding function of TIP5 abrogates the NoRC function (13). Therefore, noncoding RNA is involved in regulating rDNA heterochromatin formation.
A recent study identified a novel complex (the energy-dependent NoSC [eNoSC]) that regulates rRNA transcription in response to glucose deprivation (15). The eNoSC was identified through its binding to the H3K9 dimethylated (H3K9me2) peptide. The complex contains SirT1, SUV39H1, and a novel nucleolar protein, nucleomethylin (NML) (15). Knockdown of NML prevents the inhibition of rRNA synthesis by glucose starvation, resulting in apoptosis. Therefore, NML is important for maintenance of energy homeostasis and cell viability during starvation. NML represses rDNA transcription by promoting H3K9 methylation and establishing heterochromatin across the rDNA. NML has 456 amino acid residues with an N-terminal half that binds H3K9me2 and a C-terminal domain homologous to S-adenosylmethionine (SAM)-dependent methyltransferase (15). NML recruitment of SirT1 and SUV39H1 may be important for its function, since SirT1-SUV39H1 complex formation synergistically promotes deacetylation and methylation of histone H3K9 (16). Whether NML also has methyltransferase activity and participates in protein methylation is still unknown. A role of mammalian SirT1 in regulating rRNA transcription provides an interesting similarity to the well-established role of Saccharomyces cerevisiae yeast SIR2 in regulating rDNA recombination (17). Another sirtuin family member, SirT7, has also been shown to activate rRNA transcription (18).
In this study, we investigated the dynamic regulation of eNoSC assembly by nutrient signaling. The results showed that NML-SirT1 binding is competitively inhibited by rRNA. We propose a model in which upstream regulators such as mTOR trigger the initial change in rRNA transcription during nutrient deprivation. The nascent rRNA then functions in a signaling role to regulate eNoSC assembly and coordinate nucleolar heterochromatin formation with the nutrient level. Distinct from the classic ncRNA mechanism that promotes the assembly and recruitment of chromatin-modifying complexes, the abundant rRNA may act by mass effect to suppress eNoSC formation and provide local protection of actively transcribing rDNA chromatin.
MATERIALS AND METHODS
Cell lines and reagents.
NML cDNA was provided by Junn Yanagisawa. All constructs used in this study are of human origin. H1299 and A549 (non-small-cell lung carcinoma), U2OS (osteosarcoma), and HeLa (cervical carcinoma) cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). Transfection of H1299 cells was performed using a standard calcium phosphate precipitation protocol. For glucose starvation treatment, cells were washed twice with phosphate-buffered saline (PBS) before culturing in DMEM with 10% dialyzed FBS and 0 mM or 25 mM glucose. Rabbit polyclonal serum for NML was generated using His6 combined with NML residues 1 to 200 (His6-NML-1-200) as the antigen and affinity purified using immobilized His6-NML-1-200. Anti-FLAG polyclonal antibody and anti-Myc polyclonal antibody were purchased from Sigma. Anti-SirT1 monoclonal antibody 10E4 was previously generated in our lab (19). To knock down NML, cells were transfected with 100 nM control small interfering (siRNA) (AATTCTCCGAACGTGTCACGT) or an NML siRNA pool (Dharmacon) using RNAiMAX (Invitrogen). After 48 h of transfection, cells were glucose starved and analyzed for protein interactions or pre-rRNA synthesis.
rRNA biosynthesis assays.
HeLa cells treated with control siRNA for 48 h were cultured in medium with 25 mM or 0 mM glucose for an additional 16 h, labeled with 5 μCi [3H]uridine (38 Ci/mmol; PerkinElmer) for 30 min, and washed twice with PBS. RNA was extracted with the TRIzol reagent (Ambion). For quantification of the rRNA synthesis level, an identical amount of total RNA (10 μg for each sample) was analyzed by liquid scintillation counting to determine the incorporation of 3H. For reverse transcription-quantitative PCR (RT-qPCR) analysis of the pre-rRNA level, total cellular RNA was reverse transcribed using random hexanucleotides and the cDNAs were amplified with pre-rRNA primers (5′TGTCAGGCGTTCTCGTCTC and 5′AGCACGACGTCACCACATC). The pre-rRNA signal was normalized to the amount of actin mRNA.
Immunoprecipitation.
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail) and centrifuged for 10 min at 14,000 × g to remove the insoluble debris. The supernatant was used for immunoprecipitation and Western blotting. Treatment with RNase A (20 μg/ml) was performed during cell lysis for 20 min at room temperature. Cell lysate (200 to 1,000 μg of protein) was immunoprecipitated with 10E4 against SirT1 and protein A-agarose beads or anti-FLAG M2-agarose beads (Sigma) for 18 h at 4°C. After washing 4 times with SNNTE buffer (50 mM Tris-HCl, pH 7.4, 5 mM EDTA, 500 mM NaCl, 0.1% NP-40, 5% sucrose), the beads were boiled in sample buffer for 5 min and subjected to SDS-PAGE and Western blotting to detect the SirT1-NML interaction. To detect endogenous NML-SirT1 complex, cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate), diluted 10-fold with lysis buffer, immunoprecipitated with SirT1 antibody 10E4, washed with SNNTE buffer, and blotted with affinity-purified rabbit anti-NML antibody.
RNA immunoprecipitation.
FLAG-NML was transfected into H1299 cells for 18 h and cultured in medium containing 25 mM or 0 mM glucose for 16 h. Cells were harvested and resuspended in an equal volume of cold lysis buffer (10 mM HEPES, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol [DTT], protease inhibitors, 100 U/ml RNasin). The cells were passed through a 27-gauge needle before being frozen at −80°C. The lysate was thawed on ice and treated with DNase I for 30 min on ice (60 U/100 μl lysate). Lysate (100 μl) precleared by centrifuging at 15,000 × g for 15 min at 4°C was added to an antibody mixture including 800 μl of NT2 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), 20 μl of M2 beads, 100 U of RNasin, 10 mM DTT, and 20 mM EDTA. The mixture was incubated for 18 h at 4°C with tumbling. The beads were washed with NT2 buffer, and a portion was removed for Western blot analysis. The beads were resuspended in 100 μl of NT2 buffer and 100 μl of proteinase K buffer (20 mM Tris-HCl, pH 7.8, 10 mM EDTA, 1.0% SDS) with 30 μg of proteinase K. The mixture was incubated at 55°C for 30 min. RNA was extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol after adding 25 μg of glycogen. One-step RT-qPCR was performed by a SuperScript III platinum SYBR green one-step RT-qPCR system (Invitrogen) with primers for 5S, 5.8S, 18S, 28S, pre-rRNA, and U1snRNA: primers 5SF (GATCTCGGAAGCTAAGCAGG) and 5SR (AAGCCTACAGCACCCGGTAT), primers 5.8SF (GTGGATCACTCGGCTCGT) and 5.8SR (GCAAGTGCGTTCGAAGTGT), primers 18SF (AAACGGCTACCACATCCAAG) and 18SR (CCTCCAATGGATCCTCGTTA), primers 28SF (TCATCAGACCCCAGAAAAGG) and 28SR (GATTCGGCAGGTGAGTTGTT), primers pre-rRNAF (TGTCAGGCGTTCTCGTCTC) and pre-rRNAR (AGCACGACGTCACCACATC), and primers U1snRNAF (CTCCGGATGTGCTGACCC) and U1snRNAR (CAAATTATGCAGTCGAGTTTCCC), respectively.
GST-NML RNA binding assay.
5S and 5.8S rRNA-coding sequences were amplified by PCR from HeLa genomic DNA and subcloned into the Bluescript-KS(−) vector. The following PCR primers were used: primers 5SF (CGCGAGCTCGTCTACGGCCATACCACCCTGAAC) and 5SR (CGCGGATCCAAAGCCTACAGCACCCGGTATTCC) and primers 5.8SF (CGCGAGCTCCGACTCTTAGCGGTGGATCACTCG) and 5.8SR (CGCGGATCCAAGCGACGCTCAGACAGGCGTAGC), respectively. 5S rRNA and 5.8S rRNA were transcribed using a Riboprobe in vitro transcription kit (Promega). Glutathione S-transferase (GST)–NML was captured onto glutathione beads. After washing with PBS, the beads were incubated with or without RNase A (20 μg/ml) at 25°C for 30 min. RNase A was removed by washing 4 times with PBS and once with binding buffer containing RNasin (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 100 mM KCl, 0.1 mM DTT, 5% glycerol, 0.01 mg/ml bovine serum albumin [BSA], 40 U/ml RNasin). Beads loaded with ∼500 ng GST-NML were mixed with 2 μl of 32P-labeled RNA probes in a 30-μl mixture including 0.75 μl of BSA (50 μg/μl), 0.3 μl of 0.1 M DTT, 0.25 μl of 1 M MgCl2, 0.5 μl of RNasin (40 U/μl), 5 μg of yeast tRNA, and 16 μl of binding buffer. The mixture was incubated at 30°C for 2 h. After washing 4 times with binding buffer, the beads were boiled in Laemmli sample buffer and fractionated on a 12.5% SDS-polyacrylamide gel. The gel was directly exposed to film at −80°C for 0.5 to 6 h.
RNA binding assays.
An RNA homopolymer [poly(U)] binding assay was performed as described previously using in vitro-translated NML and SirT1 (20). For RNA electrophoretic mobility shift assay (EMSA), RNA probes were transcribed using the Riboprobe in vitro transcription kit (Promega) in the presence of α-35S-UTP (12.5 mCi/ml; PerkinElmer). GST-NML was pulled down by glutathione-agarose beads, incubated with RNase A (20 ng/μl) at 25°C for 30 min, washed 4 times with PBS, and eluted using elution buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 μM DTT, 10 mM reduced glutathione, 0.4 U/μl RNasin). Purified GST-NML (∼50 ng) was mixed with 1 μl of 35S-labeled RNA probe in a 10-μl reaction mixture containing 1.25 μg/μl BSA, 10 mM DTT, 75 mM MgCl2, 1 U/μl RNasin, 5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 75 mM KCl, and 2.5% glycerol. The mixture was incubated at 30°C for 2 h. An equal volume of loading buffer (2 mM Tris-HCl, pH 7.5, 0.2 mM EDTA, 10% glycerol, 0.1% bromophenol blue) was added, and the RNA-protein complex was resolved by electrophoresis on a 5% (wt/vol) native polyacrylamide gel containing 40 mM Tris-acetate, pH 7.8, 2.5 mM EDTA, and 1% glycerol. Gels were fixed in 30% methanol, 10% acetic acid for 30 min, incubated with enhancer solution (1 M Na salicylate, 5% glycerol) for 1 h, dried at 80°C on a vacuum drier, and exposed to film at −80°C for 6 to 18 h.
RNA ChIP.
H1299 cells (∼1 × 107) were cross-linked with 1% formaldehyde for 10 min at 23°C. The cross-linking was stopped by 0.125 M glycine for 5 min at 23°C. The cells were washed 3 times with PBS, lysed in 200 μl of buffer A with NP-40 [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 8.0, 85 mM KCl, 0.5% NP-40, protease inhibitors, 50 U/ml RNasin] on ice for 10 min, and pelleted by microcentrifugation at 5,000 rpm for 5 min. The cell pellets were washed once with 200 μl of buffer A without NP-40, resuspended in 350 to 500 μl of buffer B (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, protease inhibitors, 50 U/ml RNasin) for 10 min on ice, sonicated, and centrifuged for 10 min. The lysate was diluted 1:10 in immunoprecipitation (IP) buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, protease inhibitors, 50 U/ml RNasin) and incubated with affinity-purified rabbit anti-NML antibody for 18 h. Protein A beads (25 μl, packed) were added, and the mixture was incubated for 2 h at 4°C. The beads were washed once each with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl), LiCl buffer (250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0), and TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). The beads were eluted twice with a total of 200 μl of elution buffer (1% SDS, 100 mM NaHCO3, 50 U/ml RNasin) at 25°C for 15 min with shaking (1,000 rpm). NaCl was added to 200 mM, and the samples were de-cross-linked at 65°C for 2 h. Proteinase K (0.1 mg/ml), 10 mM EDTA, and 40 mM Tris-HCl, pH 6.8, were added, and the mixture was incubated at 42°C for 1 h. RNA was extracted with phenol-chloroform and precipitated with ethanol using 25 μg of glycogen as the carrier. The RNA was dissolved in 20 μl H2O, DNase I (0.1 mg/ml) was added, and the mixture was incubated at 37°C for 2 h before analysis by RT-qPCR. The signal of rRNA was normalized to the amount of actin mRNA. The following PCR primers were used: primers 5SF (GATCTCGGAAGCTAAGCAGG) and 5SR (AAGCCTACAGCACCCGGTAT), primers 5.8SF (GTGGATCACTCGGCTCGT) and 5.8SR (GCAAGTGCGTTCGAAGTGT), primers 18SF (AAACGGCTACCACATCCAAG) and 18SR (CCTCCAATGGATCCTCGTTA), and primers 28SF (TCATCAGACCCCAGAAAAGG) and 28SR (GATTCGGCAGGTGAGTTGTT).
RESULTS
SirT1 recruitment to rDNA during nutrient deprivation requires NML.
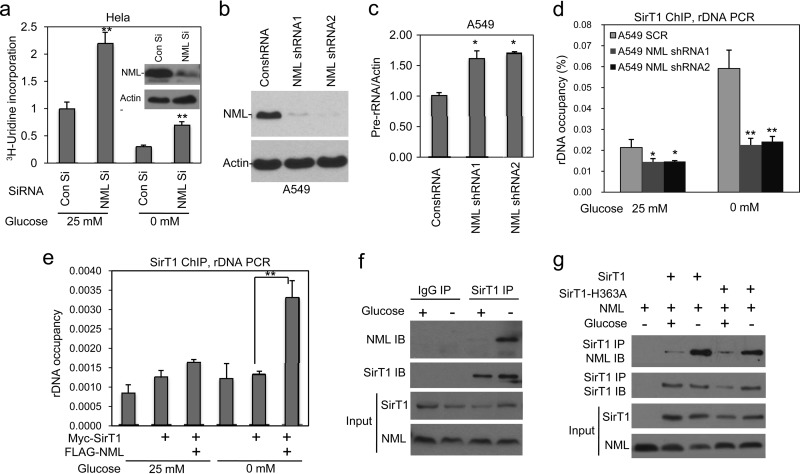
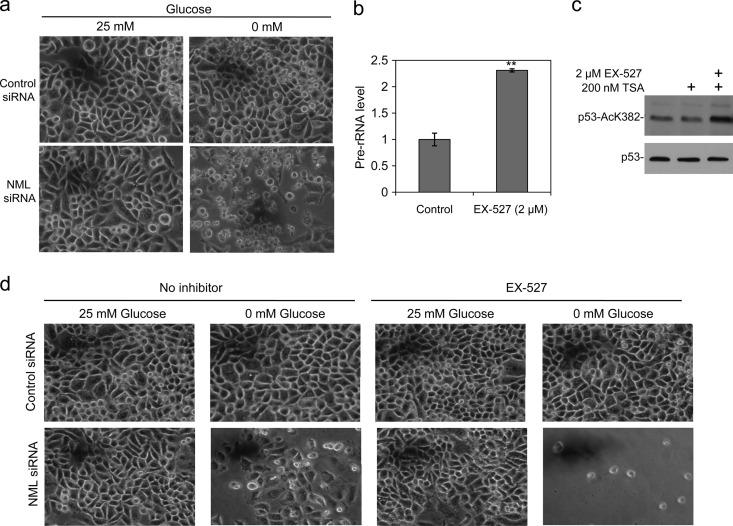
A recent study by Murayama et al. showed that the nucleolar protein NML functions as a repressor of rRNA transcription and interacts with SirT1 and SUV39H1 (15). We confirmed that transient knockdown of NML in HeLa cells stimulated rRNA synthesis in a [3H]uridine incorporation assay (Fig. 1a). NML knockdown also dampened the downregulation of rRNA transcription by glucose starvation (Fig. 1a). Furthermore, we found that stable knockdown of NML in A549 cells also increased the pre-rRNA level (Fig. 1b and c). As reported previously, knockdown of NML significantly increased cell death during glucose starvation (Fig. 2a), in which the cell death exhibited features of both apoptosis and necrosis (data not shown). Treatment with SirT1-specific inhibitor EX-527 reduced endogenous SirT1 activity (as shown by an increased p53 acetylation level) (Fig. 2c) and also increased pre-rRNA synthesis (Fig. 2b) (21). Combination of NML siRNA and EX-527 synergistically promoted cell death during glucose starvation (Fig. 2d). These results confirmed the role of NML in regulating rRNA transcription and the cooperation between NML and SirT1.
Fig 1.
SirT1 recruitment to rDNA requires NML. (a) HeLa cells treated with control and NML siRNA were glucose starved for 16 h and labeled with [3H]uridine for 30 min. An identical amount of total RNA from each sample was analyzed for the 3H incorporation level. Values are means ± SDs of triplicates. (Inset) Western blot confirming NML knockdown. Con, control. (b) A549 cells were stably infected with retrovirus expressing NML short hairpin RNAs (shRNAs), and NML knockdown in the cell lines was confirmed by Western blotting. (c) Pre-rRNA expression levels in A549 stable NML knockdown cell lines in the presence of 25 mM glucose were determined by RT-qPCR. Values are means ± SDs of triplicates. (d) SirT1 binding to rDNA loci was determined in A549 stable knockdown cell lines by ChIP-qPCR after glucose deprivation for 16 h. Values are means ± SDs of triplicates. SCR, scrambled control shRNA. (e) H1299 cells were transiently cotransfected with Myc-SirT1 and FLAG-NML and glucose starved for 16 h, followed by SirT1 chromatin IP with Myc antibody and PCR detection of rDNA. Values are means ± SDs of triplicates. (f) The interaction of endogenous SirT1 and NML in glucose-starved H1299 cells was analyzed by immunoprecipitation with SirT1 monoclonal antibody, followed by Western blotting with NML polyclonal rabbit antibody. IB, immunoblotting. (g) H1299 cells were transfected with SirT1 and FLAG-NML and treated with glucose starvation for 16 h. The interaction of ectopic SirT1 and NML was analyzed by IP-Western blotting. Statistically significant differences from the control are marked by asterisks (t test; *, P < 0.05; **, P < 0.01).
Fig 2.
NML and SirT1 cooperate to promote cell survival during nutrient deprivation. (a) HeLa cells treated with NML siRNA were glucose starved for 24 h. Cell death was shown by photography. (b) HeLa cells were glucose starved for 16 h in the absence or presence of the SirT1 inhibitor EX-527. The pre-rRNA levels were determined by RT-qPCR. (c) The effect of EX-527 in inhibiting SirT1 was confirmed by treatment of U2OS (chosen for its expression of p53) for 16 h with the indicated compounds and analysis of the p53 K382 acetylation level by Western blotting. TSA, trichostatin A. (d) HeLa cells were transfected with NML siRNA for 24 h and cultured in glucose-free medium with 2 μM EX-527 for 24 h. Cell death was shown by photography. Statistically significant differences from the control are marked by asterisks (t test; **, P < 0.01).
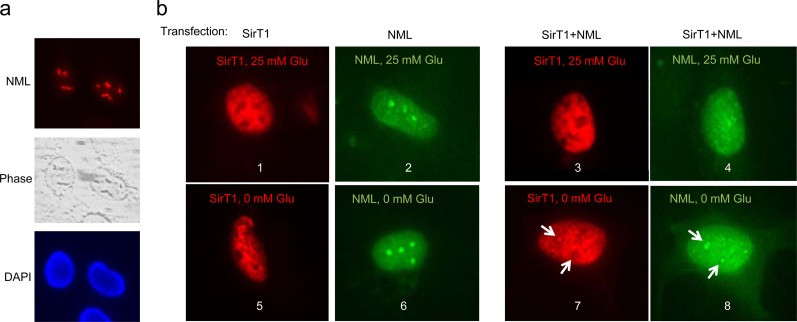
SirT1 is largely localized in the nucleoplasm (22), whereas NML is a nucleolar protein (Fig. 3a). The ability of SirT1 to regulate rRNA transcription suggests that it may be transiently recruited to the nucleolus by NML. In a chromatin immunoprecipitation (ChIP) assay, we found that endogenous SirT1 binding to rDNA was significantly increased after glucose deprivation in an NML-dependent fashion (Fig. 1d). Transfection with ectopic NML also stimulated SirT1 binding to rDNA during glucose deprivation (Fig. 1e). When U2OS cells were cotransfected with SirT1 and NML, colocalization of a small amount of SirT1 with NML in the nucleolus was also observed after glucose withdrawal (Fig. 3b). Murayama et al. showed that SirT1-NML binding was stimulated by glucose deprivation (15). We confirmed that endogenous SirT1-NML binding increased significantly after glucose starvation (Fig. 1f). The binding between transfected NML and SirT1 was also increased after glucose starvation (Fig. 1g). The SirT1 H363A catalytic mutant retained NML binding and a response to glucose deprivation (Fig. 1g). Together the results suggest that nutrient deprivation stimulates SirT1-NML binding, leading to the recruitment of SirT1 to rDNA and repression of rRNA transcription.
Fig 3.
NML recruits SirT1 to the nucleolus during glucose deprivation. (a) Endogenous NML was detected using a rabbit polyclonal antibody in U2OS cells. DAPI, 4′,6-diamidino-2-phenylindole. (b) U2OS cells were transfected with FLAG-NML and SirT1 alone or in combination, glucose starved for 16 h, and stained for SirT1 and NML using 10E4 and anti-FLAG antibodies. Panels 7 and 8 are different exposures of the same cell. NML expression recruited a small fraction of SirT1 to the nucleolus during glucose deprivation.
RNA regulates NML-SirT1 interaction.
To investigate the mechanism that regulates NML-SirT1 binding during glucose starvation, we tested NML-SirT1 binding using an in vitro assay. FLAG-NML and SirT1 were separately transfected into H1299 cells, followed by treatment with normal or low levels of glucose. Cell lysates containing NML and SirT1 were mixed, and the formation of the NML-SirT1 complex in vitro was analyzed. The result showed that NML prepared from cells cultured in glucose-free medium had an increased ability to bind SirT1 in vitro, whereas SirT1 from glucose-starved cells did not show an increased affinity for NML (Fig. 4a). This result suggests that the change in NML was responsible for promoting SirT1 binding after glucose deprivation.
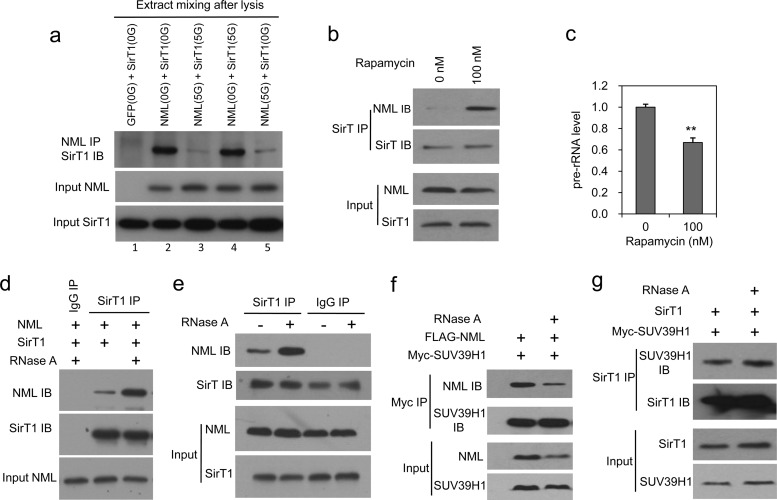
Fig 4.
RNA inhibits NML-SirT1 binding. (a) H1299 cells were transfected with SirT1 and NML separately for 24 h, followed by incubation in medium with 5 mM or 0 mM glucose (5G and 0G, respectively) for 4 h. The extracts were mixed and incubated at 23°C for 30 min, followed by IP-Western blotting to detect NML-SirT1 binding in vitro. (b) H1299 cells were treated with 100 nM the mTOR inhibitor rapamycin for 18 h and analyzed by IP-Western blotting for endogenous NML-SirT1 binding. (c) Pre-rRNA levels in rapamycin-treated H1299 cells were determined by RT-qPCR. (d) A lysate of H1299 cells transfected with NML and SirT1 was treated with 5 μg/ml RNase A at 23°C for 30 min, followed by IP-Western blotting to detect NML-SirT1 binding. (e) An H1299 cell lysate was treated with 5 μg/ml RNase A at 23°C for 30 min, followed by IP-Western blotting to detect endogenous NML-SirT1 binding. (f) A lysate of H1299 cells transfected with NML and SUV39H1 was treated with RNase A and analyzed for NML-SUV39H1 binding by IP-Western blotting. (g) A lysate of H1299 cells transfected with SirT1 and SUV39H1 was treated with RNase A and analyzed for SirT1-SUV39H1 binding by IP-Western blotting. Statistically significant differences from the control are marked by asterisks (t test; **, P < 0.01).
Next, we tested the roles of several kinases using inhibitors. The results suggested the involvement of mTOR. The activity of mTOR is suppressed by nutrient deprivation and stress signals such as oxidative stress and DNA damage (23). When H1299 cells cultured in high glucose were treated with rapamycin, pre-rRNA synthesis was downregulated and endogenous NML-SirT1 binding was stimulated (Fig. 4b and c). Therefore, mTOR may suppress NML-SirT1 binding in the presence of nutrients. A metabolic labeling experiment using [32P]orthophosphate showed that the phosphorylation level of NML was not altered by rapamycin (data not shown), thus ruling out NML as a direct phosphorylation substrate of mTOR. Analysis of immunopurified NML using antiacetyllysine antibody or mass spectrometry did not detect acetylation (data not shown).
Since pre-rRNA expression was suppressed by energy deprivation or rapamycin (Fig. 1a and 4c), it is possible that the rRNA level plays a role in regulating SirT1-NML binding. Therefore, we treated the cell extract with RNase A. The NML-SirT1 interaction was significantly increased after RNA removal (Fig. 4d and e), when assayed using transfected or endogenous proteins. RNase A treatment did not stimulate NML-SUV39H1 binding or SUV39H1-SirT1 binding (Fig. 4f and g). These results suggest that NML-SirT1 binding is specifically regulated by RNA.
NML-SirT1 binding is blocked by RNA.
Murayama et al. found that GST-NML did not interact with in vitro-translated SirT1 (15). We also found that incubation of recombinant GST-NML with H1299 extract containing endogenous SirT1 resulted in very poor binding (Fig. 5a). However, RNase A treatment significantly stimulated GST-NML binding to SirT1 (Fig. 5a), suggesting that the SirT1 binding site on GST-NML was blocked by Escherichia coli RNA.
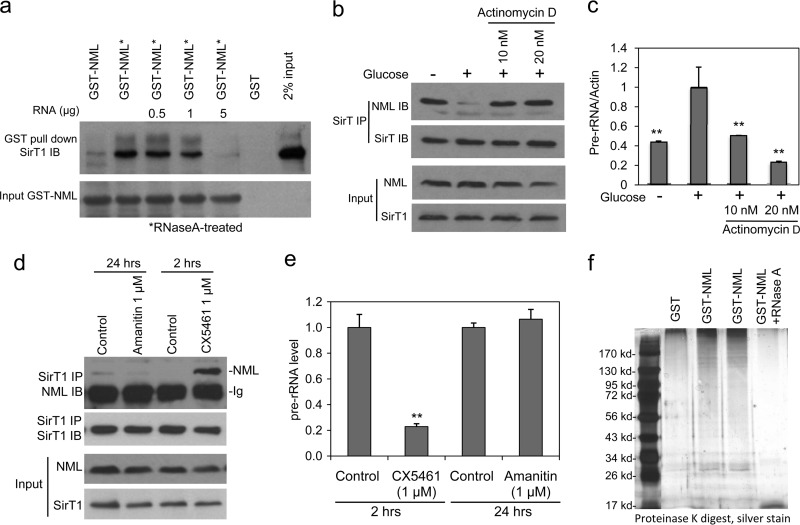
Fig 5.
rRNA inhibits NML binding to SirT1 in vitro and in vivo. (a) Glutathione beads loaded with GST-NML were treated with RNase A and RNasin and incubated with an H1299 cell lysate in the absence or presence of purified total H1299 cell RNA at the indicated amounts. The amount of SirT1 captured by GST-NML was detected by Western blotting. (b) H1299 cells were treated with actinomycin D or glucose deprivation for 8 h. Endogenous NML-SirT1 binding was detected by IP-Western blotting using the indicated antibodies. (c) Inhibition of pre-rRNA transcription by glucose deprivation and actinomycin D was confirmed by RT-qPCR. (d) H1299 cells were treated with Pol I-specific inhibitor CX-5461 or Pol II-specific inhibitor α-amanitin for the indicated times and analyzed for endogenous NML-SirT1 complex formation by IP-Western blotting. (e) H1299 cells were treated with CX-5461 or α-amanitin and analyzed for their pre-rRNA levels by RT-PCR. The pre-rRNA level was normalized to the level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA. (f) GST and GST-NML were purified from E. coli lysate using glutathione beads and digested with proteinase K, and the copurified E. coli RNA was detected by SDS-PAGE and silver staining. Statistically significant differences from the control are marked by asterisks (t test; **, P < 0.01).
To test whether E. coli-expressed GST-NML was occupied with bacterial RNA, the GST-NML preparation was digested with proteinase K and the remaining material was analyzed by gel electrophoresis and silver staining. GST-NML indeed copurified with significant amounts of E. coli RNA of various molecular masses, and the RNA was eliminated by RNase A treatment (Fig. 5f). Furthermore, total RNA purified from H1299 cells inhibited NML-SirT1 binding in vitro in a dose-dependent fashion (Fig. 5a). The interaction of recombinant His6-NML and GST-SirT1 purified from E. coli was also increased after removal of RNA and blocked by readdition of H1299 RNA (data not shown). These data suggest that RNA and SirT1 compete for binding to NML.
On the basis of the findings presented above, we hypothesized that rRNA may regulate NML-SirT1 binding in vivo. Thus, we treated H1299 cells with actinomycin D at low concentrations (10 to 20 nM) that inhibit Pol I-dependent transcription of rRNA but not Pol II-mediated transcription (Fig. 5c). As expected, actinomycin D significantly promoted the binding between endogenous NML and SirT1 (Fig. 5b), which was similar to the effect of glucose starvation. To further confirm the role of rRNA transcription in regulating NML-SirT1 binding, H1299 cells were treated with the recently developed Pol I-specific inhibitor CX-5461 (24). This compound inhibited pre-rRNA synthesis and strongly stimulated NML-SirT1 complex formation in 2 h (Fig. 5d and e). In contrast, the Pol II-specific inhibitor α-amanitin had no effect when tested for up to 24 h (to accommodate its slow uptake). These results suggest that eNoSC assembly is regulated by nascent rRNA through competitive binding of SirT1 and rRNA to NML.
NML copurifies with rRNA.
We tested whether NML is an RNA binding protein by incubating poly(U)-agarose beads with in vitro-translated NML or SirT1. The results showed that NML but not SirT1 bound to poly(U)-agarose (Fig. 6a), suggesting that NML has affinity for nucleic acid. A similar result was also obtained using poly(C)-agarose beads (data not shown). To further test whether RNA regulates NML conformation, cell extract was treated with RNase A and subjected to trypsin digestion. Endogenous NML became more resistant to trypsin digestion after RNase A treatment (Fig. 6b), suggesting that removal of RNA changed the conformation of NML or the accessibility of certain residues.
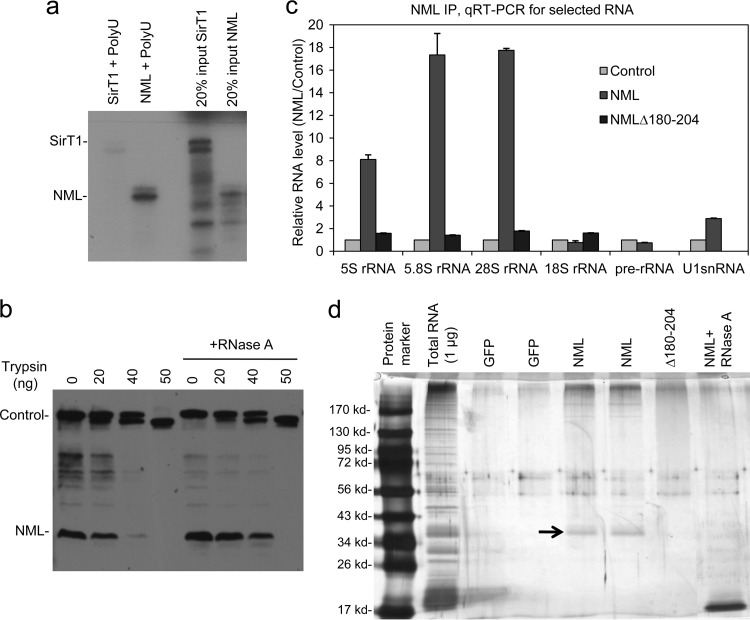
Fig 6.
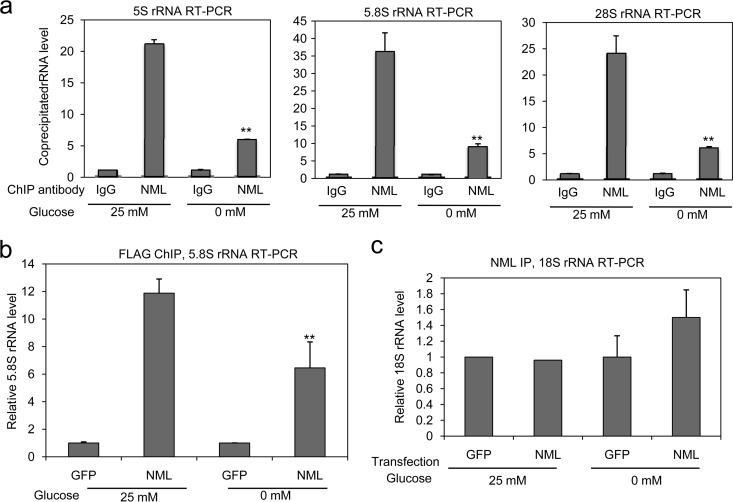
NML interacts with rRNA. (a) In vitro-translated NML was incubated with RNA-agarose beads, and the bound proteins were detected by autoradiography. SirT1 served as a negative control. (b) An H1299 cell lysate was treated with 5 μg/ml RNase A or mock treated for 30 min at 23°C, followed by trypsin digestion for 10 min at 23°C. Endogenous NML was analyzed by Western blotting. Cleavage of a background band near the top of the gel served as an internal control for trypsin dosage. (c) H1299 cells transfected with GFP, FLAG-NML, or FLAG-NMLΔ180-204 were immunoprecipitated using M2 beads. The coprecipitated RNAs were purified and analyzed by RT-qPCR using primers for the indicated RNA species. The relative rRNA level was normalized to the input amount. (d) H1299 cells were transfected with FLAG-NML, immunoprecipitated with M2 antibody, and digested with proteinase K. RNA isolated from the NML complex was subjected to 12.5% SDS-PAGE and detected by silver staining. The band marked with an arrow was found to contain predominantly 5S and 5.8S rRNA by RT-qPCR.
To determine whether NML interacts with rRNA, RNA isolated from the NML complex by RNA IP (RIP) was analyzed by RT-qPCR using validated primers. The results indicated that NML coprecipitated with 5S, 5.8S, and 28S rRNA but showed little binding to pre-rRNA and 18S rRNA (Fig. 6c). Next, the RNA samples isolated from the NML complex were fractionated on SDS-polyacrylamide gels and subjected to silver staining. NML coprecipitated with a prominent RNA band that comigrated with ∼40-kDa protein markers, as well as multiple higher-molecular-mass species (Fig. 6d). Treatment with RNase A destroyed the 40-kDa band, suggesting that it was composed of RNA. High-molecular-mass materials were also eliminated by RNase A digestion. This result further confirmed that the NML complex contains a significant amount of RNA. As a control, an NML mutant with an internal deletion of residues 180 to 204 (NMLΔ180-204) was deficient in RNA coprecipitation in these assays (this is further addressed in Fig. 11).
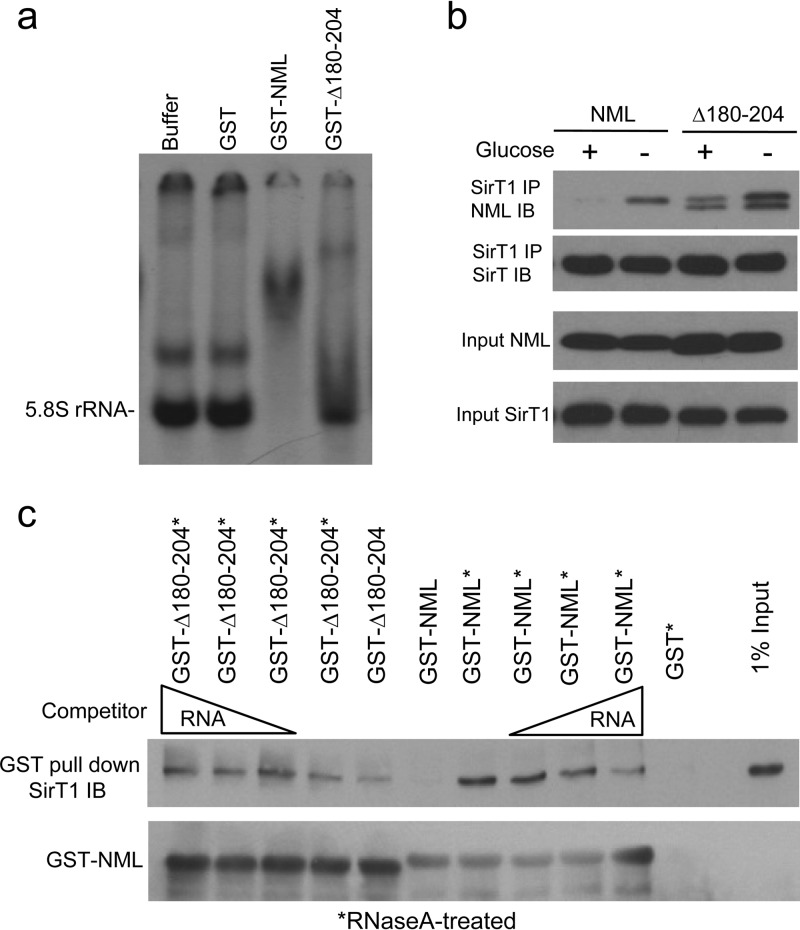
Fig 11.
Reduced NML-RNA binding correlates with increased SirT1 binding and transcription repression. (a) Identical amounts of GST-NML and GST-NMLΔ180-204 proteins were tested for RNA binding by EMSA. (b) H1299 cells cotransfected with SirT1 and NML or NMLΔ180-204 were glucose starved for 16 h. NML-SirT1 binding was detected by IP-Western blotting. (c) Glutathione beads loaded with GST-NML and GST-NMLΔ180-204 were treated with RNase A and RNasin and incubated with H1299 cell lysate in the absence or presence of purified H1299 RNA. The amount of SirT1 captured by GST-NML or GST-NMLΔ180-204 was detected by Western blotting.
The most abundant RNAs that migrate at the 17- to 40-kDa range are 5.8S rRNA, 5S rRNA, and tRNA. Therefore, the likely candidates of NML-associated RNA are 5.8S and 5S rRNA. To test this possibility, the 40-kDa band was excised and the RNA was extracted and subjected to RT-qPCR analysis using primers against 28S, 5.8S, and 5S rRNA. Additional gel slices excised at different positions were also analyzed to provide background controls. The qPCR analysis confirmed that the major components of the 40-kDa RNA band were 5.8S and 5S rRNA (data not shown). Furthermore, there was a substantial amount of 28S rRNA that copurified with NML and migrated at the higher-molecular-mass region. This result was consistent with that of the RIP–RT-qPCR analysis (Fig. 6c), suggesting that NML can bind to multiple rRNA species.
NML nonspecifically interacts with RNA in vitro.
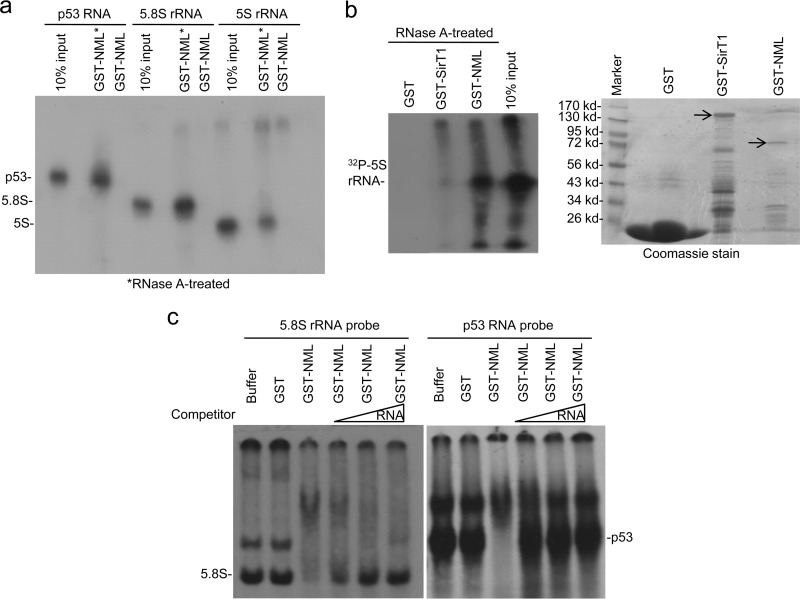
To test whether NML directly binds to rRNA, GST-NML produced in E. coli was used to pull down in vitro-transcribed, 32P-labeled 5S and 5.8S rRNA. A strong interaction between GST-NML and RNA probes was observed only after removing bacterial RNA with RNase A/RNasin pretreatment (Fig. 7a). Without pretreatment with RNase A, GST-NML captured only <1% of the RNA probe, whereas RNase A-treated GST-NML captured >20% of the 5.8S rRNA probe. However, a fragment of p53 mRNA was also pulled down by GST-NML with an efficiency comparable to that for rRNAs (Fig. 7a). GST-NML also efficiently captured RNA transcribed from a different region of p53 or from a plasmid polylinker region (data not shown), suggesting that the in vitro RNA binding by NML may not be restricted to rRNA. Consistent with the poly(U) binding result, GST-SirT1 did not bind 5S rRNA even after pretreatment with RNase A (Fig. 7b). To further test the RNA binding activity of NML, we performed an electrophoretic mobility shift assay (EMSA) using RNase A-treated GST-NML. The purified GST-NML was able to induce a mobility shift of the labeled 5.8S rRNA probe, which was blocked by the addition of H1299 RNA as a competitor (Fig. 7c). Similar results were observed using p53 RNA (Fig. 7c) or 5S rRNA probes (data not shown). These results suggest that NML can directly interact with RNA without sequence specificity in vitro. A titration experiment using EMSA showed that GST-NML bound to 5.8S rRNA with a dissociation constant (Kd) of ∼0.1 μM (data not shown). Although NML-SirT1 binding affinity has not been determined, other strong protein interactions, such as the p53-MDM2 interaction, have an affinity in a similar range (Kd, ∼0.2 μM), suggesting a potential for RNA to competitively regulate the NML interaction with other proteins, such as SirT1.
Fig 7.
NML binds to RNA in vitro. (a) GST-NML expressed in E. coli was loaded onto glutathione-agarose beads, treated with RNase A and RNasin (marked with asterisks), and incubated with in vitro-transcribed 32P-labeled 5S rRNA, 5.8S rRNA, and a fragment of p53 mRNA. The captured RNA was detected by autoradiography. (b) GST-SirT1 and GST-NML treated with RNase A and RNasin were tested for binding to 5S rRNA by pulldown assay (left). Protein levels were confirmed by Coomassie staining (right). Arrows, full-length fusion proteins. (c) GST-NML was purified, treated with RNase A and RNasin, and incubated with 35S-labeled RNA probes. RNA binding was detected by EMSA and autofluorography. Total RNA purified from H1299 cells was used as a competitor.
NML-rRNA binding is reduced after glucose deprivation.
If NML-SirT1 binding in the cell was regulated by rRNA, the NML-rRNA interaction should be reduced during starvation when NML-SirT1 binding was increased. To test whether this is the case, we performed RNA ChIP using NML antibody after in vivo chemical cross-linking. RT-qPCR analysis showed that immunoprecipitation of endogenous NML coprecipitated a significant amount of rRNA compared to control IP with IgG (Fig. 8a). The amount of coprecipitated rRNA was reduced by 2- to 3-fold after glucose deprivation (Fig. 8a), a finding correlating with reduced pre-rRNA expression. Ectopic NML binding to rRNA was also reduced after glucose deprivation in the RNA-ChIP assay (Fig. 8b). These results suggest that NML interacts with rRNA in vivo. The amount of NML-associated rRNA is determined by nutrient level, which is possibly responsible for the increased NML-SirT1 complex formation during starvation. Interestingly, NML showed negligible binding to 18S rRNA even after in vivo cross-linking (Fig. 8c), which is consistent with the finding of the RNA co-IP experiment without cross-linking (Fig. 6c). Therefore, despite its lack of sequence specificity for RNA in vitro, the in vivo RNA binding by NML may be determined by other mechanisms, such as colocalization and competition with other RNA binding proteins.
Fig 8.
The level of NML-associated rRNA is reduced after glucose deprivation. (a) H1299 cells were cultured in medium containing 25 mM or 0 mM glucose for 16 h. Endogenous NML-rRNA binding was analyzed by RNA ChIP assay using primers for the indicated RNA species. Values are means ± SDs of triplicates. (b and c) H1299 cells were transfected with FLAG-NML and cultured in medium containing 25 mM or 0 mM glucose for 16 h. NML-rRNA binding was analyzed by in vivo cross-linking and RNA ChIP assay using FLAG antibody and primers for the indicated RNA species. Values are means ± SDs for triplicates. Statistically significant differences from the control are marked by asterisks (t test; **, P < 0.01).
Overlapping SirT1 and RNA binding sites on NML.
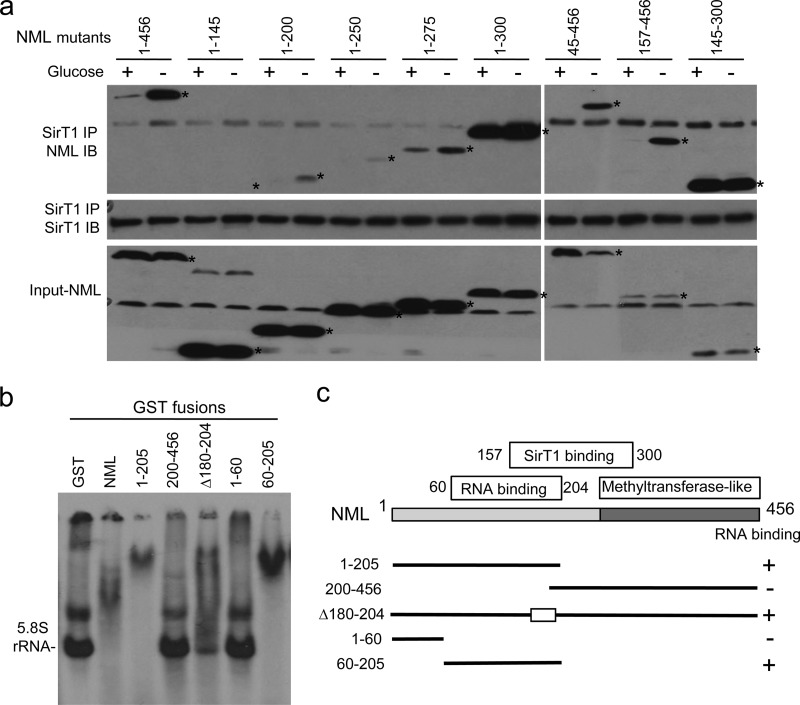
The results presented above suggest that SirT1 and RNA may have overlapping binding sites on NML, resulting in competitive binding that regulates eNoSC formation. To test this hypothesis, NML deletion mutants were analyzed for binding to SirT1. The result suggested that residues 157 to 300 of NML encode the SirT1 binding function (Fig. 9a). This region probably makes multiple contacts with SirT1, since weak SirT1 binding was still detectable with the NML fragments from residues 1 to 200 and residues 1 to 250. Most mutants capable of binding SirT1 also retained a response to glucose deprivation, but the NML fragment from residues 145 to 300 bound SirT1 constitutively, suggesting a potential loss of the RNA binding domain.
Fig 9.
Mapping of SirT1 and RNA binding sites on NML. (a) H1299 cells cotransfected with SirT1 and NML deletion mutants were cultured in normal medium or glucose-free medium for 16 h. The SirT1-NML interaction was analyzed by IP-Western blotting. (b) GST-NML fusion proteins treated with RNase A and RNasin were purified and tested for binding to in vitro-transcribed 35S-labeled 5.8S rRNA by EMSA. (c) Diagram of GST-NML mutants and summary of RNA binding analysis in panel b.
To identify the region of NML involved in RNA binding, GST-NML fusion proteins were purified from E. coli and treated with RNase A/RNasin, and similar amounts of proteins were tested for binding to RNA using EMSA. The analysis showed that residues 1 to 205 of NML had strong RNA binding, whereas the C-terminal half (residues 200 to 456) did not bind RNA (Fig. 9b). A smaller fragment of NML that contained the sequence between residues 60 and 205 also showed RNA binding activity. This region contains 25% lysine and arginine residues, has a calculated isoelectric point of 9.4, and is predicted to be partially unstructured (Fig. 10). The lack of homology to known RNA binding domains suggests that this region binds to RNA mainly through electrostatic interactions, independently of the specific RNA sequence or structure. These results suggest that the SirT1 and RNA binding sites on NML overlap partially, which may account for the competition (Fig. 9c). The result is also consistent with the unregulated SirT1 binding by the mutant consisting of the NML fragment from residues 145 to 300 (NML-145-300) (Fig. 9a), since this mutant has lost a significant part of the RNA binding site. The strong basal SirT1 binding by the NML-1-300 mutant remains unclear but may be caused by abnormal folding of this mutant.
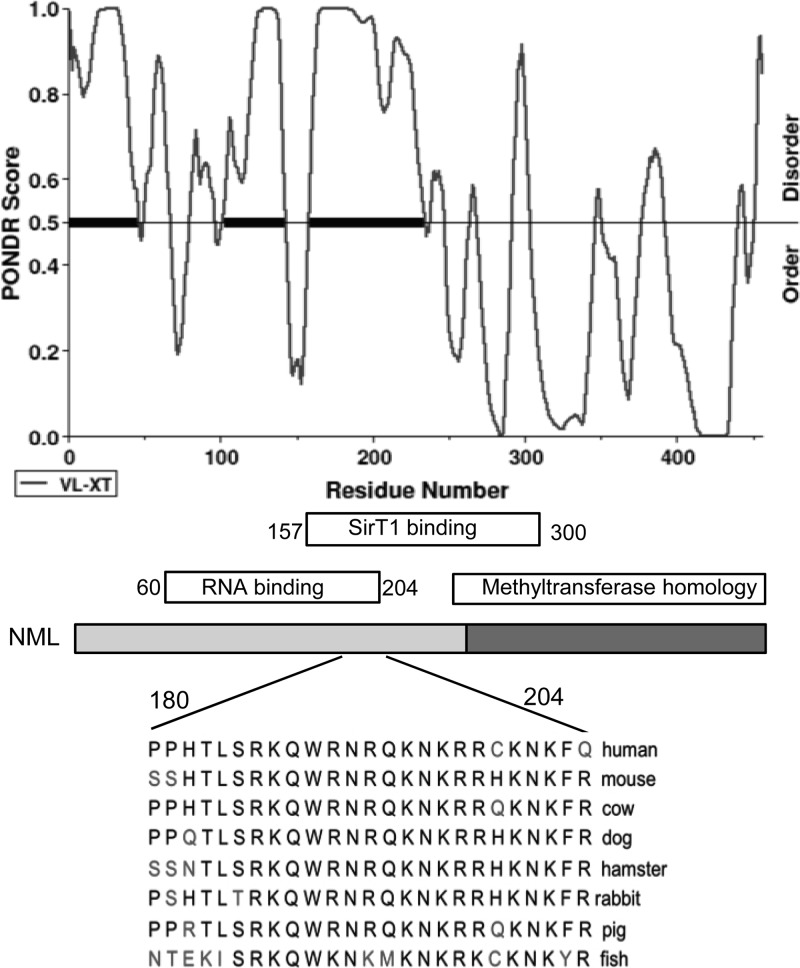
Fig 10.
The RNA binding site of NML is predicted to be unstructured. (Top) Result of analysis of NML by predictors of natural disordered regions (PONDR); (middle) NML diagram showing the relative locations of SirT1 and RNA binding regions mapped by deletion analysis; (bottom) alignment of residues 180 to 204 of NML showing the sequence conservation between species.
Reduction of RNA binding increases the repression function of NML.
Within the RNA binding site, a region from residues 180 to 204 was of particular interest because it contains 10 K/R residues (42%), and is highly conserved among different species (Fig. 10). A small internal deletion mutant, NMLΔ180-204, showed reduced binding to RNA (Fig. 9b and 11a) in EMSA and did not coprecipitate with rRNA during immunoprecipitation (Fig. 6c and d). Interestingly, the NMLΔ180-204 mutant showed stronger SirT1 binding than wild-type NML under both high- and low-glucose conditions, and the stimulation by glucose deprivation was less dramatic than that for wild-type NML (Fig. 11b). Consistent with its partial RNA binding deficiency, GST-NMLΔ180-204 showed stronger SirT1 binding than GST-NML before RNase A treatment. RNase A stimulated GST-NML binding to SirT1 but did not stimulate GST-NMLΔ180-204 binding to SirT1. Furthermore, purified RNA did not inhibit SirT1 binding by GST-NMLΔ180-204 (Fig. 11c). Therefore, this property of NMLΔ180-204 was consistent with the notion that interaction with RNA interferes with SirT1 binding.
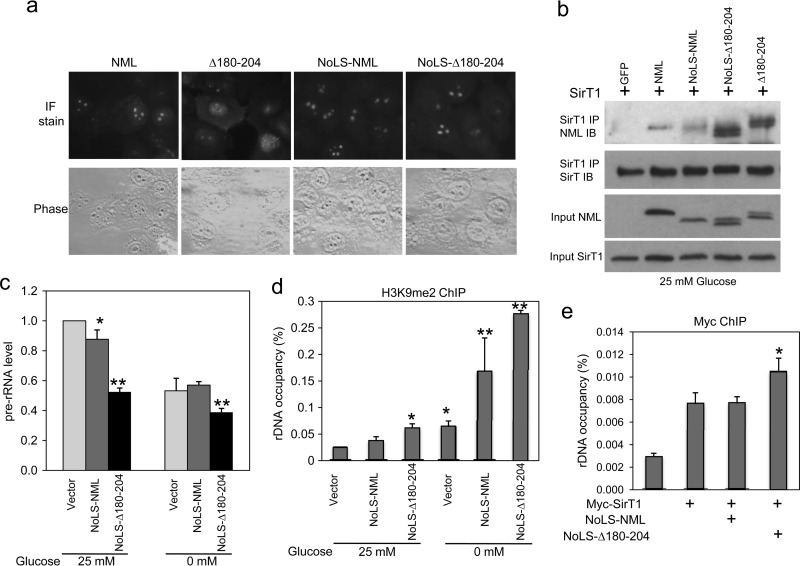
Immunofluorescence staining showed that the NMLΔ180-204 mutant was partially deficient for nucleolar localization (Fig. 12a), which may complicate analysis of its repression function in the nucleolus. To address this issue, the nucleolar localization signal sequence (NoLS; KKLKKRNK) from the MDM2 RING domain was grafted to the N terminus of NMLΔ180-204 and wild-type NML (as a control). Previous studies showed that this sequence was sufficient to target green fluorescent protein (GFP) or p53 to the nucleolus (25, 26). The NoLS fusion restored nucleolar localization of NMLΔ180-204 (Fig. 12a) while still maintaining elevated SirT1 binding compared to that for NML or NoLS-NML (Fig. 12b).
Fig 12.
Increased transcription repression by an RNA binding NML mutant. (a) H1299 cells were transfected with FLAG-tagged NML constructs fused to a nucleolar targeting signal sequence (NoLS) and stained with FLAG antibody to confirm the nucleolar localization of NoLS-NMLΔ180-204. IF, immunofluorescent. (b) NML and NMLΔ180-204 fused to the nucleolar targeting signal sequence were tested by IP-Western blotting after transfection into H1299 cells to confirm their SirT1 binding efficiency. (c) H1299 cells were transfected with the indicated plasmids and cultured in 0 mM glucose for 16 h. The pre-rRNA expression level was determined by RT-qPCR. Values are means ± SDs of triplicates. (d) H1299 cells were transfected with the indicated plasmids and cultured in 0 mM glucose for 16 h. The H3K9me2 level at the rDNA was analyzed by ChIP. Values are means ± SDs of triplicates. (e) H1299 cells were transfected with the indicated plasmids and cultured in 25 mM glucose. Myc-SirT1 recruitment to rDNA was analyzed by ChIP using anti-Myc antibody. Values are means ± SDs of triplicates. Statistically significant differences from the control are marked by asterisks (t test; *, P < 0.05; **, P < 0.01).
To test whether reduced RNA binding by NMLΔ180-204 leads to increased repression of pre-rRNA expression, H1299 cells were transfected with NoLS-NMLΔ180-204 and NoLS-NML. The NoLS-NMLΔ180-204 mutant reproducibly suppressed pre-rRNA expression more efficiently than NoLS-NML in high glucose, whereas wild-type NML had poor activity (Fig. 12c). Without the NoLS fusion, NMLΔ180-204 showed repression activity similar to that of wild-type NML, despite incomplete nucleolar localization (data not shown). ChIP analysis showed that NoLS-NMLΔ180-204 was more efficient in stimulating the H3K9me2 level at the rDNA (Fig. 12d) and recruiting SirT1 to rDNA (Fig. 12e). Therefore, reducing the RNA binding affinity of NML increased its SirT1 binding and transcription repression activities in high glucose. However, the NMLΔ180-204 mutant retained some response to glucose starvation in SirT1 binding, repression, and H3K9 methylation assays, probably due to its partial RNA binding activity (Fig. 9b and 11a). It is currently unclear whether a complete RNA binding-defective NML mutant can be created without compromising other functions. Overall, these results provide evidence that RNA binding inhibits the ability of NML to bind SirT1 and repress transcription in the nucleolus.
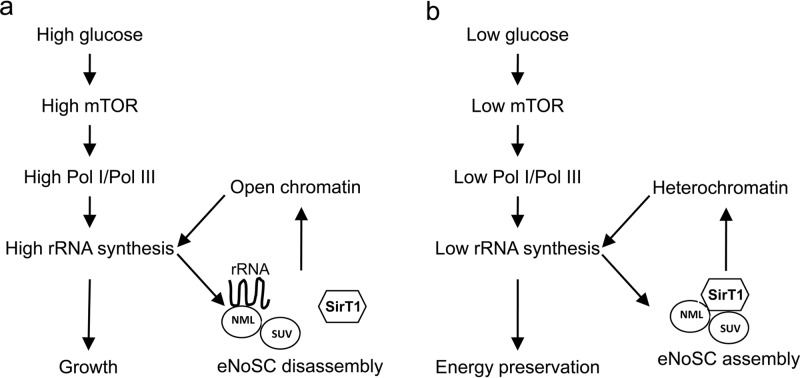
In summary, the results suggest that under nutrient-rich conditions, the assembly of eNoSC in the nucleolus is suppressed by nascent rRNA. As such, nutrient signaling that activates RNA Pol I in the nucleolus induces an RNA-mediated positive feedback to maintain the rDNA in open chromatin (Fig. 13a). Nutrient deprivation or other stresses that inhibit mTOR activity and reduce RNA Pol I transcriptional output will reduce the amount of nascent rRNA and stimulate NML-SirT1 binding, thus causing further suppression of rRNA transcription (Fig. 13b).
Fig 13.
Model of nutrient-dependent regulation of eNoSC assembly. (a) During nutrient-rich growth, mTOR stimulates RNA Pol I and Pol III synthesis of rRNA. The nascent rRNA in turn binds to NML and inhibits eNoSC assembly on rDNA, providing a positive feedback to amplify mTOR signaling. (b) During starvation, mTOR inactivation reduces Pol I and Pol III activity. The reduced nascent rRNA level enables NML to bind and recruit SirT1 to the nucleolus, promoting heterochromatin formation and silencing of the rDNA. As such, eNoSC reinforces mTOR regulation of rDNA transcription.
DISCUSSION
Ribosomal biogenesis consumes >50% of the energy supply in proliferating cells and is tightly coordinated with nutrient availability and growth signaling. Multiple stress, nutrient, and growth factor signaling pathways converge on mTOR. The ability of mTOR to simultaneously regulate rRNA and ribosomal protein biosynthesis by RNA Pol I, Pol II, and Pol III allows it to function as a central signal integrator (1, 23). Glucose deprivation causes a rapid decrease in pre-rRNA transcription through several mechanisms. A low ATP level activates AMP-activated protein kinase (AMPK), which phosphorylates Pol I-associated transcription factor TIF-1A and inhibits the assembly of the active transcription initiation complex (27). AMPK activation also inhibits mTOR. Since mTOR is a positive regulator of TIF-1A, inactivation of mTOR leads to further inhibition of pre-rRNA transcription by Pol I.
Recent identification of the SirT1- and SUV39H1-containing complex eNoSC suggests that there is an additional layer of nutrient-sensitive control at the level of rDNA heterochromatin formation. The composition of eNoSC suggests that it has an intrinsic ability to spread heterochromatin marks from inactive rDNA repeats into active repeats. NML binds to histone H3 with methylated K9, and the recruited SirT1 and SUV39H1 may act synergistically to promote deacetylation and methylation of adjacent H3K9 residues (16). NML also has structural features of a SAM-dependent methyltransferase in its C-terminal domain. Although unproven, it may methylate histones directly or in cooperation with SUV39H1. In proliferating cells, ∼50% of the rDNA repeats are thought to exist as silent heterochromatin. Therefore, the eNoSC constitutively present on the inactive rDNA repeats is poised to spread into active repeats. While the mTOR-mediated regulation of Pol I and Pol III provides a rapid mechanism to downregulate rRNA transcription, eNoSC may serve as a secondary feedback mechanism through epigenetic modification of rDNA.
Our results suggest that glucose availability regulates eNoSC assembly in the nucleolus through an rRNA-dependent mechanism. This places eNoSC under the control of upstream signal integrators, such as mTOR, and allows nucleolar heterochromatin formation to be coordinated with RNA polymerase activity. Overall, the results are consistent with the following model: under nutrient-rich conditions, mTOR and other factors stimulate RNA Pol I and Pol III synthesis of rRNA. The nascent rRNA in turn binds to NML, blocks NML interaction with SirT1, and prevents recruitment of SirT1 to the nucleolus. This mechanism provides a positive-feedback loop to mTOR signaling by protecting the active nucleolar rDNA from repression by eNoSC (Fig. 13a). During starvation, mTOR inactivation reduces Pol I and Pol III activity. The reduced level of nascent rRNA enables NML-SirT1 complex formation in the nucleolus, promoting heterochromatin spreading and silencing of the rDNA repeats (Fig. 13b). As such, eNoSC reinforces mTOR regulation of rRNA synthesis by sensing the level of Pol I and Pol III activity (nascent rRNA output).
The regulation of eNoSC assembly by rRNA suggests a unique mechanism different from the prevailing theme of ncRNA-mediated silencing. Nearly 70% of the mammalian genome produces ncRNA (28, 29). Most mammalian genes also produce antisense RNAs that regulate their expression (30). A well-established mechanism is that ncRNAs function by promoting the de novo assembly of chromatin-silencing complexes and targeting the protein complexes through siRNA base pairing with nascent ncRNA transcribing from specific target genes (31–33). In this model, a small amount of ncRNA can significantly inhibit target gene transcription. SirT1 activity is inhibited by the NAD+-dependent deacetylation product nicotinamide (34). Assembly of the SIR complex in yeast is inhibited by another product of deacetylation, O-acetyl-ADP-ribose (35). Our results add to the list of metabolic products that regulate SirT1 and suggest that rRNA also plays a role in regulating SirT1 complex assembly in the nucleolus.
The results suggest that nascent rRNA acts by a mass effect to saturate the SirT1 binding site on NML and prevents eNoSC assembly on rDNA. Such a mechanism may require high-level RNA expression and function only locally near active genes. rRNA is well suited for performing such a regulatory role because of the exceptionally high rate of pre-rRNA transcription per rDNA unit. Pre-rRNA transcription and processing occur simultaneously in the nucleolus. 5S rRNA is also transcribed near the nucleolus and delivered to the nucleolus by RPL5 (36). The local concentration of nascent 5S and 5.8S rRNA near active rDNA repeats may establish a protective zone that prevents NML-SirT1 complex formation. When rRNA synthesis starts to decline during stress, the nascent rRNA available for interaction with NML should rapidly decrease due to competition by ribosomal proteins, thus increasing NML-SirT1 binding. In addition to providing a positive-feedback mechanism to amplify nutrient signaling to the nucleolus, 5S rRNA expression outside the nucleolus may also stimulate nucleolar pre-rRNA transcription to ensure comparable output, since the rRNAs are assembled into mature ribosomes at an equal molar ratio. It remains to be determined whether similar feedback mechanisms also regulate gene expression outside the nucleolus.
Our results suggest that regulation of NML-SirT1 binding by RNA may not require sequence specificity. The E. coli RNA copurified with GST-NML strongly inhibited SirT1 binding in vitro. Several control RNAs interacted with NML as efficiently as rRNA in vitro. The region of NML important for RNA binding is rich in basic amino acids but does not have sequence homology to known RNA binding domains. It is likely that rRNAs copurify with NML mainly due to their abundance and colocalization in the nucleolus. The high concentration of rRNA in the nucleolus may be sufficient for regulating local eNoSC formation independently of sequence-specific binding. Interestingly, 18S rRNA showed negligible binding to NML in vivo, suggesting that despite its lack of sequence specificity for RNA in vitro, the in vivo RNA binding by NML may be regulated by accessibility to specific RNA species and competition with other RNA binding proteins. It is unclear whether the process of nascent 18S rRNA assembly into the preribosome precludes interaction with NML.
The sequence-nonspecific RNA binding by an unstructured region of NML is similar to the nonspecific RNA binding by heterochromatin proteins HP1swi6 and Chp1 from fission yeast (37, 38). HP1swi6 binds RNA through a positively charged flexible hinge region, whereas Chp1 binds RNA through a flexible sequence in its chromodomain. Furthermore, the RNA binding region of HP1swi6 also functions as a nuclear localization signal (38). This is similar to our observation that the positively charged region of NML from residues 180 to 204 involved in RNA binding is also needed for nucleolar localization. RNA binding has diverse functional effects on these proteins. RNA competes with the HP1swi6 chromodomain for binding to methylated histone H3 K9, stimulates Chp1 chromodomain binding to methylated H3 K9, and inhibits NML binding to SirT1. Intrinsically unstructured peptide sequences have important roles in signaling and regulation due to their ability to interact with diverse partners (39). It is possible that nonspecific RNA binding through an intrinsically unstructured region is an important feature of heterochromatin proteins necessary for their regulation at multiple genomic loci by locally produced RNA with diverse sequences.
ACKNOWLEDGMENTS
We thank Junn Yanagisawa for kindly providing the NML construct. We also thank the Moffitt Molecular Genomics Core for DNA sequence analyses.
This work is supported by grants from the National Institutes of Health (CA141244 and CA109636) and a Miles for Moffitt Milestone Award from the Moffitt Foundation to J. Chen.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 29 July 2013
REFERENCES
- 1.Mayer C, Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25:6384–6391 [DOI] [PubMed] [Google Scholar]
- 2.McStay B, Grummt I. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24:131–157 [DOI] [PubMed] [Google Scholar]
- 3.Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G. 2010. Initial genomics of the human nucleolus. PLoS Genet. 6:e1000889. 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemeth A, Langst G. 2011. Genome organization in and around the nucleolus. Trends Genet. 27:149–156 [DOI] [PubMed] [Google Scholar]
- 5.Fedoriw AM, Starmer J, Yee D, Magnuson T. 2012. Nucleolar association and transcriptional inhibition through 5S rDNA in mammals. PLoS Genet. 8:e1002468. 10.1371/journal.pgen.1002468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matera AG, Frey MR, Margelot K, Wolin SL. 1995. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J. Cell Biol. 129:1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. 2006. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 25:7577–7586 [DOI] [PubMed] [Google Scholar]
- 8.Voit R, Schafer K, Grummt I. 1997. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell. Biol. 17:4230–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhai W, Comai L. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grummt I, Pikaard CS. 2003. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4:641–649 [DOI] [PubMed] [Google Scholar]
- 11.Santoro R, Li J, Grummt I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32:393–396 [DOI] [PubMed] [Google Scholar]
- 12.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329–334 [DOI] [PubMed] [Google Scholar]
- 13.Schmitz KM, Mayer C, Postepska A, Grummt I. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24:2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer C, Neubert M, Grummt I. 2008. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 9:774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J. 2008. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133:627–639 [DOI] [PubMed] [Google Scholar]
- 16.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. 2007. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450:440–444 [DOI] [PubMed] [Google Scholar]
- 17.Gottlieb S, Esposito RE. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771–776 [DOI] [PubMed] [Google Scholar]
- 18.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. 2006. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20:1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. 2006. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 8:1025–1031 [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Marechal V, Moreau J, Levine AJ, Chen J. 1997. Proteolytic cleavage of the mdm2 oncoprotein during apoptosis. J. Biol. Chem. 272:22966–22973 [DOI] [PubMed] [Google Scholar]
- 21.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. 2006. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 26:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. 2005. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16:4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta S, Peterson TR, Sabatini DM. 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40:310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG. 2011. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 71:1418–1430 [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Chen J. 2003. MDM2-ARF complex regulates p53 sumoylation. Oncogene 22:5348–5357 [DOI] [PubMed] [Google Scholar]
- 26.Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. 2000. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2:179–181 [DOI] [PubMed] [Google Scholar]
- 27.Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, Voit R. 2009. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. U. S. A. 106:17781–17786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309:1559–1563 [DOI] [PubMed] [Google Scholar]
- 29.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488 [DOI] [PubMed] [Google Scholar]
- 30.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564–1566 [DOI] [PubMed] [Google Scholar]
- 31.Gerace EL, Halic M, Moazed D. 2010. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol. Cell 39:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 106:11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khalil AM, Rinn JL. 2011. RNA-protein interactions in human health and disease. Semin. Cell Dev. Biol. 22:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137–148 [DOI] [PubMed] [Google Scholar]
- 35.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121:515–527 [DOI] [PubMed] [Google Scholar]
- 36.Steitz JA, Berg C, Hendrick JP, La Branche-Chabot H, Metspalu A, Rinke J, Yario T. 1988. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 106:545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishida M, Shimojo H, Hayashi A, Kawaguchi R, Ohtani Y, Uegaki K, Nishimura Y, Nakayama J. 2012. Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol. Cell 47:228–241 [DOI] [PubMed] [Google Scholar]
- 38.Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Buhler M. 2012. HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol. Cell 47:215–227 [DOI] [PubMed] [Google Scholar]
- 39.Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18:756–764 [DOI] [PubMed] [Google Scholar]