Abstract
Monomethylated histone H4 lysine 20 (H4K20me1) is tightly regulated during the cell cycle. The H4K20me1 demethylase PHF8 transcriptionally regulates many cell cycle genes and is therefore predicted to play key roles in the cell cycle. Here, we show that PHF8 protein levels are the highest during G2 phase and mitosis, and we found PHF8 protein stability to be regulated by the ubiquitin-proteasome system. Purification of the PHF8 complex led to the identification of many subunits of the anaphase-promoting complex (APC) associated with PHF8. We showed that PHF8 interacts with the CDC20-containing APC (APCcdc20) primarily during mitosis. In addition, we defined a novel, KEN- and D-box-independent, LXPKXLF motif on PHF8 that is required for binding to CDC20. Through various in vivo and in vitro assays, we demonstrate that mutations of the LXPKXLF motif abrogate polyubiquitylation of PHF8 by the APC. APC substrates are typically cell cycle regulators, and consistent with this, the loss of PHF8 leads to prolonged G2 phase and defective mitosis. Furthermore, we provide evidence that PHF8 plays an important role in transcriptional activation of key G2/M genes during G2 phase. Taken together, these findings suggest that PHF8 is regulated by APCcdc20 and plays an important role in the G2/M transition.
INTRODUCTION
Proper cell division involves a highly coordinated sequence of events that is essential for genomic integrity. Failure of the cell to efficiently regulate various phases of the cell cycle leads to DNA damage, genomic instability, and, ultimately, cancer (1). Histone modifications are important players in this process, as they can directly modify chromatin and serve as a signaling platform to potentiate DNA template-based cellular events such as DNA replication, transcription, and DNA damage sensing and repair (2). Histones, through which DNA is packaged and organized, are subjected to a plethora of posttranslational modifications, such as methylation. Monomethylation of histone 4 lysine 20 (H4K20me1) is tightly regulated during the mammalian cell cycle (3). Various studies have shown the importance of this mark and the corresponding methyltransferase, PR-Set7/Set8/KMT5A, in the regulation of the cell cycle (3–6). PR-Set7 and H4K20me1 abundances are dynamically regulated during the cell cycle: they are highest during G2 phase and mitosis and lowest during G1 and S phases. H4K20me1 accumulation during late G2 phase and mitosis recruits L3MBTL1 and the condensin subunits N-CAPD3 and N-CAPG2 to chromosomes, triggering chromatin compaction and shutdown of transcription in preparation for mitosis (7, 8).
Two related histone demethylases, PHF8 and KIAA1718, have been reported to demethylate a variety of substrates, including H4K20me1 (7, 9). Both proteins bind H3K4me3 via their PHD finger, which is typically enriched at the transcription start sites (TSSs) and may therefore play a role in their recruitment to target promoters (10). PHF8 activates gene transcription primarily by demethylating H3K9me1 and H4K20me1 (7, 9). At ribosomal DNA (rDNA) loci, however, PHF8 preferentially demethylates H3K9me2 (11, 12). The importance of enzymatic demethylation mediated by PHF8 is underscored by the discovery of the link between PHF8 mutations that disrupt its enzymatic activity and X-linked intellectual disability (XLID) and craniofacial deformities (13–15).
PHF8 binds to the TSSs of 7,000 to 8,000 genes, or about one-third of the annotated genome, but affects the expression of only a small number of genes (7, 9, 16). Therefore, PHF8 is likely to be important for the regulation of gene expression in a context-dependent manner. Consistent with this hypothesis, PHF8 acts as a transcriptional coactivator for retinoic acid receptor alpha (RARα) and is recruited to target genes upon retinoic acid induction (such as in the case of all trans-retinoic acid (ATRA) treatment in acute promyelocytic leukemia) (17, 18). Notch activation also leads to the assembly of PHF8 and complex components on Notch target genes, which is important for the activation of key Notch genes, such as those for interleukin-7 receptor and DTX1 (19). Notch signaling plays an important role in human T cell acute lymphoblastic leukemia (T-ALL); consequently, a loss of PHF8 compromises Notch-mediated gene activation, leading to an inhibition of tumor formation in a mouse xenograft model (19). In HeLa cells, PHF8 interacts with E2F1, HCF-1, and SET1A to regulate E2F1-regulated promoters during G1/S phase (7). Interestingly, chromatin immunoprecipitation-sequencing (ChIP-seq) and microarray data also show that PHF8 binds to and regulates many other important cell cycle genes, including genes that are enriched at G2/M phase and are important for the G2/M transition and whose transcription is known to persist beyond G1/S phase.
In this study, we show that PHF8 plays an important role in the regulation of the G2/M transition and is regulated by the anaphase-promoting complex (APC). We show that PHF8 protein levels are highest during G2 phase and mitosis, and through various assays, we demonstrate that PHF8 protein levels are regulated by ubiquitin-mediated proteolysis mediated by the APC. The APC is an E3 ubiquitin ligase complex that is a key regulator of the cell cycle (20, 21). CDC20 and CDH1 are the primary substrate recognition subunits of the APC, with the APC containing CDC20 (APCcdc20) and APCcdh1 exhibiting distinct temporal activities and roles during the cell cycle. During mitosis, the APCcdc20 is inhibited by the spindle assembly checkpoint (SAC) and most notably targets securin for degradation, allowing sister chromatid separation during anaphase. On the other hand, the APCcdh1 is active later during mitosis and in G1 phase and is important for suppressing mitotic cyclin levels and regulating the G1/S transition. CDC20 binds proteins via conserved sequence motifs such as the KEN box or D box (22, 23). We find that PHF8 binds primarily to the APCcdc20 in a KEN- and D-box-independent manner, and we identify a novel motif, LXPKXLF, that mediates the interaction between PHF8 and CDC20. Mutation of this motif renders PHF8 resistant to polyubiquitylation and targeting for proteasomal degradation by the APCcdc20. Additionally, we show that high PHF8 protein levels at G2 phase and mitosis correlate with accumulation of PHF8 on its G2/M target genes and is important for the maintenance of low H3K9me1 and H4K20me1 levels over TSSs of these genes during G2 phase, allowing active transcription. Loss of PHF8 leads to pleiotropic cell cycle defects, most prominently an extended G2 phase and highly defective mitosis. Collectively, our results indicate that PHF8 plays a key role in the regulation of the G2/M transition and is targeted for degradation by the important cell cycle regulator APCcdc20.
MATERIALS AND METHODS
Cell culture, drug treatments, cell synchronization, and flow cytometry.
HeLa, HEK293T, and MCF7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. MG132 (Calbiochem) and cycloheximide (Sigma) were resuspended in dimethyl sulfoxide (DMSO; Sigma) and used at final concentrations of 10 μM and 100 μg/ml, respectively. Thymidine (Sigma) and nocodazole (Sigma) were resuspended in double-distilled water (ddH2O) and used at final concentrations of 2 mM and 100 ng/ml, respectively. Puromycin dichloride (Sigma) was resuspended in ddH2O and used at a final concentration of 1 to 2 μg/ml. Cells were synchronized in S phase by double-thymidine treatment including incubation in thymidine for 18 h, washing with phosphate-buffered saline (PBS) twice, and supplementation with fresh medium to release the cells for 9 h, followed by an additional thymidine treatment for 18 h. Cells were synchronized in M phase by thymidine-nocodazole treatment including incubation in thymidine for 18 h, washing with PBS twice, and supplementation with fresh medium with nocodazole for 12 h. For flow cytometry analysis, cells were fixed in cold 80% ethanol and stored at −20°C until they were ready for staining. Cells were washed twice with cold PBS and pelleted by centrifugation. The cell pellet was resuspended in PBS containing 50 μg/ml propidium iodide and 250 μg/ml RNase A and incubated at 37°C for 20 min. Cells were filtered by passage through a cell strainer cap attached to a round-bottom tube (BD Falcon). Cells were analyzed on a BD LSR Fortessa instrument using BD FACS Diva software and further analyzed with FlowJo software.
Plasmids, mutagenesis, transfection, siRNA, and shRNA knockdown.
Sequence-verified open reading frame clones were cloned into the pOZ-Flag-HA construct at the XhoI/NotI site or into pENTR-D-TOPO (Invitrogen) and recombined into the following Gateway destination vectors: MSCV-Flag-HA-IRES-PURO, CMV-Flag-HA, and CMV-MYC. The following CDC20 plasmids were obtained from Addgene: pCS2-HA-CDC20 (Addgene plasmid 11594) and pCS2-MYC-CDC20 (Addgene plasmid 11593). Point mutations were generated by using a QuikChange site-directed mutagenesis kit (Agilent). Cells were transfected with the indicated plasmids by using Lipofectamine 2000 (Invitrogen) for HeLa cells and TransIT-293 (Mirus) for HEK293T cells, according to the manufacturers' specifications. Transfected cells were typically harvested at 48 h posttransfection for analysis. Small interfering RNA (siRNA) transfection was carried out by using Lipofectamine RNAiMAX according to the manufacturer's specifications. siRNA (20 nM) was reverse transfected into cells. Twenty-four hours later, 20 nM the same siRNA was transfected into the same cells. Seventy-two hours after the first transfection, cells were harvested for analysis. Two CDC20 siRNAs targeting different regions of CDC20 were used: CDC20 siRNA 1 (catalog number D-003225-12-0005; Thermo Scientific) (5′-GCACAGUUCGCGUUCGAGA-3′) and CDC20 siRNA 2 (catalog number D-003225-10-0005; Thermo Scientific) (5′-GGGCCGAACUCCUGGCAAA-3′). Nontargeting control siRNA (catalog number D-001210-01-05) was purchased from Thermo Scientific. For short hairpin RNA (shRNA) knockdown experiments, lentiviral constructs containing shRNAs were transfected with helper plasmids into HEK293T cells to be packaged into lentiviruses, which were harvested at 48 to 72 h posttransfection. Cells were infected with lentivirus with the aid of hexadimethrine bromide (Polybrene) at a 4-μg/ml final concentration. Five PHF8 shRNAs in the pLKO.1 vector targeting different regions of PHF8 were used: PHF8 shRNA 1 (5′-CCGGGCAGGTAAATGGGAGAGGTTTCTCGAGAAACCTCTCCCATTTACCTGCTTTTTG-3′), PHF8 shRNA 2, (5′-CCGGCGACCCTGATAATAAGACCAACTCGAGTTGGTCTTATTATCAGGGTCGTTTTTG-3′), PHF8 shRNA 3 (5′-CCGGCGAACCGTACAGCTCATTAAACTCGAGTTTAATGAGCTGTACGGTTCGTTTTTG-3′), PHF8 shRNA 4 (5′-CCGGGCGAACCGTACAGCTCATTAACTCGAGTTAATGAGCTGTACGGTTCGCTTTTTG-3′), and PHF8 shRNA 5 (5′-CCGGCCCAACTGTGAAGTCTTGCATCTCGAGATGCAAGACTTCACAGTTGGGTTTTTG-3′). The control shRNA was a pLKO.1 vector containing an shRNA sequence targeting green fluorescent protein (GFP) from Thermo Scientific.
RNA extraction, reverse transcription, and qPCR.
Total RNA was extracted from cells by using TRIzol (Invitrogen), followed by chloroform extraction and isopropanol precipitation. RNA was then treated with RNase-free DNase I (Ambion), and the DNase was inactivated. Reverse transcription was carried out on 1 to 5 μg total RNA by using Superscript III (Invitrogen) and oligo(dT). cDNAs obtained were diluted accordingly and used for quantitative PCR (qPCR). Gene-specific primers were used together with SYBR green (Roche) for detection on a LightCycler 480 system (Roche). Primers used in this study include GAPDH_F (ATGCCTCCTGCACCACCAAC), GAPDH_R (GGGGCCATCCACAGTCTTCT), PHF8_F (AGGACAAGGAAAGCGTCCCAA), PHF8_R (ACACAGGAGGGCTCACAGAA), CDC20_F (CTGAACTCAAAGGTCACACAT), CDC20_R (GGTCCAACTCAAAACAGCG), CDH1_F (AACGAGCTGGTGAGCACG), CDH1_R (GGACATTGCCAGGTACAGCA), CCNB1_F (ACAAAGCACATGACTGTCAAGA), CCNB1_R (CAAGTTACACCTTTGCCACAG), NCAPG_F (GACTCTGAAAGTGATCATGAAG), NCAPG_R (CATTGAGAAATTGGGCAAGG), CENPA_F (ACAAGAGGCAGCAGAAGCAT), CENPA_R (TCCGGGCCAGTTGCACAT), E2F1_F (TGGAGCAAGAACCGCTGTTGT), E2F1_R (AAGTCCTCCCGCACATGCT), CDC25A_F (ATGAAATGCCAGTCTTACTGTGA), CDC25A_R (CTTCTTCAGACGACTGTACATC), RBL1_F (GATAGATTGTGACTTAGAAGATGC), and RBL1_R (CAGTGCAAATGACTTCACTCTTC).
Immunoprecipitation, tandem affinity purification, mass spectrometry, and silver staining.
Regular immunoprecipitation (IP) was carried out by lysing cells with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, protease, and phosphatase inhibitors) on ice for 30 min, centrifuging the cells at maximum speed for 10 min, and retrieving the supernatant (lysate). Specific antibodies or the corresponding IgG controls were added to the lysate and incubated overnight at 4°C. Protein A or G beads were then added to the tube and incubated for 1 h at 4°C. Beads were washed three times with lysis buffer, and the immunocomplexes were eluted from beads by the addition of SDS loading buffer and boiled for 5 min. The immunocomplexes could then be analyzed by immunoblotting. Tandem affinity purification (TAP) was carried out by using 20 500-cm2 dishes of cells (24). Briefly, cells underwent hypotonic treatment and Dounce homogenization, followed by centrifugation. The pellet was retained as the nuclear fraction and subjected to high-salt extraction to extract nuclear protein complexes. The nuclear extract was dialyzed (20 mM Tris-HCl [pH 7.9], 100 mM KCl, 1 mM MgCl2, 0.2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 20% glycerol) and was subjected to a first round of immunoprecipitation using Flag M2 beads (Sigma). Immunocomplexes were eluted by using Flag peptide, subjected to a second round of immunoprecipitation by using hemagglutinin (HA) beads (Santa Cruz), and eluted by using HA peptides. A portion of the elution mixture was run on a 4 to 12% Bis-Tris gel and silver stained (SilverQuest silver staining kit; Invitrogen). The rest of the elution mixture was trichloroacetic acid (TCA) precipitated, trypsin digested, loaded onto stage tips, and analyzed on a LTQ-Velos linear ion trap mass spectrometer (ThermoFinnigan).
Live-cell imaging and analysis and immunofluorescence staining.
HeLa-RFP-H2B and HeLa-RFP-ligase cells were plated onto 24-well black/clear imaging plates. Twenty-four hours later, lentiviruses with shRNAs targeting GFP or PHF8 were added to the cells. At 24 h postinfection, the imaging plate was mounted onto a motorized stage (Prior ProScan II) on a Nikon TE2000E PFS inverted microscope fitted with an incubation chamber maintained at 37°C and supplied with 5% CO2. Differential interference contrast (DIC) or red fluorescent protein (RFP) fluorescent images were taken every 12 min for 48 h by using a 20× Plan Apo 0.75-numerical-aperture (NA) objective lens with a Hamamatsu Orca cooled charge-coupled-device (CCD) camera and Nikon Elements software. TIFF files of each image were exported and analyzed manually in stacks with ImageJ (NIH). For HeLa-RFP-ligase cells, the duration of G1 phase was determined by taking the first frame after the end of mitosis (concentration of the RFP signal posttelophase) to the frame in which foci first appeared. The duration of S phase was determined by taking the first frame from the appearance of foci to the frame in which the last foci disappeared. The duration of G2 phase was determined from the first disappearance of the last foci to the frame in which the signal became diffused. The duration of M phase was determined from the first frame in which the signal was diffused throughout the cell to the frame where the signal was concentrated to the chromosomes again. An uninfected control was also tracked to document any effect of virus infection itself on the cell cycle. A separate set of cells was plated and transduced in parallel on the same day, and cells were harvested at 48 h postinfection to determine the efficiency of PHF8 knockdown by immunoblotting. Knockdown of PHF8 by the 4 independent PHF8 shRNAs was determined to be >90% each time. Immunofluorescence staining was carried out by plating cells onto coverslips, fixing cells with 3% paraformaldehyde (PFA), permeabilizing cells with PBS with 0.2% Triton X, and blocking with 1% bovine serum albumin (BSA) in PBS-Tween (PBST) before staining with antibodies at the appropriate dilutions.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was carried out as described previously (25). ChIP-qPCR primers used in this study are as follows: CCNB1_TSS_F (GTGAGAAAGAGAACTGGACG), CCNB1_TSS_R (GTTACAGAGTTGCTGGCGG), CCNB1_3′end_F (GTCTTCCAGACTGTCAAGAACA), CCNB1_3′end_R (CAAGTTTACAAGTTACACCTTTGCC), NCAPG_TSS_F (TAGACGCATGCGCCCAAAG), NCAPG_TSS_R (AGTATTTACCGCTCGAACGTTCA), NCAPG_3′end_F (GCTAGTAGCTTGCATTTGAGAAG), NCAPG_3′end_R (GCCAGTTTGTGGTACTTCCTTC), CENPA_TSS_F (CACCGCCAACCAGGCATT), CENPA_TSS_R (ACCAAGTTGTGTGACTCTGCT), E2F1_TSS_F (TTAAAGCCAATAGGAACCGCC), E2F1_TSS_R (GCAAAGTCCCGGCCACTTTTA), E2F1_3′end_F (ACCCTCCAATCTGCACTTTGAT), E2F1_3′end_R (CGAAATGTTCCCAACAGAGTC), CDC25A_TSS_F (AGCCGATGACCTGGCAGAGT), CDC25A_TSS_R (TAGTTGGCGCCAAACGGAATC), CDC25A_3′end_F (GAGGACTTCTTCCTACACGC), CDC25A_3′end_R (GGTATAATCTGAAGGCCATCC), RBL1_TSS_F (GGACAGGTCTTTCAGAATCTGA), RBL1_TSS_R (ACGTGTTGTCATCCACCGTCT), CKS1B_TSS_F (GTTGGGAGTTGCTTGGAGGTT), and CKS1B_TSS_R (ACTAACCGATACTCAAACTCCTC).
Protein purification from Sf9 cells and histone demethylation assay.
Wild-type (WT) and mutant PHF8 proteins were cloned into a baculovirus expression vector, pFastBacHT A (Invitrogen), and transfected into Sf9 cells by using the Bac-to-Bac HT Vector kit (Invitrogen). His-tagged PHF8 was produced according to the manufacturer's specifications, purified on Ni-nitrilotriacetic acid (NTA)-agarose beads (Qiagen), and eluted by imidazole (Sigma). A histone demethylation assay was carried out on calf histones at 37°C for 4 to 6 h with the following buffer: 50 mM HEPES-KOH (pH 7.9), 50 mM KCl, 1 mM MgCl2, 1 mM α-ketoglutarate, 2 mM ascorbic acid, and 100 μM ammonium iron(II) sulfate.
In vivo ubiquitylation and PHF8 degradation in mitotic Xenopus extracts.
Cells were harvested in PBS containing 10 mM the deubiquitylating enzyme inhibitor N-ethylmaleimide (NEM). Pelleted cells were lysed in a solution containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% SDS, 1 mM dithiothreitol (DTT), and 10 mM NEM and vortexed vigorously before boiling for 20 min. Denatured lysates were cooled, and about 20 volumes of a solution containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 10 mM NEM was added to dilute out the SDS in the lysate. Diluted lysates were centrifuged at maximum speed for 15 min, the supernatant was retrieved, and the amount of protein was quantitated by a bicinchoninic (BCA) protein assay (Thermo scientific). Similar amounts of lysate were used for IP, which was carried out at room temperature for 4 h. Mitotic Xenopus laevis extracts and PHF8 degradation assays were prepared as described previously (26).
Antibodies.
Antibodies used in this work include anti-PHF8 (catalog numbers ab36068 [Abcam] and A201-772A [Bethyl Laboratories]), anti-RNA polymerase II (Pol II) (CTD4H8) (catalog number 05-623; Millipore), anti-H3 (catalog number 39163; Active Motif), anti-H3K4me3 (MC315) (catalog number 04-745; Millipore), anti-H3K4me2 (CMA303) (catalog number 05-1338; Millipore), anti-H3K9me2 (catalog number ab1220; Abcam), anti-H3K9me1 (catalog number ab8896; Abcam), anti-H3K36me3 (catalog number ab9050; Abcam), anti-H4 (catalog number 39269; Active Motif), anti-H4K20me1 (catalog number ab9051; Abcam), anti-CDC27 (catalog number sc-13154; Santa Cruz), anti-CDC20 (catalog number sc-13162; Santa Cruz), anti-CDH1 (catalog number sc-56381; Santa Cruz), anti-cyclin B1 (catalog number sc-53236; Santa Cruz), anti-cyclin E (catalog number sc-198; Santa Cruz), antiactin (catalog number A2228, Sigma), anti-Flag (M2) (catalog number F1804; Sigma), anti-HA (catalog number MMS-101P; Covance), anti-MYC (catalog number sc-40; Santa Cruz), and anti-HIS (catalog number sc-8036; Santa Cruz).
RESULTS
PHF8 protein levels are regulated by the ubiquitin-proteasome pathway.
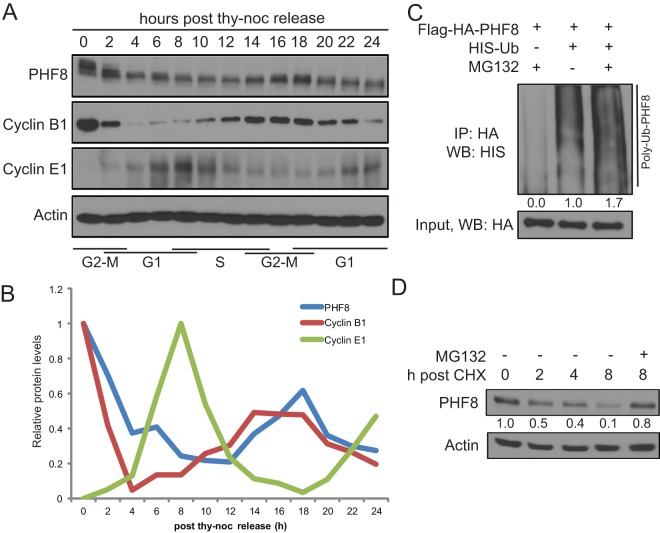
Given that previous studies suggested that PHF8 is an important regulator of the cell cycle, we wished to determine whether its expression is modulated during the cell cycle (7). HeLa cells were synchronized in mitosis (M phase) and harvested at 2-h intervals upon release over 24 h. As shown in Fig. 1A and B, PHF8 protein levels were highest in M phase, declined 3- to 4-fold in G1 phase, and reaccumulated during G2 phase. However, PHF8 mRNA levels were more or less constant throughout the cell cycle (data not shown), suggesting that PHF8 protein fluctuations during the cell cycle occur via posttranscriptional mechanisms.
Fig 1.
PHF8 protein levels are regulated by the ubiquitin-proteasome system, with the highest levels being found at G2 and M phases. (A and B) HeLa cells were synchronized at M phase and harvested at 2-h intervals upon release for 24 h. (A) Total cell lysates were probed for PHF8, the G2/M marker cyclin B1, and the G1/S marker cyclin E1. (B) ImageJ quantification of data in panel A was carried out with normalization against actin, and the highest protein level was set to 1. thy-noc, thymidine-nocodazole. (C) Flag-HA-PHF8 was transfected with the empty vector or HIS-Ub and treated with DMSO or MG132. Denatured lysates underwent HA IP and were probed for HIS-Ub. The extent of ubiquitylation in each lane was quantified by ImageJ and normalized against the input, and the quantification is presented below the HIS-Ub blot. WB, Western blotting. (D) Endogenous PHF8 half-life was assessed by cycloheximide (CHX) chase. A total of 100 μg/ml of cycloheximide and 10 μM MG132 or an equal volume of DMSO were added, and cells were harvested at 0, 2, 4, or 8 h. ImageJ was used to quantify the amount of PHF8 (normalized against actin), which is presented below the PHF8 blot.
Given that many important cell cycle-regulated proteins, such as cyclins, are regulated by cell cycle-specific E3 ligases, we investigated whether the ubiquitin-proteasome system plays a role in regulating PHF8 protein levels. Flag-HA-PHF8 and HIS-tagged ubiquitin (HIS-Ub) were cotransfected into HEK293T cells treated with DMSO or the proteasome inhibitor MG132. Cells were lysed under denaturing conditions, and anti-HA immunocomplexes were probed for HIS-Ub. As shown in Fig. 1C, PHF8 was polyubiquitylated, and the level of PHF8 polyubiquitylation increased in the presence of MG132. The half-life of PHF8 is approximately 2 to 3 h, as determined by a cycloheximide chase (Fig. 1D). Importantly, degradation of PHF8 was rescued by MG132 (Fig. 1D), indicating that PHF8 is ubiquitylated and degraded by the ubiquitin-proteasome system.
PHF8 interacts with the APCcdc20, primarily during G2/M phase.
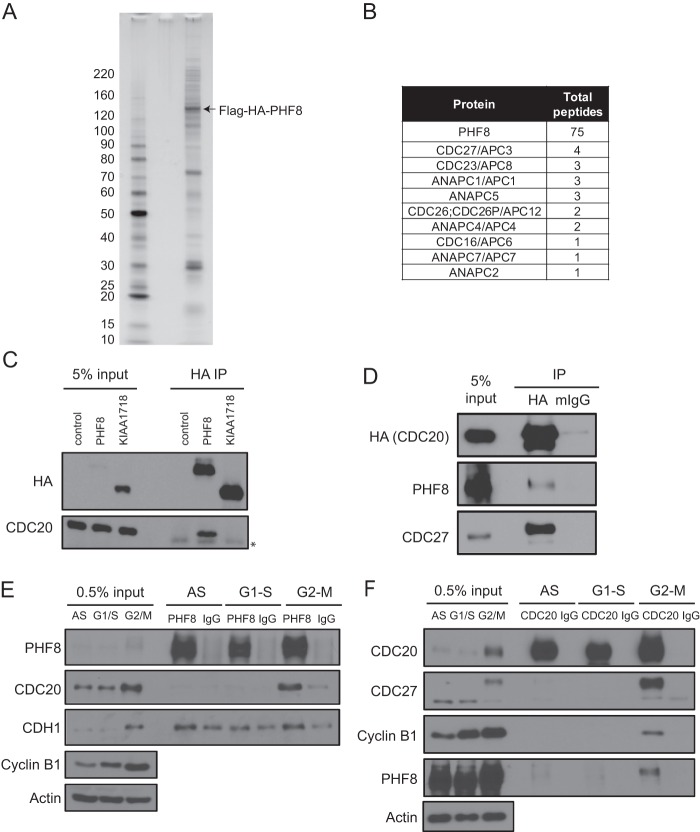
To better understand the function of PHF8, we used tandem affinity purification (TAP) to purify PHF8 from the breast cancer cell line MCF7, as breast cancer cells have been reported to overexpress PHF8 (Oncomine). Flag-HA-tagged PHF8 and its associated proteins were purified and identified by mass spectrometry (Fig. 2A and B). Consistent with its role in chromatin and transcription, many transcriptional factors, chromatin remodelers, and DNA damage proteins were found to be associated with the TAP-purified PHF8. Interestingly, we also identified multiple components of the anaphase-promoting complex (APC) (9 out of approximately 11 to 13 core APC subunit components) (Fig. 2B). Because the APC functions primarily by targeting key cell cycle proteins for proteasomal degradation during the cell cycle, we hypothesized that the APC's interaction with PHF8 may be important for the regulation of PHF8's protein abundance and function during the cell cycle.
Fig 2.
PHF8 interacts with the APCcdc20 during M phase. (A and B) MCF7-pOZ-Flag-HA-PHF8 cells were used for complex purification. Tandem affinity purification (Flag followed by HA IPs) was carried out on nuclear extracts. Immunocomplexes were subjected to silver staining (A) and mass spectrometry (B). Panel B shows a summary of components of the APC complex that was identified in the PHF8 complex and their total peptide numbers. (C) The control empty vector, Flag-HA-PHF8, or Flag-HA-KIAA1718 was transfected in HEK293T cells, and lysates were subjected to HA IP and probed for CDC20. (∗, IgG heavy chain) (D) Semiendogenous IP was carried out by transiently transfecting HA-CDC20 and carrying out HA or mouse IgG (mIgG) IP with the lysates. (E and F) Endogenous IP using PHF8 (E) or CDC20 (F) antibody was carried out by using lysates from asynchronous (AS) or S- or M-phase-synchronized HeLa cells.
To confirm the mass spectrometry data, we overexpressed Flag-HA-PHF8 in HEK293T cells and performed semiendogenous immunoprecipitation (IP) using either anti-HA antibody or mouse IgG as a negative control. We observed that approximately 3% of CDC27, a core subunit of the APC, was pulled down by HA but not by IgG antibodies (see Fig. S1 in the supplemental material). The binding specificity of the APC is generally conferred by the coactivator subunits CDC20 and CDH1, which activate the APC at distinct times of the cell cycle. Interestingly, Flag-HA-PHF8 appeared to interact primarily with CDC20 but not CDH1 (see Fig. S1 in the supplemental material). Furthermore, overexpression of Flag-HA-tagged PHF8 or its closely related demethylase KIAA1718, followed by HA semiendogenous IP, showed that the interaction with CDC20 is specific to PHF8 and does not occur with KIAA1718 (Fig. 2C).
Next, we performed reciprocal IPs, where we observed that CDC20 interacts with an approximately stoichiometric amount of CDC27, as expected, and has a weaker (<1%) but specific (compared to the control) interaction with PHF8 (Fig. 2D). We wished to determine whether this interaction could be recapitulated between endogenous PHF8 and CDC20 and whether the interaction occurs in a cell cycle-specific manner. To this end, we used asynchronous and S-phase- and M-phase-synchronized cell lysates for IP. As binding of APC with its substrates is often weak and transient, cells were treated with MG132 for 4 h before harvesting. Interestingly, we observed that PHF8 interacted most strongly with CDC20 in M phase, which is when the APCcdc20 is typically active (Fig. 2E). The reciprocal anti-CDC20 IP also clearly showed that CDC20 immunocomplexes from M-phase lysates pulled down PHF8 most strongly (Fig. 2F). The strength and cell cycle specificity of the CDC20-PHF8 interaction were similar to those of the interaction between CDC20 and cyclin B1, a substrate of the APCcdc20 (Fig. 2F).
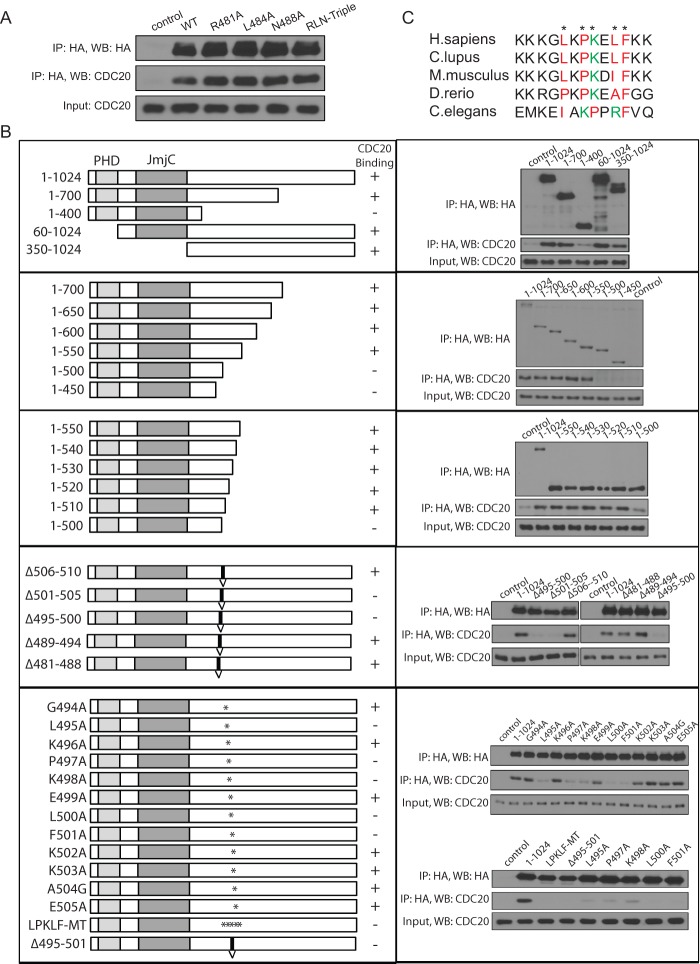
A novel motif mediates PHF8's binding to CDC20.
Several amino acid motifs are known to be important for binding to CDC20 and recruitment to the APC, the most common being the KEN box and the destruction or D box (RXXLXXXN) (22, 23). We found that PHF8 contained a putative D-box sequence between amino acids (aa) 481 and 488. However, mutation of each key amino acid residue to alanine (R481A, L484A, and N488A) and an R481A L484A N488A triple mutant (RLN-Triple) did not affect CDC20 binding (Fig. 3A). We therefore systematically mapped the region responsible for PHF8 binding to CDC20 (Fig. 3B). We transfected various truncation mutants of Flag-HA-PHF8 (aa 1 to 1024, aa 1 to 700, aa 1 to 400, aa 60 to 1024, and aa 350 to 1024) into HEK293T cells and performed anti-HA IPs. As shown in Fig. 3B (top), loss of aa 400 to 700 abolished CDC20 binding, and this region that is critical for interactions was narrowed down further to be between aa 400 and 510 by analyzing the progressive C-terminal deletion mutants (Fig. 3B, second and third panels).
Fig 3.
A novel motif on PHF8 governs its binding to CDC20. (A) The D-box motif on PHF8 was mutated in Flag-HA-PHF8 to generate mutants with an R481A, L484A, or N488A mutation or the RLN-Triple mutant (R481A L484A N488A triple mutant). The empty vector, wild-type (WT) Flag-HA-PHF8, or the various mutants were transfected in HEK293T cells, and lysates were used for HA IP and immunoblotted for CDC20. (B) Deletion mutants of Flag-HA-PHF8 were generated to map the CDC20 binding region. In the left column, constructs are shown with respect to the PHD domain and JmjC domain. Constructs were first cut at the N or C terminus (first three panels) to narrow down the region needed for binding. Next, small deletions (indicated with a “V”) were generated in the full-length protein, followed by single- or multiple-point mutations (indicated with an ∗). These constructs were transfected into HEK293T cells, subjected to HA IP, and immunoblotted for CDC20. An LXPKXLF motif from aa 495 to 501 was determined to be essential for CDC20 binding to PHF8. (C) PHF8 orthologues from various species were aligned by ClusterW2 analysis, and the LXPKXLF motif (indicated with an ∗) is highlighted in color, with red indicating hydrophobic and green indicating basic amino acids. H. sapiens, Homo sapiens; C. lupus, Canis lupus; D. rerio, Danio rerio.
To map the interaction domain further, using full-length PHF8 as the template, we generated in-frame deletion mutations surrounding aa 400 to 510 and demonstrated that aa 495 to 505 are important for CDC20 binding (Fig. 3B, fourth panel). We subsequently generated single-point mutations by mutating each amino acid from aa 494 to 505 to alanine, except for A504, which was mutated to glycine (G). Point mutations of amino acids 495 to 501 (L495A, P497A, K498A, L500A, and F501A) abolished the PHF8-CDC20 interaction. Mutations of all five key amino acids (aa 495 to 501) (LPKLF-MT) or an in-frame deletion of aa 495 to 501 (Δ495–501) also led to a similar abolishment of CDC20 binding (Fig. 3B, bottom). These analyses identified the LXPKXLF motif (aa 495 to 501) as a specific CDC20 recognition motif within PHF8. Importantly, this newly identified motif appears to be conserved in PHF8 from human to Caenorhabditis elegans (Fig. 3C).
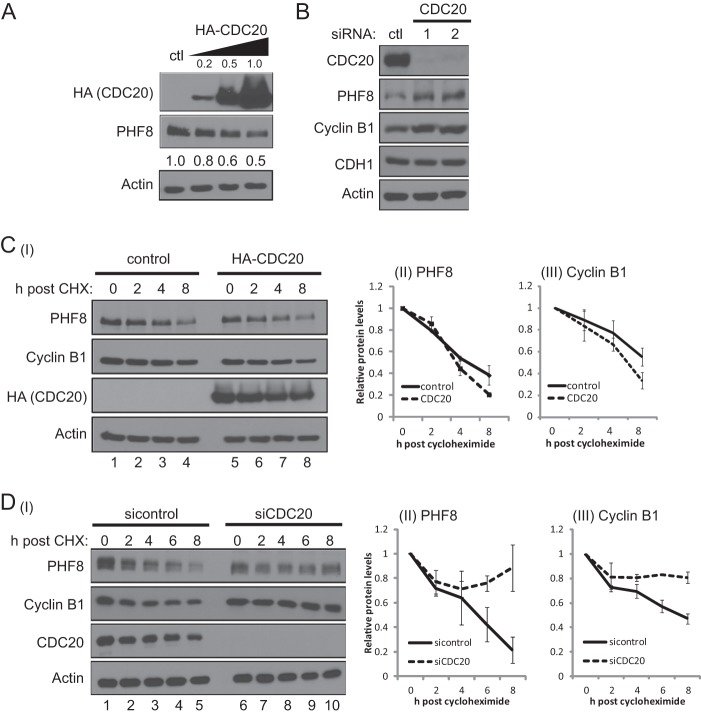
PHF8 protein levels can be modulated by changing CDC20 levels.
As the substrate recognition subunit of the APC, CDC20 is typically the limiting component of the APC in targeting substrates for proteasomal degradation. We therefore investigated whether altering CDC20 levels through overexpression or RNA interference (RNAi) had any effect on PHF8 protein levels. We first noted that PHF8 protein but not mRNA levels were inversely correlated with increasing amounts of transfected HA-CDC20 (Fig. 4A; see also Fig. S2A in the supplemental material), which is consistent with PHF8 being a substrate of APCcdc20. We next analyzed the potential impact of the loss of CDC20 on PHF8 levels. Two independent CDC20 siRNAs led to a >90% knockdown of CDC20 mRNA and protein levels but had no effect on the closely related CDH1 (Fig. 4B; see also Fig. S2BI and II in the supplemental material). RNAi of CDC20 led to an approximately 2-fold increase in PHF8 protein levels, similar to that of cyclin B1, a substrate of APCcdc20 (Fig. 4B). RNAi of CDC20 also led to a ∼40% decrease in the PHF8 mRNA level for unknown reasons (see Fig. S2BIII in the supplemental material).
Fig 4.
PHF8 protein levels can be modulated by changing CDC20 levels. (A) The empty vector control or increasing amounts of HA-CDC20 (0.2, 0.5, and 1.0 μg) were transfected into HEK293T cells, and lysates were immunoblotted for endogenous PHF8. (B) A total of 20 nM the control or two different siRNAs targeting different regions of CDC20 was transiently transfected into HeLa cells twice at 0 h and 24 h. Seventy-two hours after the first transfection, cells were harvested, lysed, and subjected to immunoblotting. (C) HEK293T cells were transfected with 0.5 μg of the empty vector control or the HA-CDC20 plasmid, treated with 100 μg/ml of cycloheximide, and harvested at 0, 2, 4, or 8 h. Lysates were immunoblotted for PHF8 and cyclin B1, and bands were quantitated by ImageJ and normalized against actin and the 0-h sample (I). Means and standard deviations of normalized PHF8 (II) and cyclin B1 (III) levels from triplicate experiments are plotted. (D) A total of 20 nM the control or CDC20 siRNA was transfected into HeLa cells. At 72 h posttransfection, cells were treated with 100 μg/ml of cycloheximide and harvested at 0, 2, 4, 6, or 8 h. Lysates were immunoblotted for PHF8, cyclin B1, CDC20, and actin, with bands quantitated as described above for panel C (I). The means and standard deviations of normalized PHF8 (II) and cyclin B1 (III) levels from triplicate experiments are plotted.
Consistently, overexpression of HA-CDC20 had the opposite effect on PHF8; i.e., HA-CDC20 overexpression led to a modest acceleration of endogenous PHF8 degradation, similar to that of the APCcdc20 substrate cyclin B1 (Fig. 4CI to III; quantitation of data from triple experiments is shown in Fig. 4CII and III), while CDC20 siRNA led to a decreased turnover rate of PHF8 protein, which was also similar to that of cyclin B1 (Fig. 4DI to III; quantitated by ImageJ and presented in graphical form in Fig. 4DII and III). Overall, these results suggest that perturbing CDC20 leads to changes in overall PHF8 protein levels and degradation rates.
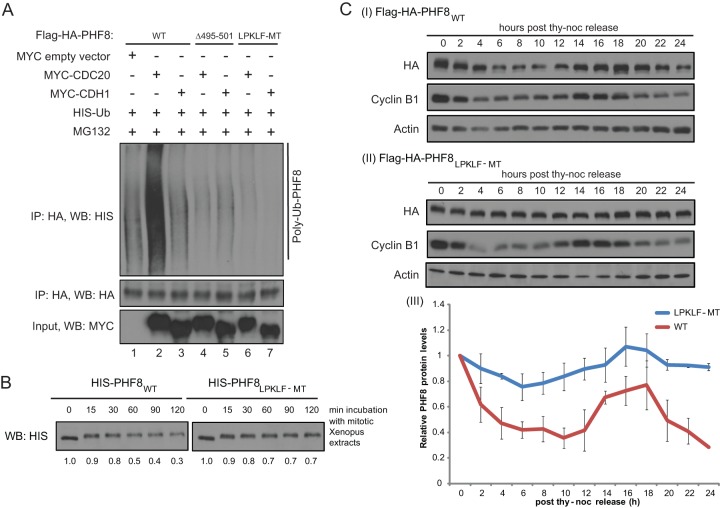
The LXPKLF mutation renders PHF8 refractory to polyubiquitylation by APCcdc20 and proteasomal degradation in vitro.
Given that mutations within the LXPKXLF motif disrupt the PHF8 interaction with CDC20, we next asked whether interaction-defective mutants of PHF8 are also resistant to APCcdc20-mediated ubiquitylation and degradation. Full-length WT, Δ495–501, or LPKLF-MT Flag-HA-PHF8 was transfected into HEK293T cells together with the empty vector, CDC20, or CDH1, respectively, and the transfected cells were subjected to MG132 treatment. Harvested cells were lysed in the presence of an inhibitor of deubiquitylating enzymes and under denaturing conditions to disrupt any protein complexes. Anti-HA IP was then carried out in order to pull down Flag-HA-PHF8. As shown in Fig. 5A, overexpression of CDC20 but not CDH1 led to an increase in PHF8 polyubiquitylation. Moreover, polyubiquitylation occurred only on the WT but not on the LPKLF-MT PHF8 protein, indicating that PHF8 is polyubiquitylated by the APC through CDC20 targeting PHF8 via the LXPKXLF motif. Consistent with this, WT PHF8 (PHF8WT) was degraded more readily than LPKLF-MT when recombinant PHF8 proteins purified from Sf9 cells were incubated with mitotic Xenopus extracts (Fig. 5B).
Fig 5.
The PHF8 LPKLF mutant resists polyubiquitylation by the APCcdc20 and proteasomal degradation but has an as-yet-undefined effect on cells. (A) WT or mutant Flag-HA-PHF8 with either the entire 7-amino-acid motif deleted (Δ495–501) or each key amino acid mutated to alanine (LPKLF-MT) was transfected into HEK293T cells together with HIS-Ub and the empty vector, MYC-CDC20, or MYC-CDH1. Forty-eight hours later, cells were treated with MG132 for 4 h and harvested. Equal amounts of denatured lysates were used for HA IP, and immunocomplexes were probed for HIS-Ub. (B) Sf9-purified full-length HIS-tagged WT or LPKLF-MT PHF8 protein was incubated with mitotic Xenopus extracts and harvested at 0, 15, 30, 60, 90, and 120 min. Samples were immunoblotted for HIS. Bands were quantitated by ImageJ and normalized against the sample from 0 min and are presented below the blot. (C) Stable HeLa cells expressing Flag-HA-tagged PHF8WT (I) or PHF8LPKLF-MT (II) were synchronized at M phase and harvested at 2-h intervals upon release for 24 h. Total cell lysates were subjected to immunoblotting. HA (PHF8) levels were quantitated by ImageJ from duplicate experiments and plotted on a graph (III).
To further understand the effect of the LPKLF mutation in vivo, we generated stable HeLa cell lines expressing either Flag-HA-PHF8WT or Flag-HA-PHF8LPKLF-MT (see Fig. S3A in the supplemental material). Immunofluorescence staining showed that the mutant localized in the nucleus and nucleoli similarly to wild-type PHF8 (see Fig. S3B in the supplemental material) and bound to a similar extent on target gene TSSs (see Fig. S3C in the supplemental material). Moreover, mutant PHF8 purified from Sf9 cells also retained the ability to demethylate histone H3K9me2/1 to a similar extent as wild-type PHF8 (see Fig. S3D in the supplemental material), indicating that the LPKLF mutations did not disrupt the catalytic activity of the protein. Importantly, while the Flag-HA-tagged wild-type PHF8 protein appears to be regulated similarly to endogenous PHF8 during the cell cycle (Fig. 1A and 5CI), with the highest protein levels at M phase, the LPKLF-MT PHF8 protein appeared to have lost the ability to be regulated during the cell cycle (Fig. 5CII and III), supporting the idea that the PHF8 protein level is regulated by the APCcdc20 during the cell cycle.
Loss of PHF8 leads to pleiotropic cell cycle effects, including a slight increase in the duration of S phase, a significantly prolonged G2 phase, and defective mitosis.
A key role of the APC in cell cycle regulation is targeting important cell cycle regulators such as cyclin A and cyclin B1, whose levels are carefully modulated during the cell cycle, and this occurs primarily through its E3 ligase function. As an APC substrate, PHF8 likely plays important roles in the cell cycle. In support of such a role, PHF8 has been shown to transcriptionally regulate a plethora of cell cycle genes (7). Moreover, loss of PHF8 is known to lead to an approximately 2-fold reduction in HeLa cell proliferation (see Fig. S4B in the supplemental material) (7, 9). It is not known, however, whether this decrease in proliferation is a result of a defect or delay in a specific cell cycle phase or programmed cell death such as apoptosis. In fact, flow cytometry data from previous studies reported only a modest 5 to 10% difference between cell cycle phases when PHF8 was knocked down (7). To better understand the effect of PHF8 knockdown on the cell cycle, we utilized HeLa-RFP-ligase cells to perform live-cell imaging (see Fig. S4CI to V in the supplemental material) (27). This cell line expresses DNA ligase I (LIG1) fused with red fluorescent protein (RFP-ligase) and is frequently used to measure the length of each cell cycle phase based on the properties of LIG1 during the cell cycle (see Fig. S4CI to V in the supplemental material). During interphase, LIG1 is present on chromatin but falls off chromatin during mitosis (appearing as a diffused signal) (see Fig. S4CIV in the supplemental material). During S phase, LIG1 forms distinct foci in the nucleus (see Fig. S4CII in the supplemental material), as it is involved in DNA replication and base excision repair. These characteristics of LIG1 allow us to monitor a single cell as it transits through different cell cycle phases and to quantitate the length of each cell cycle phase in the presence and absence of PHF8.
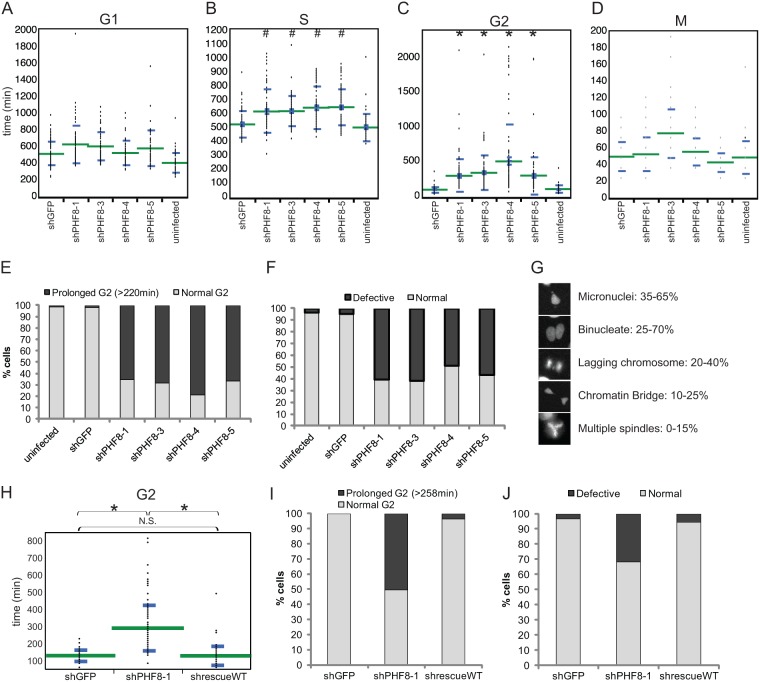
We infected HeLa-RFP-ligase cells with lentiviruses of similar titers containing a control or 4 different PHF8 shRNAs (shRNAs 1, 3, 4, and 5) (see Fig. S4A in the supplemental material). Cells were imaged starting from 24 h postinfection for 48 h, at 12-min intervals. At least 50 cells under each condition of each experiment were manually tracked as they progressed through the cell cycle, and a representative experimental set is reported here. Although the length of the G1 phase was slightly prolonged upon lentiviral infection (compare uninfected with infected cells), PHF8 RNAi (all four shRNAs) did not appear to lead to any significant differences (P > 10−4) in G1-phase length, with an average G1-phase length of about 500 to 600 min (Fig. 6A). However, S-phase length was slightly but significantly (P < 10−4) prolonged upon knockdown of PHF8 (Fig. 6B). The average S-phase length for control RNAi cells was 519 min, and loss of PHF8 consistently led to an increase of S-phase length to between 610 and 650 min. The most dramatic effect of PHF8 RNAi on the cell cycle was observed during G2 phase, when loss of PHF8 led to a very significant (P < 10−10) 3- to 5-fold increase in the average G2-phase length, from 110 min in control RNAi cells to 309 to 515 min in PHF8 RNAi cells (Fig. 6C and E). Out of the >50 cells analyzed, a large proportion (65 to 80%, as opposed to <3% in the control) of PHF8 RNAi cells exhibited a prolonged G2-phase phenotype (defined as >2× the average duration of G2 phase in control RNAi cells) (Fig. 6E). Finally, loss of PHF8 did not appear to have any significant impact on M-phase length (Fig. 6D). In sum, loss of PHF8 lengthens G2 phase most drastically but also increases S phase slightly. However, loss of PHF8 had no effect on the length of G1 and M phases of the cell cycle.
Fig 6.
Loss of PHF8 leads to prolonged G2 phase and defective mitosis. (A to D) An uninfected control or lentiviruses of similar titers containing shGFP or shPHF8-1, -3, -4, or -5 were transduced into HeLa-RFP-ligase cells. Twenty-four hours later, cells were imaged at 12-min intervals for 48 h and manually tracked as they progressed through the cell cycle. The length of each phase, G1 (A), S (B), G2 (C), and M (D), was determined based on fluorescent DNA ligase I appearance in different phases of the cell cycle, and data were plotted on a dot plot by using JMP10 software. The green lines mark the means and the blue lines mark the standard deviations of the values. Each set of PHF8 RNAi data was compared with GFP RNAi data by using Student's t test, and only S phase and G2 phase were determined to be significantly different (#, P < 10−4; ∗, P < 10−10), while G1 and M phases were not significantly different (P > 10−4). (E) Data from panel C were replotted on a histogram to show the proportion of cells under each condition that underwent a normal or prolonged G2 phase. (F and G) HeLa-RFP-H2B cells were used to observe the integrity of mitosis. (F) Data were plotted on a histogram to show the proportion of cells under each condition that underwent normal or defective mitosis. (G) Images showing examples of defective mitosis in PHF8 RNAi cells, with the range of percentages of these defects being observed across all the PHF8 shRNAs. (H and I) HeLa-RFP-ligase cells were transduced with combinations of virus, shGFP plus pOZ-Flag-HA (shGFP), shPHF8-1 plus pOZ-Flag-HA (shPHF8-1), or shPHF8-1 plus pOZ-Flag-HA-PHF8WT (shrescueWT); imaged; and quantified as described above to determine G2-phase length. Student's t test was carried out to determine the significance between each data set, and the significance is indicated (N.S., not significant; ∗, P < 10−10). (J) HeLa-RFP-H2B cells were transduced as described above for panels H and I, imaged, and quantified to determine the proportion of cells undergoing defective mitosis.
We next turned to HeLa-RFP-H2B cells, which express histone H2B fused with RFP. This fusion protein makes chromosomes fluorescent and therefore visually tractable in cells. Although the loss of PHF8 did not significantly affect M-phase length (Fig. 6D), we found mitotic events to be highly defective (Fig. 6F). Specifically, ∼50% of mitotic events showed one or more aberrant structures such as micronuclei, a chromatin bridge, multiple spindles, a lagging chromosome(s), and binucleate cells after telophase (Fig. 6G). These phenotypes were progressively severe with time, sometimes leading to cell death postmitosis.
To ensure that the cell cycle phenotypes that we observed were due to PHF8 RNAi and not due to off-target effects, we restored PHF8 expression through rescue experiments and performed live-cell imaging experiments, as detailed above. HeLa-RFP-ligase and HeLa-RFP-H2B cells were infected with a lentivirus containing shRNA targeting GFP (shGFP) or shPHF8-1, which targets the 3′ untranslated region (UTR) of PHF8, as well as a retrovirus containing either the empty vector pOZ-Flag-HA or pOZ-Flag-HA-PHF8WT (see Fig. S4D in the supplemental material). Expressing ectopic wild-type PHF8 to levels similar to endogenous PHF8 levels was sufficient to restore any defective cell cycle phenotypes. The prolonged G2-phase length (290 min) was restored to an average of 127 min in the rescue sample (shrescueWT), which is highly similar to the length of 129 min observed for the control shGFP cells (Fig. 6H). Moreover, the proportion of cells that underwent a prolonged G2 phase was restored to <5% in the PHF8 rescue sample, as opposed to the ∼50% of cells with a prolonged G2 phase observed with shPHF8-1 infection (Fig. 6I). The proportion of cells undergoing defective mitosis was also restored from ∼35% to ∼5% in the rescue sample (Fig. 6J).
PHF8 is important for keeping a subset of key G2/M target genes transcriptionally active throughout G2 phase.
PHF8 has been shown to be a transcriptional activator of many cell cycle genes (7), and we were therefore interested in determining whether high PHF8 protein levels would be especially important for the regulation of G2/M-enriched genes during this period of the cell cycle. Specifically, we wished to determine whether there were differences in PHF8 binding and histone modifications over selected PHF8 target genes that are preferentially expressed in G2/M versus G1/S phases of the cell cycle. Our selection of G1/S and G2/M genes was based on previously reported microarray data (28, 29) and was also separately verified by qPCR (CCNB1 and E2F1 mRNA levels are shown in Fig. 7BI and CI, respectively). Using ChIP-qPCR, we confirmed PHF8 binding to transcriptional start sites (TSSs) of selected G1/S-enriched genes, including the genes for E2F1, CDC25A, and RBL1, and the G2/M-enriched genes for CCNB1, NCAPG, and CENPA (7, 9) (see Fig. S5A in the supplemental material) in HeLa cells. The CKS1B gene, which is not a PHF8 target gene but is preferentially expressed in G2/M phase, was included as a negative control. Furthermore, knockdown with two independent PHF8 shRNAs (shRNAs 1 and 4) led to reduced transcription of these genes (see Fig. S5B in the supplemental material), suggesting that PHF8 is a positive regulator of their expression.
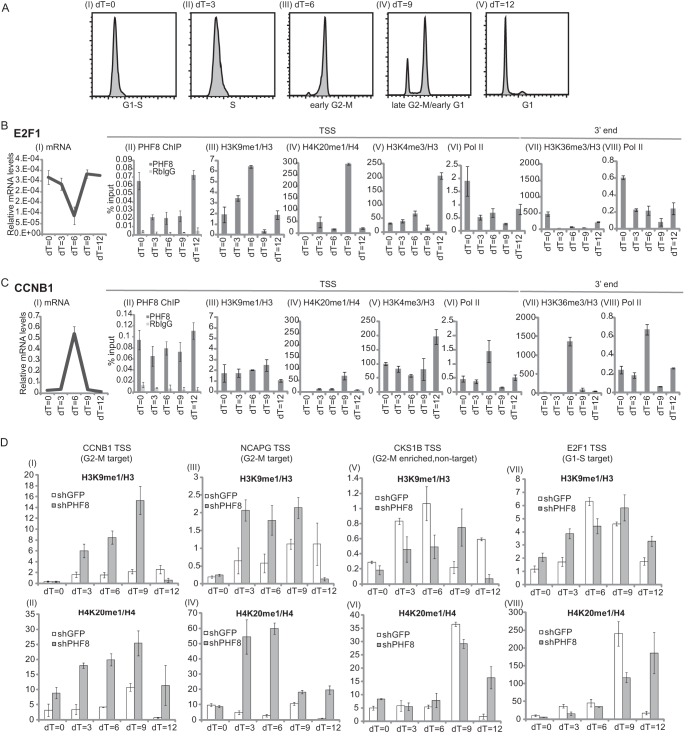
Fig 7.
PHF8 transcriptionally activates a subset of key G2/M target genes throughout G2 phase. (A to C) HeLa cells were synchronized at S phase and collected at 3-h time intervals upon release for flow cytometry analysis (A) or mRNA analysis and ChIP assays (B and C). (A) Cells from the various time points, dT of 0 h (I), dT of 3 h (II), dT of 6 h (III), dT of 9 h (IV), and dT of 12 h (V), were analyzed by flow cytometry. (B and C) Two PHF8 target genes, one enriched at G1/S phase (the E2F1 gene) and one enriched at G2/M phase (the CCNB1 gene), were used for the following analysis. All data are means and standard deviations of duplicate experiments. mRNA levels were normalized against glyceraldehyde-3-phosphate dehydrogenase (I). PHF8, rabbit IgG, and Pol II ChIPs were normalized against input DNA (II, VI, and VIII). Histone mark ChIPs are presented as a ratio over total histone ChIP (i.e., H3K9me1/H3 or H4K20me1/H4) (III to V and VII). qPCR was carried out by using gene-specific primers for E2F1 (B) or CCNB1 (C) targeting the genomic region corresponding to the TSS or 3′ end. (D) HeLa cells transduced with shGFP or shPHF8 were synchronized as described above for panels A to C. H3K9me1/H3 and H4K20me1/H4 ChIP-qPCRs were carried out over the CCNB1 TSS (I and II), NCAPG TSS (III and IV), CKS1B TSS (V and VI), and E2F1 TSS (VII and VIII).
Next, we wished to identify any changes in PHF8 binding, Pol II recruitment, and PHF8-targeted methylation marks on the G1/S- versus G2/M-enriched genes during the cell cycle. We synchronized HeLa cells by double-thymidine treatment and collected cells at 3-h intervals after release from the thymidine block (dT) (dT = 0, 3, 6, 9, and 12 h) (Fig. 7AI to V). A portion of the cells was harvested for flow cytometry analysis, which indicated that a dT of 0 h corresponds to G1/S phase (Fig. 7AI), a dT of 3 h corresponds to S phase (Fig. 7AII), a dT of 6 h corresponds to early G2/M phase (Fig. 7AIII), a dT of 9 h corresponds primarily to late G2/M phase and a smaller proportion of early-G1-phase cells (Fig. 7AIV), and a dT of 12 h corresponds to G1 phase (Fig. 7AV).
As reported previously, E2F1 and CCNB1 mRNA levels are highly regulated transcriptionally and fluctuate during the cell cycle (28, 29). E2F1 mRNA levels were highest at G1 and S phases and lowest at G2 and M phases (Fig. 7BI), while CCNB1 mRNA levels were highest at G2 and M phases and lowest at G1 and S phases (Fig. 7CI). Thus, it appears that these two genes exhibit inverse expression patterns during the cell cycle. Importantly, the level of PHF8 binding to the E2F1 TSS was highest between G1 and early S phases and was significantly lower (about 3-fold) from late S through G2 and M phases, similar to the E2F1 mRNA profile during the cell cycle and consistent with the observation that loss of PHF8 results in a significant reduction of the E2F1 mRNA level in G1/S phase (Fig. 7BI and II). PHF8 binding at the CCNB1 TSS, on the other hand, showed little difference between various cell cycle phases and was 2- to 3-fold enriched between late S and G2/M phases (dT = 3, 6, and 9 h) compared to PHF8 binding at the E2F1 TSS (Fig. 7CII). These findings suggest that PHF8 regulates E2F1 and CCNB1 at G1/S and G2/M phases of the cell cycle, respectively.
We next investigated whether PHF8-regulated histone methylation events as well as Pol II occupancy exhibit corresponding changes concomitant with PHF8 binding to E2F1 and CCNB1 promoters during different stages of the cell cycle. We observed that H3K9me1 and H4K20me1 accumulated on the TSS of the G1/S-specific E2F1 gene during G2 and M phases (dT = 6 and 9 h) (Fig. 7BIII and IV), with H4K20me1 accumulation being especially dramatic (a few hundredfold higher) at a dT of 9 h compared to other time points. This correlates well with the level of PHF8 binding to the E2F1 TSS being highest at G1/S phase and lowest in G2 phase, as discussed above. H4K20me1 accumulation has been shown to be essential during G2 and M phases, facilitating the recruitment of the transcriptional repressor L3MBTL1 and the condensin subunits NCAPD3 and NCAPG2, inducing chromatin compaction and shutdown of transcription in preparation for mitosis (7, 8, 30). On the contrary, H3K9me1 and H4K20me1 marks over the TSS of CCNB1 remained relatively constant or low during G2 and M phases (Fig. 7CIII and IV), correlating well with PHF8 occupancy over the TSS and when its protein levels are highest.
Indeed, keeping the transcriptionally repressive H3K9me1 and H4K20me1 marks low over CCNB1 TSS appears to be important for active transcription of this gene during G2 phase. Enhanced Pol II occupancy over the TSS and the 3′ end of the gene indicates that CCNB1 is actively transcribed during G2 phase (Fig. 7CVI and VIII). In addition, H3K36me3, a mark of transcriptional elongation at the 3′ end of actively transcribed genes, is enriched over CCNB1 during this time (Fig. 7CVII). In comparison, the G1/S-enriched E2F1 gene exhibits active transcription during G1/S phase (dT = 0 h), as evidenced by enriched Pol II over the TSS and 3′ end and enriched H3K36me3 over the 3′ end (Fig. 7BVI to VIII). H3K4me3 accumulation over the E2F1 TSS appears to occur at G1 phase (dT = 12 h) (Fig. 7BV) and precedes active transcription elongation.
To determine whether the lower H3K9me1 and H4K20me1 levels during G2 phase over the G2/M gene for CCNB1 (compared to the G1/S gene E2F1) is attributable to PHF8, we used HeLa cells transduced with a lentivirus containing either control shGFP or shPHF8-4 (Fig. 7DI to VIII). We examined H3K9me1/H3 and H4K20me1/H4 levels over the PHF8 G2/M target genes for CCNB1 and NCAPG (Fig. 7DI to IV). As controls, we also investigated the non-PHF8 target G2/M-enriched gene for CKS1B (Fig. 7DV and VI) and the G1/S-enriched PHF8 target gene for E2F1 (Fig. 7DVII and VIII). We observed that knockdown of PHF8 led to increased H3K9me1 and H4K20me1 levels over the G2/M genes for CCNB1 and NCAPG at the highest levels between late S and G2/M phases (Fig. 7DI to IV). However, knockdown of PHF8 did not lead to a significant increase in H3K9me1 or H4K20me1 levels over the non-PHF8-target G2/M gene for CKS1B or the PHF8 G1/S target gene for E2F1 during this time (Fig. 7DV to VIII). Overall, these results support an important role of PHF8 in the regulation of G2/M genes during G2 phase. PHF8 accumulation during G2 phase correlates with when PHF8 is functionally essential for the targeting of specific key G2/M genes such as the CCNB1 gene, demethylating H3K9me1 and H4K20me1 over their TSSs to keep these genes transcriptionally active throughout G2 phase.
DISCUSSION
As a histone demethylase, PHF8 activates its target genes through demethylation of the transcriptionally repressive marks H3K9me2/1 and H4K20me1. This study identifies PHF8 as an important regulator of the G2/M transition, and this conclusion is supported by multiple lines of evidence. First, PHF8 levels are highest at G2 and M phases, which may play a role in its access to its transcriptional targets important for G2/M transition (such as CCNB1). Second, PHF8 protein levels start to decrease as cells transition out of mitosis, possibly due to the action of the APC, which interacts with and targets PHF8 for proteasomal degradation. Third, loss of PHF8 leads to a lengthened G2 phase, indicating the importance of maintaining proper PHF8 levels in G2 phase. Lastly, our study also identifies a novel APC binding motif, which may be conserved in a number of mammalian proteins.
A novel motif on PHF8 governs its binding to CDC20, polyubiquitylation by the APC, and proteasomal degradation during mitosis.
We uncovered a previously unreported KEN-box- and D-box-independent motif (LXPKXLF) on PHF8 that is essential for interactions with CDC20 (Fig. 3A and B), polyubiquitylation by the APCcdc20, and proteasomal degradation (Fig. 5A and B). The presence of the mutation did not appear to impair PHF8's overall structure or function (see Fig. S3B to D in the supplemental material), suggesting that the main role of this motif is for PHF8's targeting by the APC for degradation. Interestingly, while the loss of this motif led to the inability of PHF8 to be regulated during the cell cycle (Fig. 5C), PHF8 still appeared to be able to be proteasomally regulated (see Fig. S3E in the supplemental material), suggesting that PHF8 is likely regulated by additional E3 ligases. Interestingly, we found that this new motif seems to be conserved among a few dozen proteins, including claspin, a known substrate of the APC. Claspin possesses a PXPKXLF motif (aa 847 to 853) that is very similar to the LXPKXLF consensus. Previously, two groups mapped different motifs and regions that are essential for claspin binding to CDH1 and degradation by the APC (31, 32). One of two key regions on claspin that is polyubiquitylated by the APC (aa 681 to 924) coincides with where the PXPKXLF motif lies (31). In addition, we also found that speriolin, a reported interactor of CDC20 and CDC27, possesses a similar PXPKXAF motif (aa 293 to 299), which falls within the region shown to be necessary for its interaction with CDC20 (aa 283 to 480) (33). The identification of this novel APC binding motif can therefore be used to predict new, potential APC interactors and substrates.
Regulation of PHF8 protein levels could be one way in which PHF8 functionally exerts itself.
Our study suggests that PHF8 protein levels can be regulated during the cell cycle posttranslationally by E3 ligases such as the APC. Regulation of PHF8 abundance appears to be an important mechanism through which PHF8 exerts its regulation over its target genes, especially G2/M genes in G2 phase. PHF8 protein levels fluctuate during the cell cycle, with the highest protein levels being found during G2 and M phases (Fig. 1A); this coincides with the cell cycle phase in which PHF8 appears to be the most functionally essential, as RNAi of PHF8 led to a prolonged G2 phase and defective mitosis (Fig. 6C and E to G). Moreover, PHF8's abundance in G2 phase was correlated with increased PHF8 binding and, importantly, decreased accumulation of the transcriptionally repressive marks H3K9me1 and H4K20me1 over the TSS of the G2/M gene for CCNB1 compared to the G1/S gene for E2F1 (Fig. 7B and C). This suggests that regulation of PHF8 protein levels, in particular accumulation of PHF8 during G2 phase, is important for transcriptional regulation of key genes, including the CCNB1 gene, that regulate the G2/M transition. Additionally, once PHF8 has fulfilled its transcriptional role in G2 phase, it is targeted for degradation by the APC, explaining the decrease in its protein levels as cells exit mitosis (Fig. 1A). The timing of degradation of PHF8 follows a trend similar to that of many important G2/M regulators, such as cyclin A and cyclin B, which are APC substrates.
In fact, ubiquitylation and proteasomal degradation have emerged as key mechanisms through which histone methylation and demethylation enzymes are regulated. The histone methyltransferases MLL and PR-Set7 and the histone demethylase KDM4A/JMJD2A are ubiquitylated and targeted for degradation by multiple E3 ligases (25, 34–42). Protein degradation of these epigenetic enzymes via the ubiquitin-proteasome system is important in the functional regulation of these enzymes and allows cells to respond rapidly to environmental signals or developmental cues. Therefore, misregulation of protein levels of these enzymes results in dramatic effects on the cell cycle, DNA damage, and transcription.
Consistent with the role of PHF8 in regulating key G2/M genes, RNAi of PHF8 leads to a lengthening of G2 phase and defects in M phase (Fig. 6C and E to G). Interestingly, while RNAi of PHF8 was reported previously to result in an approximately 2-fold-decreased proliferation rate, it did not appear to lead to major changes (only 5 to 10% changes) in the proportion of cells in each cell cycle phase (7, 9). This is probably because G2 phase takes up only about 10 to 20% of the total cell population, of which about 60 to 70% of these cells exhibit a prolonged G2 phase; thus, changes occurring in the G2/M phase would not be readily detectable by flow cytometry, as PHF8 RNAi led to only a 5% increase in the abundance of G2/M cells (7). In addition, while RNAi of PHF8 did not lead to an increase in the length of mitosis, it resulted in highly defective mitosis (Fig. 6D, F, and G). For example, the presence of binucleate cells as a result of failed cytokinesis was one of the major mitotic defects and can possibly be attributed to PHF8's regulation of cytoskeletal proteins such as RhoA, which is needed for specifying the cleavage plane for the initiation of cytokinesis (43, 44). The pleiotropic mitotic defects observed with PHF8 knockdown are consistent with a role of PHF8 in the regulation of a transcriptional cluster of G2/M genes, similar to that of FoxM1 (45). Loss of FoxM1, a transcriptional regulator of multiple G2/M genes, including those for CCNB1 and CENP-F (both are PHF8 target genes), has been shown to lead to cell defects that are similar to those with the loss of PHF8, including a prolonged G2 phase, chromosome missegregation, and cytokinesis failure (45). Despite their similar cell cycle phenotypes, we did not observe FoxM1 interacting with PHF8 based on our TAP data. Therefore, it is possible that they regulate similar G2/M genes in complementary but separate pathways.
While the loss of PHF8 has dramatic effects on the cell cycle, effects of its overexpression on the cell cycle as well as H4K20me1 levels are not overtly apparent (data not shown; see also Fig. S3A in the supplemental material). Moreover, rescue with degradation-resistant LPKLF-MT restores normal cell cycle timings similar to those of WT rescue (Fig. 6H to J and data not shown). It is possible that overexpression of PHF8 or inhibition of its degradation causes more subtle effects, such as changes in how PHF8 is targeted to specific gene loci, which would be an interesting area for experimentation in the future.
PHF8/PR-Set7 balance is important for H4K20me1 regulation during the cell cycle.
Intriguingly, the H4K20me1 methyltransferase PR-Set7, which appears to be similarly enriched in G2/M phase, is also regulated by the APC (although via a CDH1 coactivator) (46). The loss of PR-Set7 results in similar, but more severe, phenotypes compared those resulting from the loss of PHF8, including G2 arrest and defective mitosis. The disparity in the phenotype severities could be attributed to PR-Set7 being the sole H4K20me1 methyltransferase, whereas there are at least two histone demethylases that target H4K20me1 that may functionally compensate for one another. Given that PHF8 and PR-Set7 work in an opposing manner on H4K20me1, it is interesting that they demonstrate remarkable similarities in how their protein levels are regulated. PR-Set7 has been shown to be important for global accumulation of H4K20me1. Our data suggest that PHF8 is responsible for localized demethylation of H4K20me1 over specific gene loci, as changing of PHF8 levels by knockdown or overexpression does not lead to global changes in H4K20me1 (data not shown; see also Fig. S3A in the supplemental material) (9). Therefore, it appears that, at least in the context of G2/M regulation, a delicate balance of PR-Set7 and PHF8 abundance and localization is essential.
APCcdc20-mediated degradation of PHF8 may be important in development and tumorigenesis.
PHF8 is a potential oncogene in prostate cancer, acute promyelocytic leukemia, and T cell acute lymphoblastic leukemia (17, 19, 47). Different PHF8 target genes have been proposed to be essential in the initiation or progression of different cancer types. As an important transcriptional regulator of cell cycle genes, misregulation of PHF8 may increase cells' susceptibility to malignancy through genomic instability due to defective mitosis. In addition, PHF8 is likely to play a key role in brain development, as mutations in PHF8 are found in patients with XLID and craniofacial and autism spectrum disorders (13–15, 48, 49). In zebrafish, loss of PHF8 leads to apoptosis in the developing brain and jaw (9), although it is not clear if this is triggered by misregulation of the cell cycle and accumulation of DNA damage. Furthermore, although the APC is known primarily for its role in regulation of the cell cycle, recent studies have shown that APC proteins continue to be highly expressed in postmitotic neurons that have exited the cell cycle, and loss of these proteins leads to defects in neuronal growth and survival (50). It will be interesting to determine whether the APC-PHF8 interaction is conserved in postmitotic neurons and whether PHF8 protein levels are regulated by the APC in the brain and in what context.
Supplementary Material
ACKNOWLEDGMENTS
We thank Benoit Laurent for suggestions and technical assistance. We are also grateful to members of the Shi laboratory for their useful suggestions.
This work is supported by NIH grants to Y.S. (GM071004, CA118487, and MH096066) and to R.W.K. (GM66492). H.-J.L. and M.-K.M.T. are supported by predoctoral fellowships from the Agency of Science, Technology and Research (A*STAR), Singapore.
Y.S. is a cofounder of Constellation Pharmaceuticals, Inc., and a member of its scientific advisory board. The remaining authors have declared no conflicts of interest.
Footnotes
Published ahead of print 26 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00689-13.
REFERENCES
- 1.Williams GH, Stoeber K. 2012. The cell cycle and cancer. J. Pathol. 226:352–364 [DOI] [PubMed] [Google Scholar]
- 2.Badeaux AI, Shi Y. 2013. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 14:211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC. 2008. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 283:19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 16:2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. 2009. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol. Cell. Biol. 29:2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tardat M, Murr R, Herceg Z, Sardet C, Julien E. 2007. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J. Cell Biol. 179:1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466:508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trojer P, Li G, Sims RJ, III, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, Wang YH, Reinberg D. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129:915–928 [DOI] [PubMed] [Google Scholar]
- 9.Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y. 2010. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466:503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. 2010. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17:38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Z, Wang Y, Li X, Wang Y, Xu L, Wang X, Sun T, Dong X, Chen L, Mao H, Yu Y, Li J, Chen PA, Chen CD. 2010. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 20:794–801 [DOI] [PubMed] [Google Scholar]
- 12.Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. 2010. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct. Mol. Biol. 17:445–450 [DOI] [PubMed] [Google Scholar]
- 13.Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, Lespinasse J, Van Esch H, Lacombe D, Goizet C, Phan-Dinh Tuy F, van Bokhoven H, Fryns JP, Chelly J, Ropers HH, Moraine C, Hamel BC, Briault S. 2005. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 42:780–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abidi F, Miano M, Murray J, Schwartz C. 2007. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin. Genet. 72:19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koivisto AM, Ala-Mello S, Lemmela S, Komu HA, Rautio J, Jarvela I. 2007. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin. Genet. 72:145–149 [DOI] [PubMed] [Google Scholar]
- 16.Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, Helin K. 2010. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell 38:165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arteaga MF, Mikesch JH, Qiu J, Christensen J, Helin K, Kogan SC, Dong S, So CW. 2013. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell 23:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu J, Shi G, Jia Y, Li J, Wu M, Li J, Dong S, Wong J. 2010. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 20:908–918 [DOI] [PubMed] [Google Scholar]
- 19.Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. 2012. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol. Cell 48:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters JM. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 [DOI] [PubMed] [Google Scholar]
- 21.Skaar JR, Pagano M. 2009. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr. Opin. Cell Biol. 21:816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glotzer M, Murray AW, Kirschner MW. 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349:132–138 [DOI] [PubMed] [Google Scholar]
- 23.Pfleger CM, Kirschner MW. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655–665 [PMC free article] [PubMed] [Google Scholar]
- 24.Nakatani Y, Ogryzko V. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430–444 [DOI] [PubMed] [Google Scholar]
- 25.Tan MK, Lim HJ, Harper JW. 2011. SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol. Cell. Biol. 31:3687–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. 2012. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat. Cell Biol. 14:168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardoso MC, Joseph C, Rahn HP, Reusch R, Nadal-Ginard B, Leonhardt H. 1997. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell Biol. 139:579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ. 2001. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 27:48–54 [DOI] [PubMed] [Google Scholar]
- 29.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13:1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD. 2008. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27:4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao D, Inuzuka H, Korenjak M, Tseng A, Wu T, Wan L, Kirschner M, Dyson N, Wei W. 2009. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol. Biol. Cell 20:3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. 2008. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 134:256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goto M, Eddy EM. 2004. Speriolin is a novel spermatogenic cell-specific centrosomal protein associated with the seventh WD motif of Cdc20. J. Biol. Chem. 279:42128–42138 [DOI] [PubMed] [Google Scholar]
- 34.Van Rechem C, Black JC, Abbas T, Allen A, Rinehart CA, Yuan GC, Dutta A, Whetstine JR. 2011. The SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein. J. Biol. Chem. 286:30462–30470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Cheng EH, Hsieh JJ. 2007. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 21:2385–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40:9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40:22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC. 2010. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 24:2531–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, Sorensen CS. 2011. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 192:43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E. 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 12:1086–1093 [DOI] [PubMed] [Google Scholar]
- 41.Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40:364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S. 2012. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31:1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asensio-Juan E, Gallego C, Martinez-Balbas MA. 2012. The histone demethylase PHF8 is essential for cytoskeleton dynamics. Nucleic Acids Res. 40:9429–9440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Normand G, King RW. 2010. Understanding cytokinesis failure. Adv. Exp. Med. Biol. 676:27–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7:126–136 [DOI] [PubMed] [Google Scholar]
- 46.Brustel J, Tardat M, Kirsh O, Grimaud C, Julien E. 2011. Coupling mitosis to DNA replication: the emerging role of the histone H4-lysine 20 methyltransferase PR-Set7. Trends Cell Biol. 21:452–460 [DOI] [PubMed] [Google Scholar]
- 47.Bjorkman M, Ostling P, Harma V, Virtanen J, Mpindi JP, Rantala J, Mirtti T, Vesterinen T, Lundin M, Sankila A, Rannikko A, Kaivanto E, Kohonen P, Kallioniemi O, Nees M. 2012. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene 31:3444–3456 [DOI] [PubMed] [Google Scholar]
- 48.Qiao Y, Liu X, Harvard C, Hildebrand MJ, Rajcan-Separovic E, Holden JJ, Lewis ME. 2008. Autism-associated familial microdeletion of Xp11.22. Clin. Genet. 74:134–144 [DOI] [PubMed] [Google Scholar]
- 49.Nava C, Lamari F, Heron D, Mignot C, Rastetter A, Keren B, Cohen D, Faudet A, Bouteiller D, Gilleron M, Jacquette A, Whalen S, Afenjar A, Perisse D, Laurent C, Dupuits C, Gautier C, Gerard M, Huguet G, Caillet S, Leheup B, Leboyer M, Gillberg C, Delorme R, Bourgeron T, Brice A, Depienne C. 2012. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl. Psychiatry 2:e179. 10.1038/tp.2012.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Kim AH, Bonni A. 2010. The dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr. Opin. Neurobiol. 20:92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.