Abstract
DNA cytosine methylation is an epigenetic modification involved in the transcriptional repression of genes controlling a variety of physiological processes, including hematopoiesis. DNA methyltransferase 1 (Dnmt1) is a key enzyme involved in the somatic inheritance of DNA methylation and thus plays a critical role in epigenomic stability. Aberrant methylation contributes to the pathogenesis of human cancer and of hematologic malignancies in particular. To gain deeper insight into the function of Dnmt1 in lymphoid malignancies, we genetically inactivated Dnmt1 in a mouse model of MYC-induced T-cell lymphomagenesis. We show that loss of Dnmt1 delays lymphomagenesis by suppressing normal hematopoiesis and impairing tumor cell proliferation. Acute inactivation of Dnmt1 in primary lymphoma cells rapidly induced apoptosis, indicating that Dnmt1 is required to sustain T-cell lymphomas. Using high-resolution genome-wide profiling, we identified differentially methylated regions between control and Dnmt1-deficient lymphomas, demonstrating a locus-specific function for Dnmt1 in both maintenance and de novo promoter methylation. Dnmt1 activity is independent of the presence of Dnmt3a or Dnmt3b in de novo promoter methylation of the H2-Ab1 gene. Collectively, these data show for the first time that Dnmt1 is critical for the prevention and maintenance of T-cell lymphomas and contributes to aberrant methylation by both de novo and maintenance methylation.
INTRODUCTION
Cytosine methylation is an epigenetic mark that is abundant throughout intragenic and intergenic regions within the mammalian genome. The methylation of gene promoters has been associated with gene repression, while the methylation of gene bodies may promote proper transcription (1, 2). Due to its genome-wide distribution and effects on transcriptional regulation, DNA methylation plays a critical role in a wide range of physiological processes, including silencing of endogenous retroviral elements, X-chromosome inactivation, imprinting, proliferation, differentiation, and apoptosis (3, 4). The disruption of normal methylation patterns contributes to the pathogenesis of a variety of human diseases such as neurodegenerative, developmental, and autoimmune disorders (5, 6). In particular, global deregulation of cytosine methylation is apparent in cancer, where genome-wide hypomethylation is suggested to promote tumorigenesis by invoking genomic instability and upregulating oncogenes, whereas aberrant promoter hypermethylation supports tumorigenesis by silencing tumor suppressor genes (7).
Whereas the association of deregulated methylation with cancer is well established, the individual roles of the enzymes catalyzing DNA methylation, DNA methyltransferases (Dnmts), in the pathogenesis of human cancer are unclear. Three catalytically active Dnmts (Dnmt1, Dnmt3a, and Dnmt3b) are responsible for the generation and maintenance of methylation patterns in the mammalian genome. While Dnmt3a and Dnmt3b are associated with de novo methylation because of their involvement in the establishment of normal methylation patterns (8, 9), Dnmt1 is essential for maintenance of the methylation landscape due to its ability to recognize hemimethylated DNA and conserve methylation during somatic cellular division (10). However, the discrete roles of Dnmts in the formation and maintenance of the methylation landscape are more complex. Several studies have suggested that Dnmt1 may function as a de novo enzyme. For example, overexpression of Dnmt1 induced locus-specific de novo methylation in human fibroblasts that was associated with specific sequence motifs (11, 12). Additionally, knockout of Dnmt1 in embryonic stem cells suggests de novo activity for Dnmt1 at repeat elements and single-copy genes (13). Although the main function of Dnmt1 appears to be related to cytosine methylation, Dnmt1 also interacts with a large number of repressor proteins, such as histone deacetylases and DNA methyltransferase-associated protein 1 (DMAP1), to inhibit transcription in a methylation-independent manner (14–16).
Recent studies have identified somatic mutations in DNMT1 in cancer; however, they are infrequent. DNMT1 is mutated in about 3% of cases of colorectal adenocarcinoma and 1.6% of prostate cancer as well as a small subset of cases of acute myeloid leukemia (AML) (17–19). Furthermore, increased DNMT1 expression is observed in subsets of human T-cell, B-cell, and myeloid malignancies, suggesting that DNMT1 may be important for tumor maintenance (20, 21). Functional studies in mice have shown that decreased levels of Dnmt1 resulted in an increased tumor incidence at early stages of colon cancer in ApcMin/+ mice and simian virus 40 (SV40)-induced tumorigenesis in the prostate, suggesting a tumor suppressor function for Dnmt1 in these models (22, 23). Mutations in hematologic malignancies are very rare, with only one mutation found to date in AML (24). Studies in mice suggested an important role for Dnmt1 in myeloid malignant hematopoiesis. In an AML model driven by MLL-AF9 overexpression, a reduction in Dnmt1 delayed leukemogenesis and impaired leukemic stem cell self-renewal, indicating a key role for Dnmt1 in the maintenance and establishment of leukemia (25). In contrast, a severe reduction in levels of Dnmt1 clearly promotes transformation of T cells. A combination of a hypomorphic Dnmt1chip allele (expressing only 10% of Dnmt1 levels relative to the wild-type allele) with a conventional Dnmt1 knockout allele in Dnmt1chip/− mice resulted in the development of T-cell lymphomas (TCLs) by 8 months of age, despite severely impaired T-cell development (26, 27). To further elucidate the role of Dnmt1 in malignant hematopoiesis and to determine Dnmt1's contribution to aberrant DNA methylation in cancer, we used a conditional knockout approach to inactivate Dnmt1 in a mouse model of MYC-induced T-cell lymphomagenesis. Here we show that loss of Dnmt1 delayed lymphomagenesis by suppression of normal hematopoiesis and by impairing proliferation of tumor cells. Global approaches identified 730 gene promoters differentially methylated in normal thymocytes, MYC-induced T-cell lymphomas (MTCLs), and MTCLs deficient for Dnmt1, suggesting that Dnmt1 contributes to the cancer methylome by both de novo and maintenance activity. Thus, our studies not only provide a biological mechanism explaining delayed lymphomagenesis in the absence of Dnmt1 but also identify Dnmt1 target genes for the first time in the relevant in vivo setting.
MATERIALS AND METHODS
Mouse studies.
EμSRα-tTA; Teto-MYC and Dnmt12lox mice were acquired from D. W. Felsher (Stanford University, Stanford, CA) and R. Jaenisch (Whitehead Institute, Cambridge, MA), respectively. ROSA26EGFP mice (28) and Teto-Cre mice (29) were obtained from The Jackson Laboratory. All mice were back-bred for five generations into the FVB/NJ background. Standard genetic crosses were performed to generate the appropriate transgenic mice for these experiments, and the results were confirmed by PCR-based genotyping. Genomic DNA for genotyping was obtained from mouse tails. Tumor-bearing mice were carefully monitored for their overall health and harvested when they became terminally ill.
Mice used for tumor studies were EμSRα-tTA; Teto-MYC; Teto-Cre; Rosa26LOXPEGFP/EGFP; Dnmt1flox/flox and EμSRα-tTA; Teto-MYC; Rosa26LOXPEGFP/EGFP; Dnmt1flox/flox mice (referred to as MYC; Dnmt1−/− and MYC; Dnmt1flox/flox mice, respectively). Further studies were performed utilizing EμSRα-tTA; Teto-MYC; Teto-Cre; ROSA26EGFP/EGFP; Dnmt1+/+, EμSRα-tTA; Teto-MYC; Teto-Cre; ROSA26EGFP/EGFP; Dnmt3aflox/flox, and EμSRα-tTA; Teto-MYC; Teto-Cre; ROSA26EGFP/EGFP; Dnmt3bflox/flox mice (referred to as MYC; Dnmt1+/+, MYC; Dnmt3a−/−, and MYC; Dnmt3b−/− mice, respectively).
Fluorescence-activated cell sorter (FACS) analysis, proliferation, and apoptosis.
Single-cell suspensions from in vitro cultures or from the thymus, spleen, lymph node, and bone marrow were prepared and stained with the appropriate antibodies. For bromodeoxyuridine (BrdU) incorporation assays, cells were incubated with BrdU and labeled using allophycocyanin (APC)-conjugated anti-BrdU (BrdU-Flow kit; BD Biosciences) according to the manufacturer's instructions and analyzed using BD FACSDiva analysis software after processing samples on the FACSCalibur II flow cytometer. Similarly, tumor cells were stained with annexin V-APC (eBioscience) antibody and events were acquired on the FACSCalibur II flow cytometer. Cell cycle analysis was performed utilizing Vybrant DyeCycle Orange stain (Invitrogen) according to standard protocols and acquired on the LSR II cytometer. Thymi, spleens, lymph nodes, and bone marrow from developmental and tumor studies were stained with the following antibodies to analyze cell surface marker expression: CD4, CD8, CD11b, B220, CD3, TER119, Sca-1, c-kit, CD19, gamma-delta T-cell receptor (TCRγδ), and TCRβ conjugated to phycoerythrin (PE), PE-Cy5, or APC. All antibodies were obtained from eBioscience. Populations of Lin− Sca-1+ c-kit+ (LSK; negative for lineage markers CD4, CD8, CD11b, CD19, TER119, CD3, gamma-delta T-cell receptor [TCRγδ], and TCRβ and positive for Sca-1 and c-kit) cells were distinguished by gating against lineage markers (CD4, CD8, CD11b, B220, CD3, and TER119) and subsequent gating for Sca-1+ c-kit+ cells. Sorting of enhanced green fluorescent protein (EGFP)-positive CD4− CD8−, CD4+ CD8+, CD4+, and CD8+ thymocytes was performed on the FACSAria cell sorter.
MSCC and data analysis.
Methyl-sensitive cut counting (MSCC) library preparation, data collection, and repeat analysis were performed as previously described (30, 31) with the addition of a second enzyme—HpyCh4IV. The readout in counts for each individual HpaII and HpyCh4IV site is relative to the methylation status at that particular locus. The method results in output of sequencing tags, where a high number of counts correlates with low methylation and vice versa. The data in the methylation heat maps (see Fig. 5C and D and Fig. 6C) represent averages for raw counts of all statistically significant changes of 2-fold or greater within individual promoters between MYC; Dnmt1flox/flox and MYC; Dnmt1−/− mice. All statistical analysis was performed using the R programming language bioconductor package “edgeR” (32–34). The promoter is defined as at bp −1500 to +350 relative to the transcription start site. All genes considered to be hypomethylated or hypermethylated included two or more independent HpaII/HpyCh4IV sites that showed changes of 2-fold or greater with a false discovery rate (FDR) of less than 0.05.
Fig 5.
Analysis of DNA methylation in Dnmt1-deficient tumors. (A) Total 5-methylcytosine levels in normal thymocytes (Th), MYC; Dnmt1flox/flox (F/F) tumors, and MYC; Dnmt1−/− (−/−) tumors. The human colorectal carcinoma parental cell line HCT116 (H) and the HCT116 DNMT1−/−; DNMT3B−/− cell line (H13B) served as controls. (B) In silico analysis of relative methylation levels of Line-L1, Line-L2, and Sine/Alu repeats in normal thymus (Th), MYC; Dnmt1flox/flox tumors, and MYC; Dnmt1−/− tumors by MSCC. (C) A heat map analysis of MSCC data displaying 427 hypomethylated and 303 hypermethylated promoters in MYC; Dnmt1−/− tumors relative to MYC; Dnmt1flox/flox tumors (FDR < 0.05 [negative binomial]). Genes proposed to have Dnmt1 de novo (214 genes) and Dnmt1 maintenance (213 genes) activity are labeled. A color bar is shown, with black representing a high degree of methylation and yellow representing lower levels. Th indicates normal thymocytes. n represents the number of biological replicates. (D) A heat map displaying the numbers of hypermethylated (n = 337) and hypomethylated (n = 440) gene bodies in MYC; Dnmt1−/− tumors relative to MYC; Dnmt1flox/flox tumors. Normal thymus (Th) levels are also shown. Changes represent a P of <0.05 with a 2-fold or greater change.
Fig 6.
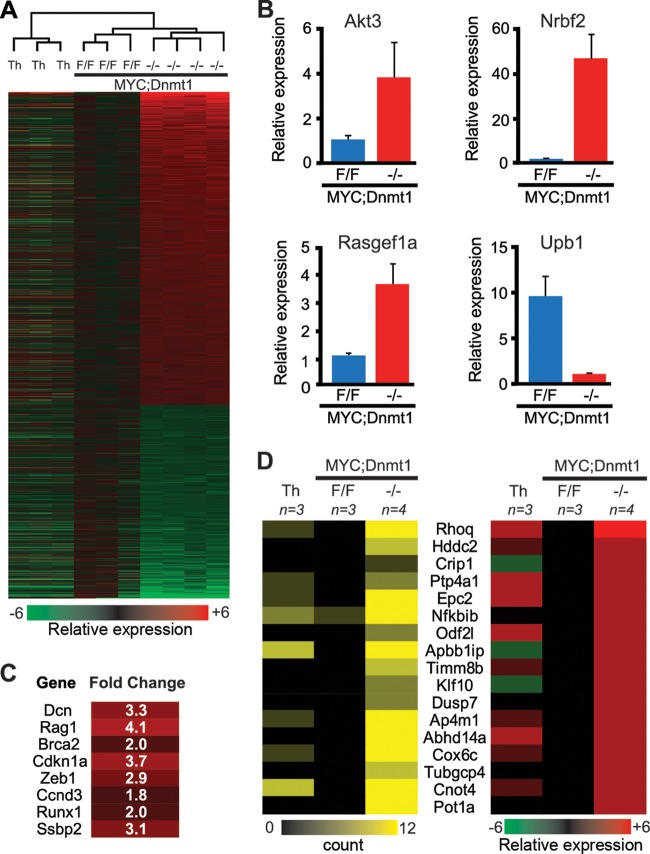
Significantly deregulated genes in MYC; Dnmt1−/− tumors. (A) A heat map derived from global transcription profiling by microarray displaying 1,260 genes that are 1.75-fold changed in MYC; Dnmt1−/− tumors relative to MYC; Dnmt1flox/flox tumors. A total of 780 genes show upregulation, while 480 show downregulation (FDR < 0.05 [Bayesian t test]). A color bar is shown to reference upregulation in red and downregulation in green. Above the heat map, an unsupervised hierarchical clustering of normal thymi (Th), MYC; Dnmt1flox/flox (F/F) tumors, and MYC; Dnmt1−/− (−/−) tumors is shown. (B) qRT-PCR displaying the relative mRNA levels of 4 differentially expressed genes in MYC; Dnmt1flox/flox and MYC; Dnmt1−/− tumors. The average results of two replicates are shown for n = 2 samples for each group. Error bars represent SEM. (C) The network tumorigenesis in thymic lymphomas derived from Ingenuity pathway analysis of 780 upregulated genes from MYC; Dnmt1−/− cells. All eight genes within the network are predicted to suppress thymic lymphomagenesis. (D) Heat maps for genes with at least a 2-fold decrease in promoter methylation as determined by MSCC (left; FDR < 0.05 [negative binomial]) and at least a 1.75-fold induction in expression as determined by microarray (right; FDR < 0.05 [Bayesian t test]) in MYC; Dnmt1−/− lymphomas. Average values from normal thymocytes (Th) (n = 3) and MYC; Dnmt1flox/flox (n = 3) and MYC; Dnmt1−/− (n = 4) tumors are shown. Red and green indicate induction and reduction of transcription, respectively, while black represents methylation and yellow stands for hypomethylation.
Tissue culture.
Mouse MYC; Dnmt1flox/flox cells were maintained in RPMI 1640 (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and 0.025 mM 2-mercaptoethanol. Cell lines were cultured at 37°C in a humidified 5% CO2 atmosphere. Cre-mediated deletion of Dnmt1 was achieved using an established MYC; Dnmt1flox/flox cell line infected with murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-puro-CreERT2 (Addgene plasmid 22776 [35]). Retroviral infections were done as described previously (30). Cre-mediated excision of Dnmt1 was induced by addition of 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich) at a concentration of 150 nM. DNA for PCR-based determinations of Dnmt1 deletion efficiency was prepared from cells harvested after 24 and 72 h. For the colony PCR genotyping analysis, cells from two primary MYC; Dnmt1−/− lymphomas were plated into 48-well culture plates in RPMI medium with 10% fetal calf serum (FCS) at dilutions of one cell or fewer per well and cultured for several weeks. DNA was extracted from individual cones and subjected to genotyping.
Total methylcytosine.
Measurement of 5-methyl-2′-deoxycytidine levels was performed at the University at Buffalo Pharmaceutical Sciences Instrumentation Facility, as described previously (36).
Affymetrix microarray analyses.
Microarray analysis was carried out and evaluated statistically using Cyber-T software as previously described (30, 37, 38). Heat maps were generated using the average levels of expression of three MYC; Dnmt1flox/flox tumors as a reference. Ingenuity pathway analysis (IPA) (Ingenuity Systems) core analysis was used to interrogate microarray data based upon their functions and association with canonical pathways. Significant gene expression changes (FDR < 0.05) between MYC; Dnmt1flox/flox and MYC; Dnmt1−/− mice were imported into IPA with a threshold of 1.75-fold change.
Statistics.
The survival data in Fig. 1B were analyzed using the Kaplan-Meier method for overall survival and the log-rank test for survival distributions. Continuous variables were compared using 2-sample Student's t tests, with error bars representing 1 standard error of the mean. Bisulfite sequencing data were analyzed using paired t tests. P values and FDR of less than 0.05 were considered statistically significant.
Fig 1.
Loss of Dnmt1 delays MYC-induced T-cell lymphomagenesis. (A) Transgene schematic depicting the EμSRα-tTA; Teto-MYC; Teto-Cre; ROSA26EGFP/EGFP; Dnmt1flox/flox genetic setting used in these studies. tTA is expressed in ∼40% of cells and drives expression at the Teto promoter, resulting in transcription of the MYC oncogene and activating Cre-lox recombination to conditionally delete Dnmt1 and activate EGFP expression. (B) Kaplan-Meier survival curves of EμSRα-tTA; Teto-MYC; ROSA26EGFP/EGFP; Dnmt1flox/flox (F/F) and EμSRα-tTA; Teto-MYC; Teto-Cre; ROSA26EGFP/EGFP; Dnmt1flox/flox (−/−) mice. Median survival rates (MS), numbers of mice (n), and P values (log-rank test) are shown. (C) PCR-based analysis of deletion efficiency of the Dnmt1 conditional knockout allele in MYC; Dnmt1−/− tumors. F and KO indicate DNA fragments derived from the floxed and knockout alleles, respectively. Dnmt1flox/flox and Dnmt1flox/− genomic DNAs served as controls. NC indicates negative control (no DNA). (D) Immunoblot analysis of Dnmt1 expression in normal thymocytes (Th) and in MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas. γ-Tubulin served as a loading control. (E) PCR-based analysis of deletion efficiency of the Dnmt1 conditional knockout allele in cellular clones from MYC; Dnmt1−/− tumors. The top and bottom panels represent two tumors with 10 clones each. F and KO indicate floxed and knockout alleles, respectively. Dnmt1flox/flox and Dnmt1flox/− genomic DNAs served as controls. NC indicates negative control (no DNA). (F) Immunoblot analysis of Dnmt3a and Dnmt3b expression in normal thymocytes (Th) and in MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas. (G) Representative examples of FACS analysis from immunophenotyping of MYC; Dnmt1−/− tumors using anti-CD4 and anti-CD8 antibodies. Examples of CD4 single-positive and CD4/CD8 double-positive tumors are shown. (H) Summary of immunophenotypes for CD4 single-positive and CD4/CD8 double-positive tumors as determined by FACS analysis using CD4 and CD8 expression in MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas.
Western blot analysis.
The following antibodies were used for Western blot analysis: Dnmt3b (52A1018; Imgenex), Dnmt1 (H-300; Santa Cruz), Dnmt3a (H-295; Santa Cruz), and γ-tubulin (H-183; Santa Cruz). Western blot analyses were carried out as described previously (30).
qRT-PCR.
cDNA was prepared from RNA using Bio-Rad iScript according to the manufacturer's protocol. cDNA was combined with SYBR green Supermix (Bio-Rad) with a final volume of 20 μl, and experiments were done in duplicate. Reaction conditions were optimized by the use of standard curves for each primer pair. Thermocycling was performed using a CFX96 system (Bio-Rad). Threshold cycle (CT) values were normalized based upon the expression of ubiquitin.
Primer sequences.
The primer sequences used in these experiments are listed below. For determinations of Dnmt1 deletion efficiency, GGGCCAGTTGTGTGACTTGG, ATGCATAGGAACAGATGTGTGC, and CTTGGGCCTGGATCTTGGGGA were used. For Akt3 quantitative real-time RT-PCR (qRT-PCR), CATCTGAAACAGACACCCGATA and GTTGTCCATGCCGTCCAT were used. For Nrbf2 qRT-PCR, TCTCTGAAGCCATGAAGCTG and GTGCTCTGCTGCGCTTTC were used. For Rasgef1a qRT-PCR, AGGACAGCTGGAAGGCACT and ATCTGTCGCAATGTCACCAA were used. For Upb1 qRT-PCR, TCTCTGCCAGCAGATCAATG and TAGTTGGGTTTGACCGCTTC were used. For H2-Ab1 combined bisulfite restriction analysis (COBRA) and bisulfite sequencing, TGGTTTTTTATTTGGGATTAATTTT and TAAACCCTACTATCTCCCCATACAC were used. For Abhd14a COBRA, GTTAAGTTTGGTTATTAGGGAAGAA and ATAAATCTTTTACACCCTTCCTAAC were used.
Bisulfite sequencing and COBRA.
Sodium bisulfite treatment of genomic DNA was carried out utilizing an EpiTect bisulfite kit (Qiagen). Primers for bisulfite PCR for both COBRA and bisulfite sequencing were designed through the use of MethPrimer (39). For COBRA, bisulfite PCR products were digested with TaqI (NEB). The resulting fragments were then loaded onto an 8% PAGE gel, separated by electrophoresis, and detected with SYBR green Gold (Invitrogen). Bisulfite PCR fragments were also processed for bisulfite sequencing by cloning into pGem-T Easy vector (Promega). DNA was purified from clones and sequenced at the University of Nebraska Medical Center High-throughput DNA Sequencing and Genotyping Core Facility.
Study approval.
This study was performed in rigorous accordance with the guidelines established by the Guide for the Care and Use of Laboratory Animals at the National Institutes of Health. All experiments involving mice were approved by the IACUC (protocol number 08-083-10-FC) at the University of Nebraska Medical Center.
RESULTS
Loss of Dnmt1 delays MYC-induced T-cell lymphomagenesis.
In this study, we utilized the genetic setting outlined in Fig. 1A and described previously (30, 40). Briefly, the EμSRα promoter is active in ∼40% of all hematopoietic lineages, including stem cells, and drives the expression of the tetracycline transcriptional transactivator (tTA). Expression of tTA drives simultaneous expression of the MYC oncogene and Cre recombinase from the Teto promoter. Activation of Cre results in the excision of the “stop cassette” located upstream of the Rosa26LOXPEGFP locus, leading to the synthesis of EGFP. Thus, EGFP allows monitoring of tTA/Cre-expressing cells by flow cytometry (fluorescence-activated cell sorter [FACS]) analysis. Importantly, Cre expression also results in the excision of the conditional knockout allele of Dnmt1 (referred to herein as Dnmt1flox).
To assess the effects of Dnmt1 loss on MYC-induced lymphomagenesis, we compared levels of tumor development in cohorts of EμSRα-tTA; Teto-MYC; Teto-Cre; Rosa26LOXPEGFP/EGFP; Dnmt1flox/flox and EμSRα-tTA; Teto-MYC; Rosa26LOXPEGFP/EGFP; Dnmt1flox/flox mice (designated MYC; Dnmt1−/− and MYC; Dnmt1flox/flox mice, respectively). As shown in Fig. 1B, lymphomagenesis was significantly delayed in the cohort of MYC; Dnmt1−/− mice relative to MYC; Dnmt1flox/flox mice (median survival, 166 and 90 days, respectively). Analysis of DNA and protein revealed that tumors arising in MYC; Dnmt1−/− mice retained ∼50% of Dnmt1 levels (Fig. 1C and D). Furthermore, analysis of individual clones grown in vitro from single cells isolated from MYC; Dnmt1−/− tumors revealed that a vast majority of cells had the Dnmt1flox/− genotype, suggesting that primary tumors mainly consisted of cells heterozygous for the Dnmt1 knockout allele (Fig. 1E). Taken together, these data suggest that Dnmt1 is required for the survival of tumor cells.
Reduced Dnmt1 levels did not affect the levels of Dnmt3a and Dnmt3b, suggesting that these enzymes are unlikely to exert compensatory effects on the process of lymphomagenesis (Fig. 1F). FACS analysis confirmed similar immunophenotypes of lymphomas developed in MYC; Dnmt1−/− and MYC; Dnmt1flox/flox mice. Specifically, MTCLs from both genetic groups consisted of cells that were either CD4+ CD8+ CD44+ CD25− or CD4+ CD8− CD44+ CD25−, suggesting that loss of Dnmt1 does not affect the immunophenotype of the tumors (Fig. 1G and H). We previously observed no measurable effects of Cre and EGFP expression on the process of lymphomagenesis in this model (30, 40). In addition, loss of Dnmt1 had no measurable effects on MYC expression (data not shown). Therefore, these results suggest that differences in survival between cohorts of MYC; Dnmt1−/− and MYC; Dnmt1flox/flox mice can be attributed to the loss of Dnmt1 and that this loss does not result in an altered immunophenotype(s) in this mouse model.
Dnmt1 is required for cellular proliferation and maintenance of tumor phenotypes in MTCLs.
To examine the underlying cellular basis for the extended survival of MYC; Dnmt1−/− mice, we measured annexin V expression and bromodeoxyuridine (BrdU) incorporation in cell lines established from lymphomas of terminally sick MYC; Dnmt1−/− and MYC; Dnmt1flox/flox mice. Flow cytometry quantification of annexin V revealed similar expression levels for MYC; Dnmt1flox/flox and MYC; Dnmt1−/− mice. However, a modest but statistically significant reduction in BrdU incorporation was observed in cells from MYC; Dnmt1−/− lymphomas (Fig. 2A and B). These data suggest that defective tumor cell proliferation may, in part, be responsible for the delayed tumorigenesis seen in MYC; Dnmt1−/− mice.
Fig 2.

Loss of Dnmt1 results in decreased proliferation and is critical for tumor cell survival. (A and B) Analysis of apoptosis by annexin V staining (A) and BrdU incorporation assay (B) of four independent T-cell lines derived from MYC; Dnmt1flox/flox and MYC; Dnmt1−/− primary MYC-induced lymphomas. Representative FACS diagrams are shown with average percentages and standard deviations in the upper right quadrant (P < 0.05 (Student's t test). Quantification of obtained results for each assay is shown as an average value with error bars representing standard errors of the means (SEM). Statistically significant differences are indicated by an asterisk. (C) A representative example of cell cycle analysis of a MYC; Dnmt1flox/flox cell line expressing Cre-ER without 4-hydroxytamoxifen (4-OHT) and after 24, 48, and 72 h of incubation with 4-OHT. The cell cycle was measured by Invitrogen Vybrant DyeCycle Orange stain. The percentage of dead cells (blue) is indicated on FACS diagrams. (D) PCR-based analysis of deletion efficiency of the Dnmt1 conditional KO allele in MYC; Dnmt1flox/flox cell lines infected with Cre-ER and treated with 4-OHT at 24 and 72 h. F and KO indicate DNA fragments derived from the floxed and knockout alleles, respectively. A Dnmt1flox/− (F/−) genomic DNA control and a negative control (NC) are shown.
To determine whether Dnmt1 is required for maintenance of the tumor phenotype, we generated two inducible MYC; Dnmt1flox/flox; Cre-ER cell lines, in which Cre-mediated excision of the Dnmt1 conditional allele is achieved by the addition of 4-hydroxytamoxifen (4-OHT) to cell cultures. Importantly, while the cellular viability of two MYC; Dnmt1+/+; Cre-ER control cell lines treated with 4-OHT was not affected (data not shown), loss of Dnmt1 resulted in death of MYC; Dnmt1flox/flox lymphoma cells within 72 h (Fig. 2C). As shown in Fig. 2D, activation of Cre-ER in vitro yielded efficient deletion of the Dnmt1 conditional allele at as early as 24 h and remained high at 72 h. Collectively, these data support the idea of a requirement for Dnmt1 in the maintenance of the tumor phenotype in vitro.
Loss of Dnmt1 impairs T-cell development.
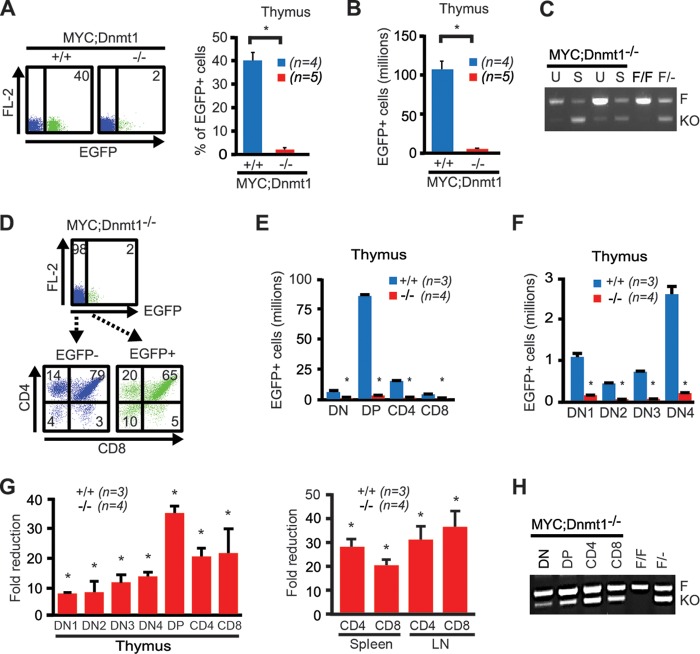
The presence of the Rosa26LOXPEGFP reporter transgene in MYC; Dnmt1−/− mice allows evaluation of biological and molecular events occurring in hematopoietic cells by FACS. To further investigate the processes responsible for delayed lymphomagenesis, we evaluated T-cell development in MYC; Dnmt1−/− mice by measuring levels of EGFP in thymi. Both percentages and numbers of EGFP-positive cells were decreased ∼20-fold in MYC; Dnmt1−/− mice relative to thymi isolated from control EμSRα-tTA; Teto-MYC; Teto-Cre; Rosa26LOXPEGFP/EGFP; Dnmt1+/+ mice (designated MYC; Dnmt1+/+ mice) (Fig. 3A and B). PCR-based genotyping revealed low levels of deletion efficiency of the conditional Dnmt1 allele in the thymi of MYC; Dnmt1−/− mice, whereas EGFP-positive thymocytes sorted by FACS analysis retained ∼50% of the conditional Dnmt1 allele (Fig. 3C). A marked decrease in cell numbers was evident at all stages of T-cell development in EGFP-positive MYC; Dnmt1−/− thymi, including DN1 to -4, CD4− CD8−, CD4+ CD8+, CD4+, and CD8+ cells (Fig. 3D to F). Dnmt1 was particularly important for the transition from CD4− CD8− double-negative (DN1 to -4) cells to the CD4+ CD8+ double-positive (DP) stage during differentiation, since CD4+ CD8+ cells showed the highest fold reduction (Fig. 3G). Consistently, the number of T cells in spleen and lymph nodes was substantially reduced (Fig. 3G). PCR-based genotyping of sorted EGFP+ DN, DP, CD4+, and CD8+ MYC; Dnmt1−/− cells showed the lowest level of Dnmt1 knockout efficiency in the DN population, with progressive increases in DP, CD4, and CD8 cells (Fig. 3H), suggesting that the requirement of Dnmt1 for cellular survival during normal thymocyte development is highest in the least-differentiated DN cells but becomes less stringent in the more-differentiated stages. This is consistent with the CD4+ CD8+ immunophenotype and deletion efficiency of Dnmt1 observed in the majority of MYC; Dnmt1−/− lymphomas (Fig. 1C and H). Altogether, these results suggest that T cells lacking Dnmt1 are selected against during T-cell development and that an efficient transition from the DN stage to the DP stage of thymocyte differentiation requires Dnmt1.
Fig 3.
Loss of Dnmt1 impairs T-cell development. (A) Representative FACS profiles (left, averages are shown within diagrams) and total percentages of EGFP expression (right). (B) Total numbers of EGFP-positive cells isolated from thymi of 24-day-old MYC; Dnmt1+/+ and MYC; Dnmt1−/− mice. (C) PCR-based analysis of deletion efficiency of the Dnmt1 conditional KO allele in unsorted (U) and EGFP-positive sorted (S) cells isolated from thymi of MYC; Dnmt1−/− mice (n = 2). F and KO indicate DNA fragments derived from the floxed and knockout alleles, respectively. Dnmt1flox/flox and Dnmt1flox/− genomic DNA served as controls. (D) FACS analysis of CD4 and CD8 expression in EGFP-positive and EGFP-negative cells from thymi of 24-day-old MYC; Dnmt1−/− mice. Percentages within each portion are shown on the flow diagram. (E) The total number of EGFP-positive T-cell populations within the thymus of MYC; Dnmt1+/+ and MYC; Dnmt1−/− mice. Double-negative (DN), CD4/CD8 double-positive (DP), CD4 single-positive (CD4), and CD8 single-positive (CD8) cell results are shown. (F) Further analysis of thymocyte development within the CD4/CD8 double-negative population (DN1 to DN4) by FACS in MYC; Dnmt1+/+ and MYC; Dnmt1−/− thymi. (G) The left bar graph shows the fold reduction of EGFP-positive double-negative (DN1 to -4), CD4/CD8 double-positive (DP), CD4 single-positive, and CD8 single-positive cells in the thymi of 24-day-old MYC; Dnmt1−/− mice relative to EGFP-positive MYC; Dnmt1+/+ cells. The right bar graph depicts fold reduction of EGFP-positive CD4 and CD8 single-positive cells in the spleen and lymph node (LN) relative to EGFP-positive MYC; Dnmt1+/+ cells. Error bars represent SEM; P < 0.05 (Student's t test). Statistically significant differences are indicated by an asterisk. n represents the number of biological replicates. (H) PCR-based analysis of deletion efficiency of the Dnmt1 conditional KO allele in FACS-sorted double-negative (DN), CD4/CD8 double-positive (DP), CD4 single-positive (CD4), and CD8 single-positive (CD8) EGFP-positive thymic cells from 24-day-old MYC; Dnmt1−/− mice. F and KO indicate DNA fragments derived from the floxed and knockout alleles, respectively. Dnmt1flox/flox and Dnmt1flox/− genomic DNA served as controls.
Ablation of Dnmt1 alters hematopoietic development.
We next investigated whether loss of Dnmt1 negatively impacted development of other hematopoietic lineages. Indeed, cell surface marker expression revealed decreased levels of EGFP-positive B -cells and myeloid cells in the spleen, lymph node, and bone marrow in MYC; Dnmt1−/− mice (Fig. 4A and B). Interestingly, the fold reduction of B220+ cells was substantially greater than that of CD11b+ cells in all three tissues, indicating that the lymphoid lineage may be more sensitive to reductions in the Dnmt1 level. This finding is consistent with previous work in which Dnmt1 was found to be essential for normal B-cell differentiation but not myeloid differentiation (27). DNA isolated from the bone marrow and spleen showed low efficiency of Dnmt1 knockout (Fig. 4C), further supporting the idea that Dnmt1 is required for normal hematopoiesis.
Fig 4.
Impaired hematopoiesis in MYC; Dnmt1−/− mice. (A) The total numbers of EGFP-positive cells within the spleen, lymph node (LN), and bone marrow (BM) of MYC; Dnmt1+/+ and MYC; Dnmt1−/− mice. (B) Fold reductions in total numbers of EGFP-positive B-lymphoid (B220) and myeloid (CD11b) cells relative to EGFP-negative B-lymphoid and myeloid cells in spleens, lymph nodes, and bone marrow of 24-day-old MYC; Dnmt1−/− mice. (C) PCR-based analysis of deletion efficiency of the Dnmt1F allele in total DNA isolated from bone marrow (BM) and spleen (SP) of 24-day-old MYC; Dnmt1−/− mice. Dnmt1flox/flox and Dnmt1flox/− genomic DNA controls as well as a negative control (NC) are shown. F and KO indicate DNA fragments derived from the floxed and knockout alleles, respectively. (D) FACS analysis of c-kit and Sca-1 expression in lineage-negative (Lin−) EGFP-negative and Lin− EGFP-positive cells (LSK cells: Lin− Sca1+ c-kit+) in the bone marrow of MYC; Dnmt1−/− mice. Average percentages of LSK cells are indicated in the FACS diagrams. (E) The total numbers of LSK cells in EGFP-positive populations in bone marrow of MYC; Dnmtflox/flox and MYC; Dnmt1−/− mice. Error bars represent SEM; P < 0.05 (Student's t test). Statistically significant differences are indicated by an asterisk. The number of samples is indicated by n.
The Eμ-tTA transgene that activates Cre-mediated excision of the conditional Dnmt1 allele is expressed in hematopoietic stem cells and early hematopoietic progenitor cells (HSC/HPCs), which are defined by a Lin− Sca-1+ c-kit+ marker profile (LSK; cells negative for lineage markers CD4, CD8, CD11b, CD19, TER119, CD3, gamma-delta T-cell receptor [TCRγδ], and TCRβ and positive for Sca-1 and c-kit). Thus, the substantial decrease in EGFP-positive cells in hematopoietic organs of MYC; Dnmt1−/− mice could be caused by an essential role of Dnmt1 in either the differentiation or the maintenance of HSC/HPCs. To assess this, we analyzed LSK populations in the bone marrow. Numbers of EGFP-positive LSK cells in MYC; Dnmt1−/− mice were significantly lower than of those isolated from MYC; Dnmt1+/+ mice (Fig. 4D and E). The relative absence of EGFP-positive MYC; Dnmt1−/− LSK cells indicated these cells have a competitive disadvantage in comparison to their EGFP-negative counterparts with respect to maintenance of their physiological levels in the bone marrow and during differentiation into hematopoietic lineages, likely due to the critical role of Dnmt1 in these processes.
Reduction of Dnmt1 in MTCLs leads to altered locus-specific methylation.
To investigate how decreased levels of Dnmt1 affect DNA methylation in tumors, we first analyzed the content of 5-methyl-2′-deoxycytosine in DNA from primary mouse MTCLs using an established mass spectrometry method (36). MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas had decreased levels of global 5-methylcytosine relative to normal thymocytes, suggesting that tumors underwent global DNA hypomethylation (Fig. 5A). No significant differences between MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas were observed. This result is consistent with the moderate effects observed in an HCT116 cell line with knockout of Dnmt1 alleles (41). To determine locus-specific effects of Dnmt1 on DNA methylation, we profiled the methylation status of HpaII and HpyCh4IV restriction sites using methyl-sensitive cut counting (MSCC) in genomic DNA isolated from MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas as described previously (30). Analysis of Line-L1, Line-L2, and Sine/Alu repeat elements revealed hypomethylation in both tumor groups relative to normal thymocytes but no significant differences in repeat element methylation between tumor groups (Fig. 5B).
We next analyzed MSCC data to determine how Dnmt1 affected the DNA methylation landscape in MYC; Dnmt1flox/flox tumors. A total of 24,236 promoters in the mouse genome have at least two restriction sites as seen from HpaII and/or HpyCh4IV digests. To rigorously assess the methylation status of promoters, we considered a change in methylation to be significant only if it occurred in two or more independent restriction sites in promoter areas at from bp −1500 to +350 relative to the transcription start site, with a 2-fold or greater change at a false discovery rate (FDR) of less than 0.05. A comparison of the methylation landscapes of MYC; Dnmt1flox/flox and MYC; Dnmt1−/− cells revealed that 730 promoters were differentially methylated between these two tumor groups (Fig. 5C; see also Data Set S1 in the supplemental material). Of these, 303 promoters showed increased levels of methylation in MYC; Dnmt1−/− lymphomas, possibly as a result of deregulation of DNA methylation machinery. Of the 427 promoters that showed decreased methylation levels in MYC; Dnmt1−/− lymphomas, 214 promoters were also hypomethylated in normal thymocytes, suggesting that these promoters might be de novo targets of Dnmt1 in tumorigenesis. The remaining 213 promoters were highly methylated in normal thymocytes and MYC; Dnmt1flox/flox lymphomas but hypomethylated in MYC; Dnmt1−/− lymphomas, suggesting that Dnmt1 is involved in the maintenance of methylation at these promoters.
Decreased levels of Dnmt1 resulted in methylation changes of 777 genes, with 440 hypomethylation and 337 hypermethylation events in gene bodies (Fig. 5D). Additionally, we have seen a relatively small amount of overlap of genes that are hypomethylated in both the promoter and the gene body (∼6%), suggesting that Dnmt1 activity is locus specific. Collectively, these data indicate that the contribution of Dnmt1 to the promoter methylation consists not only of maintenance methylation patterns but also of cancer-specific de novo activity. Since MYC; Dnmt1flox/flox tumors retain ∼50% of Dnmt1 protein levels, our data also suggest that gene promoters require higher levels of Dnmt1 than gene bodies or intragenic regions to maintain proper methylation patterns.
Deregulated transcription in Dnmt1-deficient lymphomas.
To further understand the molecular basis for increased survival of MYC; Dnmt1−/− mice, as well as the effects of Dnmt1 on gene transcription, we next compared microarray-based gene expression profiles of MYC; Dnmt1−/− lymphomas to those of normal thymocytes and MTCLs. We identified 1,260 genes whose expression levels were significantly different (1.75-fold; FDR < 0.05) between MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas (Fig. 6A; see also Data Set S2 in the supplemental material). Loss of Dnmt1 resulted in transcriptional upregulation of 780 genes, which is consistent with the function of Dnmt1 as a repressor protein. Although we cannot rule out the possibility that Dnmt1 plays a role in transcriptional activation, 480 genes downregulated in MYC; Dnmt1−/− lymphomas likely represent secondary changes rather than being a direct consequence of Dnmt1 inactivation. Real-time quantitative RT-PCR (qRT-PCR) confirmed that Akt3, Nrfb2, and Rasgef1a genes are upregulated whereas Upb1 is downregulated in MYC; Dnmt1−/− lymphomas (Fig. 6B). Unsupervised hierarchical clustering analysis using global gene transcription profiles of all genes resulted in perfect segregation of tumors in a Dnmt1-specific manner, with MYC; Dnmt1flox/flox tumors clustering closer to MYC; Dnmt1−/− lymphomas than to normal thymocytes (Fig. 6A). Tight clustering of MYC; Dnmt1−/− lymphomas suggests that Dnmt1 plays an important role in regulating the transcriptome in MTCLs, likely through specific target genes.
To gain further insight into the pathogenesis of MYC; Dnmt1−/− lymphomas, we performed IPA using 780 genes whose transcription was significantly changed (upregulated 1.75-fold or more, FDR < 0.05) relative to MYC; Dnmt1flox/flox lymphomas. The top five disease groups associated with higher expression in MYC; Dnmt1−/− lymphomas relative to control MYC; Dnmt1flox/flox lymphomas were cancer, hematological disease, developmental disorders, immunological disease, and renal and urological disease (data not shown). Of these, the most significant functional disease network was that of cancer. Eight genes (Dcn, Rag1, Brca2, Cdkn1a, Zeb1, Ccnd3, Runx1, and Ssbp2) from the hematologic disease network “tumorigenesis in thymic lymphomas” were overexpressed in Dnmt1-deficient lymphomas, and their upregulation was suggested by IPA to be involved in the suppression of T-cell lymphomagenesis (Fig. 6C). In addition, IPA showed that negative regulators of cell cycle, such as retinoblastoma (Rb1), were upregulated in Dnmt1-deficient lymphomas. Considering all these results together, these molecular events may contribute to the delayed lymphomagenesis observed in MYC; Dnmt1−/− mice.
To determine the effects of Dnmt1-dependent methylation on gene transcription, we next analyzed levels of gene expression in Dnmt1 target genes. This comparison revealed that expression of 17 of 427 hypomethylated genes correlated with promoter hypomethylation, suggesting that differential methylation affects a relatively small (∼4.0%) subset of genes (Fig. 6D).
H2-Ab1 is a target of cancer-specific de novo methylation by Dnmt1 in vivo.
The major histocompatibility class 2 gene (H2-Ab1) is involved in antigen processing and presentation (42). Its locus encodes two distinct isoforms (Fig. 7A). Our MSCC data showed that the promoter region driving expression of the longer H2-Ab1 isoform may be a target of Dnmt1, as it was consistently hypomethylated in MYC; Dnmt1−/− tumors (Fig. 7B). To determine if H2-Ab1 is a maintenance or de novo target of Dnmt1, we performed combined bisulfite restriction analysis (COBRA) and bisulfite sequencing of the −43 to +471 region of this promoter in normal thymocytes and MYC; Dnmt1flox/flox and MYC; Dnmt1−/− tumors. In both assays, MYC; Dnmt1flox/flox tumors were consistently hypermethylated relative to normal thymocytes, while Dnmt1-deficient tumors showed near-complete ablation of promoter methylation (Fig. 7C and D). These changes in methylation correlated with gene transcription of the major isoform of H2-Ab1, which was severely repressed in MYC; Dnmt1flox/flox tumors compared to normal thymocytes but was derepressed in MYC; Dnmt1−/− tumors (Fig. 7E). Importantly, this potential de novo activity of Dnmt1 is independent of the presence of Dnmt3a and Dnmt3b, as levels of methylation of the H2-Ab1 locus remained high in thymic lymphomas with genetic inactivation of either Dnmt3a or Dnmt3b (Fig. 7D). Importantly, a potential de novo activity of Dnmt1 is not limited to H2-Ab1 since the promoter of the Abhd14a gene is also hypermethylated in MYC; Dnmt1flox/flox but is unmethylated in MYC; Dnmt1−/− lymphomas (Fig. 7E). Furthermore, global methylation profiling of MYC; Dnmt1−/− and MYC; Dnmt3b−/− tumors showed an overlap of only 3% in Dnmt1 target genes (Fig. 7G, reference 30, and data not shown). In addition, only 35 of 780 (4%) Dnmt1 target genes were overexpressed in both Dnmt1−/− and Dnmt3b−/− MTCLs (Fig. 7G). Thus, Dnmt1 appears to have cancer-specific target genes whose methylation and/or expression is largely independent of the presence of Dnmt3b and perhaps Dnmt3a.
Fig 7.
H2-Ab1 is a target of Dnmt1-mediated methylation in lymphomas. (A) An illustration of the exons and introns comprising the two H2-Ab1 isoforms. Black arrows represent the transcription start site. Red arrows denote the location of qRT-PCR primers. (B) A methylation bar graph depicting average methylation levels across three CpG sites analyzed by MSCC in the promoter of H2-Ab1 for normal thymocytes (Th) and MYC; Dnmt1flox/flox and MYC; Dnmt1−/− tumors. (C) COBRA of the H2-Ab1 promoter in normal thymocytes (Th) and in MYC; Dnmt1flox/flox (F/F) and MYC; Dnmt1−/− lymphomas. PCR fragments were digested with restriction enzyme TaqI. Undigested (U) and digested (D) fragments correspond to unmethylated and methylated DNA, respectively. CpG indicates a fully methylated control. (D) Bisulfite sequencing of the H2-Ab1 promoter in normal thymocytes (Th) and in MYC; Dnmt1flox/flox, MYC; Dnmt1−/−, EμSRα-tTA; Teto-MYC; Teto-Cre; ROSA26EGFP/EGFP; Dnmt3aflox/flox (3a−/−), and EμSRα-tTA; Teto-MYC; Teto-Cre; ROSA26EGFP/EGFP; Dnmt3bflox/flox (3b−/−) lymphomas. The dashed lines denote the location of the CpG dinucleotides within the locus. Each pie denotes an individual CpG dinucleotide within the locus, and each wedge of the pie represents the sequence of an individual allele. Black denotes the percentage of methylated alleles, while white represents unmethylated alleles. The average methylation levels for two tumor samples (≥8 clones sequenced for each sample) are shown. The total methylation of the longer isoform H2-Ab1 locus was calculated by averaging the results for all clones within the group, and numbers are displayed as percentages. (E) qRT-PCR analysis of H2-Ab1 longer-isoform expression in normal thymocytes (Th), MYC; Dnmt1flox/flox tumors, and MYC; Dnmt1−/− tumors. Error bars represent SEM; P < 0.05 (Student's t test). Statistically significant differences are indicated by an asterisk. The number of samples is indicated by n. (F) COBRA of the Abhd14a promoter in normal thymocytes (Th) and in MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas. PCR fragments were digested with restriction enzyme TaqI. Undigested (U) and digested (D) fragments correspond to unmethylated and methylated DNA, respectively. CpG indicates a fully methylated control. (G) Overlap of 260 genes identified in Dnmt3b-deficient MTCLs (25) and 427 potential Dnmt1-dependent targets, as determined by MSCC, are shown in the left Venn diagram. The right Venn diagram shows 253 genes that were at least 1.75-fold upregulated in the absence of Dnmt3b in MTCLs overlaid with the 780 genes that were upregulated in MYC; Dnmt1−/− tumors. The overlapping genes are listed below each diagram.
DISCUSSION
In the present study, we used a MYC model to characterize the role of Dnmt1 in T-cell lymphomagenesis. We show that Dnmt1 plays a crucial role in the prevention and maintenance of the tumor phenotype in MTCLs as well as in normal hematopoiesis. Importantly, we identified locus-specific maintenance and de novo activity for Dnmt1. Our results suggest that Dnmt1, in addition to its involvement in maintenance of DNA methylation patterns during cellular division, also plays an irreplaceable role in promoter and gene body methylation during tumorigenesis. This report represents the first in which the cancer-specific de novo and maintenance activity of Dnmt1 in the in vivo setting is presented.
Suppressed tumorigenesis in MYC; Dnmt1−/− mice is surprising in view of previous work that has suggested that T cells are particularly sensitive to low levels of Dnmt1. Whereas a global decrease in Dnmt1 levels is achieved in all cells in Dnmt1chip/− mice, these mice almost exclusively (91%) develop T-cell lymphomas over the course of 8 months, despite the fact that this all occurs in the context of reduced numbers of HSC/HPCs and thymocytes in Dnmt1chip/− mice. This is likely occurring through the induction of genomic instability and activation of oncogenes such as Notch by intracisternal A particles (26, 27, 43).
There are at least two biological processes that may explain the increased latency of Dnmt1 tumors in our model. First, deletion of Dnmt1 in MYC; Dnmt1−/− mice results in considerable defects in hematopoietic development which may limit the pool of cells susceptible to MYC-induced lymphomagenesis. Inefficient hematopoiesis is largely a consequence of a severe reduction in the number of HSC/HPCs. The total quantity of EGFP-positive LSK cells in MYC; Dnmt1−/− mice is decreased 4-fold relative to the EGFP-negative population, suggesting that HSC/HPCs encountering loss of Dnmt1 are at a competitive disadvantage relative to EGFP-negative cells in terms of their ability to self-renew within the same microenvironment. Additionally, decreased cellularity of EGFP-positive cells of lymphoid and myeloid origins suggests that the differentiation potential of the remaining LSK cells—likely with only partial Dnmt1 expression—is also skewed. Our data are consistent with observations showing an essential role for Dnmt1 in the self-renewal and differentiation of HSCs (27, 44). c-Myc has been reported to be involved in differentiation of HSCs, as loss of c-MYC results in pooling of stem cells (45, 46). Since our data were obtained in a setting where high levels of transgenic MYC were present, it also appears that MYC overexpression is insufficient to rescue Dnmt1-driven defects in hematopoietic differentiation. Our results show an accumulation of Dnmt1-deficient T cells in the double-negative stage of T-cell differentiation, similar to what was observed in Lck-Cre; Dnmt1−/− mice, whereby Cre actively deletes Dnmt1 alleles at the DN2 stage of thymocyte differentiation and results in impaired differentiation (47). Altogether, these data indicate serious defects in hematopoiesis in the absence of Dnmt1, which ultimately diminishes the pool of hematopoietic cells available for MYC-induced transformation.
Second, a biological process that may contribute to increased survival of MYC; Dnmt1−/− mice is the decreased proliferation potential of MYC; Dnmt1−/− cells. This may reflect increases in the levels of key antiproliferative genes such as retinoblastoma (Rb) that were observed in MYC; Dnmt1−/− lymphomas. Rb is a negative regulator of cell cycle and is a tumor suppressor in hematopoietic cancers (48, 49). Another possibility is that a larger cadre of genes contributes to this phenomenon. For example, Ingenuity pathway analysis of genes upregulated in MYC; Dnmt1−/− lymphomas unveiled an eight-gene (Dcn, Rag1, Brca2, Cdkn1a, Zeb1, Runx1, Ssbp2, and Ccnd3) signature (tumorigenesis in thymic lymphomas) where all eight genes are predicted to decrease thymic lymphomagenesis. Indeed, inactivation of seven of these eight genes has been functionally linked to promotion of thymic lymphomagenesis. Loss of Ssbp2 or Dcn accelerated T-cell lymphomagenesis in p53−/− mice, while ablation of Rag1 accelerated Eμ-MYC tumorigenesis (50–52). Germ line inactivation of Brca2, Cdkn1a, Runx1, or Zeb1 is sufficient to result in hematologic malignancies in mice (53–57). Thus, these putative Dnmt1 target genes have been independently linked to their ability to suppress lymphomagenesis in mouse tumor prevention settings.
Therefore, within one cellular compartment (T cells), Dnmt1 plays a dichotomous role by preventing tumorigenesis through maintaining the integrity of the genome and silencing oncogenes (Dnmt1chip/− studies [26, 43]) and promoting lymphomagenesis by retention of cells susceptible to transformation (MYC; Dnmt1−/− studies). This is likely due to the fact that low levels of Dnmt1 allow cells to survive and accumulate genetic and epigenetic changes promoting tumorigenesis whereas complete deletion of Dnmt1 is incompatible with the viability of normal hematopoietic cells. Thus, Dnmt1 may both promote and inhibit tumorigenesis even within the same tumor type, depending on the degree of its activity. At present, we cannot rule out the possibility that germ line inactivation of one Dnmt1 allele in Dnmt1chip/− mice promotes T-cell transformation in a non-cell-autonomous way. Thus, further studies are required to determine if microenvironments deficient for Dnmt1 can enhance T-cell lymphomagenesis.
This study also uncovered an important role for Dnmt1 in the maintenance of the tumor phenotype. For example, all tumors that developed in MYC; Dnmt1−/− mice retained ∼50% of Dnmt1 levels and exhibited decreased proliferation, suggesting that Dnmt1 is crucial for the survival of tumor cells. This was further confirmed through functional inactivation of Dnmt1 in fully developed MYC; Dnmt1flox/flox lymphomas, which led to severely impaired cellular survival. In addition to being critical for the maintenance of the tumor phenotypes either in AML induced by MLL-AF9 or in B-cell leukemia induced by combined overexpression of Myc and Bcl2 (25, 27), Dnmt1 function is also fundamental for the maintenance of mouse MTCLs.
Our data further indicate that even incomplete inactivation of Dnmt1 has a profound effect on the molecular landscape of MTCLs. By comparing methylation patterns of normal thymocytes, MYC; Dnmt1flox/flox lymphomas, and MYC; Dnmt1−/− lymphomas, we identified 427 promoters whose methylation appeared to depend upon Dnmt1. Of these, 214 promoters are likely targets of Dnmt1 de novo activity, as their methylation is low in normal thymocytes and MYC; Dnmt1−/− lymphomas but is increased in MYC; Dnmt1flox/flox tumors. The remaining 213 gene promoters are methylated in normal thymocytes and MYC; Dnmt1flox/flox lymphomas but hypomethylated in MYC; Dnmt1−/− lymphomas, suggesting that Dnmt1 is involved in maintenance of the methylation at these loci throughout tumorigenesis. Furthermore, Dnmt1 target loci appear to be largely independent of Dnmt3b, as a comparison with our previously published Dnmt3b targets (30) showed only a 3% overlap with newly unveiled Dnmt1 targets in this study. Along the same lines, locus-specific de novo methylation by Dnmt1 at the H2-Ab1 target gene is independent of both Dnmt3a and Dnmt3b, as lymphomas deficient for either methyltransferase gain methylation at levels similar to those seen with MYC; Dnmt1flox/flox lymphomas. Results showing differing spectra of Dnmt1 and Dnmt3b target genes identified in MTCLs are consistent with the opposing effects of Dnmt1 and Dnmt3b on MYC-induced lymphomagenesis, during which, as our recent studies showed, Dnmt3b functions as a tumor suppressor gene (30). Our data also show increased methylation of 303 promoter regions in MYC; Dnmt1−/− lymphomas. At present, it is unclear why a decrease in Dnmt1 levels results in promoter hypermethylation. However, locus-specific increases in methylation levels have been recently observed upon loss of methyltransferase activity of Dnmt3a in mouse hematopoietic stem cells (58).
The present knowledge base of target loci of Dnmt1 is minimal. Previous studies have identified targets of Dnmt1-dependent methylation and transcription in the HCT116 colon cancer cell line (59, 60). When we compared these target genes with Dnmt1 targets identified in this study, we observed only a 2% overlap (data not shown), suggesting that Dnmt1 may have tissue- or species-specific target loci. The apparent lack of a significant overlap in Dnmt1 target genes between the HCT116 cell line and MTCLs may also stem from the different growth conditions under which the cells were maintained. For example, the spectrum of Dnmt1 target genes in HCT116 cell line may reflect the different selection pressures that cells experience during in vitro culturing. In contrast, the methylation profiles of the MTCLs are more likely to faithfully reflect physiological patterns, as they were obtained from primary lymphomas grown in vivo. In any case, in addition to Dnmt1's canonical function as a maintenance methyltransferase during DNA replication, Dnmt1 may localize to specific loci in a cell- or tissue-specific manner.
Juxtaposition of 427 Dnmt1-dependent methylation targets and 780 genes upregulated in MYC; Dnmt1−/− tumors identified only a ∼4% correlation of promoter methylation with the transcription status. This number is likely underestimated, perhaps due to limitations in MSCC analysis as well as in array-based gene transcription profiling. The use of two restriction enzymes (HpaII and HpyCh4IV) increased the coverage of DNA methylation profiling to ∼16% of all CpGs in the mouse genome but still may not have been sufficient to fully reveal the effects of Dnmt1 on the cancer methylome (61). Similarly, Affymetrix gene transcription profiling is unable to reliably distinguish between the levels of expression of various gene isoforms. For example, the Dnmt1 target gene H2-Ab1 encodes at least two isoforms whose transcripts significantly overlap. As a result, global microarray profiling has not identified differences in expression of this gene between MYC; Dnmt1flox/flox and MYC; Dnmt1−/− lymphomas. However, isoform-specific qRT-PCR clearly showed upregulation of the longer isoform which is driven by a specific promoter upon hypomethylation in Dnmt1-deficient MTCLs. Thus, future studies will focus on the use of more-sensitive methods, such as whole-genome bisulfite sequencing for methylation profiling and transcriptome sequencing (RNA-seq) for transcriptional profiling, to further analyze the relationship of DNA methylation to transcription. It is possible that inhibitory effects of DNA methylation on transcription are limited in some biological settings. Indeed, recent studies have identified a low correlation between changes in promoter methylation and differential gene expression in mouse hematopoiesis and human AML (58, 62). Thus, additional discrete studies should be performed to address this point.
Our present findings highlight the importance of Dnmt1 in the prevention and maintenance of T-cell malignancies and complex activities of Dnmt1 in the tumor methylome and transcriptome.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an Eppley Cancer Center pilot project grant (R.O.), the Nebraska Cancer and Smoking Disease Research Program NE DHHS LB506 2012-28 (R.O.), the National Center for Research Resources, and National Institutes of Health grant 5P20GM103489. R.A.H. was supported by a UNMC assistantship/fellowship.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Donna Rusjaz of the University at Buffalo Pharmaceutical Sciences Instrumentation Facility for the design and implementation of the LC-MS assay to measure 5mdC.
Footnotes
Published ahead of print 3 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00776-13.
REFERENCES
- 1.Poetsch AR, Plass C. 2011. Transcriptional regulation by DNA methylation. Cancer Treat. Rev. 37:S8–S12 [DOI] [PubMed] [Google Scholar]
- 2.Shenker N, Flanagan JM. 2012. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br. J. Cancer 106:248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCabe MT, Brandes JC, Vertino PM. 2009. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin. Cancer Res. 15:3927–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson KD. 2005. DNA methylation and human disease. Nat. Rev. Genet. 6:597–610 [DOI] [PubMed] [Google Scholar]
- 6.Portela A, Esteller M. 2010. Epigenetic modifications and human disease. Nat. Biotechnol. 28:1056–1068 [DOI] [PubMed] [Google Scholar]
- 7.Fernandez AF, Huidobro C, Fraga MF. 2012. De novo DNA methyltransferases: oncogenes, tumor suppressors, or both? Trends Genet. 28:474–479 [DOI] [PubMed] [Google Scholar]
- 8.Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257 [DOI] [PubMed] [Google Scholar]
- 9.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Saskie H, Forne T, Weber M. 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42:1093–1100 [DOI] [PubMed] [Google Scholar]
- 10.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, Macleod AR. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33:61–65 [DOI] [PubMed] [Google Scholar]
- 11.Vertino PM, Yen RW, Gao J, Baylin SB. 1996. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol. Cell. Biol. 16:4555–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. 2003. Predicting aberrant CpG island methylation. Proc. Natl. Acad. Sci. U. S. A. 100:12253–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, Walter J. 2012. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 8:e1002750. 10.1371/journal.pgen.1002750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuks F, Bergers WA, Brehm A, Hughes-Davies L, Kouzarides T. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88–91 [DOI] [PubMed] [Google Scholar]
- 15.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338–342 [DOI] [PubMed] [Google Scholar]
- 16.Rountree MR, Bachman KE, Baylin SB. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269–277 [DOI] [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasso CS, Wu Y0 Rosincon MDR, Cao X, Dhanasekaran M, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487:239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. 2001. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. U. S. A. 98:15149–15154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, Olofsson T, Rade J, Fontes M, Porwit-Macdonald A, Behrendtz M, Hoglund M, Johansson B, Fioretos T. 2007. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia 21:1198–1203 [DOI] [PubMed] [Google Scholar]
- 22.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. 2005. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 102:13580–13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinney SR, Moser MT, Pascual M, Greally JM, Foster BA, Karpf AR. 2010. Opposing roles of Dnmt1 in early- and late-stage murine prostate cancer. Mol. Cell. Biol. 30:4159–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, Mauch J, Haldemann B, Fries T, Krönke J, Kühn MW, Paschka P, Kayser S, Wolf S, Gaidzik VI, Schlenk RF, Rücker FG, Döhner H, Lottaz C, Döhner K, Bullinger L. 2012. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood 120:e83–e92 [DOI] [PubMed] [Google Scholar]
- 25.Trowbridge JJ, Sinha AU, Zhu N, Li M, Armstrong SA, Orkin SH. 2012. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev. 26:344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300:489–492 [DOI] [PubMed] [Google Scholar]
- 27.Bröske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, Nerlov C, Leutz A, Andrade-Navarro MA, Jacobsen SE, Rosenbauer F. 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat. Genet. 41:1207–1215 [DOI] [PubMed] [Google Scholar]
- 28.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. 2001. Activation of EGFP expression by cre-mediated excision in a new ROSA26 reporter mouse strain. Blood 97:324–326 [DOI] [PubMed] [Google Scholar]
- 29.Perl A, Wert SE, Nagy A, Lobe CG, Whitsett JA. 2002. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc. Natl. Acad. Sci. U. S. A. 99:10482–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hlady RA, Novakova S, Opavska J, Klinkebiel D, Peters SL, Bies J, Hannah J, Iqbal J, Anderson KM, Siebler HM, Smith LM, Greiner TC, Bastola D, Joshi S, Lockridge O, Simpson MA, Felsher DW, Wagner KU, Chan WC, Christman JK, Opavsky R. 2012. Loss of Dnmt3b function upregulates the tumor modifier Ment and accelerates mouse lymphomagenesis. J. Clin. Invest. 122:163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 27:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, Smyth GK. 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23:2881–2887 [DOI] [PubMed] [Google Scholar]
- 34.Robinson MD, Smyth GK. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9:321–332 [DOI] [PubMed] [Google Scholar]
- 35.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23:2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song L, James SR, Kazim L, Karpf AR. 2005. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal. Chem. 77:504–510 [DOI] [PubMed] [Google Scholar]
- 37.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509–519 [DOI] [PubMed] [Google Scholar]
- 39.Li LC, Dahiya R. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431 [DOI] [PubMed] [Google Scholar]
- 40.Opavsky R, Wang SH, Trikha P, Raval A, Huang Y, Wu YZ, Rodriguez B, Keller B, Liyanarachchi S, Wei G, Davuluri RV, Weinstein M, Felsher D, Ostrowski M, Leone G, Plass C. 2007. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 3:1757–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. 2000. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404:1003–1007 [DOI] [PubMed] [Google Scholar]
- 42.Stables MJ, Shah S, Camon EB, Lovering RC, Newson J, Bystrom J, Farrow S, Gilroy DW. 2011. Transcriptomic analyses of murine resolution-phase macrophages. Blood 118:e192–e208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. 2008. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27:404–408 [DOI] [PubMed] [Google Scholar]
- 44.Trowbridge JJ, Snow JW, Kim J, Orkin SH. 2009. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, Bernstein ID, Trumpp A. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3:611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. 2001. A critical role for Dnmt1 and DNA methylation in T-cell development, function, and survival. Immunity 15:763–774 [DOI] [PubMed] [Google Scholar]
- 48.Ginsberg AM, Raffeld M, Cossman J. 1991. Inactivation of the retinoblastoma gene in human lymphoid neoplasms. Blood 77:833–840 [PubMed] [Google Scholar]
- 49.Sherr CJ. 1996. Cancer cell cycles. Science 274:1672–1677 [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Klumpp S, Amin HM, Liang H, Li J, Estrov Z, Zweidler-McKay P, Brandt SJ, Agulnick A, Nagarajan L. 2010. SSBP2 is an in vivo tumor suppressor and regulator of LDB1 stability. Oncogene 29:3044–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iozzo RV, Chakrani F, Perrotti D, McQuillan DJ, Skorski T, Calabretta B, Eichstetter I. 1999. Cooperative action of germ-line mutations in decorin and p53 accelerates lymphoma tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 96:3092–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nepal RM, Zaheen A, Basit W, Li L, Berger SA, Martin A. 2008. AID and RAG1 do not contribute to lymphomagenesis in Emu c-myc transgenic mice. Oncogene 27:4752–4756 [DOI] [PubMed] [Google Scholar]
- 53.Connor F, Bertwistle D, Mee JP, Ross GM, Swift S, Grigorieva E, Tybulewicz VL, Ashworth A. 1997. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat. Genet. 17:423–430 [DOI] [PubMed] [Google Scholar]
- 54.Martín-Caballero J, Flores JM, Garcia-Palencia P, Serrano M. 2001. Tumor susceptibility of p21 (Waf1/Cip1)-deficient mice. Cancer Res. 61:6234–6238 [PubMed] [Google Scholar]
- 55.Silva FP, Morolli B, Storlazzi CT, Anelli L, Wessels H, Bezrookove V, Kluin-Nelemans HC, Giphart-Gassler M. 2003. Identification of RUNX1/AML1 as a classical tumor suppressor gene. Oncogene 22:538–547 [DOI] [PubMed] [Google Scholar]
- 56.Putz G, Rosner A, Nuesslein I, Schmitz N, Buchholz F. 2006. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene 25:929–939 [DOI] [PubMed] [Google Scholar]
- 57.Hidaka T, Nakahata S, Hatakeyama K, Hamasaki M, Yamashita K, Kohno T, Arai Y, Taki T, Nishida K, Okayama A, Asada Y, Yamaguchi R, Tsubouchi H, Yokota J, Taniwaki M, Higashi Y, Morishita K. 2008. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood 112:383–393 [DOI] [PubMed] [Google Scholar]
- 58.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JP, Godley LA, Li W, Goodell MA. 2012. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, Baylin SB. 2012. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 40:4334–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee EJ, Pei L, Srivastava G, Joshi T, Kushwaha G, Choi JH, Robertson KD, Wang X, Colbourne JK, Zhang L, Schroth GP, Xu D, Zhang K, Shi H. 2011. Targeted bisulfite sequencing by solution hybrid selection and massively parallel sequencing. Nucleic Acids Res. 39:e127. 10.1093/nar/gkr598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colaneri A, Staffa N, Fargo DC, Gao Y, Wang T, Peddada SD, Birnbaumer L. 2011. Expanded methyl-sensitive cut counting reveals hypomethylation as an epigenetic state that highlights functional sequences of the genome. Proc. Natl. Acad. Sci. U. S. A. 108:9715–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O'Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. 2010. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363:2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.