Abstract
The genus Metapneumovirus within the subfamily Pneumovirinae and family Paramyxoviridae includes only two viruses, human metapneumovirus (hMPV) and avian metapneumovirus (aMPV), which cause respiratory disease in humans and birds, respectively. These two viruses grow poorly in cell culture and other quantitation methods, such as indirect immuno-staining and immuno-fluorescent assays, are expensive, time consuming, and do not allow for plaque purification of the virus. In order to enhance research efforts for studying these two viruses, a direct plaque assay for both hMPV and aMPV has been developed. By optimizing the chemical components of the agarose overlay, it was found that both hMPV with a trypsin-independent F cleavage site and aMPV formed clear and countable plaques in a number of mammalian cell lines (such as Vero-E6 and LLC-MK2 cells) after 5 days of incubation. The plaque forming assay has similar sensitivity and reliability as the currently used immunological methods for viral quantitation. The plaque assay is also a more simple, rapid, and economical method compared to immunological assays, and in addition allows for plaque purification of the viruses. The direct plaque assay will be a valuable method for the quantitation and evaluation of the biological properties of some metapneumoviruses.
Keywords: Human metapneumovirus, avian metapneumovirus, plaque assay, quantitation
1. Introduction
Human metapneumovirus (hMPV) is a non-segmented negative-sense RNA virus belonging to the family of Paramyxoviridae, the subfamily Pneumovirinae, and the genus Metapneumovirus (van den Hoogen et al., 2001). There are at least two lineages of hMPV circulating in human population, designated A and B, which can be further divided into A1, A2, B1, and B2, based on their surface glycoproteins and antigenicity (van den Hoogen et al., 2004). Since its discovery in 2001, hMPV has been recognized worldwide as one of the leading causes of lower respiratory infections, second only to human respiratory syncytial virus (hRSV) (van den Hoogen et al., 2001; Kahn, 2006). hMPV infections are observed in all age groups with a high prevalence and severity among infants, children, the elderly, and immunocompromised patients (van den Hoogen et al., 2003; Bastien et al., 2003; Boivin et al., 2003; Falsey et al., 2003; Stockton et al., 2002; Williams et al., 2004). Clinical symptoms associated with hMPV infection are similar to those caused by hRSV, ranging from asymptomatic infection to severe bronchiolitis and pneumonia (Boivin et al., 2002; Esper et al., 2003). To date, no vaccines or antiviral drugs are available against this important pathogen.
Avian metapneumovirus (aMPV) is the only other member in the Metapneumovirus genus. Discovered in the 1970s, aMPV has been recognized as an important pathogen for the poultry industry worldwide (Buys and Preez, 1980). aMPV is the causative agent of tract reproaccute rhinotracheitis (TRT) in turkeys, and is associated with swollen head syndrome (SHS) in chickens (Buys, du Preez, and Els, 1989). It causes respiratory tract and reproductive infections with low mortality but high morbidity in turkeys and chickens, resulting in economic losses in both egg and poultry production for the turkey and chicken industries. Based on genetic and serological homology, aMPV is divided into four antigenic subtypes, namely, A, B, C, D (Bayon-Auboyer et al., 2000). Interestingly, aMPV subtype C is more closely related to hMPV than the other aMPV subtypes.
One of the major challenges in metapneumovirus research is the lack of a convenient, economic, and reliable quantitation method, which is due mainly to the fact that both hMPV and aMPV grow poorly in cell culture. It usually takes more than 10 days for hMPV to develop significant cytopathic effects (CPE) in mammalian cells. The growth of some hMPV strains is trypsin-dependent and, in general, it has been shown that aMPV grows better than hMPV (van den Hoogen et al., 2001). However, it takes more than 5 days for aMPV subtype C strains to develop typical CPE.
Currently, indirect immunostaining or an immunofluorescence assay (IFA) followed by TCID50 calculations are used to determine the titer of hMPV and aMPV (van den Hoogen et al., 2001; Herfst et al., 2004; Ebihara et al., 2005). Unfortunately, this methodology is relatively complicated and costly. Although hMPV forms small spots in immunostaining-based assays, it requires fixation of the cells followed by antibody straining. The viruses are inactivated by fixation and therefore purification of the virus post-staining cannot be executed. Agarose overlay plaque assays have been used to quantify, separate, and propagate a great number of viruses. However, no plaque assay for tittering hMPV has been reported to date. As for aMPV, only one study has shown that aMPV subtype C can form visible plaques in the Japanese quail fibrosarcoma cell line (QT-35) after 6 days incubation (Sabara and Larence, 2003). It is not known whether aMPV can form plaques in other cell lines.
In this study, an agarose overlay plaque assay is developed for titer determination and plaque purification of hMPV strains with trypsin-independent cleavage sites in F protein and aMPV using African green monkey kidney cells (Vero) and rhesus monkey kidney cells (LLC-MK2). The results indicate that plaque formation can occur in 5 days with sensitivity comparable to that of the conventional indirect immunostaining assays. In addition, the plaque assay is more convenient and economical than other immunological methods. Hence, the direct plaque assay developed in this study could provide a valuable method for detection, isolation, purification, and quantitation of aMPV and some hMPV strains with self-cleavable F protein.
2. Materials and methods
2.1.Cell culture
African green monkey kidney epithelial cells (Vero ATCC no. CCL-81 and Vero E6 ATCC no. CRL-1586) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) in a humidified incubator at 37 °C and 5% CO2. Rhesus monkeys kidney cell (LLC-MK2) cells were maintained in OPTI-MEM reduced serum medium (Invitrogen) supplemented with 2% FBS under the same incubation conditions.
2.2.Virus recovery and characterization
An infectious cDNA clone of hMPV lineage A strain NL/1/100 was kindly provided by Dr. Ron A. M. Fouchier at Department of Virology, Erasmus Medical Center, Rotterdam, The Netherlands. Recombinant hMPV (rhMPV) was isolated using a reverse genetics system (Herfst et al., 2004; Biacchesi et al., 2004). Briefly, rhMPV was recovered by co-transfection of a plasmid encoding the full-length genomic cDNA of hMPV NL/1/00 (phMPV) and support plasmids encoding viral N (pCITE-N), P (pCITE P), L (pCITE-L), and M2-1 (pCITE-M2-1) proteins into BHK.SR19T7pac cells (kindly provided by Apath LLC, Brooklyn, NY, USA) which express stably the T7 RNA polymerase. Six days post-transfection, the cells were subjected to three freeze-thaw cycles followed by centrifugation at 3000×g for 10 min. The supernatant was used subsequently to infect LLC-MK2 cells. Since hMPV requires trypsin to grow, TPCK-trypsin was added to the media to the final concentration of 0.1µg/ml at day 2 post-infection. Cytopathic effect (CPE) was observed 5 days post-infection and the recovered viruses were amplified further in LLC-MK2 cells.
The F cleavage site (99RQSR102) in the full-length genome of wild type hMPV NL/1/100 was mutated into 99RRRR102 (Fig. 1A) using the QuikChange Site-Directed Mutagenesis Kit (Strategene, La Jolla, CA, USA) with the following primers: forward, 5’- GAGAGGAGCAAATTGAAAATCCCAGACGACGTAGATTCGTTCTAGGAGCAATAGC -3’; reverse, 5’-GCTATTGCTCCTAGAACGAATCTACGTCGTCTGGGATTTTCAATTTGCTCCTCTC-3’. Recombinant hMPV carrying the trypsin-independent F cleavage site (rhMPV-F) was recovered as described above. Recombinant rhMPV-F exhibited trypsin-independent growth, and was propagated subsequently in LLC-MK2 cells in the absence of trypsin.
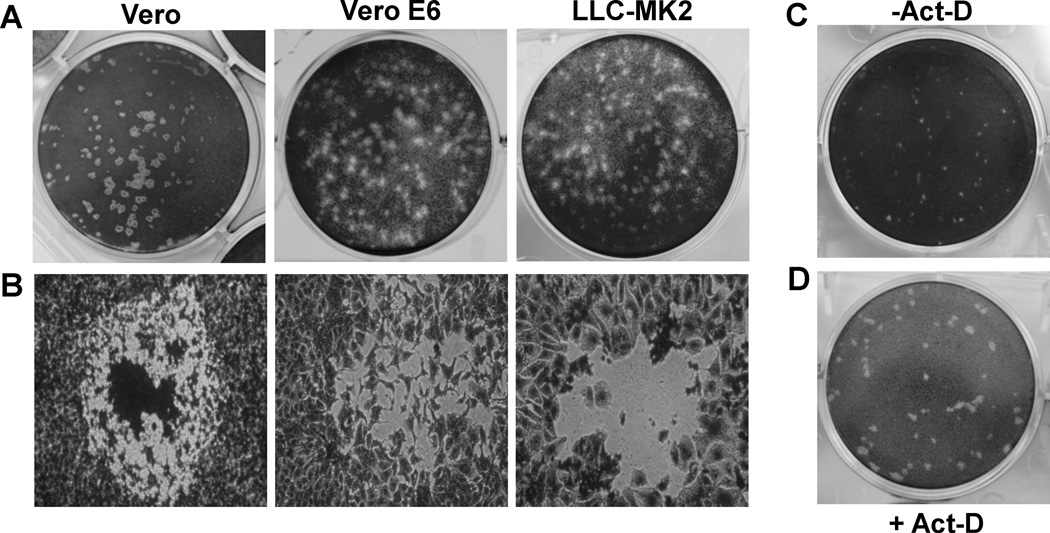
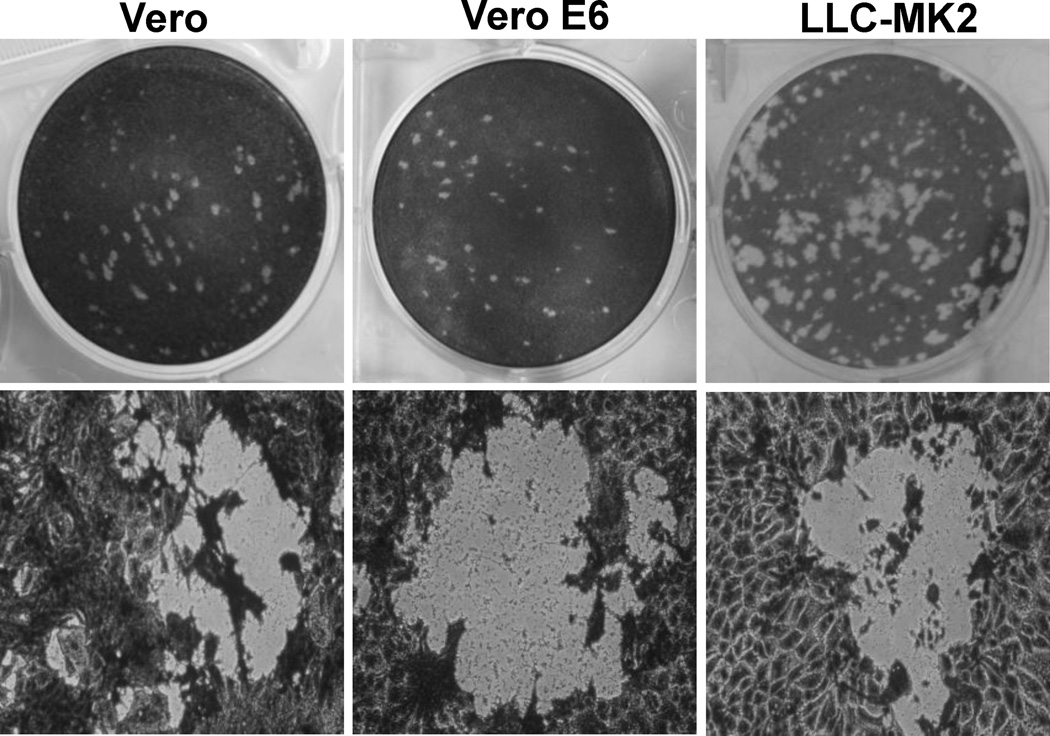
Fig.1. Plaque formation of aMPV subtype C MN strain in Vero, Vero-E6, and LLC-MK2 cells.
(A) Plaque morphology of aMPV in different cell lines. Agarose overlay plaque assay was performed in six-well plates as described in Materials and Methods. After incubation for 6 days, the plates were fixed in 10% formaldehyde, and the plaques were visualized by staining with 0.05% crystal violet. (B) Microscopic examination of viral plaques in different cell lines. Plaques were examined under at 100× magnification under light microscope. Digital photographs of plaque were taken under a Nikon TS100 inverted phase-contrast microscope mounted with a Nikon Coolpix995 camera. (C) Plaque morphology of aMPV in Vero cells without actinomycin-D. Plaque assay was performed without actinomycin-D. Plaques were developed at day 4 post-infection. (D) Plaque morphology of aMPV in Vero cells with actinomycin-D. The agarose overlay contains 0.1 μg/mL of actinomycin-D. Plaques were developed at day 4 post-infection.
aMPV subtype C Minnesota (MN) strain was a generous gift from Mo Saif at the Ohio Agricultural Research and Development Center (OARDC, Wooster, OH) and was propagated in LLC-MK2 cells.
2.3.Cytopathogenic effect(CPE)-based TCID50 assay for aMPV
LLC-MK2 cells were seeded in 96 well plates (Corning, Lowell, MA, USA) at a density of 105 cells per well and were grown at 37 °C for 18 h. Upon infection, the medium was removed and 0.2 ml of tenfold serial virus dilutions in Opti-MEM was added to each well. Eight wells containing a monolayer of cells were infected with 50 µl of each virus dilution and the cytopathogenic effect (CPE) was examined under a microscope daily for 10 days post infection. The CPE was recorded and the virus titer was calculated as the tissue culture infection dose (TCID50) using the Reed-Muench method (1938).
2.4. An indirect immunostaining assay for rhMPV
LLC-MK2 cells were seeded and then infected with serial dilutions of rhMPV or rhMPV-F as described in Section 2.3. At day 6 post-infection, the supernatant was removed and cells were fixed in a pre-chilled acetone: methanol solution (at the ratio of 3:2) at room temperature (RT) for 15 min. Cells were permeablized in a phosphate saline buffer (PBS) containing 0.4% Triton X-100 at RT for 10 min, and blocked at 37 °C for 1 h using 1% bovine serum albumin (BSA) in PBS. The cells were then labeled with an anti-hMPV N protein primary monoclonal antibody (Millipore, Billerica, MA) at dilution of 1:1,000, followed by incubation with horseradish peroxidase (HRP)-labeled rabbit anti-mouse secondary antibody (Thermo Scientific, Waltham, MA, USA) at dilution of 1:5,000. After incubation with AEC substrate chromogen (Sigma, St. Louis, MO, USA), positive cells were then visualized under the microscope. Viral titer was calculated as TCID50 using the Reed-Muench method (1938).
2.5. Direct plaque assays for rhMPV-F and aMPV
Vero or Vero-E6 cells were seeded in 6-well plates (Corning) at the density of 2×106 cells per well. After incubation for 18 h, the medium was removed and cell monolayers were infected with 400 µl of a 10-fold dilution series of each virus. After incubation at 37°C for 1 h with agitation every 10 min, the cells in each well were overlaid with 2.5 ml of Eagle minimum essential medium (MEM) containing 1% agarose, 1% FBS, 0.075% sodium bicarbonate (NaHCO3), 20mM HEPES (pH 7.7), 2 mM L-Glutamine, 12.5 mg/mL of penicillin, 4 mg/mL of streptomycin, and 4 mg/mL of kanamycin. The plates were incubated at 4 °C for 30 min to solidify the overlay media. Cells were then grown at 37 °C and 5% CO2 to allow for plaque formation. Where indicated, the overlay medium was supplemented with actinomycin-D (Sigma) (0.1 to 0.6 µg/ml) or TPCK-trypsin (0.1 to 0.6 µg/ml). After incubation for 4–10 days, the cells were fixed in 10% (v/v) formaldehyde for 2 h, and the plaques were visualized by staining with 0.05% (wt/vol) crystal violet.
2.6. Growth kinetics of rhMPV-F
Confluent LLC-MK2 cells in 35-mm dishes were infected by rhMPV-F at a multiplicity of infection (MOI) of 0.1. After 1 h of adsorption, fresh DMEM (supplemented with 2% FBS) was added, and infected cells were incubated at 37°C. At different time points post infection, the cells were harvested by three freeze-thaw cycles followed by centrifugation at 1,500 × g, RT for 15 min. Virus titer was determined using indirect immunostaining assays or direct plaque assays on LLC-MK2 cells as described above.
2.7. Statistical analyses
All experiments were carried out in triplicate. Statistical analysis was performed by one-way multiple comparisons using SPSS 8.0 statistical analysis software (SPSS Inc., Chicago, IL). A value of p <0.05 was considered statistically significant.
3. Results
3.1. aMPV, but not rhMPV, formed plaques in an agarose overlay plaque assay
First, the ability of aMPV to form plaques in Vero, Vero E6, and LLC-MK2 cells was investigated, because these cell lines are susceptible to metapneumovirus infection. As shown in Fig. 1A, aMPV subtype C MN strain formed clear and countable plaques in all three cell lines tested. Plaques produced in Vero cells were round and much clearer than those in Vero E6 or LLC-MK2 cells (Fig.1A). Further microscopic examination showed that the edge between the intact and lysed cells was clear in Vero cells (Fig. 1B). In contrast, the cells at the edge of the plaque were not completely detached in LLC-MK2 and Vero E6 cells, leading to the irregular-shaped plaques. Further optimization of the experimental conditions was established by including 0.1 µg/ml of actinomycin-D, an inhibitor of mammalian cell proliferation, in the agarose overlay media. The addition of actinomycin-D produced plaques that were visible and countable at day 4 post-infection in all three cell lines. As shown in Fig1. C and D, plaques formed in the presence of actinomycin-D were significantly larger than those in the absence of actinomycin-D. Inhibition of cell growth with actinomycin-D significantly improved the ability of aMPV to replicate in cell culture and improved the quality of the plaque assay results.
Subsequently, an attempt was made to develop a similar plaque assay for hMPV. hMPV is known to grow much poorer compared to aMPV in most of the cell lines tested. Also, some hMPV strains require trypsin for growth, so in order to optimize the plaque assay for hMPV growth the assay was conducted as described above with the following modifications. First, the overlay medium was supplemented with 0.2 µg/ml of trypsin-TPCK but no FBS. Second, the time for plaque development was extended to 3 weeks. Third, due to the high sensitivity of Vero cells to trypsin at concentrations required for hMPV growth, only Vero E6 and LLC-MK2 cells were chosen for this study. Unfortunately, no visible plaques were formed by hMPV in LLC-MK2 or Vero E6 cells after incubation for up to three weeks post-infection (data not shown). In addition, increasing the concentration of trypsin and/or including actinomycin-D in the overlay medium did not led to any observable plaques, indicating that rhMPV is not able to form plaques in Vero E6 or LLC-MK2 cells although both cell lines are permissive for hMPV replication.
3.2. Recombinant hMPV with a trypsin-independent F cleavage site (rhMPV-F) formed plaques in both Vero E6 and LLC-MK2 cells
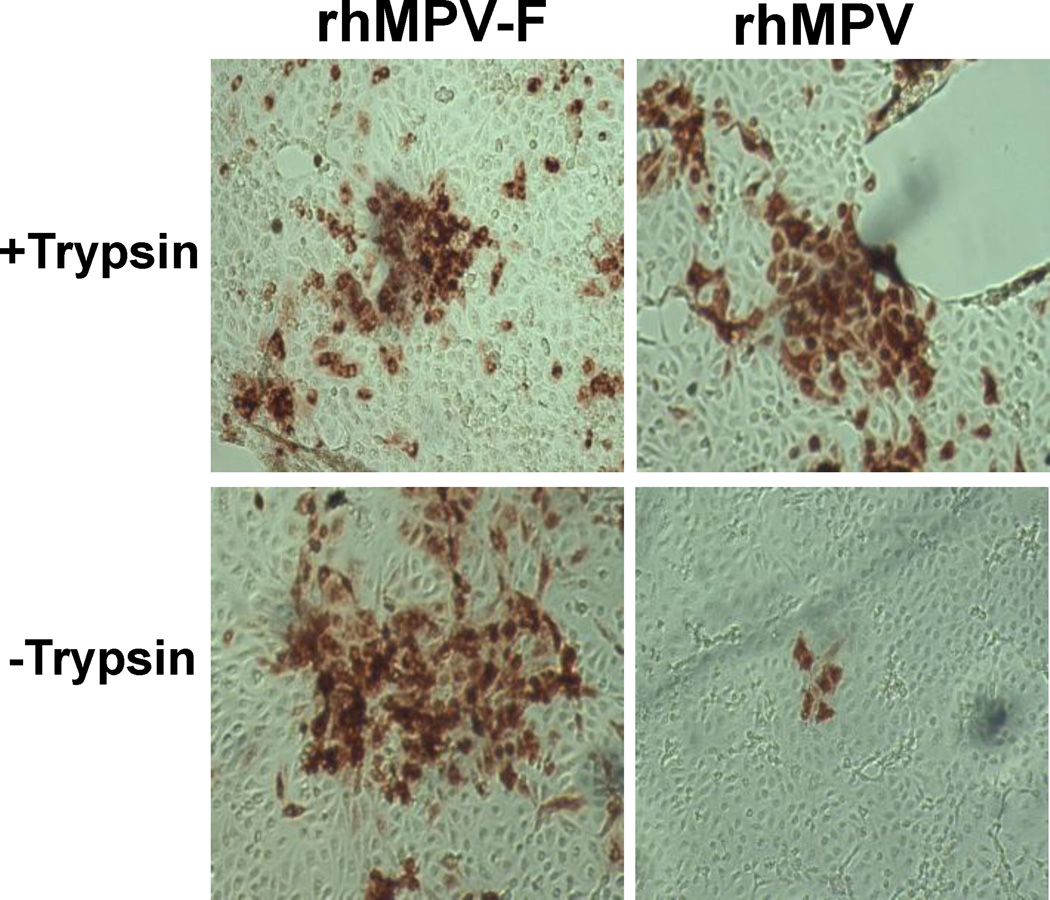
It has been reported that the growth of hMPV clinical isolates was improved in the presence of trypsin in cell culture medium. For paramyxoviruses, the F protein cleavage site sequence is the major determinant for trypsin dependence (Collins et al., 1993; Lamb 1993). The F protein of hMPV NL/1/100 strain contains a trypsin-dependent cleavage site (99RQSR102 motif). Hence, it was speculated that the inability of hMPV NL/1/100 strain to form plaques in Vero E6 and LLC-MK2 cells could be associated with its putative cleavage site as well as trypsin dependence. To address this premise, the 99RQSR102 motif was mutated to 99RRRR102 in the F cleavage site in an infectious clone of hMPV NL/1/100, and successfully recovered the recombinant hMPV carrying the trypsin-independent F cleavage site (rhMPV-F). As expected, rhMPV-F displayed a trypsin-independent growth pattern in LLC-MK2 cells (Fig. 2). Next, the evaluation of the ability of rhMPV-F to form plaques in Vero, Vero E6, and LLC-MK2 cells was carried out using an approach similar to that for aMPV. Remarkably, visible plaques were observed in Vero, Vero E6, and LLC-MK2 cells infected with rhMPV-F after 8 days incubation (Fig. 3). In addition, plaques formed in Vero and Vero E6 cells had larger and more pronounced zones of clearing and were easier to count than those in LLC-MK2 cells. Taken together, these results demonstrated trypsin-independent rhMPV-F was able to form plaques in an agarose overlay plaque assay, suggesting that a direct agarose plaque assay is applicable for some (if not all) hMPV strains with trypsin-independent F cleavage sites.
Fig.2. Trypsin dependence of rhMPV-F and rhMPV.
LLC-MK 2 cells were infected by rhMPV-F or rhMPV and incubated with DMEM with or without 0.2μg/mL of trypsin-TPCK. Indirect immunostaining assay was performed at day 6 post-infection. After removal of cell culture medium, the cells were fixed in acetone: methanol (at the ratio of 3:2) and permeablized in PBS containing 0.4% Triton X-100. The cells were then probed with anti-hMPV N monoclonal antibody, followed by incubation with HRP-conjugated rabbit anti-mouse antibody. After incubation with AEC substrate chromogen, positive cells were visualized under light microscope.
Fig.3. Plaque formation of rhMPV-F in Vero, Vero-E6, and LLC-MK2 cells.
(A) Plaque morphology of rhMPV-F in different cell lines. Agarose overlay plaque assay was performed in six-well plates as described in Materials and Methods. Plaques were developed at day 6 post-infection. (B) Microscopic examination of viral plaques in different cell lines. Plaques were examined under at 100× magnification under light microscope.
3.3. Optimization of a direct plaque assay for metapneumoviruses
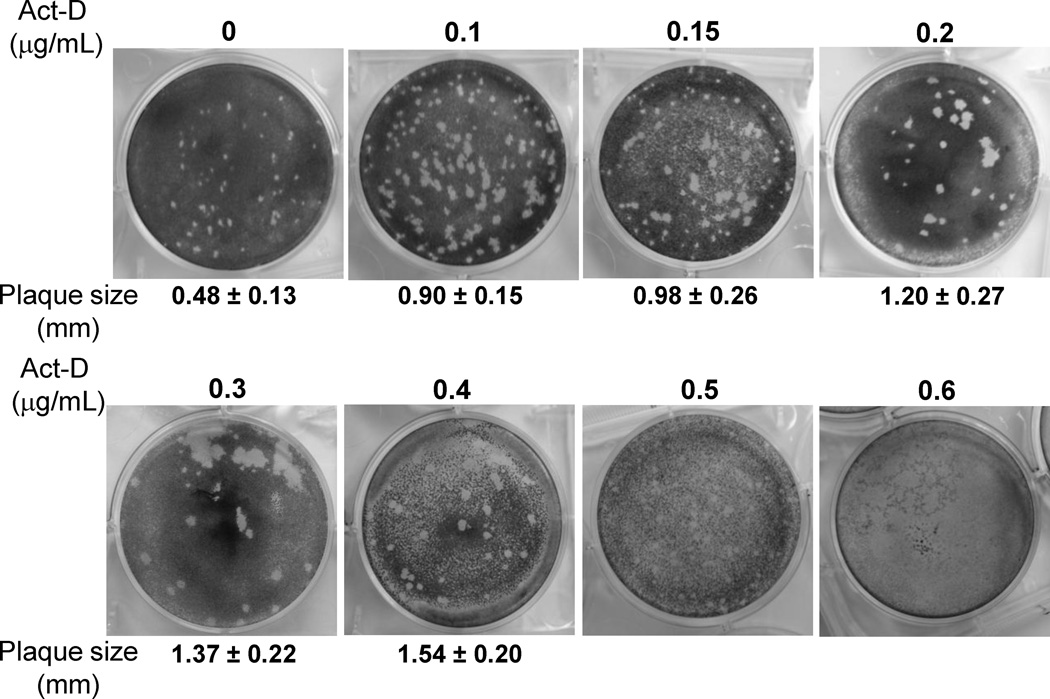
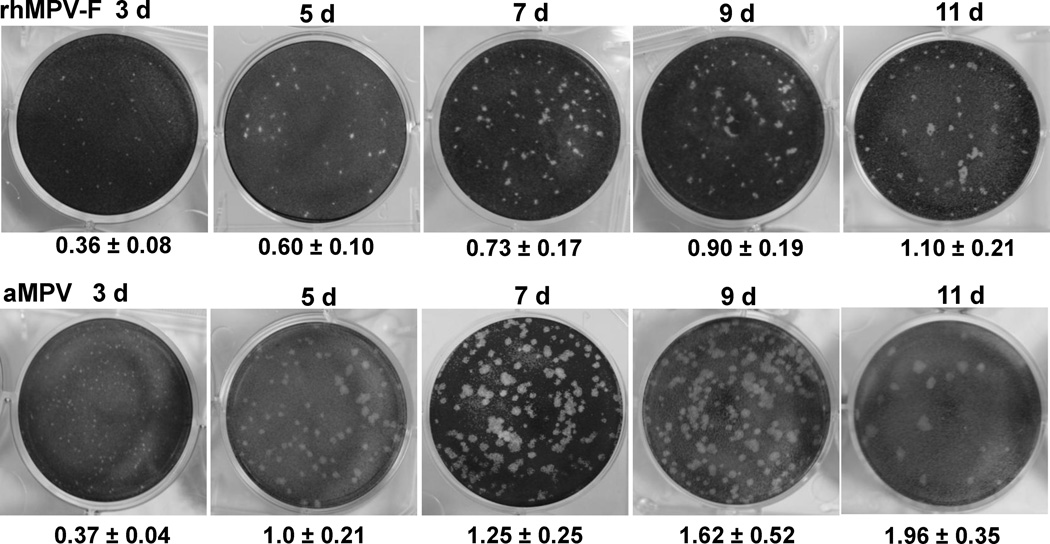
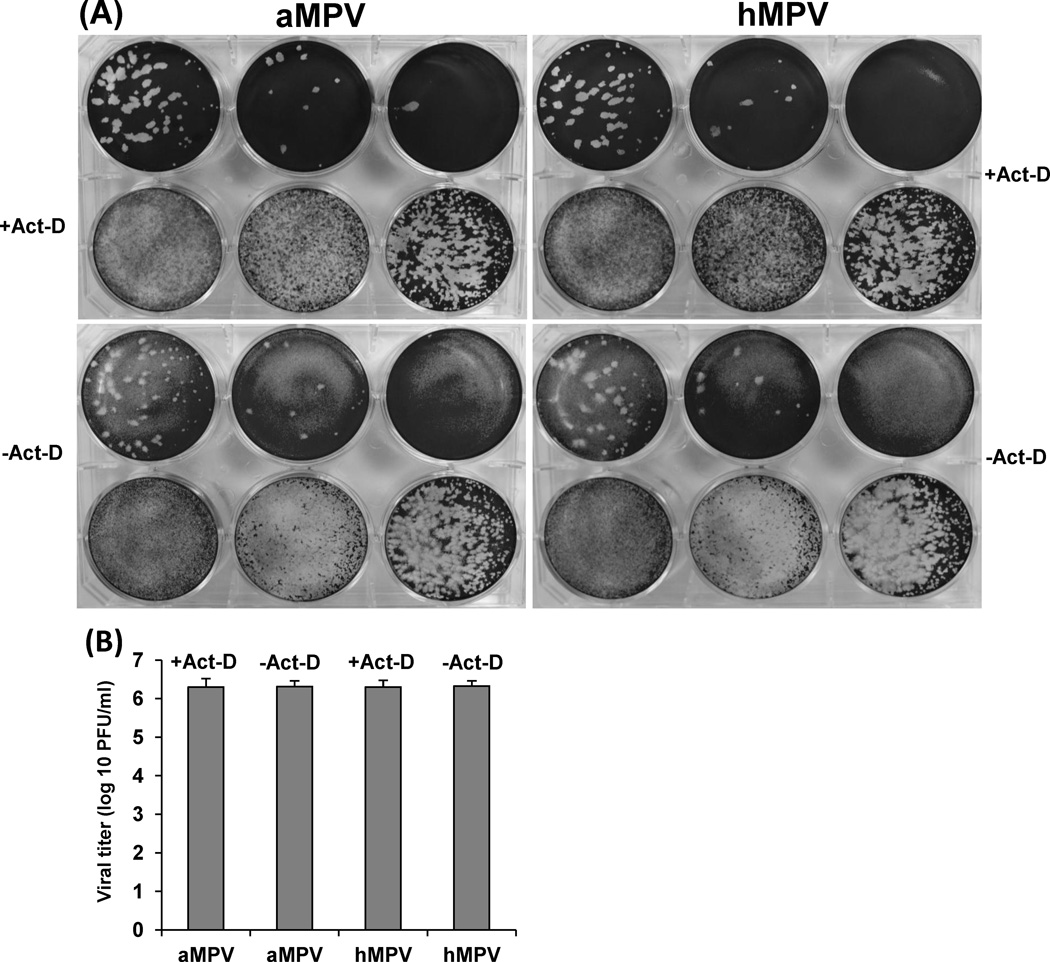
Modification of the parameters of the direct plaque assay was then optimized so that plaques could be visualized and detected in the shortest possible time. First, the effect of growth temperature on the plaque formation was evaluated. It was found that rhMPV-F formed significantly bigger plaques at 37 °C than at 30 °C after seven days of infection, indicative of a preference for the higher temperature. The average size of a total of twenty plaques formed at 37 °C at day 7 post-infection was 1.0 ± 0.12 mm, whereas the plaque size formed at 30 °C was 0.44 ± 0.11 mm. Secondly, the effect of cell confluence on the plaque formation was tested. Vero cells were seeded at different densities to achieve 50%, 80%, and 100% confluency after incubation at 37 °C for 18 h, and plaque assays were performed as described previously. There was a reduction in the size of plaques observed as the cell confluency increased. The average plaque size in cells of 80% confluency at day 7 post-infection (1.1 ± 0.2 mm) was significantly larger than that in cells of 100% confluency (0.9 ± 0.2 mm) (p < 0.05). In addition, larger plaques were observed when the cell confluency was reduced to 50%. However, cells at 50% confluency made it challenging to count the plaques due to the light background after staining with crystal violet. Therefore, it was concluded that a cell culture of 80% confluence was the most suitable for the direct plaque assay taking into account the plaque size and clear background. Thirdly, the impact of actinomycin-D on the plaque formation of rhMPV-F was investigated. The plaque assay was performed in Vero cells with 80% confluency as described previously except that the agarose overlay medium contained various amounts of actinomycin-D and the plaque was developed at day 7 after incubation. As shown in Fig. 4, actinomycin-D increased the plaque size of rhMPV-F in a dose dependent manner. However, high concentrations of actinomycin-D (0.5– 0.6 µg/mL) brought about significant damage to the monolayer of cells. In contrast, 0.1–0.2 µg/mL of actinomycin-D was found to be the optimal concentration, as it increased significantly the plaque size (P<0.05) but did not cause any noticeable damage to the cell monolayer. Fourth, the shortest time for forming visible plaques was determined, as well as the dynamics of the plaque size formation for rhMPV-F and aMPV (Fig.5). Plaque assays were carried out in the presence of 0.1 µg/mL of actinomycin-D, plates were incubated at 37°C, and fixed from days 4–10 post-infection. For both rhMPV-F and aMPV, the plaque size became bigger as the incubation time increased (Fig.5). However, aMPV appeared to be more efficient in forming plaques than rhMPV-F. For rhMPV-F, visible plaques were observed at day 5 post-infection, and became countable at day 7 post-infection. In contrast, aMPV plaques could be visualized at day 4 and became countable at day 5 post-infection. Finally, the effect of actinomycin-D on the accuracy of the direct plaque assay was evaluated. The stock of aMPV and hMPV was titrated using the direct plaque assay with or without 0.2 µg/mL of actinomycin-D. As shown in Fig. 6A, the plaques in actinomycin-D treated plates (developed at day 7) were bigger than those in non-treated plates (developed at day 10). As shown in Fig.6B, there was no significant difference between the titers determined by plaque assay with or without actinomycin-D (P>0.05). Therefore, actinomycin-D increased the size of the plaques without altering the number of the plaques.
Fig.4. The effect of actinomycin-D on plaque formation of rhMPV-F.
The plaque assay was performed in Vero cells with 80% confluency. Vero cells in six-well plates were infected with 400 µl of a 10-fold dilution series of rhMPV-F. The agarose overlay medium contained various amounts of actinomycin-D and the plaques were developed at day 7 post-infection. One representative well (dilution range from105 to107) from each six-well plate was shown. The size of individual plaque was measured by Photoshop software. The average size of a total of twenty plaques was shown.
Fig.5. Dynamics of plaque formation of rhMPV-F and aMPV in Vero cells.
Direct plaque assay were carried out in the presence of 0.1 µg/mL of actinomycin-D and plates were incubated at 37°C and fixed from day 3 to 11 post-infection. One representative well (dilution range from105 to107) from each six-well plate was shown. The average size of a total of twenty plaques was shown.
Fig.6. Effect of actinomycin-D on the number and size of viral plaques.
(A) Effect of actinomycin-D on the size of aMPV and rhMPV-F plaques. Plaque assay with or without 0.2 µg/ml of actinomycin-D was developed at days 7 and 10 post-infection, respectively. (B) Effect of actinomycin-D on the number of aMPV and rhMPV-F plaques. The titer of aMPV and rhMPV-F stocks was determined by direct plaque assay with or without 0.2 µg/ml of actinomycin-D. Titers were the average of three independent experiments.
Taken together, the optimized direct plaque assay for rhMPV-F was determined as the following: Vero or Vero E6 cells in six-well plates with 80% confluency infected by rhMPV-F; after 1 hour incubation at 37°C, the agarose overlay medium containing 0.1µg/mL of actinomycin-D is added to the cells; and the plates are incubated at 37°C for at least 5 days.
3.5. Sensitivity of the direct plaque assay
The sensitivity of the optimized direct plaque assay was compared to other conventional methods, such as TCID50 and indirect immunostaining assays. Briefly, Vero E6 cells were infected by aMPV and rhMPV-F at MOI of 1, and viruses were harvested at day 7 post-infection. The titer of aMPV and rhMPV was determined by direct plaque assay, TCID50, or indirect immunostaining assay. Overall, the virus titer from direct plaque assays was consistent with the corresponding values determined by TCID50 (aMPV) and indirect immunostaining (rhMPV-F). While the viral titer of rhMPV-F determined by the direct plaque assay is 1.7 ± 0.2×10 7 PFU/mL, the corresponding values determined by immunostaining assay is 3.8 ± 1.4×10 7 TCID50/mL. For aMPV, the viral titer determined by direct plaque assay was 2.16 ± 0.29×10 6 , the corresponding values determined by TCID50 was 3.76 ± 0.78×10 6 . The correlation in plaque formation between indirect immunostaining and direct plaque assays was also compared. Both methods were able to detect the presence of rhMPV-F at the highest virus dilution tested in this study (1:107), suggesting that the direct plaque assay has a comparable sensitivity to that of the conventional immunostaining method.
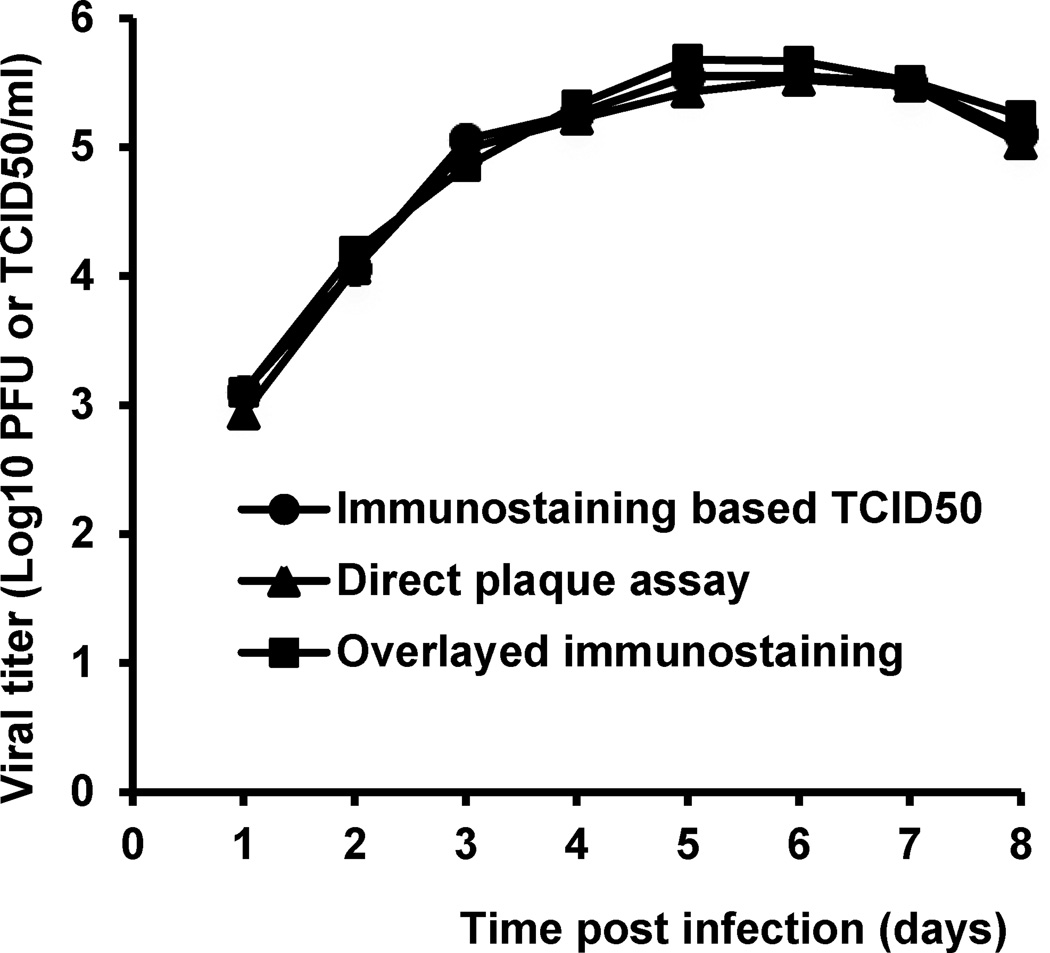
Finally, the direct plaque assays were used to study the growth kinetics of rhMPV-F in LLC-MK2 cells. As shown in Fig. 7, the results reveal an exponential phase of rhMPV-F replication until day 6 post-infection whereas the virus titer starts to drop after day 8 post-infection. The growth curve generated from direct plaque assay was indistinguishable from those of TCID50 and indirect immunostaining assays, suggesting that the direct plaque assay is as reliable as other conventional methods.
Fig.7. Growth kinetics of rhMPV-F determined by direct plaque assay and immunostaining assay.
Confluent LLC-MK2 cells in 35-mm dishes were infected with rhMPV-F at an MOI of 0.1. After a 1-h adsorption, the inoculum was removed, the cells were washed with DMEM, and fresh medium (containing 2% FBS) was added, followed by incubation at 37°C. At the indicated intervals, the cells were harvested by three freeze-thaw cycles followed by centrifugation at 1,500 × g for 15 min. Virus titers were determined by direct plaque assay, indirect immunostaining assay or TCID50 assay in Vero cells. Titers were the average of three independent experiments.
4. Discussion
aMPV and hMPV are the only two known virus members in the genus Metapneumovirus within the subfamily Pneumovirinae and family Paramyxoviridae. It has been a challenge to work with these viruses because they grow poorly in cell culture. Although aMPV was discovered forty years ago, quantitation of this virus has been limited to TCID50 assay by observation of cytopathic effect (CPE) in permissive cell lines. hMPV was first discovered in 2001 and was found to grow much poorer than aMPV in cell culture. Quantitation of hMPV is limited to indirect immunostaining assay by examination of viral antigen expression using antibody staining. In general, RNA viruses have a high mutation rate and it has been reported that both aMPV and hMPV can easily undergo gene truncation and/or mutation during passing in mammalian cells (Govindarajan et al., 2004; Agrawal et al., 2001). Therefore, there is an urgent need to develop a direct plaque assay that allows for plaque purification of virus. In this study, direct plaque assays for aMPV and trypsin-independent hMPV strains was developed. The plaque assay exhibited sensitivity and reliability comparable to conventional quantitation methods such as TCID50 and indirect immunostaining assays, but is more convenient and economical. The direct plaque assay takes only 5 days to develop plaques whereas conventional quantitation methods usually take more than 10 days to produce results. Most importantly, the direct plaque assay does not inactivate the viruses, providing a valuable tool to isolate and purify aMPV and hMPV.
Previously, it has been reported that aMPV subtypes A, B and C were able to form visible plaques in agarose overlay plaque assays using a Japanese quail fibrosarcoma (QT-35) cell line (Sabara and Larence, 2003). However, it took 13 days to develop visible plaques in QT-35 cells. Since Vero, Vero E6, and LLC-MK2 cells are typically used for propagation of aMPV and hMPV, it is beneficial to determine whether the metapneumoviruses are also able to form plaques in these cell lines. By optimizing the chemical components of the agarose overlay, it was found that aMPV and certain hMPV strains formed clear and countable plaques in these permissive cell lines after 5 days of incubation. The size of the plaques was dependent on cell confluency, and incubation temperature and time. Furthermore, the addition of actinomycin D in the agarose overlay significantly increased the size of the plaques. This compound inhibits the proliferation of cells in nonspecific ways by forming a stable complex with double-stranded DNA or causing single-strand breaks in DNA, thus inhibiting DNA-primed RNA synthesis (Mizutani et al., 2000). It is likely that addition of actinomycin D to the overlay medium inhibits cell proliferation which in turn supports viral infection and cell-cell spread. More importantly, the number of plaques is not significantly altered by actinomycin D treatment (P>0.05) (Fig.6), demonstrating that actinomycin D did not affect the accuracy and sensitivity of direct plaque assay.
The main limitation of the direct plaque assay is that it can be only applied to hMPV strains with self-cleavable F protein. The paramyxovirus F protein is synthesized as an inactive precursor (F0), which is cleaved at the F cleavage site by cellular proteases to produce a fusion-competent, disulfide-linked F2-F1 complex (Lamb 993). It is well known that F protein, especially the F protein cleavage site sequence, is the major determinant for trypsin dependence and viral virulence of paramyxoviruses, such as Newcastle Disease Virus (NDV) and Sendai virus (SeV) (Collins et al., 1993; Lamb 1993). While the trypsin-dependent hMPV strains contain a 99RQSR102 motif at the putative cleavage site of F gene, it has been reported that recombinant hMPV with a 99RRRR102 motif at the putative cleavage site (rhMPV-F) exhibits trypsin-independent growth (Schickli et al., 2005; Biacchesi et al., 2006). It was found that wild type hMPV with a 99RQSR102 cleavage site was not able to form visible plaques whereas recombinant hMPV with a 99RRRR102 cleavage site efficiently formed plaques. Despite this limitation, it should not minimize the significance of the development of the direct plaque assay for hMPV. One striking characteristic of hMPV pathogenesis is that the F cleavage site is not the major determinant of virulence (Schickli et al., 2005; Biacchesi et al., 2006). It has been shown that rhMPV-F improves significantly viral growth in cell culture but does not affect the viral replication, distribution, and pathogenesis in hamster and African green monkeys as compared to wild type hMPV (Schickli et al., 2005; Biacchesi et al., 2006). Therefore, rhMPV-F could serve as an appropriate surrogate to study the biology of hMPV. The direct plaque assay will be useful for the study of laboratory adapted hMPV strains and live vaccine candidates. Additionally, it has been documented that a number of hMPV isolates mutated spontaneously to self-cleavage F gene after multiple passages in cell culture (Schickli et al., 2005). Specifically, it was found that the F cleavage site mutation S101P was associated with trypsin-independent viral growth in vitro (Schickli et al., 2005). Interestingly, this mutation has been documented in hMPV isolated from clinical samples. Yang et al., (2009) identified two hMPV subgroup A1 clinical isolates and one subgroup B1 clinical isolate containing naturally occurring S101P mutations which have trypsin-independent F cleavage sites. Presumably, the direct plaque assay would be applicable to these clinical isolates. Finally, it should also be emphasized that self-cleavable F protein does not necessarily require all four basic amino acids (99RRRR102) in the F cleavage site. The F cleavage sites of aMPV subtypes A, B, C are 99RRRR102, 99RKKR102, and 99RKAR102 respectively, all of which possess self-cleavable characteristics (Alkhalaf et al., 2002; Govindarajan et al., 2004). Also, hMPV F protein with 99RQPR102 (S101P mutation) is self-cleavable, demonstrating that self-cleavable hMPV F protein can occur without all four basic amino acids (Schickli et al., 2005). Therefore, the direct plaque assay may be applicable to hMPV strains containing any F cleavage sequence other than 99RQSR102, which has been shown to be non-permissive to plaque formation in this study. Metapneumovirus research has been limited by the inability to plaque purify the virus and the expense of immunology-based assays for quantitation, the direct plaque assay will be a valuable tool for the quantitation and evaluation of the biological properties of metapneumoviruses.
Highlights.
A direct plaque assay for hMPV and aMPV was developed.
The direct plaque assay is highly sensitive and reliable.
The direct plaque assay is easy, rapid, more convenient, more economical, and allowed for plaque purification.
The direct plaque assay is a valuable method for quantitation and evaluation of the biological properties of metapneumoviruses.
Acknowledgements
This study was partially supported by grants from NIH (R01AI090060) and USDA Agriculture and Food Research Initiative (2010-65119-20602) to J.L. We thank Dr. Ron A. M. Fouchier for reverse genetics system of hMPV lineage A strain NL/1/100. We thank Dr. Mo Saif for aMPV subtype C Minnesota strain. We are grateful to the members of the Li laboratory for critical reviews of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal AS, Roy T, Ghosh S, Chawla-Sarkar M. Genetic variability of attachment (G) and Fusion (F) protein genes of human metapneumovirus strains circulating during 2006–2009 in Kolkata, Eastern India. Virol. J. 2011;8:67. doi: 10.1186/1743-422X-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalaf AN, Ward LA, Dearth RN, Saif YM. Pathogenicity, transmissibility, and tissue distribution of avian pneumovirus in turkey poults. Avian Dis. 2002;46:650–659. doi: 10.1637/0005-2086(2002)046[0650:PTATDO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bastien N, Ward D, Van Caeseele P, Brandt K, Lee SH, McNabb G, Klisko B, Chan E, Li Y. Human metapneumovirus infection in the Canadian population. J. Clin. Microbiol. 2003;41:4642–4646. doi: 10.1128/JCM.41.10.4642-4646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayon-Auboyer MH, Arnauld C, Toquin D, Eterradossi N. Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup. J. Gen. Virol. 2000;81:2723–2733. doi: 10.1099/0022-1317-81-11-2723. [DOI] [PubMed] [Google Scholar]
- Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. Modification of the trypsin-dependent cleavage activation site of the human metapneumovirus fusion protein to be trypsin independent does not increase replication or spread in rodents or nonhuman primates. J. Virol. 2006;80:5798–5806. doi: 10.1128/JVI.00294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S, Skiadopoulos MH, Tran KC, Murphy BR, Collins PL, Buchholz UJ. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology. 2004;321:247–259. doi: 10.1016/j.virol.2003.12.020. [DOI] [PubMed] [Google Scholar]
- Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Cote S, Peret TC, Erdman DD, Anderson LJ. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 2002;186:1330–1334. doi: 10.1086/344319. [DOI] [PubMed] [Google Scholar]
- Boivin G, De Serres G, Cote S, Gilca R, Abed Y, Rochette L, Bergeron MG, Dery P. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 2003;9:634–640. doi: 10.3201/eid0906.030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buys BS, Preez JH. A preliminary report on the isolation of a virus causing sinusitis in turkeys in South Africa and attempts to attenuate the virus. Turkeys. 1980;28:36. [Google Scholar]
- Buys SB, du Preez JH, Els HJ. The isolation and attenuation of a virus causing rhinotracheitis in turkeys in South Africa. Onderstepoort J. Vet. Res. 1989a;56:87–98. [PubMed] [Google Scholar]
- Buys SB, du Preez JH, Els HJ. Swollen head syndrome in chickens: a preliminary report on the isolation of a ossible aetiological agent. J.S. Afr. Vet. Assoc. 1989b;60:221–222. [PubMed] [Google Scholar]
- Collins MS, Bashiruddin JB, Alexander DJ. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch. Virol. 1993;128:363–370. doi: 10.1007/BF01309446. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Endo R, Ma X, Ishiguro N, Kikuta H. Detection of human metapneumovirus antigens in nasopharyngeal secretions by an immunofluorescent-antibody test. J. Clin. Microbiol. 2005;43:1138–1141. doi: 10.1128/JCM.43.3.1138-1141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F, Boucher D, Weibel C, Martinello RA, Kahn JS. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics. 2003;111:1407–1410. doi: 10.1542/peds.111.6.1407. [DOI] [PubMed] [Google Scholar]
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- Govindarajan D, Yunus AS, Samal SK. Complete sequence of the G glycoprotein gene of avian metapneumovirus subgroup C and identification of a divergent domain in the predicted protein. J. Gen. Virol. 2004;85:3671–3675. doi: 10.1099/vir.0.80400-0. [DOI] [PubMed] [Google Scholar]
- Herfst S, de Graaf M, Schickli JH, Tang RS, Kaur J, Yang CF, Spaete RR, Haller AA, van den Hoogen BG, Osterhaus AD, Fouchier RA. Recovery of human metapneumovirus genetic lineages A and B from cloned cDNA. J. Virol. 2004;78:8264–8270. doi: 10.1128/JVI.78.15.8264-8270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn JS. Epidemiology of human metapneumovirus. Clin. Microbiol. Rev. 2006;19:546–557. doi: 10.1128/CMR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- Mizutani T, Inagaki H, Tada M, Hayasaka D, Murphy M, Fujiwara T, Hamada J, Kariwa H, Takashima I. The mechanism of actinomycin D-mediated increase of Borna disease virus (BDV) RNA in cells persistently infected by BDV. Microbiol. Immunol. 2000;44:597–603. doi: 10.1111/j.1348-0421.2000.tb02539.x. [DOI] [PubMed] [Google Scholar]
- Pelletier G, Dery P, Abed Y, Boivin G. Respiratory tract reinfections by the new human Metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 2002;8:976–978. doi: 10.3201/eid0809.020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Sabara MI, Larence JE. Plaque assay for avian metapneumovirus using a Japanese quail fibrosarcoma cell line (QT-35) J. Virol. Methods. 2003;107:9–14. doi: 10.1016/s0166-0934(02)00207-0. [DOI] [PubMed] [Google Scholar]
- Schickli JH, Kaur J, Ulbrandt N, Spaete RR, Tang RS. An S101P substitution in the putative cleavage motif of the human metapneumovirus fusion protein is a major determinant for trypsin-independent growth in vero cells and does not alter tissue tropism in hamsters. J. Virol. 2005;79:10678–10689. doi: 10.1128/JVI.79.16.10678-10689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton J, Stephenson I, Fleming D, Zambon M. Human metapneumovirus as a cause of community-acquired respiratory illness. Emerg. Infect. Dis. 2002;8:897–901. doi: 10.3201/eid0809.020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo-Neto E, de Swart RL, Osterhaus AD, Fouchier RA. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 2004;10:658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 2003;188:1571–1577. doi: 10.1086/379200. [DOI] [PubMed] [Google Scholar]
- Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE., Jr Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. New Engl. J. Med. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Wang CK, Tollefson SJ, Piyaratna R, Lintao LD, Chu M, Liem A, Mark M, Spaete RR, Crowe JE, Jr, Williams JV. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virol. J. 2009;6:138–148. doi: 10.1186/1743-422X-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]