Abstract
Challenges in imaging lipid-processing events in live, intact vertebrate models have historically led to reliance on cultured cell studies, thus hampering our understanding of lipid metabolism and gastrointestinal physiology. Fluorescently-labeled molecules, such as BODIPY-labeled lipids, can reveal lipid-processing events in live zebrafish (Danio rerio) and has expanded our understanding of digestive physiology. This review will cover recent advances from the past two to three years in the use of fluorescence-based imaging techniques in live zebrafish to characterize gastrointestinal physiology in health and disease and to conduct small molecule screens to discover therapeutic compounds.
Introduction
According to a report by the Center for Disease Control, 33.8% of adults and 17% of children and adolescents were classified as obese in the United States in 2010. Defects in gastrointestinal (GI) physiology and lipid metabolism play critical roles in the development and progression of obesity and related cardiovascular disease and type II diabetes. To improve medical treatments for these conditions and reduce the societal burden of metabolic and GI diseases, a better understanding of GI physiology is needed. Imaging fluorescently-labeled molecules as they are processed in the GI tract is a powerful method to elucidate digestive function in health and diseased states. The larval zebrafish (Danio rerio) has recently emerged as an ideal model to visualize fluorescently-labeled molecules in vivo (Fig. 1) [1–3]. Here we review promising advances in imaging GI physiology and lipid processing in live zebrafish.
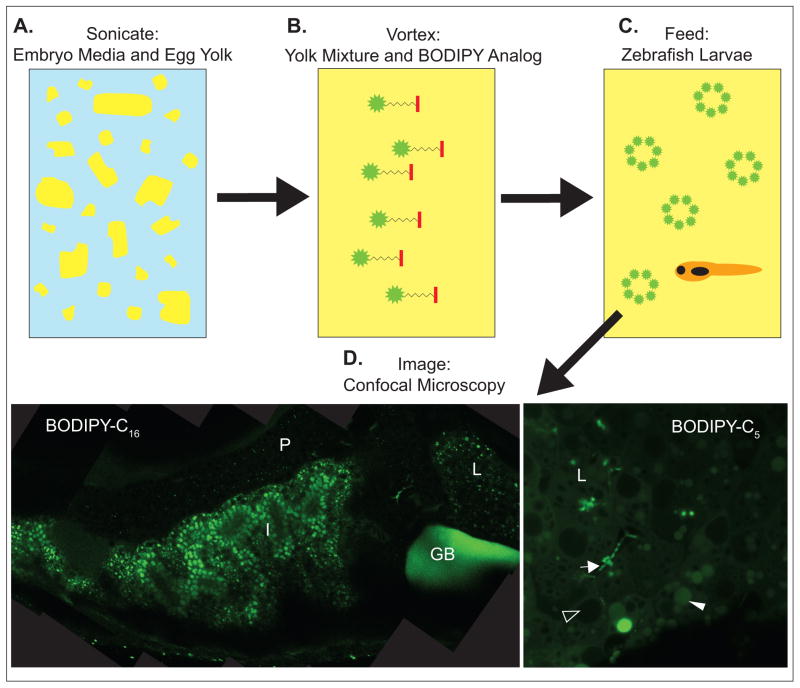
Figure 1. Feeding zebrafish fluorescently-labeled lipids in chicken egg yolk liposomes.
A. Egg yolk is added to embryo media at a concentration of 5% and sonicated to mix and form liposomes. B. Fluorescently-labeled lipid analogs are added to the yolk mixtures and incorporated into liposomes by vortexing. C. Zebrafish larvae are allowed to feed on the yolk mixture. D. Fish are imaged by confocal microscopy; representative composite image of a 6-day post fertilization (dpf) larvae fed BODIPY-C16 in egg yolk mixture and a single image of the liver of a 6-dpf larvae feed BODIPY-C5, both at 63x magnification. Gallbladder, GB; intestine, I; liver, L; pancreas, P; arrow, intrahepatic duct; filled arrowhead, lipid droplet; open arrowhead, hepatic nuclei.
Larval Zebrafish as a Model of Lipid Metabolism
Traditional methods of visualizing lipid metabolism are largely limited to cell culture systems and the use of lipophilic dyes. Although ex vivo studies provide important information regarding lipid metabolism, the complex milieus of the liver, intestine, and enterohepatic circulation cannot be completely replicated in vitro. Additionally, lipophilic dyes often produce unspecific lipid labeling and most cannot be used in live animals (reviewed in: [4]). Therefore, recent studies have undertaken the use of fluorescent lipids in live, larval zebrafish. The zebrafish GI system is similar to that of humans; it is comprised of a liver, gallbladder, exocrine and endocrine pancreas, and intestine, with multiple cell types, mucus, and resident microbiota. Inter-organ lipid transport is also conserved from fish to humans, with lipids initially absorbed in the intestine, transported to the liver via chylomicrons, exported to the periphery in low-density lipoproteins, and returned to the liver in high-density lipoproteins [5]. In addition, the rapid reproduction, ex utero embryonic development, ease of transgene expression, and optical clarity of zebrafish larvae allows fluorescently-labeled proteins and metabolites to be used to study gastrointestinal physiology and lipid metabolism [4].
Zebrafish rely on nutrients supplied by the embryonic yolk sac, including triacylglycerol (TAG) and cholesterol, for the first 4 days of development. Upon formation of the circulatory system, yolk lipids are transported from the yolk embryo interface, known as the yolk syncytial layer, to the periphery by lipoproteins [6,7]. In captivity, zebrafish are fed a lipid-rich diet (TAG, phospholipids, and sterols) with a typical fat content of at least 10% by weight [8,9]. At 8 days post fertilization (dpf), adipogenesis begins; ultimately, zebrafish develop a large visceral lipid depot and several smaller peripheral lipid depots [10]. At 6 dpf, zebrafish larvae have completely depleted their yolk supply. Because adipocytes have not yet formed, lipids must be derived from dietary sources and de novo synthesis. The techniques described in this review mainly focus on larvae at the 5 and 6 dpf developmental time points because the dietary lipids administered are the first exogenous lipids encountered by larvae and the larvae readily consume the lipids.
Imaging Zebrafish Digestive Organ Function
Lipases are largely responsible for the ability of TAG to enter the intestinal enterocytes that line the digestive tract. Synthesized in the pancreas and collected in the gall bladder, pancreatic lipases are released into the intestinal lumen where they cleave fatty acids from TAG. As emulsification in bile and cleavage by lipases are necessary for efficient fatty acid absorption by enterocytes, defects in bile dynamics or pancreatic lipase activity result in lipid malabsorption. Zebrafish ingestion of the fluorescent, self-quenched phospholipid reporter N-((6-(2,4-dinitrophenyl)amino)hexanoyl)-1-palmitoyl-2-BODIPY-FL-pentanoyl-sn-glycero-3-phosphoethanolamine (PED6) allows imaging of phospholipase A2 (PLA2) activity and subsequent phospholipid transport to be visualized in larval digestive organs [1]. PED6 is self-quenching: it only produces a fluorescent signal upon cleavage by PLA2. The photostability, strong and narrow wavelength emission, and uncharged state of the (4,4-difluoro-4-bora-3a, 4a-diaza-S-indacene) BODIPY fluorescent moiety make it an ideal fluorescent tag for zebrafish studies [11,12]. When zebrafish larvae are soaked in PED6, the reporter is ingested and labeling of the intestinal lumen and gallbladder can be observed in healthy larvae. Larvae with defects in intestinal lipase activity or hepatobiliary dysfunction display attenuated PED6 signal, making it a valuable screening tool.
Following validation of PED6 as a lipase reporter, an assay was developed to concurrently image lipase activity with PED6 and protease activity with EnzChek. In the PED6-EnzChek assay zebrafish larvae are fed a cocktail of PED6 and EnzChek (an intramolecularly quenched fluorescent reporter that emits at a wavelength unique from PED6 upon cleavage), anesthetized, mounted in 3% methylcellulose, and imaged on a fluorescence stereomicroscope at 10x magnification, which allows observation of entire digestive organs (Fig. 2) [2]. It was found that the ratio of PED6 to EnzChek ingestion was comparable across wildtype individuals and thus could be used as a measure of interindividual variation in ingestion volume. Additionally, intestinal phospholipase activity correlated highly with intestinal protease activity within individuals, but that activity varied significantly between individuals [2]. Thus, it was concluded that the ratio of phospholipase to protease activity could be used to screen for individuals with defects in exocrine pancreas function.
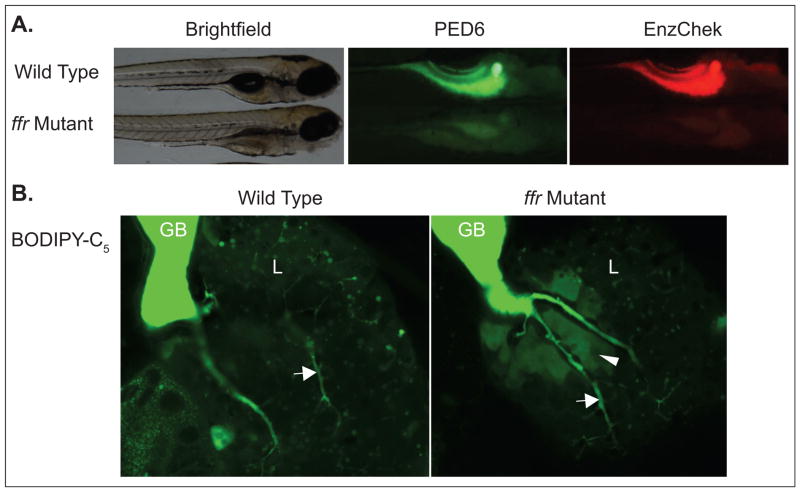
Figure 2. Use of fluorescent molecules to characterize the zebrafish fat-free (ffr) mutant.
A. The ffr mutant exhibits less PED6 and Enzchek fluorescence than wild type, indicating reduced intestinal lipase and protease activity. B. BODIPY-C5 labeling shows abnormal hepatic bile accumulation in ffr mutants; gallbladder, GB; liver, L; arrow, intrahepatic duct; arrowhead, bile accumulation.
PED6 in Physiological Screening
Several groups have taken advantage of PED6 screening to identify defects in hepatobiliary function and lipid metabolism in mutant, morpholino-treated, or experimentally-manipulated zebrafish [13–21]. For example, Hama et al. used the PED6-EnzChek cocktail to investigate zebrafish pancreatic phospholipase and protease activity during development [2]. Cholecystokinin (CCK) is a gut hormone released upon lipid ingestion that stimulates the release of pancreatic lipases and proteases to aid digestion. Hama et al. administered an agonist to the CCK receptor A to stimulate lipase and protease activity and it was found, through PED6 and EnzChek imaging, to reduce protease activity at 5 and 6 dpf, and phospholipase activity at 6 dpf, but to not have an effect on phospholipase activity at 5 dpf [2]. Therefore, it was concluded that exocrine pancreas-derived phospholipase activity increases from 5 to 6 dpf, while protease activity remains constant.
PED6 in Drug Discovery
PED6 and EnzChek have also been used to characterize mutant zebrafish and in screens to identify compounds that modulate digestive function [22]. Zebrafish fat-free (ffr) mutants display abnormal Golgi structure and vesicular trafficking, which lead to defects in lipid transport and metabolism [23]. PED6 and EnzChek were used to further characterize the ffr mutants, which had decreased intestinal phospholipase and protease activity (Fig. 2) [2]. These findings revealed the physiological consequences of the subcellular defects initially identified in the ffr mutants.
Non-steroidal anti-inflammatory drugs (NSAID) inhibit cyclooxygenases, enzymes that produce protective prostaglandins and maintain the gastrointestinal mucosa, thus leading to irritation of the GI tract [24]. GI irritation can also result from inflammatory diseases, such as inflammatory bowel disease, Crohn’s disease, and ulcerative colitis [25,26], mediated in part by group II phospholipase A2 enzymes. However, it is not clear if NSAIDs are also associated with changes in lipase or protease function. Therefore, PED6 and EnzChek were used to screen several NSAIDs to determine if changes in lipase or protease activity are part of the mechanism by which GI irritation occurs. Glafenine, a once commonly prescribed NSAID/analgesic, was found to enhance intestinal lipase activity, possibly reflecting increased inflammation, and cause sloughing off of the intestinal lining in larval zebrafish [2].
A screen of a non-biased chemical library with PED6 identified 7 novel compounds that inhibit processing of BODIPY lipid analogs [22]. This assay utilized a high-throughput technique, soaking 5 dpf larvae in compounds overnight in 96-well plates and then PED6 for 6 hours. Gallbladder fluorescence was observed on an inverted compound microscope and used as a readout of lipase production. A lack of gallbladder fluorescence was interpreted as chemical inhibition of lipid absorption due to changes in swallowing, phospholipase activity, hepatic metabolism, or biliary secretion. Thus, PED6 is a valuable tool that can be used to identify mutants with changes in digestive function and novel compounds that have the potential to treat human disease.
Imaging Subcellular Lipid Metabolism
Imaging lipids at the organ and subcellular levels in live vertebrates has been a challenge for a number of reasons (e.g., limitations of previous microscopy and fluorescent lipid technology, small size and dynamic movements of lipids, and metabolism of lipids into different metabolites) and only recent technological advancements have enabled progress in this area. Following the success of imaging digestive function at the whole organ level with PED6, we developed a powerful feeding assay for subcellular visualization of fluorescent dietary lipids as they are processed in the intestine, liver, gallbladder, and pancreas (Figs. 1 and 3) of larval zebrafish [3]. In this assay, larvae are fed BODIPY-labeled lipids suspended in liposomes in a lipid-rich chicken egg yolk meal (65% triglyceride, 30% phospholipid, 5% cholesterol; 5% in embryo media). Following feeding, larvae are anesthetized and analyzed by confocal microscopy with a 63x magnification oil immersion objective while mounted in 3% methylcellulose or with a water immersion objective while mounted in agar. An advantage of the mounting techniques is that, following imaging, the live larvae can be rinsed from methylcellulose or freed from agar and used in additional studies or raised. Depending on the time of feeding and type of fluorescently-labeled lipid fed, visualization of subcellular lipid localization (e.g., lipid droplets, bile ducts) is possible (Fig. 3) [3]. Processing of BODIPY-labeled lipids can be further analyzed by fluorescent thin layer chromatography (TLC). Presently, this feeding assay has been used to complete a comparative study of the metabolism of various dietary saturated fatty acids and to further characterize the ffr zebrafish mutant [3].
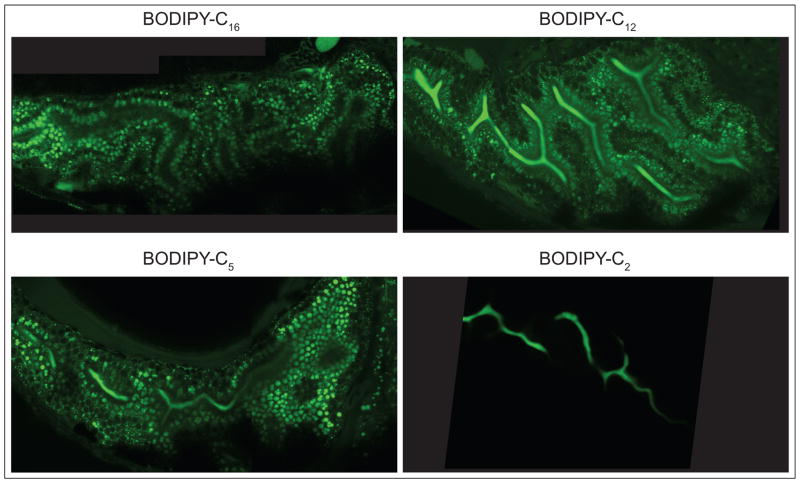
Figure 3. Differences in processing of saturated fatty acids of various chain lengths.
Representative composite images of the intestine of 6-day post fertilization larvae fed BODIPY-C16, -C12, -C5, and -C2, in egg yolk mixture at 63x magnification. BODIPY-C16 is observed in lipid droplets (LD) of enterocytes, BODIPY-C12 and -C5 in enterocyte LD and the intestinal lumen, and BODIPY-C2 in the intestinal lumen.
Visualization of Short, Medium and Long-Chain Fatty Acid Processing
Carten et al. used BODIPY labeled fatty acid analogs to investigate how the chain length of saturated dietary fatty acids affects their processing (Fig. 3) [3]. Saturated fatty acids are classified by the length of their acyl chain: short (2–6 carbons (C)), medium (8–12 C), long (14–20 C), and very long (≥22 C). The BODIPY moiety has been observed to make lipids be processed as if there are 2–3 additional C [3]. To determine how dietary saturated fatty acids of these different chain lengths are metabolized, BODIPY-C16, -C12, -C5, and -C2 were fed to 6 dpf larval zebrafish in an egg yolk meal [3]. These BOPIDY-labeled saturated fatty acid analogs had been previously studied in cell culture and invertebrates, but not in live vertebrates [27–31]. In summary, C16, C12, and C5 are incorporated into lipid droplets in enterocytes, hepatocytes, and pancreatic cells, but C2 is not. C16 is metabolized into neutral lipids (e.g., non-polar lipids such as TAG), and C12 and C5 are metabolized to neutral and complex lipids (e.g., polar lipids such as glycolipids, phospholipids). C2 is not metabolized. Both C12 and C5 appear in the intestinal lumen and hepatic ducts; C5 also labels the endosomal compartment of enterocytes and pancreatic ducts. C2 is observed in the intestinal lumen, diffusely in hepatocytes, and in hepatic and pancreatic ducts, causing speculation that it behaves as a xenobiotic in vivo. Finally, the polarized function of enterocytes can be observed in the different fluorescence patterns observed on the apical and basolateral sides of these cells following a C16 feeding, likely representing accumulation in lipid droplets and chylomicrons, respectively. A previous study, in which larvae were soaked in BODIPY-C5 or -C12, supports the finding that both are absorbed in the intestine, transported to the liver, and secreted into bile, with BODIPY-C5 producing stronger fluorescent signals in the liver and gallbladder [32]. Thus, imaging of dietary fluorescent lipids in live zebrafish larvae showed that fatty acid metabolism appears to be conserved amongst vertebrates, namely zebrafish larvae, in that they also exhibit differential metabolism of different chain length fatty acids much like humans do.
Imaging Changes in Lipid Processing in Disease
In addition to characterizing normal digestive processes, feeding zebrafish BODIPY-labeled lipids can elucidate metabolic changes in mutant and disease states. For example, an ongoing mutagenesis screen in our laboratory uses BODIPY-labeled lipids and imaging at 10 x magnification to screen for defects in GI function. More specifically, understanding of the frr mutant zebrafish was improved following BODIPY-C5 feeding (Fig. 2) [3]: ffr mutants have abnormal hepatic bile accumulation and cholestasis. Hepatic bile accumulation may be due to canalicular and ductal structural defects resulting from deficient Golgi-related vesicular trafficking of bile transporters to the plasma membrane. Thus, the reduction in intestinal lipase activity in ffr mutants previously demonstrated by PED6 studies [2] likely results from a bile secretion defect. BODIPY-labeled fatty acids can also be used to screen for defects in embryonic yolk lipid processing. BODIPY-cholesterol has been injected into the zebrafish embryonic yolk sac and tracked to the sterol rich brain, while the larvae exhibit normal development [33]. A screen has also been performed for genes involved in lipid processing prior to exogenous food intake in which BODIPY-C12 was injected into the yolk of 1 dpf embryos treated with various morpholinos [34]. At 3 dpf, yolk lipids were analyzed for defects in lipid processing by fluorescent TLC and it was discovered that apolipoprotein C2 is essential for normal yolk lipid processing.
Future Directions
There is tremendous potential for the use of fluorescently-labeled molecules, such as PED6 and BODIPY analogs, in live zebrafish to improve our understanding of GI and lipid function in health and disease. Zebrafish models of lipid processing and GI diseases have been developed that include diet-induced obesity [35], hypercholesterolemia [36], atherosclerosis [37,38], and hepatic steatosis [39]. The possibility of further studying these disease models with fluorescently-tagged proteins, which are readily expressed in zebrafish, and/or BODIPY analogs, is very promising. In fact, Stoletov et al. fed cholesteryl-BODIPY-conjugated fatty acid ester with a high-cholesterol diet in their zebrafish model of atherosclerosis and found that it brightly labeled lipid deposits in blood vessels [36]. Recently, an in vitro study demonstrated that the efflux of BODIPY-labeled cholesterol from macrophages can be measured more sensitively than the traditional method that measures efflux of radio-labeled cholesterol [40]; it will be exciting to apply this technique to study macrophages in live zebrafish.
There is immense potential for the use of fluorescent lipid feeding assays in high-throughput screens to identify compounds or mutations affecting GI physiology, lipid metabolism, and developmental processes at the organ and/or subcellular levels. High-throughput screens of uncharacterized small molecules are often conducted in ex vivo systems, with activity often translating poorly to subsequent in vivo models. However, the fluorescent lipids described in this review allow initial small molecule screens to bypass the ex vivo step, conserving time, money, and additional resources.
Conclusions
Limitations in the ability to image lipid processing in a live, vertebrate animal model and the subsequent reliance on cultured cells has hampered our understanding of the physiological function of these important metabolites. The use of fluorescently labeled lipids to image lipid metabolism and GI physiology in live zebrafish has the potential to expand our understanding of key cell biological processes that occur throughout the GI system, embryonic yolk, and vasculature. These powerful assays can be used to characterize lipid metabolism in health and disease and to conduct high-throughput, in vivo screens of small molecules for drug discovery.
Acknowledgments
The authors thank and Juliana Carten and Jennifer Anderson for critical readings of the manuscript. Funding was provided by the National Institutes of Health (R56DK093399 and RO1GM63904), the Carnegie Institution for Science endowment, and the G. Harold and Leila Y. Mathers Charitable Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jessica P. Otis, Email: otis@ciwemb.edu.
Steven A. Farber, Email: farber@ciwemb.edu.
Literature Cited
- 1.Farber SA, Pack M, Ho SY, Johnson ID, Wagner DS, Dosch R, Mullins MC, Hendrickson HS, Hendrickson EK, Halpern ME. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292 (5520):1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- ••2.Hama K, Provost E, Baranowski TC, Rubinstein AL, Anderson JL, Leach SD, Farber SA. In vivo imaging of zebrafish digestive organ function using multiple quenched fluorescent reporters. Am J Physiol Gastrointest Liver Physiol. 2009;296(2):G445–453. doi: 10.1152/ajpgi.90513.2008. Using the PED6 and EnzChek fluorescent reporters of lipase and protease activity, the authors imaged gastrointestinal function in live zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••3.Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev biol. 2011;360(2):276–285. doi: 10.1016/j.ydbio.2011.09.010. The authors introduce a method using dietary BODIPY-labeled fatty acids to image subcellular details of lipid processing in the liver, intestine, gallbladder, and pancreas of larval zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson JL, Carten JD, Farber SA. Zebrafish lipid metabolism: from mediating early patterning to the metabolism of dietary fat and cholesterol. Method cell biol. 2011;101:111–141. doi: 10.1016/B978-0-12-387036-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheridan MA. Lipid dynamics in fish: aspects of absorption, transportation, deposition and mobilization. Comp Biochem Phys B. 1988;90 (4):679–690. doi: 10.1016/0305-0491(88)90322-7. [Review] [99 refs] [DOI] [PubMed] [Google Scholar]
- 6.Babin PJ, Thisse C, Durliat M, Andre M, Akimenko MA, Thisse B. Both apolipoprotein E and A-I genes are present in a nonmammalian vertebrate and are highly expressed during embryonic development. PNAS. 1997;94 (16):8622–8627. doi: 10.1073/pnas.94.16.8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marza E, Barthe C, Andre M, Villeneuve L, Helou C, Babin PJ. Developmental expression and nutritional regulation of a zebrafish gene homologous to mammalian microsomal triglyceride transfer protein large subunit. Dev Dyn. 2005;232 (2):506–518. doi: 10.1002/dvdy.20251. [DOI] [PubMed] [Google Scholar]
- 8.Enzler L, Smith V, Lin JS, Olcott HS. The lipids of Mono Lake, California, brine shrimp (Artemia salina) J agr food chem. 1974;22 (2):330–331. doi: 10.1021/jf60192a017. [DOI] [PubMed] [Google Scholar]
- 9.Spence R, Gerlach G, Lawrence C, Smith C. The behaviour and ecology of the zebrafish, Danio rerio. Biol Rev Camb Philos Soc. 2008;83 (1):13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- 10.Flynn EJ, 3rd, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) J lipid res. 2009;50 (8):1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagano RE, Martin OC, Kang HC, Haugland RP. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol. 1991;113 (6):1267–1279. doi: 10.1083/jcb.113.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monsma FJ, Jr, Barton AC, Kang HC, Brassard DL, Haugland RP, Sibley DR. Characterization of novel fluorescent ligands with high affinity for D1 and D2 dopaminergic receptors. J neurochem. 1989;52 (5):1641–1644. doi: 10.1111/j.1471-4159.1989.tb09220.x. [DOI] [PubMed] [Google Scholar]
- 13.Matthews RP, Eauclaire SF, Mugnier M, Lorent K, Cui S, Ross MM, Zhang Z, Russo P, Pack M. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53 (3):905–914. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136 (3):1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews RP, Plumb-Rudewiez N, Lorent K, Gissen P, Johnson CA, Lemaigre F, Pack M. Zebrafish vps33b, an ortholog of the gene responsible for human arthrogryposis-renal dysfunction-cholestasis syndrome, regulates biliary development downstream of the onecut transcription factor hnf6. Development. 2005;132 (23):5295–5306. doi: 10.1242/dev.02140. [DOI] [PubMed] [Google Scholar]
- 16.Matthews RP, Lorent K, Russo P, Pack M. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev biol. 2004;274 (2):245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev dynam. 2008;237 (1):124–131. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- 18.Cui S, Capecci LM, Matthews RP. Disruption of planar cell polarity activity leads to developmental biliary defects. Dev biol. 2011;351 (2):229–241. doi: 10.1016/j.ydbio.2010.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullinane AR, Straatman-Iwanowska A, Zaucker A, Wakabayashi Y, Bruce CK, Luo G, Rahman F, Gurakan F, Utine E, Ozkan TB, et al. Mutations in VIPAR cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nat genet. 2010;42 (4):303–312. doi: 10.1038/ng.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev biol. 2006;297 (2):374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Daroczi B, Kari G, Ren Q, Dicker AP, Rodeck U. Nuclear factor kappaB inhibitors alleviate and the proteasome inhibitor PS-341 exacerbates radiation toxicity in zebrafish embryos. Mol Cancer Ther. 2009;8 (9):2625–2634. doi: 10.1158/1535-7163.MCT-09-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22.Clifton JD, Lucumi E, Myers MC, Napper A, Hama K, Farber SA, Smith AB, 3rd, Huryn DM, Diamond SL, Pack M. Identification of novel inhibitors of dietary lipid absorption using zebrafish. PLoS One. 2010;5(8):e12386. doi: 10.1371/journal.pone.0012386. An excellent example of how fluorescent reporters, such as PED6 and BODIPY-labeled lipids, can be used to conduct in vivo, high-throughput screens of compounds that can be developed to inhibit lipid absorption. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho SY, Lorent K, Pack M, Farber SA. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 2006;3 (4):289–300. doi: 10.1016/j.cmet.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parente L, Perretti M. Advances in the pathophysiology of constitutive and inducible cyclooxygenases: two enzymes in the spotlight. Biochem pharmacol. 2003;65 (2):153–159. doi: 10.1016/s0006-2952(02)01422-3. [DOI] [PubMed] [Google Scholar]
- 25.Haapamaki MM, Gronroos JM, Nurmi H, Alanen K, Kallajoki M, Nevalainen TJ. Gene expression of group II phospholipase A2 in intestine in ulcerative colitis. Gut. 1997;40 (1):95–101. doi: 10.1136/gut.40.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami T, Tojo H, Shinomura Y, Matsuzawa Y, Okamoto M. Increased group II phospholipase A2 in colonic mucosa of patients with Crohn's disease and ulcerative colitis. Gut. 1994;35 (11):1593–1598. doi: 10.1136/gut.35.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furlong ST, Thibault KS, Morbelli LM, Quinn JJ, Rogers RA. Uptake and compartmentalization of fluorescent lipid analogs in larval Schistosoma mansoni. J lipid res. 1995;36 (1):1–12. [PubMed] [Google Scholar]
- 28.Huang H, Starodub O, McIntosh A, Atshaves BP, Woldegiorgis G, Kier AB, Schroeder F. Liver fatty acid-binding protein colocalizes with peroxisome proliferator activated receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry-US. 2004;43 (9):2484–2500. doi: 10.1021/bi0352318. [DOI] [PubMed] [Google Scholar]
- 29.Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, Mak HY. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. PNAS. 2010;107 (10):4640–4645. doi: 10.1073/pnas.0912308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat genet. 2006;38 (3):363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 31.Karsenty J, Helal O, de la Porte PL, Beauclair-Deprez P, Martin-Elyazidi C, Planells R, Storch J, Gastaldi M. I-FABP expression alters the intracellular distribution of the BODIPY C16 fatty acid analog. Mol cell biochem. 2009;326 (1–2):97–104. doi: 10.1007/s11010-008-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlegel A, Stainier DY. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry-US. 2006;45 (51):15179–15187. doi: 10.1021/bi0619268. [DOI] [PubMed] [Google Scholar]
- 33.Holtta-Vuori M, Uronen RL, Repakova J, Salonen E, Vattulainen I, Panula P, Li Z, Bittman R, Ikonen E. BODIPY-cholesterol: a new tool to visualize sterol trafficking in living cells and organisms. Traffic. 2008;9 (11):1839–1849. doi: 10.1111/j.1600-0854.2008.00801.x. [DOI] [PubMed] [Google Scholar]
- 34.Pickart MA, Klee EW, Nielsen AL, Sivasubbu S, Mendenhall EM, Bill BR, Chen E, Eckfeldt CE, Knowlton M, Robu ME, et al. Genome-wide reverse genetics framework to identify novel functions of the vertebrate secretome. PLoS ONE. 2006;1:e104. doi: 10.1371/journal.pone.0000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •35.Oka T, Nishimura Y, Zang L, Hirano M, Shimada Y, Wang Z, Umemoto N, Kuroyanagi J, Nishimura N, Tanaka T. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010;10:21. doi: 10.1186/1472-6793-10-21. This manuscript describes a zebrafish model of diet-induced obesity, which mirrors many of the changes in human obesity, and can thus be used in the future to develop pharmaceutical strategies to treat obesity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoletov K, Fang L, Choi SH, Hartvigsen K, Hansen LF, Hall C, Pattison J, Juliano J, Miller ER, Almazan F, et al. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ Res. 2009;104 (8):952–960. doi: 10.1161/CIRCRESAHA.108.189803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang YP, Lin YK, Su YH, Fang JY. Tryptanthrin-loaded nanoparticles for delivery into cultured human breast cancer cells, MCF7: the effects of solid lipid/liquid lipid ratios in the inner core. Chem Pharm Bull (Tokyo) 2011;59 (2):266–271. doi: 10.1248/cpb.59.266. [DOI] [PubMed] [Google Scholar]
- •38.Fang L, Harkewicz R, Hartvigsen K, Wiesner P, Choi SH, Almazan F, Pattison J, Deer E, Sayaphupha T, Dennis EA, et al. Oxidized cholesteryl esters and phospholipids in zebrafish larvae fed a high cholesterol diet: macrophage binding and activation. J biol chem. 2010;285(42):32343–32351. doi: 10.1074/jbc.M110.137257. The authors use a zebrafish atherosclerosis model to study lipid oxidation during this disease process. High cholesterol feeding increased oxidized lipids and proatherogenic macrophages in the zebrafish larvae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passeri MJ, Cinaroglu A, Gao C, Sadler KC. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49 (2):443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sankaranarayanan S, Kellner-Weibel G, de la Llera-Moya M, Phillips MC, Asztalos BF, Bittman R, Rothblat GH. A sensitive assay for ABCA1-mediated cholesterol efflux using BODIPY-cholesterol. J lipid res. 2011;52 (12):2332–2340. doi: 10.1194/jlr.D018051. [DOI] [PMC free article] [PubMed] [Google Scholar]