Summary
Variability of regenerative potential among animals has long perplexed biologists1. Based on their amazing regenerative abilities, planarians have become important models for understanding the molecular basis of regeneration2; however, planarian species with limited regenerative abilities are also found3,4. Despite the importance of understanding the differences between closely related, regenerating and non-regenerating organisms, few studies have focused on the evolutionary loss of regeneration5, and the molecular mechanisms leading to such regenerative loss remain obscure. Here we examine Procotyla fluviatilis, a planarian with restricted ability to replace missing tissues6, utilizing next-generation sequencing to define the gene expression programs active in regeneration-permissive and regeneration-deficient tissues. We found that Wnt signaling is aberrantly activated in regeneration-deficient tissues. Remarkably, down-regulation of canonical Wnt signaling in regeneration-deficient regions restores regenerative abilities: blastemas form and new heads regenerate in tissues that normally never regenerate. This work reveals that manipulating a single signaling pathway can reverse the evolutionary loss of regenerative potential.
Unlike more commonly studied planarians2, representatives of the family Dendrocoelidae regenerate heads only when amputated within the anterior third of the body, yet retain full posterior regeneration ability along the antero-posterior axis (Fig. 1a,b and Supplementary Fig. 1)3,4. After amputation in regeneration-deficient tissues, P. fluviatilis fails to produce a blastema and never regenerates anterior structures (Fig. 1b and Supplementary Fig. 1). The stage at which regenerative processes fail in this animal is unknown. Planarian regeneration encompasses several processes, including: wound healing to cover exposed tissues and allow signaling between the wound epidermis and underlying mesenchymal cells7; apoptosis8; and stem cell (neoblast) proliferation9. Following amputation, neoblast proliferation occurs in two waves: a systemic response shortly after amputation and a later burst near the wound site9. In addition, regenerating tissues re-establish proper axial polarity, using a number of conserved signaling pathways10–14. Subsequently, tissue outgrowth occurs as an undifferentiated mass of cells (the regeneration blastema) differentiates to replace lost structures15.
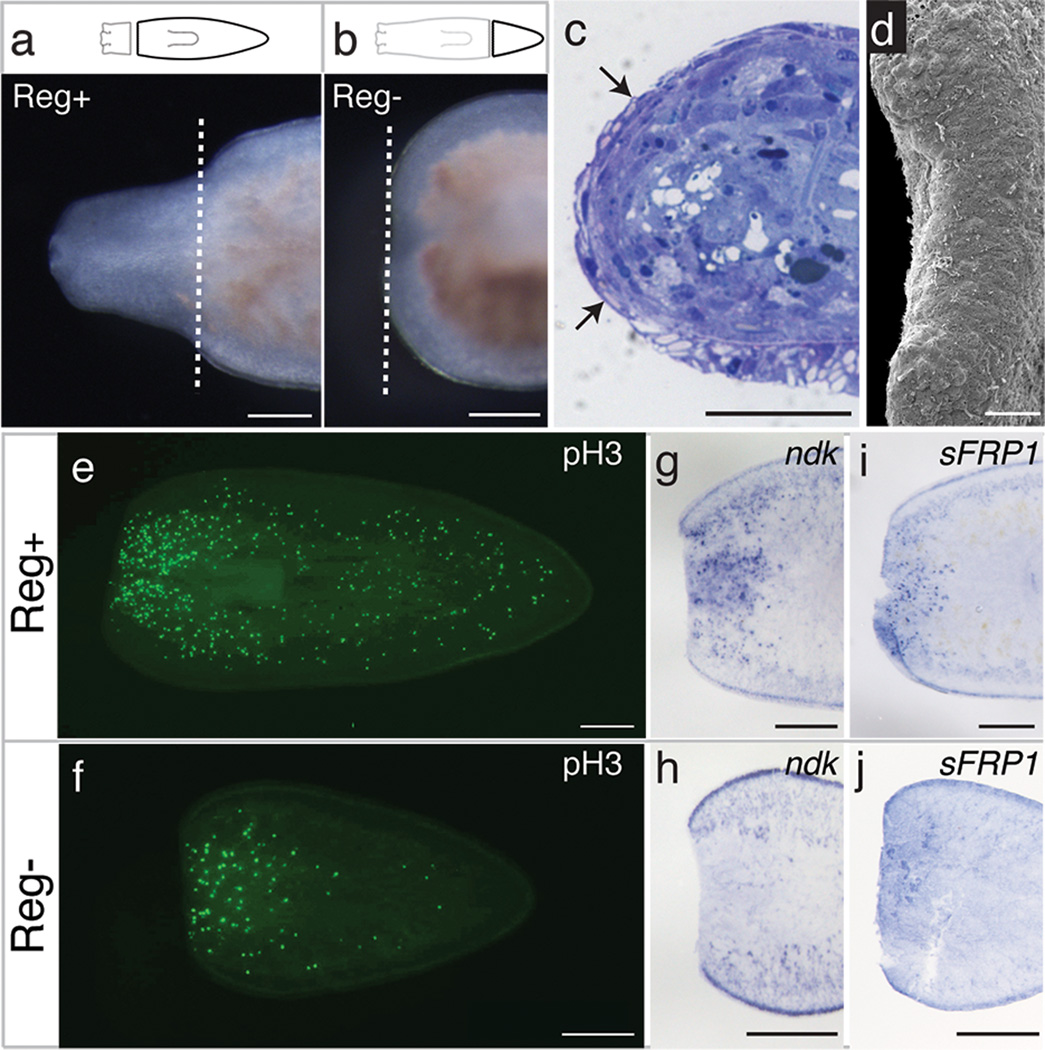
Figure 1. Procotyla fluviatilis fails to regenerate heads after amputation in posterior body regions.
a–b, Proficient and failed head regeneration 10 days after amputation in Reg+ regions Reg− regions respectively (n=25/25). Dashed lines indicate amputation planes. c–d, Complete wound epithelium (arrows; n=8/9) and full wound closure (n=7/8) 48 hours after amputation in Reg− tissue. Scale bar in d, 100 µm. e–f, Mitotic activity 4 days after amputation in Reg+ and Reg− tissues (n=10/treatment). g–j, Pf-ndk and Pf-sFRP1 expression 24 hours after amputation in Reg+ and Reg− tissues (n=8/treatment). Anterior is to the left. Scale bars, 250 µm unless otherwise noted.
To identify the nature of regeneration failure in P. fluviatilis, we characterized several of these early regenerative processes following amputation in both regeneration-proficient (Reg+) and regeneration-deficient (Reg−) tissues. Histological staining and electron microscopy revealed that wound healing occurs properly following amputation in all tissues, regardless of regenerative potential (Fig. 1c,d). After amputation in Reg− tissues, biphasic cell division occurs in both Reg+ and Reg− tissues (Fig. 1e,f and Supplementary Fig. 2). In addition, gut tissues appear to remodel16 in fragments that fail to regenerate (Supplementary Fig. 1). These data show that early phases of the regenerative response occur, although Reg− tissues fail to form a blastema.
To assess whether axial polarity is re-established properly following amputation in Reg− tissues of P. fluviatilis, we characterized the spatio-temporal expression of homologs of nou-darake (ndk) and sFRP1, genes expressed specifically at the anterior of other planarian species10,11,17. Both Pf-ndk and Pf-sFRP1 are expressed at the anterior wound site shortly after amputation in Reg+ tissue (Fig. 1g,i). However, the expression of these genes was reduced in Reg− tissue following injury (Fig. 1h,j), suggesting that the initial failure of regeneration occurs at or upstream of axial re-polarization.
Since these polarity markers are not expressed appropriately following amputation in Reg− fragments, we sought to identify gene expression differences between Reg+ and Reg− tissue after amputation. We generated a de novo P. fluviatilis transcriptome and used RNA sequencing (RNAseq) to characterize transcripts from excised tissue fragments in Reg+ and Reg− body regions 24 hours post-amputation (Fig. 2a). We performed parallel analyses on tissues excised from intact animals at identical body regions to account for regional differences in transcripts, thereby identifying changes resulting from amputation (Fig. 2a). Analysis of amputated versus intact tissues revealed that 10.7% of the assembled contigs (16,026/149,594) were significantly altered ≥ 2-fold (p < 0.05) after amputation in either Reg+ or Reg− tissues. After collapsing contigs likely representing the same transcript based upon blast similarity, we focused our analysis on 15,742 contigs that appear to be expressed differentially after amputation (based on the large number of contigs, many individual transcripts are still likely represented by multiple contigs). While a small number of contigs were simultaneously over- or under-represented in both conditions (74/15,742), many were over- or under-represented exclusively in either Reg+ or Reg− fragments (14,288/15,742). Other contigs were over-represented in Reg+ tissue and under-represented in Reg− tissue (537/15,742) or vice versa (842/15,742) (Supplementary Table 1). Upon close examination of transcripts over-represented in Reg− tissues and under-represented in Reg+ tissues, we found that several represented genes were involved in Wnt signaling. Given the importance of Wnt signaling in defining anteroposterior polarity in other planarian species10,11,14, we focused on genes involved in this pathway.
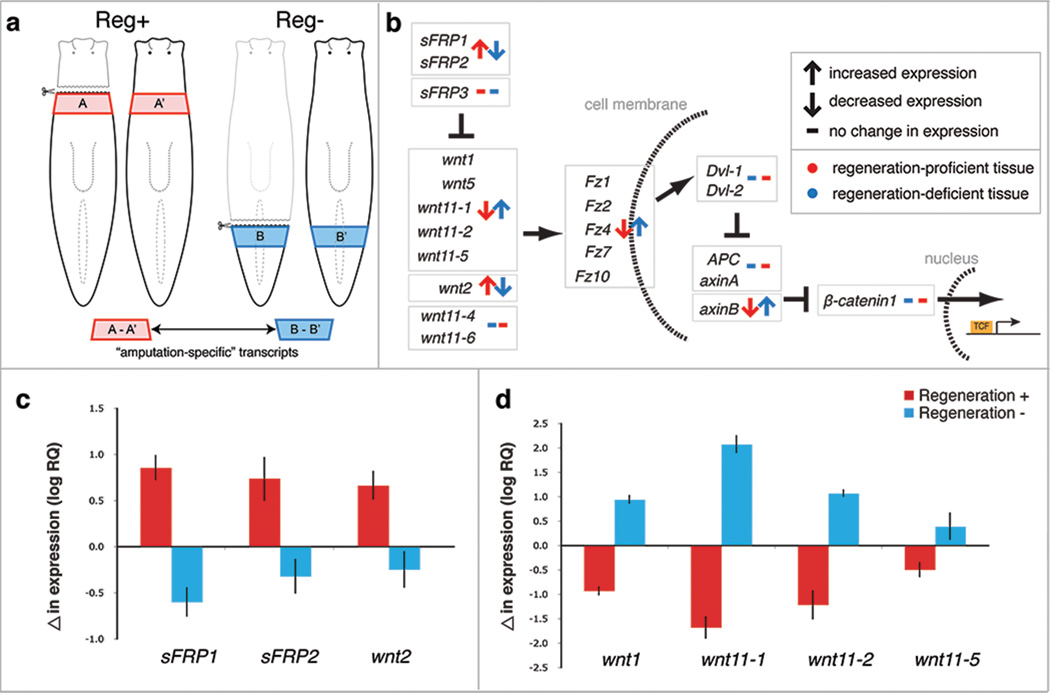
Figure 2. Comparative transcriptomics reveal differentially expressed genes following amputation in Reg+ and Reg− tissues.
a, Experimental strategy to identify transcripts responsive to amputation in Reg+ (red) and Reg− (blue) tissues. b, Alteration in expression levels of Wnt signaling components after amputation in Reg+ (red) and Reg− (blue) tissues measured by RNAseq. c–d, Changes in transcript levels of selected anterior- and posterior-specific Wnt signaling components after amputation in Reg+ (red) and Reg− (blue) tissues relative to uncut controls measured by qRT-PCR. Error bars represent standard deviations.
RNAseq revealed significant over-representation of many transcripts encoding Wnt ligands and receptors in Reg− tissues after amputation (Fig. 2b), with some transcripts, such as Pf-wnt11-1, upregulated as much as ~400-fold relative to intact controls. These same transcripts were downregulated in Reg+ tissues relative to their position-adjusted intact controls (Fig. 2b). In addition, homologs of Wnt inhibitors, such as sFRP1 and sFRP218, were downregulated in Reg− tissues and upregulated in Reg+ tissues (Fig. 2b). These patterns of gene expression were confirmed by quantitative RT-PCR (qRT-PCR) (Fig. 2c,d). Pf-β-catenin1, the intracellular effector of Wnt signaling, was not expressed differentially following amputation in either tissue region (Fig. 2b), indicating that Pf-β-catenin1 expression is not responsive to wounding in either Reg+ or Reg− tissues.
Since axial repolarization fails in Reg− tissues after amputation, we hypothesized that upregulation of posteriorly expressed genes, including wnt ligands, may inhibit signals that lead to proper anterior-posterior patterning and, thus, block regeneration. To test this hypothesis, we disrupted Wnt signaling using RNA-interference (RNAi) to target Pf-β-catenin1, the intracellular Wnt signaling effector. Remarkably, Pf-β-catenin1 RNAi resulted in blastema formation and regeneration of a complete head and brain in Reg− fragments as assayed by regeneration of the photoreceptors (n=64/71) (Fig. 3b,f), while control(RNAi) animals, injected with dsRNA from a ccdB and camR-containing bacterial sequence, failed to form a blastema or neural structures (Fig. 3a,e). Knocking down wnts individually or in combination did not rescue Reg− tissue (Supplementary Table 2). Pf-β-catenin1(RNAi) animals with rescued regeneration demonstrated anteriorly directed movements within 15 days after amputation (Supplementary Video 1), suggesting complete and functional regeneration of the head. Rescue of regeneration in Pf-β-catenin1(RNAi) animals reveals that Reg− tissues are competent to express the head regeneration program, but either lack signals required for re-establishment of axial polarity or these signals are inhibited.
Figure 3. Disruption of Wnt signaling via RNAi rescues regeneration in Reg− tissues.
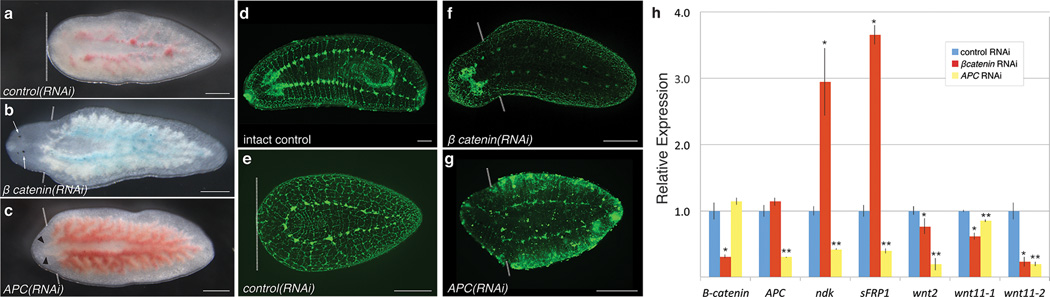
a-c, Posterior fragments 14 days after amputation in Reg− regions of control(RNAi), Pf-β-catenin1(RNAi), and APC(RNAi) animals. Dashed lines indicate amputation planes; white arrows indicate eyespots; black arrowheads indicate gut branches indicative of posterior fate. d–g, Nervous system in uncut and Reg− body regions of control(RNAi), Pf-β-catenin1(RNAi), and APC(RNAi) animals 28 days after amputation. Anterior is to the left. Scale bars, 250 µm. h, Relative transcript levels in Reg− tissue from Pf-β-catenin1(RNAi), Pf-APC(RNAi), and control(RNAi) animals 48 hours after amputation. Significant differences between control and experimental samples based on p-values (< 0.05) from two-tailed unpaired t-test are shown. (*, Pf-β-catenin1(RNAi) versus control(RNAi); **, Pf-APC(RNAi) versus control(RNAi)). Error bars represent standard deviations.
In contrast, knockdown of APC, an inhibitor of β-catenin and Wnt signaling, also resulted in blastema formation, but led to regeneration of tails at anterior-facing wounds (n=14/53, Fig. 3c,g), as observed in S. mediterranea10. These data suggest that altering gene expression to create either an anterior or posterior polarity cue within Reg− tissues can perpetuate downstream steps in regeneration, thereby allowing blastema formation and regeneration of heads or tails at anterior-facing wounds. Notum has recently been identified as a wnt inhibitor expressed at anterior facing wounds in the planarian S. mediterranea19. However, Pf-notum RNAi failed to initiate a similar posterior-regeneration program. To assay alterations in gene expression following regeneration rescue, we used qRT-PCR to characterize expression patterns of polarity genes following Pf-β-catenin1 RNAi and Pf-APC RNAi (Fig. 3h). After amputation in Pf-β-catenin1(RNAi) animals, both Pf-sFRP1 and Pf-ndk were significantly upregulated, suggesting that a latent anterior regeneration program is reactivated in Reg− tissues following Pf-β-catenin1 RNAi.
Our data provide important clues about mechanisms regulating regeneration. In a related Dendrocoelid species with reduced regenerative capacity, anterior regeneration ability in posterior fragments was rescued via grafting irradiated anterior tissue onto Reg− tissue20, suggesting that signals from differentiated anterior cells were sufficient to allow regeneration. Our results suggest that such signals are involved in re-establishing anterior-posterior polarity. Recent efforts have identified many requirements for the regenerative response to wounding, including, proliferation9, apoptosis8, and cell signaling11,12,21, but the inter-relationships between these processes are not yet well understood. Our data confirm that increased neoblast proliferation following amputation occurs independently of the re-establishment of anterior-posterior polarity (Fig. 1e,f and Supplemental Fig. 2)14. We have also shown that subsequent regenerative processes are inhibited until polarity is re-established. These observations suggest a checkpoint in the regeneration program that must be satisfied before downstream developmental processes can occur. Such a checkpoint would act as a vulnerable stage at which evolutionary modifications could alter regenerative potential.
Loss of regenerative ability would seem to carry a selective disadvantage, and possible causes for such losses have been the subject of much speculation1,5. Identification of aberrant Wnt signaling following amputation in Reg− tissues of P. fluviatilis suggests one mechanism limiting regeneration ability in planarians; however, we can only speculate about the driving force leading to regeneration loss. Unlike most planarians, P. fluviatilis is semelparous, reproducing only once, then dying within a single season6. Life-history studies have shown that semelparous species invest more in reproduction22, possibly at the expense of other developmental programs. Whereas highly regenerative, iteroparous planarian species resorb reproductive structures during periods of starvation23 and following amputation24, semelparous species do not resorb testes after such events23,25. We speculate that the signals maintaining the reproductive system in semelparous planarians may perturb the proper re-establishment of polarity after amputation, providing a selective advantage for later reproductive potential. As a consequence, anterior regeneration ability has been limited.
Next-generation sequencing technologies and functional analyses facilitate research on understudied, yet biologically informative, non-model organisms. Expanding use of these technologies will help elucidate causes for limited regeneration in other animals, potentially identifying inhibitory signals that must be overcome to elicit a regenerative response after wounding. Given that perturbation of a single gene’s function can rescue an entire regenerative program, identifying additional regeneration-inhibiting signals will increase our understanding of the evolution of regeneration loss and provide the intriguing prospect of restoring regenerative abilities in regeneration-deficient animals.
Methods
Animal collection & culture
Procotyla fluviatilis was collected from streams at Blockhouse Point Conservation Park (Maryland) or the LaRue Pine Hills/Otter Pond Research Natural Area (Illinois). Animals were maintained in the laboratory at 18°C in Montjuïch salts31 and fed bi-weekly. Planarians were starved for 1 week before use.
In situ hybridization
In situ hybridizations were performed using the formaldehyde-based fixation procedure as described previously26 with the following modifications. Planarians were killed in 8% n-acetyl cysteine for 5 min, fixed in 4% formaldehyde in PBSTx (PBS + 0.3% Triton X-100) for 25 mins, and incubated in reduction solution for 5 mins at room temperature. Samples were bleached in 6% H2O2 for 1 hour. Samples were imaged with a Leica M205A stereoscope.
Immunostaining
Immunostaining was conducted as described previously27 using methacarn fixative. Primary antibody incubation was performed overnight at 4°C at the following concentrations: anti-synapsin mouse monoclonal antibody (1:75 [Developmental Studies Hybridoma Bank, 3C11]), anti-phospho-histone H3 (Ser10) rabbit monoclonal antibody (1:1000 [Cell Signaling Technology, 3377]). Animals were incubated in secondary antibody (goat anti-mouse Alexa-488, 1:400 or goat anti-rabbit, Alexa-488, 1:500 [Molecular Probes, Invitrogen, A11029]) overnight at 4°C. Animals were mounted in Vectashield. Images were captured with Zeiss SteREO LumarV12, Zeiss AxioZoomV16, and Zeiss LSM710 confocal microscopes.
Histology & Scanning Electron Microscopy
Animals were fixed and prepared as previously described28. For histology, ethanol was gradually replaced with acetone, followed by infiltration with Araldite/Embed 812 (Electron Microscopy Sciences). Sections (1 µm) were collected on glass slides, dried over a heating plate, and stained with 1% Toluidine Blue for 10 seconds. Slides were mounted with Cytoseal 60 (Thermo Scientific) and imaged on a Nikon Eclipse TE200 inverted microscope. For Scanning Electron Microscopy, once the samples reached 100% ethanol, they were critical-point dried using a Tousimis Samdri-PVT-3D, mounted on aluminum stubs, coated with Au/Pd using a Denton Desk II TSC turbo-pumped sputter coater and imaged on a Philips XL30 ESEM-FEG.
Transcriptome sequencing and RNAseq
For sequencing the reference transcriptome, RNA from 4 random, intact P. fluviatilis adults was isolated using TRIzol (Invitrogen), DNAse-treated, purified with RNA Clean and Concentrator kit (Zymo, Irvine, CA), and submitted to the W. M. Keck Center for Comparative and Functional Genomics for Roche 454 pyrosequencing. Reads were assembled de novo using iterations of SeqMan NGen (DNASTAR, Madison, WI) and CLC Genomics (CLCbio, Cambridge, MA). For RNAseq experiments, RNA was isolated from tissue fragments of 5 worms, each excised ~2 mm posterior to the amputation sites. Control RNA was purified from corresponding control fragments excised from intact animals at equivalent body regions as described above. Samples were submitted to the W. M. Keck Center for Comparative and Functional Genomics for Illumina sequencing. Reads were mapped to the reference transcriptome using CLC Genomics and compared as per Marioni et al29. Contigs with ≥2-fold change and p-value < 0.05 (from two-sided unpaired t-tests) were used for detailed analysis. Selected contigs were screened to identify redundant contigs by using BLAST homology to the NCBI nr database to merge contigs with top hits to the same gene.
Cloning
To generate riboprobes, candidate genes were PCR amplified from cDNA generated from total RNA (iScript cDNA Synthesis Kit, Bio-Rad, Hercules, CA). For cDNA preparations, RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA). For cloning, 2–3 µL of PCR product was ligated with 70 ng of Eam1105I-digested pJC53.2 (Rapid DNA Ligation Kit, Roche, Mannheim, Germany)30 and used to transform DH5α. In vitro transcriptions with the appropriate RNA polymerase were performed using standard approaches with the addition of Digoxigenin-11-UTP (Roche, Mannheim, Germany). All primers used to amplify candidate genes are included in Supplemental Table 3.
RNAi
To generate dsRNA, templates cloned into pJC53.230 were amplified with a modified T7 oligonucleotide (GGATCCTAATACGACTCACTATAGGG), purified using a DNA Clean & Concentrator kit (Zymo Research, Orange, CA), and eluted in 15 µL of water. 10.5 µL of each PCR product (Supplemental Fig. 5) was used as template for in vitro transcription in a reaction containing 5 µL 100 mM mix of rNTPs (Promega), 1 µL high-yield transcription buffer (0.4 M Tris pH 8.0, 0.1 M MgCl2, 20 mM spermidine, 0.1 M DTT), 1 µL thermostable inorganic pyrophosphatase (New England Biolabs, Ipswich, MA), 0.5 µL Optizyme recombinant ribonuclease inhibitor (Fisher Scientific, Pittsburg, PA), and 2 µL T7 RNA polymerase. Samples were incubated at 37°C for 12 h and then treated with RNase-free DNase (Fisher Scientific, Pittsburgh, PA, FP2231) and cleaned/concentrated via ammonium acetate precipitation. Synthesized RNA was then annealed by heating at 95°C, 75°C, and 50°C each for 3 min. dsRNA solution was mixed with dye and 65nL (~1ug/uL) was microinjected into the gut of randomized adult planarians 3 times over the course of 1 week prior to amputation using a Nanoject II micromanipulator (Drummond Scientific). As a negative control, animals were injected with dsRNA synthesized from the ccdB and camR-containing insert of pJC53.230. Live (RNAi) animals were imaged and videos were captured with a Leica M205A.
Quantitative RT-PCR
To examine transcript levels following amputation in regeneration-proficient and regeneration-deficient tissues, RNA was extracted using Trizol Reagent from 5 random tissue fragments identical to those used for RNA sequencing. Following DNase treatment (DNA-free RNA Kit, Zymo Research, Orange, CA), reverse transcription was performed (iScript cDNA Synthesis Kit) and quantitative PCR was conducted using Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK) and a 7900HT real-time PCR system (Applied Biosystems). Three biological replicates were performed and all samples were measured in triplicate to account for pipetting error. Absolute quantities of each transcript were determined for each gene and normalized to the level of Pf-actin in each sample. The mean value for each amputated treatment was then normalized to the intact tissue fragments extracted from an identical axial position to determine the relative changes in expression due to amputation. For qPCR of genes following RNAi experiments, the mean values of control(RNAi) and experimental(RNAi) samples were graphed independently without normalization to intact fragments. All primers used for these studies are included in Supplemental Table 3.
Supplementary Material
Acknowledgments
We thank Newmark lab members for comments, A. Vieira for technical assistance, A. Hernandez and the W. M. Keck Center for Comparative and Functional Genomics for sequencing assistance, and the National Forest Service, Illinois Department of Natural Resources, and Montgomery County (MD) Department of Parks for field-collection permits. We also thank A. Boney, J. Brubacher, T. Chong, M. Issigonis, H. Iyer, and B. Lambrus for assistance in field collections. This work was supported by National Institute of General Medicine Sciences fellowship F32GM097921 (J.M.S.). P.A.N. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. Both authors contributed to the design of the experimental strategy. J.M.S. conducted all experiments, analyzed the data, and drafted the manuscript, which was critically reviewed and revised by P.A.N. Both authors discussed the results and commented on the final version of the manuscript.
Author Information. Sequence read archive (SRA) data reported in this paper were deposited at NCBI under project accession PRJNA205293. The assembled transcriptome has been deposited at NCBI under TSA Bioproject 205293. RNA-seq analyses have been deposited in the NCBI Gene Expression Omnibus under accession GSE48497. The authors declare no competing financial interests.
References
- 1.Bely AE. Evolutionary loss of animal regeneration: pattern and process. Integr. Comp. Biol. 2010;50:515–527. doi: 10.1093/icb/icq118. [DOI] [PubMed] [Google Scholar]
- 2.Newmark PA, Sánchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nat. Rev. Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- 3.Lillie FR. Notes on regeneration and regulation in planarians. Am. J. Physiol. 1901;6:129–141. [Google Scholar]
- 4.Morgan TH. Notes on Regeneration. Biol. Bull. 1904;6:159–172. [Google Scholar]
- 5.Bely AE, Nyberg KG. Evolution of animal regeneration: re-emergence of a field. Trends Ecol. Evol. 2010;25:161–170. doi: 10.1016/j.tree.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Kenk R. Biota of Freshwater Ecosystems: Identification Manual 1. Washington, DC: Environmental Protection Agency; 1972. Freshwater planarians (Turbellaria) of North America. [Google Scholar]
- 7.Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Gene. Dev. 2012;26:988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellettieri J, et al. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurley KA, Rink JC, Sánchez Alvarado A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen CP, Reddien PW. Smed-Bcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- 12.Yazawa S, Umesono Y, Hayashi T, Tarui H, Agata K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc Natl Acad Sci USA. 2009;106:22329–22334. doi: 10.1073/pnas.0907464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rink JC, Gurley KA, Elliott SA, Sánchez Alvarado A. Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science. 2009;326:1406–1410. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurley KA, et al. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 16.Forsthoefel DJ, Park AE, Newmark PA. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev. Biol. 2011;356:445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cebria F, et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- 18.Mii Y, Taira M. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signaling range. Development. 2009;136:4083–4088. doi: 10.1242/dev.032524. [DOI] [PubMed] [Google Scholar]
- 19.Petersen CP, Reddien PW. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science. 2011;332:852–855. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephan-Dubois F, Gilgenkrantz F. Transplantation et regeneration chez la planaire Dendrocoelum lacteum. J. Embryol. Exp. Morph. 1961;9:642–649. [PubMed] [Google Scholar]
- 21.Tasaki J, et al. ERK signaling controls blastema cell differentiation during planarian regeneration. Development. 2011;138:2417–2427. doi: 10.1242/dev.060764. [DOI] [PubMed] [Google Scholar]
- 22.Calow P. Life Cycles. London: Chapman and Hall; 1978. [Google Scholar]
- 23.Romero R, Baguna J. Quantitative cellular analysis of life-cycle strategies of iteroparous and semelparous triclads. Forstchr. Zool. 1988;36:283–289. [Google Scholar]
- 24.Fedecka-Bruner B. Etudes sur la regeneration des organes genitaux chez la planaire Dugesia lugubris I. Regeneration des testicules apres destruction. Bull. Biol. Fr. Belg. 1967;101:255–319. [PubMed] [Google Scholar]
- 25.Grasso M, Gardenghi G. The involvement of cellular elements other than neoblasts in Dendrocoelum lacteum regeneration. Boll. Zool. 1978;45:365–368. [Google Scholar]
- 26.Pearson BJ, et al. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong T, Stary JM, Wang Y, Newmark PA. Molecular markers to characterize the hermaphroditic reproductive system of the planarian Schmidtea mediterranea. BMC Dev. Biol. 2011;11:69. doi: 10.1186/1471-213X-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouhana L, Vieira AP, Roberts-Galbraith RH, Newmark PA. PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development. 2012;139:1083–1094. doi: 10.1242/dev.076182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNAseq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins JJ, et al. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- 31.Cebria F, Newmark PA. Planarian homologues of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.