Abstract
PRDM9 directs human meiotic crossover hotspots to intergenic sequence motifs, whereas budding yeast hotspots overlap low nucleosome density regions in gene promoters. To investigate hotspots in plants, which lack PRDM9, we used coalescent analysis of Arabidopsis genetic variation. Crossovers increase towards gene promoters and terminators, and hotspots are associated with active chromatin modifications, including H2A.Z, histone H3K4me3, low nucleosome density and low DNA methylation. Hotspot-enriched A-rich and CTT-repeat DNA motifs occur upstream and downstream of transcriptional start respectively. Crossovers are asymmetric around promoters and highest over CTT-motifs and H2A.Z-nucleosomes. Pollen-typing, segregation and cytogenetic analysis show decreased crossovers in the arp6 H2A.Z deposition mutant, at multiple scales. During meiosis H2A.Z and DMC1/RAD51 recombinases form overlapping chromosomal foci. As arp6 reduces DMC1/RAD51 foci, H2A.Z may promote formation or processing of meiotic DNA double-strand breaks. We propose that gene chromatin ancestrally designates hotspots within eukaryotes and PRDM9 is a derived state within vertebrates.
In fungi and mammals the majority of meiotic recombination occurs in narrow (1-2 kilobase) hotspots1-3. Human and mouse hotspots are targeted to DNA sequence motifs by the zinc finger domain protein PRDM94-11. PRDM9-dependent crossovers occur mainly in intergenic regions and introns, with the lowest recombination in exons9,12. PRDM9 also contains a SET domain with histone H3K4 trimethyltransferase activity and targets this modification to hotspot chromatin during meiosis11,13-15. In contrast, hotspots in the budding yeast, Saccharomyces cerevisiae are not sequence-dependent, show polarity within genes and occur predominantly at regions of low nucleosome density in gene promoters3,16-21. However, S.cerevisiae hotspots are also closely associated with H3K4 trimethylation (H3K4me3), which is required for wild type patterns of recombination22-26. Therefore, mammalian and yeast recombination hotspots are specified to varying degrees by genetic and epigenetic information. Although recombination rate varies extensively within plant genomes27-33, the control of meiotic crossover hotspots in plants is poorly understood. We therefore sought to map fine-scale recombination rates in Arabidopsis thaliana, which lacks PRDM9, and investigate the contribution of DNA sequence and chromatin to the control of plant hotspot locations.
Coalescent analysis of Arabidopsis genetic variation
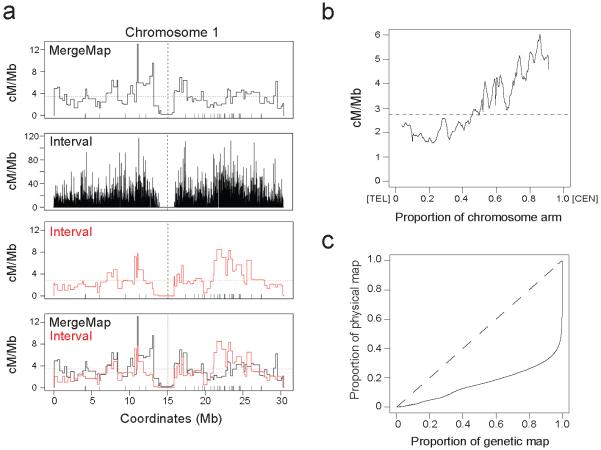
To generate a map of crossover frequency and hotspots in Arabidopsis we applied coalescent theory to a large single nucleotide polymorphism (SNP) dataset generated from 80 Eurasian accessions34. We used the Interval program from the LDhat package to estimate the population-scaled recombination rate between pairs of SNPs2. After conditioning on diallelic SNPs in unique sequence, we analysed a total of 2,112,845 SNPs (17.7 SNPs/kb) (Supplementary Table 1). We validated crossover frequencies estimated by Interval by comparing them to a consensus genetic map from 17 F2 populations33, which was generated previously using the MergeMap program35. We observed correlation between historical and experimental crossover frequencies for all chromosomes (eg 0.44 −0.55 at the 500 kb scale), although there are regions of substantial divergence (Fig. 1a, Supplementary Fig. 1, Supplementary Tables 2-3). Structural genetic variation between accessions may contribute to differences in recombination rate measurements. For example, megabase (Mb) inversions on chromosome 3 between Col and Sha accessions33,36, and the short arm of chromosome 4 between Col and Ler accessions37, cause crossover suppression. Population genetic forces, such as mutation, selection and drift, will also contribute to differences between historical and experimental recombination rates. As reported previously the disease resistance gene (R-gene) dense regions on chromosomes 1 and 5 show elevated historical crossover frequency relative to experimental measurements38 (Fig. 1a and Supplementary Figs 1-2). This may reflect balancing selection on R-genes, variable recombination or mutation rates or chance effects in previous generations.
Figure 1. Meiotic crossover frequency in the Arabidopsis genome.
(a) Plots show crossover frequency (cM/Mb) along chromosome 1 estimated by MergeMap (black), Interval (black), Interval plotted using MergeMap markers (red) and the overlay of MergeMap and Interval maps. MergeMap was generated previously from analysis of genotype data from 17 × F populations33 using the MergeMap program35,84. The ‘bin’ widths are variable because they are determined by intermarker distances. Horizontal dashed lines represent mean crossover frequency, vertical dashed lines represent centromeres and vertical ticks above the x-axis indicate disease resistance gene (R-gene) positions. (b) Mean crossover frequency (cM/Mb) estimated by Interval as a proportion along the length of the chromosome arms, orientated with the telomere (TEL) at 0 on the x-axis and the centromere (CEN) at 1. (c) The proportion of the crossovers estimated by Interval plotted against the proportion of physical sequence (solid black line). The dashed line represents a uniform relationship between the genetic and physical maps.
We observed Mb-scale variation in crossover frequency along the chromosomes, with an increase from telomere to centromere (Fig. 1b and Supplementary Fig. 3), although the centromere itself is crossover suppressed27,29,35. To investigate variation in recombination rates we plotted the proportion of crossovers against the proportion of physical sequence and observed a non-linear relationship, with 80% of crossovers occurring in 26% of the sequence (Fig. 1c and Supplementary Table 4). This is comparable to humans and chimpanzees and provides evidence for the presence of hotspots12,39. To identify crossover hotspots we analysed the same SNP data as used for Interval with an approximate marginal likelihood method called SequenceLDhot40. Missing data necessitated dropping a small proportion of SNPs (1.66%) relative to those analysed by Interval (Supplementary Table 1). We optimized SequenceLDhot run parameters using comparisons to the 3a hotspot, which we previously defined experimentally using pollen-typing35. SequenceLDhot identified 8,448 hotspots that correspond to 3.55% of the sequence and contain 14.73% of crossovers identified by Interval (ratio 14.73/3.55=4.15) (Supplementary Table 4). Therefore, our recombination maps show evidence for substantial variation in Arabidopsis crossover frequency at both domain and hotspot scales.
Gene chromatin at Arabidopsis promoter hotspots
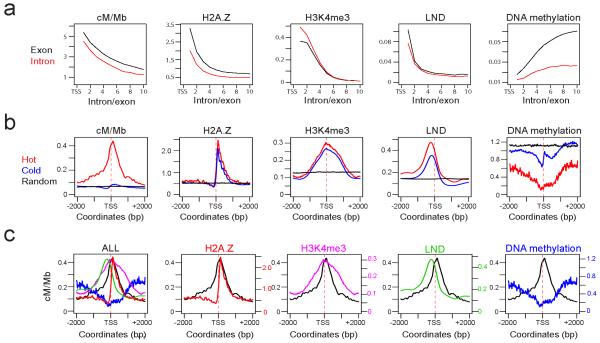
Because the 3a crossover hotspots overlapped with gene transcriptional start (TSS) and termination sites (TTS)35, we tested for overlap between hotspots and TSS/TTS35,41. Hotspots identified by SequenceLDhot overlapped with 5.75% (1,565) of TSS and 4.14% (1,127) of TTS (Supplementary Table 5), which was significantly more than expected by chance (Bickel’s block bootstrap42, P<1×10−15). 1 kb windows centered on hotspot-associated TSS correspond to 1.33% of the sequence and contain 10.07% of crossovers (ratio 10.07/1.33=7.57), whereas windows centered on all TSS correspond to 22.86% of the sequence and 64.39% of crossovers (ratio 64.39/22.86=2.82). Gradients of recombination rate have been observed in yeast genes3,17,19, so we analysed the mean crossover frequency of introns and exons in relation to their proximity to TSS and TTS. We observed gradients of increasing crossover frequency towards TSS and TTS, with a stronger effect for TSS (Fig. 2a and Supplementary Fig. 5a). Therefore, Arabidopsis crossovers are associated with genes and increase towards promoters and terminators.
Figure 2. Chromatin landscape at hot and cold promoters.
(a) Plots show the mean value of the variable printed above at exons (black) and introns (red) at increasing position numbers relative to the transcriptional start site (TSS). (b) Plots showing summed, normalized values for the variable printed above across +/− 2 kilobase (kb) windows centered on hot TSS (red), cold TSS (blue) or random (black) positions. All hot and cold distributions are significantly different by Wilcoxon signed rank test, P<1×10−15. (c) Plots are as for hot TSS plots in (b) but with the indicated variable H2A.Z45 (red), low nucleosome density47 (green), H3K4me3 44 (purple) and DNA methylation48 (blue) overlaid with cM/Mb (black). The final panel shows an overlay of all variables.
TSS/TTS are associated with specific chromatin features that facilitate RNA Pol-II transcriptional regulation43. We tested whether these patterns were different between hot, cold and random positions by analyzing 4 kb windows centered on hotspot overlapping (“hot”) TSS/TTS, non-hotspot overlapping (“cold”) TSS/TTS and the same number of random positions as for cold TSS/TTS (Fig. 2b and Supplementary Fig. 5b). Crossover frequency is highly elevated in proximity to hot TSS and shows an asymmetric peak relative to the TSS, shifted to the +1 nucleosome position (Fig. 2b and Supplementary Fig. 6). TSS/TTS are flanked by low nucleosome density (LND) regions upstream (TSS) or downstream (TTS) respectively, which facilitate RNA pol-II transcriptional initiation and termination43,44. The LND peaks occur on the opposite side of TSS/TTS to the peak of hotspot crossover frequency (Fig. 2b-c and Supplementary Fig. 5b-c). We observed higher LND at hot versus cold TSS/TTS (Wilcoxon signed rank test P<1×10−15), consistent with increased accessibility promoting recombination (Fig. 2b and Supplementary Fig. 5b). H3K4me3 occurs at the 5′ end of genes and is associated with mammalian and yeast recombination hotspots11,14,15,22-25. We observed a H3K4me3 peak overlapping the hotspot crossover frequency peak44 (Fig. 2b and Supplementary Fig. 5b), consistent with a conserved role for H3K4me3 in promoting plant hotspots11,14,15,22-25. Although H3K4me3 levels were significantly higher at hot TSS relative to cold (Wilcoxon signed rank test P<1×10−15), the difference is small relative to the difference in cM/Mb (Fig. 2b).
The histone variant H2A.Z occupies highly positioned +1 nucleosomes at TSS (Fig. 2b), where it facilitates transcriptional regulation45,46. We observed that the hotspot crossover frequency peak closely overlaps H2A.Z-containing nucleosomes (Fig. 2b-c and Supplementary Fig. 6). H2A.Z levels are significantly higher at hot TSS/TTS relative to cold TSS/TTS (Wilcoxon signed rank test P<1×10−15) (Fig. 2b and Supplementary Fig. 5b), consistent with a role for this histone variant in crossover formation. However, similar to H3K4me3 the difference in H2A.Z levels between hot and cold promoters was small relative to the difference in crossover frequency (Fig. 2b). Because crossover frequency exhibits gradients within genes, we tested whether chromatin follows similar patterns. H2A.Z, LND, H3K4me3 and crossover frequency show similar decreases as distance from TSS increases44,45,47 (Fig. 2a). This is again consistent with H2A.Z, H3K4me3 and LND playing a role in recombination at hotspot promoters.
DNA methylation is known to inhibit RNA pol-II transcriptional initiation and TSS/TTS show low DNA methylation48 (Fig. 2b and Supplementary Fig. 5b). We observe that hot TSS/TTS are DNA hypomethylated relative to cold (Wilcoxon signed rank test P<1×10−15) (Fig. 2b and Supplementary Fig. 5b). This is consistent with DNA methylation inhibiting meiotic recombination, as is known for Ascobolus immersus49. DNA methylation increases in genes as distance to the TSS increases and is higher in exons compared to introns, which reflects gene body methylation48 (Fig. 2a). Of the epigenetic marks analysed DNA methylation is the most different between hot and cold TSS/TTS, suggesting that this may be an important determinant of hotspot location and activity (Fig. 2b and Supplementary Fig. 5b). Therefore, Arabidopsis crossover hotspots associate with chromatin patterns that promote RNA pol-II transcription, with highest recombination rates over +1 H2A.Z-containing, H3K4me3-modified nucleosomes at DNA hypomethylated gene promoters. Although these chromatin modifications are correlated with crossover frequency, none alone is specific to hotspot promoters, suggesting they interact quantitatively to influence hotspot identity. It is also important to note that the chromatin datasets analysed were generated from non-meiotic cells44,45,47,48.
A-rich and CTT-repeat DNA motifs are enriched at hotspots
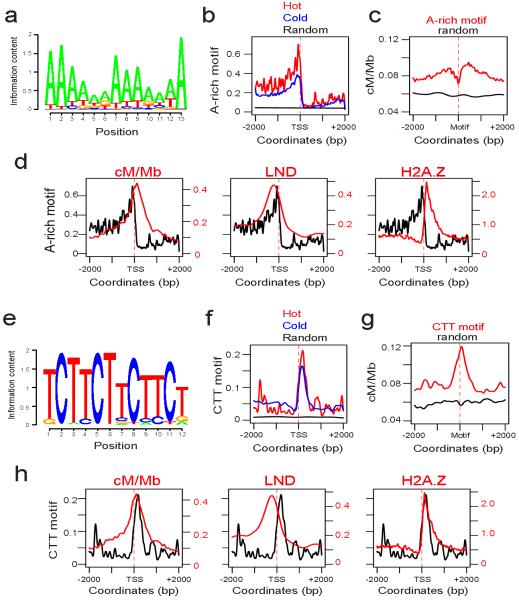
Human hotspots are determined by specific DNA sequence motifs recognized by the PRDM9 zinc finger protein4-11. Therefore, we tested whether Arabidopsis hotspots associate with specific DNA sequence motifs. We used three de novo DNA motif search algorithms, MEME/COSMO50,51, SOMBRERO52 and WEEDER53, to test for motifs enriched within 1 kb windows around hotspot-associated TSS compared with cold TSS. All three methods identified A-rich and CTT-repeat motifs as enriched at hotspot promoters (for example, Fig. 3a and 3e). This is consistent with previous work that demonstrated an association between A-rich motifs and crossover frequency in Arabidopsis38. The hotspot-enriched A-rich motifs were between 6-30 bp and the CTT-motifs were between 6-21 bp in length. Hot and cold promoters share both motifs, but they are significantly higher around hotspot TSS (Fig. 3b and f). The A-rich motifs are located upstream of TSS and overlap with regions of low nucleosome density (Fig. 3b and d), consistent with work in S.cerevisiae demonstrating that homopolymeric A and T tracts define nucleosome depleted regions54. Crossover frequency is significantly higher in +/− 2 kb windows around A-rich motifs compared with random positions (Wilcoxon signed rank test P<1×10−15) (Fig. 3c). Recombination is highest in regions flanking the A-rich motifs, which reflects positioning of these motifs upstream of TSS and the recombination rate peak (Fig. 3c-d). In contrast, the CTT-repeat motifs are located downstream of TSS and overlap the crossover frequency peak and H2A.Z nucleosomes (Fig. 3f and h). Crossover frequency is significantly higher in +/− 2 kb windows around CTT-motifs compared with random positions (Wilcoxon signed rank test P<1×10−15), and crossovers are elevated on the motifs themselves (Fig. 3g). Because the A-rich and CTT-repeat motifs overlap with low nucleosome density and H2A.Z respectively, they may contribute to nucleosome positioning or chromatin organization at promoters, with consequences for recombination. Alternatively, they may directly recruit factors that promote crossovers, analogous to PRDM94-11. Interestingly, the CTT-motifs we identified are characterized by a three bp C periodicity that is reminiscent of PRDM9 target motifs4-11 (Fig. 3e).
Figure 3. A-rich and CTT-repeat DNA sequence motifs at hot and cold promoters.
(a) Logo plot for a 13 bp A-rich motif identified as enriched at hotspot-associated promoters. (b) Enrichment of the A-rich motif shown in (a) at hot (red) and cold (blue) promoters in +/− 2 kb windows centered on TSS, and permutated (random, black) promoter sequences. (c) Crossover frequency (cM/Mb) in +/− 2 kb windows centered on the start coordinate of matches to the motif shown in (a) (red), compared to the same number of random positions (black), which were significantly different by a Wilcoxon rank sum test P<1×10−15. (d) Enrichment of the A-rich motif (black) shown in (a) at hotspot-associated promoters, overlaid with crossover frequency (cM/Mb, red), low nucleosome density47 (red) and H2A.Z45 (red) over +/− 2 kb windows centered on TSS. (e) Logo plot for a 12 bp CTT-repeat motif identified as enriched at hotspot promoters. (f) Enrichment of the CTT-repeat motif shown in (e) at hot (red) and cold (blue) promoters in +/− 2 kb windows centered on TSS and permutated promoter sequences (random, black). (g) Crossover frequency (cM/Mb) in +/− 2 kb windows centered on the start coordinate of matches to the motif shown in (e) (red), compared to the same number of random positions (black), which were significantly different by a Wilcoxon rank sum test P<1×10−15. (h) Enrichment of the CTT-repeat motif (black) shown in (e) at hotspot promoters, overlaid with crossover frequency (cM/Mb, red), low nucleosome density47 (red) and H2A.Z45 (red) over +/− 2 kb windows centered on TSS.
Experimental validation of hotspots using pollen-typing
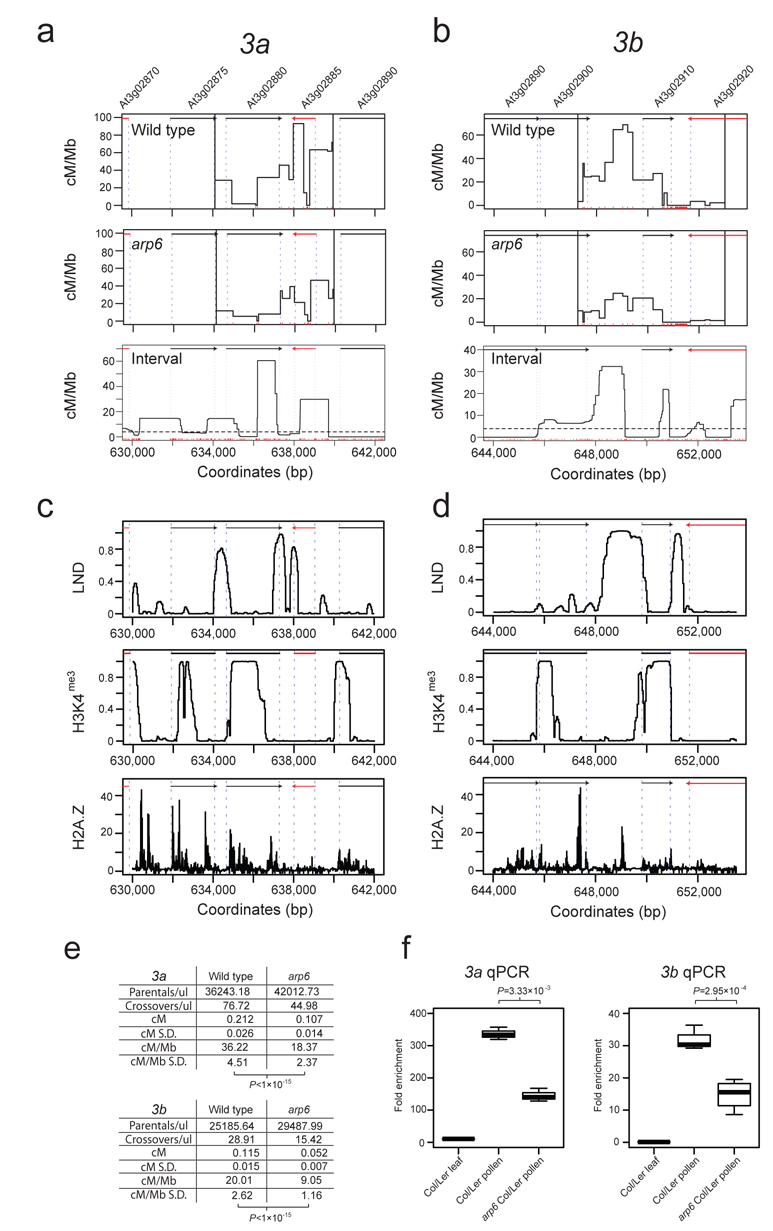
We next used pollen-typing to test whether crossover hotspots defined by coalescent analysis could be detected experimentally35,41. We previously used genetic mapping to identify a high crossover frequency interval on chromosome 3 between 632,767 and 660,906 bp that contained the 3a hotspot35. We searched within this window for additional hotspots using the Interval genetic map and identified a hotspot adjacent to 3a that we call 3b, which overlaps the TTS of At3g02900 and TSS of At3g02910 (Fig. 4a-b and Supplementary Table 6). Both 3a and 3b high crossover frequency intervals overlap with H2A.Z peaks as measured by ChIP-seq45 and ChIP-qPCR analysis (Fig. 4c-d and Supplementary Fig. 7). We designed Col/Ler allele-specific primers flanking 3b and amplified crossover and parental molecules from Col/Ler F1 pollen DNA35,41. The 3b crossover frequency is 20.01 cM/Mb, which is lower than 3a (36.22 cM/Mb) (Fig. 4e, Supplementary Tables 6-7). This is consistent with lower 3b recombination rates measured by Interval relative to 3a (Fig. 4a-b). Sequencing of 3b crossover molecules revealed a hotspot in the At3g02900/At3g02910 intergenic region, with a peak rate of 68.81 cM/Mb (male chromosome average=4.77 cM/Mb) that overlaps with a peak in crossover rate estimated by Interval (Fig. 4b and Supplementary Table 6).
Figure 4. The arp6 mutant has decreased crossover frequency at the 3a and 3b hotspots.
(a-b) Plots showing crossover frequency (cM/Mb) estimated by pollen-typing in wild type or arp6, and Interval across the 3a (a) and 3b (b) hotspots on chromosome 3. Vertical black lines in the pollen-typing plots indicate inner allele-specific primer positions. Red vertical ticks above the x-axis indicate SNP positions. Black and red arrows represent forward and reverse strand genes respectively. Associated gene numbers are printed above the arrows. Vertical, dashed blue lines indicate TSS and TTS positions. Horizontal dotted lines indicate the chromosome average recombination rate. (c-d) Low nucleosome density (LND) ChIP-chip47, histone 3 lysine 4 trimethylation (H3K4me3) ChIP-chip44 and H2A.Z ChIP-seq45 data plotted across the 3a and 3b hotspots, with plot annotations as for (a-b). (e) Crossover frequency for hotspots 3a and 3b calculated by single molecule pollen-typing of crossover and parental molecules in wild type and arp6 and associated standard deviations (SD). Crossover rates are significantly lower in arp6 (t-test 3a P<1×10−15, 3b P<1×10−15). (f) Crossover frequency for hotspots 3a and 3b measured by pollen-typing quantitative PCR (qPCR) are significantly lower in arp6 (t-test 3a P=3.33×10−3, 3b P=2.95×10−4).
Analysis of crossover frequency within the 3a pollen-typing amplicon shows three hotspots separated by at least one interval of 0 cM/Mb (3a-1 634,109-636,119 bp, 3a-2 636,199-638,633 bp and 3a-3 638,779-639,934) and that overlap TSS/TTS35 (Fig. 4a and Supplementary Table 7). The position of the central 3a-2 hotspot identified by pollen-typing is wider compared with Interval recombination rates estimates (Fig. 4a and Supplementary Table 7). To investigate the difference between LD-based and direct estimates of crossover rate we considered the effect of using different variant sets for analysis by Interval. Using a high confidence set of variants we find that the locations of inferred crossover hotspots are robust, but the relative height of peaks is altered (Supplementary Fig. 8a). This suggests that low levels of error may be influencing the exact crossover rate estimates but not the overall location of hotspots. We also note that SNP and deletion distributions are non-uniform across 3a and there are a number of deletions in the 3a-2 and 3a-3 regions (Supplementary Fig. 8b). Because local sequence polymorphisms are known to alter the recombination topology of mammalian hotspots55-57, variation in polymorphism patterns may also contribute directly to differences between historical and experimental crossover frequencies at 3a.
Decreased hotspot crossover frequency in arp6
H2A.Z-containing nucleosomes possess biophysical properties that contribute to transcriptional regulation at TSS/TTS46. Given the overlap between H2A.Z-containing nucleosomes and crossover frequency at hotspot promoters, we next tested the functional relationship between H2A.Z and crossovers. To do this we repeated pollen-typing in the actin related protein6 (arp6) mutant58-60. ARP6 is a component of the SWR1 nucleosome remodeling complex required for H2A.Z deposition58-61. We crossed Col (suf3-158) and Ler (esd1-262) arp6 alleles to generate arp6 Col/Ler F1s and repeated pollen-typing. The 3a and 3b regions show reduced crossover frequency in arp6 relative to wild type (t-test 3a P<1×10−15, 3b P<1×10−15), though with related hotspot topology (Fig. 4e, Supplementary Tables 6-7). We also developed a pollen-typing quantitative PCR (qPCR) assay and again observed that 3a and 3b crossovers are significantly reduced in arp6 (t-test 3a P=3.33×10−3, 3b P=2.95×10−4) (Fig. 4f and Supplementary Table 8). This experimentally demonstrates that H2A.Z deposition promotes crossovers at Arabidopsis hotspots.
Decreased domain and chromosome scale crossovers in arp6
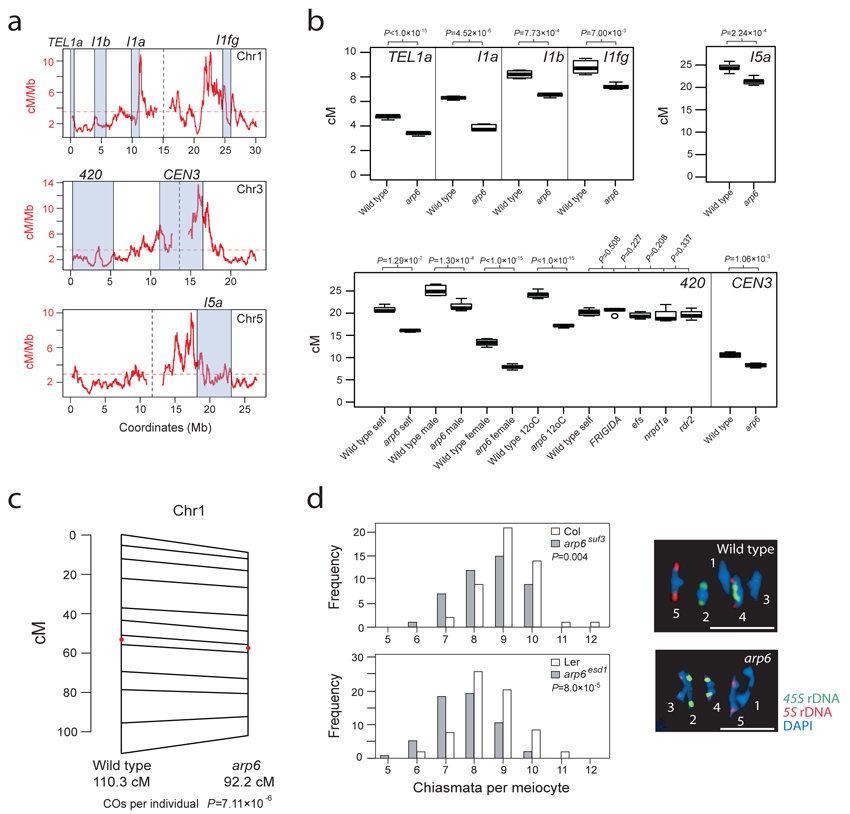
Because we observed decreased hotspot activity in arp6, we next tested crossover frequency at the domain scale (>1 Mb), via segregation of linked, heterozygous T-DNA insertions35,63. The FTL system measures segregation of T-DNAs expressing distinct colors of fluorescent protein from the pollen-specific LAT52 promoter63. FTL intervals I1a, I1b and I1fg each span >1 Mb of gene-dense sequence on chromosome 1 and show significantly reduced genetic distance in arp6 (chi square test P=4.52×10−6, P=7.73×10−4 and P=7.00×10−3 respectively) (Fig. 5a-5b and Supplementary Tables 9-11). We also observed significant reductions in arp6 crossover frequency using intervals on chromosomes 5 (I5a) and 3 (CEN3) (chi square test P=2.24×10−4 and P=1.06×10−3 respectively) (Fig. 5a-b and Supplementary Tables 12-13), demonstrating decreased crossover frequency in arp6 on different chromosomes.
Figure 5. The arp6 mutant has decreased crossover frequency at domain and whole chromosome scales.
(a) Crossover frequency estimated by Interval (red, cM/Mb) plotted along chromosomes 1, 3 and 5. Physical intervals in which we measured genetic distance are labeled and indicated by grey shading. Vertical, dotted lines indicate centromeres and horizontal dotted lines indicate the chromosome mean crossover frequency. (b) Genetic distances (cM) for intervals TEL1a, I1a, I1b, I5a, I1fg, 420 and CEN3 in wild type and arp6. test Chi square P-values comparing wild type and arp6 are printed above the plots. All temperatures are 21°C, apart from those listed as 12°C. (c) F2 genetic map length for chromosome 1 is reduced in arp6 relative to wild type. The number of crossover s per F2 individual was significantly lower in arp6 (chi square test P=1.06×10−7). Wild type and arp6 genetic maps are connected at SSLP marker positions and intervals containing the centromeres are marked with red dots. (d) Histograms for chiasma numbers per meiocyte for wild type and arp6, using alleles in Col (suf3) or Ler (esd1) accessions. There were significantly fewer chiasmata in arp6 (Conway-Maxwell-Poisson regression testing, Col P=0.004, Ler P=8.0×10−5). Representative images of metaphase-I meiocytes DAPI-stained and labeled by FISH against 45S (green) and 5S (red) rDNA to identify specific chromosomes. Chromosomes are labeled with their number. In wild type all bivalents except chr2 are ‘rings’ with >1 chiasma, whereas in arp6 all bivalents except chr2 are ‘rods’ with 1 chiasma. Scale bars represent 10 μM.
The DART method measures trans-segregation of linked insertions carrying different antibiotic or herbicide resistance genes, via double selection in backcross progeny64 (Supplementary Fig. 9). We performed DART using three insertion pairs located adjacent to the telomere of chromosome 1 termed TEL1a, TEL1b and TEL1c, which again showed reduced genetic distance in arp6 (chi square test P<1×10−15, P=0.0171 and P=0.10 respectively) (Fig. 5a-b and Supplementary Table 14). To demonstrate that reduced arp6 crossover frequency was not specific to T-DNA markers we measured segregation of simple sequence length polymorphisms (SSLPs). We generated 768 individual F2 populations from wild type and arp6 Col/Ler F1 plants and genotyped them for 12 SSLPs along chromosome 1, with an average intermarker distance of 2.75 Mb that together span 99.4% of the chromosome’s physical length (Fig. 5c). Chromosome 1 crossover number per F2 individual is significantly lower in arp6 (chi square test P=7.11×10−6), and genetic map length is reduced (wild type=110.3 cM, arp6=92.2 cM) (Fig. 5c). Crossover number per F2 individual is significantly different from the Poisson expectation in both populations (goodness-of-fit test, wild type P=2.07×10−13, arp6 P=4.97×10−15) (Supplementary Fig. 10a-b), which suggests that crossover interference is still acting in arp665.
As a further test of domain-scale recombination rates we used the 420 system, which measures segregation of T-DNAs expressing fluorescent proteins from the seed-specific NapA promoter35,66. The 420 interval is 5.1 Mb, located sub-telomerically on chromosome 3 and contains the 3a and 3b hotspots (Fig. 5a)35,66. A specific advantage of 420 is that reciprocal crossing can be used to measure male and female crossover rates separately, and 420 has been shown to have higher crossover frequency in male meiosis35,66. Sex-averaged, male and female 420 crossover frequency were all significantly reduced in arp6 (chi square test P=1.29×10−7, P=1.30×10−4 and P<1×10−15 respectively) (Fig. 5b and Supplementary Table 15). Since H2A.Z occupancy has been shown to decrease with increasing ambient temperature60, we hypothesized that crossover frequency would increase at lower temperatures. Consistent with this, we observed that wild type 420 recombination is significantly increased at 12°C compared to 21°C (chi square test P=1.78×10−4), and that arp6 was again significantly reduced relative to wild type at 12°C (chi square test P<1×10−15) (Fig. 5b and Supplementary Table 15). To test whether crossover decreases in arp6 are specific, we crossed 420 with genetic backgrounds that are late-flowering (FRIGIDA) or mutated in other epigenetic pathways, including small RNA biogenesis (rdr2, nrpd1a) and histone modification (efs). No significant difference in crossover frequency was observed in these backgrounds relative to wild type (chi square test P=0.51, P=0.21, P=0.34 and P=0.23 respectively) (Fig. 5b and Supplementary Table 15), demonstrating that decreased arp6 crossover frequency is specific.
RAD51, DMC1, MLH1 foci and chiasmata are reduced in arp6
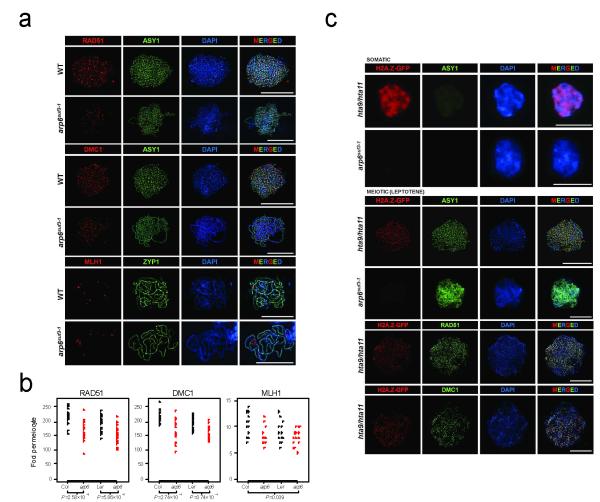
To further investigate the effect of arp6 on meiotic recombination we used immunocytology (Fig. 5d, Fig. 6 and Supplementary Fig. 11). Meiotic DSB sites can be estimated by immunostaining leptotene-stage meiotic chromosomes for RAD51 and DMC1, which are recombinases that mediate single-strand DNA invasion of the homologous chromosome after DSB formation and resection67,68. RAD51 and DMC1 accumulate as foci along chromosome axes, which can be detected using an antibody against the HORMA domain protein ASY169 (Fig. 6a). We observed significantly fewer RAD51 and DMC1 foci per meiocyte in arp6 in both Col (Wilcoxon rank sum test RAD51 P=2.58×10−5, DMC1 P=5.86×10−5) and Ler (Wilcoxon rank sum test RAD51 P=2.74×10−4, DMC1 P=3.74×10−4) accessions (Fig. 6a-b, Supplementary Fig. 11 and Supplementary Tables 16-17). This is consistent with H2A.Z promoting DSB formation, resection and/or strand invasion during meiotic prophase-I.
Figure 6. Immunolocalization of H2A.Z and meiotic proteins in wild type and arp6.
(a)The strand-invasion recombinases RAD51 and DMC1 (red) localise as foci to ASY1 (green) labeled, DAPI-stained (blue) leptotene chromosomes in wild type and arp6. MLH1 (red), a marker for crossover sites, localises to ZYP1 (green) labeled, DAPI-stained (blue) pachytene chromosomes in wild type and arp6. (b) Plots showing RAD51, DMC1 and MLH1 foci number per meiocyte in wild type and arp6. Wilcoxon rank sum test P-values are indicated for RAD51 and DMC1 counts per meiocyte between wild type and arp6 in either Col or Ler accessions. As MLH1 count data was underdispersed compared to the expectation based on the Poisson distribution, we used Conway-Maxwell-Poisson regression testing. MLH1 foci were significantly reduced in arp6 (P=0.039) across Col and Ler accessions. (c) Immunodetection of H2A.Z-GFP (red) in somatic and meiotic (leptotene) nuclei in complementing HTA11:GFP hta9 hta11 lines60. During meiosis H2A.Z-GFP (red) forms discrete foci on ASY1 labeled (green), DAPI-stained (blue) chromosomes, which overlap with RAD51 and DMC1 foci (green), but are not detected in arp6. All scale bars represent 10μM.
In Arabidopsis more DSBs are generated than mature into crossovers, for example we observed ~214 DMC1/RAD51 foci per meiocyte in wild type (Fig. 6a-b and Supplementary Tables 16-17), which are repaired into approximately 10 crossovers per meiosis33,70. Crossovers can be measured cytologically when Arabidopsis bivalents segregate at metaphase-I and adopt either rod or ring chiasma configurations, representing single or multiple crossovers respectively71 (Fig. 5d). We observed significantly fewer chiasmata in arp6 using both Col (Conway-Maxwell-Poisson regression test72, P=0.004) and Ler (Conway-Maxwell-Poisson regression test72, P=8.0×10−5) alleles (Fig. 5d and Supplementary Tables 18-19). Meiocytes with fewer than one chiasma per chromosome in arp6 were not observed, suggesting that obligate crossovers are maintained65 (Fig. 5d and Supplementary Table 18). We also immunostained pachytene meiocytes for MLH1, which marks class-I interfering crossovers, and ZYP1, which marks the synaptonemal complex73 (Fig. 6a and Supplementary Fig. 11). We observed significantly fewer MLH1 foci in arp6 comparing across both Col and Ler alleles (Conway-Maxwell-Poisson regression test72, P=0.039) (Fig. 6a-b and Supplementary Table 20), consistent with our chiasma data. MLH1 foci numbers per meiocyte are significantly different from the Poisson expectation in both wild type and arp6 (goodness-of-fit test, wild type P=3.47×10−3, arp6 P=5.79×10−4) (Supplementary Fig. 10c-d), again indicating that crossover interference is acting in arp665. Together this demonstrates that arp6 reduces the frequency of RAD51, DMC1 and MLH1 foci and chiasmata at whole chromosome scale.
Axis-associated H2A.Z foci during meiotic prophase-I
To investigate the localization of H2A.Z on meiotic chromosomes we performed immunostaining using a complementing GFP-tagged line HTA11-GFP hta9 hta11 (hereafter referred to as H2A.Z-GFP)60. Consistent with previous work we observed diffuse H2A.Z-GFP staining in somatic nuclei, and lower signal in the densely DAPI-staining heterochromatic chromocentres59-61 (Fig. 6c). Immunostaining of leptotene meiocytes for H2A.Z-GFP and ASY1 revealed ~250 axis-associated H2A.Z foci that overlapped RAD51 (85%) and DMC1 (93%) significantly more than randomized controls (42% and 48% respectively) (chi square test RAD51 P<1×10−15, DMC1 P<1×10−15) (Fig. 6c). We observed an absence of H2A.Z-GFP signal in the arp6 mutant (Fig. 6c). These data further support a role for H2A.Z during early meiotic recombination steps, including DSB formation and strand invasion. We propose that decreased recombination activity of individual hotspots in the absence of H2A.Z deposition leads to reduced crossover frequency at domain and whole chromosome scales.
DISCUSSION
We demonstrate extensive variation in crossover frequency throughout the Arabidopsis genome at both fine and broad scales. At fine scale (kb) crossovers are concentrated in hotspots associated with the start, and to a lesser extent, the end of genes. Hotspot promoters have chromatin marks associated with active RNA Pol-II transcription, including low nucleosome density, H2A.Z, H3K4me3 and low DNA methylation. Hotspot promoter crossovers are highest over CTT-repeat DNA motifs and +1 H2A.Z-nucleosomes. To test the importance of promoter chromatin for recombination we analysed the H2A.Z deposition mutant arp6 and observed reduced crossovers at hotspot, domain and whole chromosome scales. Although H2A.Z and H3K4me3 levels are significantly higher at hotspot promoters the differences are small relative to the difference in crossover frequency. Similarly, budding yeast H3K4me3 levels are a poor predictor of hotspot activity, despite this chromatin modification being required for normal meiotic recombination22-26. As none of the chromatin variables analysed here uniquely identifies hotspot promoters they are likely to act jointly to influence crossover frequency. The most divergent chromatin modification between hot and cold promoters is DNA methylation, consistent with this epigenetic modification inhibiting crossovers. Suppression of crossovers in the densely DNA methylated Arabidopsis centromeric regions provides further evidence for a negative relationship27,29,33,35. The chromosome arms show Mb scale variation in recombination activity, which may relate to similar patterns observed in yeast and mammalian genomes2,3,12,74-76. We propose that crossover frequency in plant genomes is determined by quantitative interactions between chromatin, DNA sequence and meiotic chromosome organisation.
H2A.Z functions in somatic DSB repair, where it is recruited to break sites77,78. Furthermore, a second H2A variant H2A.X has roles in both somatic and meiotic DSB repair. H2A.X is phosphorylated (ϒ-H2A.X) in 1-2 Mb domains surrounding DSBs79-81, and forms SPO11- dependent foci during meiosis69. Our data further demonstrate the involvement of histone H2A variants in DNA repair, specifically a role for H2A.Z in promoting Arabidopsis meiotic crossovers. In this context H2A.Z may function analogously to H3K4me3 in yeast. During meiosis, yeast Spp1, a subunit of the COMPASS complex, binds to both H3K4me3 and the Mer2 recombination protein, which is located on the meiotic chromosome axis23,25, consistent with the tethered loop axis model82. We have demonstrated that H2A.Z accumulates in foci on leptotene-stage meiotic chromosomes that colocalise with RAD51 and DMC1 recombinase foci. As RAD51/DMC1 foci are reduced in arp6 we propose that H2A.Z promotes the formation or processing of meiotic DNA DSBs. However, as crossovers are reduced in arp6, despite an estimated ~150 remaining DSBs, H2A.Z may also play additional downstream roles in crossover formation. Therefore, multiple elements of promoter chromatin, including H2A.Z, are likely to contribute to recruitment of hotspot sequences to the meiotic axis and promotion of DNA repair and recombination.
Arabidopsis hotspots most closely resemble budding yeast DSB hotspots, which overlap with low nucleosome density regions in gene promoters and H3K4me3 3,16-25. In contrast, mouse and human hotspots are positioned to intergenic sequence motifs by the PRDM9 zinc-finger histone methyltransferase4-10,14,15. However, in the absence of PRDM9 mouse DSB hotspots revert to the +1 nucleosome position at TSS15. We therefore propose that gene chromatin is an ancestral hotspot designation mechanism within eukaryotes and that PRDM9 evolved more recently in animals83.
ONLINE METHODS
Estimating crossover frequency using LDhat
From single nucleotide polymorphism (SNP) data of 80 Eurasian Arabidopsis thaliana accessions34 we selected diallelic positions with <10% missing data (coded as Z, N, M or X), >10% minimum allele frequency and no deletions (coded as D or −). Repetitive sequences are polymorphic between accessions and can be difficult to map to the TAIR10 genome assembly85,86. Therefore, we masked SNPs within (i) TAIR10 transposable element annotation, (ii) RepeatMasker output87, (iii) Tandem Repeats Finder output88, (iv) Inverted Repeats Finder output89 and (v) centromeric regions showing CO suppression27,29,33.
The Interval program from the LDhat package2,90 was used to estimate the population-scaled recombination rate (ρ/kb, ρ=4Ner, where Ne is the effective population size and r is the per generation recombination rate) between pairs of SNPs2. SNP data was split into 5,000 SNP blocks with 500 SNP overlaps and formatted using the Convert program. Block recombination rates were joined at overlap position 250. We used the Complete program to generate two likelihood lookup tables for 80 individuals with a grid size of 100 and using a population-scaled mutation rate per site of θ=0.1, or 0.0012,90 (θ=4Neμ, where μ is the per generation mutation rate). As A.thaliana is predominantly self-fertilizing and SNPs are highly homozygous within accessions, we treated the data as haploid and phased34. The crossover model was used with either the θ=0.1 or 0.001 lookup table and varying block penalty from 1 to 15. We used 60,000,000 iterations, sampling every 40,000 updates and discarded the first 500 samples as burn-in.
To compare Interval crossover estimates with MergeMap data33,35 we converted ρ/kb to ρ by multiplying by interval widths. ρ was summed over MergeMap marker intervals and divided by interval width (Mb). ρ/Mb was regressed onto MergeMap cM/Mb and the regression coefficient used to rescale ρ/Mb to cM/Mb. Interval and MergeMap recombination rates at different physical scales were compared using Spearman’s rank correlation. We smoothed cM/Mb over 10,000 site windows using the rollmean function from the R package zoo91. After smoothing chromosome arms were orientated with the lowest base pair coordinates at the telomeres and crossover rates calculated along the chromosome as a proportion of total arm length. Mean values across the 8 long chromosome arms were taken at regular intervals. To calculate the proportion of the genetic map in the physical map, cM and bp values for each interval were calculated as a proportion of their total chromosome map lengths. Values were then ranked according to decreasing proportion of ρ and mean values taken at regular intervals for the 5 chromosomes.
Identifying crossover hotspots using SequenceLDhot
To identify hotspots we used SequenceLDhot40, which applies an approximate marginal likelihood method to generate likelihood ratio statistics for hotspot positions40. SNP data was split into 2,500 site blocks with no overlaps. We used 15,000 runs with a minimum of 300 runs per hotspot, θ=0.1 and a starting value for ρ of 2. To exclude artefactual hotspot calls, we specified that all hotspot recombination rates be between 12 and 80 ρ/kb. Windows of 500 bp containing at least 7 SNPs were tested for the presence of a hotspot every 250 bp from the starting coordinate. Background recombination rates were specified using median ρ/kb over 50 kb windows from the Interval genetic map. A subset of SNP blocks had a missing data structure that caused SequenceLDhot to fail, which we removed, resulting in the analysis of a total of 2,077,613 SNPs (17.4 SNPs/kb).
Chromatin analysis
Transcription start sites (TSS) and termination sites (TTS) from TAIR10 were divided into those overlapping SequenceLDhot hotspots (hot) and the remainder (cold). We defined control positions by randomly selecting the same number of sites as for cold TSS/TTS from a uniform distribution for each chromosome. Hot, cold and random positions were used to define +/− 2,000 bp windows. Hot and cold TSS/TTS windows were orientated so that the direction of transcription is from left to right. For each position in a window values from datasets of interest; (i) Interval cM/Mb, (ii) DNA cytosine methylation from bisulfite sequencing48, (iii) nucleosome density data from ChIP and array hybridization47, (iv) H2A.Z ChIP-seq data45 and (v) H3K4me3 ChIP and array hybridization data44, were summed across all windows and divided by the total number of windows.
DNA sequence motif analysis
To search for hotspot-enriched DNA sequence motifs we used three de novo search algorithms; MEME/COSMO50,51, SOMBRERO52 and WEEDER53. Protein-coding, representative gene models from TAIR10 were divided into those with TSS overlapping SequenceLDhot hotspots (hot) and the remainder (cold). Genes were ranked by mean crossover frequency over 1 kb windows centered on each TSS. Background sequence for use with all programs except WEEDER was generated using 1 kb windows around 1,500 cold TSS randomly chosen from the lowest cM/Mb quartile. MEME searches for motifs using a probabilistic model and maximum likelihood estimation50. The maximum data length MEME can analyse is 100,000 (eg 100 × 1,000 bp windows). Therefore, we selected the 99 TSS with highest crossover rates as a training set. To generate a background model for MEME, COSMO and SOMBRERO we used SOMBRERO (Markov model, order 3-6) with the 1,500 cold TSS. We used MEME to search for motifs of lengths 6-30 bp on both strands allowing zero or more motifs per sequence (ANR model). For the other programs, we selected the 500 hot TSS with highest crossover frequency as the training set. We used COSMO, which is an implementation of MEME in the R language that uses an improved likelihood-profile method51. COSMO does not have the 100,000 maximum data length limit and so we used 500 hot TSS as the training set. SOMBRERO searches for motifs using a self-organising map of position weight matrices52. We used a 50 × 25 grid size with the SOMBRERO default options to search for motifs of lengths 6-30 bp. WEEDER performs motif searches using exhaustive enumeration53. For the WEEDER background set we generated frequency files of expected 6-mers and 8-mers using the 1,500 cold TSS. For the training set we used the 500 hot TSS and searched for motifs of lengths 6, 8, 10 and 12 bp, using the ‘extra’ run parameter, the S and M run settings, which consider both strands and allow multiple motifs matches per sequence and the Tn output setting was 20.
To analyse motif distributions we matched motif position weight matrices, using the Biostrings function matchPWM, to +/− 2 kb windows around hot and cold TSS. Motif match start coordinates were summed over hot or cold promoters and divided by the number of promoters analysed. For random we compared to permutated hot and cold sequences. To analyse crossover frequency at the motifs we summed cM/Mb estimates over +/− 2 kb windows centered on motif-match start positions and divided by the number of windows analysed. For controls we compared to the same number of randomly chosen positions.
Pollen-typing
Pollen-typing for hotspot 3a was performed as described35,41. We followed the same methodology to design allele-specific primers for hotspot 3b, which were optimised for allele-specificity and amplification efficiency35,41 (Supplementary Table 21). 3b primer combinations used in nested amplification were;
1st Crossover amplification = KC156_LeF + KC166_CoR
1st Parental amplification = KC167_CoF + KC166_CoR
Amplification conditions for the first round of PCR were {94°C for 2′30″}, then 8 cycles of {94°C for 30″, 58°C (decrease by 0.5°C per cycle) for 45″, 68°C for 6′30″}, then 26 cycles of {94°C for 20″, 54°C for 30″, 68°C for 6′30″}, then {65°C for 6′30″}. Amplification products from the first reaction were diluted 20-fold with 5 mM Tris pH 8.0 and 1μl used for input in the second amplification. All amplifications were performed using Ex-taq (Takara).
2nd Crossover amplification = KC160_LeF + KC152_CoR
2nd Parental amplification = KC168_CoF + KC152_CoR
Amplification conditions for the second round of PCR were {94°C for 2′30″}, then 8 cycles of {94°C for 30″, 57°C (decrease by 0.5°C per cycle) for 45″, 68°C for 6′00″}, then 26 cycles of {94°C for 20″, 53°C for 30″, 68°C for 6′00″}, then {65°C for 6′00″}.
Amplification products from single crossover molecules were diluted 20-fold with 5 mM Tris pH 8.0 and re-amplified using 2nd nested amplification primers KC160_LeF + KC152_CoR. PCR products were then directly sequenced using primers KC248, KC239, KC240, KC236 and KC237 to identify internal crossover locations (Supplementary Table 21).
Pollen-typing quantitative PCR (qPCR) assay
Genomic DNA from Col/Ler F1 leaves, pollen and arp6 F1 Col/Ler pollen was extracted and crossover molecule concentrations estimated by single molecule amplification35,41. DNA inputs were adjusted such that wild type Col/Ler pollen F1 DNA contained approximately 30 amplifiable crossover molecules and equal ng amounts of Col/Ler leaf and arp6 Col/Ler pollen DNA were used. Amplifications were performed as for the first round of single molecule amplifications, with a reaction volume of 20 μl and 16 cycles. PCR reactions were diluted 20-fold for crossovers and 20,000-fold for parental molecules with 5 mM Tris pH 8.0. Real time PCRs were then performed using universal primers (Supplementary Table 21, 3a=6376+6377; 3b=KC137+KC138) and SYBR green (Invitrogen) to estimate DNA concentrations. Ct values for parental and crossover amplifications from the same DNA template were used to calculate relative enrichment using the Delta Delta Ct method. Each sample was analysed with three technical replicates, and the whole experiment repeated with three biological replicates.
H2A.Z-GFP chromatin immunoprecipitation (ChIP)
ChIP was performed using HTA11-GFP hta11 hta9 lines and Col wild type controls60. Nuclei were extracted from formaldehyde cross-linked meiotic-stage flower buds and chromatin digested with 0.05 units of mirococcal nuclease (MNase) (NEB) at 37 °C for 15 minutes. ChIP was performed using a GFP polyclonal antibody (Santa Cruz, sc-8334). The relative enrichment of immunoprecipitated 3a and 3b hotspot DNA from the HTA11-GFP hta11 hta9 versus Col was analysed by qPCR (Supplementary Tables 22 and 23).
Fluorescent pollen-tetrad and seed-based crossover scoring
Scoring of fluorescent protein expression in seed and pollen was performed as reported previously35,92.
Simple Sequence Length Polymorphism (SSLP) genotyping
DNA was extracted from wild type and arp6 Col/Ler F2 individuals using CTAB (Sigma). We used Ler polymorphism data93 to screen for SSLPs between 28-262 bp (Supplementary Table 24). Primers were designed to amplify both Col and Ler templates spanning SSLPs. Amplification products were separated electrophoretically through 3% agarose-TBE gels to genotype F2 individuals. Genotype data were analysed using Rqtl94 and genetic maps constructed using the Kosambi mapping function. Total chromosome 1 crossover numbers per F2 were compared between wild type and arp6 using a t-test. These data were also compared to the Poisson distribution using the R package vcd95 and goodfit function.
Chiasma counting and immunocytology
We performed chiasma analysis on DAPI-stained metaphase-I anther meiocytes and identified chromosomes via 45S and 5S rDNA FISH71. Chiasma numbers were underdispersed compared with the Poisson expectation and there was heterogeneity between the chromosomes, with chromosomes 2 and 4 having fewer chiasma. As Conway-Maxwell-Poisson regression outperforms other methods for underdispersed counts72, the data were modelled using version 0.3.4 of the COMPoissonReg package96. In addition to genotype, dummy variables were constructed to model chromosome effects. Variables were entered in a stepwise manner and the final model included effects for genotype, chromosome 2 and chromosome 4 (Supplementary Table 18 and 19).
Immunolocalisation was carried out as described97. Antibodies were used at concentrations of 1:500: anti-ASY1 (rat), anti-ZYP1 (rat), anti-RAD51 (rabbit), anti-DMC1 (rabbit), anti-MLH1 (rabbit) and anti-GFP (Santa Cruz Biotechnology, Catalog no. 8334). Microscopy was performed using a Nikon 90i Fluorescence Microscope (Tokyo, Japan). Deconvolution of images by the ‘Mexican Hat’ function (Nikon NIS elements software) was used to count foci as described98. Once foci overlap was scored, the green channel was rotated 180° relative to the red channel and rescored to obtain randomized measurements. Overlapping (H2A.Z versus DMC1/RAD51) versus randomized counts were compared using Wilcoxon rank sum tests. The MLH1 foci counts were underdispersed compared with the Poisson expectation and these data were also modeled using the COMPoissonReg package96.
Supplementary Material
ACKNOWLEDGEMENTS
We kindly thank Steve Jacobsen (University of California, Los Angeles) and Daniel Zilberman (University of California, Berkeley) for chromatin data, Detlef Weigel (Max Plank Institute for Developmental Biology, Tübingen) for genetic polymorphism data, Paul Fearnhead (University of Lancaster) for helpful advice running SequenceLDhot, Philip Wigge (The Sainsbury Laboratory, University of Cambridge) for the HTA11-GFP hta11 hta9 line and comments and Peter Shaw (John Innes Centre, Norwich) for the HTA11-GFP arp6 line. Work in the Henderson laboratory is supported by a Royal Society University Research Fellowship, Gatsby Charitable Foundation Grant 2962, the Isaac Newton Trust and BBSRC grant BB/K007882/1. KC was supported by an EMBO Long Term Fellowship EMBO LTF-807,2009. PAZ is supported by grant 605/MOB/2011/0 from the Polish Ministry of Science and Higher Education. GM is supported by Wellcome Trust Core Award 090532/Z/09/Z and OV by Wellcome Trust studentship 086786/Z/08/Z. Work in the Franklin laboratory is supported by the BBSRC. GPC is supported by grant MCB-1121563 from the NSF. This paper is dedicated to Simon Chan.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nature reviews. Genetics. 2004;5:413–24. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 2.McVean GAT, et al. The fine-scale structure of recombination rate variation in the human genome. Science (New York, N.Y.) 2004;304:581–4. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 3.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–31. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science (New York, N.Y.) 2010;327:836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science (New York, N.Y.) 2010;327:876–9. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg IL, et al. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12378–83. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nature genetics. 2010;42:859–63. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–5. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 10.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science (New York, N.Y.) 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–8. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science (New York, N.Y.) 2005;310:321–4. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–8. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 14.Grey C, et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS biology. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–5. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. The EMBO journal. 1994;13:5754–63. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu TC, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science (New York, N.Y.) 1994;263:515–8. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 18.Fan QQ, Petes TD. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Molecular and cellular biology. 1996;16:2037–43. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–9. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 20.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:5213–8. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome research. 2009;19:2245–57. doi: 10.1101/gr.096297.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. The EMBO journal. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sommermeyer V, Béneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Molecular cell. 2013;49:43–54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Sollier J, et al. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. The EMBO journal. 2004;23:1957–67. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Acquaviva L, et al. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science (New York, N.Y.) 2013;339:215–8. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- 26.Tischfield SE, Keeney S. Scale matters: the spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell cycle (Georgetown, Tex.) 2012;11:1496–503. doi: 10.4161/cc.19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Copenhaver GP, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science (New York, N.Y.) 1999;286:2468–74. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 28.Dooner HK. Genetic Fine Structure of the BRONZE Locus in Maize. Genetics. 1986;113:1021–36. doi: 10.1093/genetics/113.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS genetics. 2011;7:e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gore MA, et al. A first-generation haplotype map of maize. Science (New York, N.Y.) 2009;326:1115–7. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 31.Mayer KFX, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–6. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 32.Saintenac C, et al. Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.) Genetics. 2009;181:393–403. doi: 10.1534/genetics.108.097469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salomé PA, et al. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity. 2012;108:447–55. doi: 10.1038/hdy.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nature genetics. 2011;43:956–63. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 35.Yelina NE, et al. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS genetics. 2012;8:e1002844. doi: 10.1371/journal.pgen.1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 37.Fransz PF, et al. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell. 2000;100:367–76. doi: 10.1016/s0092-8674(00)80672-8. [DOI] [PubMed] [Google Scholar]
- 38.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nature genetics. 2012;44:212–6. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auton A, et al. A fine-scale chimpanzee genetic map from population sequencing. Science (New York, N.Y.) 2012;336:193–8. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics (Oxford, England) 2006;22:3061–6. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 41.Drouaud J, Mézard C. Characterization of meiotic crossovers in pollen from Arabidopsis thaliana. Methods in molecular biology (Clifton, N.J.) 2011;745:223–49. doi: 10.1007/978-1-61779-129-1_14. [DOI] [PubMed] [Google Scholar]
- 42.Bickel PJ, Boley N, Brown JB, Huang H, Zhang NR. Subsampling methods for genomic inference. Annals of Applied Statistics. 2010;4:1660–1697. [Google Scholar]
- 43.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Critical reviews in biochemistry and molecular biology. 2009;44:117–41. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Bernatavichute YV,, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome biology. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS genetics. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deal RB, Henikoff S. Histone variants and modifications in plant gene regulation. Current opinion in plant biology. 2011;14:116–22. doi: 10.1016/j.pbi.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS biology. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maloisel L, Rossignol JL. Suppression of crossing-over by DNA methylation in Ascobolus. Genes & development. 1998;12:1381–9. doi: 10.1101/gad.12.9.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bembom O, Keles S, van der Laan MJ. Supervised detection of conserved motifs in DNA sequences with cosmo. Statistical applications in genetics and molecular biology. 2007;6 doi: 10.2202/1544-6115.1260. Article8. [DOI] [PubMed] [Google Scholar]
- 52.Mahony S, Golden A, Smith TJ, Benos PV. Improved detection of DNA motifs using a self-organized clustering of familial binding profiles. Bioinformatics (Oxford, England) 2005;21(Suppl 1):i283–91. doi: 10.1093/bioinformatics/bti1025. [DOI] [PubMed] [Google Scholar]
- 53.Pavesi G, Mauri G, Pesole G. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics (Oxford, England) 2001;17(Suppl 1):S207–14. doi: 10.1093/bioinformatics/17.suppl_1.s207. [DOI] [PubMed] [Google Scholar]
- 54.Field Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS computational biology. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baudat F, de Massy B. Cis- and trans-acting elements regulate the mouse Psmb9 meiotic recombination hotspot. PLoS genetics. 2007;3:e100. doi: 10.1371/journal.pgen.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cole F, Keeney S, Jasin M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Molecular cell. 2010;39:700–10. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Human molecular genetics. 2005;14:2277–87. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- 58.Choi K, et al. SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. The Plant cell. 2005;17:2647–60. doi: 10.1105/tpc.105.035485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. The Plant cell. 2007;19:74–83. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–47. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–9. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin-Trillo M, et al. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development (Cambridge, England) 2006;133:1241–52. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- 63.Francis KE, et al. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3913–8. doi: 10.1073/pnas.0608936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barth S, Melchinger AE, Devezi-Savula B, Lübberstedt T. A high-throughput system for genome-wide measurement of genetic recombination in Arabidopsis thaliana based on transgenic markers. Functional & integrative genomics. 2000;1:200–6. doi: 10.1007/s101420000030. [DOI] [PubMed] [Google Scholar]
- 65.Jones GH, Franklin FCH. Meiotic crossing-over: obligation and interference. Cell. 2006;126:246–8. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 66.Melamed-Bessudo C, Yehuda E, Stuitje AR, Levy AA. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 2005;43:458–66. doi: 10.1111/j.1365-313X.2005.02466.x. [DOI] [PubMed] [Google Scholar]
- 67.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation. Cell. 1992;69:439–56. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 68.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–70. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 69.Sanchez-Moran E, Santos J-L, Jones GH, Franklin FCH. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes & development. 2007;21:2220–33. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Copenhaver GP, Browne WE, Preuss D. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:247–52. doi: 10.1073/pnas.95.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanchez-Moran E, Armstrong SJ, Santos JL, Franklin FCH, Jones GH. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics. 2002;162:1415–22. doi: 10.1093/genetics/162.3.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Francis RA, et al. Characterizing the performance of the Conway-Maxwell Poisson generalized linear model. Risk analysis: an official publication of the Society for Risk Analysis. 2012;32:167–83. doi: 10.1111/j.1539-6924.2011.01659.x. [DOI] [PubMed] [Google Scholar]
- 73.Jackson N, et al. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. The EMBO journal. 2006;25:1315–23. doi: 10.1038/sj.emboj.7600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 75.Panizza S, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–83. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 76.Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11383–90. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morillo-Huesca M, Clemente-Ruiz M, Andújar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PloS one. 2010;5:e12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–13. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. The EMBO journal. 2010;29:1446–57. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Savic V, et al. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Molecular cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meier A, et al. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. The EMBO journal. 2007;26:2707–18. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–94. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 83.Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS genetics. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Y, Close TJ, Lonardi S. On the accurate construction of consensus genetic maps. Computational systems bioinformatics/Life Sciences Society. Computational Systems Bioinformatics Conference. 2008;7:285–96. [PubMed] [Google Scholar]
- 85.Borevitz JO, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12057–62. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science (New York, N.Y.) 2007;317:338–42. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 87.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996 [Google Scholar]
- 88.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic acids research. 1999;27:573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome research. 2004;14:1861–9. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Auton A, McVean G. Recombination rate estimation in the presence of hotspots. Genome research. 2007;17:1219–27. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Achim Z, Grothendieck G. zoo: S3 infrastructure for regular and irregular time series. Journal of Statistical Software. 2005;14:1–27. [Google Scholar]
- 92.Berchowitz LE, Copenhaver GP. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nature protocols. 2008;3:41–50. doi: 10.1038/nprot.2007.491. [DOI] [PubMed] [Google Scholar]
- 93.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–23. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arends D, Prins P, Jansen RC, Broman KW. R/qtl: high-throughput multiple QTL mapping. Bioinformatics (Oxford, England) 2010;26:2990–2. doi: 10.1093/bioinformatics/btq565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meyer D, Achim Z, Hornick K. vcd: Visualizing Categorical Data. 2012 [Google Scholar]
- 96.Sellers K, Lotze T. COMPoissonReg: Conway-Maxwell Poisson (COM-Poisson) regression. 2011 [Google Scholar]
- 97.Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes & development. 2004;18:2557–70. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ferdous M, et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS genetics. 2012;8:e1002507. doi: 10.1371/journal.pgen.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.