Abstract
The Tup1-Cyc8 (Ssn6) complex is a well characterized and conserved general transcriptional repressor complex in eukaryotic cells. Here, we report the identification of the Tup1 (TupA) homolog in the filamentous fungus Aspergillus niger in a genetic screen for mutants with a constitutive expression of the agsA gene. The agsA gene encodes a putative alpha-glucan synthase, which is induced in response to cell wall stress in A. niger. Apart from the constitutive expression of agsA, the selected mutant was also found to produce an unknown pigment at high temperatures. Complementation analysis with a genomic library showed that the tupA gene could complement the phenotypes of the mutant. Screening of a collection of 240 mutants with constitutive expression of agsA identified sixteen additional pigment-secreting mutants, which were all mutated in the tupA gene. The phenotypes of the tupA mutants were very similar to the phenotypes of a tupA deletion strain. Further analysis of the tupA-17 mutant and the ΔtupA mutant revealed that TupA is also required for normal growth and morphogenesis. The production of the pigment at 37°C is nitrogen source-dependent and repressed by ammonium. Genome-wide expression analysis of the tupA mutant during exponential growth revealed derepression of a large group of diverse genes, including genes related to development and cell wall biosynthesis, and also protease-encoding genes that are normally repressed by ammonium. Comparison of the transcriptome of up-regulated genes in the tupA mutant showed limited overlap with the transcriptome of caspofungin-induced cell wall stress-related genes, suggesting that TupA is not a general suppressor of cell wall stress-induced genes. We propose that TupA is an important repressor of genes related to development and nitrogen metabolism.
Introduction
The fungal wall is an essential organel. It forms a strong structural barrier that offers protection against mechanical damage, helps to withstand the internal turgor pressure, and maintains and determines the shape of the cell. Developmental stages or dimorphic switches strongly affect the composition of the cell wall, both in structure as well as in the type of cell wall mannoproteins that are incorporated into the cell wall [1]–[4]. The cell wall also contributes to invasion of sturdy substrates, and the formation of multi-cellular structures. The structural components of the wall mainly consist of polysaccharides, such as polymers of glucose (β-1,3- and β-1,6-glucan, and chitin, which consists of β-1,4-linked N-acetyl-glucosamine residues [5], [6]. In addition, filamentous fungal walls including those of Aspergillus species often contain α-glucans, β-1,3-1,4-glucan, galactomannan, galactosaminogalactan and galactomannoproteins [7]–[9]. The actual cell wall composition not only depends on the fungal species, but its composition is also highly dependent on environmental factors and developmental stages [10].
Many (pathogenic) fungi are able to switch from yeast to filamentous growth. This is accompanied by major changes in cell wall composition. The dimorphic switch has been extensively studied in Candida albicans and this has shown that the expression of cell wall genes is highly dynamic during the yeast to hyphal transition [11], [12]. Moreover, in pathogenic dimorphic fungi like Histoplasma capsulatum, Cryptococcus neoformans, Blastomyces dermatitidis, and Paracoccidioides brasiliensis, the virulent yeast form contains substantial levels of α-glucan (35–46% of total cell wall carbohydrates) in comparison to dramatically decreased levels in the avirulent mycelial form [13]–[17]. In addition, cell wall stress conditions have been reported to induce important changes in cell wall composition by the specific induction of cell wall remodeling genes or cell wall proteins [18]–[20]. The Cell Wall Integrity (CWI) pathway, a conserved signaling pathway, is one of the most important pathways mediating this response (see [21]–[23] for reviews). In A. niger, activation of the CWI pathway results in a strong transcriptional induction of agsA, an alpha-glucan synthase-encoding gene [20], [24]. The induced transcription of agsA in response to cell wall stress is mediated via a highly conserved Rlm1p-like MADS-box transcription factor protein, called RlmA [24].
The Tup1-Cyc8(Ssn6) complex is a general transcriptional co-repressor complex that controls the expression of genes involved in various processes. This complex is especially well studied in the yeast Saccharomyces cerevisiae, and mutational and genome-wide expression studies have shown that Tup1 is responsible for the repression of over 180 genes, including gene sets regulated by glucose, DNA damage, mating type, and oxygen availability, and gene sets involved in osmotic stress responses, flocculation, and dimorphism [25]–[28]. Recent studies have shown that the repressor function of Tup1-Cyc8 is caused by the interaction of the complex with a specific DNA-binding domain, thereby preventing the recruitment of transcriptional co-activators [29], [30]. The important role of Tup1/TupA in pathogenic fungi has received further attention because of its important role in dimorphism and pathogenicity. Although the role of Tup1 in fungal dimorphism is conserved, the way it controls the switch differs between fungi [12], [31]–[33].
We previously reported about the isolation of UV-mutants showing a constitutive high expression of the agsA gene by selection for improved growth on acetamide as sole nitrogen source and for the presence of GFP-labeled, fluorescent nuclei [34]. For this, a dual reporter strain was used that contained a construct with the amdS sequence (coding for an acetamidase) and the Histone2B-GFP sequence both cloned behind an agsA promoter region. In this study, we describe a mutant with a constitutive expression of the agsA gene and show that the mutant is mutated in the A. niger TupA homolog. The tupA (An15g00140) mutant in A. niger displays in addition to induced expression of agsA a strongly reduced radial growth rate, increased branching, and abundant secretion of an unknown pigment into the medium. We present further genome-wide transcriptomic consequences of the mutation in the co-repressor complex and focus on the impact of tupA on the transcriptional control of cell wall biosynthetic genes in Aspergillus niger. The genome-wide study combined with phenotypic analysis of the tupA strains also suggests that TupA is an important repressor of genes related to nitrogen metabolism, which might explain the important role of TupA in relation to dimorphic switching in dimorphic fungi.
Materials and Methods
Strains, Plasmids, Cosmids, and Growth Conditions
The A. niger strains used in this study are listed in Table 1. Strains were grown on minimal medium (MM) [35] containing 1% (w v−1) glucose or on complete medium (CM), containing 0.5% (w v−1) yeast extract and 0.1% (w v−1) casamino acids in addition to MM-glucose. When required, plates or medium were supplemented with 10 mM uridine, SDS (50 µg/ml), Calcofluor White (50–400 µg/ml), caspofungin (0.2–1.5 µg/ml), or with sorbitol (1.2 M) to assay growth. MM agar plates containing acetamide as sole nitrogen source were made as described [36].
Table 1. Strains used in this study.
| Strain | Description | Reference |
| N402 | cspA1 derivative of ATCC9029 | [89] |
| AB4.1 | pyrG − derivative of N402 | [90] |
| MA169.4 | kusA::DR-amdS-DR pyrG − | [43] |
| RD15.8 | pPagsA-H2B-GFP-TtrpC-pyrG* and pPagsA-amdS-TamdS/pAN7.1 | [34] |
| RD15.8#36 | pPagsA-H2B-GFP-TtrpC-pyrG* and pPagsA-amdS-TamdS/pAN7.1 | [34] |
| DSC12 | pyrG − derivative of RD15.8#36 | this study |
| DSC13 | DSC12 containing pMA172-An15g00140 (tupA) | this study |
| MA245.1 | ΔtupA::pyrG in MA169.4 | this study |
| MA246.1 | ΔtupA::pyrG in RD15.8 pyrG − | this study |
| SM2.36 | ΔtupA::pyrG AB4.1 pyrG − | this study |
Targeted integration of constructs at the pyrG locus using the pyrG* allele was done as described [37]. E. coli DH5α strains were transformed by electroporation for propagation and amplification of the cosmids. Amplification of plasmid DNA was performed using the XL1-Blue strain, which was transformed using the heat-shock protocol as described by [38]. Transformation of A. niger was performed as described by Meyer et al. [39] using 40 mg lysing enzyme (L-1412, Sigma, St. Louis) per gram wet weight of mycelium. A. niger genomic DNA (including cosmid DNA) was isolated as described previously [39]. [α-32P]dCTP-labeled probes were synthesized using the Rediprime II DNA labeling system (Amersham Pharmacia Biotech, Piscataway, NJ) according to the instructions of the manufacturer. All molecular techniques were carried out as described [40]. Sequencing was performed by Macrogen Europe (Amsterdam, The Netherlands).
Bioreactor Cultivations
Bioreactor cultivations were carried out as described previously. Fermentation medium (FM) adjusted to pH 3, is composed of 0.75% glucose, 0.45% NH4Cl, 0.15% KH2PO4, 0.05% KCl, 0.05% MgSO4, 0.1% trace element solution and 0.003% yeast extract as described [41]. Freshly harvested conidia (5×109) from strain N402 and RD15.8#36 were used to inoculate 5 liters of FM. Cultivations were performed in a BioFlo3000 bioreactor (New Brunswick Scientific), where the temperature, pH (set to 3), and agitation speed were controlled online using the program NBS Biocommand. The cultivation program consisted of two consecutive phases: (i) 30°C, agitation speed of 250 rpm, and headspace aeration, for the first 5 h; (ii) 30°C, agitation speed of 750 rpm, and sparger aeration during the second phase. Mycelial samples were taken after specific time points for microarray, metabolic, and microscopic analyses.
Identification and Cloning of tupA
RD15.8#36 was made pyrG − by selecting 5-fluoroorotic acid (5-FOA)-resistant mutants as described [39]. One of the generated pyrG − mutants was transformed with a genomic cosmid library in an AMA1-containing pyrG-based self-replicating vector [40]. Cosmids from transformants that show complementation of the growth-deficient phenotype at 30°C were isolated. To re-isolate the complementing cosmid, DNA from transformants was isolated and used for transformation to E. coli DH5α by electroporation. Fresh ampicillin (amp)-resistant transformants were transferred to 50 ml LB-amp medium and grown overnight at 37°C, and cosmid DNA was isolated using a small-scale isolation protocol essentially as described (Sambrook et al., 1989). Primers cosT7 and cosUL were used for sequencing the borders of the insert [34]. Subclones were generated by digestion of the cosmids with various enzymes, and fragments were ligated into properly digested pBluescriptSK(+). The generated subclones were co-transformed with pAB4.1 to RD15.8#36 pyrG −. The 10.1-kb HindIII subclone giving complementation was subject to further analysis and sequencing.
For complementation studies the An15g00140 locus, including approximately 2.3-kb promoter and 1.4-kb terminator regions, was PCR-amplified using N402 genomic DNA as template and the NotI site-containing primers P1_0140_For and P2_0140_Rev (Table 2). The fragments were cloned into pJET1.2 (Fermentas), sequenced, released from pJET1.2 via NotI restriction and cloned into NotI-linearized pMA172 [43]. Respective plasmids (pMA172-0140 and pJET-0140) were then transformed into RD15.8#36 pyrG −. Primary transformants containing the complementation plasmid were isolated on MM without uridine supplementation and further analyzed by Southern blot. To allow pMA172 plasmid loss, spores were streaked on MM containing 10 mM uridine. The DNA sequence of the An15g00140 gene in the parental strain (RD15.8), in the wild-type strain (N402) and in the mutant strains was determined by sequencing. The An15g00140 locus with a 0.7-kb promoter and a 0.7-kb terminator region was amplified with primers P3_0140_For and P8_0140_Rev (Table 2), using genomic DNA of the respective three strains as template DNA. PCR products were directly sequenced with appropriate primers (Table 2).
Table 2. Overview of the primers used in this study.
| Primer | Sequence 5′-3′ | used for | remark |
| P1_0140_For | aaggaaaaaagcggccgcTGAAGTGCCAGCCAGTAGTGG | amplifying locus | NotI underlined |
| P2_0140_Rev | aaggaaaaaagcggccgcTGGGTGATCGTGACTTTACCGCGGTAAAGTCACGATCACCCA | amplifying locus and generating deletion construct 3′ | NotI underlined |
| P3_0140_intI | CGGTCACACTAAGCGCCGTA | sequencing tupA alleles | |
| P4_0140_intII | CGACACAAATCTTTCGCGCTA | sequencing tupA alleles | |
| P5_0140_intIII | TTGCCTGACCTCTGACCTCG | sequencing tupA alleles | |
| P6_0140_intIV | GCTGGCAATGGTCGGTACA | sequencing tupA alleles | |
| P7_0140_intV | CGGAAATGCGCAGATGATG | sequencing tupA alleles | |
| P8_0140_intVI | CGAGAGATTGCATGGCAGC | sequencing tupA alleles | |
| P9_0140_ For | cccaagcttACATGATTTGCTGGCTCCGAC | generating deletion construct | HindIII underlined |
| P10_0140_ Rev | ccgctcgagAGCTGAGGCTGAAGGAGGAG | generating deletion construct | XhoI underlined |
| P11_0140_ For | ggggtaccTGAAGTGCCAGCCAGTAGTGG | generating deletion construct | KpnI underlined |
Generation of a tupA Deletion Strain
An An15g00140 gene disruption cassette, (ΔtupA::pyrG), was prepared by a three-way ligation. 5′ and 3′ regions flanking the coding region were amplified by PCR using the primers listed in Table 2. Fragments were cloned into pJet2.1. The 0.5-kb 5′ region KpnI/XhoI fragment, the 0.5-kb 3′ HindIII/NotI fragment and a 1.7-kb HindIII/XhoI fragment from pAO4-13 [44] containing the A. oryzae pyrG gene, were cloned into the pBluescript-SK+ backbone prepared by digestion with KpnI and NotI to give ΔtupA::pyrG.
Using ΔtupA::pyrG as a template, a PCR with primers P1_0140_For and P11_0140_Rev was performed and the linear deletion fragment was transformed to MA169.4 pyrG − (ku70::DR-amdS-DR pyrG −). Uridine prototrophic transformants were purified twice on MM and subjected to Southern blot analysis [39]. DNA extraction was carried out from mycelia that had been collected from liquid MM cultures containing 0.003% yeast extract.
Microarray and Northern Analysis
(i) RNA isolation and quality control
Culture broth samples (10 ml each) obtained from the above-described bioreactor cultures were quickly harvested and filtered, and the mycelial samples were immediately frozen using liquid nitrogen. Total RNA for Northern and microarray analyses was isolated from frozen, ground mycelium by Trizol extraction according to the manufacturer’s instructions. Following extraction, RNA was purified on NucleoSpin RNA II columns (Machery-Nagel), including a DNase I digestion step. RNA was eluted in 60 µl of MilliQ water. RNA quantity and quality were determined on a Nanodrop spectrophotometer, and integrity was tested on an Agilent 2100 Bioanalyser. The spectrum generated by the Agilent Bioanalyser was visually inspected for possible RNA degradation and contamination with genomic DNA to ensure good sample quality.
Microarray Analysis and Bioinformatics
Microarray analyses for N402 and RD15.8#36 were performed on mycelia obtained during the exponential growth phase from two independent bioreactor cultivations (biological duplicates), when 75% of glucose had been consumed. Probe synthesis and fragmentation were performed at ServiceXS (Leiden, Netherlands) according to the GeneChip Expression Analysis Technical Manual (Affymetrix Inc. 2002). DSM (Delft, Netherlands) proprietary A. niger gene chips were hybridized, washed, stained, and scanned as described in the Gene-Chip Expression Analysis Technical Manual (Affymetrix Inc. 2002). The generated transcriptomic data set and description of the Affymetrix gene chip used are deposited at the Gene Expression Omnibus database and can be accessed via their accession numbers GSE50523 and GPL6785, respectively. For transcriptomic data analysis, the statistical programming language R as used including open source and open development packages of the Bioconductor project [45]. Affymetrix probe-level data from CEL files were preprocessed using the Robust Multi-Array average (RMA) [46] algorithm as implemented in the Affy package [47]. For transcripts targeted by multiple probe sets, average expression values were computed prior to the identification of differentially expressed genes with the limma package [48]. The proportion of false positives was controlled by calculating the false discovery rate (FDR) according to the method of Benjamini and Hochberg [49] and controlled at 0.005 without a minimal fold change criterion. Applying a critical FDR of 0.05, Gene Ontology (GO) [50] enrichment analysis for differentially expressed gene sets was performed using Fisher’s exact Test Gene Ontology annotation tool (FetGOat) [51] including its most recent GO annotation.
Results
Phenotypic Analysis of A. niger Mutant RD15.8#36
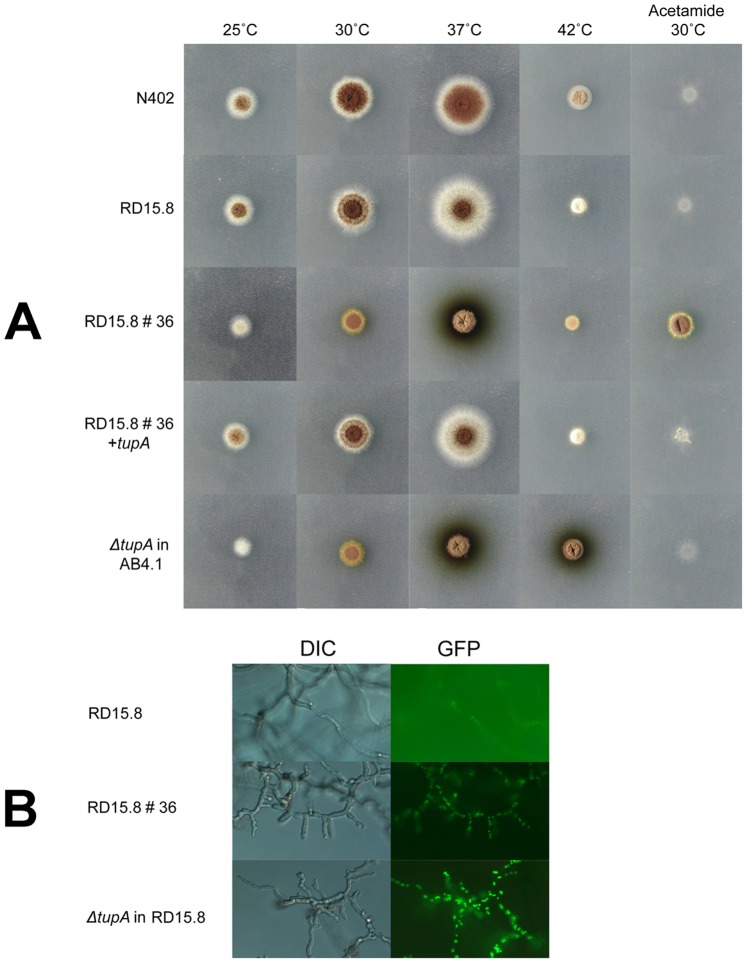
We previously reported the use of a genetic screen to isolate mutants with an induced expression of agsA, a gene encoding a putative α-glucan synthase. In this screen, a reporter strain is used that contains two reporter constructs: the agsA promoter fused to the A. nidulans acetamidase (amdS) gene and the agsA promoter fused to H2B-GFP [34]. Since agsA is specifically induced in response to cell wall stress conditions [20], this screen is expected to yield cell wall mutants with a constitutively activated cell wall integrity pathway resulting in agsA expression. The agsA-amdS reporter allows direct selection for mutants that can grow on acetamide and the second reporter was included to check for cis-acting mutations in the agsA-amdS promoter region and for mutants in which the expression of endogenous acetamidase was deregulated. In comparison to the parental strain (RD15.8), mutant RD15.8#36 shows clear induction of the two agsA reporter constructs, which results in strongly improved growth on acetamide medium at 30°C and nuclei that show enhanced green fluorescence, consistent with increased expression of the agsA gene (Figure 1a and 1b). The mutants showed several growth-related phenotypes including retarded spore germination (germination of RD15.8#36 occurred about 12 h later than in the parental strain (data not shown), and a strongly reduced radial growth rate (Figure 1a). Interestingly, at 37°C, the mutant secreted an enhanced amount of dark green- to brown-colored pigment in the medium (Figure 1a). We did not observe increased sensitivity of the mutant towards the cell wall-perturbing compounds caspofungin and Calcofluor White (CFW) or to the cell membrane-perturbing agents SDS and fenpropimorph, an inhibitor of ergosterol biosynthesis (data not shown), which would have been indicative of defects in cell wall synthesis [52], [53]. Attempts to improve the growth of the RD15.8#36 mutant by supplementing the culture medium with 1.2 M sorbitol were only partly successful (data not shown). This suggests that despite the induced expression of agsA the mechanical strength of the mutant wall was not significantly affected.
Figure 1. Phenotypic characterization of the tupA mutants.
(a) Ten thousand spores of UV mutant RD15.8#36, the parental strain RD15.8, the complemented mutant RD15.8#36/pAn14g00140, the full deletion mutant ΔtupA and wild-type N402, were spotted and growth was monitored on MM-glucose-nitrate plates or MM/glucose-acetamide plates acetamide (first vertical column or MM-glucose plates (if not stated differently) at the indicated temperature for 3 days. (b) DIC- and fluorescent pictures of parental strain (RD15.8), the tupA mutant, and the tupA deletion strain in the RD15.8 background after growth for 20 hours at 30°C in MM with casamino acids.
The A. niger Homolog of the General Transcriptional Repressor TupA Complements A. niger Mutant RD15.8#36
To identify the mutated gene causing the phenotypes of the RD15.8#36 mutant, a pyrG − derivative was made and transformed with a pyrG-based genomic cosmid library [42]. Initially, circa 1,000 uridine-prototrophic transformants were directly selected for enhanced growth at 30°C and one hundred of those were purified and retested for wild-type growth at 30°C on acetamide and minimal medium. Cosmids were isolated from ten transformants with a strong complementation phenotype and EcoRI restriction of the cosmids revealed the isolation of identical cosmid clones that could complement the RD15.8#36 phenotypes. Sequence analysis of the insert borders revealed the presence of two different chromosomal fragments, which complicated identification of the responsible gene. Hence, the isolated cosmid was restricted with various enzymes and individual fragments were co-transformed with pAB4.1 to RD15.8#36pyrG −. A 10.1-kb HindIII fragment that complemented the reduced growth rate was subsequently cloned and sequenced. The HindIII subclone harbored a single full-length predicted open reading frame, namely, An15g00140. This gene was amplified via PCR and ligated into the autonomously replicating vector pMA172 [43] and the resulting plasmid was transformed to RD15.8#36pyrG −. As shown in Figure 1a, An15g00140 complemented the different phenotypes of the RD15.8#36. This indicates that a single gene is responsible for these phenotypes.
Comparison of the protein sequence generated from the An15g00140 gene revealed that the protein displays strong sequence similarity to the transcriptional repressor RcoA of A. nidulans and to Tup1 of S. cerevisiae, and we will refer to this gene as tupA.
Characterization of tupA Mutants in the Cell Wall Mutant Collection
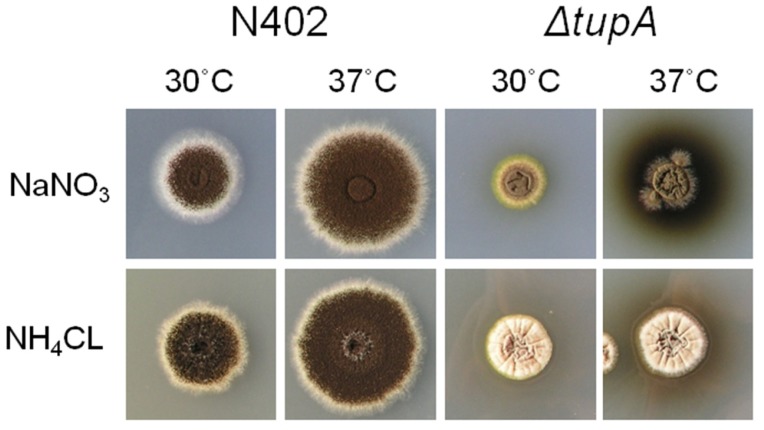
The 240 cell wall mutants of A. niger that were isolated via a genetic screen for high agsA expression [34] and yielded the identification of TupA as described above, was screened for pigment-producing mutants at high temperature. Apart from mutant RD5.18#36, sixteen more mutants were identified that secreted the pigment at high temperature in a nitrogen source-dependent manner (data not shown). To verify whether these 17 mutants were all mutated in tupA or otherwise affected in agsA expression, the tupA locus of each of the mutant was PCR amplified and sequenced. All mutants were mutated in the tupA gene (Table 3). We were unable to PCR amplify the tupA locus from mutant tupA-14 indicating rearrangement of the tupA locus in this strain. A variety of mutations including point mutations, insertions, deletions, missense mutations and mutations affecting intron splicing were detected. Tup1 is known to interact with Cyc8/Ssn6 and deletion of either tupA or cys8/ssn6 leads to similar phenotypes [26]. Because of the saturating high number of tupA mutants identified by screening and the lack of any other mutant with a similar phenotype but not tupA, we suspect that mutations in the Cyc8/Ssn6 homolog in A. niger (An02g03940) do not lead to a strong induction of the agsA promoter. The sequencing of the mutant alleles also confirms that in our original mutants the tupA locus was mutated and that tupA was not acting as a suppressor gene in our complementation studies. To investigate the consequences of a loss of function of the tupA gene in A. niger, a gene deletion vector (pΔtupA::pyrG) was constructed, in which the ORF was replaced by the A. oryzae pyrG gene. After transformation, primary transformants were purified twice on MM lacking uridine, and stable transformants were obtained. Proper disruption of the tupA gene in several transformants was confirmed by Southern blot analysis (data not shown). The phenotypes of the ΔtupA deletion mutant were similar to tupA mutant (RD15.8#36) (Figure 1a and b). The phenotype of the tupA deletion strain was characterized on glucose medium containing either nitrate or ammonium as a nitrogen source. The phenotype was characterized by a reduced radial growth rate (approximately 50% reduced after 7 days of incubation) and a strong reduction (∼80%) in conidiation in the presence of ammonium but not in the presence of nitrate (Table 4). We also noticed that the production of the pigment was highest when the tupA mutant grew at high temperatures (37°C) on nitrate. Equivalent amounts on ammonium (10 mM) strongly repressed pigment formation (Figure 2).
Table 3. Overview of mutations in A niger tupA mutants.
| Strain | Allel | Position of mutation inORF (including introns) | Consequence of the mutation (s) | Protein sequence(WT: 1–590) | |
| RD6.47#28 | tupA-1 | GA deletion (61, 62) | out of frame after aa 21 | truncation | 1–21 |
| RD6.13#22 | tupA-2 | TA to AT (1561, 1562) | ATT to ATA (silent I remains I);AAG to TAG (K to stop) | truncation | 1–358 |
| RD6.13#53 | tupA-3 | A to C (487) | mutation in 3′ intron splice siteTAG to TCG | truncation | 1–60 |
| RD15.4#2 | tupA-4 | G to C (2351) | TGG to TCG (W to S) | point mutation | 1–590 (W to S at 575) |
| RD15.4#3 | tupA-5 | G to A (889) | mutation in 3′ intron splice siteCAG to CAA | truncation | 1–170 |
| RD15.4#24 | tupA-6 | extra T (1355) | Out of frame after aa 316 | truncation | 1–316 |
| RD15.4#26 | tupA-7 | extra T (786) | Out of frame after aa 160 | truncation | 1–160 |
| RD15.4#27 | tupA-8 | C to T (2128) | TCT to TTT (S to F) | point mutation | 1–590 (S to F at 489) |
| RD15.4#29 | tupA-9 | A to T (1772) | AAG to TAG (K to stop) | truncation | 1–473 |
| RD15.4#37 | tupA-10 | C to T and C deletion(669, 670) | GGC to GGT (Silent G remains G);out of frame after aa 122 | truncation | 1–122 |
| RD15.4#47 | tupA-11 | TG to GA (1664, 1665) | TGG to GAT (W to D) | point mutation | 1–590 (W to D at 393) |
| RD15.4#49 | tupA-12 | C to T (43) | CAA to TAA (Q to stop) | truncation | 1–14 |
| RD15.4#52 | tupA-13 | T to C (2350) | TTG to CCG (W to R) | point mutation | 1–590 W to R at 575) |
| RD15.4#60 | tupA-14 | no PCR product obtained | |||
| RD15.8#13 | tupA-15 | C to T (1856) | TCG to TTG (S to L) | point mutation | 1–590 (S to L at 457) |
| RD15.8#27 | tupA-16 | G to A (2125) | GGT to GAT (G to D) | point mutation | 1–590 (G to D at 488) |
| RD15.8#36 | tupA-17 | G to A (2144) | TGG to TGA (W to stop) | truncation | 1–519 |
Table 4. Growth and sporulation analysis of wild-type (N402) and ΔtupA (tupA::pyrG in MA169.4) strains after 7 days of growth.
| Strain | N-source | Temp.(°C) | Colonydiameter (cm) | Spores percolony | Spores/cm2 * | Relativegrowth | Relativesporulation |
| WT | ammonium | 30 | 10.0 | 2.0×108 | 2.5×106 | 100% | 100% |
| WT | ammonium | 37 | 14.0 | 6.8×108 | 4.4×106 | 100% | 100% |
| WT | nitrate | 30 | 9.8 | 2.0×108 | 2.6×106 | 100% | 100% |
| WT | nitrate | 37 | 14.9 | 3.0×108 | 1.7×106 | 100% | 100% |
| ΔtupA | ammonium | 30 | 5.8 | 0.16×108 | 0.61×106 | 58.0% | 24.4% |
| ΔtupA | ammonium | 37 | 7.3 | 0.33×108 | 0.79×106 | 52.1% | 18.0% |
| ΔtupA | nitrate | 30 | 5.2 | 0.68×108 | 3.2×106 | 53.1% | 123% |
| ΔtupA | nitrate | 37 | 5.5 | 0.49×108 | 2.1×106 | 36.9% | 124% |
Number of spores of the colony/((Radius of the colony)2×π).
Figure 2. The effects of growth temperature and nitrogen source on pigment formation in the tupA mutant.
High temperatures (37°C) and nitrate are required for optimal production of the pigment. Note also the strong reduction of conidiospores on top of the vegetative mycelium in the tupA mutant grown on ammonium as nitrogen source. Pictures taken after 3 days.
Microarray Analysis of Bioreactor-grown Wild-type and Mutant Strains
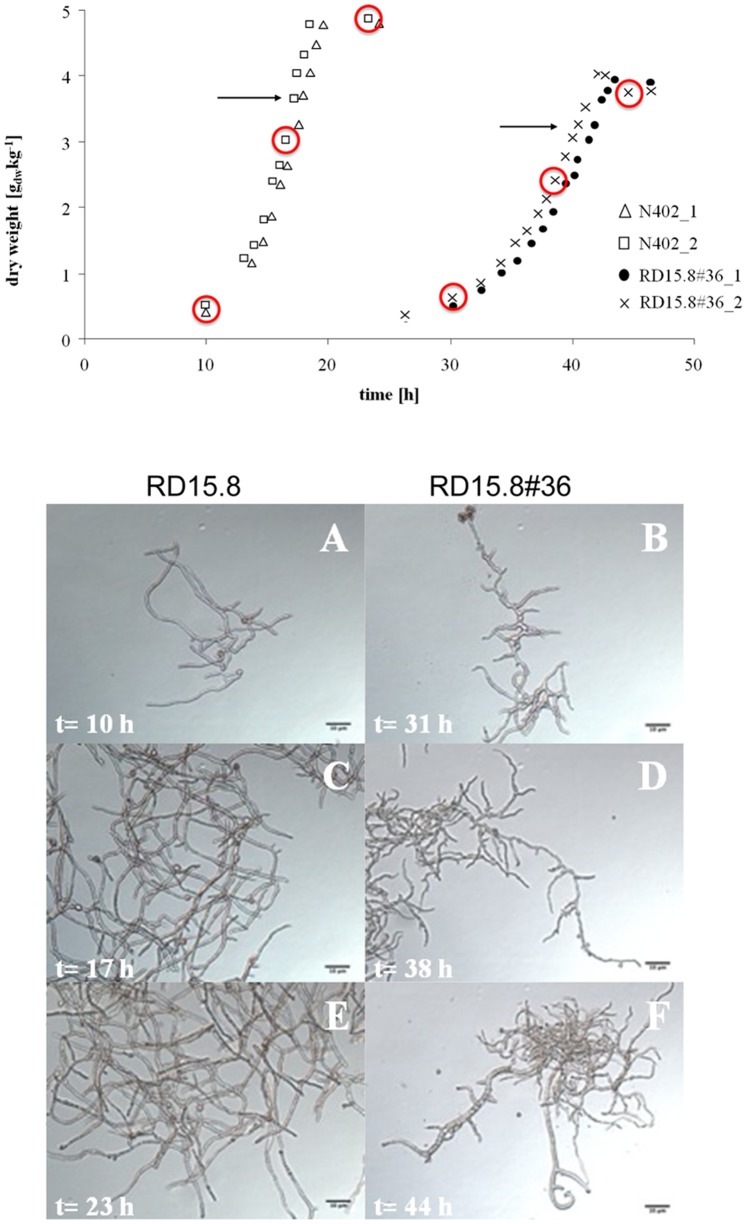
To identify genes in A. niger that are differentially expressed in the tupA mutant compared to the wild-type strain, RNA samples were extracted from the wild-type (N402) and the tupA mutant strain (RD15.8#36). To ensure controlled and reproducible growth conditions, both N402 and RD15.8#36 were cultivated in a bioreactor (see Materials and Methods). As shown in Figure 3, initial spore germination of the tupA mutant was delayed compared to the wild-type strain. The dry weight measurements were used to determine the maximal specific growth rate of both strains under the given growth conditions and this showed that the growth rate of the tupA mutant strain (0.16 h−1) was substantially lower than that of the wild-type strain N402 (0.25 h−1) (data not shown), consistent with the substantial decrease in the radial growth rate when cultured on a solid surface (Figure 1 and 2). The final growth yields (4.0 gDW kg−1 for the mutant and 4.8 gDW kg−1 for N402) differed only moderately and were highly reproducible. At the end of the exponential growth phase the supernatant of the mutant culture turned dark-green to brown whereas the wild-type broth remained colorless. The green-brownish color at this time point was not due to the presence of melanized conidiospores as microscopic observation did not indicate the presence of spores. Conceivably, the metabolic expenditure required for the formation and secretion of these secondary metabolites by the mutant strain might explain its lower biomass yield. As shown in Figure 3 (panels A-F), also under these growth conditions the tupA mutant showed morphological aberrations, which included increased branching, increased variability in hyphal diameters, and curled hyphae. In addition, RD15.8#36 sporadically formed swollen, abnormally shaped, branched hyphae (Figure 3. These morphologic differences might also result in less efficient metabolism and therefore reduced biomass yield.
Figure 3. Physiological and morphological of A. niger RD15.8#36 in comparison to wild-type N402.
(Top figure) Biomass accumulation in the duplicate cultures and the maximum specific growth rate (µmax). The arrows indicate the time point when 75% of glucose was consumed and mycelia were harvested for transcriptomic analysis. (Lower figure (A-F) Morphology of the mutant RD15.8#36 in bioreactor cultures in comparison to the wild-type N402 both grown on FM. Mycelium from the exponentially growing culture was harvested at the indicated time points. Scale bar, 10 µm.
RNA was isolated from both cultures when 75% of the carbon source (glucose) was converted into biomass (3.2 gDW kg−1 for RD15.8#36, 3.7 gDW kg−1 for N402). Two biological replicates for each strain were performed corresponding to four microarray analyses in total.
Genome-wide Expression Analysis of the tupA Mutant in Comparison to the Wild-type Strain
To identify the processes in A. niger affected by the tupA mutation, we first confirmed that the biological reproducibility of gene transcription for the biological duplicates was high, as evidenced by a mean relative standard deviation of 0.05 for both the wild-type and mutant, respectively, and similar to values reported previously [54], [55]. Two thousand and eight out of 14,165 A. niger genes (using a low corrected P-value of <0.005) were differentially expressed (as defined in Materials and Methods). The large size of this set of genes (∼14%) corroborates the importance of tupA as a general transcriptional repressor. Of the 2008 differentially expressed genes 1053 were higher expressed in the tupA mutant compared to wild-type (tupAUP) and 955 genes were down-regulated (tupADOWN). A comprehensive list of all differentially expressed genes including statistical significance and transcript ratios is presented in.
For more insight into the processes that are affected in the tupA mutant, we performed GO-enrichment analysis of the genes that were significantly higher expressed in the tupA mutant using FetGOat [51]. The GO-terms (Biological Process) overrepresented in the tupA mutant include 28 GO terms. The GO-terms are given in Table S2. Among those 28 BP terms, 9 terminal nodes were present which are summarized in Table 5. The presence of GO term ´related to carbon metabolic procesś (acetate and pentose metabolic processes), ´related to differentiation and development´ (conidium formation, reproductive processes and sporulation), ´related to stress responsé (oxidative stress) suggest conservation of TupA function among fungi in repressing gene sets under non-inducing conditions.
Table 5. GO terms (Biological Process) enriched among up-regulated genes in the tupA strain.
| Description | FDR | ||
| GO:0006083 | acetate metabolic process | BP | 7,13E-04 |
| GO:0044282 | small molecule catabolic process | BP | 7,46E-03 |
| GO:0015695 | organic cation transport | BP | 1,95E-02 |
| GO:0075307 | positive regulation of conidium formation | BP | 2,65E-02 |
| GO:2000243 | positive regulation of reproductive process | BP | 3,03E-02 |
| GO:0019321 | pentose metabolic process | BP | 3,36E-02 |
| GO:0006979 | response to oxidative stress | BP | 3,95E-02 |
| GO:0045881 | positive regulation of sporulation resulting in formation of a cellular spore | BP | 3,95E-02 |
| GO:0033609 | oxalate metabolic process | BP | 4,29E-02 |
About half of the differentially expressed genes were down-regulated in the tupA mutant and also for these genes a GO enrichment analysis was performed. Sixty-seven GO terms (BP-terms) were enriched, which are summarized in Table S3. Among the sixty-seven enriched GO-terms, 19 terminal nodes were present, which are summarized in Table 6. Most of the terms are associated with metabolic and biosynthetic process, which may be caused by the slower growth rate of the tupA mutant. The inactivation of TupA not only affects the expression of a large number of genes, but the fold-changes of many of these genes were also remarkable. Seventy-five genes were higher expressed in the tupA strain with a FC>10 and 77 genes were down-regulated with a FC>10 (Tables S4 and S5).
Table 6. GO terms (Biological Process) enriched among down-regulated genes in the tupA strain.
| Description | FDR | ||
| GO:0009108 | coenzyme biosynthetic process | BP | 3,56E-03 |
| GO:0046355 | mannan catabolic process | BP | 4,92E-03 |
| GO:0006534 | cysteine metabolic process | BP | 8,41E-03 |
| GO:0006879 | cellular iron ion homeostasis | BP | 9,53E-03 |
| GO:0019184 | nonribosomal peptide biosynthetic process | BP | 1,10E-02 |
| GO:0042364 | water-soluble vitamin biosynthetic process | BP | 1,64E-02 |
| GO:0006544 | glycine metabolic process | BP | 1,99E-02 |
| GO:0006066 | alcohol metabolic process | BP | 2,16E-02 |
| GO:0071577 | zinc ion transmembrane transport | BP | 2,92E-02 |
| GO:0006733 | oxidoreduction coenzyme metabolic process | BP | 2,92E-02 |
| GO:0043156 | chromatin remodeling in response to cationstress | BP | 2,92E-02 |
| GO:0019662 | non-glycolytic fermentation | BP | 2,92E-02 |
| GO:0009082 | branched chain family amino acidbiosynthetic process | BP | 3,19E-02 |
| GO:0000301 | retrograde transport, vesicle recyclingwithin Golgi | BP | 3,36E-02 |
| GO:0006555 | methionine metabolic process | BP | 4,12E-02 |
| GO:0015812 | gamma-aminobutyric acid transport | BP | 4,12E-02 |
| GO:0008219 | cell death | BP | 4,12E-02 |
| GO:0010106 | cellular response to iron ion starvation | BP | 4,45E-02 |
| GO:0015893 | drug transport | BP | 4,92E-02 |
In the following sections we will focus on genes related to synthesis of the fungal cell wall and to development and highlight some of the most significant changes in gene expression (Tables 7–10).
Table 7. Cell wall biosynthetic genes up-regulated in the tupA mutant.
| Gene number | Gene name | Description | tupA | WT | FC | P-value |
| An08g03580 | bgtA | Putative beta-1,3-glucanosyltransferase (GH17-family) | 1669 | 37 | 44.6 | 2.28E-07 |
| An18g04100 | exgA | Putative exo-beta-1,3-glucanase (GH5-family) | 907 | 45 | 20.1 | 1.46E-06 |
| An02g03980 | kslA | Putative transglycosidase required for beta-1,6 glucan biosynthesis; ScKre6-like | 179 | 32 | 5.5 | 3.21E-06 |
| An01g06500 | dfgD | Putative endo-mannanase with a possible role in GPI-CWP incorporation; ScDfg5-like(GH76-family) | 152 | 29 | 5.2 | 1.02E-06 |
| An16g02850 | crhF | Putative transglycosidase involved in cell wall biosynthesis (GH16-family) | 673 | 135 | 5.0 | 5.78E-06 |
| An02g09050 | gelG | GPI- anchored beta-1,3-glucanosyltransferase | 362 | 78 | 4.6 | 2.21E-05 |
| An05g00130 | knlA | Putative transglycosidase required for beta-1,6 glucan biosynthesis; ScKre9-like | 1008 | 219 | 4.6 | 1.31E-06 |
| An15g07800 | agtC | GPI-anchored alpha-glucanotransferase | 85 | 21 | 4.1 | 9.14E-06 |
| An07g04650 | bgtB | Putative beta-1,3-glucanosyltransferase (GH17-family) | 381 | 120 | 3.2 | 5.76E-06 |
| An15g07810 | agsB | Putative catalytic subunit alpha-glucan synthase complex | 82 | 31 | 2.7 | 5.40E-04 |
| An01g12450 | bxgA | Putative exo-beta-1,3-glucanase (GH55-family) | 1122 | 473 | 2.4 | 5.87E-05 |
| An08g09030 | cfcB | Putative ClassV Chitinase (GH18) | 57 | 24 | 2.4 | 1.01E-04 |
| An07g07530 | crhB | Putative transglycosidase involved in cell wall biosynthesis (GH16-family) | 1589 | 753 | 2.1 | 9.72E-04 |
| An10g00400 | gelA | GPI- anchored beta-1,3-glucanosyltransferase | 2918 | 1435 | 2.0 | 1.02E-04 |
| An09g04010 | chsB | Putative chitin synthase Class III | 1574 | 819 | 1.9 | 4.51E-04 |
| An02g10490 | – | Putative endo-1,3(4)-beta-glucanase | 457 | 262 | 1.7 | 5.18E-04 |
| An03g05260 | csnA | similarity to chitosanase csnA - Aspergillus oryzae | 72 | 44 | 1.6 | 9.15E-04 |
| An01g04560 | mlgA | strong similarity to mixed-linked glucanase precursor MLG1 - Cochliobolus carbonum | 194 | 125 | 1.6 | 4.33E-03 |
| An02g13180 | bgxB | Putative exo-beta-1,3-glucanase (GH55-family) | 38 | 25 | 1.5 | 1.56E-03 |
| An07g01540 | rotA | Protein with putative role in beta-1,6 glucan biosynthesis | 735 | 506 | 1.5 | 2.39E-03 |
Table 10. Predicted GPI-anchored cell wall protein-encoding genes down-regulated in the tupA mutant.
| Gene number | Gene name | Description | tupA | WT | FC | P-value |
| An07g05670 | Putative cell wall protein | 196 | 342 | 1.7 | 6.45E-04 | |
| An12g07750 | Putative cell wall protein serine threonine rich | 141 | 1076 | 7.6 | 1.89E-03 | |
| An16g07950 | Putative cell wall protein serine threonine rich | 109 | 7139 | 65.4 | 6.98E-08 |
Differential Expression of Cell Wall Biosynthesis and Remodeling Genes
The A. niger tupA mutant was identified in a plate screen for mutants with an increased expression of agsA. We therefore looked specifically if the agsA gene was also induced in the microarray data set. Surprisingly, the agsA gene was not induced in the tupA mutant under the growth conditions used in the bioreactor (normalized expression of agsA in the tupA mutant: 294 vs normalized expression wild-type: 254; FC 1.2; FDR 2.45E-01). Indeed, Northern analysis of additional samples taken from the bioreactor runs confirmed that agsA was not induced during the bioreactor cultivation (data not shown), whereas the expression of agsA on plate was clearly induced. We investigated whether nitrogen source was influencing the expression of agsA as the plate screen was carried out on minimal medium containing acetamide as a nitrogen source whereas the bioreactor cultivation was carried out in minimal medium containing ammonium as a carbon source. To also examine the effect of the pH, spores of the tupA mutant containing the PagsA-H2B-GFP reporter were allowed to germinate in nitrate (pH3.0 and pH6.0) and ammonium (pH3.0 and pH6.0) medium and the fluorescence was analyzed. Under all condition, we observed fluorescent nuclei to a similar strength, indicating that neither the low pH nor the nitrogen source explained for the absence of agsA expression in the bioreactor (Figure S1). It is important to note that the bioreactor condition represents only a single condition and thus only a small fraction of the genes affected by the tupA deletion will be revealed. For example transcriptomic consequences in relation to the loss of TupA such as conidiation, carbon stress, nitrogen stress, effects of pH and DNA damage conditions will not be detected. Further experiments are required to understand in detail the effects of loss of TupA repression in general and of agsA expression in particular. Several factors, including the physiology of the fungus (exponential growth in the bioreactor and a mix of exponential growing mycelium (edge) and stationary phase mycelium (central parts of the colony) [56], [57], or different expression dependent on the water content [58]–[59] could account for these differences. The morphological differences of the tupA strain still raised the question whether the transcription of other cell wall formation-associated genes was altered in the tupA strain. To examine this, the cell wall-related genes that were annotated as such [60] were examined. This analysis identified 20 up-regulated genes involved in the biosynthesis of cell wall polysaccharides (Table 7). Especially, genes related to β-glucan processing were significantly up-regulated, but several genes involved in chitin and α-glucan synthesis were up-regulated in the tupA mutant as well (Table 7). Two genes (bgt1/An08g03580 and exgA/An18g04100), both enzymes involved in β-glucan synthesis, were very highly expressed in the tupA mutant. In addition, the expression of genes predicted to encode structural (GPI-anchored) cell wall proteins [60] were analyzed (Table 8). Fifteen cell wall protein-encoding genes were higher expressed indicating that the cell wall protein composition has changed in the tupA mutant. One of these proteins, CwpA, has been shown to encode a GPI-anchored cell wall protein that is expressed during stationary phase, but before conidiation markers such as brlA and rodA [61]. The higher expression of bgt1 and cwpA in the tupA mutant was also confirmed by Northern blot analysis (Figure S2).
Table 8. Predicted (GPI-anchored) cell wall protein encoding genes up-regulated in the tupA mutant.
| Genenumber | Genename | Description | PredictedGPI-anchor | tupA | WT | FC | P-value |
| An15g07790 | putative cell wall protein; serine/threonine rich | yes | 2412 | 32 | 74.4 | 4.40E-07 | |
| An11g02730 | putative cell wall protein; serine/threonine rich | yes | 743 | 23 | 32.5 | 1.68E-07 | |
| An02g09010 | putative cell wall protein; serine/threonine rich | yes | 1464 | 48 | 30.3 | 2.75E-07 | |
| An14g02100 | cwpA | hydrogen fluoride extractable GPI-anchored cell wall protein | yes | 1433 | 97 | 14.7 | 1.75E-06 |
| An16g07920 | putative cell wall protein | yes | 5261 | 403 | 13.1 | 9.09E-07 | |
| An16g01780 | putative cell wall protein | yes | 9574 | 808 | 11.8 | 3.41E-06 | |
| An06g01000 | putative cell wall protein; serine/threonine rich | yes | 4951 | 699 | 7.1 | 1.47E-05 | |
| An12g00140 | putative cell wall protein; serine/threonine rich | yes | 763 | 113 | 6.8 | 9.53E-07 | |
| An18g06360 | putative cell wall protein; serine/threonine rich | yes | 689 | 137 | 5.0 | 9.96E-07 | |
| An07g04620 | putative cell wall protein | yes | 2155 | 493 | 4.4 | 3.28E-04 | |
| An01g05230 | putative cell wall protein; serine/threonine rich | yes | 6536 | 1940 | 3.4 | 7.03E-06 | |
| An07g06210 | putative cell wall protein; serine/threonine rich | yes | 738 | 231 | 3.2 | 4.29E-05 | |
| An14g01820 | phiA | strong similarity to cell wall protein binB - Aspergillus nidulans | no | 1966 | 611 | 3.2 | 1.02E-04 |
| An02g00120 | putative cell wall protein serine/threonine rich | yes | 771 | 403 | 1.9 | 1.69E-03 | |
| An18g03730 | putative cell wall protein serine/threonine rich | yes | 3086 | 1895 | 1.6 | 1.24E-03 |
In addition to higher expressed cell wall-related genes, 19 genes encoding cell wall biosynthetic enzymes and three genes encoding GPI-anchored cell wall proteins were lower expressed in the tupA mutant (Table 9 and 10 respectively). Most dramatically down-regulated are a putative endo-mannanase of the DFG family, a GPI-anchored chitinase, a putative alpha-glucanase (mutA) and a GPI-anchored cell wall protein of unknown function. The differential expression of mutA was confirmed by Northern blot analysis (Figure S2). Collectively, the observations presented in Tables 7, 8, 9, 10 strongly indicate that TupA plays an important role in regulating the formation and remodeling of the cell wall.
Table 9. Cell wall biosynthetic genes down-regulated in the tupA mutant.
| Genenumber | Genename | Description | tupA | WT | FC-down | P-value |
| An11g01240 | dfgH | Putative endo-mannanase (GH76-family) with a possible role in GPI-CWP incorporation | 42 | 503 | 12.0 | 1.67E-07 |
| An09g06400 | ctcA | Predicted GPI-anchored protein. Putative ClassIII Chitinase (GH18) | 161 | 1852 | 11.5 | 3.26E-07 |
| An08g09610 | agnD | Putative alpha-1,3-glucanase GH71 | 324 | 2187 | 6.7 | 8.15E-07 |
| An06g00360 | dfgF | Putative endo-mannanase (GH76-family) with a possible role in GPI-CWP incorporation | 131 | 753 | 5.8 | 8.84E-07 |
| An07g01160 | crhC | Predicted GPI-anchored protein. Putative transglycosidase of GH16-family | 153 | 584 | 3.8 | 8.69E-06 |
| An08g07350 | gelB | Predicted GPI-anchored protein. Putative 1,3-beta-glucanosyltransferase GH72 | 224 | 713 | 3.2 | 1.78E-05 |
| An09g00670 | gelD | Predicted GPI-anchored protein. Putative 1,3-beta-glucanosyltransferase GH72 | 1424 | 4169 | 2.9 | 8.65E-06 |
| An04g04670 | cfcC | Putative ClassV Chitinase (GH18) | 232 | 609 | 2.6 | 3.08E-05 |
| An13g02510 | crhE | Predicted GPI-anchored protein. Putative transglycosidase of GH16-family | 83 | 213 | 2.6 | 4.88E-05 |
| An01g11010 | crhD | Predicted GPI-anchored protein. Putative transglycosidase of GH16-family | 256 | 641 | 2.5 | 6.91E-05 |
| An12g10380 | chsE | Putative chitin synthase ClassIII | 1203 | 2556 | 2.1 | 3.43E-04 |
| An16g08090 | dfgE | Putative endo-mannanase (GH76-family) with a possible role in GPI-CWP incorporation | 281 | 597 | 2.1 | 1.44E-04 |
| An12g02450 | agsC | Putative catalytic subunit alpha-glucan synthase complex | 161 | 323 | 2.0 | 1.63E-04 |
| An08g05290 | chsG | Putative chitin synthase ClassVI | 28 | 55 | 2.0 | 8.78E-05 |
| An02g07020 | cfcA | Putative ClassV Chitinase (GH18) | 132 | 258 | 2.0 | 5.38E-04 |
| An11g07660 | exgB | Putative exo-1,3-beta-glucanase | 144 | 272 | 1.9 | 2.36E-04 |
| An09g03070 | agsE | Putative catalytic subunit alpha-glucan synthase complex | 376 | 699 | 1.9 | 1.88E-03 |
| An09g06260 | agnC | Putative alpha-1.3-glucanase GH71 | 355 | 571 | 1.6 | 9.68E-04 |
| An12g02460 | agtB | GPI-anchored alpha-glucanotransferase | 328 | 483 | 1.5 | 2.48E-03 |
| Cell wall signaling | ||||||
| An04g10140 | mltB | Putative plasma membrane sensor required for cell wall integrity signalling | 47 | 235 | 5.0 | 3.27E-05 |
Differential Expression of Asexual Development and Secondary Metabolite Production Related Genes
GO enrichment analysis identified the differential expression of asexual development-related genes. In Table 11 the genes and a short description of the gene products are presented. Four transcription factors (flbC, flbD, brlA, and proA) are significantly up-regulated in the tupA mutant during exponential growth. The first three are positive transcription factors required for conidiation in A. nidulans (see [62], [63] for reviews) and for brlA we confirmed that this gene is required for conidiation in A. niger [64]. The ProA transcription factor is homologous to the Pro1/NosA transcription factor which is required for sexual development in Sordaria macrospora and a repressor of sexual development in Aspergillus nidulans [65], [66]. The genes ppoA and ppoC encode two fatty acid oxygenases that are required for the production of oxylipins called psi-factors. In A. nidulans, psi factors have been shown to alter the ratio of asexual to sexual sporulation. The lack of synthesis of psi-factor in ppo disruption strains increased and misregulated the activation of sexual development [67], [68] and has been shown to affect brlA expression levels.
Table 11. Developmental genes up-regulated in the tupA mutant.
| Genenumber | Genename | Description | TupA* | WT* | FC | P-value |
| Regulation of Development | ||||||
| An02g05420 | flbC | Putative regulator containing two zinc-finger motifs | 672 | 221 | 3.0 | 1.04E-04 |
| An01g04830 | flbD | strong similarity to myb-like DNA binding protein, required for conidiation | 1916 | 83 | 23.2 | 3.20E-07 |
| An01g10540 | brlA | BrlA C2H2 Zn (II) finger transcription factor required for conidiation | 158 | 22 | 7.1 | 6.61E-06 |
| An04g07400 | proA | Zn(II)2Cys6- transcriptional activator similar to Pro1 | 597 | 135 | 4.4 | 1.68E-04 |
| An05g00480 | stuA | APSES-transcription factor (spatial expression of abaA) | 2202 | 870 | 2.5 | 7.07E-04 |
| An17g01580 | steA | Transcriptional Activator containing homeodomain DNA binding; STE12-LIKE | 427 | 231 | 1.8 | 1.80E-04 |
| An02g09610 | nsdD | GATA-transcription factor, light regulation | 199 | 115 | 1.7 | 4.03E-04 |
| An01g13660 | abr2 | Putative laccase possible role in pigment biosynthesis | 891 | 28 | 31.7 | 7.97E-08 |
| An04g05880 | ppoA | Fatty acid oxygenase for Psi factor production | 211 | 86 | 2.5 | 4.18E-04 |
| An02g07930 | ppoC | Fatty acid oxygenase for Psi factor production | 661 | 90 | 7.4 | 1.72E-06 |
| An12g00710 | esdC | Required for sexual development in A. nidulans negative regulation of conidium formation, positive regulation of sexual sporulation resulting in formation of a cellular spore | 3989 | 804 | 5.0 | 3.94E-05 |
| An14g01820 | phiA | Strong similarity to hypothetical cell wall protein binB; caspofungin induced | 1966 | 611 | 3.2 | 1.02E-04 |
| An08g05100 | veA | Velvet activator induces sexual reproduction A. nidulans | 319 | 124 | 2.6 | 4.15E-05 |
| An12g03660 | CAAX-prenyl cysteine carboxymethyltransferase; a-factor modification; STE14-LIKE | 272 | 114 | 2.4 | 7.99E-05 | |
| An02g03160 | flbA | Regulator of G-protein signalling | 167 | 83 | 2.0 | 3.02E-04 |
| An18g06110 | rgsA | Regulator of G-protein signalling | 180 | 96 | 1.9 | 4.91E-04 |
| An14g02970 | fphA | Red light phytochrome An14g02970 AN9008.2 89.m01927 20173.m00405 | 83 | 46 | 1.8 | 3.85E-03 |
| Hydrophobins | ||||||
| An15g03800 | hypF | Putative hydrophobin | 948 | 22 | 42.7 | 5.52E-08 |
| An07g03340 | hypE | Putative hydrophobin | 1726 | 136 | 12.7 | 2.34E-07 |
| An01g10940 | hypA | Putative hydrophobin | 125 | 43 | 2.9 | 2.62E-05 |
| An04g08500 | rodA | Hydrophobin: strong similarity to rodletless protein rodA - Aspergillus nidulans | 185 | 88 | 2.1 | 1.34E-03 |
| Melanin biosynthesis | ||||||
| An09g05730 | fwnA | polyketide synthase required for melanin synthesis A. niger | 54 | 128 | 0.4 | 3.94E-04 |
| An14g05350 | olvA | strong similarity to hypothetical yellowish-green 1 ayg1 - Aspergillus fumigatus | 363 | 85 | 4.3 | 3.42E-05 |
| An14g05370 | brnA | strong similarity to cell surface ferroxidase precursor Fet3 - Saccharomyces cerevisiae | 52 | 34 | 1.5 | 1.91E-03 |
| Growth and Morphology | ||||||
| An16g03740 | pkaR | protein kinase A regulatory subunit | 469 | 286 | 1.6 | 1.59E-03 |
| An01g02320 | rasA | RAS protein, small GTP binding protein | 1938 | 1217 | 1.6 | 7.34E-04 |
Several hydrophobin genes were up-regulated in the tupA mutant (Table 11, Figure S2). However, the hydrophobin gene that is induced during conidiation in response to carbon starvation during zero growth conditions (An03g02360) [41] was not induced in the tupA mutant. Finally, we noticed that two genes that are required for spore-related melanin production in A. niger (olvA and brnA) were up-regulated. However, the polyketide synthase (fwnA), which is required for the melanin production [69], was not higher expressed. As described above, the loss of repression of transcription due to the tupA mutation has an important effect on the expression of genes that regulate and coordinate asexual development. However, not all the genes that are induced during asexual development were induced. We therefore suggest that TupA assists in repression of these genes under non-inducing conditions. For other genes, like fwnA, a specific activator is probably required to induce expression. It is important to note that we did not observe formation of asexual structures (conidiospores) during the exponential growth phase of the tupA mutant when cultivated in the bioreactor, indicating that not the entire asexual developmental program was turned on in the tupA mutant during exponential growth. Also noticeable is the up-regulation of two genes related to cAMP signaling (RasA and PkaR), which hints to an increased activation of cAMP synthesis in the tupA mutant.
Differentiation is closely linked to the production of secondary metabolites [70] and therefore the expression of genes and gene clusters potentially encoding secondary metabolite synthesis was also analyzed. To do so, the list of 376 secondary metabolite-related genes in A. niger as published by Pel and co-workers was used [60]. Fifteen genes were found to be up-regulated (Table S6). One of them does not belong to any known gene cluster (An02g00840) and is predicted to encode a non-ribosomal protein synthase (NRPS). The 14 remaining up-regulated genes belong to predicted gene clusters. In all cases, only a limited number of the genes in an annotated gene cluster were induced. The gene cluster bordered by genes An08g03730 and An08g03820 includes 6 induced genes of the 10 genes in total in the cluster. An08g03770, which belongs to this cluster and is strongly (31-fold) induced, is predicted to encode a Zn(II)2Cys6 transcription factor. It remains to be elucidated which secondary metabolite is produced by this cluster. In total 34 secondary metabolite genes were down-regulated in the tupA mutant, belonging to 17 clusters and three down-regulated genes were not clustered. Similarly, as observed for the tupA-induced secondary metabolite genes, not all genes from a cluster were down-regulated. All five genes in the cluster bordered by An03g03520 and An03g03560 showed a strong down-regulation in the tupA mutant (Table S6).
TupA is Involved in Controlling Expression of Extracellular Proteases
The genes pepA and pepB, both encoding extracellular proteases in A. niger [42], are highly expressed in the tupA mutant during exponential growth on glucose and ammonium, whereas their expression is low in the wild-type strain. Their respective fold-changes are 224 and 99 (Table S7). Expression of pepA and pepB requires the Zn2Cys6 transcription factor called PrtT [42]. Thirteen PrtT targets (including pepA and pepB) have been described in patent application US 2008/0108105 A1. These PrtT-dependent proteases were identified because their expression is dependent on a functional prtT gene; in the prtT knock out strain, these proteases are significantly lower expressed. Of the thirteen PrtT targets, 6 genes (including pepA and pepB) were up-regulated in the tupA mutant. The expression of the remaining four genes was less dramatically different between the tupA strain and the wild-type strain (Table S7).
Discussion
It is well established that the Tup1/Cyc8 complex functions as an important repressor complex in eukaryotic cells. Transcriptional analysis of a A. niger tupA mutant reveals the important role for TupA in controlling gene expression as about 14% of the genes are differentially expressed (up- or down-regulated) in the tupA mutant. Some of these differences could be indirectly caused by the slower growth rate of the tupA mutant compared to the wild-type, but the detailed analysis of the genes differentially expressed in the tupA mutants (Tables 7, 8, 9, 10, 11, and Tables S6 and S7) suggest that TupA is also important in regulating some specific processes related to cell wall biosynthesis, development, secondary metabolism, and nitrogen regulation.
The tupA mutant identified in this study was isolated in a cell wall mutant screen. The selection is based on the observation that agsA is induced in response to cell wall stress [71]. This screen has been successful in establishing that galactomannan biosynthesis and functional vacuolar ATPase activity are required for cell wall biosynthesis [34], [72]. The mutants identified (ugmA and vmaD, respectively) showed several cell wall-related phenotypes such as increased sensitivity towards CFW and SDS. The mutant selected for this study showed a relative strong induction of the agsA reporters as observed by its ability to grow relatively well on acetamide, and displayed a relatively strong GFP fluorescence signal compared to other mutants (data not shown). However, the mutant did not show increased sensitivity towards cell wall- or cell membrane-perturbing compounds, suggesting that cell wall integrity was not significantly affected.
Caspofungin, an inhibitor of beta-1,3-glucan synthase, induces the cell wall integrity pathway. Genome-wide expression analysis of A. niger treated with sub-lethal concentrations of caspofungin resulted in induced expression of 166 genes [20]. These genes are considered to represent the cell wall stress-responsive genes in A. niger. To examine whether loss of tupA function results in derepression of the cell wall stress-responsive genes we determined the overlap between caspofungin-induced and tupA-induced genes. Of the 166 genes induced by caspofungin (CA) only 47 genes (∼28%) were also induced in the tupA strain (Table S8). Eighty-six (∼52%) of the CA-induced genes were not differentially expressed in the tupA strain. In addition, a considerable number of CA-induced genes (33 genes; ∼20%) had lower expression levels in the tupA mutant. Although the overlap is substantial, this indicates that loss of tupA function under the growth conditions used here does not simply lead to derepression of cell wall stress-induced genes and suggests that TupA does not function as a repressor of cell wall stress-induced genes under non-stressed conditions.
Interestingly, the two strongest induced cell wall-related genes in the tupA mutant, bgtA and exgA, are not expressed under normal growth conditions. We noticed that these two genes are very highly expressed during sclerotia formation in the A. niger sclA-1 mutant strain [73], based on RNA analysis extracted from sclerotia (Jørgenson and Ram, unpublished results). In several filamentous fungi, Tup1 functions as a global repressor which regulates genes associated with morphological differentiation, sexual and asexual reproduction, and pathogenicity [31]–[33]. Possibly, TupA of A. niger also has a repressing role with respect to the expression of sclerotia-associated genes under conditions in which sclerotia formation is normally repressed.
An additional interesting phenotype of the tupA mutant is the secretion of a currently unknown pigment in the medium when grown at high temperatures and with nitrate as a nitrogen source (Fig 1). Both replacement of nitrate by ammonium (Figure 2) as well as the addition of yeast extract and casamino acids to nitrate-containing minimal medium (not shown) reduce pigment production. This suggests that production of the pigment is normally repressed in the presence of nitrate, but that nitrate-controlled repression is lost in the tupA mutant. In the tupA mutant however, the synthesis of the pigment can still be repressed by more preferred nitrogen sources such as ammonium. In Penicillium marneffi the lack of tupA also results in pigment production [32], but the effect of nitrogen sources has not been analyzed. Attempts to identify the nature of the pigment secreted in the tupA mutant have not been conclusive. It probably has an elemental composition of C10H14O4, but further analysis is required to identify the compound (Kristian F. Nielsen, unpublished results). Intriguingly, the tup1 deletion strain of C. albicans also secretes excessive amounts of a (yellow-green) pigment into the medium [74].
In S. cerevisiae, it is well established that Tup1, together with Ssn6 (Cyc8), is involved in carbon repression. In S. cerevisiae Mig1p, the repressor responsive to the carbon status of the cell, is known to recruite the Tup1/Ssn6 complex to enable its repressor function [75]. Also in the yeasts Schizosaccharomyces pombe and Candida albicans Tup1 homologs have been found to be required for carbon repression [76]–[77]. However, in Aspergillus nidulans it has been shown that the Tup1 homolog, in this species designated as RcoA, is not involved in carbon repression [78]–[79].
Delmas et al. recently proposed a model by which starvation leads to the expression of genes that encode extracellular enzymes [80]. These enzymes are normally repressed by CreA and it was proposed that these enzymes have a sensing role for the presence of alternative substrates. These genes were identified in the study of Delmas et al., because the genes encoding these extracellular enzymes were rapidly induced under carbon starvation conditions. The authors show that the gene encoding cellobiohydrolase (An01g11660; cbhB) is induced upon C-starvation in a XlnR-independent way and that the expression is higher in a creA mutant strain, strongly suggesting that the induced expression of cbhB upon starvation is mediated via carbon catabolite derepression. As cbhB (An01g11660) is not higher expressed in the tupA mutant, this offers further support for the notion that similar to A. nidulans tupA of A. niger is not required for carbon catabolite repression.
The expression of the two major extracellular proteases in A. niger (pepA and pepB) has been shown to be under carbon catabolite repression [81]. The involvement of CreA in mediating this repression was shown by [82] as the expression of pepA and pepB was higher in the creA strain under repressing conditions. The latter observation seems to contradict the conclusion the TupA is not mediating carbon repression since pepA and pepB are highly expressed in the tupA mutant (Table S7). To explain these observations, it is important to note that pepA and pepB are also under nitrogen repression control. The presence of a preferred nitrogen source such as ammonium strongly represses the expression of pepA and pepB [80]. Additional support for a connection between TupA and nitrogen repression comes from the observation that the formation of the pigment in the tupA mutant strain was found to be nitrogen source-dependent and repressed by ammonium.
TupA has been implicated in several fungi to have a role in dimorphism in the transition from yeast to filamentous growth [31]–[33], [83]. The role of Tup1 in fungal dimorphism might well be linked to nitrogen metabolism as nitrogen availability has been shown to be an important factor in fungal dimorphism [84]–[88]. We suggest that the link to nitrogen metabolism and TupA is important to understand the involvement of TupA in developmental processes and dimorphic switches in fungi.
Supporting Information
Spores of the MA246.1 (ΔtupA in RD15.8) were inoculated in MM-glucose containing 10 mM ammonium or 10 mM nitrate at pH3.0 or pH5.7. Pictures were taken after 16 of incubation at 30°C. The fluorescence detected under all conditions shows that agsA expression in germinating spores of the tupA mutant is not affected by pH or nitrogen source.
(TIF)
Northern blot analysis of selected differentially expressed genes of RNA samples that were used for the microarrays of the wild-type strain (wt) or the tupA mutant. Gene identifiers are indicated as well as the gene name (when available). Behind the gene identified the fold change in expression (tupA vs wild-type) is given based on the microarray data.
(TIFF)
Expression data WT and tupA mutant.
(XLS)
GO-terms of up-regulated genes in tupA mutant.
(XLS)
GO-terms of down-regulated genes in tupA mutant.
(XLS)
TupA up-regulated gene with Fold change >10.
(XLSX)
TupA down-regulated gene with Fold change >10.
(XLSX)
Expression analysis of all secondary metabolite genes.
(DOCX)
Expression analysis of PrtT target genes.
(DOCX)
Comparison of caspofungin treated and TupA differentials.
(XLSX)
Acknowledgments
We thank Peter Punt and Frank Schuren for the A. niger genomic library. Doreen Schachtschabel is grateful for a postdoctoral fellowship by the Deutsche Forschungsgemeinsschaft (DFG). We thank Joohae Park for his help in preparing the figures.
Funding Statement
The research group of A.F.J. Ram is part of the Kluyver Centre for Genomics of Industrial Fermentation (http://www.kluyvercentre.nl), which is supported by the Netherlands Genomics Initiative (http://www.genomics.nl/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bastos KP, Bailao AM, Borges CL, Faria FP, Felipe MS, et al. (2007) The transcriptome analysis of early morphogenesis in Paracoccidioides brasiliensis mycelium reveals novel and induced genes potentially associated to the dimorphic process. BMC Microbiol 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, et al. (2011) Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157: 2297–2307. [DOI] [PubMed] [Google Scholar]
- 3. Morales-Vargas AT, Dominguez A, Ruiz-Herrera J (2012) Identification of dimorphism-involved genes of Yarrowia lipolytica by means of microarray analysis. Res Microbiol 163: 378–387. [DOI] [PubMed] [Google Scholar]
- 4. Robledo-Briones M, Ruiz-Herrera J (2013) Regulation of genes involved in cell wall synthesis and structure during Ustilago maydis dimorphism. FEMS Yeast Res 13: 74–84. [DOI] [PubMed] [Google Scholar]
- 5. Klis FM, Boorsma A, De Groot PW (2006) Cell wall construction in Saccharomyces cerevisiae . Yeast 23: 185–202. [DOI] [PubMed] [Google Scholar]
- 6. Gastebois A, Clavaud C, Aimanianda V, Latge JP (2009) Aspergillus fumigatus: cell wall polysaccharides, their biosynthesis and organization. Future Microbiol 4: 583–59. [DOI] [PubMed] [Google Scholar]
- 7. Damveld RA, vanKuyk PA, Arentshorst M, Klis FM, van den Hondel CA, et al. (2005) Expression of agsA, one of five 1,3-alpha-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet Biol 42: 165–177. [DOI] [PubMed] [Google Scholar]
- 8. Levdansky E, Kashi O, Sharon H, Shadkchan Y, Osherov N (2010) The Aspergillus fumigatus cspA gene encoding a repeat-rich cell wall protein is important for normal conidial cell wall architecture and interaction with host cells. Eukaryot Cell 9: 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fontaine T, Delangle A, Simenel C, Coddeville B, van Vliet SJ, et al. (2011) Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus . PLoS Pathog 7: e1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Latge JP (2010) Tasting the fungal cell wall. Cell Microbiol 12: 863–872. [DOI] [PubMed] [Google Scholar]
- 11. Sohn K, Urban C, Brunner H, Rupp S (2003) EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol Microbiol 47: 89–102. [DOI] [PubMed] [Google Scholar]
- 12. Braun BR, Johnson AD (2000) TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans . Genetics 155: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hogan LH, Klein BS (1994) Altered expression of surface alpha-1,3-glucan in genetically related strains of Blastomyces dermatitidis that differ in virulence. Infect Immun 62: 3543–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reese AJ, Doering TL (2003) Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol 50: 1401–1409. [DOI] [PubMed] [Google Scholar]
- 15. Rappleye CA, Engle JT, Goldman WE (2004) RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1,3)-glucan in virulence. Mol Microbiol 53: 153–165. [DOI] [PubMed] [Google Scholar]
- 16. San-Blas F, San-Blas G, Cova LJ (1976) A morphological mutant of Paracoccidioides brasiliensis strain IVIC Pb9. Isolation and wall characterization. J Gen Microbiol 93: 209–218. [DOI] [PubMed] [Google Scholar]
- 17. San-Blas G, Vernet D (1977) Induction of the synthesis of cell wall alpha-1,3-glucan in the yeastlike form of Paracoccidioides brasiliensis strain IVIC Pb9 by fetal calf serum. Infect Immun 15: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, et al. (2004) The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J Biol Chem 279: 15183–15195. [DOI] [PubMed] [Google Scholar]
- 19. Boorsma A, de Nobel H, ter Riet B, Bargmann B, Brul S, et al. (2004) Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21: 413–427. [DOI] [PubMed] [Google Scholar]
- 20. Meyer V, Damveld RA, Arentshorst M, Stahl U, van den Hondel CA, et al. (2007) Survival in the presence of antifungals: genome-wide expression profiling of Aspergillus niger in response to sublethal concentrations of caspofungin and fenpropimorph. J Biol Chem 282: 32935–32948. [DOI] [PubMed] [Google Scholar]
- 21. Levin DE (2005) Cell wall integrity signaling in Saccharomyces cerevisiae . Microbiol Mol Biol Rev 69: 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shapiro RS, Robbins N, Cowen LE (2011) Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75: 213–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levin DE (2011) Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Damveld RA, Arentshorst M, Franken A, vanKuyk PA, Klis FM, et al. (2005) The Aspergillus niger MADS-box transcription factor RlmA is required for cell wall reinforcement in response to cell wall stress. Mol Microbiol 58: 305–319. [DOI] [PubMed] [Google Scholar]
- 25. Green SR, Johnson AD (2005) Genome-wide analysis of the functions of a conserved surface on the corepressor Tup1. Mol Biol Cell 16: 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith RL, Johnson AD (2000) Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25: 325–330. [DOI] [PubMed] [Google Scholar]
- 27. Kobayashi Y, Inai T, Mizunuma M, Okada I, Shitamukai A, et al. (2008) Identification of Tup1 and Cyc8 mutations defective in the responses to osmotic stress. Biochem Biophys Res Commun 368: 50–55. [DOI] [PubMed] [Google Scholar]
- 28. Braun BR, Johnson AD (1997) Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277: 105–109. [DOI] [PubMed] [Google Scholar]
- 29. Wong KH, Struhl K (2011) The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev 25: 2525–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parnell EJ, Stillman DJ (2011) Shields up: the Tup1-Cyc8 repressor complex blocks coactivator recruitment. Genes Dev 25: 2429–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee H, Chang YC, Varma A, Kwon-Chung KJ (2009) Regulatory diversity of TUP1 in Cryptococcus neoformans. . Eukaryot Cell 8: 1901–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Todd RB, Greenhalgh JR, Hynes MJ, Andrianopoulos A (2003) TupA, the Penicillium marneffei Tup1p homologue, represses both yeast and spore development. Mol Microbiol 48: 85–94. [DOI] [PubMed] [Google Scholar]
- 33. Elias-Villalobos A, Fernandez-Alvarez A, Ibeas JI (2011) The general transcriptional repressor Tup1 is required for dimorphism and virulence in a fungal plant pathogen. PLoS Pathog 7: e1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Damveld RA, Franken A, Arentshorst M, Punt PJ, Klis FM, et al. (2008) A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett JW, Lasure LL (1991) Growth media. In: Bennett JW, Lasure LL (Eds.). More Gene Manipulations in Fungi. Academic Press, San Diego, 441–458. [Google Scholar]
- 36. Kelly JM, Hynes MJ (1985) Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans . EMBO J 4: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Gorcom RF, van den Hondel CA (1988) Expression analysis vectors for Aspergillus niger . Nucleic Acids Res 16: 9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inoue H, Nojima H, Okayama H (1990) High efficiency transformation of Escherichia coli with plasmids. Gene 96: 23–28. [DOI] [PubMed] [Google Scholar]
- 39.Meyer V, Ram AFJ, Punt PJ (2010) Genetics, genetic manipulation and approaches to strain improvement of filamentous fungi. In: Demain AL, Davis J (eds) Man. Indust. Microbiol. Biotech. Wiley, NY, 3rd edn., 318–329. [Google Scholar]
- 40.Sambrook J, Fritsch EF, and Maniatis T (1989) Molecular Cloning: a Laboratory Manual, Cold Spring Harbor Laboratory Press, Plainview NY. [Google Scholar]
- 41. Jorgensen TR, Nitsche BM, Lamers GE, Arentshorst M, van den Hondel CA, et al. (2010) Transcriptomic insights into the physiology of Aspergillus niger approaching a specific growth rate of zero. Appl Environ Microbiol 76: 5344–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Punt PJ, Schuren FH, Lehmbeck J, Christensen T, Hjort C, et al. (2008) Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet Biol 45: 1591–1599. [DOI] [PubMed] [Google Scholar]
- 43. Carvalho ND, Arentshorst M, Jin Kwon M, Meyer V, Ram AF (2010) Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl Microbiol Biotechnol 87: 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Ruiter-Jacobs YM, Broekhuijsen M, Unkles SE, Campbell EI, Kinghorn JR, et al. (1989) A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae . Curr Genet 16: 159–163. [DOI] [PubMed] [Google Scholar]
- 45. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 47. Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315. [DOI] [PubMed] [Google Scholar]
- 48. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 49. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 57: 289–300. [Google Scholar]
- 50. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nitsche BM, Crabtree J, Cerqueira GC, Meyer V, Ram AF, et al. (2011) New resources for functional analysis of omics data for the genus Aspergillus . BMC Genomics 12: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ram AF, Arentshorst M, Damveld RA, vanKuyk PA, Klis FM, et al. (2004) The cell wall stress response in Aspergillus niger involves increased expression of the glutamine:fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology 150: 3315–3326. [DOI] [PubMed] [Google Scholar]
- 53. de Groot PW, Ruiz C, Vazquez de Aldana CR, Duenas E, Cid VJ, et al. (2001) A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae . Comp Funct Genomics 2: 124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wösten HA, Moukha SM, Sietsma JH, Wessels JG (1991) Localization of growth and secretion of proteins in Aspergillus niger. J Gen Microbiol. 137: 2017–2023. [DOI] [PubMed] [Google Scholar]
- 55. Levin AM, de Vries RP, Conesa A, de Bekker C, Talon M, et al. (2007) Spatial differentiation in the vegetative mycelium of Aspergillus niger . Eukaryot Cell 6: 2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. te Biesebeke R, van Biezen N, de Vos WM, van den Hondel CA, Punt PJ (2005) Different control mechanisms regulate glucoamylase and protease gene transcription in Aspergillus oryzae in solid-state and submerged fermentation. Appl Microbiol Biotechnol 67: 75–82. [DOI] [PubMed] [Google Scholar]
- 57. Kobayashi A, Sano M, Oda K, Hisada H, Hata Y, et al. (2007) The glucoamylase-encoding gene (glaB) is expressed in solid-state culture with a low water content. Biosci Biotechnol Biochem 71: 1797–1809. [DOI] [PubMed] [Google Scholar]
- 58. Jorgensen TR, Goosen T, Hondel CA, Ram AF, Iversen JJ (2009) Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Piper MD, Daran-Lapujade P, Bro C, Regenberg B, Knudsen S, et al. (2002) Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae . J Biol Chem 277: 37001–37008. [DOI] [PubMed] [Google Scholar]
- 60. Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25: 221–231. [DOI] [PubMed] [Google Scholar]
- 61. Damveld RA, Arentshorst M, VanKuyk PA, Klis FM, van den Hondel CA, et al. (2005) Characterisation of CwpA, a putative glycosylphosphatidylinositol-anchored cell wall mannoprotein in the filamentous fungus Aspergillus niger . Fungal Genet Biol 42: 873–885. [DOI] [PubMed] [Google Scholar]
- 62. Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Muller WH, et al. (2013) Development in Aspergillus . Stud Mycol 74: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adams TH, Wieser JK, Yu JH (1998) Asexual sporulation in Aspergillus nidulans . Microbiol Mol Biol Rev 62: 35–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krijgsheld P, Nitsche BM, Post H, Levin AM, Muller WH, et al. (2013) Deletion of flbA results in increased secretome complexity and reduced secretion heterogeneity in colonies of Aspergillus niger. J Proteome Res. 12: 1808–1819. [DOI] [PubMed] [Google Scholar]
- 65. Masloff S, Poggeler S, Kuck U (1999) The pro1(+) gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vienken K, Scherer M, Fischer R (2005) The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics 169: 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP (2004) Endogenous lipogenic regulators of spore balance in Aspergillus nidulans . Eukaryot Cell 3: 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tsitsigiannis DI, Zarnowski R, Keller NP (2004) The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans . J Biol Chem 279: 11344–11353. [DOI] [PubMed] [Google Scholar]
- 69. Jorgensen TR, Park J, Arentshorst M, van Welzen AM, Lamers G, et al. (2011) The molecular and genetic basis of conidial pigmentation in Aspergillus niger . Fungal Genet Biol 48: 544–553. [DOI] [PubMed] [Google Scholar]
- 70. Calvo AM, Wilson RA, Bok JW, Keller NP (2002) Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev 66: 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Damveld RA, vanKuyk PA, Arentshorst M, Klis FM, van den Hondel CA, et al. (2005) Expression of agsA, one of five 1,3-alpha-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet Biol 42: 165–1. [DOI] [PubMed] [Google Scholar]
- 72. Schachtschabel D, Arentshorst M, Lagendijk EL, Ram AF (2012) Vacuolar H(+)-ATPase plays a key role in cell wall biosynthesis of Aspergillus niger . Fungal Genet Biol 49: 284–293. [DOI] [PubMed] [Google Scholar]
- 73. Jorgensen TR, Nielsen KF, Arentshorst M, Park J, van den Hondel CA, et al. (2011) Submerged conidiation and product formation by Aspergillus niger at low specific growth rates are affected in aerial developmental mutants. Appl Environ Microbiol 77: 5270–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Knight SA, Lesuisse E, Stearman R, Klausner RD, Dancis A (2002) Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 148: 29–40. [DOI] [PubMed] [Google Scholar]
- 75. Treitel MA, Carlson M (1995) Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci U S A 92: 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Janoo RT, Neely LA, Braun BR, Whitehall SK, Hoffman CS (2001) Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 157: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Murad AM, d’Enfert C, Gaillardin C, Tournu H, Tekaia F, et al. (2001) Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol Microbiol 42: 981–993. [DOI] [PubMed] [Google Scholar]
- 78. Hicks J, Lockington RA, Strauss J, Dieringer D, Kubicek CP, et al. (2001) RcoA has pleiotropic effects on Aspergillus nidulans cellular development. Mol Microbiol. 39: 1482–1493. [DOI] [PubMed] [Google Scholar]
- 79. Garcia I, Mathieu M, Nikolaev I, Felenbok B, Scazzocchio C (2008) Roles of the Aspergillus nidulans homologues of Tup1 and Ssn6 in chromatin structure and cell viability. FEMS Microbiol Lett 289: 146–154. [DOI] [PubMed] [Google Scholar]
- 80. Delmas S, Pullan ST, Gaddipati S, Kokolski M, Malla S, et al. (2012) Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing. PLoS Genet 8: e1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jarai G, Buxton F (1994) Nitrogen, carbon, and pH regulation of extracellular acidic proteases of Aspergillus niger . Curr Genet 26: 238–244. [DOI] [PubMed] [Google Scholar]
- 82.van den Hombergh JPTW (1996) An analysis of the proteolytic system in Aspergillus in order to improve protein production. PhD thesis, Wageningen Agriculture University, Wageningen. [Google Scholar]
- 83. Lee H, Chang YC, Kwon-Chung KJ (2005) TUP1 disruption reveals biological differences between MATa and MATalpha strains of Cryptococcus neoformans . Mol Microbiol 55: 1222–1232. [DOI] [PubMed] [Google Scholar]
- 84. Mitchell LH, Soll DR (1979) Commitment to germ tube or bud formation during release from stationary phase in Candida albicans. . Exp Cell Res 120: 167–179. [DOI] [PubMed] [Google Scholar]
- 85. Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR (1992) Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090. [DOI] [PubMed] [Google Scholar]
- 86. Ruiz-Herrera J, Sentandreu R (2002) Different effectors of dimorphism in Yarrowia lipolytica . Arch Microbiol 178: 477–483. [DOI] [PubMed] [Google Scholar]
- 87. Szabo R, Stofanikova V (2002) Presence of organic sources of nitrogen is critical for filament formation and pH-dependent morphogenesis in Yarrowia lipolytica . FEMS Microbiol Lett 206: 45–50. [DOI] [PubMed] [Google Scholar]
- 88. Heintz-Buschart A, Eickhoff H, Hohn E, Bilitewski U (2013) Identification of inhibitors of yeast-to-hyphae transition in Candida albicans by a reporter screening assay. J Biotechnol 164: 137–142. [DOI] [PubMed] [Google Scholar]
- 89. Bos CJ, Debets AJ, Swart K, Huybers A, Kobus G, et al. (1988) Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger . Curr Genet 14: 437–443. [DOI] [PubMed] [Google Scholar]
- 90. van Hartingsveldt W, Mattern IE, van Zeijl CM, Pouwels PH, van den Hondel CA (1987) Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet 206: 71–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spores of the MA246.1 (ΔtupA in RD15.8) were inoculated in MM-glucose containing 10 mM ammonium or 10 mM nitrate at pH3.0 or pH5.7. Pictures were taken after 16 of incubation at 30°C. The fluorescence detected under all conditions shows that agsA expression in germinating spores of the tupA mutant is not affected by pH or nitrogen source.
(TIF)
Northern blot analysis of selected differentially expressed genes of RNA samples that were used for the microarrays of the wild-type strain (wt) or the tupA mutant. Gene identifiers are indicated as well as the gene name (when available). Behind the gene identified the fold change in expression (tupA vs wild-type) is given based on the microarray data.
(TIFF)
Expression data WT and tupA mutant.
(XLS)
GO-terms of up-regulated genes in tupA mutant.
(XLS)
GO-terms of down-regulated genes in tupA mutant.
(XLS)
TupA up-regulated gene with Fold change >10.
(XLSX)
TupA down-regulated gene with Fold change >10.
(XLSX)
Expression analysis of all secondary metabolite genes.
(DOCX)
Expression analysis of PrtT target genes.
(DOCX)
Comparison of caspofungin treated and TupA differentials.
(XLSX)