Abstract
The red leaf coloration of Empire Red Leaf Cotton (ERLC) (Gossypium hirsutum L.), resulted from anthocyanin accumulation in light, is a well known dominant agricultural trait. However, the underpin molecular mechanism remains elusive. To explore this, we compared the molecular biological basis of anthocyanin accumulation in both ERLC and the green leaf cotton variety CCRI 24 (Gossypium hirsutum L.). Introduction of R2R3-MYB transcription factor Rosea1, the master regulator anthocyanin biosynthesis in Antirrhinum majus, into CCRI 24 induced anthocyanin accumulation, indicating structural genes for anthocyanin biosynthesis are not defected and the leaf coloration might be caused by variation of regulatory genes expression. Expression analysis found that a transcription factor RLC1 (Red Leaf Cotton 1) which encodes the ortholog of PAP1/Rosea1 was highly expressed in leaves of ERLC but barely expressed in CCRI 24 in light. Ectopic expression of RLC1 from ERLC and CCRI 24 in hairy roots of Antirrhinum majus and CCRI 24 significantly enhanced anthocyanin accumulation. Comparison of RLC1 promoter sequences between ERLC and CCRI 24 revealed two 228-bp tandem repeats presented in ERLC with only one repeat in CCRI 24. Transient assays in cotton leave tissue evidenced that the tandem repeats in ERLC is responsible for light-induced RLC1 expression and therefore anthocyanin accumulation. Taken together, our results in this article strongly support an important step toward understanding the role of R2R3-MYB transcription factors in the regulatory menchanisms of anthocyanin accumulation in red leaf cotton under light.
Introduction
Red leaf cotton is an important genetic resource. The red color produced by anthocyanins is found in most tissues of the plant, except in the cotton fibers; the leaves, stems, petals, and bolls are all red. Accumulation of anthocyanins confers resistance to insect pests such as bollworm, whiteflies, and the boll weevil [1], [2]. Previous genetic studies found that three anthocyanin-related loci – R1, R2, and Rd – are responsible for anthocyanin accumulation in red-colored plant species [3]. The R1 locus affects red leaf pigmentation in red leaf cotton including ERLC, the R2 locus is responsible for the red petal spots in green leaf cotton, and the Rd locus is involved in dwarf red cotton species [3], [4]. Anthocyanin accumulation in red leaf cotton (ERLC) is sensitized to the light environment such that the red coloration is reduced when plants are grown in shade, whereas plants of the same genotype grown in the field in the summer are solid red in appearance [4]. Red leaf color is a morphological trait that could potentially be useful in cotton breeding. However, the molecular basis of the anthocyanin pigment synthesis mechanism in red leaf cotton remains elusive.
Anthocyanins are flavonoids that are produced in response to a range of environmental and developmental signals. They protect plants against UV light damage, pathogen invasion, and insect attack [5]–[7]. The genetics and biochemistry of the anthocyanin biosynthetic pathway have been well characterized in plants [8]. In Arabidopsis, genes participating in the anthocyanin biosynthetic pathway are divided into two groups, early biosynthetic genes (EBGs) including those for chalcone synthase (CHS), chalcone isomerase (CHI), flavonol 3-hydroxylase (F3H), and flavonol 3′-hydroxylase (F3′H), and late biosynthetic genes (LBGs) including those for dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and anthocyanidin reductase (ANR). Each of the two groups has been found to be co-regulated by different regulators [9]–[11]. This gene regulation pattern is similar to that seen in other dicot plants such as Antirrhinum and Petunia. Anthocyanin biosynthetic pathway genes are predominantly regulated at the transcriptional level, both developmentally and/or in response to various biotic and abiotic stress factors. Many regulatory genes have been identified in different species, for example AN1 and AN2 in Petunia [12], [13], C1 and P1 in Zea mays [14], [15], Rosea1 and Delila in Antirrhinum [16], [17], PAP1 and TTG8 in Arabidopsis [18], [19], MYB1 and MdMYB10 in apples [20], [21] and VvMYBA in grape [22].
The MYB transcription factors constitute the largest transcriptional factor gene family in plants and can be classified into three subfamilies based on the number of highly conserved imperfect repeats in the DNA-binding domain. R3-MYB has one repeat, R2R3-MYB has two repeats, and R1R2R3-MYB has three repeats [23], [24]. Of these, the R2R3-MYB transcription factors have been shown to play important roles in tissue-specific anthocyanin accumulation in many plants; these include AN2 in Petunia [12], Rosea1, Rosea2, and Venosa in Antirrhinum [17], C1 and P1 in Maize [14], [15], and PAP1 in Arabidopsis [18]. Some mutant varieties display different pigment intensities, also caused by variations in the R2R3-MYB regulators. For instance, a mini-satellite-like structure insertion in the MYB10 promoter activates its expression resulting in anthocyanin accumulation in the flesh and folium in apples [25]. Mutations in two adjacent MYB regulators, VvMYBA1 and VvMYBA2, inhibit their expression and change the skin color of grape (Vitis vinifera) from red to white [26]. Several anthocyanin repressors are also MYB transcription factors, including an R2R3-MYB repressor from strawberry FaMYB1 [27], AtMYB4 and AtMYB6 from Arabidopsis [28], AmMYB308 from Antirrhinum [29], and a single repeat MYB gene AtMYBL2 from Arabidopsis [30].
The basic helix-loop-helix (bHLH) transcription factors are another group of regulatory genes involved in anthocyanin biosynthesis in plants. The insertion of a transposon into a bHLH regulatory gene was found to alter the flower color of morning glory [31], and a transposon insertion altered flower color in Antirrhinum [16]. bHLH factors also regulate several unrelated biosynthetic pathways, including the development of trichomes and seed coat mucilage [32], [33].
In plants, regulation of the anthocyanin biosynthetic pathway is controlled by a transcriptional activation complex consisting of R2R3-MYB, bHLH transcription factors, and WD40 family proteins [34], [35]. In Arabidopsis, four R2R3-MYB regulators (PAP1, PAP2, MYB113, and MYB114), three bHLH regulators (GL3, EGL3, and TT8), and a WD40 regulator (TTG1) participate in anthocyanin synthesis regulation by forming a MYB-bHLH-WD40 complex [33], [35]. This complex mainly regulates the expression of LBGs in conjunction with their promoters [36]–[39].
In this study, we evidence functional characterization of an R2R3-MYB transcription factor RLC1 from Empire Red Leaf Cotton (ERLC). Transcriptional analysis in various tissues indicated that RLC1 was preferentially expressed in the hypocotyl (stem) and leaves. This expression was influenced by light conditions and correlated with anthocyanin accumulation in the leaves of ERLC. Ectopic expression of RLC1in Antirrhinum and green leaf cotton CCRI 24 significantly upregulated the expression of genes in the anthocyanin pathway and resulted in anthocyanin accumulation in the hairy roots. Transformation analysis of 35S::RLC1a (from the start codon to the stop codon in the DNA sequence of CCRI 24) in Antirrhinum revealed anthocyanin accumulation in the transformed hairy roots, suggesting that promoter differentiation might be an effect of the gene transcript in CCRI 24. Transient expression analysis of promoter activity revealed that the RLC1 promoter in ERLC, which contains two 228-bp fragments forming a direct tandem repeat, normally promoted anthocyanin accumulation when introduced into the leaves of CCRI 24; however, the promoter of CCRI 24, which contains only a single 228-bp fragment in the RLC1 promoter region did not result in anthocyanin accumulation in the leaves. Therefore, the 228-bp fragment tandem repeat is an important feature of the RLC1 promoter in cotton. Taken together, our results strongly suggest that the RLC1 transcription factor is induced by light is a major regulatory gene controlling anthocyanin accumulation in cotton leaves.
Materials and Methods
Plant Material and Growth Conditions
Seeds of Antirrhinum majus JI 7 (provided by Professor Enrico Coen, John Innes Centre, Norwich, UK) and cotton seeds of the ERLC and CCRI 24 varieties (from the Cotton Research Institute, Chinese Academy of Agricultural Science, Anyang, China) were used in this study. The seed coats of the cotton seeds were removed and the seeds of A. majus JI7 were surface-sterilized with a brief rinse in 70% (v/v) ethanol followed by a wash in a 2% (v/v) solution of sodium hypochlorite for 10 min, and germinated on solid MS medium in a growth room at 25°C with a 16-h light/8-h dark photoperiod.
For light/shading experiments, 20 10-day old seedlings of both ERLC and CCRI 24 germinated on MS medium were divided into two groups. One group was transferred to a greenhouse in direct light conditions, and other group was transferred to a greenhouse shadowed by a double layer of black shading net. After 30 days in culture, the upper fully opened mature leaves were used to isolate RNA and carry out anthocyanin analysis.
Quantification of Anthocyanins
The leaves were ground to a fine powder in liquid nitrogen, and 0.3 g of the powder was extracted with 1 mL acidic methanol (1% hydrochloric acid, w/v) at room temperature for 12 h with moderate shaking. After centrifugation at 12,000 rpm for 10 min, 800 µL of the supernatant was added to 4 mL of acidic methanol. The absorbance at 530 and 657 nm was determined using a spectrophotometer (UV757CRT, Shanghai), and the relative level of anthocyanin was calculated using the equation A530 – (0.25×A650) [40]. Each sample was tested three times. Error bars indicate the standard deviation (SD) values of the average anthocyanin content.
Isolation of Full-length RLC1 cDNA and Phylogenetic Analysis
Total RNA was isolated from 7-day-old hypocotyl tissues (0.3 g) of ERLC using the SV Total RNA Isolation System combined with RNase-free DNase (Promega, Madison, WI). First-strand cDNA was synthesized from 2 µg of total RNA with the Superscript III First Strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA) in combination with a PCR library kit (Takara, Shiga, Japan). For the isolation of the anthocyanin-specific MYB transcription factor, a set of degenerate primers (Table S1) were designed from plant MYB cDNA and the conserved DNA-binding domain of R2R3-MYBs was amplified by PCR. Amplified fragments of about 240 bp were subsequently ligated into the pMD19-T cloning vector (Takara) and the insert sequenced using vector M13 primers. Nucleotide sequences were translated into amino acid sequences using DNAMAN (version 6) and compared using the BLAST network service from the National Center for Biotechnology Information (NCBI). To isolate the full-length cDNA of the candidate MYB genes, the 3′-ends were amplified using a combination of the gene-specific primer MYB3SP and the RP primer, and the 5′-end was amplified using the specific primer MYB15SP1 and the CA primer (PCR library kit, Takara,) (Table S1). The 3′- and 5′-RACE PCR products were ligated into the pMD19-T vector and sequenced. Additionally, based on the sequence obtained by RACE-PCR, a set of gene-specific primers, Ghi2-F2 and Ghi2-R2, were designed to amplify the full-length coding sequence using Pfu DNA polymerase (Promega). The amplified fragment was cloned into a pEASY-Blunt vector (named pEASY-RLC1) (TransGen Biotech, Beijing, China) and confirmed as the cDNA sequence. The amplified 744-bp full-length cDNA was named RLC1 (Red Leaf Cotton 1). This identified coding region of RLC1 was aligned with published MYB gene sequences involved in the anthocyanin biosynthetic pathway from different plants and a phylogenetic tree constructed using ClustalW (version 2.0) [41] and MEGA version 4.0 [42]. The GenBank accession numbers of these proteins are as follows: AmROSEA1 (ABB83826), AtMYB75 (AAG42001), VvMYBA1 (BAD1897), AtMYB90 (AAG42002), NtAN2 (ACO52470), CsRuby (AFB73909), AmROSEA2 (ABB83827), AmVENOSA1 (ABB83828), AtMYB90 (AAG42002), AtMYB113 (NP_176811), AtWER (NP_196979), AtMYB0 (NP_189430), GhMyb10 (CAD87010), MdMYB10 (ABB84753), MdMYB1 (ABK58136), PhAN2 (AAF66727), PhPHZ (ADW94951), PhDPL (ADW94950), VvMYBA1 (BAD18977), VvMYBA2 (BAD18978), and NtAN2 (ACO52470).
Construction of pBI35S::RLC1 and Transformation of A. majus and CCRI 24
The full length RLC1 cDNA coding region was amplified from the pEASY-RLC1 plasmid using an Sma I-Ghi2-F2 and Sac I-Ghi2-R2 gene-specific primer set (Table S1) in combination with Pfu DNA polymerase (Promega), and then sub-cloned into a pEASY-Blunt vector for confirmation by sequencing. Using the Sma I and Sac I restriction sites, RLC1 was substituted for the GUS gene in the pBI121 vector to produce the construct pBI35S::RLC1, containing the RLC1 coding region between the 35S promoter and the CaMv terminator. The pBI35S::RLC1 binary vector was electroporated into A. rhizogenes AR1193 cells [43]. AR1193/pBI121 as a negative control, AR1193/pBI35S::ROSEA1 as a positive control, and AR1193/pBI35S::RLC1 were used to transformation of 7-day-old CCRI 24 and 4-week-old A. majus JI 7 hypocotyls according to the method described by Cui [44].
Expression Analysis of RLC1 and Structural Genes of the Anthocyanin Pathway
Total RNA was extracted from 0.1-g samples of roots, 7-day-old hypocotyls, leaves, and mature petals of both ERLC and CCRI 24 varieties, and transformed hair roots of A. majus and CCRI 24 using the SV Total RNA Isolation System in combination with RNase-free DNase (Promega). First-strand cDNA was synthesized from 2 µg of total RNA with a Superscript III First Strand cDNA Synthesis Kit (Invitrogen). Semi-quantitative RT-PCR for the genes of involved in this work were carried out using a ABI 2720 machine with 10 ng first cDNA and gene-specific primers (Table S2). The following PCR conditions were used: preliminary denaturation for 5 min at 95°C followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55–60°C for 30 s, and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. The genes AmUBI of A. majus and UBI 7 of cotton were used as positive controls [45]. The RT-PCR experiments were repeated at least three times independently and the PCR products confirmed by sequencing.
Isolation of RLC1 Promoter Region
The promoters for ERLC and CCRI 24 were isolated using the Genome Walking kit (Takara) with the gene-specific primers SP1, SP2, and SP3 (Table S1). The PCR-amplified 2.3-kb fragment from ERLC and 2.08-kb fragment from CCRI 24 were ligated to the pMD19-T vector (the resulting constructs were named pMD-Rpro and pMD-Gpro, respectively), and M13 primers were used for sequencing. To isolate the DNA sequence between the start and stop codons, PCR was carried out using two primer sets, GhDNA-F1/GhDNA-R1 for exon 1 and exon 2, and GhDNA-F2/GhDNA-R2 for exon 2 and exon 3, and combined with the LA Tag polymerase (Takara). The products were ligated into the pMD19-T vector and sequenced. The full-length RLC1 genomic sequence was obtained by compiling partial sequences based on overlapping regions. Analysis of the promoter sequence was performed using the PLACE database [46].
Transient Analysis of RLC1 Promoter Region
To compare the promoter activities of R−pro of ERLC and G−pro of CCRI 24, the two promoters were amplified using Pfu DNA polymerase (Promega) and the primers Hind III-ProC-F and Sma I-ProC-R. The PCR product was double-digested with the restriction enzymes Hind III/Sma I, and the 35S promoter on pBI121 and pBI35S::RLC1 was replaced with R−pro and G−pro to construct the expression vectors R−pro::RLC1, G−pro::RLC1, R−pro::GUS, and G−pro::GUS. These vectors were electroporated into A. tumefaciens strain GV3101 and used for the transient assay.
To determine the promoter activity of RLC1, transient assays were performed using mature leaves of green leaf cotton CCRI 24 by infiltration [47]. After infiltration, the leaves were cultured in a growth chamber at 25°C with a 16-h light/8-h dark photoperiod, illuminated with fluorescent light at an intensity of 100 µmol m−2s−1. After three days of culture, the leaves were histochemically stained for GUS, or color accumulation was observed by microscopy (Leica S8 APO).
Histochemical GUS Assay
Infiltrated young leaves with GV3101/R−pro::GUS, GV3101/G−pro::GUS and GV3101/pBI121 were stained for GUS according to the method described by Jefferson [48].
Results
Phenotypic Characterization of Empire Red Leaf Cotton (ERLC)
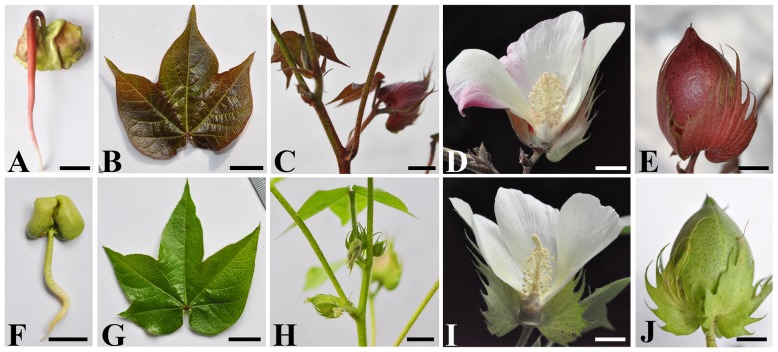
The ERLC plant exhibits red pigment accumulation throughout its growing period, displaying red coloration in most tissues except the fibers, including seedlings, stems, leaves, calyces, flowers, and bolls when grown in light conditions (Fig. 1A–E). The red color is more intense in the older tissues of the leaves, stems, calyces, and bolls (data not shown); however, the visible color is not apparent in tissues and organs grown in shaded conditions. Additionally, red pigmentation is not observed in fibers in any growth conditions (data not shown). In contrast, the green-leaved cotton cultivar CCRI 24 did not display any color accumulation in seedlings, stems, leaves, calyces, bolls, or fibers, regardless of light conditions (Fig. 1F–J).
Figure 1. Phenotypic comparison between Empire red leaf cotton (ERLC) (upper panel) and green leaf cotton CCRI 24 (bottom panel) cultivars grown in light.
A, A 7-day-old young seedling of ERLC;B, A young leaf from a 4-week-old ERLC plant; C, stems of ERLC; D, A mature flower of ERLC; E, A boll of ERLC; F, A 7-day-old seedling of CCRI 24; G, A young leaf from a 4-week-old plant of CCRI 24; H, stems of CCRI 24; I, A mature flower of CCRI 24; J, A boll of CCRI 24. Scale bar indicates 1 cm.
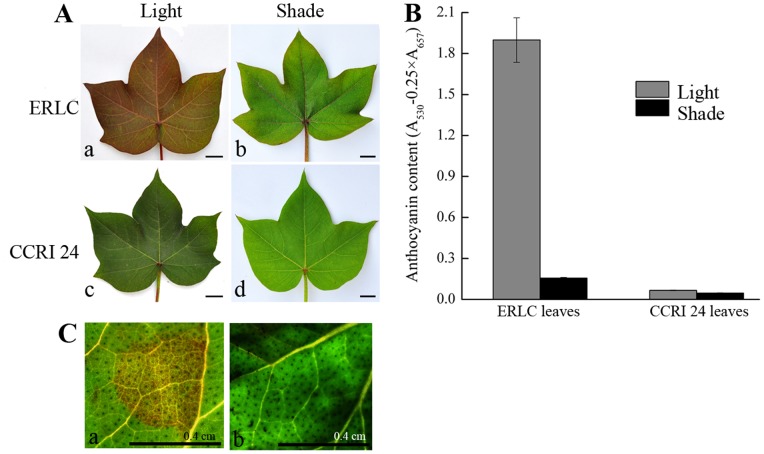
Previous research found that the red color produced by anthocyanins was distributed throughout the plant in several red leaf cotton cultivars and that the accumulation of this pigment was affected by light conditions [4]. To determine whether the color was indicative of anthocyanin accumulation and whether light conditions affects this accumulation in ERLC, the total anthocyanin content in the leaves of both ERLC and CCRI 24 plants grown in light and shaded conditions was measured using a UV spectrometer (Fig. 2A, 2B). High concentrations of anthocyanins were detected in the leaves of ERLC plants grown in light (Fig. 2A, panel a). However, a lower concentration of anthocyanin was detected in the leaves of ERLC plants grown in shade, and in the leaves of CCRI 24 plants grown in light (Fig. 2A, panels b and c and Fig. 2B). Anthocyanins were barely detectable in the leaves of CCRI 24 plants grown in shaded conditions (Fig. 2A, panel d, and Fig. 2B). Anthocyanin content correlated with light conditions in ERLC plants, with the concentration in light being 11-fold higher than that in shade. However, no significant differences in anthocyanin content were detected between the leaves of CCRI 24 plants grown in light and shade. These results suggest that the light environment strongly affects the anthocyanin accumulation process in ERLC, but that the anthocyanin biosynthesis pathway is not influenced by light conditions in CCRI 24.
Figure 2. Comparison of leaf colors and analysis of total anthocyanin concentrations of leaves from ERLC and CCRI 24 cultivars grown in light and shade conditions.
A, Comparison of leaf colors. The cultivars are indicated on the left and conditions are indicated above. The bar indicates 1; b,A mature leaf of ERLC grown in shade; c, A mature leaf of CCRI 24 grown in light; d, A mature leaf of CCRI 24 grown in shade. B, Total anthocyanin extracted from three fully opened young leaves of each cultivar, respectively, measured using a UV spectrometer. Means of three replicates with error bars indicating standard error (± SD). C, Transient analysis was performed on the leaves of CCRI 24, the Agrobacterium strain GV3101/pBI35S::ROSEA1 (left), and the negative control GV3101/pBI121 (right). The treated cotton leaves were cultured at 25°C in, 16 h light for three days, and observed for color accumulation by microscopy. Scale bar is 0.4 cm.
Recent studies have detected the expression of structural genes involved in the flavonoid (including anthocyanin) synthetic pathway, such as CHS, CHI, F3H, DFR, ANS, and ANR, in the cotton fibers of several cotton cultivars [49], [50]. Despite no visible anthocyanin accumulation, the expression of structural genes was detected in the leaves of CCRI 24 plants grown in light conditions in our study (data not shown). These results indicate that the expression of structural genes may not directly respond to light and affect anthocyanin accumulation in cotton. Therefore, anthocyanin synthesis in leaves of cotton might be affected by light-induced regulatory genes.
To confirm our prediction, we performed a transient analysis using a combination of mature green young leaves of CCRI 24 and the Agrobacterium strain GV3101/pBI35S::ROSEA1, harboring the R2R3-MYB transcriptional factor ROSEA1, (verified to be a regulatory gene controlling the anthocyanin synthetic pathway in Antirrhinum), under the control of the 35S promoter, by infiltration [47]. Red anthocyanin accumulation was detected in treated leaves of CCRT 24 (Fig. 2C, panel a), but not in GV3101/pBI121, a negative control (Fig. 2C, panel b). This result shows that the expression of ROSEA1-like MYB genes affects anthocyanin biosynthesis in the leaves of cotton, and that its expression might be controlled by light.
Isolation of RLC1 and Phylogenetic Analysis
Several studies have reported that the R2R3-MYB genes regulate the anthocyanin biosynthetic pathway and that their expression is controlled by light [51], [52]. In this study, we observed color accumulation in the leaves of ERLC in response to light. We designed a degenerate primer set (Table S1) based on the conserved regions in the R2R3 domain of anthocyanin-related MYB transcription factors and performed degenerate PCR to isolate R2R3-MYB transcription factor genes from the cDNA of 7-day-old pigmented hypocotyls of ERLC (Fig. 1A). A 240-bp cDNA fragment was isolated and showed 77% DNA sequence identity with the R2R3 domain of the MYB transcription factor ROSEA1, which has been verified to regulate the anthocyanin biosynthetic pathway in Antirrhinum [17]. Full-length cDNAs were determined using 5′/3′-RACE PCR techniques. The isolated 744-bp candidate cDNA encodes a protein of 247 amino acids. We named this gene RLC1 (Red Leaf Cotton 1).
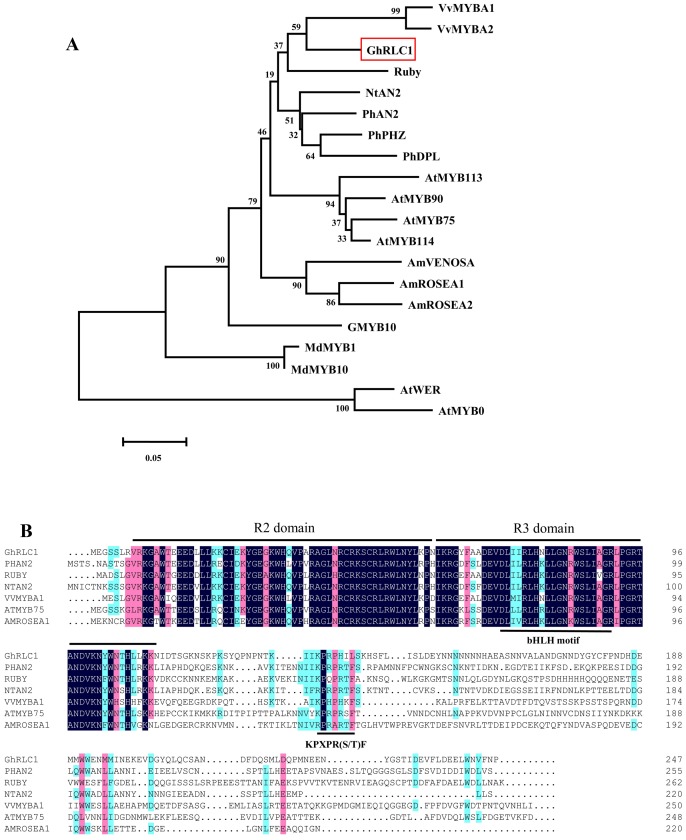
Phylogenetic analysis showed that RLC1 belonged to the same cluster as VvMYB1, VvMYB2, and Ruby, which have been shown to regulate the anthocyanin synthesis pathway in grape species and in Citrus sinensis (Fig. 3A). The deduced amino acid sequence of RLC1 shared 85% and 86% identity with VvMYB1 and Ruby, respectively, in the R2R3-MYB DNA-binding domain. The completed protein sequence of RLC1 was 44% identical to VvMYB1 of grape, and 46% identical to Ruby of Citrus sinensis. The alignment of the RLC1 protein sequence with some known R2R3-MYB regulators from different plant species revealed a very conserved R2R3 repeat domain and a [DE]Lx2[RK]x3Lx6Lx3R motif for interaction with bHLH proteins (Fig. 3B). Additionally, another signature motif, KPXPR(S/T)F, which is specific to MYB proteins that activate anthocyanin biosynthesis, was found [53]. Thus, sequence analysis indicated that RLC1 is an R2R3-MYB like gene involved in the regulation of the anthocyanin synthetic pathway in ERLC.
Figure 3. Comparison of the deduced amino acid sequence of RLC1 with verified MYB genes of other plant species.
A, Phylogenetic tree of RLC1 and selected R2R3-MYBs from other plant species. The multiple sequence alignment was performed with the R2R3 domain of MYB proteins. The tree was constructed using the neighbor-Joining method using MEGA software. Numbers along the branches indicate bootstrap support determined from 1,000 trials, and the bar indicates an evolutionary distance of 0.05%. B, Alignment of deduced amino acid sequences of RLC1 with MYB transcriptional regulators. The R2 and R3 repeat domains are indicated by lines above, and the conserved region of the bHLH interacting motif ([DE]Lx2[RK]x3Lx6Lx3R) and the conserved KPRPR[S/T]F motif are underlined.
Functional Analysis of RLC1
To determine the function of RLC1, an expression binary vector pBI35S::RLC1, harboring the full-length RLC1 cDNA under the control of the cauliflower mosaic virus 35S promoter, was transformed into hypocotyl segments of A. majus JI7 by A. rhizogenes-mediated transformation [44]. pBI35S::ROSEA1 as a positive control and pBI121(35S::GUS) as a negative control were also transformed into A. majus. Two weeks after infection, transformed non-pigmented hairy roots emerged from the wounded end of hypocotyl segments of A. majus co-cultivated with pBI121, pBI35S::CRL1, and pBI35S::ROSEA1 (Fig. 4A). Highly pigmented hairy roots emerged from hypocotyl segments transformed with pBI35S::RLC1 and pBI35S::ROSEA1 later than non-pigmented hairy roots. Pigmented hair roots were also found to elongate slower than non-pigmented hairy roots (Fig. 4B, 4C). The callus, hairy roots, and emerged shoots of segments transformed with pBI121 showed no pigmentation. Additionally, we obtained 11 and 14 independent pigmented hairy roots of pBI35S::RLC1 and pBI35S::ROSEA1, respectively, after four weeks of culture. Transformed shoots from pigmented hairy roots consistently showed pale or dark red pigmentation in the leaves and stem (Fig. 4D).
Figure 4. Phenotypic comparison of the hairy roots of A. majus transformed with pBI121, pBI35S::ROSEA1, and pBI35S::RLC1, and expression analysis of genes involved in the anthocyanin pathway in A. majus by RT-PCR.

Hairy roots transformed with A, pBI121; B, pBI35S::RLC1; C, pBI35S::ROSEA1. D, A shoot regenerated from hairy roots transformed with pBI35S::RLC1. Anthocyanin pigmentation is apparent in the leaves and stems. E, RT-PCR analysis of gene expression in the hairy roots of A. majus transformed with pBI121 (negative control), pBI35S::ROSEA 1 (positive control), and pBI35S::RLC1, respectively.
To confirm that RLC1 activates genes of the anthocyanin biosynthetic pathway, we investigated the expression levels of the structural genes CHS, F3H, DFR, and ANS in the hairy roots of A. majus of transformed with pBI121, pBI35S::RLC1 or pBI35S::ROSEA1 by semi-quantitative reverse transcription (RT)-PCR. No visible expression of the investigated structural genes was detected in negative control hairy roots transformed with pBI121. However, the expression of structural genes was dramatically up-regulated in hair roots transformed with both pBI35S::RLC1 and pBI35S::ROSEA1 (Fig. 4E). RLC1 and ROSEA1 activated the expression levels of these structural genes to very similar extents, and this correlated with anthocyanin accumulation in hairy roots transformed with pBI35S::RLC1 and pBI35S::ROSEA1 (Fig. 4). These results indicate that RLC1 has similar functions to ROSEA1 of A. majus and that it is commonly involved in regulating the anthocyanin synthetic pathway in ERLC.
Analysis of RLC1 Expression Patterns between ERLC and CCRI 24
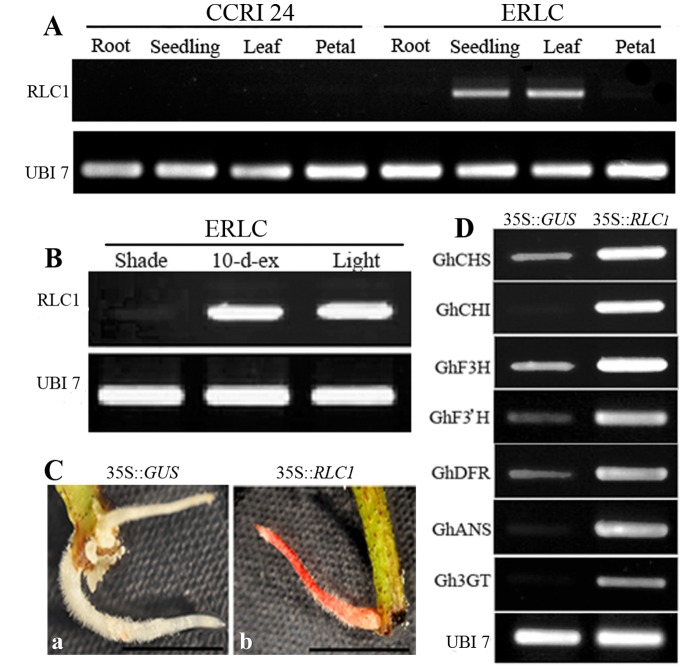
To investigate and compare RLC1 expression between the ERLC and CCRI 24 varieties, semi-quantitative RT-PCR analysis was performed using total RNA isolated from roots, seedlings, leaves, and mature petals. Both ERLC and CCRI 24 plants were grown in light conditions. The expression pattern of RLC1 was compared in ERLC and CCRI 24 (Fig. 5A). Strong expression of RLC1 was detected in the seedlings and leaves of ERLC, but RLC1 was barely expressed in the roots and petals of ERLC. Slight expression of RLC1 was detected in the leaves of CCRI 24, but expression of RLC1 was silenced in the roots and seedlings of CCRI 24.
Figure 5. Analysis of RLC1 and gene expression levels in cotton by RT-PCR. UBI 7 was used as a positive control.
A, RLC1 expression analysis was performed in roots, seedlings, leaves, and mature petals of ERLC and CCRI 24 cultivars grown in light by semi-quantitative RT-PCR. B, Comparison of the expression levels of RLC1 in mature leaves of ERLC grown in shade, light, and combined conditions. C, Comparison of color accumulation in transformed hairy roots of CCRI 24. a, Hairy root transformed with pBI121; b, Hairy root transformed with pBI35S::RLC1. Scale bar indicates 1 cm. D, Expression analysis of structural genes in hairy roots of transformed CCRI 24 with the negative controls pBI121 and pBI35S::RLC1, respectively.
To investigate the effect of light conditions on the expression level of RLC1, RT-PCR analysis was performed using total RNA isolated from the leaves of ERLC plants grown in light, those grown in the shade, and those transferred to light after 10 days in the shade (Fig. 5B). Slight expression of RLC1 was detected in the leaves of plants grown in the shade. Remarkably up-regulated expression of RLC1 was detected in the leaves of plants 10 days after being transferred from shade to light. The expression level was very similar to that in leaves grown in light (Fig. 5B). The RLC1 transcript level coincided with anthocyanin content and coloration in ERLC and CCRI 24 cultivars (Fig. 2A, 2B; Fig. 5B). These results indicate that light-induced RLC1 regulates anthocyanin accumulation in the leaves of ERLC plants. Furthermore, reduced expression of RLC1 may cause the absence of pigmentation in the leaves of CCRI 24 plants.
To test our hypothesis, pBI35S::RLC1 and the negative control plasmid pBI121 were introduced into 7-day-old hypocotyls of CCRI 24 by A. rhizogenes–mediated transformation. After six weeks of infection, five pigmented hairy roots were obtained from 30 hypocotyl segments transformed with pBI35S::RLC1. Hairy roots from plants transformed with pBI121 did not show any colors (Fig. 5C). To investigate the role of RLC1 in the anthocyanin biosynthetic pathway in CCRI 24, we analyzed the expression levels of the structural genes CHS, CHI, F3H, F3′H, DFR, ANS, and 3GT in hairy roots transformed with pBI12 and pBI35S::RLC1 by semi-quantitative RT-PCR. All the investigated structural genes were expressed in negative control hairy roots transformed with pBI121; in particular, CHI and 3GT were only slightly expressed. In pigmented hairy roots transformed with pBI35S::RLC1, RLC1 clearly activated the expression of the investigated structural genes, with CHI and 3GT expression being significantly up-regulated (Fig. 5D). These findings clearly indicate that all the structural genes involved in the anthocyanin biosynthetic pathway in CCRI 24 were normal. The expression level of RLC1 influences the transcription of structural genes and affects anthocyanin accumulation in the leaves of CCRI 24 plants. Differential expression patterns of RLC1 between ERLC and CCRI 24 might be due to differences between alleles.
Comparison of RLC1 Alleles between ERLC and CCRI 24
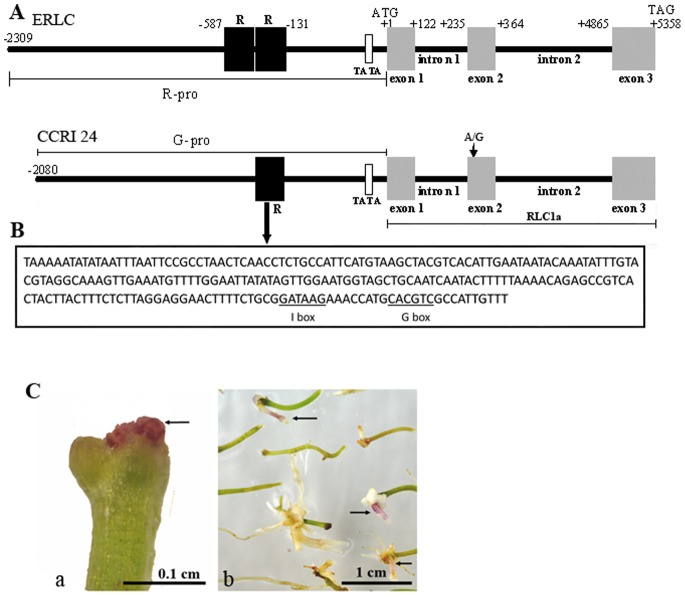
To test this idea, a 7-kb DNA fragment was isolated from ERLC genomic DNA using a DNA Walking Kit. This fragment included a 2.3-kb promoter region, three exons, and two intron regions (Fig. 6A, top). Based on the 7-kb DNA sequence of ERLC, a corresponding DNA fragment was amplified from the CCRI 24 genomic DNA by PCR (Fig. 6A, bottom). On comparing the DNA coding regions of ERLC and CCRI 24, we found that the CCRI 24 sequence also showed the same structural features as that of ERLC, with the same lengths for exon 1, exon 2, exon 3, intron 1, and intron 2. We found thirteen nucleotide polymorphisms in the introns (data not shown) and 1 nucleotide difference in exon 2 where an adenine (A) instead of a guanine (G) in exon 2 resulted in an Arg-to-His change at the R2 repeat domain of the deduced amino acid sequence (Fig. 6A, bottom). Except for an amino acid change, the coding region can be encoded as a full-length RLC1 cDNA with 99.6% identity between ERLC and CCRI 24. A major difference between ERLC and CCRI 24 was determined in promoter regions. The 2.3-kb promoter region of ERLC (R−pro) has a putative TATA box at position −89 and two 228-bp length repeat fragments (R) located in sequence between positions −131 and −587. However, the promoter region of CCRI (G−pro) is shorter than R−pro by 228 bp and lacks one of the repeats (R) at position −359 to −588. We also analyzed the 228-bp fragment sequence in the Plant Cis-acting Regulatory DNA Elements (PLACE) database [46], and found a putative I-box and a G-box that have been identified by several groups as a cis-acting element involved in the activation of light-regulated plant genes [54], [55]. However, we did not find other putative cis-acting elements or structural candidates in the sequence of the 228 bp-fragment.
Figure 6. Comparison of the RLC1 alleles between ERLC and CCRI 24.
A, The coding sequence is shown in gray boxes and the non-coding sequence is shown as a black line. Location of the putative TATA box is shown by an empty square. Repeat fragments (R) are indicated by black squares. The exons, introns, and promoter region are labeled. The location of a nucleotide change site is also indicated in exon 2 of CCRI 24. Numbers refer to the position relative to the first nucleotide of the start codon. B, DNA sequence of the 228-bp fragment, with the location of putative I-box and G-box (underlined). C, Functional analysis of the RLC1a region of CCRI 24 (Fig. 6A, bottom) in the hairy roots of A. majus by A. rhizhogenes-mediated transformation. The pBI35S::RLC1a expression vector contained the RLC1a region on pBI121 driven by the cauliflower mosaic virus 35S promoter. a, a red pigmented mass developed on the end of hypocotyl segment of A. majus two weeks after infection; b, red pigmented hairy root developed from the ends of hypocotyl segments four weeks after infection. The pigmented mass (a) and hairy roots (b) are indicated by small arrows.
The Two 228-bp Fragments Form an Important Motif in the RLC1 Promoter
To detect the effects of the thirteen polymorphisms in the introns and the amino acid change for RCL1 transcription in CCRI 24, we created an expression vector pBI35S::RLC1a, harboring a 5-kb DNA fragment including the three exons and two introns of CCRI 24(Fig. 6A, bottom), under the control of the cauliflower mosaic virus 35S promoter. This expression vector was transformed into hypocotyls of A. majus by A. rhizogenes-mediated transformation. Two weeks after infection, we observed a red colored mass growing from the end of the hypocotyl, and several red-pigmented hairy roots were obtained four weeks after culture (Fig. 6C). This result indicated that the amino acid change and thirteen polymorphisms in the introns did not affect RLC1 transcription in CCRI 24. This suggests that the differences in the promoter region between ERLC and CCRI 24 might affect RLC1 expression (Fig. 6A).
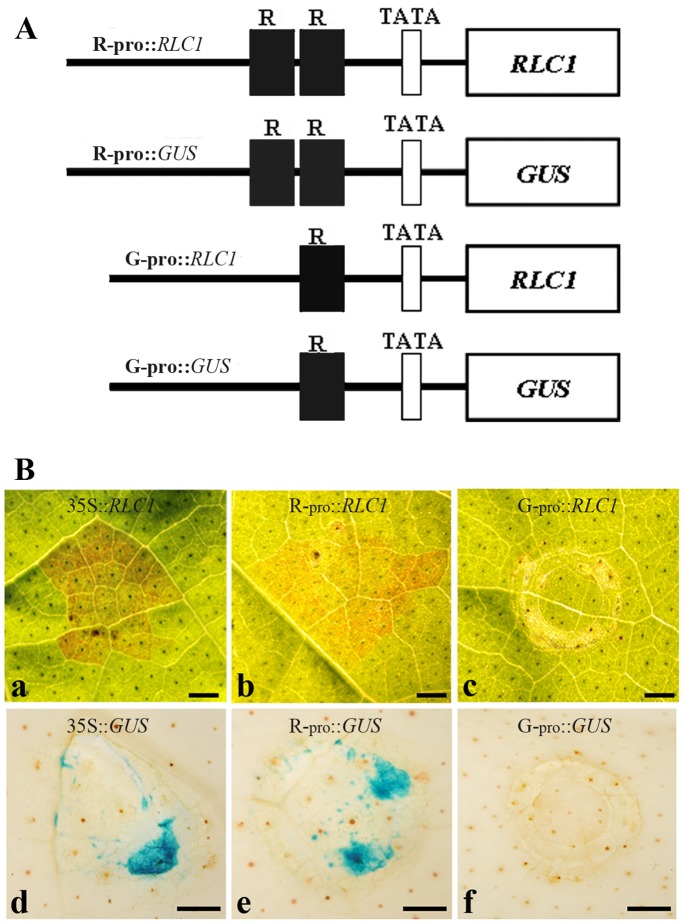
To confirm our expectation, we created several expression vectors R−pro::RLC1, R−pro::GUS, G−pro::RLC1, and G−pro::GUS (Fig. 7A) and performed transient analysis using young mature green leaves of CCRI 24 with an infiltration method [47]. At three days post-infiltration, red color was observed at the infiltrated points in cotton leaves with R−pro::RLC1 and 35S::RLC1 respectively (Fig. 7B, panels a and b). However, the red color was not observed in leaves infiltrated with G−pro::RLC1 (Fig. 7B, panel c). Similar results were also observed in the transient analysis using the β-glucuronidase (GUS) gene as the reporter, driven by the R−pro or G−pro promoter (Fig. 7B, panels d–f). These experiments strongly support the hypothesis that the repeat structure is necessary as a putative donor motif on the promoter to regulate RLC1 expression in cotton in light conditions.
Figure 7. Analysis of RLC1 promoter activity by infiltration.
A, Diagrams of constructs for the analysis of RLC1 promoter activity. R−pro: 2300-bp promoter region of RLC1 of ERLC; G−pro: 2080-bp promoter region of RLC1 of CCRI 24 (Fig. 6). B, promoter activity tests were performed using mature young leaves of CCRI 24 using the combination of expression vectors described above. Treated cotton leaves were cultured at 25°C with 16-h light periods for three days, and the leaves were used for color observation or GUS staining. a, A leaf infiltrated with 35::RLC1 and cultured for three days in light; b, A leaf infiltrated with R−pro::RLC1 and cultured for three days in light; c, A leaf infiltrated with G−pro::RLC1 and cultured for three days in light; d, A GUS-stained leaf infiltrated with pBI121 and cultured for three days in light; e, A GUS stained leaf infiltrated with R−pro::GUS and cultured for three days in light; f, A leaf infiltrated with G−pro::GUS and cultured for three days in light. Scale bar is 0.1 cm.
Discussion
RLC1 is an R2R3-MYB Transcription Factor
In this study, using degenerate PCR based on a R2R3-MYB domain-specific sequence, we isolated a transcriptional factor, named RLC1, from ERLC. RLC1 encoded a R2R3-MYB protein clearly placed in a clade with many anthocyanin-regulating MYB transcription factors from different species such as PhAN2 [12], Rosea1 [17], PAP1 [18], MdMYB1 [51], Ruby [56], VvMYBAs [22], [26], [57] and NtAN2 [58]. Alignment of the amino acid sequence of RLC1 showed significant similarity to structures with Ruby and PhAN2, which harbored very conserved R2R3 domains, a conserved ([DE]Lx2[RK]x3-Lx6Lx3R) motif thought to be responsible for the interaction between bHLH and MYB proteins (Fig. 3) [59], and a KPRPR[S/T]F motif specific to R2R3-MYB regulators in anthocyanin synthesis [53]. Genomic DNA sequence analysis also showed that RLC1 has three exons. The R2 domain spans exons 1 and 2, and the R3 domain spans exons 2 and 3. The genomic organization and R2R3 domain distribution of RLC1 was similar to that of other R2R3-MYBs [12], [18], [58]. These structural similarities in the DNA and amino acid sequences suggest that RLC1 is likely to have similar functions to the anthocyanin -specific MYB regulators and a common evolutionary origin.
RLC1 is PAP1/ROSEA1/Ortholog Gene
In this study, we confirmed that RLC1 is a positive regulator of anthocyanin synthesis through the transformation of Antirrhinum and green leaf cotton CCRI 24. Overexpression of RLC1 resulted in anthocyanin accumulation in the transformed hairy roots of Antirrhinum and cotton.
In the hairy roots of Antirrhinum, 35S::GUS did not affect expression of the endogenous anthocyanin synthesis genes Am CHS, AmF3H, AmDFR, and AmANS, and the hairy roots lacked coloration. However, the expression of Am CHS, AmF3H, AmDFR, and AmANS was significantly upregulated by 35S::RLC1, and strong anthocyanin accumulation was observed in the hairy roots in contrast to those of plants transformed with 35S::GUS, which lacked anthocyanin. 35S::RLC1-activated expression levels of anthocyanin synthetic genes showed patterns similar to those activated by 35S::ROSEA1 in transformed hairy roots (Fig. 4). These results suggest that RLC1 is an ortholog of ROSEA1 in Antirrhinum, and is mainly involved in the regulation of anthocyanin synthesis in cotton.
In cotton, despite the negative control in hairy roots transformed with 35S::GUS that lacked anthocyanin accumulation, the endogenous structural genes GhCHS, GhCHI, GhF3H, GhF3′H, GHDFR, GhANS, and Gh3GT in the anthocyanin pathway showed detectable expression. Recent studies have detected the expression of these structural genes in white fibers [49], [50]. Therefore, we determined which structural genes are commonly expressed in most tissues and organs of green leaf cotton, and whether they might be involved in the production of other flavonoids, such as proanthocyanidins. We also found that the structural genes GhCHI and Gh3GT were significantly upregulated in the transformed hairy roots of 35S::RLC1 as well; these hairy roots showed notable anthocyanin accumulation (Fig. 5C, 5D). Therefore, the expression levels of GhCHI, an EBG, and Gh3GT, an LBG, may affect anthocyanin biosynthesis and which genes expression is controlled by RLC1 expression. Although the R2R3-MYB genes mostly regulate LBG expression in the flavonoid pathway [17], similar disagreements regarding the regulation of EBGs and LBGs have been reported in the over-expression of PAP1 in transgenic Arabidopsis and NtAN2 in tobacco. Over-expression of the PAP1 and NtAN2 MYB regulators results in the up-regulation of genes across the entire phenylpropanoid pathway in Arabidopsis and tobacco [35], [58]. Therefore, RLC1 and PAP1 may redundantly regulate the transcription of EBGs in a manner similar to MYB regulators in Arabidopsis and cotton. Additionally, the elevated expression of EBGs may be related to a metabolite feedback phenomenon, induced by the up-regulation of LBGs.
Since the transcription of structural genes in the anthocyanin pathway was affected, the genetic basis of cotton color is probably due to the activity of a common regulator of the RLC1 gene. This would be analogous to PAP1 in Arabidopsis and ROSEA1 in Antirrhinum [17], [18].
Promoter Differentiation Affects RLC1 Expression between ERLC and CCRI 24
In this study, we investigated RLC1 alleles between red leaf cotton ERLC and green leaf cotton CCRI 24. We found a 228-bp fragment spanning positions −131 to −359 bp in the RLC1 promoter of CCRI 24 (G−pro), and two 228-bp fragments were detected from position −131 to −587 bp forming a near-perfect direct tandem repeat in the RLC1 promoter of ERLC (R−pro). Further, through transient expression analysis of RLC1 promoter activity in both ERLC and CCRI 24, we found out that the promoter of ERLC (R−pro) normally drove fused genes (Fig. 7). These results suggested that the repeat structure is a putative donor motif of the promoter region that acts to promote RLC1 gene expression in leaves of cotton.
Light Stimulation Causes RLC1 Expression in ERLC
Environmental conditions control the anthocyanin biosynthetic pathway in plants. In Arabidopsis, maize, apple, and grape, anthocyanin biosynthesis is enhanced by sunlight stimulation [39], [53], [60]–[63]. In mature grape plants, light and sugar content affect MYB regulation of flavonoid biosynthesis [61]. Furthermore, in apple, MYB activation of anthocyanin synthesis is affected by light. When the fruit are bagged, MdMYB1 expression is prevented in the red-skinned apple “Cripps Pink” [51]. High light exposure also upregulates the Arabidopsis R2R3-MYB activators PAP1 and PAP2 and their bHLH partner TT8 [39], [64]. Moreover, it has been previously reported that the expression of specific maize MYB genes is strongly light induced and is closely correlated with the light induction of structural genes and anthocyanin biosynthesis [60], [64].
In this study, we have shown that anthocyanin biosynthesis in the leaves of cotton plant is also controlled by light. The expression level of RLC1 and anthocyanin accumulation in leaves of ERLC was affected by light conditions (Fig. 2; Fig. 5B). When grown in light, RLC1 was strongly expressed in leaves and seedlings. However, when grown in shaded conditions, RLC1 was scarcely expressed in leaves, but when plants were transferred to light, expression of RLC1 recovered to normal levels and the leaves developed a deep red color (Fig. 5B). The RLC1 expression pattern also correlated with the expression of other anthocyanin biosynthetic genes.
In addition, we also analyzed the 228-bp fragment sequence against the PLACE database [46], and found several putative cis elements. Among them, we detected a putative I-box and G-box identified by several groups as a cis-acting element involved in the activation of light-regulated plant genes [54], [55], [65]. Therefore, a light signal might interact with a light-responsive element in the promoter region of the RLC1 gene, and enhance the expression of RLC1, which would then regulate the expression of anthocyanin structural genes and the anthocyanin pathway in cotton. Although most genes in the anthocyanin biosynthesis pathway are known, it is not known how the MYB genes regulate anthocyanin biosynthesis in cotton. We have shown that the R2R3-MYB gene RLC1 regulates anthocyanin structural genes in response to light induction. However, the influence of other bHLH and WD40 cofactors required for anthocyanin synthesis have not been determined and so their role in light regulation remains to be determined.
In summary,in ERLC the anthocyanin pathway is constitutively regulated but is also responsive to light. We have identified an R2R3-MYB transcriptional factor RLC1 that has an expression pattern correlating with that of anthocyanin accumulation in the leaf and in response to light. The MYB regulator RLC1 can enhance the expression of structural genes in transformed hairy roots of Antirrhinum and cotton. This expression is due to a motif of two 228-bp fragments forming tandem repeats more than −131 bp upstream of the promoter region of RLC1. These results suggest that RLC1 regulates by light induction structural genes across the anthocyanin pathway in red leaf cotton. This provides the potential to modulate leaf color by altering the expression of this regulatory gene. It will be of great interest to determine how the response to light mediated through this key regulator controls anthocyanin accumulation in cotton leaf.
Supporting Information
Primers used for gene cloning in this study.
(DOC)
Primers used for quantitative RT-PCR analysis in this study.
(DOC)
Acknowledgments
The authors thank Professor Enrico Coen from John Innes Centre, United Kingdom for providing the Antirrhinum majus seeds.
Funding Statement
This work was supported by the Chinese Academy of Sciences (grant no. KSCX3-EW-N-03), and the Yunnan Provincial Science and Technology Department (grant no. 2012IB011) to M.L.C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zafar Y, Asif M, Kausar A, Riaz S (2009) Niaz (2009) Development of genetic linkage map of leaf red colour in cotton (Gossypium hirsutum) using DNA markers. Pak J Bot 41: 1127–1136. [Google Scholar]
- 2. Fitt GP (1994) Cotton pest management: part 3. An Australian perspective. Annu Rev Entomol 39: 543–562. [Google Scholar]
- 3. Stephens S (1974) Geographic and taxonomic distribution of anthocyanin genes in New World cottons. J Genet 61: 128–141. [Google Scholar]
- 4. Killough D, Horlacher W (1933) The inheritance of virescent yellow and red plant colors in cotton. Genetics 18: 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55: 481–504. [DOI] [PubMed] [Google Scholar]
- 6. Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780. [DOI] [PubMed] [Google Scholar]
- 7. Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749. [DOI] [PubMed] [Google Scholar]
- 8. Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinum majus . Plant J 1: 37–49. [DOI] [PubMed] [Google Scholar]
- 9. Pelletier MK, Shirley BW (1996) Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings (Coordinate regulation with chalcone synthase and chalcone isomerase). Plant Physiol 111: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pelletier MK, Murrell JR, Shirley BW (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis (Further evidence for differential regulation of “early” and “late” genes). Plant Physiol 113: 1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, et al. (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spelt C, Quattrocchio F, Mol J, Koes R (2002) ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 14: 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grotewold E, Athma P, Peterson T (1991) Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc Natl Acad Sci USA 88: 4587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sainz MB, Grotewold E, Chandler VL (1997) Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9: 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68: 955. [DOI] [PubMed] [Google Scholar]
- 17. Schwinn K, Venail J, Shang YJ, Mackay S, Alm V, et al. (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, et al. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, et al. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chagné D, Lin-Wang K, Espley RV, Volz RK, How NM, et al. (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982–982. [DOI] [PubMed] [Google Scholar]
- 23. Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46: 74–83. [DOI] [PubMed] [Google Scholar]
- 24. Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41: 577–585. [DOI] [PubMed] [Google Scholar]
- 25. Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, et al. (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49: 772–785. [DOI] [PubMed] [Google Scholar]
- 27. Aharoni A, De Vos C, Wein M, Sun Z, Greco R, et al. (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332. [DOI] [PubMed] [Google Scholar]
- 28. Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, et al. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. The EMBO J 19: 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, et al. (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967. [DOI] [PubMed] [Google Scholar]
- 31. Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, et al. (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49: 641–654. [DOI] [PubMed] [Google Scholar]
- 32. Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- 34. Ramsay NA, Glover BJ (2005) MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70. [DOI] [PubMed] [Google Scholar]
- 35. Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827. [DOI] [PubMed] [Google Scholar]
- 36. Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, et al. (2003) Proanthocyanidin -accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, et al. (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . Plant J 39: 366–380. [DOI] [PubMed] [Google Scholar]
- 38. Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, et al. (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana . Planta 224: 96–107. [DOI] [PubMed] [Google Scholar]
- 39. Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, et al. (2008) Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 165: 886–894. [DOI] [PubMed] [Google Scholar]
- 40. Rabino I, Mancinelli AL (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 42. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 43. Shen WJ, Forde BG (1989) Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res 17: 8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cui M, Takayanagi K, Kamada H, Nishimura S, Handa T (2001) Efficient shoot regeneration from hairy roots of Antirrhinum majus L. transformed by the rol type MAT vector system. Plant Cell Rep 20: 55–59. [DOI] [PubMed] [Google Scholar]
- 45. Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, et al. (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18: 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956. [DOI] [PubMed] [Google Scholar]
- 48. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao YH, Zhang ZS, Yin MH, Luo M, Li XB, et al. (2007) Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem Biophys Res Commun 358: 73–78. [DOI] [PubMed] [Google Scholar]
- 50. Li YJ, Zhang XY, Wang FX, Yang CL, Liu F, et al. (2013) A comparative proteomic analysis provides insights into pigment biosynthesis in brown colored fiber. J proteomics 78: 374–388. [DOI] [PubMed] [Google Scholar]
- 51. Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, et al. (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142: 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, et al. (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65: 771–784. [DOI] [PubMed] [Google Scholar]
- 53. Kui LW, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, et al. (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYBI is a member of a novel class of Myb-like proteins. Plant J 20: 641–652. [DOI] [PubMed] [Google Scholar]
- 55. Agius F, Amaya I, Botella MA, Valpuesta V (2005) Functional analysis of homologous and heterologous promoters in strawberry fruits using transient expression. J Exp Bot 56: 37–46. [DOI] [PubMed] [Google Scholar]
- 56. Butelli E, Licciardello C, Zhang Y, Liu JJ, Mackay S, et al. (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24: 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215: 924–933. [DOI] [PubMed] [Google Scholar]
- 58. Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, et al. (2010) Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231: 1061–1076. [DOI] [PubMed] [Google Scholar]
- 59. Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34. [DOI] [PubMed] [Google Scholar]
- 60. Piazza P, Procissi A, Jenkins GI, Tonelli C (2002) Members of the c1/pl1 regulatory gene family mediate the response of maize aleurone and mesocotyl to different light qualities and cytokinins. Plant Physiol 128: 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Matus JT, Loyola R, Vega A, Peña-Neira A, Bordeu E, et al. (2009) Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera . J Exp Bot 60: 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Page M, Sultana N, Paszkiewicz K, Florance H, Smirnoff N (2011) The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: further evidence for redox control of anthocyanin synthesis. Plant Cell Environ 35: 388–404. [DOI] [PubMed] [Google Scholar]
- 63. Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, et al. (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cone KC, Cocciolone SM, Burr FA, Burr B (1993) Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Giuliano G, Pichersky E, Malik V, Timko M, Scolnik P, et al. (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for gene cloning in this study.
(DOC)
Primers used for quantitative RT-PCR analysis in this study.
(DOC)