Summary
The regrowth of amputated limbs and the distal tips of digits represent models of tissue regeneration in amphibians, fish, and mice. For decades it had been assumed that limb regeneration derived from the blastema, an undifferentiated pluripotent cell population thought to be derived from mature cells via dedifferentiation. Here we show that a wide-range of tissue stem/progenitor cells contribute to restore the mouse distal digit. Genetic fate mapping and clonal analysis of individual cells revealed that these stem cells are lineage restricted, mimicking digit growth during development. Transplantation of CFP expressing hematopoietic stem cells, and parabiosis between genetically marked mice, confirmed that the stem/progenitors are tissue resident, including the cells involved in angiogenesis. These results, combined with those from appendage development/regeneration in lower vertebrates, collectively demonstrate that tissue stem cells rather than pluripotent blastema cells are an evolutionarily conserved cellular mode for limb regeneration after amputation.
Keywords: blastema, digit tip, limb development, regeneration, stem cells
Introduction
While whole body regeneration from blood and vascular structures can occur in protochordate colonial tunicates1-3, regeneration of body parts is highly restricted in vertebrates, especially in mammals4. Therefore, digit tip [fingertip or toetip] regrowth in mice5-9 and humans10-12, present unique model systems for mammalian organ regeneration. Several hypothesis have been used to explain the mechanisms contributing to regeneration13. One hypothesis is that circulating precursors, perhaps of the mesenchymal or hematopoietic lineages, can enter damaged tissues and transdifferentiate (lineage conversion of a defined cell into another cell type) into the lost cell types14-17. Another concept is that the residual local mature cells of diverse types can dedifferentiate to form a blastema, a pluripotent class of cells (cells that may differentiate into various types of specialized cells) that can use pathways other than used in development to form limbs and digit structures18-21; this idea has recently been emphasized as an alternative to tissue specific stem cells playing an important role in regeneration22. A third idea is that tissue homeostasis and regeneration derive from the kinds of tissue-specific stem cells that originally were responsible for their embryonic development23-26.
Mammalian digit tips exhibit morphological variations that contributed to the evolutionary diversification27 of mammals. The digit tip is derived from multiple and distinct embryonic origins, and includes the distal bone with associated marrow cavity and hematopoietic cells, ventral (flexor) and dorsal (extensor) tendons, sweat glands with myoepithelial and luminal secreting cells and associated neurons for innervation, dermis with resident melanocytes and dendritic cells, mesenchyme with resident fibroblasts, skin epidermis with hair follicles, a nail organ composed of six specific parts (the root, nail bed, nail plate, eponychium (cuticle), perionychium, and hyponychium) and is highly vascularised28. Each of these distal structures has a specific function, damage to which may result in an abnormal digit.
Newborn and adult mice are able to regrow forelimb [finger] and hindlimb [toe] digit tips after their amputation through the distal interphalangeal joint5-9. Regeneration of the digit tip involves the integrated regrowth of multiple tissues within 2-3 months, reaching an external morphology that is cosmetically and functionally similar to normal digits. Most importantly, regeneration of the mouse distal digit shares morphological similarities with clinical cases documenting regrowth of missing distal portions of fingers in both children and adults10-12.
Here we explore the cellular source for mouse digit tip regeneration and provide evidence that regrowth is a cumulative effort of distinct stem cells and their daughter progenitors which are germ layer and lineage restricted.
Results and Discussion
Amputations that removed most structures of the distal digit consistently failed to regenerate (Supplementary Fig. 1, a, b). Consistent with previous observations7-9, amputations with residual bone, sweat glands and nail organ remaining (amputation plane 1 and 2, AP1/2), showed partial to complete regrowth of distal structures after 70 days (Supplementary Fig. 1, a-c, Supplementary table 1), with histological indications of mesenchyme cells at the digit apex (Supplementary Fig. 1, d-d″). Short pulse regimes of BrdU revealed local proliferations within defined sites of the distal digit (Supplementary Fig. 1, e-g, white arrowheads). These results implied that digit regrowth resulted from the combined activities of tissue resident stem cells. We tested this hypothesis via genetic fate mapping and lineage tracing of the major tissues within the digit.
Developmental restriction of ectoderm
The contribution of the epidermis to the regenerating digit was analyzed using Keratin-14-CreER (K14CreER) transgenic mice29. K14CreER mice were crossed with mTmG, a double-fluorescent reporter mouse that replaces the expression of tomato red with green fluorescent protein (GFP) following Cre-mediated excision30.
K14CreERmTmG mice were injected with tamoxifen, and then amputated along AP1/2 (see methods) and digits were processed for histology after three months. GFP expression within digits was confined to ectodermal tissues (Fig. 1, a-e), including dorsal and ventral epidermis, nail plate (with eponychium and hyponychium), hair follicles, and secreting portions of sweat-glands. The GFP expression was sustained, almost certainly implicating epithelial stem cells as the original tamoxifen induced cells, since several skin turnover times had occurred over the 3-month interval. GFP expression was absent from mesoderm tissues, including bone and marrow cavity, blood vessels, tendon, nail bed, dermis, and surrounding mesenchyme of the digit (Fig. 1, d, e).
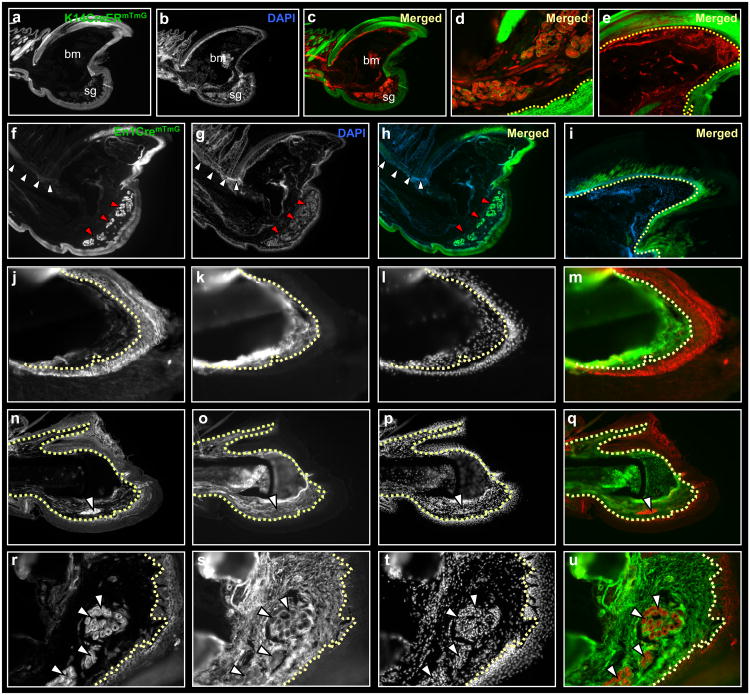
Figure 1.
Germ layer restriction of ectoderm/mesoderm during digit tip regeneration. Sections through a distal digit of K14CreERmTmG (a-e), En1CremTmG (f-i) and Prx1CremTmG (j-u) transgenic mice, following three months post-amputation. Ectoderm contributes to epidermis, nail and sweat glands and fails to contribute to mesoderm tissues (a-i). Dashed line outlines the border between epidermis/dermis (d) and nail plate/matrix (e, i). Segregation of ectoderm in En1CremTmG into dorsal and ventral fates; ventral ectoderm contributes to ventral epidermis and sweat glands (f-h, red arrowheads) with no contributions to the dorsal epidermis or hair follicles (f-h, white arrowheads). A partial contribution to nail reveals dorsal and ventral chimeric origins to the nail plate (i). This boundary is shifted compared to the published lineage mapping study, using En1CreER. Lineage tracing of Prx1Cre shows restricted GFP expression to bone, tendon and mesenchyme, with no contribution to ectoderm. Keratin-14 (K14) expression in Prx1CremTmG digits is mutually exclusive from GFP expression. High magnifications of nail (j-m), bone (n-q) and sweat glands (r-u). Dashed lines outline borders between nail plate/matrix (j-m) and epidermis/dermis (n-u). White arrowheads (r-u) shows sweat glands within ventral mesenchyme are GFP negative. bm, bone marrow; sg, sweat glands.
A similar ectoderm-specific expression of GFP emerged in regenerated digits of Keratin-5-CreERmTmG (K5CreERmTmG)31 transgenic mice (Supplementary Fig. 2, a-e). Again, the sustained expression of GFP implies that keratin 5, like keratin 14, is expressed in self-renewing stem cells.
Engrailed-1-Cre transgenic mice (En1Cre) were used to lineage trace the ventral ectoderm32, in a similar method. GFP expression was present within the ventral epidermis (Fig. 1, f-h), duct and secreting portions of sweat glands (Fig. 1, f-h, red arrowheads) and within a subset of cells of the nail plate (compare Fig. 1e to Fig. 1i). GFP expression was absent from the dorsal epidermis and its associated hair follicles (Fig. 1, f-h, white arrowheads) and from all mesodermal tissues (Fig. 1, h, i). Engrailed expression appears to be in a subset of more lineage restricted stem cells, limited to development of ventral ectoderm and to a subset of the nail plate. These results collectively show that germ-layer restriction is maintained by classes of ectodermal stem cells during digit tip regeneration, with further restriction of ectoderm into dorsal and ventral fates and chimeric contributions to the nail plate.
Developmental restriction of mesoderm
The contribution of mesoderm to the regenerating digit was analyzed using Prx1Cre33 transgenic mice, in a similar method. Within regenerated digits of Prx1CremTmG mice, GFP expression was confined to mesodermal tissues, including bone, tendons, nail bed, dermis and mesenchyme (Supplementary Fig. 2, f-j) and was absent from all ectodermal tissues (Supplementary Fig. 2, i, j).
The possibility of germ layer conversion (transdifferentiation) of mesoderm to ectoderm during digit tip regeneration was further analyzed by immunoassaying sections of regenerated digits from Prx1CremTmG mice for K14 protein. K14 protein was expressed in all ectoderm lineages of the digit and was mutually exclusive from GFP expression (Supplementary Fig. 2, k-n). The K14/GFP border distinguished between nail plate and bed (Fig. 1, j-m), between surrounding epidermis (dorsal & ventral) to digit bone and dermis (Fig. 1, n-q) and between sweat glands (duct & secreting portions) to the surrounding mesenchyme (Fig. 1, r-u), demonstrating that germ-layer restriction of mesoderm and ectoderm by their respective stem and progenitor cells is maintained during regeneration of the distal digits.
We used Sox9Cre transgenic mice to lineage trace bone/cartilage precursors34, in a similar method. In line with Sox9 expression in stem cells of the skin35, GFP was expressed within all epidermal lineages of the digits (Fig. 2, a-d; Supplementary Fig. 3). Within the mesoderm, GFP expression was restricted to the bone (Fig. 2, a-d, dotted line; Supplementary Fig. 3, a″, a‴) and was absent from tendons, nail bed, dermis, vasculature and the mesenchyme surrounding the sweat glands (Supplementary Fig. 3, b-c‴). As the Sox9 promoter drives Cre in these mice, and not Cre-ER, and also because we do not know the lifespan of osteocytes, we could not conclude whether the GFP+ bone cells derived from stem or progenitor cells.
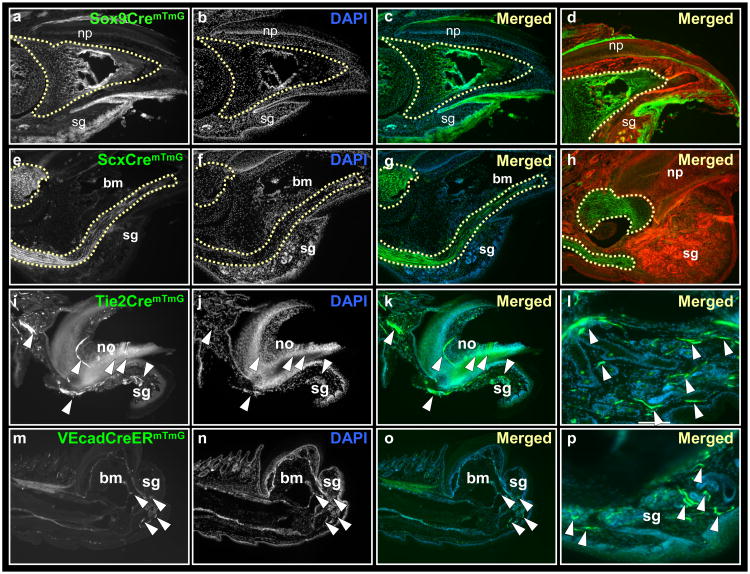
Figure 2.
Lineage restriction of bone, tendons and endothelium during digit tip regeneration. Sections through Sox9CremTmG (a-d), ScxCremTmG (e-h), Tie2CremTmG (i-l) and VEcadherinCreERmTmG (m-p) transgenic mice, following three months post amputations. Lineage tracing of Sox9 shows expression within all epidermal lineages. Within mesoderm, GFP expression is restricted to the distal digit bone (a-d, outlined by a dashed line). Lineage tracing of Scleraxis shows restriction of GFP to tendons (e-h, outlined by a dashed line). Lineage tracing of Tie2 and VEcadherin shows restriction of GFP in blood cells and blood vessels of the distal digit (i-p, white arrowheads). bm, bone marrow; np, nail plate, no, nail organ; sg, sweat glands.
Likewise, we used ScleraxisCre (ScxCre) transgenic mice crossed to the mTmG reporter to lineage trace the tendon precursors36. Within regenerated digits, GFP expression was restricted to dorsal and ventral tendons (Fig. 2, e-h, dotted line), with some GFP staining also present in a distinct group of cells within the hair follicles as previously described for developing limbs36. GFP was absent from bone, blood vessels, and surrounding mesenchyme and from ectodermal tissues (Fig. 2, g, h).
The contribution of endothelium to the regenerating digits was assayed similarly by crossing mTmG reporter mice with Tie2Cre37 and VECadherinCreER38 transgenic mice. In line with published work31, GFP+ blood vessels were abundant within the distal regenerated digits of Tie2CremTmG mice. GFP expression was restricted to blood cells and blood vessels within nail bed, the dermal/hypodermal border and surrounding the sweat glands (Fig.2, i-l, white arrowheads). GFP expression was absent from other mesodermal tissues, and all ectodermal tissues (Fig. 2, k, l). A restricted expression of GFP to blood vessels, also emerged within regenerated digits of VE-CadherinCreERmTmG mice treated with tamoxifen prior to amputation (Fig. 2, m-p, white arrowheads).
Specificity of the GFP to endothelium was confirmed by immunoassaying sections of regenerated digits from Tie2CremTmG mice for the endothelial markers Platelet Endothelial Cell Adhesion Molecule 1 (CD31; Supplementary Fig. 4, a-a‴, white arrowheads) and Pan Endothelial Cell Antigen (PEA; Supplementary Fig. 4, b-b‴, white arrowheads). Both antibodies marked blood vessels within the nail bed, sweat glands and surrounding mesenchyme and co-localized with that of GFP. We then looked at ectoderm markers of epidermis and neural fates by immunoassaying for Keratin14 (K14; Supplementary Fig. 4, c-c‴) and Myelin Basic Protein (MBP; Supplementary Fig. 4, d-d‴), respectively, and found they were mutually exclusive from that of GFP expression. Likewise, in digits from VEcadCreERmTmG mice, GFP expression was separate from mesenchyme/fibroblast-associated (CD105, CD90, S100A4) and hematopoietic (CD45) markers (Supplementary Fig. 5), indicating absence of endothelial contribution to these cell populations.
A similar tissue restricted pattern was apparent during the regrowth timepoint (10 days post amputation) associated with the appearance of a blastema39 (Supplementary Fig. 6), collectively, showing that germ-layer and lineage restriction of ectoderm and mesoderm including bone, tendon and blood vessel lineages is maintained throughout regeneration of the distal digits.
Circulating cells do not contribute to major tissues of the regenerating digits
To address hematopoietic contributions to the regenerating digits, hematopoietic stem cells were isolated from transgenic mice expressing the cyan fluorescent protein (CFP-HSCs), on the basis of the following surface expressions40: Lin- (CD3- CD4- CD8- B220- Mac- Gr-1- Ter119-) CD34- Flk2- Sca1+ c-Kit+ Slamf1+ (Fig. 3a). Wild type mice were reconstituted with 20 CFP-HSCs (n=4). CFP-HSC injected mice reached full blood chimerism (Supplementary Fig. 7), at which time digits were amputated at AP1/2, and analyzed 3 months later. HSC derived CFP cells were observed within the marrow cavity, the dermis and mesenchyme of the distal digit, and expressed the hematopoietic antigen CD45, ubiquitously (Fig. 3, b-g, white arrowheads). We did not find any contributions of CFP-HSC derived cells to the major tissues of the digit, including bone, tendons, nail organ, sweat glands, epidermis or blood vessels (Fig. 3, d, g).
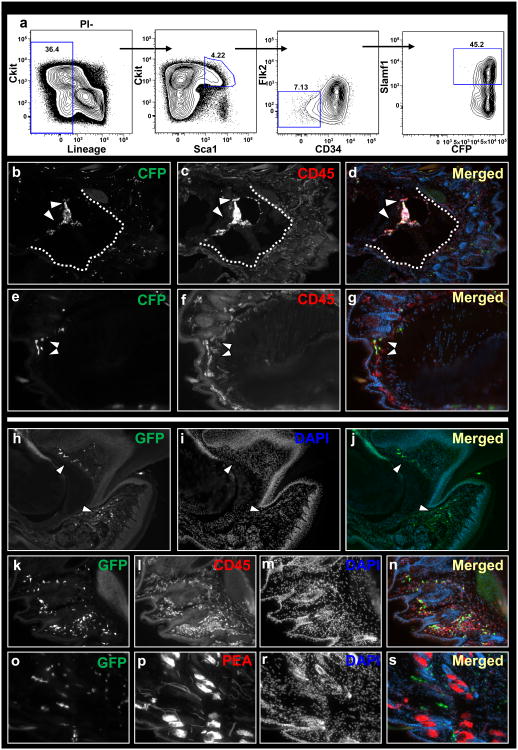
Figure 3.
Circulating cells do not contribute to regenerating tissues of the digit tip. Flow chart showing gating of HSCs on the basis of Lin− (CD3− CD4− CD8− Mac− Gr-1− B220− Ter119−) Flk2− CD34− Sca+ Slamf1+ surface expressions (a). Sections through the digit of a host mice that was infused with HSCs following three months post-amputation. HSC-derived cells within bone marrow (b-d) and dermis (e-g). Dotted line outlines bone/epidermis border. Parabiosis between wild-type and genetically marked (GFP) littermates (h-r). Circulating cells within nail organ, mesenchyme surrounding sweat glands and dermis of regenerated digits (h-j, white arrowheads). Circulating cells within the regenerated digit express the hematopoietic marker CD45 (k-n) but not the endothelial marker PEA (o-r).
To further address the possibility of a circulatory contribution to the regenerating digits, we created pairs of genetically marked parabiotic mice that have a shared anastomosed circulatory system41. Wild-type mice were surgically conjoined to mice expressing GFP under the chicken β-actin promoter. Mice were left parabiosed for one month for full chimerism to be established within the hematopoietic system. Then digits from wild type mice were amputated at AP1/2 and mice were sacrificed 3 months later. Donor derived GFP cells were present within numerous tissues of the distal digit, including its marrow cavity, epidermis, nail matrix and mesenchyme surrounding the sweat glands (Fig. 3, h-j, white arrowheads). All donor derived GFP cells expressed the hematopoietic antigen CD45 within all sites examined (Fig. 3, k-n), and failed to contribute to the major tissues of the digit. We then checked to see if donor derived GFP cells would contribute to new blood vessel formation following parabiosis. Donor derived GFP cells in the digits were closely associated with blood vessels but were mutually exclusive from PEA expression (Fig. 3, o-r). Thus, the angiogenesis that results in vascularized digit tips must derive from local, non-hematopoietic or any other circulating endothelial precursors, consistent with our previous results in tumor angiogenesis26. These results also demonstrate that circulating cells (of any origin) do not contribute to the major tissues within the digit, and are consistent with early irradiation experiments in salamanders by Butler and O'Brien (1942) that showed appendage regeneration requires a local cellular source42.
Clonal analysis of the regenerating digits
We looked at emerging clones within regenerating digit tips, using ‘Rainbow’ mice, a multi-color Cre-dependent marker system which harbor a four-color reporter construct (red, yellow, green, blue; see supplementary methods section). Clonal patterns were visualized within all tissues by crossing ‘Rainbow’ mice with mice harboring an inducible Cre under the promoter of the actin gene (ActinCreER). ActinCreERRainbow offspring were injected with tamoxifen and digits were amputated at AP1/2. Mice were sacrificed 3 months afterward, when digits were processed for histology. Clonal domains appeared within all regenerated tissue types (Fig. 4), with multiple clones occupying each tissue. A pattern of clonal stripes emerged from the nail bed contributing cumulatively multiple clones to the nail plate (Fig. 4, a-a′). Within the epidermis, clones expanded laterally to the basal epidermis, outwards into the stratified layer of the epidermis and inwards into dermal pegs close to the epidermis/dermis border (Fig. 4, b-b′), revealing that this new epidermis is derived from multiple expanding keratinocytes, not from hair follicles44. Within sweat glands, several clones contributed separately to the regenerating gland (Fig. 4, c-c′, white arrowheads). Blood vessels in association with the sweat glands derived from separate clones (Fig. 4, d-d′, white arrowheads), confirming our previous genetic fate mapping, and within bone, clones emerged with a restricted pattern to the proximal epiphysis, contributing laterally to new bone (Fig. 4, e-e′, white arrowheads). Although we are unable at this point to indicate the contributions of fibroblasts, or rule out a pre-determined multipotent cell that contributes to bone/tendon and fibroblasts, our work with isolated bone/cartilage progenitors44 and fibroblast precursors from mouse, indicate that both cell types maintain lineage restriction in-vivo and in-vitro (Y.R. personal observations).
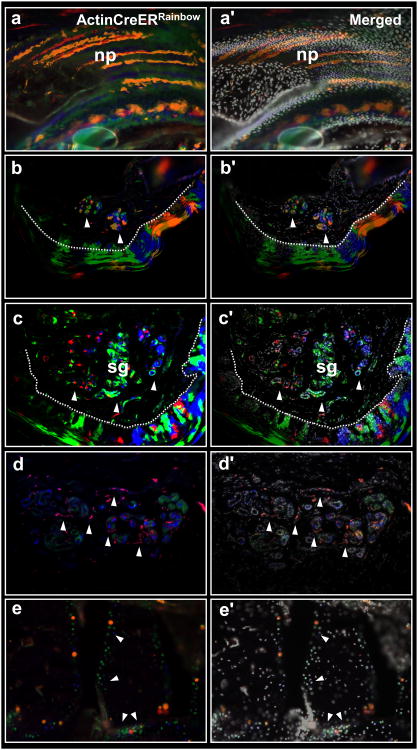
Figure 4.
Multiple lineage-restricted clones contribute to digit tip regeneration. Sections through regenerated digits of ActinCreERRainbow mice. Expanding clones within nail (a-a′), epidermis (b-b′) and sweat glands (c-c′). Dashed line (b & c) outlines the epidermis/dermis border; white arrowheads show clones within ventral sweat glands. Sweat glands (d-d′) and the surrounding blood vessels (d-d′, white arrowheads) are derived from separate clones. Expanding clones within the digit bone (e-e′, white arrowheads). np, nail plate, sg, sweat glands.
Early work by Gargiolo and Slack on xenopus tail regeneration, revealed a similarly limited range of potential to the different stem/progenitors of the xenopus tail45. In the salamander Ambystoma mexicanum, Kragl and Knapp46 recently shown that transplanted embryonic cells destined to form specific limb tissues, maintain their embryonic fates following the amputation and subsequent regeneration of the salamander limbs. More recently, a similar fate restriction was documented in developing and regenerating zebrafish fins47 Collectively, our findings in the mouse, with that in appendage development/regeneration of lower vertebrates attest to lineage restriction of stem/progenitor cells as an evolutionarily conserved cellular mode, in which change in cell fate or dedifferentiation to pluripotency is uncommon and negligible to regeneration, outcomes carrying significant implications for regenerative medicine.
Methods
Mice
Mice were derived and maintained at the Stanford University Research Animal Facility in accordance with Stanford University guidelines. All the animals were housed in sterile micro-insulators and given water and rodent chow ad libitum.
K5CreERT2, K14CreERT2, En1Cre,VE-cadherinCreERT2 and Tie2Cre transgenic mice were obtained from The Jackson Laboratory (Sacramento, CA) and a breeding colony was established. Sox9Cre and Prx1Cre mice were a gift from Dr. Alejandro Sweet-Cordero (Stanford University, California). ScxCre mice were a gift from Dr. Ronen Schweitzer (Shriners Hospital, Portland Oregon). mTmG reporter mice were a gift from Dr. Liqun Luo (Stanford University, California).
Digit amputations
Amputations were conducted under the guidelines of Stanford University Administrative Panel on Laboratory Animal Care (APLAC), Protocol #21621. Mice (post-weaning age) were anesthetized with i.p. injection of Nembutal (Pentobarbital; 50 mg/kg body weight) or Ketamine/Xylazine (80 mg and 8 mg per kg body weight, respectively). Digits 1-5 were amputated at proximal or distal levels using a No. 11 scalpel and allowed to heal without suturing for 2-3 months.
Generation of Rosa26-Rainbow mice
Lox2272-loxN-loxp-NheI-EcoRI sequence was introduced into the EcoRI site of pCAGGS. Fluorescent cDNAs, EGFP, mCherry, mOrange and mCerulean (Addgene, Cambridge, MA) were subcloned into an expression vector, pCAGGS. The fluorescent cDNAs together with 3′ globin enhancer and polyA sequences of the pCAGGS vector were PCR amplified and loxp (for mCherry), loxN (for mOrrange) or lox2272 (for mCerulean) sequences at 5′ non-coding region of the cDNA, and NheI at 5′ and XbaI at 3′ sites were attached to the fragments. The resulting NheI-XbaI fragments were subcloned into the NheI site of the lox2272-loxN-loxp-pCAGGS sequentially in the following order (loxp-mCherry, loxN-mOrange, lox2272-mCerulean, and EGFP). The whole construct from the CAG promoter to the polyA sequence downstream of the mCherry was cut out and subcloned into the Rosa26-1 neo vector. The vector was introduced into R1 ES cells and the knockin ES clone was established, and mice were generated.
Mice genotyping
The following primers and PCR conditions were used for genotyping: Cre; CGGTCGATGCAACGAGTGATGAGG and CCAGAGACGGAAATCCATCGCTCG. 94°C for 10 min, 94°C for 30 sec, 56°C for 1:30 min, 72°C for 1:30 min, repeat 35 cycles, 72°C for 8 min. mTmG; CTCTGCTGCCTCCTGGCTTCT, CGAGGCGGATCACAAGCAATA and TCAATGGGCGGGGGTCGTT. 94°C for 3 min, 94°C for 30 sec, 61°C for 1 min, 72°C for 1 min, repeat 35 cycles, 72°C for 2 min.
Tamoxifen injections
4-Hydroxytamoxifen (Sigma) was prepared by dissolving in absolute ethanol to a stock concentration of 100mg/ml using extensive vortexing. Tamoxifen stock solution was further emulsified in corn oil (Sigma) at 1:10. Prior to injection, the tamoxifen/oil suspension was sonicated in a sterile glass vial in ultrasonic bath sonicator. 1-2.5mg tamoxifen was injected i.p. with a tuberculin syringe and 25-gauge needle.
HSC transplantation
Transplantation and engraftment of HSCs into mice was carried as previously published42.
Histology and tissue analysis
For fixation, tissues were placed in 2% paraformaldehyde for 12-16h at 4°C, then decalcified in 0.4 M EDTA in PBS (pH 7.2) at 4 °C for 2 weeks. Samples were prepared for embedding by soaking in 30% sucrose in PBS at 4°C for 24h. Samples were removed from the sucrose solution and tissue blocks were prepared by embedding in Tissue Tek O.C.T (Sakura Finetek) under dry ice to freeze the samples within the compound. Frozen blocks were mounted on a MicroM HM550 cryostat (MICROM International GmbH) and 5-8 micron thick sections were transferred to Superfrost/Plus adhesive slides (Fisher brand). Representative sections were stained with Movat's modified pentachrome stain, hematoxylin and eosin stain and aniline blue stain as previously described45.
Immunohistochemistry
Immunostaining was performed using the following primary antibodies KI67 (Abcam), CD31 (Abcam), CD90 (eBioscience), CD105 (eBioscience), PEA (BD Pharmingen), cytokeratin 5 (Abcam), cytokeratin 14 (Covance), CD45 (Biolegend), MBP (Abcam), S100A4 (Abcam).
Briefly, slides were blocked for 30min in 10% BSA with 2% goat serum followed by incubation with primary antibody for 12-16 hours. For immunoassaying on sections from mTmG mice, Alexa Fluor 647 conjugated antibody was used as secondary 1:1000 for 1 hour (Invitrogen), and were visualized in the far-red channel (Cy5). Fluorescent and bright-field images were taken with a Leica DM4000B microscope (Leica Microsystems) and RETIGA 2000R camera (QImaging Scientific Cameras).
Supplementary Material
Acknowledgments
We thank A. Mosley for assistance with animal care and parabiosis experiments, Dr. Alejandro Sweet-Cordero (Stanford University, California) for providing Sox9Cre and Prx1Cre mice, Dr. Ronen Schweitzer (Shriners Hospital, Portland Oregon) for providing ScxCre mice, and Dr. Liqun Luo (Stanford University, California) for providing mTmG reporter mice. We thank Daniel Montoro for assistance with histochemical stains and Guy Paz for assistance with figure preparations. This work was supported in part by a grant from the California Institute of Regenerative Medicine (RC1 00354) and from the Smith Family Trust (to ILW), the Oak Foundation and the Hagey Laboratory for Pediatric Regenerative Medicine (to MTL). Y.R. is supported by the Human Frontier Science Program (HFSP) Long Term Fellowship, and the Machiah Foundation Fellowship.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints.
Author Contributions: Y.R. and I.L.W. designed the experiments. Y.R. performed the regeneration experiments, imaged and analyzed the data from all regeneration experiments. H.U. provided the ‘Rainbow’ reporter mice for part of the experiments. Y.R. and P.L. performed the HSC transplantations and analyzed the data. Y.R. and I.L.W. wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Rinkevich B, Shlemberg Z, Fishelson L. Whole-body protochordate regeneration from totipotent blood cells. Proc Natl Acad Sci U S A. 1995;92:7695–7699. doi: 10.1073/pnas.92.17.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinkevich Y, Paz G, Rinkevich B, Reshef R. Systemic bud induction and retinoic acid signaling underlie whole body regeneration in the urochordate Botrylloides leachi. PLoS Biol. 2007;5:e71. doi: 10.1371/journal.pbio.0050071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voskoboynik A, et al. Striving for normality: whole body regeneration through a series of abnormal generations. FASEB J. 2007;21:1335–44. doi: 10.1096/fj.06-7337com. [DOI] [PubMed] [Google Scholar]
- 4.Sánchez Alvarado A. Regeneration in the metazoans: why does it happen? Bioessays. 2000;22:578–590. doi: 10.1002/(SICI)1521-1878(200006)22:6<578::AID-BIES11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Han M, Yang X, Lee J, Allan CH, Muneoka K. Development and regeneration of the neonatal digit tip in mice. Dev Biol. 2008;315:125–135. doi: 10.1016/j.ydbio.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schotte OE, Smith CB. Wound healing processes in amputated mouse digits. Biol Bull. 1959;117:546–561. [Google Scholar]
- 7.Borgens RB. Mice regrow the tips of their foretoes. Science. 1982;217:747–750. doi: 10.1126/science.7100922. [DOI] [PubMed] [Google Scholar]
- 8.Reginelli AD, Wang YQ, Sassoon D, Muneoka K. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development. 1995;121:1065–1076. doi: 10.1242/dev.121.4.1065. [DOI] [PubMed] [Google Scholar]
- 9.Neufeld DA, Zhao W. Phalangeal regrowth in rodents: postamputational bone regrowth depends upon the level of amputation. Prog Clin Biol Res. 1993;383A:243–52. [PubMed] [Google Scholar]
- 10.Illingworth CM. Trapped fingers and amputated fingertips in children. J Pediatr Surg. 1974;9:853–858. doi: 10.1016/s0022-3468(74)80220-4. [DOI] [PubMed] [Google Scholar]
- 11.Rosenthal LJ, Reiner MA, Bleicher MA. Nonoperative management of distal fingertip amputations in children. Pediatrics. 1979;64:1–3. [PubMed] [Google Scholar]
- 12.Vidal P, Dickson MG. Regeneration of the distal phalanx. A case report. J Hand Surg Br. 1993;18:230–233. doi: 10.1016/0266-7681(93)90116-w. [DOI] [PubMed] [Google Scholar]
- 13.Morgan TH. Regeneration. New York: Macmillan; 1901. [Google Scholar]
- 14.Bratincsák A, et al. CD45-positive blood cells give rise to uterine epithelial cells in mice. Stem Cells. 2007;25:2820–2826. doi: 10.1634/stemcells.2007-0301. [DOI] [PubMed] [Google Scholar]
- 15.Faustman DL, et al. Comment on papers by Chong et al., Nishio et al., and Suri et al. on diabetes reversal in NOD mice. Science. 2006;314:1243. doi: 10.1126/science.1129918. [DOI] [PubMed] [Google Scholar]
- 16.Djouad F, Bouffi C, Ghannam S, Noël D, Jorgensen C. Mesenchymal stem cells: innovative therapeutic tools for rheumatic diseases. Nat Rev Rheumatol. 2009;5:392–399. doi: 10.1038/nrrheum.2009.104. [DOI] [PubMed] [Google Scholar]
- 17.Zaidi N, Nixon AJ. Stem cell therapy in bone repair and regeneration. Ann N Y Acad Sci. 2007;1117:62–72. doi: 10.1196/annals.1402.074. [DOI] [PubMed] [Google Scholar]
- 18.Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Ohgo S, Yokoyama H. Limb blastema cell: a stem cell for morphological regeneration. Dev Growth Differ. 2010;52:89–99. doi: 10.1111/j.1440-169X.2009.01144.x. [DOI] [PubMed] [Google Scholar]
- 20.Christen B, Robles V, Raya M, Paramonov I, Belmonte JC. Regeneration and reprogramming compared. BMC Biol. 2010;20:8, 5. doi: 10.1186/1741-7007-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsonis PA. Stem cells and blastema cells. Curr Stem Cell Res Ther. 2008;3:53–4. doi: 10.2174/157488808783489408. [DOI] [PubMed] [Google Scholar]
- 22.http://www.nytimes.com/2010/11/09/science/09wade.html; http://www.nytimes.com/2010/11/09/health/09stem.html; http://www.nytimes.com/2010/08/06/science/06cell.html; http://tierneylab.blogs.nytimes.com/2009/07/02/rumsfeldian-stem-cells; http://www.nytimes.com/2009/03/10/science/10lab.html
- 23.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 24.Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science. 2002;297:2256–2259. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
- 25.Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 116:639–648. doi: 10.1016/s0092-8674(04)00208-9. [DOI] [PubMed] [Google Scholar]
- 26.Purhonen SJ, et al. Bone marrow-derived circulating endothelial precursors do not contribute to vascular endothelium and are not needed for tumor growth. Proc Natl Acad Sci U S A. 2008;105:6620–6625. doi: 10.1073/pnas.0710516105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamrick MW. Development and evolution of the mammalian limb: adaptive diversification of nails, hooves, and claws. Evol Dev. 2001;3:355–363. doi: 10.1046/j.1525-142x.2001.01032.x. [DOI] [PubMed] [Google Scholar]
- 28.Said S, Parke W, Neufeld DA. Vascular supplies differ in regenerating and nonregenerating amputated rodent digits. Anat Rec A Discov Mol Cell Evol Biol. 2004;278:443–449. doi: 10.1002/ar.a.20034. [DOI] [PubMed] [Google Scholar]
- 29.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 31.Indra AK, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimmel RA, et al. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- 33.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 34.Akiyama H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 37.Forde A, Constien R, Gröne HJ, Hämmerling G, Arnold B. Temporal Cre-mediated recombination exclusively in endothelial cells using Tie2 regulatory elements. Genesis. 2002;33:191–7. doi: 10.1002/gene.10117. [DOI] [PubMed] [Google Scholar]
- 38.Monvoisin A, et al. VE-cadherin-CreERT2 transgenic mouse: a model for inducible recombination in the endothelium. Dev Dyn. 2006;235:3413–3422. doi: 10.1002/dvdy.20982. [DOI] [PubMed] [Google Scholar]
- 39.Fernando WA, et al. Wound healing and blastema formation in regenerating digit tips of adult mice. Dev Biol. 2011;350:301–10. doi: 10.1016/j.ydbio.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forsberg EC, Bhattacharya D, Weissman IL. Hematopoietic stem cells: expression profiling and beyond. Stem Cell Rev. 2006;2:23–30. doi: 10.1007/s12015-006-0005-z. [DOI] [PubMed] [Google Scholar]
- 41.Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 42.Liversage RA. In: A History of Regeneration Research: Milestones in the Evolution of a Science. Dinsmore CE, editor. Cambridge University Press; 1991. [Google Scholar]
- 43.Ito M, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–20. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 44.Chan CK, et al. Endochondral ossification is required for haematopoietic stem-cell niche formation. Nature. 2009;457:490–494. doi: 10.1038/nature07547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gargioli C, Slack JM. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–79. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- 46.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 47.Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell. 2011;20:725–32. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.